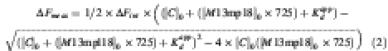Abstract
Helicases are transferred to replication origins by helicase loading factors. The Escherichia coli DnaC and eukaryotic Cdc6/18 helicase loaders contain ATP sites and are both members of the AAA+ family. One might expect that ATP is required for helicase loading; however, this study on DnaC illustrates that ATP is not actually needed for DnaC to load helicase onto single-strand DNA (ssDNA). In fact, it seems to be a paradox that after transfer of helicase to DNA, DnaC–ATP inhibits helicase action. In addition, ATP is required for DnaC function at an early step in oriC replication in which ATP stimulates ssDNA binding by DnaC, leading to expansion of the ssDNA bubble at the origin. Two cofactors, ssDNA and DnaB, trigger hydrolysis of ATP, converting DnaC to the ADP form that no longer inhibits DnaB. These observations have led to the idea that DnaC is a ‘dual’ switch protein, where both the ATP and the ADP forms are sequentially required for replication. This dual switching process may underlie the sensitivity of DnaB to even small fluctuations in DnaC levels.
Keywords: ATP/ADP switch protein/DnaB/DnaC/helicase loader/oriC
Introduction
In both prokaryotes and eukaryotes, replication origins are activated by one or more origin binding proteins, leading to a structure upon which helicases can be assembled to initiate replication forks (Baker and Bell, 1998). In Escherichia coli, DnaA binds the origin and melts a region within three AT-rich 13mer direct repeats (Fuller et al., 1984; Bramhill and Kornberg, 1988; Gille and Messer, 1991). Two DnaB hexameric helicases are then assembled onto the single-strand DNA (ssDNA) bubble to form bi-directional replication forks (Baker et al., 1987; Funnell et al., 1987; Bramhill and Kornberg, 1988; Fang et al., 1999). The DnaC helicase loader is required in this process, but dissociates from DnaB after loading it onto DNA (Funnell et al., 1987; Fang et al., 1999). Each of these proteins binds and/or hydrolyzes ATP (Reha-Krantz and Hurwitz, 1978; Arai and Kornberg, 1981; Sekimizu et al., 1987; Wahle et al., 1989a).
Similar events occur in other organisms. In eukaryotes, the origin binding protein is the heterohexameric origin recognition complex (ORC) (Bell and Stillman, 1992; Dutta and Bell, 1997; Kelly and Brown, 2000). The Mcm2–7 complex, analogous to a DnaB hexamer, is believed to act as a helicase at replication forks (Baker and Bell, 1998). The Cdc6 (cdc18) initiation factor (along with Cdt1) acts early in the process of forming a pre-replication complex (preRC) and is thought to behave analogously to DnaC to assemble the heterohexameric Mcm2–7 complex into the preRC at origins (Donovan et al., 1997). Like DnaC, Cdc6 is only needed in the preRC assembly process and does not remain bound to the origin with ORC and Mcm2–7 (Weinreich et al., 1999).
Several of these replication initiation factors are members of the AAA+ family of ATPases (ATPases associated with a variety of cellular activities) including DnaA, Orc1, Orc4, Orc5, DnaC, Cdc6 and the Mcm2–7 proteins (Koonin, 1992; Neuwald et al., 1999). The mechanisms of these proteins, and the function of ATP in each of them, are topics of current interest. DnaA utilizes ATP to destabilize the AT-rich region in the origin, forming a ssDNA bubble, although the mechanism of this reaction is unclear (Bramhill and Kornberg, 1988). In budding yeast, the Orc1 subunit uses ATP for origin binding and a functional ATP site is essential for cell viability (Klemm and Bell, 2001). Mutations in Orc5p that interfere with ATP binding interfere with cell growth (Klemm et al., 1997). In contrast, Orc4 does not seem to bind ATP (Klemm et al., 1997). Mcm2–7 proteins are thought to couple ATP to helicase action (Ishimi, 1997). The ATP site of Cdc6 is important for function, as mutants in the ATP site are not viable, and do not appear to be capable of forming the preRC in vivo (Perkins and Diffley, 1998; Weinreich et al., 1999). Likewise, E.coli DnaC protein is required to load DnaB helicase onto oriC, and ATP has generally been assumed to play a role in this helicase loading process (Kobori and Kornberg, 1982; Wahle et al., 1989b).
This report uses the E.coli system to examine the mechanism of DnaC in helicase loading and origin activation. Several conclusions can be derived from the study. We find that DnaC is a dual ATP/ADP switch protein, where ATP hydrolysis by DnaC is triggered by DnaB and ssDNA. Surprisingly, ATP is not required for DnaC to load DnaB onto ssDNA. In fact, after loading DnaB onto ssDNA (which DnaC can do with ATP, ADP or even with no nucleotide) the DnaC–ATP form inhibits DNA unwinding by DnaB. However, ATP does play a crucial role in DnaC action at oriC. Results presented herein indicate that the role of ATP is to strengthen DnaC interaction with ssDNA, thereby expanding the ssDNA bubble at the origin. After transfer of DnaB to oriC, the inhibitory effect of DnaC–ATP must be switched off. We find that ATP hydrolysis by DnaC is triggered by the combined presence of DnaB and ssDNA. Hence, after assembly of DnaB onto the ssDNA bubble at oriC, DnaC hydrolyzes bound ATP leading to the DnaC–ADP form, which no longer inhibits the helicase. These findings lead to a model of DnaC action as a dual ATP/ADP switch, in which DnaC–ATP loads the helicase onto oriC and DnaC–ADP relieves inhibition of the DnaB helicase.
Results
DnaC ATPase requires two effectors
DnaC is a member of the AAA+ family of ATPases, but has not been demonstrated to hydrolyze ATP. Some members of this family lack a consensus ATP binding site and do not bind or hydrolyze ATP. For example, the δ and δ′ subunits of the E.coli γ complex clamp loader are AAA+ proteins, but they do not interact with ATP (Naktinis et al., 1995; Neuwald et al., 1999; Jeruzalmi et al., 2001). In these proteins, the ATP site has been mutated during evolution, preventing it from binding ATP. However, binding of adenine nucleotide to DnaC has been observed (Wahle et al., 1989a; Galletto et al., 2000), which prompted us to take a closer look at DnaC for ATPase activity.
Although DnaC does not hydrolyze ATP by itself, it seemed a reasonable expectation that this activity may require certain cofactors, such as ssDNA, or even binding to its partner protein, DnaB. In the event that DnaC requires DnaB for ATPase activity, an activity originating from DnaC may have been obscured in previous studies by the very active DNA-dependent ATPase intrinsic to DnaB (Reha-Krantz and Hurwitz, 1978; Arai and Kornberg, 1981). To test whether DnaB causes ATP hydrolysis within DnaC, we used a DnaB active site mutant (K236A), which binds ATP but does not hydrolyze it (Fang et al., 1999).
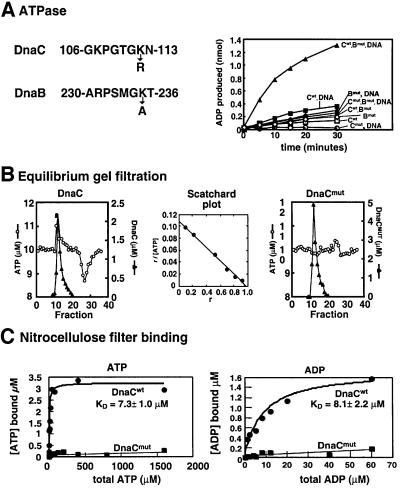
Study of DnaC in the presence of the DnaB mutant revealed a weak ATPase activity (0.6 ATP/min/DnaC) (Figure 1A; Table I). Slight ATPase activity by DnaC was also observed in the presence of ssDNA (0.9 ATP/min/DnaC). Maximal activity required the presence of both DnaB and ssDNA (3.5 ATP/min/DnaC). To examine more definitively whether the observed ATPase is intrinsic to DnaC, we constructed a mutant of DnaC in the putative ATP site (K112R; see Figure 1A) and tested it for ATP binding and hydrolysis activity. The mutant DnaC lacked ATPase activity whether ssDNA and mutant DnaB were present or not (Figure 1A). In Figure 1B, we determined the Kd value of wild-type and mutant DnaC for ATP using the equilibrium gel filtration technique. In this experiment, DnaC is passed through a gel filtration column equilibrated with [α-32P]ATP. If DnaC binds ATP, then a peak of [32P]ATP will comigrate with DnaC through the gel filtration column. In addition, a corresponding trough in ATP concentration elutes later from the gel filtration column and is caused by loss of [32P]ATP from the buffer due to the [32P]ATP bound to DnaC. An example of a positive result is shown in the left panel of Figure 1B. Performing the experiment at several different concentrations of ATP yields the information for a Scatchard plot shown in the middle panel of Figure 1B (see Materials and methods for details). Whereas DnaC binds to ATP (Kd ∼9 µM), consistent with previous reports (Wahle et al., 1989a; Galletto et al., 2000), ATP binding to the DnaC mutant is not detected (Figure 1B, right panel).
Fig. 1. Nucleotide binding and hydrolysis by wild-type and mutant DnaC. (A) The sequences of the Walker A motifs in DnaB and DnaC are shown (left panel). The sites of the DnaB alanine mutation and the DnaC arginine mutation are indicated. ATP hydrolysis by DnaC (Cwt) and DnaC mutant (Cmut) in the presence of M13mp18 ssDNA (DNA) and mutant DnaB (Bmut) were measured as described in Materials and methods. When present, 16 pmol DnaC or DnaCmut, 4.6 pmol DnaBmut hexamer (27.6 pmol as monomer) and 165 ng M13mp18 ssDNA were used in 25 µl of reaction buffer with 100 µM [α-32P]ATP. Assays contained DnaC only (open squares), DnaB mutant only (open diamonds), DnaC with DnaB mutant (open triangles), DnaC with ssDNA (filled squares), DnaC mutant with ssDNA (open circles), DnaB mutant with ssDNA (filled diamonds), DnaC with DnaB mutant and ssDNA (filled triangles), and DnaC mutant, DnaB mutant and ssDNA (filled circles). (B) ATP binding by DnaC (left panel) and DnaC mutant (right panel) was measured using equilibrium gel filtration. The concentration of ATP (open circles) and DnaC (filled triangles) was determined for each fraction. In the left panel, 4.4 nmol of DnaC was used in the presence of 10 µM [32P]ATP. In the right panel, 9.9 nmol of DnaC mutant was used in the presence of 10 µM [32P]ATP. A Scatchard curve analysis of ATP binding data by wild-type DnaC from these experiments in shown in the middle panel. (C) Nucleotide binding was also determined for both wild-type (filled circles) and mutant (filled squares) DnaC using nitrocellulose filter retention assays as described in Materials and methods. Binding to ATP (left panel) and ADP (right panel) was measured. With wild-type protein a Kd was determined by fitting with equation 1 (see Materials and methods). With mutant DnaC, virtually no binding was detected.
Table I. Effects of various cofactors and substrates on nucleotide hydrolysis by DnaC.
| (d)NTP | Nucleic acid | DnaC, DnaBmut [pmol (d)NDP/min] |
|---|---|---|
| ATP | None | 11.4 |
| ATP | M13mp18 ssDNA | 63.8 |
| ATP | φX174 ssDNA | 58.9 |
| ATP | poly(dA) oligo dT | 61.1 |
| ATP | pUC18 | 11.3 |
| ATP | pUC18oriC | 12.7 |
| UTP | M13mp18 ssDNA | 4.4 |
| GTP | M13mp18 ssDNA | 2.4 |
| CTP | M13mp18 ssDNA | 3.2 |
| dATP | M13mp18 ssDNA | 67.7 |
| dTTP | M13mp18 ssDNA | 2.8 |
| dGTP | M13mp18 ssDNA | 7.1 |
| dCTP | M13mp18 ssDNA | 2.4 |
Nucleotide hydrolysis was performed as described in Materials and methods, except that the 25 µl reactions contained 18 pmol DnaC, 4.2 pmol mutant DnaB and 165 ng DNA. After 15 min at 37°C, reactions were quenched and analyzed by thin-layer chromatography.
Characterization of the DnaC ATPase activity shows it is specific to ATP/dATP; other (deoxy)ribonucleoside triphosphates are not hydrolyzed (Table I). This specificity mirrors the nucleotide binding specificity of DnaC (Galletto et al., 2000), but contrasts with the specificity of DnaB. DnaB hydrolyzes all four ribonucleoside tri phosphates equally well, but has poor activity with deoxy ribonucleosides (Reha-Krantz and Hurwitz, 1978; Arai and Kornberg, 1981). Of the various ssDNA and double strand DNA cofactors examined, ssDNA was the most effective as an effector of the DnaC ATPase (Table I).
ATP is not needed for interaction with DnaB
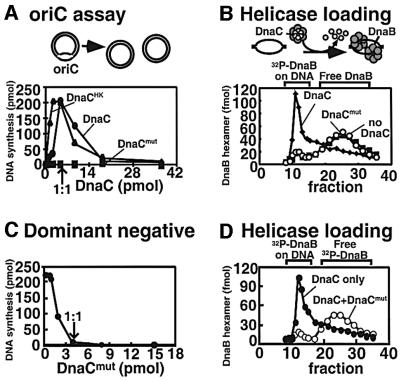
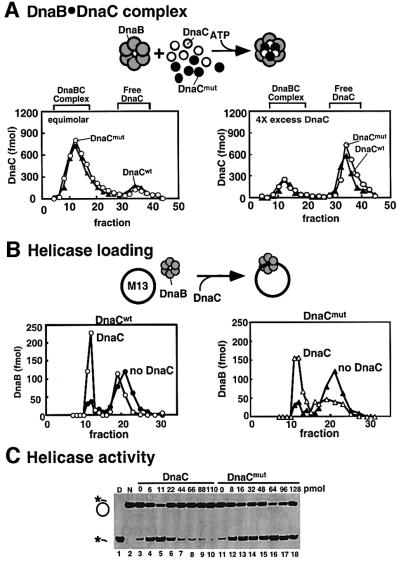
We next compared the abilities of wild-type DnaC and mutant DnaC to bind DnaB. To perform this study, we radiolabeled wild-type DnaC with tritium and placed a kinase site on the DnaC mutant for labeling with 32P. Neither modification interferes with DnaC function in replication of an oriC plasmid (Fang et al., 1999; see also Figure 3A). A mixture of the wild-type [3H]DnaC and mutant [32P]DnaC was used to analyze their relative ability to interact with DnaB through a gel filtration column (Figure 2A). At a 4-fold molar excess of DnaC1 to DnaB1, approximately equal amounts of wild-type and mutant DnaC assemble into the large DnaB6–DnaC6 complex (fractions 10–18); the excess DnaC monomer elutes later (fractions 30–40). When the experiment was repeated under conditions of equimolar DnaC1 to DnaB1, mutant and wild-type DnaC still assembled into the DnaB6–DnaC6 complex with equal efficiency. These observations indicate that ATP binding to DnaC is not needed for DnaB–DnaC complex formation, since the DnaC mutant does not bind ATP.
Fig. 3. DnaC in oriC replication assays. (A) The activity of wild-type and mutant DnaC in oriC replication. Increasing amounts of wild-type (filled circles), N-terminally tagged (filled triangles) or mutant (filled squares) DnaC were titrated into oriC replication assays (see Materials and methods). DNA synthesis was measured by the pmol incorporation of [α32P]dTMP into pUC18oriC. (B) Assembly of DnaB onto oriC. Wild-type (filled diamonds), mutant (open circles) or no DnaC (filled squares) was added to DnaA, HU, ATP, Mg2+, [32P]DnaB and supercoiled pUC18oriC, and incubated at 30°C. The samples were then applied to 5 ml BioGel A-15m columns equilibrated in buffer C containing 100 mM NaCl. Fractions were analyzed as described in Materials and methods. The early excluded fractions contained DnaB assembled onto DNA and the later included fractions contained free DnaB. (C) The DnaC mutant is dominant-negative to wild-type DnaC. Increasing amounts of DnaC mutant were added into an oriC replication assay that contained 4.4 pmol of DnaC. The arrow marks the point that contains equal amounts of wild-type and mutant DnaC. (D) DnaC mutant is dominant-negative for DnaB assembly onto oriC. DnaB (4.4 pmol) assembly was performed in the presence of 25 pmol wild-type DnaC and 50 pmol DnaC mutant (open circles) or 75 pmol wild-type DnaC (filled circles).
Fig. 2. Interaction between DnaB and mutant DnaC. The ability of the DnaC mutant to interact with, assemble and inhibit DnaB was examined. In (A) the association of [32P]DnaC mutant (open circles) and wild-type 3H-DnaC (filled triangles) with DnaB was compared. DnaB (10 pmol as monomer in the right panel and 40 pmol as monomer in the left panel) was mixed with mutant and wild-type DnaC (20 pmol of each, for a total of 40 pmol DnaC) in 125 µl reaction buffer containing 0.1 mM ATP and incubated for 10 min at 26°C. Complex was separated from free DnaC by gel filtration through a Superose-6 column equilibrated in buffer C with 100 mM NaCl and 0.1 mM ATP. The amounts of wild-type and mutant DnaC in each fraction were determined by scintillation counting. In (B), the ability of mutant (right panel) and wild-type DnaC (left panel) to stimulate DnaB assembly onto M13mp18 ssDNA was determined. Each reaction contained 1.2 pmol DnaB hexamer, 139 fmol M13mp18 ssDNA and, when present, 16 pmol DnaC or DnaC mutant. Assays in the presence (open circles) or absence (filled circles) of DnaC were performed in 100 µl reaction buffer containing 5 mM ATP, and then incubated for 10 min at 30°C before separating over a 5 ml BioGel A15-m column equilibrated in buffer C containing 100 mM NaCl. Fractions were collected and then 20 µl of each fraction was analyzed by western blot for the presence of DnaB as described in Materials and methods. (C) The DnaC mutant is unable to inhibit DNA unwinding by DnaB. Helicase assays were performed as described in Materials and methods. The amounts of DnaC (wild type or mutant) added to each reaction are indicated. The migrations of native substrate (lane N) and denatured substrate (lane D) are shown.
ATP is not needed for helicase loading onto ssDNA
The results above demonstrate that the DnaC mutant binds to DnaB. Can it load DnaB onto DNA? This was tested using DnaB and circular M13mp18 ssDNA (Figure 2B). Upon assembly of DnaB onto ssDNA, the large DnaB–ssDNA complex can be resolved from unbound DnaB by gel filtration analysis on a large pore resin, such as BioGel A15m. Column fractions were assayed for the presence of DnaB by quantitative western blot analysis. The result demonstrates that the DnaC mutant is as proficient a helicase loader as wild-type DnaC, indicating that ATP is not required for helicase loading onto ssDNA. Perhaps the DnaC mutant cannot finish the entire cycle of helicase loading and inhibits DnaB helicase. Next we examined the effect of mutant DnaC on helicase activity.
ATP induces DnaC inhibition of helicase
To monitor the effect of DnaC on DnaB helicase activity we annealed a short 32P end-labeled DNA oligonucleotide to M13mp18 ssDNA. Helicase activity dislodges the oligonucleotide, which then migrates ahead of the slow-migrating M13mp18 ssDNA in a non-denaturing polyacrylamide gel. Although DnaB functions optimally at a forked junction (LeBowitz and McMacken, 1986; Kaplan, 2000), we observe that DnaB can melt short regions of fully duplex DNA (Fang et al., 1999), and the following assays were performed using a flush DNA 30mer fully annealed to M13mp18 ssDNA. The results, shown in Figure 2C, demonstrate that DnaB alone displays weak helicase activity (lane 3), and that DnaC stimulates DnaB as much as 10-fold (lane 5). However, DnaB helicase is inhibited when DnaC is added in excess of DnaB (lanes 6–10). This behavior is consistent with the known inhibitory effect of DnaC on DnaB ssDNA-dependent ATPase and helicase activity (Wahle et al., 1989b).
A parallel study of DnaB helicase using the DnaC mutant in place of wild-type DnaC is shown in Figure 2C, lanes 11–18. The DnaC mutant still stimulates DnaB, consistent with its ability to load DnaB onto ssDNA. However, unlike wild-type DnaC, mutant DnaC does not inhibit DnaB helicase at even the highest concentration added to the assay. Thus we conclude that DnaC requires ATP to inhibit DnaB.
DnaC requires ATP for function at oriC
The results thus far indicate that DnaC does not require ATP to load DnaB onto ssDNA, and that in fact ATP binding to DnaC inhibits DnaB helicase activity. Assuming DnaC plays no additional roles at the origin, one would predict that the DnaC ATP site mutant should be capable of loading DnaB onto oriC. The activities of wild-type and mutant DnaC in replication of an oriC-containing plasmid are compared in the experiments shown in Figure 3. As observed previously, DnaC is required for replication, but DnaC inhibits the reaction when present in excess over DnaB. Contrary to our predictions, however, the DnaC mutant is completely inactive in this assay (Figure 3A).
To determine whether the DnaC mutant loads DnaB onto the origin, [32P]DnaB was used to assess directly whether DnaB is attached to the oriC plasmid by gel filtration analysis (Figure 3B). The control reaction using wild-type DnaC demonstrates attachment of [32P]DnaB helicase to oriC (Figure 3B, diamonds). However, [32P]DnaB does not become attached to oriC in the reaction using the DnaC mutant (Figure 3B, circles). It remained possible that mutant DnaC was inactive by virtue of not binding DnaB under these particular assay conditions. This can be tested easily, due to the multimeric nature of the DnaB6–DnaC6 complex. If mutant DnaC binds DnaB, but is otherwise inactive at oriC, then mutant DnaC will interfere with wild-type DnaC when the two are mixed in the assay. The results shown in Figure 3C demonstrate that activity is lost completely at 1:1 stoichiometry of mutant to wild-type DnaC. Next, [32P]DnaB and equimolar wild-type DnaC were used in the presence of an inhibitory amount of mutant DnaC (2-fold over wild-type DnaC) to determine whether [32P]DnaB was placed onto oriC but held in an inactive state (i.e. due to incomplete dissociation of DnaC). The gel filtration analysis demonstrates that [32P]DnaB is simply not loaded onto oriC by the mutant/wild-type DnaC mixture (Figure 3D). These results indicate that mutant DnaC is efficient in binding DnaB, but is inactive in some early step that requires ATP at oriC.
ATP strengthens DnaC–ssDNA binding
Inactivity of the DnaC mutant in transfer of DnaB onto oriC contrasts with its ability to assist DnaB onto ssDNA, although it is important to note that these are not directly comparable assays. Nonetheless, these divergent results suggest that DnaC does something more at the origin. It has been suggested that DnaC may help recruit DnaB to ssDNA by direct contact between DnaC and ssDNA, although DnaC–ssDNA interaction in the absence of DnaB has not been observed (Learn et al., 1997). Next, we developed a sensitive assay to determine whether DnaC interacts directly with ssDNA and, if so, whether ATP has an effect on this interaction.
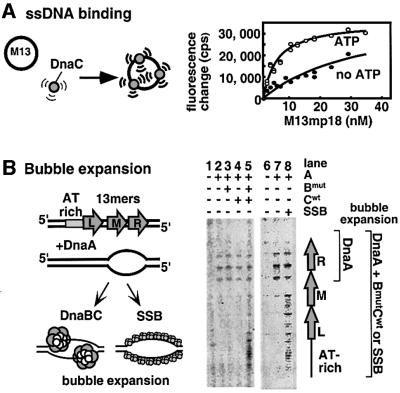
In the experiment of Figure 4A, we utilized the intrinsic fluorescence of DnaC as a tool to examine DnaC for direct interaction with ssDNA. Titration of M13mp18 ssDNA into a solution of DnaC resulted in an enhancement of fluorescence, indicating that DnaC binds ssDNA in a fashion that increases the intrinsic fluorescence of one or more of its three tryptophan residues. When the experiment was repeated in the presence of 100 µM ATP the rise in fluorescence was much steeper. Hence, ATP binding to DnaC increases the affinity of DnaC for ssDNA. Curve fitting, assuming a simple association reaction, indicates that ATP increases the affinity of DnaC for ssDNA ∼6- to 8-fold. It seems likely that DnaC–ssDNA interaction, strengthened by ATP, may underlie the ATP requirement for DnaC function at oriC.
Fig. 4. ssDNA binding by DnaC. (A) Equilibrium association of DnaC and M13mp18 ssDNA was monitored by fluorescence emission at 335 nm in the absence of ATP (filled circles), and in the presence of 100 µM ATP (open circles). The reactions were carried out as described in Materials and methods. The data points represent the fluorescence change at 335 nm after subtraction of the signal from free M13mp18 in counts per second (cps); the solid curves represent the best fits to equation 2 (see Materials and methods). (B) Bubble expansion by SSB and DnaB–DnaC in the presence of DnaA was measured by potassium permanganate footprinting at oriC, performed as described in Materials and methods. On the right is depicted the region of the origin under examination. The 13mer repeats are indicated by the gray arrows. The changes in the origin sensitivity to potassium permanganate are summarized in cartoon form as well as the protein interactions that cause them (left panel). PhosphorImager scans of the experiments with DnaB mutant and DnaC as well as with SSB, both in the presence of DnaA, are shown on the right. When present, 460 ng DnaA, 125 ng DnaB mutant, 200 ng DnaC and/or 800 ng SSB were used in the reactions.
One possible role for ssDNA binding by DnaC at oriC is to promote expansion of the DnaA-induced bubble at oriC. At oriC, DnaA (along with HU protein) induces strand opening of two of the three direct repeats (R and M; see Figure 4B). Addition of DnaB–DnaC complex further expands this bubble to include all three repeats and an AT-rich area just beyond the L repeat. As described in a previous study, DnaB encircles the R and L 13mers on this strand, protecting them from modification (Fang et al., 1999). The helicase activity of DnaB is not needed for bubble expansion, as these experiments are performed using the inactive DnaB mutant. If ssDNA binding by DnaC is sufficient for bubble expansion, then addition of only DnaC (and DnaA) may be necessary. However, as demonstrated in Figure 4B, lanes 3 and 4, DnaB or DnaC alone is insufficient to promote bubble expansion (with DnaA). Presumably, the combined ssDNA binding strength of both DnaC and DnaB are required to expand the bubble at oriC. If ssDNA binding by the DnaB–DnaC complex truly underlies the mechanism for bubble expansion, then a strong ssDNA binding protein, such as E.coli SSB, may substitute for DnaB–DnaC complex in this expansion function. The result shows that addition of SSB causes a similar extent of bubble expansion as observed using the DnaB–DnaC complex (Figure 4B, lane 8). Neither DnaB–DnaC nor SSB caused unwinding of oriC in the absence of DnaA (not shown), consistent with results of a previous study (Krause and Messer, 1999). Interestingly, although SSB can bind to the bubble in the origin, it does not seem to inhibit DnaB–DnaC function at oriC (Fang et al., 1999). The difference may lie in the ability of DnaA to interact with DnaB and direct DnaB–DnaC to the oriC bubble (Marszalek and Kaguni, 1994).
The fact that SSB promotes bubble expansion at oriC supports the idea that ssDNA binding by DnaC within the DnaB–DnaC complex is involved in bubble expansion at oriC. In summary, the role of ATP in DnaC action at oriC may be to enhance the ssDNA binding affinity of DnaB–DnaC complex, leading to bubble expansion, thereby providing sufficient ssDNA for DnaC to load DnaB onto the open origin. DnaC within the DnaB–DnaC complex may perform this role directly, by interacting with ssDNA, or indirectly, by modifying DnaA interaction with the origin (or both).
DnaC is an ATP/ADP switch protein
The dependence of the DnaC ATPase on ssDNA and DnaB (Figure 1A) is reminiscent of GTP switch proteins. For example, the prototypical switch protein, Ras, is active when bound to GTP and inactive when bound to GDP (Bourne et al., 1990). The switch from the active/GTP form to the inactive/GDP form is dependent on GAPs (GTPase activating proteins) (Sprang, 1997). Here, ssDNA and DnaB are acting in a manner analogous to GAPs; DnaC cannot hydrolyze ATP without them.
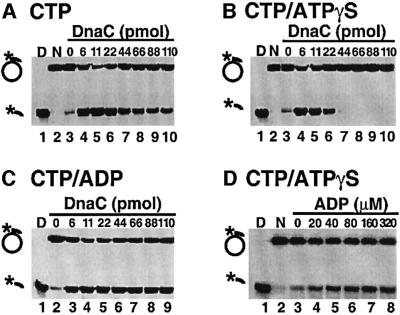
If DnaC acts as an ATP/ADP switch protein, it should have two states, DnaC–ATP and DnaC–ADP, each with distinct properties. Our prediction from the results obtained herein is that the ATP bound form of DnaC is inhibitory to DnaB helicase, and that the ADP form is not. To begin to test this prediction, we designed experiments to determine whether ATP binding to DnaC is sufficient to inhibit DnaB and whether exchange of ADP for ATP converts DnaC back to a stimulatory state. Essential to these next few experiments is the fact that DnaB functions with CTP (Arai and Kornberg, 1981), but DnaC does not (Table I; Galletto et al., 2000). Use of high concentrations of CTP for DnaB helicase activity afforded us the ability to test the effect of ATP, ATP-γS and ADP on DnaC action. In the assays that follow (Figure 5A–C), CTP is present at 5 mM, and adenine nucleotide is at 40 µM. Thus DnaB will predominantly be bound to CTP; DnaC will only bind the adenine nucleotide since it does not interact with CTP.
Fig. 5. DnaC is an ATP/ADP switch protein. Helicase assays were performed using M13mp18 ssDNA to which a 32P end-labeled oligonucleotide was annealed as a substrate, as described in Materials and methods. Substrate, the indicated nucleotide(s), DnaB (4.4 pmol hexamer, when present) and DnaC (indicated amounts) were incubated at 37°C before separating free oligonucleotide from intact substrate by electrophoresis through a 15% polyacrylamide gel (in Tris–borate). The left-most lane in each panel is substrate that has been boiled before electrophoresis to indicate the migration of free oligonucleotide in the gel (lanes D). Lanes N contain neither DnaC nor DnaB. DNA unwinding by DnaB with increasing amounts of DnaC with (A) 5 mM CTP, (B) 5 mM CTP and 40 µM ATP-γS, and (C) 5 mM CTP and 40 µM ADP. In (D), ADP was titrated into helicase reactions that contain an inhibitory amount of DnaC, 5 mM CTP and 40 µM ATP-γS.
First, we performed a control experiment (Figure 5A) in which wild-type DnaC (DnaCwt) is titrated into the reaction using CTP and no ATP. Since ATP is not present and DnaC does not bind CTP, the results should mimic those using mutant DnaC (DnaCmut; in the presence or absence of ATP), and in fact they essentially do. DnaC stimulates DnaB helicase even when present at a 10-fold molar excess over DnaB (Figure 5A, lane 10), although helicase action is diminished ∼2-fold compared with the use of stoichiometric DnaC. Use of ATP-γS, not hydrolyzed by DnaC, should mimic the inhibitory DnaC–ATP state, and the result shown in Figure 5B shows that indeed DnaC inhibits DnaB using ATP-γS when DnaC is present in excess. The low concentration of ATP-γS did not significantly affect unwinding by DnaB alone (compare lanes 3 in Figure 5A and B). This result indicates that ATP binding, but not hydrolysis, induces a conformation of DnaC that inhibits helicase activity when it remains associated with DnaB. Consistent with this idea, inhibition in the presence of ATP (Figure 2C) is not as strong as in the presence of ATP-γS (Figure 5B).
The ATP/ADP switch hypothesis for DnaC makes the further prediction that the ADP-bound form of DnaC should not inhibit DnaB. In fact, ADP should reverse the DnaC–ATP inhibited state by competing out bound ATP. As a control leading up to a test of this, we examined the effect of ADP on DnaC with DnaB–CTP. The result demonstrates that DnaC–ADP is unable to inhibit DnaB, as predicted by the switch model (Figure 5C). However, an alternative explanation for the lack of helicase inhibition in the presence of ADP is the lack of ADP binding by DnaC under these conditions. We tested DnaC for ADP binding using a nitrocellulose filter retention assay with the result that DnaC binds ADP with an approximate Kd of 8 µM (Figure 1C).
To confirm that ADP places DnaC in a conformation that does not inhibit DnaB, we titrated ADP into a DnaB helicase assay that was already inhibited by DnaC– ATP-γS. The result shows that as the ADP concentration is increased, the inhibition of DnaB by DnaC is relieved (Figure 5D). We presume in this experiment that ADP competes out ATP-γS from DnaC to yield the reactivated DnaC–ADP. Based on these observations we conclude that DnaC has two different states that depend on which nucleotide it binds; the DnaC–ATP state inhibits DNA unwinding by DnaB, whereas the DnaC–ADP state does not inhibit DnaB. Curiously, the DnaC–ATP state is required for both DnaB loading onto oriC and helicase inhibition. Therefore, rather than having ‘on’ and ‘off’ states like prototypical switch proteins, DnaC possesses dual states, each with its own positive role in replication. The DnaC–ATP state promotes assembly of DnaB onto oriC and the DnaC–ADP state relieves inhibition of DnaB.
Discussion
DnaC as a dual ATP/ADP switch protein
Nucleotide switch proteins have two different states, dependent on the bound nucleotide (Bourne et al., 1990). Switching from one state to the other is often facilitated by accessory factors that either affect nucleotide hydrolysis or release of the bound nucleotide (Sprang, 1997). Like most switch proteins, DnaC exists in two states: DnaC–ATP, which expands the bubble, and DnaC–ADP, which allows DnaB to unwind DNA. The transition from the ATP-bound to the ADP-bound state (hydrolysis) is dependent on the substrates, ssDNA and DnaB. However, unlike most other switch proteins, the two states are not ‘on’ and ‘off’. Each of the two states is required in turn for replication of oriC plasmids. DnaC–ATP is required to bind ssDNA for DNA melting at oriC, and DnaC–ADP is required to relieve the inhibition exerted on DnaB by DnaC–ATP.
This dual switch may also be found in a subset of other switch proteins. For example, the yeast protein Bud1 is a GTP/GDP switch protein in which the GDP and GTP forms interact with different proteins (Park et al., 1997). Both of these interactions are crucial for Bud1 function, determining the localization of the bud site. In fact, several switching cycles are required for Bud1 to perform its function. Another dual switch protein is the P1 plasmid partition protein ParA. ParA is an ATP/ADP switch protein in which the ATP form participates in the partition complex required for proper plasmid segregation and the ADP form is a transcriptional repressor (Bouet and Funnell, 1999). Both forms exert an overall positive effect on partition.
DnaC is not the only nucleotide switch protein controlling replication initiation (Lee and Bell, 2000). A second E.coli replication protein, DnaA, is also an ATP/ADP switch protein. DnaA–ATP supports partial unwinding of the AT-rich 13mer repeats in oriC, whereas DnaA–ADP does not (Kornberg and Baker, 1992). Recent work has shown that switching from the active ATP-bound form to the inactive ADP-bound form is stimulated by replicating polymerase (Katayama et al., 1998). The early events in replication initiation are therefore controlled by the sequential action of nucleotide switch proteins. Interestingly, DnaA shares sequence homology with DnaC, particularly in and around the motifs required for ATP binding and hydrolysis. DnaA, like DnaC, is also a member of the AAA+ family.
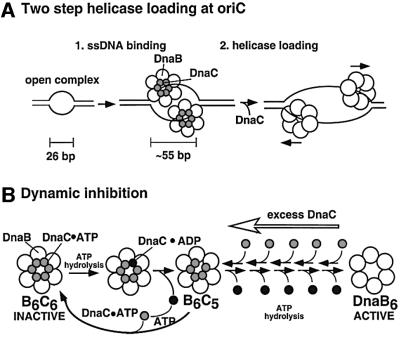
A model for DnaC action during helicase assembly onto oriC
The observations herein on DnaC, and its behavior with ATP, ADP, ssDNA, DnaB and oriC, lead to a model in which the sequential interaction of these components with DnaC controls assembly of proteins to achieve full activation of the origin (Figure 6A). The first step in loading DnaB onto the origin is the formation of the DnaB–DnaC complex. Given an intracellular ATP concentration 12-fold higher than ADP (Neuhard and Nygaard, 1987), and the fact that DnaC binds these two nucleotides with similar affinity, most intracellular DnaC will be in the ATP-bound state. At this stage, DnaC is unable to hydrolyze ATP, because it has not yet associated with ssDNA. Next, the DnaB–DnaC complex is directed to the DnaA-induced open complex at oriC, presumably by DnaB interaction with DnaA already bound at the origin (Marszalek and Kaguni, 1994). When mutant DnaB is assembled onto the origin by DnaC, the unwound region expands from 26 to ∼55 nucleotides (Fang et al., 1999). The DnaB mutant is inactive as a helicase, and thus the observed bubble expansion is not due to helicase action. We suggest, therefore, that DnaC within the DnaB–DnaC complex enlarges the bubble. For example, DnaB–DnaC-dependent bubble expansion may occur either by DnaB– DnaC modification of DnaA function and/or by direct binding of DnaC to ssDNA. Consistent with the latter suggestion that non-specific ssDNA binding underlies bubble expansion, Figure 4B illustrates that SSB also expands the DnaA induced bubble at oriC. We demonstrate here that ATP induces DnaC to bind ssDNA tighter than in the absence of ATP. Thus, we propose that ssDNA binding by DnaC is the step in DnaC function at oriC that requires ATP. The hexameric DnaB is also needed for bubble expansion, either as a scaffold to bring six molecules of DnaC to the task, or to assist directly in binding ssDNA. In a second step, DnaC may then assemble DnaB onto the expanded bubble in the same manner as it assembles DnaB onto ssDNA. Since DnaA is known to interact with DnaB (Marszalek et al., 1994), it is possible that this interaction may also play an important role in DnaB assembly onto oriC. Interestingly, N-terminal deletion mutants of DnaA do not support DnaC-dependent assembly of DnaB onto DNA, even though these truncated DnaAs are able to interact with DnaB in solution and still support bubble unwinding at oriC (Sutton et al., 1998). In this current study we show that DnaB–DnaC interaction with ATP and ssDNA are important for DnaB–DnaC function; however, DnaB– DnaC interaction with other factors, such as DnaA, may also be critical for helicase loading at oriC.
Fig. 6. Models of DnaC action. The models are discussed in greater detail in the text. (A) The two steps of helicase loading at oriC are ssDNA binding by the DnaB–DnaC complex and then helicase loading so that the helicase encircles ssDNA. (B) Dynamic inhibition of DnaB by DnaC is controlled by the dual ATP/ADP switch in DnaC and explains the sensitivity of DnaB to small fluctuations in DnaC.
A snapshot of DnaB taken in this moment would show two assembled, inactive DnaB hexamers, each bound to six molecules of DnaC–ATP. While DnaC is in the ATP form and bound to DnaB, DnaB cannot unwind DNA. It may be important for DnaC to keep helicase activity inert during its assembly onto the origin so that the process of assembling each of the two helicases can be coordinated. We propose that DnaC may recognize when its two effector molecules, DnaB and ssDNA, have reached their proper geometry in which ssDNA passes through the DnaB ring, and only then is ATP hydrolysis permitted. Once DnaC bound to DnaB is in the ADP form, DnaB is no longer inhibited.
Why does DnaC–ATP inhibit DnaB?
This report demonstrates that neither ATP nor ADP is needed for DnaC to load DnaB onto ssDNA, and in fact, ATP bound to DnaC inhibits the helicase activity of DnaB. If DnaC is inhibitory to DnaB helicase, why did DnaC evolve to interact with ATP in the first place? As shown in this study, DnaC uses ATP to regulate its affinity for ssDNA. Interaction with ssDNA may help load DnaB into sites that are internal to double strand DNA, such as oriC, by aiding strand separation. However, the act of binding ssDNA may inhibit translocation of DnaB along ssDNA, thereby preventing helicase action. Thus, in order for DnaC to bind ssDNA but not inhibit helicase, it appears to have evolved a mechanism to separate its actions into two distinct steps that act in sequential fashion; first to bind ssDNA for melting and second to decrease its affinity for ssDNA to allow helicase progression. This mechanism utilizes nucleotide binding and hydrolysis; first, by using ATP binding to strengthen DnaC interaction with ssDNA, and then decreasing its grip on ssDNA by hydrolyzing ATP to yield DnaC–ADP.
Is the ATP switch regulatory?
Inhibition of DnaB by DnaC is surprisingly sensitive to DnaC concentration, suggesting that helicase action could possibly be regulated in vivo by slight fluctuations in the ratios of DnaC to DnaB. For example, at 1:1 DnaC1: DnaB1, DnaC stimulates DnaB rather than inhibiting it. But at 2:1 DnaC1:DnaB1, DnaC strongly inhibits DnaB. Inhibition by DnaC occurs after the helicase is assembled onto the DNA, not before, thus helicase loading is not impaired (Allen and Kornberg, 1991). Under inhibitory conditions of excess DnaC, DnaC remains associated with DnaB, preventing movement on ssDNA. Consistent with the idea that DnaC could possibly act in a regulatory fashion, experiments in which DnaC is expressed from an inducible plasmid demonstrate that DnaC can shut down ongoing replication in vivo when supplied in excess over the amount normally present in growing cells (Skarstad and Wold, 1995).
Normally, DnaC is not stuck tightly to DnaB loaded onto DNA and readily dissociates during gel filtration, freeing DnaB for helicase action (Kobori and Kornberg, 1982; Funnell et al., 1987; Fang et al., 1999). Furthermore, simply adding more DnaB to a reaction with inhibitory amounts of DnaC, such that the concentration of DnaB matches the elevated DnaC concentration, relieves the inhibition (Allen and Kornberg, 1991). Therefore, inhibition of DnaB by excess DnaC is a freely reversible reaction. Indeed, this rapid on and off action of DnaC appears to underlie the mechanism of inhibition. As ATP is hydrolyzed, DnaC–ADP may dissociate from DnaB, but another DnaC–ATP can take its place, thus maintaining the inhibited state (Figure 6B).
The stoichiometry of six DnaCs per DnaB hexamer probably explains the narrow window of DnaC activation and inhibition of helicase. If six (or nearly six) DnaCs are required to activate DnaB, the activity of DnaB will be rather steep in response to DnaC concentration. Likewise, if DnaB activity requires several of the DnaC molecules to dissociate, then the presence of even a little extra DnaC–ATP in solution will take the place of some DnaC–ADP as it dissociates from DnaB6, resulting in a dynamic maintenance of the inhibited state (Figure 6B).
The modulation of DnaB activity by the DnaC:DnaB ratio prompts us to consider DnaC as a regulatory molecule at the level of a moving replication fork. For example, when a condition arises in the cell that is inconsistent with DNA synthesis, such as breaks in the DNA, the cell may set into motion processes that prevent ongoing replication, and stop new reinitiation events at replication origins. DnaC seems ideally suited in this task for several reasons. First, only a subtle increase in concentration of DnaC is needed to produce the inhibited state. Secondly, inhibition is achieved at moving replication forks and at the origin. Thirdly, inhibition and reactivation can be achieved by modulation of either DnaC or DnaB concentration (or both). Finally, since DnaB remains on DNA while inhibited by DnaC, the replication fork machinery should remain intact, and would not need to be reassembled after inhibition is relieved. Reactivation of replication could be achieved by production of extra DnaB to soak up the DnaC, or the removal of DnaC by proteolysis. Consistent with DnaC regulation of moving replication forks, there exist DnaC mutants that interfere with replication elongation rather than initiation (Carl, 1970; Wechsler and Gross, 1971; Schubach et al., 1973; Wechsler, 1975). We are currently testing the idea that DnaC may play a regulatory role in DNA replication.
It is not known at this time whether all bacteria contain DnaC. Direct sequence searches do not detect homologs in some bacteria (Caspi et al., 2001; e.g. Gram-positive DnaI protein is a weak homolog of E.coli DnaC). However, given the small size, and possibilities for convergent evolution, recognition of functional homologs to DnaC may be impossible by sequence comparison methods alone. For example, the prokaryotic and eukaryotic DNA sliding clamps, β and proliferating cell nuclear antigen (PCNA), share no sequence homology but have the same chain fold and three-dimensional structure (Kelman and O’Donnell, 1995).
Cdc6 as a DnaC homolog
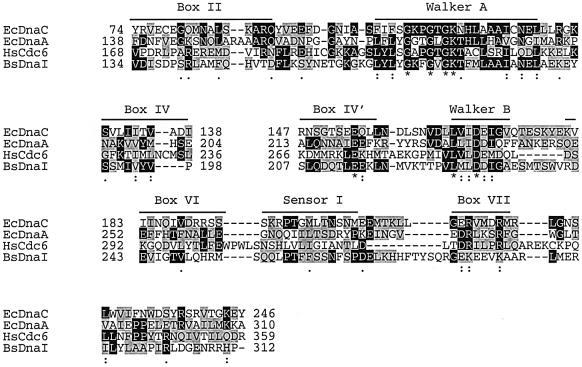
The eukaryotic functional equivalent of E.coli DnaC is thought to be the Cdc6/Cdc18 protein (Baker and Bell, 1998; Davey and O’Donnell, 2000). Eukaryotic Cdc6 and E.coli DnaC are homologous to one another, and the Cdc6 protein, like DnaC, is a member of the AAA+ family (Koonin, 1992; Neuwald et al., 1999) (Figure 7). The most notable experiments linking Cdc6 to DnaC action are a series of cell biology studies. These reports demonstrate that Cdc6 acts at origins, and that an intact ATP site in Cdc6 is required for assembly of Mcm2–7 onto origin sequences to form the preRC (Donovan et al., 1997). After Mcm2–7 is assembled onto the origin, the Cdc6 protein, like DnaC, appears to dissociate and is not needed again during S phase (Weinreich et al., 1999). It should be noted that Mcm2–7 complex interaction with replication origins is likely more complicated than DnaB loading. For example, at least a second protein, Cdt1, is required for Mcm2–7 loading (Maiorano et al., 2000; Nishitani et al., 2000). This and other differences reflect the increased complexity of eukaryotic cells.
Fig. 7. Alignment of DnaA, DnaC, DnaI and Cdc6. Homo sapiens Cdc6 (HsCdc6; DDBJ/EMBL/GenBank accession no. NP001245), Bacillus subtilis DnaI (BsDnaI; accession no. NP390776), E.coli DnaC (EcDnaC; accession no. NP418781) and E.coli DnaA (EcDnaA; accession no. NP418157) were aligned using CLUSTAL_X. The alignment was refined manually using previous alignments between DnaA, DnaI and DnaC (Koonin, 1992), as well as between DnaA and Cdc6 (Neuwald et al., 1999). An asterisk indicates positions that have a fully conserved residue, a colon indicates positions that have fully conserved strongly similar residues and a period indicates positions that have fully conserved residues with weaker similarity. Black boxes indicate residues that are identical and gray boxes indicate similar residues at each position.
Also included in the AAA+ subclass are proteins involved in the initiation of DNA replication in eukaryotic cells (Neuwald et al., 1999). These proteins include some of the subunits of the ORC, and the six subunits of the Mcm2–7 complex. Hence, like prokaryotes, eukaryotes also use multiple AAA+ proteins, at least some of which probably function as ATP/ADP switches like DnaA and DnaC.
Materials and methods
Reagents and buffers
Sources for reagents were as follows: radioactive nucleotides, New England Nuclear; unlabeled nucleotides, Pharmacia; ATP-γS, Sigma; and DNA modification enzymes, New England Biolabs. Protein concentrations were determined by Bradford (Bio-Rad) using the manufacturer’s directions with bovine serum albumin (BSA) as a standard. Reaction buffer is 40 mM HEPES-NaOH pH 7.4, 10 mM MgCl2, 5 mM dithiothreitol (DTT) and 100 µg BSA/ml. Cell lysis buffer is 20 mM Tris–HCl pH 8.0 and 100 mM NaCl. Buffer A is 25 mM HEPES–NaOH pH 7.4, 1 mM EDTA, 2 mM DTT and 15% glycerol. Buffer B is 50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 mM DTT and 10% glycerol. Buffer C is 20 mM Tris–HCl pH 7.5, 0.1 mM EDTA, 5 mM MgCl2 and 10% glycerol.
Proteins and antibodies
The following replication proteins were prepared as described previously: DnaA, DnaB mutant and [3H]DnaC (Fang et al., 1999), HU (Parada and Marians, 1991), DnaB, SSB and [32P]DnaB (Yuzhakov et al., 1996). Polyclonal antibodies against DnaB and DnaC were raised in rabbits by BabCo (Berkley, CA). The antibodies were affinity purified using pure DnaB or DnaC immobilized on nitrocellulose (Sambrook et al., 1989).
DNAs
M13mp18 ssDNA was phenol-extracted from phage that had been banded twice in cesium chloride density gradients as described previously (Turner and O’Donnell, 1995). DNA oligonucleotides were purchased from Oligos etc. pUC18oriC is a version of pUC18 that contains the oriC sequence (Fang et al., 1999). pET11-dnaC expresses DnaC under control of the T7 RNA polymerase promoter and was constructed by PCR amplification of dnaC from the E.coli genome using primers that produce an NdeI restriction site at the start codon of the dnaC coding sequence [primer 1: 5′-d(GGAGATATACATATGAAAAACGTTGGA)-3′] and a BamHI site after the stop codon [primer 2: 5′-d(GTTAGCAGCCGGAT CCGGACCGCCAGA)-3′]. After restriction with NdeI and BamHI, the dnaC fragment was inserted into the NdeI–BamHI sites of pET11a (Novagen). In the same manner, a version of DnaC with six histidine residues and a protein kinase A (PKA) phosphorylation site at the amino terminus was formed by inserting the same DnaC NdeI–BamHI fragment into the NdeI–BamHI sites of pHK (Kelman et al., 1995) (pHK-dnaC). The DnaC coding region in both plasmids was sequenced. Consistent with previous reports, the predicted amino acid at codon six is aspartate, not alanine (Ludlam et al., 2001).
Mutation of the DnaC ATP binding motif
A mutation was introduced into the ATP binding site of the tagged DnaC using mutagenic oligonucleotides and PCR. The dnaC gene was amplified in two parts: using primer 1 from above and a mutagenic oligonucleotide that contains a 3 bp mismatch (underlined) [5′-d(AGC CAGGAACCGGCCGTAACCATCTGGCGGC)-3′], the upstream portion of DnaC was amplified. This mismatch changes the conserved lysine at position 112 of DnaC to arginine. The rest of DnaC was amplified using primer 2 from above and a second mutagenic oligonucleotide [5′-d(GCCGCCAGATGGTTACGGCCGGTTCCTGGCT)-3′]. Amplific ation was for 30 cycles using Vent polymerase. The amplified products were purified away from primers and mixed for a second PCR with primers 1 and 2, to amplify the entire coding sequence. The reaction was heated to 95°C for 2.5 min, cooled to 37°C over 10 min and allowed to extend for 5 min at 72°C before proceeding with standard PCR. Following PCR, the resulting fragment was restricted with NdeI and BamHI, purified and then inserted into the NdeI–BamHI sites of pHK to yield pHK-dnaCK112R. The modified sequence was confirmed by sequencing the entire dnaC gene.
Purification of His-tagged DnaC and DnaC mutant
Four liters of the E.coli strain BL21(λDE3) [Novagen; F– ompT hsdSB (rB– mB–) gal dcm (λDE3)] harboring pHK-dnaC or pHKdnaCK112R were grown at 37°C in LB media containing 100 µg ampicillin/ml. When the cells reached a density of A600 = 0.3, they were shifted to 25°C. IPTG was added to a final concentration of 0.4 mM when the culture reached an A600 of 0.6. After further growth at 25°C for 4 h, the cells (∼4 g) were collected by centrifugation and resuspended in 50 ml cell lysis buffer containing 20 mg lysozyme. The cells were homogenized with a glass Dounce and incubated on ice for 1 h. Only the purification of wild-type DnaC will be described, as the purification of the mutant DnaC was essentially identical.
After cell lysis, the cell debris was removed by centrifugation for 1 h at 18 000 r.p.m. in an SS-34 rotor. The clarified supernatant (Fr I, 391 mg in 38 ml) was applied to a 10 ml TALON Metal Affinity column (Clontech) that had been equilibrated with cell lysis buffer. After the sample was loaded on the column, the column was washed with 20 mM Tris–HCl pH 8.0, containing 20 mM imidazole and 100 mM NaCl. Protein was eluted in a linear gradient from 20 mM imidazole and 100 mM NaCl to 100 mM imidazole and 400 mM NaCl in 20 mM Tris–HCl pH 8.0. Peak fractions (identified by SDS–PAGE) were pooled and dialyzed against buffer C. During dialysis, a portion of the protein precipitated, the majority of which was not DnaC. The insoluble material was removed by centrifugation and the clarified supernatant contained most of the DnaC (Fr II, 22 mg in 34 ml). Fr II was applied to a 5 ml DEAE-Fast Flow Sepharose column equilibrated in buffer C. DnaC was recovered in the flowthrough, then aliquoted and stored at –80°C (Fr III, ∼20 mg in 30 ml).
ATP hydrolysis assay
ATPase assays were in 25 µl of reaction buffer containing 0.1 mM [α32P]ATP (3 × 106 c.p.m.) and 165 ng M13mp18 ssDNA. The DnaC and/or DnaB mutant proteins were added to the reactions on ice and the tubes were shifted to 30°C. At the indicated times (see Figure 1), 3 µl were removed from the reaction and quenched by the addition of 40 mM EDTA and 1% SDS (3 µl). Quenched reactions were spotted (1 µl) onto thin-layer chromatography sheets coated with polyethyleneimine cellulose (PEI-Cellulose F; EM Science) and developed in 0.6 M potassium phosphate pH 3.4. ADP and ATP migrated differently and were quantitated using a PhosphorImager and ImageQuant software (Molecular Dynamics).
Nucleotide binding by DnaC
Nitrocellulose filter binding assays were performed as described previously (Hingorani and O’Donnell, 1998). Briefly, 4 µM wild-type or mutant tagged DnaC was mixed with the indicated concentrations of [α32P]ATP or [2,8 3H]ADP in 15 µl of reaction buffer. The nucleotide binding isotherms were fit to a quadratic solution for 1:1 binding of a ligand to a macromolecule:
(ML) = 1/2 {(Kd + LT + MT) – [(Kd + LT + MT)2/4 – (MTLT)]0.5}(1)
where Kd is the dissociation constant, LT is the total ligand (ATP) concentration, MT is the total protein (DnaC monomer) concentration, and ML is the molar amount of ligand bound to protein.
Equilibrium gel filtration was performed as described previously (Davey and Funnell, 1997). Briefly, a 5 ml Sephadex G100 column was equilibrated in column buffer (20 mM Tris–HCl pH 7.5, 0.1 mM EDTA, 5 mM DTT, 4% glycerol, 0.1 mg BSA/ml and 8 mM MgCl2) with the indicated concentration of [α32P]ATP. Either 140 µg (4.4 nmol) of wild-type DnaC or 313 µg (9.9 nmol) of mutant DnaC was mixed an equal volume of 2× column buffer. The samples were applied to the columns and 250 µl fractions were collected. A portion of each fraction was analyzed by liquid scintillation counting for ATP concentration. Another portion of the fraction was used to determine the protein concentration by Bradford.
Helicase activity
DNA unwinding by DnaB was measured by displacement of a 30mer DNA oligonucleotide annealed to M13mp18 as modified from Shrimankar et al. (1992). The substrate was prepared by annealing 30 pmol of the 5′-end 32P-labeled 30mer (complementary to map positions 6817 through 6846 of M13mp18) to 5 pmol of M13mp18 circular ssDNA in a total volume of 70 µl as described previously (Studwell and O’Donnell, 1990). The annealed oligonucleotide was separated from free oligonucleotide by gel filtration through a 5 ml BioGel A-15m column equilibrated with 10 mM Tris–HCl pH 7.5, 1 mM EDTA and 100 mM NaCl. The helicase reactions contained 4.4 pmol DnaB hexamer, 6–128 pmol of mutant or wild-type DnaC, and ∼40 fmol of the purified partial duplex substrate in 25 µl of reaction buffer containing 5 mM CTP and either 40 µM ADP or ATP-γS as indicated. Reactions were incubated at 37°C for 10 min, quenched with 0.2% SDS and 20 mM EDTA (final concentration) and analyzed on a 15% polyacrylamide gel in Tris–borate buffer at 25°C.
oriC replication
Replication of oriC plasmid DNA was performed as described previously (Fang et al., 1999). The substrate was pUC18 into which the oriC sequence had been inserted (Fang et al., 1999). Replication assays contained 63 fmol pUC18oriC plasmid, 22 pmol SSB, 4.6 pmol DnaA, 1.1 pmol (as hexamer) DnaB, 4.4 pmol DnaC (unless noted otherwise), 3 pmol DnaG, 0.9 pmol HU protein, 0.5 pmol DNA gyrase, 0.5 pmol β, 370 fmol DNA polymerase III*, 5 mM ATP, 0.5 mM each GTP, CTP and UTP, 40 µM each dATP, dGTP and dCTP, 40 µM [32P]TTP (3000–6000 c.p.m./pmol), 5 mM creatine phosphate, and 0.5 mg of creatine kinase in 25 μl reaction buffer. Reactions were assembled at 0°C and then incubated at 30°C for 15 min, within the linear range of the assay. At this time, half of the plasmid substrate is replicated using wild-type proteins in the amounts specified above. Replication products were quantitated by spotting the reactions onto DE81 filters. After washing and drying, the filters were counted in a liquid scintillation counter (Wallac).
DnaC and DnaB interaction with ssDNA
DnaC interaction with ssDNA was measured in the presence and absence of ATP using the intrinsic fluorescence of DnaC. All spectroscopic measurements were performed in 20 mM Tris–HCl buffer pH 7.5, 0.5 mM EDTA, 5 mM DTT, 8 mM MgCl2 and 120 mM NaCl, at 25°C, in the presence or absence of 100 µM ATP. Intrinsic fluorescence emission spectra (305–420 nm) of Trp were recorded on a PTI spectrofluorimeter, using an excitation wavelength of 295 nm and excitation/emission slits of 2 and 3 mm, respectively. Increasing amounts of M13mp18 ssDNA (as indicated) were added to a 1 µM solution of DnaC. Controls of M13mp18 in the reaction buffer were recorded under the same experimental conditions. To determine the difference in affinity for ssDNA of DnaC with and without ATP, the data were fit to an equation that yields an apparent Kd. To perform this fit, one must assume a site size for DnaC. For the calculation below, we assumed a site size of 10 nucleotides for DnaC, yielding 725 sites per M13mp18 ssDNA molecule. However, since the number of sites per M13mp18 ssDNA is not known, a true or absolute Kd value cannot be obtained. Nevertheless, the ratio of the apparent Kds with and without ATP accurately reflects the difference in affinity of DnaC to ssDNA in the presence and absence of ATP. The measured fluorescence change of DnaC in response to M13mp18 ssDNA was fit to the following equation:
where (C)0 is the total concentration of DnaC protein (constant), (M13mp18)0 is the concentration of M13mp18 ssDNA, and ΔFint is the difference in fluorescence between free and bound DnaC.
For DnaB assembly onto ssDNA, each reaction contained 1.2 pmol DnaB, 16 pmol of wild-type or mutant DnaC, 139 fmol M13mp18 ssDNA and 5 mM ATP in 100 µl reaction buffer. The reactions were incubated at 30°C for 10 min and then were applied to 5 ml BioGel A15-m columns equilibrated in buffer C containing 100 mM NaCl. Fractions were collected and were analyzed for the presence of DnaB by western blot analysis. Proteins were transferred to nitrocellulose by spotting 20 µl of each fraction onto BA-S 83 nitrocellulose membrane using a dot-blot apparatus. The membrane was probed for the presence of DnaB using affinity-purified antibody and standard western blotting procedures. The amount of DnaB in each fraction was determined by comparison to known amounts of DnaB on the same blot. Autoradiographs of the blot were quantified using a laser densitometer (Molecular Dynamics).
DnaB assembly at oriC
DnaB assembly at oriC was performed as described previously (Fang et al., 1999). Assembly was measured using a version of DnaB that has a PKA phosphorylation site at the amino terminus, allowing labeling with 32P (Yuzhakov et al., 1996). Briefly, 252 fmol of supercoiled pUC18oriC was mixed with 88 pmol SSB, 18 pmol DnaA, 4.4 pmol (as hexamer) of [32P]DnaB, 20 pmol DnaC (wild type or mutant), 3.6 pmol HU and 5 mM ATP in 100 µl reaction buffer. After 30 min at 30°C, the reactions were placed in a tabletop centrifuge for 30 min at 4°C. The reaction was then analyzed by gel filtration through a 5 ml BioGel A-15m (Bio-Rad) column equilibrated with buffer C containing 100 mM NaCl at 25°C. Fractions of ∼200 µl were collected and 150 µl of each reaction was analyzed in a liquid scintillation counter (Wallac).
KMnO4 modification
Reactions contained 240 fmol pUC18oriC and 5 mM ATP in 25 µl reaction buffer lacking DTT. The indicated amounts of DnaA, DnaB mutant, DnaC and/or SSB were added and reactions were incubated for 30 min at 37°C. KMnO4 was then added to a final concentration of 40 mM and after a further 4 min at 37°C, the reaction was quenched with 1.3 M 2-mercaptoethanol. DNA was purified by phenol/chloroform extraction in the presence of 0.5% SDS and 20 mM EDTA, then passed through a Sephadex G25 spin column in water followed by ethanol precipitation using 10 µg yeast tRNA as carrier. Primer extension was performed using DNA polymerase I Klenow fragment as described previously (Frappier and O’Donnell, 1992) using 5′-d(CCCGTGGATTCTACTCAAC) as a primer. Primer extension was stopped upon addition of formamide and dye and the samples were analyzed by electrophoresis through a 6% polyacrylamide 50% urea gel. After electrophoresis the gel was dried on DE81 paper and exposed to a Phosphor storage screens (Molecular Dynamics).
Acknowledgments
Acknowledgements
We are grateful to the NIH (GM38839) and HHMI for supporting this project.
References
- Allen G.C. Jr and Kornberg,A. (1991) Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J. Biol. Chem., 266, 22096–22101. [PubMed] [Google Scholar]
- Arai K. and Kornberg,A. (1981) Mechanism of dnaB protein action. II. ATP hydrolysis by dnaB protein dependent on single- or double-stranded DNA. J. Biol. Chem., 256, 5253–5259. [PubMed] [Google Scholar]
- Baker T.A. and Bell,S.P. (1998) Polymerases and the replisome: machines within machines. Cell, 92, 295–305. [DOI] [PubMed] [Google Scholar]
- Baker T.A., Funnell,B.E. and Kornberg,A. (1987) Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J. Biol. Chem., 262, 6877–6885. [PubMed] [Google Scholar]
- Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Bouet J.Y. and Funnell,B.E. (1999) P1 ParA interacts with the P1 partition complex at parS and an ATP–ADP switch controls ParA activities. EMBO J., 18, 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H.R., Sanders,D.A. and McCormick,F. (1990) The GTPase superfamily: a conserved switch for diverse cell functions. Nature, 348, 125–132. [DOI] [PubMed] [Google Scholar]
- Bramhill D. and Kornberg,A. (1988) Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell, 52, 743–755. [DOI] [PubMed] [Google Scholar]
- Carl P.L. (1970) Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol. Gen. Genet., 109, 107–122. [DOI] [PubMed] [Google Scholar]
- Caspi R., Pacek,M., Consiglieri,G., Helinski,D.R., Toukdarian,A. and Konieczny,I. (2001) A broad host range replicon with different requirements for replication initiation in three bacterial species. EMBO J., 20, 3262–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M.J. and Funnell,B.E. (1997) Modulation of the P1 plasmid partition protein ParA by ATP, ADP and P1 ParB. J. Biol. Chem., 272, 15286–15292. [DOI] [PubMed] [Google Scholar]
- Davey M.J. and O’Donnell,M. (2000) Mechanisms of DNA replication. Curr. Opin. Chem. Biol., 4, 581–586. [DOI] [PubMed] [Google Scholar]
- Donovan S., Harwood,J., Drury,L.S. and Diffley,J.F. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A. and Bell,S.P. (1997) Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell. Dev. Biol., 13, 293–332. [DOI] [PubMed] [Google Scholar]
- Fang L., Davey,M.J. and O’Donnell,M. (1999) Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell, 4, 541–553. [DOI] [PubMed] [Google Scholar]
- Frappier L. and O’Donnell,M. (1992) EBNA1 distorts oriP, the Epstein–Barr virus latent replication origin. J. Virol., 66, 1786–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R.S., Funnell,B.E. and Kornberg,A. (1984) The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell, 38, 889–900. [DOI] [PubMed] [Google Scholar]
- Funnell B.E., Baker,T.A. and Kornberg,A. (1987) In vitro assembly of a prepriming complex at the origin of the Esherichia coli chromosome. J. Biol. Chem., 262, 10327–10334. [PubMed] [Google Scholar]
- Galletto R., Rajendran,S. and Bujalowski,W. (2000) Interactions of nucleotide cofactors with the Escherichia coli replication factor DnaC protein. Biochemistry, 39, 12959–12969. [DOI] [PubMed] [Google Scholar]
- Gille H. and Messer,W. (1991) Localized DNA melting and structural pertubations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J., 10, 1579–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani M.M. and O’Donnell,M. (1998) ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J. Biol. Chem., 273, 24550–24563. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D., Yurieva,O., Zhao,Y., Young,M., Stewart,J., Hingorani,M., O’Donnell,M. and Kuriyan,J. (2001) Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell, 106, 417–428. [PubMed] [Google Scholar]
- Kaplan D.L. (2000) The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol., 301, 285–299. [DOI] [PubMed] [Google Scholar]
- Katayama T., Kubota,T., Kurokawa,K., Crooke,E. and Sekimizu,K. (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell, 94, 61–71. [DOI] [PubMed] [Google Scholar]
- Kelly T.J. and Brown,G.W. (2000) Regulation of chromosome replication. Annu. Rev. Biochem., 69, 829–880. [DOI] [PubMed] [Google Scholar]
- Kelman Z. and O’Donnell,M. (1995) Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res., 23, 3613–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z., Yao,N. and O’Donnell,M. (1995) Escherichia coli expression vectors containing a protein kinase recognition motif, His6-tag and hemagglutinin epitope. Gene, 166, 177–178. [DOI] [PubMed] [Google Scholar]
- Klemm R.D. and Bell,S.P. (2001) ATP bound to the origin recognition complex is important for preRC formation. Proc. Natl Acad. Sci. USA, 98, 8361–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm R.D., Austin,R.J. and Bell,S.P. (1997) Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell, 88, 493–502. [DOI] [PubMed] [Google Scholar]
- Kobori J.A. and Kornberg,A. (1982) The Escherichia coli dnaC gene product III. Properties of the dnaB–dnaC protein complex. J. Biol. Chem., 257, 13770–13775. [PubMed] [Google Scholar]
- Koonin E.V. (1992) DnaC protein contains a modified ATP-binding motif and belongs to a novel family of ATPases including also DnaA. Nucleic Acids Res., 20, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A. and Baker,T.A. (1992) DNA Replication. W.H. Freeman and Co., New York, NY.
- Krause M. and Messer,W. (1999) DnaA proteins of Escherichia coli and Bacillus subtilis: coordinate actions with single-stranded DNA-binding protein and interspecies inhibition during open complex formation at the replication origins. Gene, 228, 123–132. [DOI] [PubMed] [Google Scholar]
- Learn B.A., Um,S.-J. and McMacken,R. (1997) Cryptic single-stranded-DNA binding activities of the phage λ P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc. Natl Acad. Sci. USA, 94, 1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBowitz J.H. and McMacken,R. (1986) The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem., 261, 4738–4748. [PubMed] [Google Scholar]
- Lee G.L. and Bell,S.P. (2000) ATPase switches controlling DNA replication initiation. Curr. Opin. Cell Biol., 12, 280–285. [DOI] [PubMed] [Google Scholar]
- Ludlam A.V., McNatt,M.W., Carr,K.M. and Kaguni,J.M. (2001) Essential amino acids of E. coli DnaC protein in an N-terminal domain interact with DnaB helicase. J. Biol. Chem., 276, 27345–27353. [DOI] [PubMed] [Google Scholar]
- Maiorano D., Moreau,J. and Mechali,M. (2000) XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature, 404, 622–625. [DOI] [PubMed] [Google Scholar]
- Marszalek J. and Kaguni,J.M. (1994) DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem., 269, 4883–4890. [PubMed] [Google Scholar]
- Naktinis V., Onrust,R., Fang,L. and O’Donnell,M. (1995) Assembly of a chromosomal replication machine: Two DNA polymerases, a clamp loader and sliding clamps in one holoenzyme particle. J. Biol. Chem., 270, 13358–13365. [PubMed] [Google Scholar]
- Neuhard J. and Nygaard,P. (1987) Purines and pyrimidines. In Neidhardt,F.C., Ingraham,J.L., Low,K.B., Magasanik,B., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, Vol. 1. American Society for Microbiology, Washington, DC, pp. 445–473.
- Neuwald A.F., Aravind,L., Spouge,J.L. and Koonin,E.V. (1999) AAA+: A class of chaperone-like ATPases associated with the assembly, operation and disassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- Nishitani H., Lygerou,Z., Nishimoto,T. and Nurse,P. (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature, 404, 625–628. [DOI] [PubMed] [Google Scholar]
- Parada C.A. and Marians,K.J. (1991) Mechanism of DNA A protein-dependent pBR322 DNA replication. DNA A protein-mediated trans-strand loading of the DNA B protein at the origin of pBR322 DNA. J. Biol. Chem., 266, 18895–18906. [PubMed] [Google Scholar]
- Park H.O., Bi,E., Pringle,J.R. and Herskowitz,I. (1997) Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl Acad. Sci. USA, 94, 4463–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G. and Diffley,J.F. (1998) Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell, 2, 23–32. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L.J. and Hurwitz,J. (1978) The dnaB gene product of Escherichia coli. II. Single stranded DNA-dependent ribonucleoside triphosphatase activity. J. Biol. Chem., 253, 4051–4057. [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schubach W.H., Whitmer,J.D. and Davern,C.I. (1973) Genetic control of DNA initiation in Escherichia coli. J. Mol. Biol., 74, 205–221. [DOI] [PubMed] [Google Scholar]
- Sekimizu K., Bramhill,D. and Kornberg,A. (1987) ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell, 50, 259–265. [DOI] [PubMed] [Google Scholar]
- Shrimankar P., Stordal,L. and Maurer,R. (1992) Purification and characterization of a mutant DnaB protein specifically defective in ATP hydrolysis. J. Bacteriol., 174, 7689–7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K. and Wold,S. (1995) The speed of the Escherichia coli fork in vivo depends on the DnaB:DnaC ratio. Mol. Microbiol., 17, 825–831. [DOI] [PubMed] [Google Scholar]
- Sprang S.R. (1997) G proteins, effectors and GAPs: structure and mechanism. Curr. Opin. Struct. Biol., 7, 849–856. [DOI] [PubMed] [Google Scholar]
- Studwell P.S. and O’Donnell,M. (1990) Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J. Biol. Chem., 265, 1171–1178. [PubMed] [Google Scholar]
- Sutton M.D., Carr,K.M., Vicente,M. and Kaguni,J.M. (1998) Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem., 273, 34255–34262. [DOI] [PubMed] [Google Scholar]
- Turner J. and O’Donnell,M. (1995) Cycling of Escherichia coli DNA polymerase III from one sliding clamp to another: model for lagging strand. Methods Enzymol., 262, 442–449. [DOI] [PubMed] [Google Scholar]
- Wahle E., Lasken,R.S. and Kornberg,A. (1989a) The dnaB–dnaC replication protein complex of Escherichia coli. I. Formation and properties. J. Biol. Chem., 264, 2463–2468. [PubMed] [Google Scholar]
- Wahle E., Lasken,R.S. and Kornberg,A. (1989b) The dnaB–dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J. Biol. Chem., 264, 2469–2475. [PubMed] [Google Scholar]
- Wechsler J.A. (1975) Genetic and phenotypic characterization of dnaC mutations. J. Bacteriol., 121, 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J.A. and Gross,J.D. (1971) Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol. Gen. Genet., 113, 273–284. [DOI] [PubMed] [Google Scholar]
- Weinreich M., Liang,C. and Stillman,B. (1999) The Cdc6p nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc. Natl Acad. Sci. USA, 96, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzhakov A., Turner,J. and O’Donnell,M. (1996) Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell, 86, 877–886. [DOI] [PubMed] [Google Scholar]