Abstract
We have investigated the expression and function of a novel protein encoded by open reading frame (ORF) K7 of Kaposi’s sarcoma-associated herpesvirus (KSHV). Computational analyses revealed that K7 is structurally related to survivin-ΔEx3, a splice variant of human survivin that protects cells from apoptosis by an undefined mechanism. Both K7 and survivin-ΔEx3 contain a mitochondrial-targeting sequence, an N-terminal region of a BIR (baculovirus IAP repeat) domain and a putative BH2 (Bcl-2 homology)-like domain. These suggested that K7 is a new viral anti-apoptotic protein and survivin-ΔEx3 is its likely cellular homologue. We show that K7 is a glycoprotein, which can inhibit apoptosis and anchor to intracellular membranes where Bcl-2 resides. K7 does not associate with Bax, but does bind to Bcl-2 via its putative BH2 domain. In addition, K7 binds to active caspase-3 via its BIR domain and thus inhibits the activity of caspase-3. The BH2 domain of K7 is crucial for the inhibition of caspase-3 activity and is therefore essential for its anti-apoptotic function. Furthermore, K7 bridges Bcl-2 and activated caspase-3 into a protein complex. K7 therefore appears to be an adaptor protein and part of an anti-apoptotic complex that presents effector caspases to Bcl-2, enabling Bcl-2 to inhibit caspase activity. These data also suggest that survivin-ΔEx3 might function by a similar mechanism to that of K7. We denote K7 as vIAP (viral inhibitor-of-apoptosis protein).
Keywords: apoptosis/caspase-3/human survivin/K7/Kaposi’s sarcoma-associated herpesvirus
Introduction
Apoptotic cell death is a major antiviral cellular response to block the propagation of viruses (Tschopp et al., 1998b; Boya et al., 2001). Two apoptotic pathways in mammalian cells are employed to destroy virus-infected cells: the extrinsic death-receptor pathway and the intrinsic mitochondrial pathway. Both pathways converge to activate caspases, in particular the effector caspase-3 (Thornberry and Lazebnik, 1998). In the extrinsic pathway, death receptors belonging to the tumour necrosis factor (TNF) receptor gene superfamily, such as Fas/CD95 or TNFR1, utilize protein interaction modules to assemble receptor-signalling complexes that recruit and activate procaspases 8 and 10 (Ashkenazi and Dixit, 1998; Wallach et al., 1999). Activated caspase-8 and -10 then activate a cascade of caspases, which finally results in the activation of downstream effector caspases (such as caspase-3, -6 and -7), resulting in dismantling and removal of the cell (Thornberry and Lazebnik, 1998; Hengartner, 2000). This is one of the pathways activated by cytotoxic T-lymphocytes to kill virus-infected cells (Harty et al., 2000; Guidotti and Chisari, 2001).
The intrinsic protection mechanism involves the participation of mitochondria, which release caspase-activating proteins. The transcriptional activation of Bcl-2 homology domain 3 (BH3) containing pro-apoptotic proteins of the Bcl-2 family (such as Bax) results in alternations in mitochondrial membrane potential, leading to the release of pro-apoptotic molecules such as cytochrome c (Adams and Cory, 1998; Green and Reed, 1998). Cytochrome c activates caspases by binding to and activating Apaf-1, inducing it to associate with procaspase-9, thereby triggering procaspase-9 activation and initiating effector caspases. When an infected cell senses the unscheduled activation of the cell cycle by viral proteins or DNA damage induced by viral replication, the cell initiates this intrinsic pathway (Evan and Littlewood, 1998; Vousden, 2000).
The activation of effector caspases by either pathway is blocked by cytoplasmic proteins, the inhibitor-of-apoptosis proteins (IAPs). IAPs contain at least one BIR (baculovirus IAP repeat) domain that binds to procaspase-9 and activated effector caspase-3 and -7 to inhibit their activity (Deveraux and Reed, 1999; Miller, 1999). The BIR core includes a C2HC motif chelating a zinc ion that plays an essential role in inhibiting apoptosis (Deveraux et al., 1997; Roy et al., 1997; Vucic et al., 1998; Hinds et al., 1999). IAPs can also suppress apoptosis through caspase-independent mechanisms, which involve transcription factors such as NF-κB (Chu et al., 1997; You et al., 1997). The mammalian prototype of the IAP family is survivin, which is the smallest known IAP family protein (∼16 KDa) and contains a single BIR domain with which it binds caspases and prevents caspase-induced apoptosis (Tamm et al., 1998; Verhagen et al., 2001). Altered expression of survivin appears to be a common event associated with the pathogenesis of human cancer; survivin is overexpressed in many transformed cell lines and in common cancers, such as those of the lung, colon, liver, prostate and breast (Ambrosini et al., 1997; Ito et al., 2000). Reduced survivin expression causes apoptosis and sensitization to anticancer drugs, suggesting that survivin expression is important for cell survival or chemoresistance of certain carcinomas (Ambrosini et al., 1998; Olie et al., 2000; Satyamoorthy et al., 2001).
An alternative anti-apoptotic mechanism is mediated by members of the Bcl-2 anti-apoptotic family, which possess two conserved motifs known as Bcl-2 homology domains 1 and 2 (BH1 and BH2) (Adams and Cory, 1998; Chao and Korsmeyer, 1998). Coalescence of the α-helices in the BH1 and BH2 regions create an elongated hydrophobic cleft to form homo- or heterodimers with other proteins in this family (Muchmore et al., 1996; Sattler et al., 1997). The E1B-19K protein of adenovirus is the only known protein of this family that contains only a BH1, but no BH2 domain (Adams and Cory, 1998). However, no protein with only a BH2 domain has yet been described. Bcl-2 family members act by forming or controlling pores on the outer membrane of mitochondria, through which the release of cytochrome c and other intermembrane pro-apoptotic proteins is regulated. In addition, the anti-apoptotic Bcl-2 members might function directly to regulate caspase activities by binding to an adaptor molecule (Hengartner, 2000). These adaptors either bridge Bcl-2/Bcl-XL and Apaf-1, thereby inhibiting Apaf-1 activation, or bind procaspase-8 to Bcl-2/Bcl-XL, preventing its activation (Ng et al., 1997; Chau et al., 2000; Zhang et al., 2000). No effector caspase-binding adaptor has yet been identified.
Viruses employ an arsenal of proteins to interfere with pro-apoptotic signalling pathways, affording a selective advantage for them to maintain a persistent infection or to prolong the survival of lytically infected cells, allowing maximum virus progeny production (O’Brien, 1998; Tschopp et al., 1998b). Many viruses express anti-apoptotic proteins, including IAP caspase inhibitors, Bcl-2 homologues and death-effector-domain-containing proteins termed vFLIPs [viral FLICE (Fas-associated death-domain-like IL-1 β-converting enzyme)-inhibitory proteins]. The degenerated caspase homologue vFLIP blocks the death-receptor pathway and the viral Bcl-2 homologues protect cells from death by maintaining mitochondrial integrity (Tschopp et al., 1998a; Boya et al., 2001). Studies of viral apoptosis inhibitors provide a valuable insight into the biochemical processes involved in the execution of cell death (Mosialos et al., 1995; O’Brien, 1998). Viral FLIPs led to the identification of cellular homologues (cFLIPs) (Irmler et al., 1997), while the BIR-containing caspase inhibitor (p35) of baculovirus guided the identification of the mammalian IAP family (Seshagiri et al., 1999).
KSHV (Kaposi’s sarcoma-associated herpesvirus; human herpesvirus-8) is an oncogenic virus closely associated with the pathogenesis of Kaposi’s sarcoma and certain lymphoproliferations such as primary effusion lymphoma (PEL) and multicentric Castleman’s disease (Boshoff and Weiss, 2001; Moore and Chang, 2001). Two KSHV anti-apoptotic proteins have been identified: the vBcl-2 encoded by open reading frame (ORF) 16 and the vFLIP encoded by ORF K13. However, no IAP homologue has yet been identified in KSHV or any other known herpesvirus. The K7 protein of KSHV is encoded by the putative ORF K7, which is unique to KSHV. K7 has no obvious homologue in other γ2 herpesviruses, including the close relative rhesus rhadinovirus (McGeoch and Davison, 1999; Alexander et al., 2000). This suggests that KSHV may have acquired this specific gene from its human host genome relatively recently. Here we report that the K7 protein is structurally and functionally related to a recently identified spliced variant of human survivin, survivin-ΔEx3 (Mahotka et al., 1999; Krieg et al., 2002). Both proteins protect cells from apoptosis and possess similar functional domains, therefore K7 is a new viral anti-apoptotic protein and survivin-ΔEx3 appears to be its cellular homologue.
Results
K7 similarity to human survivin-ΔEx3
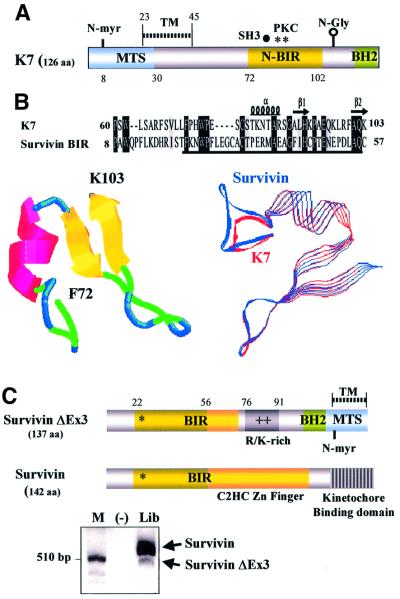
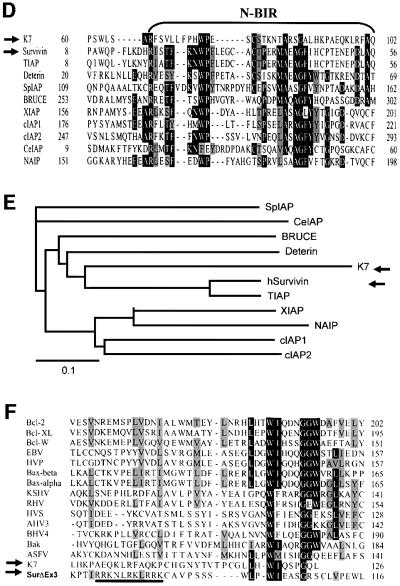
By searching the public databases, K7 was found to harbour a plant-form mitochondrial-targeting signal (MTS) and a transmembrane domain (Figure 1A), which suggested that K7 might be a mitochondrial protein. Several putative post-translational modification sites were found in K7, including an N-glycosylation site, a myristoylation site and several phosphorylation sites (Figure 1A). Hydropathy plots predicted K7 to be a single-transmembrane protein that is highly hydrophobic in the N-terminus (data not shown). Furthermore, by using different fragments of K7 protein as bait probe, we identified a BIR domain in K7 (Figure 1B). The BIR domain is essential and sufficient for IAPs to protect cells from apoptosis (Vucic et al., 1998). The IAP family member most similar to K7 is human survivin (E = 0.014; Figure 1B, upper panel). Sequence alignment and phylogenic analyses of BIR domains also revealed that K7 and survivin display the greatest similarity (Figure 1D and E). The K7 and survivin BIR domains are also comparable in three-dimensional (3D) structure predictions (Figure 1B) (Chantalat et al., 2000; Muchmore et al., 2000; Verdecia et al., 2000). However, K7 possesses the N-terminal half of a BIR motif and lacks the C-terminal-half C2HC Zn-binding fold of other BIR domains, and in this respect K7 is most similar to a naturally occurring splice variant of survivin, survivin-ΔEx3 (Figure 1C), which has been shown to be expressed in cancer cells and to be a functional anti-apoptotic protein (Mahotka et al., 1999; Krieg et al., 2002).
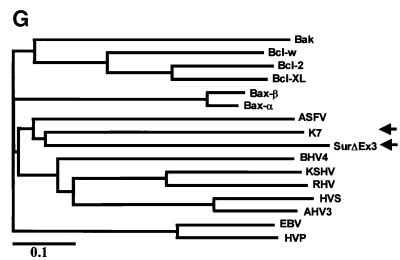
Fig. 1. K7 has sequence and predictive structural similarities to survivin-ΔEx3. (A) Schematic representation of the structure of K7. The MTS, the transmembrane (TM) domain, the BIR-like domain (N-BIR) and the putative BH2 domain of K7 are indicated. Several putative post-translational modification sites of K7, such as the N-myristoylation site (N-myr), SH3 motif, protein kinase C phosphorylation sites (PKC) and the N-glycosylation site (N-Gly), are also shown. (B). Comparison between K7 and survivin BIR domains. Sequence alignment of K7 and survivin BIR domains. (Upper panel) The BIR domain of survivin is underlined. (Bottom left) Ribbon representation of the K7 BIR domain. The α-helices, β-strands and turns are represented as red coils, yellow arrows and blue loops, respectively, and the corresponding amino acids are shown in the upper panel. (Bottom right) Superimposed image of K7 and survivin BIR domains. K7 is represented by red strands and survivin by blue strands. (C) Endogenous expression of survivin and survivin-ΔEx3, and the structures of these two isoforms. The kinetochore-binding domain of survivin, which is essential for chromosome segregation and cytokinesis, is indicated. R/K-rich, Arg/Lys-rich region. PCR analysis for cDNAs of survivin isoforms in a human fetal cDNA library is also shown (bottom panel). (D) Alignment of different BIR domains. The species designations used for the alignment, and the primary accession number or database entry number for each sequence are as follows, with the indicated accession numbers: K7, AAC57096. Human BIR domain proteins: survivin, 2315863; XIAP (X-linked IAP), P98170; cIAP1, NP_001157; cIAP2, NP_001156. Mouse: BRUCE (BIR repeat containing ubiquitin-conjugating enzyme), CAA76720; TIAP, BAA28266; NAIP (neuronal apoptosis inhibitory protein), AAB69223. Drosophila: Deterin, XP_081836. Schizosaccharomyces pombe BIR protein SpIAP, NP_587866. Caenorhabditis elegans BIR protein CeIAP, NP_505949. Numbers to the left of the sequences indicate the positions of the amino acids in each protein. Residues conserved in the majority of the sequences are in black boxes, whereas similar sequences are in grey. (E) Phylogenic relationship of different BIR domains. The alignment data shown in (D) was used to construct a Phylogenic tree, which was derived by maximum likelihood searching by the TreeView program. The divergence scale of the across-page branches is indicated. (F) Alignment of the BH2 domain of Bcl-2 family members with K7 and survivin-ΔEx3. The following human and viral proteins are used in this alignment: Bax-β (AAA03620), Bax-α (AAA03619), Bcl-2 (AAA35591), Bcl-XL (CAA80661), Bcl-W (NP_004041), EBV (Epstein–Barr virus Bcl-2; CAA01638), HVP (herpesvirus papio Bcl-2; AAF99596), KSHV (KSHV Bcl-2; AAB62596), RHV (Rhesus herpesvirus Bcl-2; AAD21342), HVS (herpesvirus simiri Bcl-2; CAA45639), AHV3 (Ateline herpesvirus 3 Bcl-2; NP_047987), BHV4 (bovine herpesvirus 4 Bcl-2; AAD34361), Bak (Q16611), and ASFV (African swine fever virus Bcl-2; NP_042735). The R/K-rich region in survivin-ΔEx3 is underlined. (G) Phylogenic tree for BH2 domain-containing apoptotic regulators. The same BH2 domains shown in Figure 13F were used to construct this tree.
survivin-ΔEx3 mRNA is a product of alternative splicing and lacks the exon 3 of survivin (Mahotka et al., 1999). The skipping of exon 3 in survivin-ΔEx3 mRNA results in an interrupted BIR domain followed by an additional frame shift in exon 4. Consequently, the survivin-ΔEx3 ORF ends in the 3′ untranslated region of survivin and generates a novel 63-amino-acid-long C-terminal tail. survivin-ΔEx3 has been shown to be present in cancer tissues (Mahotka et al., 1999; Krieg et al., 2002), and when we attempted to clone survivin from a human fetal tissue library, we amplified an additional smaller product (Figure 1C). Direct sequencing of this amplicon revealed that the sequences of survivin-ΔEx3 and survivin in the fetal tissue library are identical to those described in cancer cells (data not shown). To gain further insight into survivin-ΔEx3, we performed protein domain analysis and secondary structure predictions. We found that in the new C-terminus of survivin-ΔEx3, a transmembrane domain and a novel potential plant-form MTS are created, similar to the N-terminal part of K7 (Figure 1A and C). In addition, there is a putative BH2 domain on both K7 and survivin-ΔEx3 (Figure 1A and C). Comparison of the BH2 domain sequences of K7 and survivin-ΔEx3 with other known Bcl-2 family members indicated that they share limited similarity to the BH2 domains of known apoptotic regulators (Figure 1F), and that both proteins clustered together in phylogenetic analysis (Figure 1G). Neither K7 nor survivin-ΔEx3 has a BH1 domain, which normally cooperates with the BH2 domain for anti-apoptotic function (Adams and Cory, 1998; Chao and Korsmeyer, 1998). The highly structural similarities between K7 and human survivin-ΔEx3, together with the finding of putative BH2 and BIR domains on K7 (Figure 1A and C), suggest that K7 is an anti-apoptotic protein.
Expression of K7 gene in PEL cells
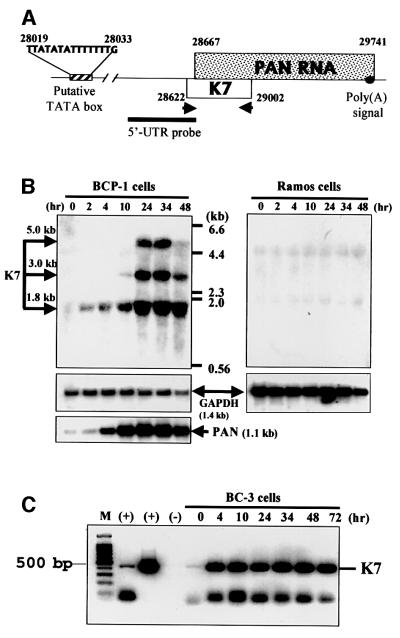
Microarray analyses have shown previously that K7 is either expressed in latency or is highly inducible upon n-butyrate induction of KSHV latently infected lymphoma cells (Jenner et al., 2001; Paulose-Murphy et al., 2001). However, the probes used in these array studies could have cross-hybridized with the PAN (polyadenylated nuclear) RNA gene, which is highly expressed in the virus lytic cycle and overlaps with the K7 ORF (Figure 2A). We performed northern blot analysis to detect K7-specific RNA in KSHV-positive PEL cell lines by using a K7-specific probe (5′-UTR probe; Figure 2A). Three K7-specific RNAs were identified in KSHV-positive BCP-1 PEL cells (Figure 2B, left) and two K7-specific transcripts in another PEL cell line BC-3 (1.8- and 5-kb species; data not shown). These RNA species could not be detected in KSHV-negative B-cell lines such as Ramos (Figure 2B, right) and DG75 cells (not shown). The 1.8-kb RNA transcript was detectable within 2 h after 12-O-tetradecaylphorbol 13-acetate (TPA) induction, followed by the 3 and 5 kb RNA species (Figure 2B). These RNAs increased with time and reached a peak 24 h post-treatment (Figure 2B). To control for loading and integrity of the mRNA, the same blots were re-probed with GAPDH (Figure 2B). Hybridizing the same blots with a probe to a KSHV lytic gene product, PAN (Sun et al., 1996, 1999), confirmed successful induction of the viral lytic cycle (Figure 2B).
Fig. 2. K7 is a lytic viral gene. (A) Schematic representation of the structure of the K7 gene. The cDNA probe used in northern blot analysis and the K7-specific primers used in RT–PCRs are shown. (B) Northern blot analyses of K7 RNAs. The KSHV latently infected PEL cell line BCP-1 and the KSHV-negative Ramos cell line were lytically induced by TPA, and total RNA was extracted at the indicated time points. Blots were first probed against K7 mRNA, then reprobed with a GAPDH probe or with another KSHV gene, PAN, to indicate the induction of viral lytic cycle. (C) RT–PCR analysis for K7 transcripts. RNA from TPA-treated, KSHV-infected PEL cells (BC3) was collected at the indicated time points. M, 100-bp DNA fragment markers. (+), RNA from TPA-treated BCP-1 cells (lane 2) and cDNA of K7 (lane 3) were used as positive controls.
We also used RT–PCR to detect K7 RNA and to confirm that K7 is expressed only during the lytic viral cycle. The sense primer used in PCR is upstream of the PAN gene but within the K7 ORF, thus avoiding amplification of PAN (Figure 2A). In TPA-untreated latent PEL cells, a small amount of K7 RNA could be detected (Figure 2C), and this possibly arises from the 2–3% of lytic cells present in untreated cells (Dupin et al., 1999). We conclude that the K7 gene is a lytic gene expressed in PEL cells.
Expression and characterization of K7 protein in transfected cells
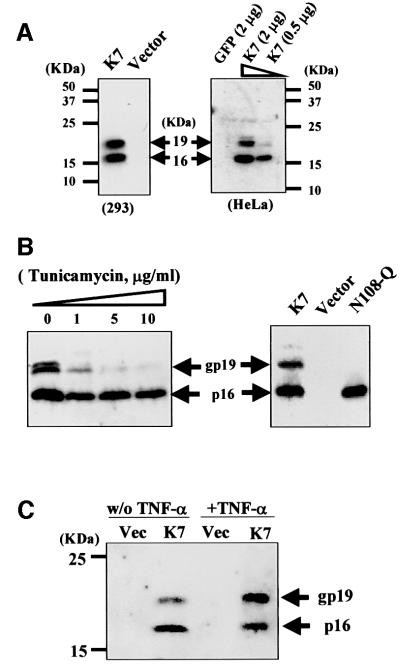
We cloned K7 ORF cDNA from BC3 cDNA library and tagged it at both the N- and C-termini with the influenza virus haemagglutinin (HA) epitope. HeLa and 293 cells were transiently transfected with this construct and western blotting revealed two K7-specific signals (Figure 3A). The predicted molecular mass of tagged K7 is 15.8 kDa, correlating to the 16 kDa band detected (Figure 3A). In addition, a 19 kDa band was also detected (Figure 3A). To determine whether this band was due to post-translational modification, we treated K7-transfected HeLa cells with tunicamycin, a potent inhibitor of N-linked glycosylation, and this led to a dose-dependent loss in the appearance of processed K7 (gp19; Figure 3B, left). Mutation in the N-glycosylation site of K7 (mutant N108Q) totally removed the higher modified form (Figure 3B, right), confirming that the 19 kDa band was due to the N-glycosylation of K7. The N-glycosylation of K7 was enhanced after TNF-α treatment (Figure 3C), further implying an important physiological role of this modification.
Fig. 3. K7 expressed as a predicted 16 kDa protein and as a 19 kDa glycoprotein. Cells were transfected with the indicated plasmids, and protein extracts were collected 48 h after transfection and probed by western blots with an anti-HA mAb. (A) K7 expression pattern. Two different forms of K7 were detected in transfected cells. (B) The 19 kDa form of K7 is a glycoprotein. (Left panel) HeLa cells transfected with K7 were treated with different doses of tunicamycin to inhibit N-glycosylation for 24 h before extraction. (Right panel) A point mutation (N108Q) on the N-glycosylation site of K7 could prevent K7 from modification. (C) The N-glycosylation of K7 is increased after TNF-α treatment. HeLa cells transfected with a K7-expressing plasmid or with vector alone were exposed 24 h post-transfection to 10 ng/ml TNF-α plus 1 µg/ml CHX. (D) K7 has a short half-life. K7-transfected HeLa cells were treated with 10 µg/ml CHX at 48 h post-transfection to block protein synthesis. Cell extracts were collected at indicated time points. (Lower panel) Same blot stained with Coomassie Blue to show equal loading of protein in each lane. (E) K7 forms homodimers. HeLa cell extracts, which contain transfected K7, were incubated with GST, GST-K7-CT or GST-K7-BIR. Specifically bound proteins were eluted from beads with SDS–PAGE sample buffer before analysis.
To determine the half-life of K7, transfected cells were treated with 10 µg/ml of the protein synthesis inhibitor cycloheximide (CHX). At this concentration, new cellular translation stops and only the existing proteins can be detected (Akgul et al., 2000). Forty-eight hours after transfection, the transfected HeLa cells were treated with CHX for the indicated periods before cell extracts were collected and analysed. K7 was undetectable within 2 h of treatment, indicating that K7 is a labile, short half-life protein, similar to some other apoptotic proteins (Figure 3D) (Somia et al., 1999; Akgul et al., 2000; Duriez et al., 2000). We next tested whether K7, like Bcl-2 family members and similar to human survivin (Adams and Cory, 1998; Chantalat et al., 2000; Muchmore et al., 2000), can associate with itself to form homodimers. The C-terminal (CT) 56 amino acids of K7, which contains the putative BIR and BH2 domains, and the BIR domain of K7 were fused to a glutathione S-transferase (GST) protein domain, expressed in Escherichia coli to give GST-K7-CT/GST-K7-BIR recombinant proteins, and then tested for interaction with full-length K7 by in vitro GST pull-down assays. Figure 3E shows that both GST-K7-CT and GST-K7-BIR recombinant proteins precipitate full-length K7.
K7 localizes to mitochondria, endoplasmic reticulum and the nuclear membrane
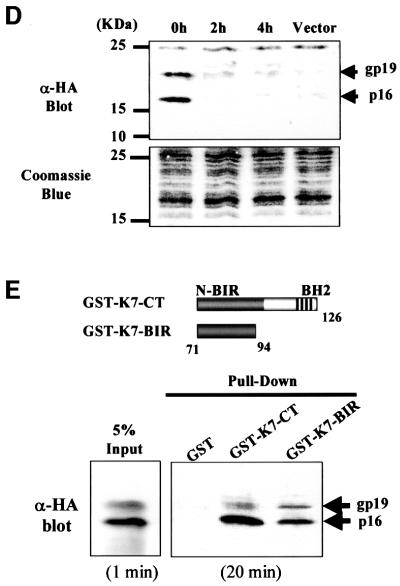
We examined the subcellular localization of K7 in cells by immunofluorescence assay. K7 is a cytosolic protein with a nuclear membrane staining pattern (Figure 4A). Expression of K7 in cells revealed a punctate cytoplasmic and lacy reticular staining pattern (Figure 4A), which is similar to that of mitochondria, but distinct from Golgi, and lysosome staining (data not shown). Staining of K7-expressing cells with the mitochondria-specific dye MitoTracker Red verified that K7 colocalized with mitochondria (Figure 4B). However, not all K7 proteins in cells are distributed to mitochondria; some green K7 signals did not overlap with the red mitochondrial staining (Figure 4B). Cellular fractionation experiments confirmed that K7 was located in mitochondria and nuclear membrane fractions, and also showed that K7 resided in the endoplasmic reticulum (ER) (Figure 4C). In the ER, only the unglycosylated 16 kDa form of K7 was present (Figure 4C). The distribution pattern of K7 was similar to that of Bcl-2, but was distinct from that of Bax (Figure 4C), which is known to reside in the cytoplasm and only translocates to mitochondria in response to death signals (Iwahashi et al., 1997; Wolter et al., 1997; Gross et al., 1998).
Fig. 4. K7 localizes to the mitochondria and ER. (A) Confocal microscopy of HeLa cells transfected with HA-tagged K7. Transfected cells were stained with an anti-HA mAb at 48 h post-transfection. (B) Colocalization of K7 and mitochondria. K7-transfected HeLa cells were stained with antibody to HA (green), and mitochondria were stained with CMXRos (red). The cell nucleus was stained with Hoechst 33258 dye (blue). Partial colocalization is seen upon overlay of these images. (C) Subcellular fractionation of HeLa cells transfected with K7. Transfected cells were separated into nucleoplasmic (NP), cytoplasmic (C), mitochondria (M) and ER fractions. The fractions were probed by western blots with antibodies to HA (for tagged K7), human Bcl-2 and Bax. (D) The N-terminal 70 amino acids of K7 is sufficient and essential for mitochondrial targeting. HeLa cells transfected with the indicated plasmids were subjected to subcellular fractionation analysis. The fractions were probed by western blots with a polyclonal antibody to GST to detect each fusion protein.
Computational analyses suggested a putative mitochondrial-targeting domain in the K7 N-terminus (Figure 1A). We next investigated whether this domain is functional. Full-length K7, as well as the N-terminus (amino acids 1–70) and C-terminus (amino acids 71–126), were fused with a GST protein domain in a eukaryotic expression vector, transfected into cells and then examined for intracellular localization. Immunoblot analysis of subcellular fractions demonstrated that the ectopically expressed GST–K7 fusion protein was located in the mitochondria and ER fractions, whereas the GST protein itself was cytosolic (Figure 4D). The N-terminus of K7 localized GST to the mitochondria and ER (Figure 4D, GST-N70), but the C-terminus did not (Figure 4D, GST-C56).
K7 can protect cells from apoptosis induced by various stimuli
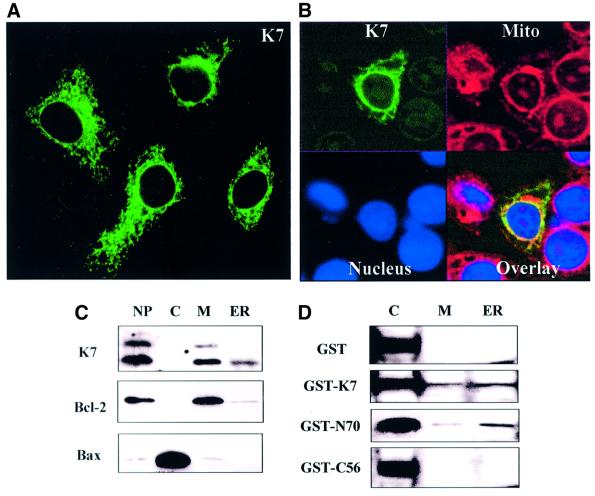
The putative functions of K7 were first explored by transient transfection assays. We tested the cytoprotective function of K7 on apoptosis induced by different stimuli (Figure 5). In Figure 5A, apoptosis was induced by TNF-α or anti-Fas monoclonal antibody (mAb) in the presence of 1 µg/ml of CHX, and apoptotic cells were stained by Annexin V–fluorescein isothiocyanate/propidium iodide (FITC/PI) double staining and counted by flow cytometry. In the presence of K7, apoptosis was reduced to 30–50% (Figure 5A), whereas GFP or LacZ protein did not protect transfected cells from apoptosis (not shown).
Fig. 5. K7 inhibits apoptosis. (A) K7 protects cells from death receptor mediated apoptosis. HeLa cells transfected with a K7-expressing plasmid or with vector alone were exposed 24 h post-transfection to 10 ng/ml TNF-α or 100 ng/ml anti-Fas mAb plus 1 µg/ml CHX. Apoptotic cells were double stained with Annexin V–FITC/PI and counted by flow cytometry. Results are expressed as the mean ± SD of three independent experiments. (B) K7 rescues Bax-induced apoptosis. HeLa cells were transfected with 100 ng of Bax-expression plasmid plus 400 ng (4:1), 900 ng (9:1) or 1800 ng (18:1) of the indicated constructs. Forty-eight hours post-transfection, all cells, both adherent and in suspension, were pooled, washed and stained with the mitochondrial membrane-specific dye CMXRosamine to assess the mitochondrial membrane potential (ΔΨm), and were then analysed by fluorescence activated cell sorting (FACS). Inhibition percentage was calculated as follows: (% apoptosis in Bax-transfected cells) – (% apoptosis in the indicated DNA-cotransfected cells)/(% apoptosis in Bax-transfected cells), where % apoptosis is the percentage of apoptotic cells relative to transfected cells. (C) Both the putative BH2 and the BIR domains of K7 appear to be essential for inhibition of TNF-α induced cell death. HeLa cells transfected with the indicated expression plasmids were treated with TNF-α plus CHX. Twenty-four hours after treatment, cell viability was monitored by PI staining and by FACS counting (top panel). (Bottom panel) The expression of wild or mutant types is shown.
We next tested whether K7 can interfere with Bax-induced apoptosis. HeLa cells were cotransfected with Bax- and K7-expressing plasmids, and transfected cells were analysed by staining with CMXRosamine (CMXRos) to measure loss of mitochondrial membrane potential (ΔΨm), which is an indicator of mitochondrial integrity (Castedo et al., 1996; Marshall et al., 1999; Oda et al., 2000). A decrease in CMXRos staining indicates a loss of mitochondrial integrity, which is mediated by the opening of the mitochondrial permeability transition pores and is an early apoptotic event (Bossy-Wetzel et al., 1998; Green and Reed, 1998). As positive controls, Bcl-2 and Bcl-XL were also cotransfected with Bax (Figure 5B). Coexpressing K7 suppressed Bax-induced apoptosis in a dose-dependent manner that was comparable to Bcl-2 (Figure 5B).
We next generated deletion mutants and examined their anti-apoptotic function by counting cell viability. Transfected cells were treated with TNF-α (Figure 5C) or anti-Fas mAb (data not shown) plus CHX. The deletion of the C-terminal BH2 domain (mutants ΔC11 and ΔC32) or the conserved amino acids in the BIR domain (mutant Δ73–82) resulted in a loss of anti-apoptotic activity (Figure 5C).
K7 bridges Bcl-2 and active caspase-3 to inhibit caspase-3 activity
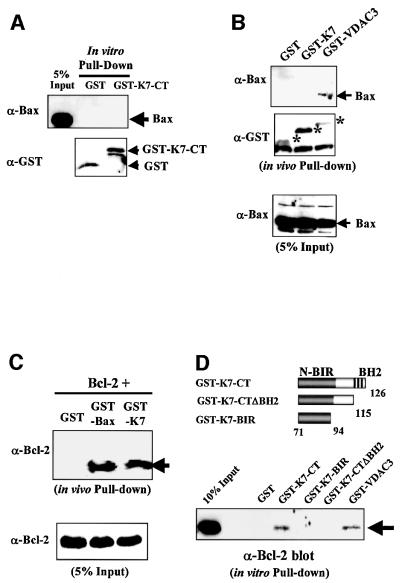
Since K7 could inhibit apoptosis and has a BH2 and a BIR domain, we used in vivo and in vitro GST pull-down experiments to investigate whether K7 interacts with Bcl-2 family members or with caspases (Figure 6). Because K7 could inhibit Bax-induced apoptosis, we tested whether K7 binds to Bax. Neither in vitro nor in vivo binding assays indicated Bax interaction with K7, whereas the positive control VDAC3 (Sampson et al., 1998; Rahmani et al., 2000) did precipitate Bax (Figure 6B).
Fig. 6. K7 interacts with Bcl-2 and activated caspase-3 via its BH2 and BIR domains, respectively. (A) K7 and Bax do not bind in vitro. HeLa cell extracts, which contain endogenous Bax, were incubated with GST or GST-K7-CT recombinant proteins as described in Figure 33E. Specifically retained proteins were blotted with either anti-Bax Ab (upper panel) or anti-GST Ab (bottom panel). (B) Bax does not bind to K7 in vivo. HeLa cells were transfected with Bax plus the indicated expression plasmids and then subjected to GST pull-down assays 48 h post-transfection. Asterisks indicate the expressed GST fusion proteins. (C) K7 heterodimerizes with Bcl-2 in vivo. HeLa cells were cotransfected with equal amounts of Bcl-2 and the indicated plasmids. Transfected cells were harvested and GST pull-down assays were performed to detect coprecipitated Bcl-2. (D) The C-terminal BH2 domain of K7 is essential for Bcl-2 interaction. HeLa cell extracts were subjected to in vitro GST pull-down assays with the indicated K7 mutants. Specifically bound Bcl-2 was detected by an anti-Bcl-2 mAb. The GST–VDAC3 recombinant protein served as a positive control. (E) K7 interacts with active caspase-3. HEK 293 cells were treated for 8 h with 10 ng/ml TNF-α plus 1 µg/ml CHX, and cell lysates were then mixed with the indicated recombinant proteins. Bounded caspase-3 was analysed by an anti-caspase-3 mAb, which recognizes both pro- and activated caspase-3. (F) K7 and active caspase-3 interact in vivo through the BIR domain. Cell extracts were made 48 h post-transfection from HeLa cells cotransfected with Bax and wild-type or BIR mutant of K7. Anti-HA mAb and isotype control were used to pull down K7, followed by western blot analysis with anti-caspase-3 mAb. (G) Bcl-2 precipitates active caspase-3 in the presence of K7. HeLa cells were cotransfected with 15 µg each of plasmids encoding K7 (or empty vector) and Bax. Cell extracts were prepared 2 days later and immunoprecipitations were performed using anti-Bcl-2 mAb. Immune complexes were analysed by western blot with anti-caspase-3 mAb. (H) K7 is an inhibitor of activated caspase-3. Fluorescent substrate (Sub) and recombinant activated caspase-3 (rCas3) were mixed with RIPA buffer or with protein extracts from K7-transfected or non-transfected HeLa cells. Reaction mixtures were incubated for 1 h at 37°C before determinion of fluorescence. A caspase-3 inhibitor was used as a positive control.
When GST–K7 and Bcl-2 were cotransfected into cells, in vivo pull-down of GST–K7 resulted in the coprecipitation of Bcl-2 (Figure 6C). The interaction was specific, as in a parallel reaction, under the same binding conditions, wild-type GST alone failed to pull-down Bcl-2. In this assay, the interaction of K7 with Bcl-2 was as efficient as that of Bax with Bcl-2 (Figure 6C). To map the site of interaction between K7 and Bcl-2, in vitro GST pull-down assays were performed. Bcl-2 bound to the C-terminus of K7 (Figure 6D, GST-K7-CT), and deletion of the BH2 domain of K7 abolished this interaction (GST-K7-CTΔBH2 and GST-K7-BIR).
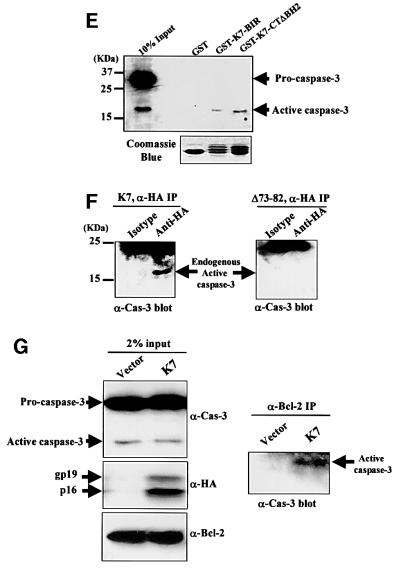
Since the BIR domains on other IAPs have been shown to be able to interact with activated caspase-3 and -7, we tested whether K7 could bind to caspase-3. The K7 BIR domain alone was sufficient to bind to active caspase-3 (Figure 6E, GST-K7-BIR and GST-K7-CTΔBH2). This interaction was specific, since K7 did not pull-down the predominant procaspase-3 (Figure 6E). Further evidence that K7 and active caspase-3 interact in vivo through the BIR domain were provided by in vivo coimmunoprecipitation experiments from HeLa cells cotransfected with Bax and either wild-type or BIR-deleted mutant K7 (Δ73–82; Figure 6F). Cell extracts were immunoprecipitated with anti-HA mAb and blotted for caspase-3; only wild-type K7 could pull down endogenous active caspase-3 (Figure 6F).
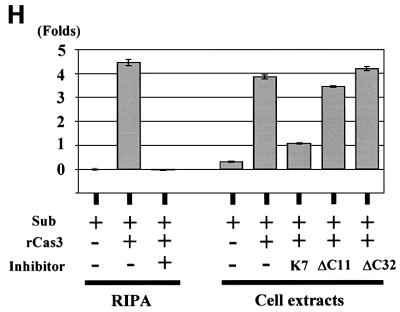
Since K7 is capable of interacting with both Bcl-2 and active caspase-3, we next tested whether K7 could bridge these two proteins together in a complex. Accordingly, K7 was transiently expressed in cells and coimmunoprecipitation was performed using anti-Bcl-2 mAb and K7-positive or -negative cell lysates. Active caspase-3 can be precipitated in the same immune complexes only when K7 is present in lysates (Figure 6G). In an in vitro caspase activity assay, endogenously expressed K7 inhibited the activity of recombinant active caspase-3, and the deletion of the BH2 domain abolished this inhibitory ability (Figure 6H).
Discussion
The recent growth in biological databases and rapid development of novel computational methods have facilitated the identification of the roles of many new genes (Eisenberg et al., 2000; Thornton, 2001). Here we report an example of this, where we used the available online databases and programs to predict and then identify the function of an unknown protein, the K7 protein of KSHV. K7 is one of the few KSHV ORFs that is not present in viruses closely related to KSHV (Albrecht et al., 1992; Russo et al., 1996; Alexander et al., 2000). We therefore hypothesized that K7 was more recently acquired from the host genome than other KSHV genes with cellular homologues, and that K7 should therefore have an identifiable cellular counterpart. Database searches first identified putative BH2-like and BIR domains present in K7 (Figure 1A). Further analyses identified a plant-form MTS, a putative N-myristoylation site and an N-glycosylation site (Figure 1A). Our computational data thus suggested that K7 is an anti-apoptotic protein, which is related in particular to a spliced variant of human survivin, survivin-ΔEx3 (E = 0.014; Figure 1C).
survivin-ΔEx3 is a spliced variant that lacks exon 3 of wild-type survivin; this results in an interrupted BIR domain and generates a frame shift resulting in a novel 63-amino-acid-long C-terminus (Figure 1C) (Mahotka et al., 1999). survivin-ΔEx3 has been shown to be expressed in cancer tissues and to have anti-apoptotic properties, similar to survivin (Mahotka et al., 1999; Krieg et al., 2002). When we used specific primers to amplify the survivin transcript from a human fetal cDNA library, survivin-ΔEx3 was also amplified (confirmed by sequencing) (Figure 1C). survivin-ΔEx3 is therefore not only expressed in certain cancers, but is also expressed in fetal tissues, similar to survivin (Ambrosini et al., 1997; Adida et al., 1998). This protein may therefore be another oncofetal protein detectable in cancers (Warnes and Smith, 1987). Our computational data also showed that survivin-ΔEx3, like KSHV K7, contains a BH2-like domain, a putative N-myristoylation site and a plant-form MTS (Figure 1C). Structural prediction programs and phylogenetic analyses showed that the K7 putative BIR and BH2 domains are most similar to those found in survivin-ΔEx3 (Figure 1B, E and G), further demonstrating the similarity between these two proteins. However, the putative BH2-like domains in K7 and survivin-ΔEx3 show only weak sequence similarity to other known BH2 motifs (Figure 1F).
KSHV encodes an array of cellular homologues, including cyclin, Bcl-2, IL-6, chemokine, FLIP and GPCR (G protein-coupled receptor) homologues (Moore et al., 1996; Russo et al., 1996; Neipel et al., 1998). The mechanisms by which a DNA virus that does not integrate into the host genome acquires cellular genes are not clearly understood. The fact that K7 represents a spliced variant of a cellular gene (survivin) and the observation that the KSHV vFLIP is similar to the smaller spliced variant of cellular FLIP (Irmler et al., 1997) suggests that the virus incorporated cDNA of spliced cellular mRNA, not host genomic DNA, into its genome.
We first tested whether K7 is expressed in KSHV-infected PEL cells during the latent phase or only during the lytic phase of infection. Both northern blot and RT–PCR data indicated that K7 transcripts are present upon lytic induction (Figure 2). Like vBcl-2, K7 may therefore prolong the lytic phase allowing maximum viral progeny production (Sun et al., 1999). Three K7-specific RNAs, of 1.8, 3 and 5 kb in length, could be detected in KSHV-positive PEL cells (Figure 2B). The 1.8 kb K7 RNA is probably transcribed from the putative TATA box upstream of K7 ORF to the polyadenylated signal downstream of the PAN RNA gene (Figure 2A) (Sun et al., 1996). The expression pattern of the 1.8 kb K7 RNA, which appears as early as 2 h post-induction of the lytic cycle, correlates with other immediate-early genes of KSHV (Sun et al., 1999). Furthermore, the existence of multiple K7 transcripts at different time points indicates differential regulation of the transcripts from the K7 gene locus.
In transfected cells, the observed K7 protein is similar in size to the predicted 16 kDa, and an additional 19 kDa protein is also present (Figure 3). Only the 16 kDa form locates to the ER, suggesting that the 19 kDa form is due to post-translational modification occurring at the post-ER stage (Figure 4C). The treatment of transfected cells with tunicamycin, a potent inhibitor for N-glycosylation, and a point mutation in the N-glycosylation site of K7 resulted in the disappearance of the modified protein, confirming that the 19 kDa form of K7 is a glycoprotein (Figure 3B). In cells, N-glycosylation allows the efficient targeting of neoglycoproteins to subcellular organelles, helps protein folding and controls protein degradation (Helenius and Aebi, 2001). The glycosylation of K7 can be enhanced by TNF-α treatment (Figure 3C), indicating an important role of N-glycosylation in the anti-apoptotic function of K7. This modification, together with other modifications such as phosphorylation, might prolong the half-life of K7 or serve as a recognition tag for K7-interacting partners. However, the glycosylation of K7 is not essential for K7 to form homodimers or to form heterodimers with Bcl-2, since the GST-K7-CT/GST-K7-BIR recombinant proteins expressed from E.coli can still interact with K7 or Bcl-2 in HeLa cell extracts (Figures 3E and 6D).
Immunofluorescence assays with a HA-tagged K7 showed predominant mitochondrial localization (Figure 4B). Cell fractionation assays confirmed that K7 localizes not only to mitochondria but also to the ER, similar to the cellular distribution of Bcl-2 (Figure 4C). Deleting the putative MTS of K7 abolished its mitochondrial localization (Figure 4D). Comparing the fractionation data of wild-type and GST–K7 fusion proteins suggests that the GST domain interferes with the localization of K7 to its target organelles (Figure 4C and D). Therefore, in Figure 4D, the mitochondrial targeting by the N70 region or full-length K7 is not as efficient as is seen with the wild-type form of K7 (Figure 4C). However, the N-terminal 70 amino acids of K7 alone is sufficient to bring the GST domain into mitochondria and ER (Figure 4D). Computational analyses revealed that K7 possesses an unusual plant-form MTS. It has been shown that a plant mitochondrial matrix protein, manganese superoxide dismutase, is efficiently imported and correctly processed by yeast mitochondria, suggesting a highly conserved mitochondrial-importing mechanism between plants and yeast (Bowler et al., 1989). Also, the mammalian translocases on the mitochondrial inner membrane show a sequence similarity to a channel-forming amino acid transporter in the outer envelope of chloroplasts, and to LivH, a component of a prokaryotic amino acid permease (Rassow et al., 1999; Rassow and Pfanner, 2000). Thus, some components in the mitochondrial transport system are evolutionarily conserved, and some plant MTSs are still functional in animal cells and vice versa. The human cytomegalovirus (CMV) also encodes a mitochondria-localized anti-apoptotic protein, vMIA (viral mitochondrial inhibitor-of-apoptosis) (Goldmacher et al., 1999). Like K7, vMIA contains a single transmembrane domain and an N-terminal plant-form mitochondria-targeting domain, which is sufficient for mitochondrial targeting (Goldmacher et al., 1999; Hayajneh et al., 2001). Why these viral mitochondrial proteins have plant MTSs are unknown. It might indicate that a selective advantage is obtained with this sequence, making the mitochondrial translocation of these viral proteins more efficient.
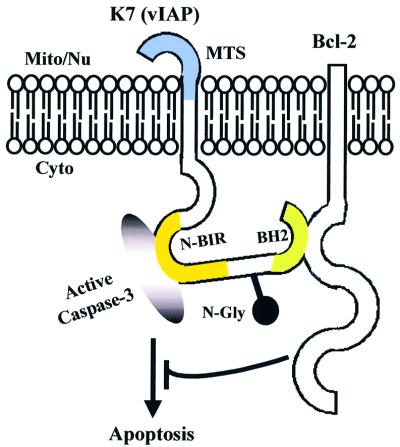
In transfected cells, K7 blocks TNF-α, anti-Fas mAb and Bax-induced apoptosis (Figure 5). This anti-apoptotic function of K7 was abolished when the putative BH2 or BIR domain was deleted (Figure 5C). Mechanistic studies showed that K7 interacts with both Bcl-2 and active caspase-3 (Figure 6C–F). survivin and other mammalian IAPs (XIAP, c-IAP-1 and c-IAP-2) have been shown to directly bind to and inhibit the terminal effectors caspase-3 and -7 in vitro and in vivo (Deveraux et al., 1998; Tamm et al., 1998; Jiang et al., 2001; Shin et al., 2001). However, none of these IAPs has been shown to interact with Bcl-2 family members. The functional BH2 motif on K7 is responsible for the interaction of K7 with Bcl-2 and the BIR domain for caspase-3 interaction (Figure 6C–F). These interactions are specific since K7 does not bind to Bax (Figure 6A and B) or procaspase-3 (Figure 6E). Thus, K7 appears to be a BH2-only protein that can bind to Bcl-2 without a BH1 domain. Although the important C-terminal end of the BH2 domain in Bcl-2 (amino acids 193–202) (Borner et al., 1994; Yin et al., 1994) is absent in K7, and the similarity between the BH2 domain of K7 and other BH2 motifs is not statistically significant (Figure 1F), our data still show that it is essential for Bcl-2 binding (Figure 6C and D). By performing an in vivo coimmunoprecipitation assay, we have further shown that in the presence of K7, endogenous Bcl-2 can pull down endogenous active caspase-3 (Figure 6G). To test whether K7 can interfere directly with caspase-3 activity, we used a cell-free system in which recombinant active caspase-3 was added together with cytosolic extracts derived from control or K7-transfected cells. K7 was able to reduce caspase-3 activity and the deletion of the K7 BH2 domain abolished its caspase-3 inhibitory ability (Figure 6H, ΔC11 and ΔC32), suggesting that the BH2 domain of K7 is not only essential for its anti-apoptotic function but is also crucial for the inhibition of caspase-3 activity. This suggests that K7 is an adaptor protein, linking the Bcl-2 family members directly to the effector caspases, thereby enabling Bcl-2 to inhibit the activities of activated effector caspases (Figure 7).
Fig. 7. Proposed model of K7 (vIAP) function. One function of K7 (vIAP) may be as a molecular adaptor to present activated caspases to anti-apoptotic proteins, such as Bcl-2.
The direct regulation of some caspases by Bcl-2 family proteins via adaptor molecules has been described in Caenorhabditis elegans: CED-9 blocks apoptosis by binding to and neutralizing CED-4 (an adaptor), an essential activator of CED-3 caspase (Frade and Michaelidis, 1997). In mammalian cells, the CED-9 homologues Bcl-2 and Bcl-XL also block apoptosis by interfering with the activation of CED-3-like caspases. Bcl-XL and Boo, a novel anti-apoptotic member of the Bcl-2 family, have been reported to bind to the CED-4 homologue Apaf-1, through which they may inhibit the association of Apaf-1 with procaspase-9 and thereby prevent caspase-9 activation (Hu et al., 1998; Pan et al., 1998; Song et al., 1999). Although some other reports argue about this (Moriishi et al., 1999; Conus et al., 2000; Newmeyer et al., 2000), several adaptor proteins such as BAR (Zhang et al., 2000), Aven (Chau et al., 2000) and the ER-localized protein Bap31 (Ng et al., 1997; Nguyen et al., 2000) have been described to bridge initiator caspases (such as procaspase-8) or Apaf-1 to Bcl-2 family members to prevent apoptosis, and form complexes with Bcl-2/Bcl-XL anti-apoptotic proteins, but not with Bax/Bak pro-apoptotic regulators (Ng et al., 1997; Chau et al., 2000; Zhang et al., 2000). Our data suggest that K7 is the first identified protein linking Bcl-2 to effector caspases, thus acting as an adaptor molecule. Furthermore, because of the structural and functional similarities between K7 and survivin-ΔEx3, survivin-ΔEx3 might be the cellular homologue of K7 and functions as an endogenous adaptor protein.
In adults, the expression of survivin is limited to active proliferating cells such as bone marrow CD34+ precursor cells and proliferating mature endothelial cells (Tran et al., 1999; Fukuda and Pelus, 2001; Mesri et al., 2001). KSHV has been shown to infect CD34+ precursor cells and mature endothelial cells in vivo (Boshoff et al., 1995; Dupin et al., 1999; Henry et al., 1999). In vitro studies also showed that KSHV could infect these two cell types (Flore et al., 1998; Panyutich et al., 1998; Renne et al., 1998; Ciufo et al., 2001; Mikovits et al., 2001; Lagunoff et al., 2002). It is therefore possible that survivin-ΔEx3 is also expressed in these cell types, where an ancestral KSHV pirated the cellular transcript/cDNA. Further analysis of the tissue distribution pattern of survivin-ΔEx3 may help to elucidate the natural host cells of KSHV and the cell types of KSHV-associated tumours. Overall, our data indicate that K7 is a functional homologue of survivin-ΔEx3 and this is the first herpesviral IAP-family member identified. We therefore designate K7 as vIAP (viral inhibitor-of-apoptosis protein).
Materials and methods
Computational analysis
Homologue searches for K7 were performed by using the PSI-BLAST protein database (Altschul et al., 1997; Schaffer et al., 2001) at The National Center for Biotechnology Information (NCBI; National Institutes of Health, Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST) and by the SSEARCH algorithm at the Pole Bio-Informatique Lyonnais (http://pbil.ibcp.fr). Amino acid sequences were aligned using the ClustalW program from the European Bioinformatics Institute (EBI; http://www.ebi.ac.uk/clustalw) (Thompson et al., 1994) then modified online by the Boxshade program from the Pasteur Institute (France; http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html). TreeView software downloaded from the Taxonomy and Systematics server at the University of Glasgow (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used to draw a phylogenic tree for aligned proteins. The MatInspector (Quandt et al., 1995) and PIX (protein identification of unknown sequences) algorithms at the UK Human Genome Mapping Project (HGMP) Resource Centre (http://www.hgmp.mrc.ac.uk) were used to deduce the potential TATA box(es) of the K7 gene and functional motifs in the K7 protein. The domains and motifs in K7 were also found by the iPSORT Prediction and MITOPORT programs (http://www.HypothesisCreator.net/iPSORT/) at the Expasy Molecular Biology Server (http://www.expasy.ch/). The New Geno3D service (at http://geno3d-pbil.ibcp.fr) was used for the modelling of proteins’ 3D structure, and the RasMol software downloaded from the University of Massachusetts (http://www.umass.edu) was then used to view the resulting files.
Plasmids
Plasmids expressing Bcl-2 (pBabe-Bcl2) and Bcl-XL (pcDNA-Bcl-XL) were gifts from Dr Eric Lam (Ludwig Institute, London, UK). The constructs expressing recombinant GST–VDAC3 (pGVDAC3) and HA-tagged VDAC3 protein (pCMV4HA-VDAC3) were kindly provided by Dr Aleem Siddiqui (Department of Microbiology, University of Colorado Health Sciences Center) (Rahmani et al., 2000). The K7 ORF, 5′-UTR of K7 mRNA, PAN RNA, Bax and GAPDH cDNAs were cloned by RT–PCR as described previously (Wang et al., 1997). Total RNA from the PEL cell line BC-3 was extracted using the total RNA isolation kit (Qiagen, Crawley, UK) and amplified by RT–PCR using the following primer pairs: for K7, 5′-GCCACCATGGCATACCCATACGACGTC CCAGACTACGCTGGAACACTGGAGATAAAAGGG (predicted start codon in bold and the HA epitope sequence underlined) and 3′-CGCCTCGAGCTAAGCGTAGTCTGGGACGTCGTATGGGTACAA CTGGCCTGGAGATTGAATCC (predicted stop codon in bold, one XhoI site in italic and underlined, and the HA epitope sequence underlined); 5′-UTR of K7 mRNA, 5′-CGACTGGTTGCGGAAGT ATTCGC and 3′-GCTAAACTGACTCAAGCTGGC; PAN RNA, 5′-ACTGGGACTGCCCAGTCACC and 3′-GCGCCTCGAGCTACAAC TGGCCTGGAGATTG; Bax, 5′-GCAGATCATGGACGGGTCCGGG GAGCAG (start codon in bold) and 3′-GAGTTAGCCCATCTTCTT CCAGATGG (stop codon in bold); and for GAPDH, 5′-ATG GGGAAGGTGAAGGTCGG and 3′-GGTCTTACTCCTTGGAGGC. The amplified fragments were TA cloned into a eukaryotic expression vector pCR3.1-TOPO/V5-His (Invitrogen, Groningen, The Netherlands) according to the manufacturer’s instructions. The primers used to amplify survivin or survivin-ΔEx3 cDNA from human fetal RNA were as described previously (Mahotka et al., 1999).
The deletion and point mutations introduced in the pCR3.1-K7 were created using the QuickChange mutagenesis kit (Stratagene, Amsterdam, The Netherlands), as per the manufacturer’s instructions, and the following mutation primer pairs (all mutated nucleotides are shown in bold): for ΔC11 (amino acid Leu116 is mutated to a stop codon), 5′-CA GTCACCCCTTGCGGGTAATTGCATTGGATTCAATC and 3′-GA TTGAATCCAATGCAATTACCCGCAAGGGGTGACTG; for ΔC32 (Glu95 is mutated to a stop codon), 5′-GCTCTCCATAAGCCCGCA TAACAAAAGCTGCGATTTGC and 3′-GCAAATCGCAGCTT TTGT TATGCGGGCTTATGGAGAGC; for Δ73–82, 5′-CCTGGCTTTCGGC TAGGTTTTCCGTCCTACTTTTCAACACCGCGCGGTCTGGAGCT CTCCATAAGCCCGC and 3′-GCGGGCTTATGGAGAGCTCCAGA CCGCGCGGTGTTGAAAAGTAGGACGGAAAACCTAGCCGAAA GCCAGG; for N108Q (Asn108 is mutated to Gln), 5′-GCCCA AAAACCTTGCCATGGCCAATATACAGTCACCCCTTGCGG and 3′-CCGCAAGGGGTGACTGTATATTGGCCATGGCAAGGTTTTTG.
Constructs pGST-K7, pGST-K7-C56 and pGST-K7-N70, which could express a series of GST–K7 fusion proteins in eukaryotic cells, were created by PCR amplification of the full-length (pGST-K7) C-terminal 56 amino acids (pGST-K7-C56), or the N-terminal 70 amino acids (pGST-K7-N70) from pCR3.1-K7 using the following primer pairs: for full-length K7 cloning, 5′-GCCGGATCCACCATGGCAGGAACACTGGAGATAAAAGGG (predicted start codon in bold and BamHI site underlined) and 3′-CCGGAATTCCTACAACTGGCCTGGAGATTG (EcoRI site underlined); for K7 C-terminus cloning, 5′-CGC GGATCCCTTTTCCCACATTGGCCTG (BamHI site underlined) and the 3′primer was the same as that used in pGST-K7 cloning; and for the K7 N-terminal fragment, the 5′ primer was the same as that used for pCR3.1-K7 cloning and 3′-CCGGAATTCTAGGACGGAAAACCACC (EcoRI site underlined). These PCR products were then digested with BamHI and EcoRI and in-frame ligated into the similarly cut vector pGST, which contains a CMV immediate-early promoter followed by a GST domain.
pGEX6P-1-K7-CT was used to produce recombinant GST-K7-CT protein and was constructed as follows: cDNA encoding the C-terminus of K7 (amino acids 71–126) was PCR amplified from the full-length K7 ORF cDNA with the primer pairs used in the construction of pGST-K7-C56. The PCR product was purified and restriction enzyme digested with BamHI and EcoRI, and ligated in-frame into similarly cut pGEX6P-1 vector (Amersham Pharmacia Biotech, Buckinghamshire, UK). pGEX6P-1-K7-CTΔBH2 and pGEX6P-1-K7-BIR were used to express recombinant GST-K7-CTΔBH2 and GST-K7-BIR proteins, respectively. Both plasmids were constructed by introducing a point mutation at amino acids 116 or 95 in pGEX6P-1-K7-CT, using the same mutagenesis kit and primers as those used in the construction of pCR3.1-K7-ΔC11 and pCR3.1-K7-ΔC32.
Cell cultures, transfection and induction of viral replication
Cell cultures were performed as described previously (Sharp et al., 2002). For transient transfections, 4 × 105 of human embryonic kidney (HEK) 293 or HeLa cells were plated in a 25 cm2 flask, and the following day were transfected by FuGENE 6 according to the manufacturer’s instructions (Roche Diagnostics, Mannheim, Germany). Tunicamycin treatment of K7 transfected cell lines was performed by adding tunicamycin to the culture medium, to a final concentration of between 1 and 10 µg/ml for 24 h before cell harvest.
Prior to induction of virus replication in PEL cells, dead cells were removed from the cultures by density centrifugation through Lymphoprep (Nycomed, Roskilde, Denmark). Cells were then resuspended in fresh medium and viral replication was induced by the addition of TPA (Sigma, St Louis, MO) at 20 ng/ml. Cells were harvested at the indicated time points, washed once with phosphate-buffered saline (PBS), then extracted using a total RNA isolation kit (Qiagen, Crawley, UK) before freezing at –80°C.
Immunofluorescent assay and subcellular fractionation
Subcellular localization was performed as described previously (La Bella et al., 2000). For immunofluorescent assay, subconfluent layers of HeLa cells (1 × 105) were grown on glass coverslips and transfected with FuGENE 6. Forty-eight hours after transfection, cells were washed three times with PBS and fixed for 20 min at –20°C with methanol/acetone (1:1), and then washed again with PBS. Cells were then blocked for 1 h with blocking buffer [3% bovine serum albumin (BSA) in PBS with 1 mM MgCl2 and 1 mM CaCl2], before 1–2 h of incubation with primary antibody [anti-HA (1:500, mouse monoclonal antibody; BabCo, Cambridge, UK), anti-Bax (1:1000, rabbit polyclonal antibody; a gift from Mary Collins), anti-Bcl-2 (1:250, mouse monoclonal antibody; BD Pharmingen, Basingstoke, UK), anti-Bcl-XL (1:500, rabbit polyclonal antibody; Sigma) and anti-GST (1:1000, rabbit polyclonal antibody; Transduction Labs, Lexington, KY)] diluted in washing buffer (0.3% BSA in PBS with 1 mM MgCl2 and 1 mM CaCl2). After washing, the cells were incubated with the indicated fluorescent secondary antibodies [1:200; donkey anti-mouse IgG congugated to FITC or AMCA (purchased from DAKO diagnostics), and donkey anti-rabbit IgG coupled to FITC (Jackson Immunochemicals, West Grove, PA)] for 1 h at 20°C, then mounted on glass slides with 20 µl of glycerol. In some experiments, cells were incubated further with 20 µM of MitoTracker Red CMXRos (Molecular Probes Europe, Rijnsburgerweg, The Netherlands) at 20°C for 10 min to stain mitochondria, then washed three times with PBS. In colocalization experiments, cellular nuclei were counterstained with Hoechst 33258 (Sigma) (Negri et al., 1997). Control slides were stained with secondary antibody alone. Images of protein localizations or cell organelles were obtained using a laser-scanning confocal microscope (Leica TSC Confocal Systems, Milton Keynes, UK).
Induction of apoptosis, apoptotic assay and flow cytometry
For TNF-α and anti-Fas mAb-induced apoptosis, transfected HeLa cells were cultured in the presence of 10 ng/ml of TNF-α (Sigma) or 100 ng/ml anti-Fas IgM mAb (clone CH11; Upstate Biotechnology, UK) plus 1 µg/ml of CHX (Sigma) for 16 h. For Bax cotransfection experiments, 4 × 105 HeLa cells were transfected with the indicated expression plasmids and incubated at 37°C for 48 or 24 h. The annexin V–FITC/PI double staining assay was performed according to the manufacturer’s instructions (BD Pharmingen). To measure mitochondrial transmembrane potential (ΔΨm), 5 × 105 transfected HeLa cells were incubated with 500 µM of CMXRos added to the culture media at 37°C for 30 min and then detached by trypsin. To evaluate cell viability, cells were trypsinized and resuspended in PI solution (2 µg/ml PI in PBS with 1% BSA and 0.01% azide) at 20°C for 20 min. Treated cells were washed once in PBS then analysed for fluorescence using FACSCalibur™ flow cytometry together with the CellQuest software (Becton Dickinson, Franklin Lakes, NJ). At least 20 000 cells were counted in each experiment, and the numbers reported represent the average and standard deviation (SD) of at least three independent experiments.
In vitro caspase-3 activity assays
The measurements of caspase-3 activity in cell lysates or in RIPA buffer were performed using the Active Caspase-3 (CPP32) SET kit (BD Pharmingen) according to the manufacturer’s instructions. Transfected HeLa cell extract (1 ml) from one 35-mm dish was mixed with 10 µl of fluorogenic substrate and 50 ng of purified active caspase-3, and incubated for 1 h at 37°C. Liberated fluorescence was measured using the STORM spectrofluorometer (Amersham Pharmacia Biotech) with an excitation wavelength of 400 nm and an emission wavelength of 505 nm. In a separate reaction, 10 µl of the inhibitor peptide was added into the 1 ml assay mixture as a positive control for inhibition.
Miscellaneous methods
The methods used for northern blot analysis were as described previously (Wang et al., 1997). The expression of GST–K7 recombinant fusion proteins, GST pull-down assays, in vivo coimmunoprecipitation and western blots were also performed as described (Sharp et al., 2002).
Acknowledgments
Acknowledgements
We thank Mary Collins, Robin Weiss, Yoshio Endo and Daniel Hollyman for helpful discussions. We also thank Genetica (Boston, MA) for providing human fetal cDNA. This work was supported by Cancer Research UK, The Medical Research Council and GlaxoSmithKline.
References
- Adams J.M. and Cory,S. (1998) The Bcl-2 protein family: arbiters of cell survival. Science, 281, 1322–1326. [DOI] [PubMed] [Google Scholar]
- Adida C., Crotty,P.L., McGrath,J., Berrebi,D., Diebold,J. and Altieri, D.C. (1998) Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am. J. Pathol., 152, 43–49. [PMC free article] [PubMed] [Google Scholar]
- Akgul C., Moulding,D.A., White,M.R. and Edwards,S.W. (2000) In vivo localisation and stability of human Mcl-1 using green fluorescent protein (GFP) fusion proteins. FEBS Lett., 478, 72–76. [DOI] [PubMed] [Google Scholar]
- Albrecht J.C. et al. (1992) Primary structure of the herpesvirus saimiri genome. J. Virol., 66, 5047–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander L., Denekamp,L., Knapp,A., Auerbach,M.R., Damania,B. and Desrosiers,R.C. (2000) The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi’s sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol., 74, 3388–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller, W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G., Adida,C. and Altieri,D.C. (1997) A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med., 3, 917–921. [DOI] [PubMed] [Google Scholar]
- Ambrosini G., Adida,C., Sirugo,G. and Altieri,D.C. (1998) Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J. Biol. Chem., 273, 11177–11182. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A. and Dixit,V.M. (1998) Death receptors: signaling and modulation. Science, 281, 1305–1308. [DOI] [PubMed] [Google Scholar]
- Borner C., Olivier,R., Martinou,I., Mattmann,C., Tschopp,J. and Martinou,J.C. (1994) Dissection of functional domains in Bcl-2 α by site-directed mutagenesis. Biochem. Cell Biol., 72, 463–469. [DOI] [PubMed] [Google Scholar]
- Boshoff C. and Weiss,R.A. (2001) Epidemiology and pathogenesis of Kaposi’s sarcoma-associated herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci., 356, 517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff C., Schulz,T.F., Kennedy,M.M., Graham,A.K., Fisher,C., Thomas, A., McGee,J.O., Weiss,R.A. and O’Leary,J.J. (1995) Kaposi’s sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med., 1, 1274–1278. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E., Newmeyer,D.D. and Green,D.R. (1998) Mitochondrial cytochrome c release in apoptosis occurs upstream of DEVD-specific caspase activation and independently of mitochondrial transmembrane depolarization. EMBO J., 17, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Alliotte,T., Van den Bulcke,M., Bauw,G., Vandekerckhove,J., Van Montagu,M. and Inze,D. (1989) A plant manganese superoxide dismutase is efficiently imported and correctly processed by yeast mitochondria. Proc. Natl Acad. Sci. USA, 86, 3237–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P., Roques,B. and Kroemer,G. (2001) Viral and bacterial proteins regulating apoptosis at the mitochondrial level. EMBO J., 20, 4325–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M., Hirsch,T., Susin,S.A., Zamzami,N., Marchetti,P., Macho,A. and Kroemer,G. (1996) Sequential acquisition of mitochondrial and plasma membrane alterations during early lymphocyte apoptosis. J. Immunol., 157, 512–521. [PubMed] [Google Scholar]
- Chantalat L., Skoufias,D.A., Kleman,J.P., Jung,B., Dideberg,O. and Margolis,R.L. (2000) Crystal structure of human survivin reveals a bow tie-shaped dimer with two unusual α-helical extensions. Mol. Cell, 6, 183–189. [PubMed] [Google Scholar]
- Chao D.T. and Korsmeyer,S.J. (1998) BCL-2 family: regulators of cell death. Annu. Rev. Immunol., 16, 395–419. [DOI] [PubMed] [Google Scholar]
- Chau B.N., Cheng,E.H., Kerr,D.A. and Hardwick,J.M. (2000) Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol. Cell, 6, 31–40. [PubMed] [Google Scholar]
- Chu Z.L., McKinsey,T.A., Liu,L., Gentry,J.J., Malim,M.H. and Ballard,D.W. (1997) Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-κB control. Proc. Natl Acad. Sci. USA, 94, 10057–10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciufo D.M., Cannon,J.S., Poole,L.J., Wu,F.Y., Murray,P., Ambinder, R.F. and Hayward,G.S. (2001) Spindle cell conversion by Kaposi’s sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol., 75, 5614–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conus S., Rosse,T. and Borner,C. (2000) Failure of Bcl-2 family members to interact with Apaf-1 in normal and apoptotic cells. Cell Death Differ., 7, 947–954. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L. and Reed,J.C. (1999) IAP family proteins—suppressors of apoptosis. Genes Dev., 13, 239–252. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L., Takahashi,R., Salvesen,G.S. and Reed,J.C. (1997) X-linked IAP is a direct inhibitor of cell-death proteases. Nature, 388, 300–304. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L., Roy,N., Stennicke,H.R., Van Arsdale,T., Zhou,Q., Srinivasula,S.M., Alnemri,E.S., Salvesen,G.S. and Reed,J.C. (1998) IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J., 17, 2215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin N. et al. (1999) Distribution of human herpesvirus-8 latently infected cells in Kaposi’s sarcoma, multicentric Castleman’s disease and primary effusion lymphoma. Proc. Natl Acad. Sci. USA, 96, 4546–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duriez P.J., Wong,F., Dorovini-Zis,K., Shahidi,R. and Karsan,A. (2000) A1 functions at the mitochondria to delay endothelial apoptosis in response to tumor necrosis factor. J. Biol. Chem., 275, 18099–18107. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Marcotte,E.M., Xenarios,I. and Yeates,T.O. (2000) Protein function in the post-genomic era. Nature, 405, 823–826. [DOI] [PubMed] [Google Scholar]
- Evan G. and Littlewood,T. (1998) A matter of life and cell death. Science, 281, 1317–1322. [DOI] [PubMed] [Google Scholar]
- Flore O., Rafii,S., Ely,S., O’Leary,J.J., Hyjek,E.M. and Cesarman,E. (1998) Transformation of primary human endothelial cells by Kaposi’s sarcoma-associated herpesvirus. Nature, 394, 588–592. [DOI] [PubMed] [Google Scholar]
- Frade J.M. and Michaelidis,T.M. (1997) Origin of eukaryotic programmed cell death: a consequence of aerobic metabolism? BioEssays, 19, 827–832. [DOI] [PubMed] [Google Scholar]
- Fukuda S. and Pelus,L.M. (2001) Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34+ cells by hematopoietic growth factors: implication of survivin expression in normal hematopoiesis. Blood, 98, 2091–2100. [DOI] [PubMed] [Google Scholar]
- Goldmacher V.S. et al. (1999) A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl Acad. Sci. USA, 96, 12536–12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.R. and Reed,J.C. (1998) Mitochondria and apoptosis. Science, 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Gross A., Jockel,J., Wei,M.C. and Korsmeyer,S.J. (1998) Enforced dimerization of BAX results in its translocation, mitochondrial dysfunction and apoptosis. EMBO J., 17, 3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti L.G. and Chisari,F.V. (2001) Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol., 19, 65–91. [DOI] [PubMed] [Google Scholar]
- Harty J.T., Tvinnereim,A.R. and White,D.W. (2000) CD8+ T cell effector mechanisms in resistance to infection. Annu. Rev. Immunol., 18, 275–308. [DOI] [PubMed] [Google Scholar]
- Hayajneh W.A., Contopoulos-Ioannidis,D.G., Lesperance,M.M., Venegas, A.M. and Colberg-Poley,A.M. (2001) The carboxyl terminus of the human cytomegalovirus UL37 immediate-early glycoprotein is conserved in primary strains and is important for transactivation. J. Gen. Virol., 82, 1569–1579. [DOI] [PubMed] [Google Scholar]
- Helenius A. and Aebi,M. (2001) Intracellular functions of N-linked glycans. Science, 291, 2364–2369. [DOI] [PubMed] [Google Scholar]
- Hengartner M.O. (2000) The biochemistry of apoptosis. Nature, 407, 770–776. [DOI] [PubMed] [Google Scholar]
- Henry M., Uthman,A., Geusau,A., Rieger,A., Furci,L., Lazzarin,A., Lusso,P. and Tschachler,E. (1999) Infection of circulating CD34+ cells by HHV-8 in patients with Kaposi’s sarcoma. J. Invest. Dermatol., 113, 613–616. [DOI] [PubMed] [Google Scholar]
- Hinds M.G., Norton,R.S., Vaux,D.L. and Day,C.L. (1999) Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat. Nat. Struct. Biol., 6, 648–651. [DOI] [PubMed] [Google Scholar]
- Hu Y., Benedict,M.A., Wu,D., Inohara,N. and Nunez,G. (1998) Bcl-XL interacts with Apaf-1 and inhibits Apaf-1-dependent caspase-9 activation. Proc. Natl Acad. Sci. USA, 95, 4386–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler M. et al. (1997) Inhibition of death receptor signals by cellular FLIP. Nature, 388, 190–195. [DOI] [PubMed] [Google Scholar]
- Ito T. et al. (2000) survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology, 31, 1080–1085. [DOI] [PubMed] [Google Scholar]
- Iwahashi H., Eguchi,Y., Yasuhara,N., Hanafusa,T., Matsuzawa,Y. and Tsujimoto,Y. (1997) Synergistic anti-apoptotic activity between Bcl-2 and SMN implicated in spinal muscular atrophy. Nature, 390, 413–417. [DOI] [PubMed] [Google Scholar]
- Jenner R.G., Alba,M.M., Boshoff,C. and Kellam,P. (2001) Kaposi’s sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol., 75, 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Wilford,C., Duensing,S., Munger,K., Jones,G. and Jones,D. (2001) Participation of survivin in mitotic and apoptotic activities of normal and tumor-derived cells. J. Cell. Biochem., 83, 342–354. [DOI] [PubMed] [Google Scholar]
- Krieg A. et al. (2002) Expression of different survivin variants in gastric carcinomas: first clues to a role of survivin-2B in tumour progression. Br. J. Cancer, 86, 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBella V., Kallenbach,S. and Pettmann,B. (2000) Expression and subcellular localization of two isoforms of the survival motor neuron protein in different cell types. J. Neurosci. Res., 62, 346–356. [DOI] [PubMed] [Google Scholar]
- Lagunoff M., Bechtel,J., Venetsanakos,E., Roy,A.M., Abbey,N., Herndier,B., McMahon,M. and Ganem,D. (2002) De novo infection and serial transmission of Kaposi’s sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol., 76, 2440–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahotka C., Wenzel,M., Springer,E., Gabbert,H.E. and Gerharz,C.D. (1999) survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res., 59, 6097–6102. [PubMed] [Google Scholar]
- Marshall W.L., Yim,C., Gustafson,E., Graf,T., Sage,D.R., Hanify,K., Williams,L., Fingeroth,J. and Finberg,R.W. (1999) Epstein–Barr virus encodes a novel homolog of the bcl-2 oncogene that inhibits apoptosis and associates with Bax and Bak. J. Virol., 73, 5181–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D.J. and Davison,A.J. (1999) The descent of human herpesvirus 8. Semin. Cancer Biol., 9, 201–209. [DOI] [PubMed] [Google Scholar]
- Mesri M., Morales-Ruiz,M., Ackermann,E.J., Bennett,C.F., Pober,J.S., Sessa,W.C. and Altieri,D.C. (2001) Suppression of vascular endothelial growth factor-mediated endothelial cell protection by survivin targeting. Am. J. Pathol., 158, 1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikovits J., Ruscetti,F., Zhu,W., Bagni,R., Dorjsuren,D. and Shoemaker,R. (2001) Potential cellular signatures of viral infections in human hematopoietic cells. Dis. Markers, 17, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.K. (1999) An exegesis of IAPs: salvation and surprises from BIR motifs. Trends Cell Biol., 9, 323–328. [DOI] [PubMed] [Google Scholar]
- Moore P.S. and Chang,Y. (2001) Molecular virology of Kaposi’s sarcoma-associated herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci., 356, 499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.S., Boshoff,C., Weiss,R.A. and Chang,Y. (1996) Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science, 274, 1739–1744. [DOI] [PubMed] [Google Scholar]
- Moriishi K., Huang,D.C., Cory,S. and Adams,J.M. (1999) Bcl-2 family members do not inhibit apoptosis by binding the caspase activator Apaf-1. Proc. Natl Acad. Sci. USA, 96, 9683–9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosialos G., Birkenbach,M., Yalamanchili,R., VanArsdale,T., Ware,C. and Kieff,E. (1995) The Epstein–Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell, 80, 389–399. [DOI] [PubMed] [Google Scholar]
- Muchmore S.W. et al. (1996) X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature, 381, 335–341. [DOI] [PubMed] [Google Scholar]
- Muchmore S.W. et al. (2000) Crystal structure and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol. Cell, 6, 173–182. [PubMed] [Google Scholar]
- Negri C., Donzelli,M., Bernardi,R., Rossi,L., Burkle,A. and Scovassi,A.I. (1997) Multiparametric staining to identify apoptotic human cells. Exp. Cell Res., 234, 174–177. [DOI] [PubMed] [Google Scholar]
- Neipel F., Albrecht,J.C. and Fleckenstein,B. (1998) Human herpesvirus 8—the first human Rhadinovirus. J. Natl Cancer Inst. Monogr., 23, 73–77. [DOI] [PubMed] [Google Scholar]
- Newmeyer D.D., Bossy-Wetzel,E., Kluck,R.M., Wolf,B.B., Beere,H.M. and Green,D.R. (2000) Bcl-xL does not inhibit the function of Apaf-1. Cell Death Differ., 7, 402–407. [DOI] [PubMed] [Google Scholar]
- Ng F.W., Nguyen,M., Kwan,T., Branton,P.E., Nicholson,D.W., Cromlish,J.A. and Shore,G.C. (1997) p28 Bap31, a Bcl-2/Bcl-XL- and procaspase-8-associated protein in the endoplasmic reticulum. J. Cell Biol., 139, 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M., Breckenridge,D.G., Ducret,A. and Shore,G.C. (2000) Caspase-resistant BAP31 inhibits Fas-mediated apoptotic membrane fragmentation and release of cytochrome c from mitochondria. Mol. Cell Biol., 20, 6731–6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien V. (1998) Viruses and apoptosis. J. Gen. Virol., 79, 1833–1845. [DOI] [PubMed] [Google Scholar]
- Oda E., Ohki,R., Murasawa,H., Nemoto,J., Shibue,T., Yamashita,T., Tokino,T., Taniguchi,T. and Tanaka,N. (2000) Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science, 288, 1053–1058. [DOI] [PubMed] [Google Scholar]
- Olie R.A., Simoes-Wust,A.P., Baumann,B., Leech,S.H., Fabbro,D., Stahel,R.A. and Zangemeister-Wittke,U. (2000) A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res., 60, 2805–2809. [PubMed] [Google Scholar]
- Pan G., O’Rourke,K. and Dixit,V.M. (1998) Caspase-9, Bcl-XL and Apaf-1 form a ternary complex. J. Biol. Chem., 273, 5841–5845. [DOI] [PubMed] [Google Scholar]
- Panyutich E.A., Said,J.W. and Miles,S.A. (1998) Infection of primary dermal microvascular endothelial cells by Kaposi’s sarcoma-associated herpesvirus. AIDS, 12, 467–472. [DOI] [PubMed] [Google Scholar]
- Paulose-Murphy M. et al. (2001) Transcription program of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus). J. Virol., 75, 4843–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani Z., Huh,K.W., Lasher,R. and Siddiqui,A. (2000) Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3 and alters its transmembrane potential. J. Virol., 74, 2840–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J. and Pfanner,N. (2000) The protein import machinery of the mitochondrial membranes. Traffic, 1, 457–464. [DOI] [PubMed] [Google Scholar]
- Rassow J., Dekker,P.J., van Wilpe,S., Meijer,M. and Soll,J. (1999) The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol., 286, 105–120. [DOI] [PubMed] [Google Scholar]
- Renne R., Blackbourn,D., Whitby,D., Levy,J. and Ganem,D. (1998) Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J. Virol., 72, 5182–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N., Deveraux,Q.L., Takahashi,R., Salvesen,G.S. and Reed,J.C. (1997) The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J., 16, 6914–6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo J.J. et al. (1996) Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl Acad. Sci. USA, 93, 14862–14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson M.J., Ross,L., Decker,W.K. and Craigen,W.J. (1998) A novel isoform of the mitochondrial outer membrane protein VDAC3 via alternative splicing of a 3-base exon. Functional characteristics and subcellular localization. J. Biol. Chem., 273, 30482–30486. [DOI] [PubMed] [Google Scholar]
- Sattler M. et al. (1997) Structure of Bcl-xL–Bak peptide complex: recognition between regulators of apoptosis. Science, 275, 983–986. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K., Bogenrieder,T. and Herlyn,M. (2001) No longer a molecular black box—new clues to apoptosis and drug resistance in melanoma. Trends Mol. Med., 7, 191–194. [DOI] [PubMed] [Google Scholar]
- Schaffer A.A., Aravind,L., Madden,T.L., Shavirin,S., Spouge,J.L., Wolf, Y.I., Koonin,E.V. and Altschul,S.F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res., 29, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshagiri S., Vucic,D., Lee,J. and Dixit,V.M. (1999) Baculovirus-based genetic screen for antiapoptotic genes identifies a novel IAP. J. Biol. Chem., 274, 36769–36773. [DOI] [PubMed] [Google Scholar]
- Sharp T.V., Wang,H.W., Koumi,A., Hollyman,D., Endo,Y., Ye,H., Du,M.Q. and Boshoff,C. (2002) K15 protein of Kaposi’s sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J. Virol., 76, 802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Sung,B.J., Cho,Y.S., Kim,H.J., Ha,N.C., Hwang,J.I., Chung, C.W., Jung,Y.K. and Oh,B.H. (2001) An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry, 40, 1117–1123. [DOI] [PubMed] [Google Scholar]
- Somia N.V., Schmitt,M.J., Vetter,D.E., Van Antwerp,D., Heinemann, S.F. and Verma,I.M. (1999) LFG: an anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc. Natl Acad. Sci. USA, 96, 12667–12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., Kuang,Y., Dixit,V.M. and Vincenz,C. (1999) Boo, a novel negative regulator of cell death, interacts with Apaf-1. EMBO J., 18, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Lin,S.F., Gradoville,L. and Miller,G. (1996) Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl Acad. Sci. USA, 93, 11883–11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Lin,S.F., Staskus,K., Gradoville,L., Grogan,E., Haase,A. and Miller,G. (1999) Kinetics of Kaposi’s sarcoma-associated herpesvirus gene expression. J. Virol., 73, 2232–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I., Wang,Y., Sausville,E., Scudiero,D.A., Vigna,N., Oltersdorf,T. and Reed,J.C. (1998) IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases and anticancer drugs. Cancer Res., 58, 5315–5320. [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput. Appl. Biosci., 10, 19–29. [DOI] [PubMed] [Google Scholar]
- Thornberry N.A. and Lazebnik,Y. (1998) Caspases: enemies within. Science, 281, 1312–1316. [DOI] [PubMed] [Google Scholar]
- Thornton J.M. (2001) From genome to function. Science, 292, 2095–2097. [DOI] [PubMed] [Google Scholar]
- Tran J., Rak,J., Sheehan,C., Saibil,S.D., LaCasse,E., Korneluk,R.G. and Kerbel,R.S. (1999) Marked induction of the IAP family antiapoptotic proteins survivin and XIAP by VEGF in vascular endothelial cells. Biochem. Biophys. Res. Commun., 264, 781–788. [DOI] [PubMed] [Google Scholar]
- Tschopp J., Irmler,M. and Thome,M. (1998a) Inhibition of fas death signals by FLIPs. Curr. Opin. Immunol., 10, 552–558. [DOI] [PubMed] [Google Scholar]
- Tschopp J., Thome,M., Hofmann,K. and Meinl,E. (1998b) The fight of viruses against apoptosis. Curr. Opin. Genet. Dev., 8, 82–87. [DOI] [PubMed] [Google Scholar]
- Verdecia M.A., Huang,H., Dutil,E., Kaiser,D.A., Hunter,T. and Noel,J.P. (2000) Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol., 7, 602–608. [DOI] [PubMed] [Google Scholar]
- Verhagen A.M., Coulson,E.J. and Vaux,D.L. (2001) Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol., 2, REVIEWS3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden K.H. (2000) p53: death star. Cell, 103, 691–694. [DOI] [PubMed] [Google Scholar]
- Vucic D., Kaiser,W.J. and Miller,L.K. (1998) A mutational analysis of the baculovirus inhibitor of apoptosis Op-IAP. J. Biol. Chem., 273, 33915–33921. [DOI] [PubMed] [Google Scholar]
- Wallach D., Varfolomeev,E.E., Malinin,N.L., Goltsev,Y.V., Kovalenko, A.V. and Boldin,M.P. (1999) Tumor necrosis factor receptor and Fas signaling mechanisms. Annu. Rev. Immunol., 17, 331–367. [DOI] [PubMed] [Google Scholar]
- Wang H.W., Wu,H.L., Chen,D.S. and Chen,P.J. (1997) Identification of the functional regions required for hepatitis D virus replication and transcription by linker-scanning mutagenesis of viral genome. Virology, 239, 119–131. [DOI] [PubMed] [Google Scholar]
- Warnes T.W. and Smith,A. (1987) Tumour markers in diagnosis and management. Baillieres Clin. Gastroenterol., 1, 63–89. [DOI] [PubMed] [Google Scholar]
- Wolter K.G., Hsu,Y.T., Smith,C.L., Nechushtan,A., Xi,X.G. and Youle,R.J. (1997) Movement of Bax from the cytosol to mitochondria during apoptosis. J. Cell Biol., 139, 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X.M., Oltvai,Z.N. and Korsmeyer,S.J. (1994) BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature, 369, 321–323. [DOI] [PubMed] [Google Scholar]
- You M., Ku,P.T., Hrdlickova,R. and Bose,H.R.,Jr (1997) ch-IAP1, a member of the inhibitor-of-apoptosis protein family, is a mediator of the anti-apoptotic activity of the v-Rel oncoprotein. Mol. Cell. Biol., 17, 7328–7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Xu,Q., Krajewski,S., Krajewska,M., Xie,Z., Fuess,S., Kitada,S., Godzik,A. and Reed,J.C. (2000) BAR: an apoptosis regulator at the intersection of caspases and Bcl-2 family proteins. Proc. Natl Acad. Sci. USA, 97, 2597–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]