Abstract
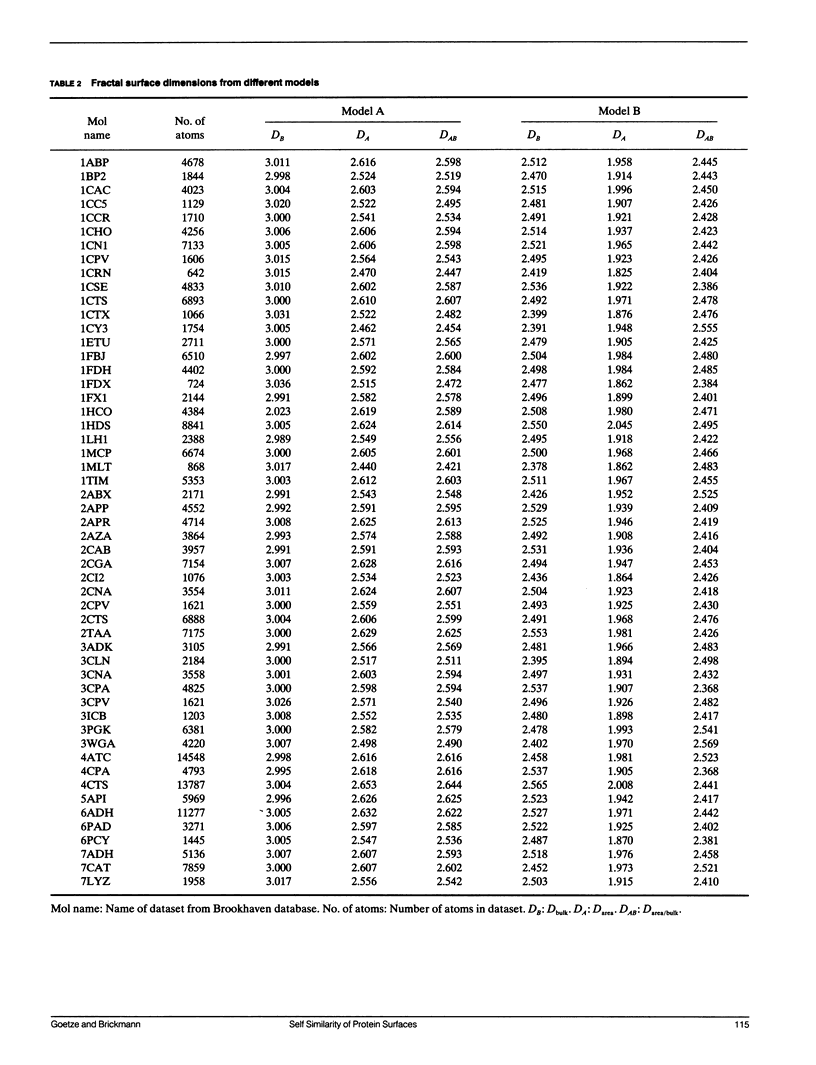
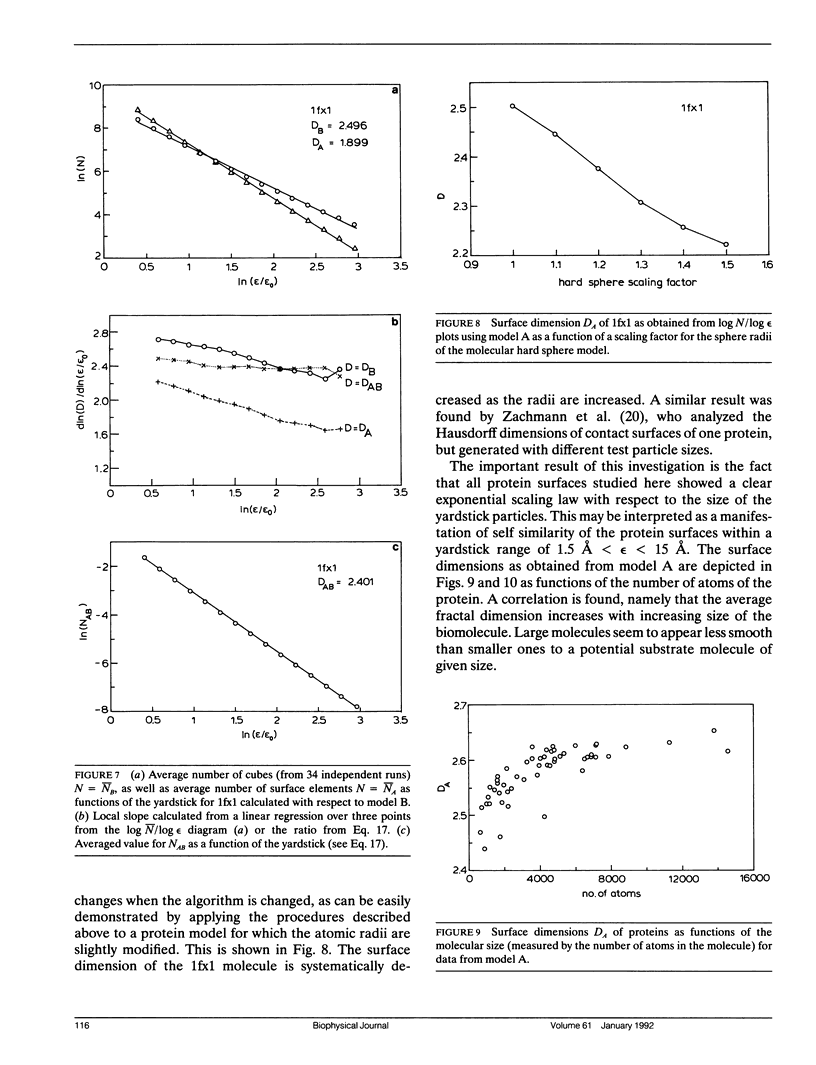
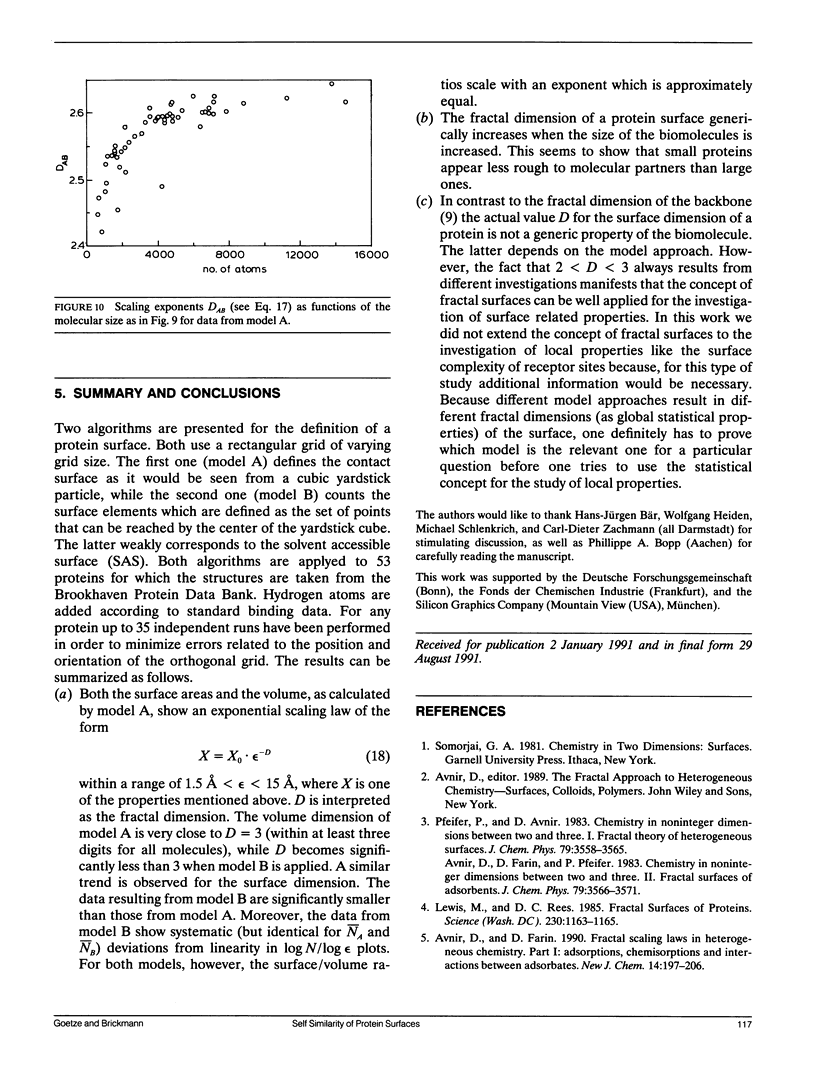
Scaling properties of the surfaces of 53 proteins are studied on the basis of experimentally determined 3-D structures of these biological macromolecules. It is found that the surfaces show self similarity within a yardstick range of 1.5 A less than epsilon less than 15 A. The self similarity is measured by the fractal dimension D of the surface. Two different algorithms for the determination of the fractal dimension are applied, both based on cubic yardstick particles. One is related to the contact surface (CS), which was first introduced by Connolly (17), while the other corresponds to the solvent accessible surface (SAS) of Richards (9). The fractal dimensions of both are different. While the CS type approach leads to relatively high values of D in the range 2.5 to 2.6 the SAS approach gives fractal dimensions of D approximately 2.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fushman D. Surface fractality of proteins from theory and NMR data. J Biomol Struct Dyn. 1990 Jun;7(6):1333–1344. doi: 10.1080/07391102.1990.10508569. [DOI] [PubMed] [Google Scholar]
- Lewis M., Rees D. C. Fractal surfaces of proteins. Science. 1985 Dec 6;230(4730):1163–1165. doi: 10.1126/science.4071040. [DOI] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]


