Abstract
Background
The alarming rise of antimicrobial resistance (AMR), especially in Klebsiella pneumoniae, demands urgent development of novel therapeutic strategies. Recently, probiotics and their metabolites, known as postbiotics, have gained attention due to their potential to enhance antibiotic efficacy and combat resistant pathogens.
Methods
In this study, 88 native Lactobacillus spp. isolates, were screened for antimicrobial activity against a highly resistant K. pneumoniae ATCC 7881 strain with a minimum inhibitory concentration ≥ 2048 µg/mL. Based on screening results, four strains—Lactiplantibacillus plantarum RP155, 403, 225 and Ligilactobacillus salivarius RP317—were selected to produce postbiotics. Antimicrobial activity of these postbiotics, alone and combined with sub-inhibitory concentrations of amoxicillin and imipenem, was evaluated using the broth microdilution method. Gene expression of key antibiotic resistance determinants (blaNDM, blaCTX, blaTEM, blaSHV) was assessed by qRT-PCR following co-culture treatments.
Results
All postbiotics exhibited inhibitory effects on the growth of multidrug-resistant K. pneumoniae when applied in combination with low-dose antibiotics. Postbiotic concentrations ranging from 25 to 100 mg/mL combined with 1–4 µg/mL of amoxicillin or imipenem resulted in complete bacterial eradication. Molecular analyses revealed differential regulation of resistance genes: some postbiotics alone (notably L. plantarum RP225 and L. salivarius RP317) increased expression of specific resistance genes, whereas combinations with antibiotics significantly suppressed or completely silenced the expression levels of genes investigated.
Conclusion
Our findings demonstrate that native postbiotics, particularly in synergy with antibiotics, exert potent antimicrobial activity against multidrug-resistant K. pneumoniae and effectively downregulate critical resistance genes; highlighting the potential of postbiotics as adjunctive therapeutic agents in the battle against AMR.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-025-04474-7.
Keywords: Postbiotic, Lacobacillus, Klebsiella pneumoniae, AMR
Introduction
Antimicrobial resistance (AMR), which is usually caused by the misuse of antibiotics or, in a broader context, by neglecting the One Health approach, has become a major problem in the 21 st century [1, 2]. According to the World Health Organization (WHO), bacterial antimicrobial resistance (AMR) was directly responsible for 1.27 million deaths worldwide in the year 2019 and contributed to nearly 5 million deaths overall during the same year, making AMR one of the biggest global threats to public health [3]. Traditional antibiotics, while initially effective, now face significant challenges due to rapid resistance development and environmental persistence [4]. Specifically, antibiotic residues entering environments through agricultural runoff, pharmaceutical waste, and incorrect disposal accumulate in rivers and soil, producing selective pressure that drives AMR evolution in environmental bacteria [5]. The environmental reservoir of antibiotic resistance genes acts as a source that can transfer resistant genes to clinically relevant pathogens through horizontal gene transfer (HGT) methods such as plasmids, creating a vicious cycle of resistance amplification [6]. Therefore, the ongoing prevalence and dissemination of multidrug-resistant strains in medical environments highlight the critical demand for improved antimicrobial stewardship and innovative treatment approaches.
As antimicrobial resistance rates continue to rise, it has become increasingly clear that traditional approaches are no longer sufficient to address this growing threat. There is an urgent need to re-evaluate and shift our current strategies by incorporating a broader array of diverse and innovative compounds. Such a transition is essential for effectively preventing the further spread of microbial resistance and ensuring the continued efficacy of antimicrobial therapies in both clinical and community settings [7]. Importantly, the One Health approach highlights that antimicrobial resistance is a multifaceted issue requiring coordinated efforts across human, animal, and environmental health sectors [8]. Effective reduction of resistance in patients depends on comprehensive management of resistance reservoirs beyond clinical settings, including agriculture and the environment [9, 10]. Given the critical importance of addressing antimicrobial resistance from a One Health perspective, it is essential to explore supportive or alternative agents to antibiotics that can enhance treatment efficacy and help curb resistance development.
Recent approaches that health care settings have chosen to deploy as alternative treatment approaches can be classified into three types: plant-, metal-, and nanotechnology-based antimicrobials [4, 11, 12]. Besides these agents, using microbial strategies like probiotics could be helpful in controlling antimicrobial resistance. Probiotics could exert antimicrobial effects by creating antibacterial chemicals, such as bacteriocins, competitive exclusion of microorganisms, and improving the immune system [13]. According to International Scientific Association for Probiotics and Prebiotics (ISAPP) in 2021, postbiotic is a preparation of inanimate (non-living) microorganisms and/or their components that confers a health benefit on the host. Aside from probiotics, their derivatives, especially postbiotics, could have positive effects, such as antibacterial effects. This includes whole inactivated microbial cells or fragments such as cell walls, membranes, proteins, and metabolites produced by the microbes, which may contribute to the beneficial outcomes [14, 15]. Due to the non-living nature of postbiotics, using these agents instead of live probiotics may offer certain advantages, especially for patients with immunocompromised conditions [16].
Klebsiella pneumoniae is one of the common resistant bacteria for which using novel approaches is essential to effectively combat. AMR poses a global health risk, especially with K. pneumoniae known for its multidrug resistance and links to hospital infections [17]. K. pneumoniae frequently leads to infections acquired in hospitals and has demonstrated a growing resistance to last-line antibiotics like carbapenems, making treatment more challenging and heightening the risk of mortality [18]. Among the diverse mechanisms that contribute to antibiotic resistance, the existence of resistance genes, such as Extended Spectrum Beta-Lactamases (ESBL) genes (e.g., blaCTX, blaTEM, and blaSHV), along with blaNDM, which encodes a carbapenemase, plays a crucial role. In recent years, CTX-M betalactamases have exhibited a rapid dissemination among Enterobacteriaceae, becoming the most common ESBLs in numerous regions across the globe [19]. The SHV and TEM enzymes, as additional ESBL types, are significant contributors to betalactam antibiotic resistance, particularly in K. pneumoniae [20, 21]. Furthermore, the rise of blaNDM−1 K. pneumoniae as a source of outbreaks is alarming, as once integrated into drug-resistant plasmids, virulence factors make these strains exceedingly resistant, virulent, and easily transmissible [22]. These genes are incredibly significant in driving a substantial shift in antimicrobial resistance. As a result, in recent years, the healthcare community has switched toward employing novel techniques, including antimicrobial peptides, phages, conventional medicine, antimicrobial nanoparticle technology, and cell-free supernatant generated from probiotics to combat resistant K. pneumoniae [23, 24].
In the current study, we aimed to introduce a novel approach to combat microbial resistance via using compounds that could enhance the effects of antibiotics. Indeed, in addition to the classic properties of the probiotics and their derivatives as an antimicrobial agent, we think that these agents could have special effects in increasing the antibiotics function. Specifically, the use of postbiotics derived from novel, native probiotic strains offer an innovative method to boost the effectiveness of antibiotics such as amoxicillin and imipenem against resistant Klebsiella strains. Moreover, this approach has the potential to effectively reduce the expression of resistance genes, thereby contributing to better control of antimicrobial resistance.
Materials and methods
Minimum inhibitory concentrations (MIC) of probiotic strains and K. pneumoniae ATCC 7881
In the current investigation, to obtain postbiotics effective against K. pneumoniae ATCC 7881, it was first necessary to screen probiotic strains for antimicrobial activity. Here, we used 88 native Lactobacillus spp. probiotic isolates collected in our previous study [25]. The evaluation of probiotic resistance to amoxicillin and imipenem was conducted to ensure the persistence and survival of our native probiotic strains in the presence of these antibiotics. For this purpose, the minimum inhibitory concentration (MIC) values were determined using the broth microdilution method on all Lactobacillus strains using 150 µL of bacterial suspension adjusted to various optical densities (OD) corresponding to 0.5, 1, and 2 McFarland standards, as well as dilutions of 1:20 and 1:100 of 0.5 McFarland. Additionally, 150 µL of antibiotic solutions with serial dilutions ranging from 2048 µg/ml to 2 µg/ml were added to each well. The use of different OD values aimed to determine the optimal concentration and density of our native probiotic strains for subsequent experimental steps.
To ensure the selection of a resistant strain, the MIC test was done on K. pneumoniae ATCC 7881 to the amoxicillin and imipenem. For this purpose, 150 µl of K. pneumoniae ATCC 7881 suspension and then 150 µl of the antibiotic, at serial dilutions ranging from 2048 µg/ml to 2 µg/ml, were added to each well.
Probiotic selection strategy
In this study, the target bacterium was co-cultured with probiotic strains in both the presence and absence of amoxicillin and imipenem antibiotics. Following incubation, bacterial growth was quantified using colony counting methods. Based on the results, the probiotic strain demonstrating the greatest enhancement of antibiotic efficacy was selected for subsequent analyses.
To carry out this step, 150 µL of K. pneumoniae ATCC 7881 suspension (1:100 dilutions of 0.5 McFarland standard) and 150 µL of our native probiotic strains adjusted to an optical density equivalent to 2 McFarland were added to each well. To assess the synergistic effects of our native probiotic strains and antibiotic, the antibiotic was first serially diluted in this stage, with concentrations ranging from 2048 µg/ml to 2 µg/ml utilizing 100 µL of the antibiotics. Next, 50 µL of K. pneumoniae ATCC 7881 (at a concentration of 1:100 of a 0.5 McFarland standard) and 50 µL of the probiotic (2 McFarland) were applied to each well. Note that culture medium was present in column 12, which was used as a negative control to look for contamination. Following a 24-hour incubation period at 37 °C, 100 µL of each well was diluted 1:100. To measure the bacterial growth and determine the decrease in colony counts, 100 µL of this 1:100 dilutions was then cultivated on growth medium. It should be mentioned that postbiotics were made from the probiotic strains that were chosen for their ability to reduce the colony number of K. pneumoniae ATCC 7881.
Postbiotic preparation
Initially, for the primary preculture, 200 µl of probiotic bacteria (2 McFarland) were grown in 15 ml MRS broth and incubated at 37 °C for 24 h. Subsequently, for the postbiotic preparation, 15 mL bacterial suspension was inoculated to 285 ml MRS broth and incubated again at 37 °C for 36 h. Next, the cultures were centrifuged at 3500 rpm at 4 °C, and the resulting supernatant was filtered through 0.22 μm filters. The obtained suspensions were then frozen at − 20 °C. After freezing, the samples were lyophilized in a freeze-dryer (Martin Christ, Germany).
MIC of our native postbiotic and K. pneumoniae ATCC 7881
To determine the MIC of our native postbiotics against K. pneumoniae ATCC 7881, various concentrations of the postbiotics, including 100, 50, 25, 12.5, 6.25, and 3 mg/ml, were tested against K. pneumoniae ATCC 7881 at a concentration of 1:100 of a 0.5 McFarland standard.
Evaluation of the synergistic effects of our native postbiotic with antibiotic
In this step, the MIC test was performed using 50 µL of postbiotic at concentrations of 100, 50, 25, and 12.5 mg/ml, along with 50 µL of K. pneumoniae ATCC 7881 (at a concentration of 1:100 of a 0.5 McFarland standard) and 50 µL of antibiotic with serial dilutions ranging from 2048 µg/ml to 2 µg/ml. The plates were incubated at 37 °C for 24 h, after which the wells were examined for bacterial growth.
Evaluation of fractional inhibitory concentration (FIC)
Fractional Inhibitory Concentration (FIC) index is widely used in checkerboard assays and other combination studies to assess the potential enhancement or inhibition of antimicrobial activity when drugs are combined [26]. This method was calculated using the formula:
FIC = MIC of drug A in combination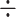 MIC of drug A alone + MIC of drug B in combination
MIC of drug A alone + MIC of drug B in combination MIC of drug B alone.
MIC of drug B alone.
Here: Drug A is our native postbiotic and drug B is our antibiotics (imipenem or amoxicillin).
Interpretation of FICI values is generally as follows:
FICI ≤ 0.5 indicates synergistic interaction.
0.5 < FICI ≤ 1 indicates additive effect.
1 < FICI < 4 indicates no interaction (indifference).
FICI ≥ 4 indicates antagonism.
The efficacy of postbiotics on the expression of blaSHV, blaCTX, blaTEM, and blaNDM−1 genes
To assess the efficacy of postbiotics on the expression of the genes blaSHV, blaCTX, blaTEM, and blaNDM−1, samples were collected after co-culturing this agent simultaneously with K. pneumoniae ATCC 7881. RNA was extracted from the samples, and following cDNA synthesis, real-time PCR was performed using primers specific to each gene. The list of primers used in the current study is provided in Table 1.
Table 1.
The primers used in the current study
| Gene | Primer Sequence (5’ >3’) |
|---|---|
| bla CTX−M |
F: 5-GAAAGCGAACCGAATCT-3 R: 5-GACATCGTCCCATTGAC-3 |
| bla TEM |
F:5-AGTATTCAACATTTCCGTGT-3 R:5- TAATCAGTGAGGCACCTATCTC-3 |
| bla SHV |
R:5- TAATCAGTGAGGCACCTATCTC-3 R:5- TAATCAGTGAGGCACCTATCTC-3 |
| bla NDM−1 |
F: 5′-CGCAACACAGCCTGACTTT-3′ R: 5′-TCGATCCCAACGGTGATATT-3′ |
Statistical analysis
The housekeeping gene gapdh and 16 S rRNA was utilized for normalization purposes. The relative quantification of targeted gene expression was determined using the 2−ΔΔCt method. The statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software Inc, CA, USA). The one-way test ANOVA followed by Tukey’s post hoc test was used for normal data, while the Kruskal-W allis test was used for non-normal data. The results are presented as the mean ± standard error. A P-value of less than 0.05 was considered to be statistically significant.
Results
Resistance and antibacterial effects of probiotic strains
A total of 77 native probiotic strains demonstrated complete resistance to amoxicillin and imipenem at 2048 µg/mL. Among these, four strains-Lactiplantibacillus plantarum RP155, RP403, RP225, and Ligilactobacillus salivarius RP317-were selected for postbiotic preparation due to their ability to fully inhibit the growth of K. pneumoniae ATCC 7881 at low antibiotic concentrations. Based on these results, these four strains were selected as candidates for postbiotic production.
The effectiveness of our native postbiotic on the growth of K. pneumoniae ATCC 7881
The colony counts of K. pneumoniae ATCC 7881 were evaluated at different concentrations of four postbiotics (L. plantarum RP155, 403, 225 and L. salivarius RP317). For Postbiotic L. plantarum RP155, no colonies were detected at concentrations of 100, 50, and 25 mg/ml, while at 12.5 mg/ml the count was 7200, increasing to 8100 at 6.25 mg/ml and 10,000 at 3 mg/ml. Postbiotic L. plantarum MR225 completely inhibited colony formation at concentrations of 100, 50, 25, and 12.5 mg/ml; however, colony counts increased to 5000 and 6800 at 6.25 and 3 mg/ml, respectively. For postbiotic L. salivarius MR317, zero colonies were observed at concentrations of 100, 50, and 25 mg/ml, while at 12.5 mg/ml there were 8500 colonies, rising to 9200 at 6.25 mg/ml and 12,000 at 3 mg/ml. Finally, postbiotic L. plantarum MR403 showed no bacterial colonies at 100 and 50 mg/ml, but colonies appeared at 8600 at 25 mg/ml and increased to 12,000, 12,800, and 13,300 at concentrations of 12.5, 6.25, and 3 mg/ml, respectively. These results indicate varying inhibitory effects of the postbiotics on bacterial growth depending on concentration (Fig. 1). Indeed, as shown in Fig. 1, the MIC of our postbiotics were 25 mg/ml for L. plantarum RP155 and L. salivarius MR317, 12.5 mg/ml for L. plantarum MR225, and 50 mg/ml for L. plantarum MR403.
Fig. 1.
Colony count of K. pneumoniae ATCC 7881 after coculture with our native postbiotics, including L. plantarum RP225, L. plantarum RP403, L. plantarum RP155, and L. salivarius RP317
The effectiveness of postbiotics in improving the performance of amoxicillin and imipenem
The results of the colony count for K. pneumoniae ATCC 7881, with postbiotic treatment in the presence of antibiotics, are presented in Fig. 2. The data presented in this figure show the colony counts of K. pneumoniae ATCC 7881 when exposed to combinations of four native postbiotics (L. plantarum MR155 at 25 mg/ml, L. plantarum MR225 at 12.5 mg/ml, L. salivarius MR317 at 25 mg/ml, and L. plantarum MR403 at 50 mg/ml) with varying concentrations of two antibiotics: amoxicillin and imipenem. For amoxicillin, complete inhibition of bacterial growth was observed at higher concentrations (2048 to 512 µg/ml) across all postbiotics, while lower concentrations showed increasing colony counts, especially with postbiotic L. plantarum MR225, which reached up to 250 colonies at 1 µg/ml. In contrast, the combination with imipenem showed complete inhibition with zero colony counts at all tested concentrations (2048 to 1 µg/ml) regardless of the postbiotic used. These results indicate a strong antibacterial effect of postbiotics when combined with imipenem and a concentration-dependent additive or partial inhibitory effect with amoxicillin.
Fig. 2.
Colony count of K. pneumoniae ATCC 7881 after coculture with our native postbiotics, including L. plantarum RP225, L. plantarum RP403, L. plantarum RP155, and L. salivarius RP317 along with imipenem and amoxicillin
Evaluation of fractional inhibitory concentration (FIC)
The FIC indices were calculated for the combinations of postbiotics and antibiotics against K. pneumoniae ATCC 7881. For the postbiotics L. plantarum MR155, L. plantarum MR225, L. salivarius MR317, and L. plantarum MR403, the FIC values were 1.0, 1.25, 0.75, and 0.02, respectively, indicating additive effect for L. plantarum MR155 and L. salivarius MR317, indifference for L. plantarum MR225, and synergism for L. plantarum MR403. In contrast, the combinations of the same postbiotics with imipenem resulted in FIC values of 0.04, 0.08, 0.04, and 0.02, for L. plantarum MR155, L. plantarum MR225, L. salivarius MR317, and L. plantarum MR403, respectively, demonstrating strong synergistic interactions across all tested strains. These results suggest that while some postbiotic-antibiotic combinations show only additive or indifferent effects, others, particularly those with imipenem, exhibit significant synergy, highlighting the potential of postbiotics to enhance antibiotic efficacy against resistant K. pneumoniae ATCC 7881. It should be noted that the MIC of postbiotic could be seen in Fig. 1, MIC of our native postbiotic in combination of imipenem and amoxicillin could be seen in Fig. 2, and the checker board could be found in Supplementary file 1.
The effect of postbiotics on the resistant genes’ expression
Analysis of the results regarding the effect of postbiotics on the expression of selected genes, both alone and in combination with antibiotics, revealed that our native postbiotics were able to reduce the expression of the evaluated genes (see Fig. 2). However, the effects of all postbiotics were shown to be variable. Also, it should be noted that the concentration of the postbiotic was carefully adjusted because of its strong inhibitory effect on K. pneumoniae; thus, to obtain reliable 16S rRNA (as reference gene) amplification and accurate gene expression results, the postbiotic concentration was reduced to 3 mg/mL.”
A. blaNDM expression
The expression of the blaNDM gene in K. pneumoniae ATCC 7881 exhibited distinct responses to different treatment regimens. Notably, treatment with postbiotic L. plantarum RP225 or postbiotic L. salivarius RP317 alone led to a marked increase in blaNDM expression compared to the untreated control. In contrast, postbiotics L. plantarum RP403 and L. plantarum RP155 alone showed levels comparable lower than the control (p < 0.0001). Importantly, when any of the postbiotics were combined with imipenem, blaNDM expression was dramatically reduced, with several combinations resulting in nearly undetectable levels of gene expression. These findings suggest that while certain postbiotics alone may upregulate blaNDM, their combination with imipenem exerts a strong suppressive effect on this carbapenem resistance gene (see Fig. 3A).
Fig. 3.
Effects of native postbiotics with and without antibiotic combinations on the expression of resistance genes in K. pneumoniae ATCC7881. Relative fold change in the expression levels of (A) blaNDM, (B) blaCTX, (C) blaTEM, and (D) blaSHV genes in K. pneumoniae ATCC7881 following treatment with different native postbiotics (225, 403, 317, and 155), alone or in combination with antibiotics (imipenem or amoxicillin), as compared to untreated control groups (K. pneumoniae ATCC7881 in combination with antibiotic, imipenem/amoxicillin). Gene expression was quantified by qRT-PCR and normalized to the reference gene (16 S rRNA). Data are presented as fold change relative to control. Bars represent mean values; “0” indicates complete suppression of gene expression. Significant differences compared to control are indicated. (••p < 0.01; •••p < 0.001; ••••p < 0.0001). It should be noted that K.p 7881 stands for K. pneumoniae ATCC7881, and Postbiotic 225, 403, 155, and 317 stands for L. plantarum RP225, L. plantarum RP403, L. plantarum RP155, and L. salivarius RP317, respectively
B. blaCTX expression
The expression of the blaCTX gene in K. pneumoniae ATCC 7881 was significantly influenced by the different treatment regimens. In the majority of treatment groups, blaCTX expression was completely suppressed, with fold change values reduced to zero. However, in three specific treatments—postbiotic L. plantarum RP225 alone, postbiotic L. salivarius RP317 alone, and postbiotic L. plantarum RP155 combined with antibiotic (amoxicillin)—the expression of blaCTX was not entirely eliminated, though these groups still showed a statistically significant reduction compared to the untreated control (p < 0.0001). This indicates that all postbiotic treatments exert a significant inhibitory effect on blaCTX gene expression, with most treatments completely suppressing it to zero, and even in cases where expression was not fully eliminated, the reduction was statistically significant (see Fig. 3B).
C. blaTEM expression
The expression of the blaTEM gene in K. pneumoniae ATCC 7881 was differentially affected by the various treatments. Treatment with postbiotic L. plantarum RP225 combined with amoxicillin, as well as postbiotics L. salivarius RP317 and L. plantarum RP155 alone, led to a reduction in blaTEM expression compared to the control, although the expression was not completely suppressed to zero. In contrast, all other treatments-including postbiotic L. plantarum RP403 alone or in combination with amoxicillin, amoxicillin combined with postbiotics L. salivarius RP317 and L. plantarum RP155 completely abolished blaTEM expression, resulting in a fold change of zero. Notably, postbiotic L. plantarum RP225 alone was the only treatment that resulted in an increase in blaTEM expression relative to the control, highlighting its undesirable effect when used in isolation. This underscores the importance of carefully selecting postbiotic regimens, as some may not only fail to suppress resistance gene expression but may actually enhance it when administered alone (see Fig. 3C).
D. blaSHV expression
The expression of the blaSHV gene in K. pneumoniae ATCC 7881 was significantly reduced in the groups treated with postbiotic L. plantarum RP225 alone, postbiotic L. salivarius RP317 alone, postbiotic L. plantarum RP155 alone, and combined with amoxicillin. In all other treatment groups-including postbiotic L. plantarum RP403 alone or in combination with amoxicillin, and amoxicillin combined with postbiotics L. plantarum RP225 or L. salivarius RP317 the expression level of blaSHV was completely suppressed to zero. (see Fig. 3D).
Discussion
According to World Health Organization (WHO) antimicrobial resistance is one of the most critical topics that has gained significant attention, especially in recent years, due to its high mortality rate-1.27 million deaths in 2019 alone. In addition to the global death, the economic cost of AMR is also a major concern. According to the World Bank, AMR could result in an additional US$1 trillion in healthcare costs by 2050 [27]. As antibiotic resistance increases worldwide, there has also been a lack of success in discovering new antibacterial drugs. However, by learning from previous antibiotic research and recent advances in our understanding of how antibiotics work and how bacteria function, there is hope for developing innovative therapies that can effectively combat infections in this challenging era of resistance [28]. In this study, we sought to present a novel strategy for combating microbial resistance by utilizing compounds that enhance antibiotic efficacy. Beyond the well-known antimicrobial properties of probiotics and their derivatives, we propose that these agents may exert unique effects in potentiating the action of antibiotics.
The present study was conducted in two phases, focusing on both phenotypic and genotypic analyses. Initially, to evaluate the antimicrobial activity of our native postbiotics, the K. pneumoniae strain 7881 was selected, and the minimum inhibitory concentration (MIC) against amoxicillin and imipenem was determined. Our findings revealed that K. pneumoniae ATCC 7881 shown to be highly resistant to amoxicillin, a beta-lactam antibiotic, and imipenem, a carbapenem antibiotic, at concentrations up to 2048 µg/ml. Despite this high level of resistance, the first phase demonstrated that treatment with our native postbiotic agents along with antibiotics significantly reduced the bacterial colony count, ultimately leading to the complete eradication of K. pneumoniae ATCC 7881. Also, the calculated FIC indices provide valuable insight into the interactions between postbiotics and antibiotics against K. pneumoniae ATCC 7881. The results demonstrate that combinations involving L. plantarum MR155 and L. salivarius MR317 mainly exert additive effects, which implies that their combined antibacterial activity corresponds closely to the sum of their individual effects without significant enhancement or suppression. Conversely, the indifference observed with L. plantarum MR225 suggests that this particular postbiotic neither enhanced nor diminished antibiotic efficacy, indicating a neutral interaction. Notably, L. plantarum MR403 exhibited a pronounced synergistic effect, as evidenced by an FIC value well below 0.5, signifying a substantial increase in antibacterial activity when combined with the antibiotic. The most striking finding is the consistent strong synergy observed when all tested postbiotics were combined with imipenem, evidenced by their low FIC values (0.02 to 0.08). This synergy indicates a more than additive effect, where the postbiotics could potentiate the bactericidal activity of imipenem, possibly through mechanisms such as enhanced membrane permeability or interference with bacterial resistance pathways. These interactions highlight the potential therapeutic advantage of using specific postbiotic-antibiotic combinations, particularly with carbapenem-class antibiotics, to combat antibiotic-resistant K. pneumoniae. It is also important to consider that FIC index interpretations have inherent limitations, such as not distinguishing directional effects between compounds; nonetheless, these findings strongly suggest that selected postbiotics can serve as effective adjuvants to conventional antibiotic therapy. The antimicrobial properties of postbiotics can be linked to their varied characteristics, which generally encompass a range of bioactive compounds, including organic acids, bacteriocins, peptides, and other metabolites [29]. These substances demonstrate antimicrobial effects by modifying the properties of bacterial cells; for instance, organic acids decrease intracellular pH [30], compromising membrane integrity, while bacteriocins form pores in bacterial membranes, resulting in cell lysis [31].
Some studies support the antimicrobial efficacy of postbiotics, consistent with the findings of the present study. For example, Banakar et al. reported that postbiotics derived from Lactobacillus rhamnosus GG significantly reduced the colony-forming unit (CFU) count of Streptococcus mutans, which is one of the most important bacterial species involved in dental plaque formation through biofilm production [32]. In a separate investigation carried out by Tong et al., postbiotics sourced from Bacillus amyloliquefaciens J and Lactobacillus plantarum SN4 demonstrated remarkable antimicrobial effectiveness against Escherichia coli, Staphylococcus aureus, and Salmonella typhimurium by disrupting bacterial cell membranes, leading to the leakage of cellular contents. Their findings highlighted organic acids, especially lactic acid and acetic acid, as the primary antibacterial agents responsible for this antimicrobial effect [33].
The antibacterial effects of native probiotics not only shown as phenotypically but also at the gene expression level. The summary of observed effects of our native postbiotics on resistant gene expression level in K. pneumoniae ATCC 7881 is provided in Table 2. Our data showed that Postbiotic L. plantarum RP403 demonstrated significant suppression (zero expression) of all four resistance genes, both alone and with amoxicillin/imipenem. Postbiotic L. plantarum RP225 exhibited mixed results, including increases in blaNDM and blaTEM but reductions in blaCTX and blaSHV. In combination with imipenem/amoxicillin, postbiotic L. plantarum RP225 completely suppressed blaNDM, blaCTX, and blaSHV, with a mild reduction in blaTEM. Postbiotic L. salivarius RP317 increased blaNDM expression severely but reduced other genes; its combination with amoxicillin/imipenem achieved complete suppression of blaCTX, blaTEM, and blaSHV, and reduced blaNDM significantly. Postbiotic L. plantarum RP 155 notably decreased blaNDM and blaSHV and completely suppressed blaCTX, with mild reductions blaTEM; its combination with amoxicillin/imipenem further enhanced suppression, especially of blaNDM and blaTEM. The evaluation of the antimicrobial effectiveness of postbiotics at the gene expression level has been examined in other studies. As noted by Nezhadi et al., postbiotics derived from Lactobacillus plantarum resulted in a significant reduction in the expression levels of resistance genes, such as ermB (P = 0.007) and blaKPC (P = 0.02) [34]. Furthermore, Kunishima et al. reported that the supernatants from various gut microbiota, including C. butyricum, C. difficile, C. perfringens, E. faecium, and L. plantarum, were able to lower the expression levels of blaCTX in ESBL-producing E. coli [35].
Table 2.
Summary of observed effects of our native postbiotics on resistant gene expression level in K. pneumoniae ATCC 7881
| Treatment Group | blaNDM Expression | blaCTX Expression | blaTEM Expression | blaSHV Expression |
|---|---|---|---|---|
| Postbiotic 225 alone | Moderate increase (undesirable) | Mild reduction | Severe increase (undesirable) | Severe reduction |
| Amoxicillin + Postbiotic 225 | Complete suppression (zero) | Complete suppression (zero) | Mild reduction | Complete suppression (zero) |
| Postbiotic 403 alone | Complete suppression (zero) | Complete suppression (zero) | Complete suppression (zero) | Complete suppression (zero) |
| Amoxicillin + Postbiotic 403 | Complete suppression (zero) | Complete suppression (zero) | Complete suppression (zero) | Complete suppression (zero) |
| Postbiotic 317 alone | Severe increase (undesirable) | Severe reduction | Mild reduction | Severe reduction |
| Amoxicillin + Postbiotic 317 | Severe reduction | Complete suppression (zero) | Complete suppression (zero) | Complete suppression (zero) |
| Postbiotic 155 alone | Severe reduction | Complete suppression (zero) | Mild reduction | Severe reduction |
| Amoxicillin + Postbiotic 155 | Complete suppression (zero) | Severe reduction | Complete suppression (zero) | Severe reduction |
It should be noted that one undesirable observation emerged in the current study. According to our results, certain postbiotics, specifically L. plantarum RP225 and L. salivarius RP317, when used alone, led to an increase in resistance gene expression. This phenomenon, however, does not necessarily result in adverse effects. The transient upregulation of resistance genes induced by postbiotics alone may stem from factors such as alterations in basal metabolism, cellular stress responses, or other related mechanisms. Importantly, in the absence of antibiotic treatment, this increased gene expression poses little risk, as there is no antibiotic pressure to exert selective disadvantage. Moreover, the postbiotics themselves remain unharmed and continue their antimicrobial activity through alternative aforementioned mechanisms. Conversely, when postbiotics are combined with antibiotics, a pronounced decrease in resistance gene expression is observed. This synergistic effect significantly diminishes the expression of resistance enzymes that would otherwise compromise antibiotic efficacy. This suppression not only limit bacterial defense mechanisms but also enhances the overall antimicrobial activity.
Such a mechanism holds profound implications for One Health strategies. By modulating resistance gene expression and boosting antibiotic effectiveness, postbiotics can serve as valuable adjuncts in combating antibiotic resistance, particularly in regions where antibiotics are extensively used without prescription. Furthermore, their use may help mitigate the spread of resistance driven by antibiotic residues in agriculture and poultry production. Ultimately, this combined approach can reduce resistance gene transfer, preserve antibiotic utility, and contribute substantially to integrated antimicrobial stewardship efforts. Collectively, these findings underscore the potential of postbiotic-antibiotic combinations to improve treatment outcomes and address the global challenge of multidrug resistance from a One Health perspective.
Conclusion
In summary, this research emphasizes the encouraging potential of native postbiotics as supplementary antimicrobial agents against multidrug-resistant K. pneumoniae. Although certain postbiotics independently exhibited inconsistent effects on resistance gene expression, their combination with antibiotics, especially amoxicillin, led to a marked reduction of essential resistance genes and successful bacterial elimination. These results highlight the synergistic relationship between postbiotics and antibiotics, likely facilitated by bioactive metabolites that compromise bacterial defenses and improve antibiotic effectiveness. In light of the worldwide challenge posed by antimicrobial resistance and the difficulties in developing new antibiotics, postbiotics offer an innovative and safe therapeutic pathway to enhance the effectiveness of existing treatments. Additional research and clinical validation are necessary to thoroughly investigate their use in fighting resistant infections.
Supplementary Information
Acknowledgements
The authors would like to thank Pasteur Institute of Iran as funding agency (with grant number 2283).
Authors’ contributions
Performed the experiments: ZH, FS, EHAGhKh, Data analysis: ShA, writing of the manuscript: ShA, ZH, Revised manuscript: FB, MR and Conceived and designed the experiments: ShA, MR.
Funding
The current research was done in support of Pasteur Institute of Iran supported as a funding agency (with grant number 2283).
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The experimental protocols were established following the Declaration of Helsinki and approved by the ethics committee of Pasteur Institute of Iran (IR.PII.AEC.1402.018).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shadi Aghamohammad and Fatemeh Sakaki contributed as co-first authors.
Contributor Information
Farzad Badmasti, Email: fbadmasti2008@gmail.com.
Mahdi Rohani, Email: kia.rohani1979@gmail.com.
References
- 1.Ahmed SK, Hussein S, Qurbani K, Ibrahim RH, Fareeq A, Mahmood KA, et al. Antimicrobial resistance: Impacts, challenges, and future prospects. J Med Surg Public Health. 2024;2:100081. [Google Scholar]
- 2.Velazquez-Meza ME, Galarde-López M, Carrillo-Quiróz B, Alpuche-Aranda CM. Antimicrobial resistance: one health approach. Veterinary World. 2022;15(3):743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plague AM. Antimicrobial Resistance. 2025.
- 4.Salam MA, Al-Amin MY, Salam MT, Pawar JS, Akhter N, Rabaan AA, et al. editors. Antimicrobial resistance: a growing serious threat for global public health. Healthcare: Multidisciplinary Digital Publishing Institute; 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsson DJ, Flach C-F. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022;20(5):257–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L-G, Zhang T. Plasmid-mediated antibiotic resistance gene transfer under environmental stresses: insights from laboratory-based studies. Sci Total Environ. 2023;887:163870. [DOI] [PubMed] [Google Scholar]
- 7.Breijyeh Z, Karaman R. Design and synthesis of novel antimicrobial agents. Antibiotics. 2023;12(3):628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mudenda S, Chabalenge B, Daka V, Mfune RL, Salachi KI, Mohamed S, et al. Global strategies to combat antimicrobial resistance: a one health perspective. Pharmacol Pharm. 2023;14(8):271–328. [Google Scholar]
- 9.Cocker D, Birgand G, Zhu N, Rodriguez-Manzano J, Ahmad R, Jambo K, et al. Healthcare as a driver, reservoir and amplifier of antimicrobial resistance: opportunities for interventions. Nat Rev Microbiol. 2024;22(10):636–49. [DOI] [PubMed] [Google Scholar]
- 10.Ifedinezi OV, Nnaji ND, Anumudu CK, Ekwueme CT, Uhegwu CC, Ihenetu FC, et al. Environmental antimicrobial resistance: implications for food safety and public health. Antibiotics. 2024;13(11):1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ermini ML, Voliani V. Antimicrobial nano-agents: the copper age. ACS Nano. 2021;15(4):6008–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo L, Huang W, Zhang J, Yu Y, Sun T. Metal-based nanoparticles as antimicrobial agents: a review. ACS Appl Nano Mater. 2024;7(3):2529–45. [Google Scholar]
- 13.Fijan S. Probiotics and Their Antimicrobial Effect. Microorganisms. 2023; 11(2):528. [DOI] [PMC free article] [PubMed]
- 14.Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EM, et al. The international scientific association of probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Reviews Gastroenterol Hepatol. 2021;18(9):649–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinderola G, Sanders ME, Cunningham M, Hill C. Frequently asked questions about the ISAPP postbiotic definition. Front Microbiol. 2024;14:1324565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji J, Jin W, Liu S, Jiao Z, Li X. Probiotics, prebiotics, and postbiotics in health and disease. MedComm, 2023; 4 (6): e420. [DOI] [PMC free article] [PubMed]
- 17.Lodhi MMI, Lakshminarayana S, Aaftab G. Occurrence of nosocomial multi-drug resistant Klebsiella pneumoniae in india: A systemic review and meta-analysis. IP Int J Med Microbiol Trop Dis. 2024;10:17–23. [Google Scholar]
- 18.Walsh TR, Gales AC, Laxminarayan R, Dodd PC (2023) Antimicrobial Resistance: Addressing a Global Threat to Humanity. PLoS Med 20(7): e1004264. [DOI] [PMC free article] [PubMed]
- 19.Ghenea AE, Zlatian OM, Cristea OM, Ungureanu A, Mititelu RR, Balasoiu AT, et al. TEM, CTX-M, SHV genes in ESBL-producing Escherichia coli and Klebsiella pneumoniae isolated from clinical samples in a County clinical emergency hospital Romania-predominance of CTX-M-15. Antibiotics. 2022;11(4):503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Kumar S, Zhang L, Wu H, Wu H. Characteristics of antibiotic resistance mechanisms and genes of Klebsiella pneumoniae. Open Med. 2023;18(1):20230707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quintelas M, Silva V, Araújo S, Tejedor-Junco MT, Pereira JE, Igrejas G, et al. Klebsiella in wildlife: clonal dynamics and antibiotic resistance Profiles, a systematic review. Pathogens. 2024;13(11):945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magobo RE, Ismail H, Lowe M, Strasheim W, Mogokotleng R, Perovic O, et al. Outbreak of NDM-1–and OXA-181–producing Klebsiella pneumoniae bloodstream infections in a neonatal unit, South Africa. Emerg Infect Dis. 2023;29(8):1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Kumar S, Zhang L. Mechanisms of antibiotic resistance and developments in therapeutic strategies to combat Klebsiella pneumoniae infection. Infect Drug Resist. 2024; 19(17):1107-1119 [DOI] [PMC free article] [PubMed]
- 24.Do AD, Quang HP, Phan QK. Probiotic cell-free supernatant as effective antimicrobials against Klebsiella pneumoniae and reduce antibiotic resistance development. Int Microbiol. 2024; 28(4):623-632. [DOI] [PubMed]
- 25.Rohani M, Noohi N, Talebi M, Katouli M, Pourshafie MR. Highly heterogeneous probiotic Lactobacillus species in healthy Iranians with low functional activities. PLoS ONE. 2015;10(12):e0144467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fatsis-Kavalopoulos N, Sánchez-Hevia DL, Andersson DI. Beyond the FIC index: the extended information from fractional inhibitory concentrations (FICs). J Antimicrob Chemother. 2024;79(9):2394–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Organization WH. WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance, to guide research, development, and strategies to prevent and control antimicrobial resistance. World Health Organization; 2024.
- 28.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529(7586):336–43. [DOI] [PubMed] [Google Scholar]
- 29.Isaac-Bamgboye FJ, Mgbechidinma CL, Onyeaka H, Isaac-Bamgboye IT, Chukwugozie DC. Exploring the potential of postbiotics for food safety and human health improvement. J Nutr Metabolism. 2024;2024(1):1868161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prajapati N, Patel J, Singh S, Yadav VK, Joshi C, Patani A, et al. Postbiotic production: Harnessing the power of microbial metabolites for health applications. Front Microbiol. 2023;14:1306192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Che J, Shi J, Fang C, Zeng X, Wu Z, Du Q, et al. Elimination of pathogen biofilms via postbiotics from lactic acid bacteria: A promising method in food and biomedicine. Microorganisms. 2024;12(4):704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banakar M, Pourhajibagher M, Etemad-Moghadam S, Mehran M, Yazdi MH, Haghgoo R, et al. Antimicrobial effects of postbiotic mediators derived from Lactobacillus rhamnosus GG and Lactobacillus reuteri on Streptococcus mutans. Front Bioscience-Landmark. 2023;28(5):88. [DOI] [PubMed] [Google Scholar]
- 33.Tong Y, Abbas Z, Zhang J, Wang J, Zhou Y, Si D et al. Antimicrobial activity and mechanism of novel postbiotics against foodborne pathogens. LWT. 2025; (17):117464.
- 34.Nezhadi J, Ahmadi A. Assessing the efficacy of postbiotics derived from Lactobacillus plantarum on antibiotic resistance genes in nosocomial pathogens such as Enterococcus faecalis and Pseudomonas aeruginosa. Lett Appl Microbiol. 2024;77(12):ovae127. [DOI] [PubMed] [Google Scholar]
- 35.Kunishima H, Ishibashi N, Wada K, Oka K, Takahashi M, Yamasaki Y, et al. The effect of gut microbiota and probiotic organisms on the properties of extended spectrum beta-lactamase producing and carbapenem resistant Enterobacteriaceae including growth, beta-lactamase activity and gene transmissibility. J Infect Chemother. 2019;25(11):894–900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.





