Abstract
The Ku70/80 heterodimer is a critical component of the non-homologous end-joining (NHEJ) pathway and of the telomere cap in yeast and mammals. We report the molecular characterization of the KU70 and KU80 genes in Arabidopsis and describe the consequences of a Ku70 deficiency. Arabidopsis KU70/80 genes are ubiquitously expressed and their products form stable heterodimers in vitro. Plants harboring a T-DNA insertion in KU70 exhibit no growth or developmental defects under standard growth conditions. However, mutant seedlings are hypersensitive to γ-irradiation-induced double-strand breaks. Unexpectedly, we found that mutants are hypersensitive to methyl methanosulfonate during seed germination, but lose this sensitivity in seedlings, implying that the requirement for NHEJ varies during plant development. Lack of Ku70 results in a dramatic deregulation of telomere length control, with mutant telomeres expanding to more than twice the size of wild type by the second generation. Furthermore, in contrast to the situation in mammals, chromosome fusions are not associated with a Ku deficiency in Arabidopsis. These findings imply that Ku may play a different role in capping plant and animal telomeres.
Keywords: double-strand break repair/genome stability/methyl methanosulfonate/non-homologous end-joining/telomerase
Introduction
Double-strand DNA breaks (DSBs) are lethal DNA lesions that can be induced by external assaults such as ionizing radiation and radiomimetic drugs, or by endogenous metabolic processes including DNA replication. Because a single unrepaired DSB can lead to cell-cycle arrest (Sandell and Zakian, 1993), efficient DSB repair mechanisms are essential. In eukaryotes, broken ends can be processed in two pathways: non-homologous end-joining (NHEJ) and homologous recombination (HR) (Critchlow and Jackson, 1998; Hiom, 1999; Thompson and Schild, 2001). In both cases, the break is repaired by ‘resealing’ the free DNA ends to generate a contiguous molecule.
Ku is a heterodimeric complex of 70 and 80 kDa sub units, critical for the NHEJ pathway. Ku binds with high affinity to broken DNA ends to prevent degradation and facilitate alignment (Walker et al., 2001) for subsequent ligation by DNA ligase IV. DNA-dependent protein kinase and the ligase IV-associated protein XRCC4 are also required for NHEJ, but their precise roles are unclear (Critchlow and Jackson, 1998). In yeast lacking functional Ku, NHEJ is severely impaired (Boulton and Jackson, 1998; Baumann and Cech, 2000), and mutants show enhanced sensitivity to radiomimetic agents in a rad52 background (Mages et al., 1996). Mammalian cells or animals deficient in either the Ku70 or Ku80 subunit are similarly radiosensitive and are defective in V(D)J recombination (Featherstone and Jackson, 1999). In addition, Ku-deficient mice exhibit impaired growth and the early onset of senescence (Nussenzweig et al., 1996; Vogel et al., 1999).
While NHEJ is believed to be the major DNA repair pathway in higher eukaryotes, this mechanism is inherently error prone (Kirik et al., 2000). Recombinational repair, in contrast, provides an error-free route for DSB repair and is the preferred pathway in lower eukaryotes (Hiom, 1999). Recent studies suggest that HR also plays an important role in DSB repair in higher eukaryotes during S and G2, when the homologous sequence from a sister chromatid can be utilized for accurate repair (reviewed in Thompson and Schild, 2001). Moreover, cell cycle-dependent employment of both NHEJ and HR has been reported in vertebrates (Takata et al., 1998), with NHEJ playing the dominant role during G1, and HR in late S and G2.
Relatively little is known about DNA repair in plants (Vonarx et al., 1998; Britt, 1999). Two mutants deficient in DSB repair have been described in Arabidopsis thaliana. Inactivation of MIM, a plant homolog of Rad18, reduces somatic recombination rates and increases sensitivity to genotoxic stress (Mengiste et al., 1999). Similarly, dis ruption of RAD50 increases sensitivity to methyl methanosulfonate (MMS) and renders mutants sterile (Gallego et al., 2001). To date, no mutants deficient in NHEJ have been described, although Arabidopsis orthologs of ligase IV and XRCC4 have been characterized (West et al., 2000).
The natural ends of linear eukaryotic chromosomes are masked from DNA repair and checkpoint machinery by telomeres, higher order nucleoprotein caps. Telomeres consist of short G-rich repeat arrays associated with specific non-nucleosomal telomere-binding proteins (Collins, 2000) that protect the ends from fusions (van Steensel et al., 1998) and control the length of the telomere tract (Smogorzewska et al., 2000). Replication of telomeres is mediated by telomerase, a ribonucleoprotein reverse transcriptase that uses its RNA subunit as a template for addition of telomere repeat sequences by the catalytic subunit, TERT (Nugent and Lundblad, 1998). Proteins involved in DSB repair and checkpoint response are also closely associated with telomeres, and are essential for telomere maintenance. For example, mutations in the MRT2 gene, which encodes a Caenorhabditis elegans homolog of the yeast DNA checkpoint protein Rad17, result in telomere shortening and chromosome fusions (Ahmed and Hodgkin, 2000). In addition, the DSB repair complex Mre11/Rad50/Xrs2 (Nbs2) is required for telomere maintenance in yeast and humans (Nugent et al., 1998; Ranganathan et al., 2001), and recent studies reveal cell cycle-regulated association of this complex with the human telomere protein TRF2 (Zhu et al., 2000).
The Ku heterodimer also plays a critical role in telomere length homeostasis and, while these proteins are known to interact directly with TRF1, TRF2 and the telomerase RNA subunit (Hsu et al., 2000; Song et al., 2000; Peterson et al., 2001), their contribution to the telomere cap remains enigmatic. Inactivation of Saccharomyces cerevisiae YKU70 or YKU80 leads to a temperature-sensitive phenotype characterized by shorter telomeres with extended terminal G overhangs (Boulton and Jackson, 1998; Gravel et al., 1998; Polotnianka et al., 1998). Silencing of telomere-proximal genes is also dramatically reduced and telomeres are delocalized from the nuclear periphery (Laroche et al., 1998). The temperature sensitivity of yku cells is attributed to a lethal alteration in telomere structure at elevated temperature (Fellerhoff et al., 2000). The telomeres of fission yeast that harbor ku70 mutations are shorter than wild type and exhibit extensive subtelomeric rearrangements (Baumann and Cech, 2000). However, unlike budding yeast, Schizosaccharomyces pombe ku70–/– cells are not temperature sensitive (Manolis et al., 2001). Mammalian cells deficient in Ku display frequent end-to-end chromosome fusions (Bailey et al., 1999; Hsu et al., 2000; Samper et al., 2000; d’Adda di Fagagna et al., 2001). The impact on telomere dynamics in mammals is still unclear as both telomere shortening and lengthening have been reported (Samper et al., 2000; d’Adda di Fagagna et al., 2001).
As in other eukaryotes, telomeres in plants are maintained by telomerase, and Arabidopsis mutants harboring a T-DNA insertion in the TERT gene are characterized by gross cytogenetic and developmental defects when telomeres become critically shortened (Fitzgerald et al., 1999; Riha et al., 2001). A role for the Mre11/Rad50/Xrs2 DNA repair complex at telomeres is implied from analysis of the Arabidopsis Rad50 homolog. Plants lacking Rad50 protein are sterile, but show no telomere defects. However, cell cultures derived from these mutants exhibit dramatic telomere shortening and massive cell death. Survivors harbor telomeres that are significantly longer than wild-type (Gallego and White, 2001). Here we investigate the role of NHEJ in plants and describe the consequences of a Ku70 deficiency in Arabidopsis. We demonstrate an essential role for Ku in the DNA damage response, and also provide evidence that the NHEJ pathway is not the sole mechanism for repairing DSBs in plants. Finally, we show that although telomere length homeostasis is severely perturbed in mutants lacking Ku, the protective cap on the chromosome terminus remains intact.
Results
Molecular characterization of Ku70 and Ku80 homologs in Arabidopsis
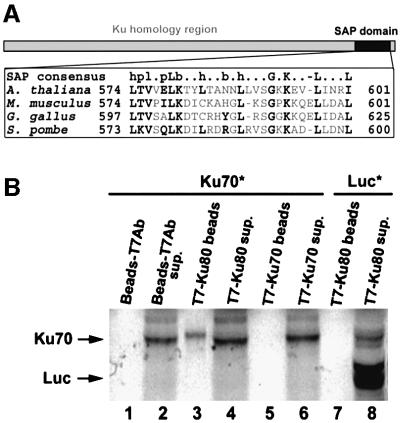
A TBLASTN search of the DNA database revealed a likely Arabidopsis homolog for Ku70 on chromosome 1 (BAC clone F6I1; DDBJ/EMBL/GenBank accession No. AC051629), which displayed ∼30% identity to human Ku70. To obtain a full-length cDNA for the Arabidopsis gene, we searched the Arabidopsis expressed sequence tag (EST) non-redundant database and found a cDNA clone, FB059c01F (DDBJ/EMBL/GenBank accession No. AV533293; Asamizu et al., 2000), with identity to the 3′ end of the AtKU70 gene. The remainder of the KU70 cDNA was obtained by RT–PCR and 5′ RACE using primers designed from the predicted coding sequence. Our KU70 cDNA sequence matches a sequence reported in the DNA database during preparation of this manuscript (DDBJ/EMBL/GenBank accession No. AF283759). Over all, the predicted Ku70 protein exhibits 29% identity (45% similarity) to the human Ku70 protein and 20% identity (33% similarity) to the S.cerevisiae Yku70. The C-terminus of AtKu70 contains a SAP domain (Aravind and Koonin, 2000), a DNA-binding motif conserved in all characterized Ku70 orthologs except S.cerevisiae Yku70 (Figure 1A).
Fig. 1. Identification of Ku 70/80 proteins in Arabidopsis. (A) Schem atic diagram of Ku70 and alignment of the C-terminal DNA-binding domain (SAP) (Aravind and Koonin, 2000). (B) Dimerization of Ku70/80 proteins in vitro. Co-immunoprecipitation experiments using recombinant T7-tagged Ku80 or Ku70 and 35S-labeled (asterisk) Ku70 or luciferase are shown. See Materials and methods for details.
Recently, the complete mRNA sequence of an Arabidopsis Ku80-like protein was reported in the DNA database (DDBJ/EMBL/GenBank accession No. AF283758; Y.Adachi, K.Oguchi, K.Tamura and H.Takahashi, unpublished data). AtKU80 codes for a predicted protein with 22% identity (43% similarity) to human Ku86 and 24% identity (41% similarity) to S.cerevisiae Yku80. We used RT–PCR to obtain a full-length cDNA of AtKU80.
Since Ku70 and Ku80 exist as a heterodimeric complex in other organisms, we tested whether the putative Ku70 and Ku80 proteins from Arabidopsis interact in vitro using co-immunoprecipitation experiments. The AtKU70 and AtKU80 coding regions were inserted into a pET28a expression vector to produce recombinant Ku70 and Ku80 proteins either with or without an N-terminal T7 tag. As shown in Figure 1B, untagged 35S-labeled Ku70 protein was precipitated in the presence of unlabeled T7-tagged Ku80 using a T7 antibody (lane 3). In control reactions, no interaction was seen between T7 antibodies and untagged Ku70 (Figure 1B, lane 1), or between Ku80 and luciferase (Figure 1B, lane 7). Furthermore, as reported for mammalian Ku proteins (Osipovich et al., 1997; Cary et al., 1998), we found no evidence for AtKu70 homodimerization (Figure 1B, lane 5).
Northern blot analysis of KU70 and KU80 transcripts from total RNA samples detected RNAs of the expected size (∼2.3 kb) in all tissues examined (Figure 2; see also Figure 3B). The ubiquitous expression profile of KU genes in Arabidopsis resembles the profile in mammals (Cai et al., 1994; Koike et al., 1996). In non-photosynthesizing tissues, a slightly higher molecular weight species of KU80 mRNA was detected (Figure 2). RT–PCR indicated that the two KU80 RNA species are not products of alternative splicing (data not shown). Instead, we suspect that they result from distinct 3′ end mRNA processing events (Hartung and Puchta, 2000), since two putative cleavage/polyadenylation signals are present 100 bp apart in the KU80 3′-untranslated region.
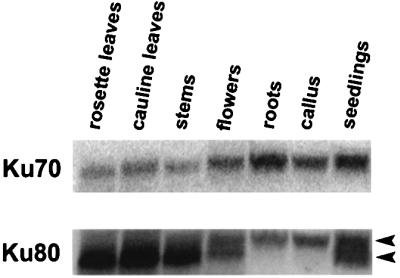
Fig. 2. Expression analysis of Ku70/80 mRNA. RNA isolated from a variety of organs was analyzed by northern blot hybridization using a probe derived from the entire Ku70 cDNA. The blot was stripped and rehybridized with a Ku80 probe. Arrowheads denote two Ku80 RNA species that may arise from different 3′ end processing events.
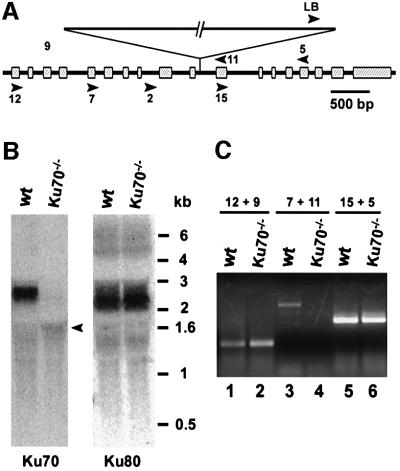
Fig. 3. Disruption of the Arabidopsis KU70 gene by T-DNA insertion. (A) Schematic diagram of the AtKU70 gene. The position of a T-DNA insertion is indicated. Boxes = exons; arrowheads = primers used for RT–PCR and genotyping. (B) Northern blot analysis of KU70/80 expression in mutant and wild-type plants. The same membrane was used for sequential hybridizations with Ku70 and Ku80 probes. The arrowhead marks a non-specific hybridization signal. (C) RT–PCR analysis of the disrupted ku70 allele. Thirty-five cycles of PCR were performed. Under semi-quantitative conditions (30 PCR cycles, 250 ng of RNA), products corresponding to the Ku70 N-terminus were barely detectable in mutants, while the wild-type signal was strong (data not shown).
Identification of an Arabidopsis ku70 mutant
To examine the role of the Ku complex in plants, we screened a collection of 60 480 Arabidopsis T-DNA insertion lines available at the University of Wisconsin Arabidopsis Knock-out Facility (Krysan et al., 1999) and identified a line harboring a disruption in the KU70 gene (Figure 3A). Southern blot analysis revealed a single site of T-DNA insertion that maps to the 10th intron of KU70. The disruption appears to be comprised of three adjacent T-DNA insertions (data not shown).
Northern blot analysis of mutants homozygous for the T-DNA insertion failed to detect KU70 mRNA (Figure 3B; the arrowhead indicates non-specific hybridization). The level of KU80 was the same as in wild-type. We confirmed that expression of the full-length transcript was abolished by the T-DNA insertion by performing RT–PCR using primers that flank the T-DNA insertion site. No product was obtained after 35 cycles of PCR amplification (Figure 3C, lane 4). This finding excludes the possibility that splicing out the entire T-DNA generates an intact full-length Ku70 transcript. RT–PCR of the KU70 5′ and 3′ coding regions flanking the T-DNA insertion revealed the presence of low abundance transcripts undetectable by northern blotting (Figure 3C, lanes 2 and 6). Sequence analysis of the 3′ transcript expressed from a cryptic promoter within the T-DNA (Mengiste et al., 1999) demonstrated the lack of a consensus AUG codon in a reading frame appropriate for translation of the Ku70 C-terminus (data not shown). Taken together, these data argue that Ku function in the mutants is likely to be severely compromised, and possibly null.
Plants homozygous for the T-DNA insertion were propagated for two generations. In striking contrast to mammals deficient in Ku (Nussenzweig et al., 1996), Arabidopsis ku70 mutants were phenotypically indistinguishable from their heterozygous and wild-type siblings. No defects in vegetative or generative growth and development were detected under standard growth conditions, and mutants were fertile.
Response of Ku70-deficient Arabidopsis to genotoxic stress
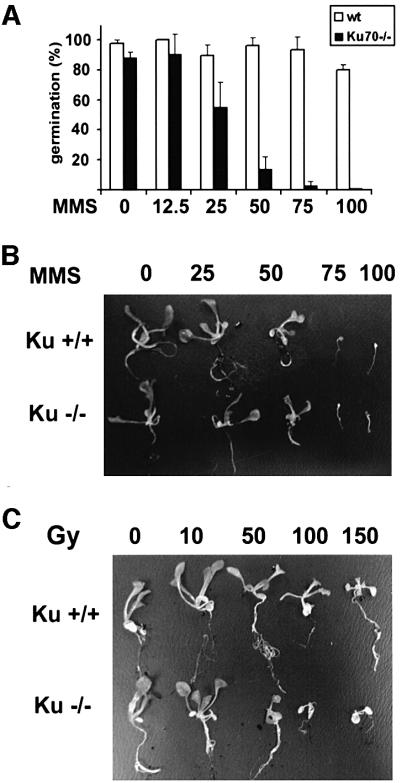
To determine whether disruption of KU70 alters the response to DNA damage, we germinated seeds from ku70–/– and wild-type plants on solid media supplemented with various concentrations of MMS. The germination efficiency of wild-type dropped by ∼20% in the presence of 100 p.p.m. MMS. In contrast, the ratio of germinating to non-germinating mutant seeds decreased steadily with increasing concentration of MMS. In 100 p.p.m. MMS, germination was completely abolished (Figure 4A). Surprisingly, a subset of the mutants that germinated in the presence of 75 p.p.m. MMS grew at a rate similar to wild type (data not shown). This observation strongly suggests that Ku-deficient plants are hypersensitive to MMS only during germination. Previous studies demonstrate that seedlings defective in DNA repair or damage checkpoint response are hypersensitive to MMS (Mengiste et al., 1999; Ulm et al., 2001). Therefore, we examined the sensitivity of Ku-deficient plants to MMS treatment. Five-day-old wild-type and mutant seedlings germinated and grown in the absence of MMS were transferred into a liquid medium supplemented with MMS. In this assay, mutant seedlings showed the same response to MMS as wild type (Figure 4B). In our experiments, the toxicity of MMS was higher in liquid media such that growth of both wild-type and mutant plants was completely inhibited in 75 p.p.m. MMS. However, we detected no difference in the growth of wild type and mutants in 50 p.p.m. MMS (Figure 4B). In addition, 8-day-old mutant seedlings transferred to solid media containing 100 p.p.m. MMS grew as well as the wild type (data not shown). Thus, the MMS hypersensitivity of Ku-deficient mutants is confined to a specific stage in development.
Fig. 4. Response of ku70 mutants to genotoxic stress. (A) Germination efficiency of wild-type and mutant seeds in media supplemented with various concentrations of MMS. Results from four independent experiments are shown. Bars denote the standard deviation. Concentration units are in p.p.m. (B) Response of seedlings to MMS. Five-day-old seedlings were incubated in media containing different concentrations of MMS (in p.p.m.). (C) Response of seedlings to γ-irradiation. Five-day-old seedlings were subjected to increasing doses of radiation as indicated. For both experiments, plants were photographed 10 days after genotoxic treatment.
In mammals, Ku-deficient mutants are hypersensitive to ionizing radiation. We examined the response of Arabidopsis ku70 mutants to ionizing radiation by exposing 5-day-old seedlings to γ-irradiation (Figure 4C). A dose of 50 Gy retarded the growth of mutants, while 100 Gy strongly suppressed development of true leaves and roots. In contrast, wild-type plants treated with this higher dose displayed only minimal growth defects. From these data, we conclude that Ku-deficient seedlings respond differently to genotoxic stresses, as they exhibit wild-type sensitivity to MMS and hypersensitivity to ionizing radiation.
Telomere status and genome stability in Ku-deficient Arabidopsis
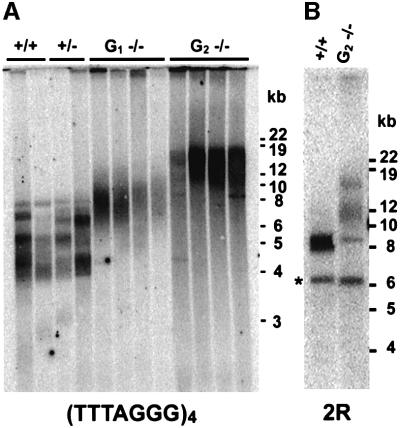
We next examined the role of Ku70 in telomere length homeostasis by determining telomere length in individual plants from the first two generations of mutants. Terminal restriction fragments (TRFs) were generated by cleavage with Tru9I and analyzed by Southern hybridization with a telomeric probe. Telomeres in the wild-type Wassilewskija (WS) ecotype and plants heterozygous for the ku mutation ranged in size from 4 to 8 kb, and were twice the size of telomeres in the Colombia ecotype (Fitzgerald et al., 1999). This finding was not unanticipated, since telomere lengths vary by ∼2-fold among different Arabidopsis ecotypes (K.Davis and T.McKnight, personal communication). Surprisingly, however, telomeres in first-generation ku70 homozygous mutants (G1) extended from 6 to 12 kb (Figure 5A) and, by the second generation (G2), approached ∼20 kb. Thus, telomeres in ku70 mutants are subject to continual extension over successive plant generations.
Fig. 5. Telomere length analysis of Ku70-deficient plants. (A) TRF analysis of DNA from individual wild-type, heterozygous and first- (G1) and second- (G2) generation mutants. Note that the sharp bands in the wild type and heterozygotes are replaced by smears in mutants. (B) Single telomere analysis of wild-type and G2 ku70 mutants using a telomere-associated probe derived from the right arm of chromosome 2. The asterisk denotes interstitial telomeric DNA.
Two types of experiments were performed to verify that the telomere elongation phenotype was associated with the T-DNA insertion in KU70. First, co-segregation analysis was conducted using 70 progeny derived from plants heterozygous for the T-DNA insertion. A total of 19/19 homozygous mutants displayed elongated telomeres, while 51/51 heterozygous and wild-type plants exhibited the wild-type telomere profile (data not shown). Secondly, the telomere phenotype was complemented by ectopic expression of wild-type Ku70. Two complementation constructs, one harboring a full-length genomic clone containing 1.6 kb of putative promoter sequence and the second containing Ku70 cDNA expressed from the cauliflower mosaic virus (CaMV) 35S promoter, were used for complementation of the ku70 mutant. The two constructs were introduced into plants heterozygous for the T-DNA insertion in KU70. In T1, four individuals were identified (two plants for each complementation construct) that were homozygous for the T-DNA disruption in Ku70 and which ectopically expressed wild-type Ku70 mRNA. In each case, ectopic expression of KU70 prevented the dramatic telomere extension (data not shown). Taken together, these data demonstrate that the telomere phenotype is linked to a disruption in KU70.
The discrete banding profile of TRFs in wild-type plants (Figure 5A) implies that telomeres are tightly regulated in the WS ecotype. Probes specific for individual chromosome ends indicated that telomere tracts are maintained within a 1 kb range (Figure 5B; data not shown). Indi vidual telomere tracts are maintained in the same size range in the Colombia ecotype (Riha et al., 2001; K.Riha and D.Shippen, unpublished data). In contrast, telomeres in ku70 mutants were extremely heterogeneous (Figure 5A). This heterogeneity was especially apparent when TRFs were visualized using probes specific for individual chromosome ends (Figure 5B; data not shown). The 2R probe, for instance, showed a discrete band of 8–9 kb in wild-type plants, and a heterogeneous signal spreading from 8 to ∼18 kb in the mutants (Figure 5B).
Tru9I recognizes the sequence TTAA, and cleaves immediately adjacent to the terminal telomere repeat tract. Hence, the longer terminal restriction fragments that arise from the mutants are comprised almost exclusively of TTTAGGG repeats. A preliminary analysis of subtelomeric regions indicated that there was no gross DNA rearrangement (data not shown). Instead, only the terminal telomere repeat tract appears to be modified in Ku-deficient plants.
Inactivation of Ku in mammals results in a high frequency of end-to-end chromosome fusions (Bailey et al., 1999; Samper et al., 2000; d’Adda di Fagagna et al., 2001). Cytogenetic analysis of >700 anaphases in G1 and G2 ku70 mutants failed to reveal any fusion events. In contrast, in late-generation telomerase-deficient mutants analyzed by the same protocol, up to 45% of the anaphases examined harbored bridges (Riha et al., 2001). We con clude from these data that despite dramatic deregulation of telomere length control, telomeres in Ku70-deficient Arabidopsis are still functional and protect chromosomes from end-to-end fusions.
Discussion
Plants face a constant risk of DNA damage due to continuous exposure to a variety of environmental factors, and thus very efficient repair mechanisms must operate to maintain genome integrity. Here we report establishment of a plant model deficient in the NHEJ DNA repair pathway. The Ku70 mutant we characterized carries a T-DNA insertion in the 10th intron located in the middle of the gene. While we could detect no full-length transcripts from the disrupted gene by northern blot analysis or RT–PCR, a low abundance transcript from the N-terminus was detected by RT–PCR. It is unlikely that a truncated protein translated from this RNA would be functional as studies in yeast show that deletion of the extreme C-terminus of Ku70 is essential for both NHEJ and telomere maintenance (Driller et al., 2000). However, we cannot rule out the possibility that a protein derived from the Ku70 N-terminus interferes with some biological function in plants. A binding site for the telomere protein TRF2 is located in the N-terminal half of human Ku70 (Song et al., 2000) and, hence, the presence of a truncated Ku protein could potentially contribute to the unique telomere phenotype associated with ku70 mutants (see below).
Requirement for the NHEJ pathway varies during plant development
Both HR and NHEJ are implicated in repair of DSBs in eukaryotes, but NHEJ has been assumed to be the major pathway in plants based on the random integration of a foreign DNA into plant genomes (Britt, 1999; Mengiste and Paszkowski, 1999). We directly investigated the contribution of the NHEJ pathway to DBS repair in Arabidopsis by exposing plants deficient in Ku to two different genotoxic stresses, γ-irradiation and MMS. Surprisingly, we found that mutant seedlings are hypersensitive to γ-irradiation, but relatively insensitive to MMS. We postulate that these distinct responses reflect the mechanism by which the break is introduced. Ionizing radiation leads to DSBs (as well as a broad spectrum of other DNA lesions) immediately after exposure. In seed lings, the majority of plant cells are in G1, and unrepaired DSBs induced by γ-irradiation would prevent cells from entering S phase. The failure of Arabidopsis ku70 mutants to produce true leaves following a relatively low dose of γ-irradiation is consistent with a crucial role for NHEJ in DSB repair during G1.
MMS is a monofunctional alkylating agent that produces abasic lesions and single-strand nicks (Helbig and Speit, 1997; Glaab et al., 1999). The damaged site subsequently is converted into a DSB during DNA replication in S phase and leads to fork arrest. Thus, in contrast to γ-irradiation, the DSBs introduced by MMS are S-phase specific. Vertebrate studies indicate that the cell cycle context of a DSB determines whether NHEJ or HR is employed to repair it (Takata et al., 1998). DSBs that arise during DNA replication, such as those introduced by MMS, are repaired through HR (Marians, 2000; Michel, 2000). Consistent with this idea, Drosophila mutants defective in HR (rad54–/–) are more sensitive to MMS treatment than ku70–/– flies (Kooistra et al., 1999). Further more, MMS-treated Ku-deficient hamster cells have the same rate of DNA break rejoining as control cells (Helbig and Speit, 1997).
Recent studies support the notion that HR contributes to DSB repair in plants. MIM and RAD50 are predicted to play a role in HR (Mengiste et al., 1999; Signon et al., 2001), and plants deficient in MIM and RAD50 are sensitive to MMS treatment (Mengiste et al., 1999; Gallego et al., 2001). These results together with data presented here argue that both NHEJ and HR are utilized to repair DSBs in plants and that the cell cycle context of the DSB strongly influences which pathway is chosen for its repair.
One surprising result from our study is that in contrast to Ku-deficient seedlings, mutant seeds are hypersensitive to MMS and germination is reduced dramatically even at very low levels of MMS. We interpret this to mean that the requirement for NHEJ varies throughout plant development. In support of this idea, mouse studies demonstrate that the relative contributions of HR and NHEJ in the repair of radiation-induced DNA damage change during development (Essers et al., 2000). During dehydration and dry storage, plant seeds are subject to extensive DNA damage, and pre-replicative repair occurs immediately after imbibition (reviewed in Vonarx et al., 1998; Whittle et al., 2001). Although germination of Ku-deficient seeds is unaffected under normal conditions, our data suggest that the additional damage incurred by MMS treatment cannot be repaired in the absence of Ku. Since seed viability and DNA integrity decrease during prolonged storage, it will be interesting to examine germination efficiency in the aging seeds of Ku mutants.
Arabidopsis Ku is required for telomere length homeostasis, but not for chromosome stability
The Ku heterodimer is involved in two seemingly paradoxical processes: it mediates resealing of broken DNA ends as a part of NHEJ and it localizes to telomeres, which protect chromosome ends from fusions. A Ku deficiency in mammals leads to massive genome instability and chromosome fusions, which are thought to cause stunted growth and accelerated aging. In contrast, the genome of Ku-deficient Arabidopsis is stable, and we could detect no chromosome fusions in >700 anaphases examined. This observation is consistent with the normal growth and development of mutant plants and argues that the telomere capping function in Ku-deficient plants is intact.
The absence of Ku affects telomere homeostasis in yeast to reset telomere tracts to a shorter but stabilized length (Boulton and Jackson, 1998; Gravel et al., 1998; Polotnianka et al., 1998; Baumann and Cech, 2000). However, in Arabidopsis ku mutants, telomere length control is deregulated, and telomere tracts steadily increase to over twice the size of wild type by the second generation. Monitoring telomere status in subsequent generations of the mutants will be necessary to determine if and when equilibrium is reached. The loss of telomere length homeostasis in ku70 mutants is remarkable, since wild-type Arabidopsis telomeres are very tightly controlled (Fitzgerald et al., 1999; Zentgraf et al., 2000; Riha et al., 2001; this study). Interestingly, inactivation of telomerase has the opposite effect, reducing telomere length polymorphism and leading to a very slow rate of telomere shortening (Fitzgerald et al., 1999; Riha et al., 2001).
The longer telomeres in Ku-deficient Arabidopsis could result from a perturbation in telomere architecture that increases access of recombination machinery or telomerase. Elevated recombination rates at telomeric and subtelomeric regions are associated with a Ku deficiency in yeast (Polotnianka et al., 1998; Baumann and Cech, 2000). Alternatively, Ku could act as a negative regulator of telomerase through direct interactions with telomerase (Grandin et al., 2000; Peterson et al., 2001) or telomere-binding proteins such as TRF1 and TRF2 (Hsu et al., 2000; Smogorzewska et al., 2000; Song et al., 2000). Intriguingly, recent studies suggest that Ku is a positive regulator of telomerase in yeast (Grandin et al., 2000). The longer telomeres and the absence of chromosome fusions in Ku-deficient plants argue that the function of Ku at telomeres differs among eukaryotes. Analysis of Arabidopsis tert/ku70 double mutants should not only provide further insight into the role of the Ku complex, but may also help to define the relative contribution of telomerase and recombination to telomere maintenance in higher eukaryotes. These studies are underway.
Materials and methods
Plant growth and assays for sensitivity to genotoxic treatment
Arabidopsis thaliana plants (ecotype Wassilewskija) were grown at 23°C in an environmental chamber under a 16/8 h light/dark photoperiod. Seeds for the germination assay were sterilized in 50% bleach and plated on solid 0.5 BM medium (Mathur and Koncz, 1998) supplemented with 0–100 p.p.m. (v/v) MMS (Aldrich). Five-day-old seedlings were transferred to separate wells of a 24-well plate containing liquid 0.5 BM medium with 0–100 p.p.m. MMS and incubated in a shaker with constant light. To test sensitivity to ionizing radiation, 5-day-old seedlings were exposed to increasing time periods of γ-irradiation from a 60Co source (Theratron 780; Atomic Energy Systems, Canada), for absorbed doses of 10, 50, 100 or 150 Gy. Following irradiation, seedlings were transferred to individual wells of a 24-well plate containing liquid 0.5 BM medium, and incubated in a shaker with constant light.
cDNA synthesis
Total mRNA was extracted from 0.1–0.5 g of plant tissue using Tri Reagent solution (Sigma). To obtain the 5′ terminus of AtKU70, 5′ RACE was performed using the 5′ RACE System (Gibco) and nested primers Ku70-8 (5′-TGCGTTTATCTGCTGTC-3′) and Ku70-9 (5′-TGAGCCTTGAGTGACTGAGCAAT-3′) according to the manufacturer’s protocol. Ku70 and Ku80 cDNAs were synthesized from total leaf RNA using Superscript II reverse transcriptase (Gibco): 5 pmol of Ku70-4 (5′-AAACTCCACCATAAAGTTAGAGCCAGAT-3′) or Ku80-2 (5′-GAA CAAAAAGGGTTGTTTCTGA-3′) primers were incubated with 2 µg of total RNA in the supplied buffer at 65°C for 5 min. Reverse transcription was carried with 100 U of Superscript II at 42°C for 50 min. RNA was degraded with RNase H (USB). The coding regions of KU70 and KU80 were amplified with Ex-Taq polymerase (Takara) from the primer pairs: Ku70-12 (5′-CATATGGCTAGCATGGAATTGGACCCAGATGATGT-3′)/Ku70–6 (5′-CTTTTGTAGCTGAGGCAAAACGAT-3′) and Ku80-1 (5′-TCAGGTTCCATGGCACGAAATCG GGAGGGTT-3′)/Ku80-3 (5′-CGGTACCGTTGTCGACAACACAGTTACAA-3′). PCR products were cloned into a pCR2.1-TOPO vector (Invitrogen).
Expression analysis of Ku70 and Ku80
A 15 µg aliquot of total RNA was separated in a 1.2% formaldehyde gel and blotted onto a nylon membrane (Sambrook and Russell, 2001). Hybridization with 32P-labeled cDNA probes labeled by random priming was carried out as described (Church and Gilbert, 1984). Expression analysis of the regions flanking the T-DNA insertion in the KU70 gene was performed using 1 µg of total RNA and the Access RT–PCR system (Promega) (Fitzgerald et al., 1999). The following primer sets were used: Ku70-12/Ku70-9 specific for the N-terminus; Ku70-15 (5′-CTGTGGAAGAGCTATCCCAAGTA-3′)/Ku70-5 (5′-ACGTCGCATTAGAGCTGATGCTT-3′) for the C-terminus and Ku70-7 (5′-GGGAGAATTCCCTTTATAGT-3′) and Ku70-11 (5′-ACTTGGGATAGCTCTTCCACAGTA-3′) flanking the T-DNA insertion (see Figure 3A).
Dimerization assay
Full-length KU80 cDNA was inserted into the BamHI–SalI restriction sites in the expression vector pET28a (Novagen) to produce a Ku80 fusion protein with an N-terminal His-T7 tag. KU70 cDNA was cloned in either the NheI–HindIII or BamHI–HindIII restriction sites of pET28a, resulting in His-Ku70 or His-T7-Ku70 fusions, respectively. All three constructs and the control vector encoding untagged luciferase were transcribed and translated separately in a TnT-coupled rabbit reticulocyte lysate (Promega) with or without [35S]l-methionine (Amersham). Translation was stopped with cycloheximide (4 ng/µl) before 35S-labeled proteins were mixed with T7-tagged unlabeled proteins in a ratio of 3:1 and incubated at 30°C for 15 min. Proteins were immunoprecipitated with T7 antibodies as described (Bryan et al., 2000). Precipitate and supernatant fractions were analyzed by SDS–PAGE and autoradiography.
Identification of a ku70 mutant
The α population of T-DNA insertion lines at the University of Wisconsin Arabidopsis Knock-out Facility was screened using primers Ku70-1 (5′-ACAACCACTGGCTTATTGACAGCTTTGTT-3′) and Ku70-4 according to a protocol available at http://www.biotech.wisc.edu/Arabidopsis/. The progeny of a plant heterozygous for a T-DNA insertion in KU70 were genotyped by PCR using the primers Ku70-2 (5′-TACTACACC AGACAAAGCCGTGATGGTT-3′), Ku70-11 and LB-CD6 (5′-GAA CATCGGTCTCAATGCA-3′) (Fitzgerald et al., 1999). A 939 bp product was generated from the wild-type allele and a 639 bp product from the disrupted allele. Plants homozygous for the ku70 disruption (referred to as G1) were self-pollinated to obtain second-generation mutants (G2).
Telomere analysis
DNA from individual plants was extracted as described (Cocciolone and Cone, 1993). TRF analysis was performed with Tru9I (Promega) restriction enzyme and 32P 5′ end-labeled (T3AG3)4 oligonucleotide as a probe (Fitzgerald et al., 1999). Single telomere analysis for the right arm of chromosome 2 (2R) was performed as follows: 1 µg of genomic DNA was digested with PvuII, and DNA was separated by electrophoresis in a 0.7% agarose gel and blotted onto a nylon membrane. A DNA sequence derived from telomere-adjacent region 2R was used as a probe for hybridization (Riha et al., 2001). Anaphase spreads were prepared from pistils as described previously (Riha et al., 2001).
Complementation analysis
Two complementation constructs were prepared. First, a 6.7 kb genomic DNA fragment of the KU70 gene containing 1.6 kb of putative promoter sequence was amplified by PCR using primers Ku18 (5′-GCCTTT CACCACGTGCATCAT-3′) and Ku21 (5′-ACGTTCTCAGGGATC CTTCAA-3′), cloned into a pDR PCR cloning vector (Qiagen) and then subcloned into a binary vector pCB302 (Xiang et al., 1999). The second complementation construct was prepared by transferring a Ku70 cDNA fragment (see above) into a binary vector, pCBK05 (K.Riha and D.E.Shippen, unpublished data), to allow expression from a 35S CaMV promoter. Both vectors carry the bar gene as a selectable marker. Complementation vectors were introduced in Agrobacterium tumefaciens strain GV3101. Plants heterozygous for the T-DNA disruption in KU70 were transformed by the in planta method (Bechtold et al., 1993). T1 transformants were selected on 0.5 BM medium supplemented with 20 mg/l of phosphinotricine (Crescent Chemical), genotyped and analyzed for Ku70 mRNA expression and telomere length as described above.
Acknowledgments
Acknowledgements
We thank Michael Walker for help in irradiating plants, the Kazusa DNA Research Institute for providing the AtKU70 cDNA clone, and the Arabidopsis Knock-out Facility for screening T-DNA insertion pools. We are also indebted to Jeff Chen, Tom McKnight and Jeff Kapler for insightful comments on the manuscript. This work was supported by an NSF grant (MCB9982499) to D.E.S.
References
- Ahmed S. and Hodgkin,J. (2000) MRT-2 checkpoint protein is required for germline immortality and telomere replication in C.elegans. Nature, 403, 159–164. [DOI] [PubMed] [Google Scholar]
- Aravind L. and Koonin,E.V. (2000) SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci., 25, 112–114. [DOI] [PubMed] [Google Scholar]
- Asamizu E., Nakamura,Y., Sato,S. and Tabata,S. (2000) A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res., 7, 175–180. [DOI] [PubMed] [Google Scholar]
- Bailey S.M., Meyne,J., Chen,D.J., Kurimasa,A., Li,G.C., Lehnert,B.E. and Goodwin,E.H. (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl Acad. Sci. USA, 96, 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. and Cech,T.R. (2000) Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell, 11, 3265–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Ellis,J. and Pelletier,G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. Paris, Life Sci., 316, 1194–1199. [Google Scholar]
- Boulton S.J. and Jackson,S.P. (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J., 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt A.B. (1999) Molecular genetics of DNA repair in higher plants. Trends Plant Sci., 4, 20–25. [DOI] [PubMed] [Google Scholar]
- Bryan T.M., Goodrich,K.J. and Cech,T.R. (2000) A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem., 275, 24199–24207. [DOI] [PubMed] [Google Scholar]
- Cai Q.Q., Plet,A., Imbert,J., Lafage-Pochitaloff,M., Cerdan,C. and Blanchard,J.M. (1994) Chromosomal location and expression of the genes coding for Ku p70 and p80 in human cell lines and normal tissues. Cytogenet. Cell Genet., 65, 221–227. [DOI] [PubMed] [Google Scholar]
- Cary R.B., Chen,F., Shen,Z. and Chen,D.J. (1998) A central region of Ku80 mediates interaction with Ku70 in vivo. Nucleic Acids Res., 26, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocciolone S.M. and Cone,K.C. (1993) Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics, 135, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. (2000) Mammalian telomeres and telomerase. Curr. Opin. Cell Biol. 12, 378–383. [DOI] [PubMed] [Google Scholar]
- Critchlow S.E. and Jackson,S.P. (1998) DNA end-joining: from yeast to man. Trends Biochem. Sci., 23, 394–398. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Hande,M.P., Tong,W., Roth,D., Lansdorp,P.M., Wang,Z. and Jackson,S.P. (2001) Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol., 11, 1192–1196. [DOI] [PubMed] [Google Scholar]
- Driller L., Wellinger,R.J., Larrivee,M., Kremmer,E., Jaklin,S. and Feldmann,H.M. (2000) A short C-terminal domain of Yku70p is essential for telomere maintenance. J. Biol. Chem., 275, 24921–24927. [DOI] [PubMed] [Google Scholar]
- Essers J., van Steeg,H., de Wit,J., Swagemakers,S.M., Vermeij,M., Hoeijmakers,J.H. and Kanaar,R. (2000) Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J., 19, 1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone C. and Jackson,S.P. (1999) Ku, a DNA repair protein with multiple cellular functions? Mutat. Res., 434, 3–15. [DOI] [PubMed] [Google Scholar]
- Fellerhoff B., Eckardt-Schupp,F. and Friedl,A.A. (2000) Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics, 154, 1039–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M.S., Riha,K., Gao,F., Ren,S., McKnight,T.D. and Shippen,D.E. (1999) Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA, 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M.E. and White,C.I. (2001) RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl Acad. Sci. USA, 98, 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M.E., Jeanneau,M., Granier,F., Bouchez,D., Bechtold,N. and White,C.I. (2001) Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J., 25, 31–41. [DOI] [PubMed] [Google Scholar]
- Glaab W.E., Tindall,K.R. and Skopek,T.R. (1999) Specificity of mutations induced by methyl methanesulfonate in mismatch repair-deficient human cancer cell lines. Mutat. Res., 427, 67–78. [DOI] [PubMed] [Google Scholar]
- Grandin N., Damon,C. and Charbonneau,M. (2000) Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol., 20, 8397–8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- Hartung F. and Puchta,H. (2000) Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res., 28, 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig R. and Speit,G. (1997) DNA effects in repair-deficient V79 Chinese hamster cells studied with the comet assay. Mutat. Res., 377, 279–286. [DOI] [PubMed] [Google Scholar]
- Hiom K. (1999) DNA repair: Rad52—the means to an end. Curr. Biol., 9, 446–448. [DOI] [PubMed] [Google Scholar]
- Hsu H.L. et al. (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev., 14, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A., Salomon,S. and Puchta,H. (2000) Species-specific double-strand break repair and genome evolution in plants. EMBO J., 19, 5562–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M., Matsuda,Y., Mimori,T., Harada,Y.N., Shiomi,N. and Shiomi,T. (1996) Chromosomal localization of the mouse and rat DNA double-strand break repair genes Ku p70 and Ku p80/XRCC5 and their mRNA expression in various mouse tissues. Genomics, 38, 38–44. [DOI] [PubMed] [Google Scholar]
- Kooistra R., Pastink,A., Zonneveld,J.B., Lohman,P.H. and Eeken,J.C. (1999) The Drosophila melanogaster DmRAD54 gene plays a crucial role in double-strand break repair after P-element excision and acts synergistically with Ku70 in the repair of X-ray damage. Mol. Cell. Biol., 19, 6269–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P.J., Young,J.C. and Sussman,M.R. (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell, 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T., Martin,S.G., Gotta,M., Gorham,H.C., Pryde,F.E., Louis,E.J. and Gasser,S.M. (1998) Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol., 8, 653–656. [DOI] [PubMed] [Google Scholar]
- Mages G.J., Feldmann,H.M. and Winnacker,E.L. (1996) Involvement of the Saccharomyces cerevisiae HDF1 gene in DNA double-strand break repair and recombination. J. Biol. Chem., 271, 7910–7915. [DOI] [PubMed] [Google Scholar]
- Manolis K.G., Nimmo,E.R., Hartsuiker,E., Carr,A.M., Jeggo,P.A. and Allshire,R.C. (2001) Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J., 20, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marians K.J. (2000) Replication and recombination intersect. Curr. Opin. Genet. Dev., 10, 151–156. [DOI] [PubMed] [Google Scholar]
- Mathur J. and Koncz,C. (1998) Callus culture and regeneration. In Martinez-Zapater,J. and Salinas,J. (eds), Arabidopsis Protocols. Humana Press Inc., Totowa, NJ, Vol. 82, pp. 31–34. [DOI] [PubMed]
- Mengiste T. and Paszkowski,J. (1999) Prospects for the precise engineering of plant genomes by homologous recombination. Biol. Chem., 380, 749–758. [DOI] [PubMed] [Google Scholar]
- Mengiste T., Revenkova,E., Bechtold,N. and Paszkowski,J. (1999) An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J., 18, 4505–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. (2000) Replication fork arrest and DNA recombination. Trends Biochem. Sci., 25, 173–178. [DOI] [PubMed] [Google Scholar]
- Nugent C.I. and Lundblad,V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev. 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Bosco,G., Ross,L.O., Evans,S.K., Salinger,A.P., Moore,J.K., Haber,J.E. and Lundblad,V. (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol., 8, 657–660. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A., Chen,C., da Costa Soares,V., Sanchez,M., Sokol,K., Nussenzweig,M.C. and Li,G.C. (1996) Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature, 382, 551–555. [DOI] [PubMed] [Google Scholar]
- Osipovich O., Durum,S.K. and Muegge,K. (1997) Defining the minimal domain of Ku80 for interaction with Ku70. J. Biol. Chem., 272, 27259–27265. [DOI] [PubMed] [Google Scholar]
- Peterson S.E., Stellwagen,A.E., Diede,S.J., Singer,M.S., Haimberger,Z.W., Johnson,C.O., Tzoneva,M. and Gottschling,D.E. (2001) The function of a stem–loop in telomerase RNA is linked to the DNA repair protein Ku. Nature Genet., 27, 64–67. [DOI] [PubMed] [Google Scholar]
- Polotnianka R.M., Li,J. and Lustig,A.J. (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol., 8, 831–834. [DOI] [PubMed] [Google Scholar]
- Ranganathan V. et al. (2001) Rescue of a telomere length defect of Nijmegen breakage syndrome cells requires NBS and telomerase catalytic subunit. Curr. Biol., 11, 962–966. [DOI] [PubMed] [Google Scholar]
- Riha K., McKnight,T.D., Griffing,L.R. and Shippen,D.E. (2001) Living with genome instability: plant responses to telomere dysfunction. Science, 291, 1797–800. [DOI] [PubMed] [Google Scholar]
- Sambrook J. and Russell,D.W. (2001) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Samper E., Goytisolo,F.A., Slijepcevic,P., van Buul,P.P. and Blasco,M.A. (2000) Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO rep., 1, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L.L. and Zakian,V.A. (1993) Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell, 75, 729–739. [DOI] [PubMed] [Google Scholar]
- Signon L., Malkova,A., Naylor,M.L., Klein,H. and Haber,J.E. (2001) Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol., 21, 2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., van Steensel,B., Bianchi,A., Oelmann,S., Schaefer,M.R., Schnapp,G. and deLange,T. (2000) Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol., 20, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Jung,D., Jung,Y., Lee,S.G. and Lee,I. (2000) Interaction of human Ku70 with TRF2. FEBS Lett., 481, 81–85. [DOI] [PubMed] [Google Scholar]
- Takata M., Sasaki,M.S., Sonoda,E., Morrison,C., Hashimoto,M., Utsumi,H., Yamaguchi-Iwai,Y., Shinohara,A. and Takeda,S. (1998) Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J., 17, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L.H. and Schild,D. (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res., 477, 131–153. [DOI] [PubMed] [Google Scholar]
- Ulm R., Revenkova,E., di Sansebastiano,G., Bechtold,N. and Paszkowski,J. (2001) Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev., 15, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska,A. and deLange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Vogel H., Lim,D.S., Karsenty,G., Finegold,M. and Hasty,P. (1999) Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl Acad. Sci. USA, 96, 10770–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarx E.J., Mitchell,H.L., Karthikeyan,R., Chatterjee,I. and Kunz,B.A. (1998) DNA repair in higher plants. Mutat. Res., 400, 187–200. [DOI] [PubMed] [Google Scholar]
- Walker J.R., Corpina,R.A. and Goldberg,J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature, 412, 607–614. [DOI] [PubMed] [Google Scholar]
- West C.E., Waterworth,W.M., Jiang,Q. and Bray,C.M. (2000) Arabidopsis DNA ligase IV is induced by γ-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J., 24, 67–78. [DOI] [PubMed] [Google Scholar]
- Whittle C.A., Beardmore,T. and Johnston,M.O. (2001) Is G1 arrest in plant seeds induced by a p53-related pathway? Trends Plant Sci., 6, 248–251. [DOI] [PubMed] [Google Scholar]
- Xiang C., Han,P., Lutziger,I., Wang,K. and Oliver,D.J. (1999) A mini binary vector series for plant transformation. Plant Mol. Biol., 40, 711–177. [DOI] [PubMed] [Google Scholar]
- Zentgraf U., Hinderhofer,K. and Kolb,D. (2000) Specific association of a small protein with the telomeric DNA–protein complex during the onset of leaf senescence in Arabidopsis thaliana.Plant Mol. Biol., 42, 429–438. [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Kuster,B., Mann,M., Petrini,J.H. and deLange,T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nature Genet., 25, 347–352. [DOI] [PubMed] [Google Scholar]