Abstract
Disruption of ypuG and ypuH open reading frames in Bacillus subtilis leads to temperature-sensitive slow growth, a defect in chromosome structure and formation of anucleate cells. The genes, which were named scpA and scpB, were found to be epistatic to the smc gene. Fusions of ScpA and ScpB to the fluorescent proteins YFP or CFP showed that both proteins co-localize to two or four discrete foci that were present at mid-cell in young cells, and within both cell halves, generally adjacent to chromosomal origin regions, in older cells. ScpA and ScpB foci are associated with DNA and depend on the presence of SMC and both Scps. ScpA and ScpB are associated with each other and with SMC in vivo, as determined using the FRET technique and immunoprecipitation assays. Genes similar to scpA and scpB are present in many bacteria and archaea, which suggests that their gene products form a condensation complex with SMC in most prokaryotes. The observed foci could constitute condensation factories that pull DNA away from mid-cell into both cell halves.
Keywords: chromosome/condensation/Scp proteins/segregation/SMC protein
Introduction
Chromosome segregation occurs in an ordered and highly efficient manner in bacteria, with an error rate for loss of chromosomes of <10–5 (Ireton et al., 1994). At one point during the cell cycle, and soon after initiation of replication, origin regions of the chromosome separate rapidly towards the cell poles, independent of cell wall growth (Glaser et al., 1997; Lin et al., 1997; Webb et al., 1997; Niki and Hiraga, 1998). Terminus regions of chromosomes are also separated in a dynamic manner, but later in the cell cycle, and, additionally, are not separated as far as origin regions (Webb et al., 1998). This process establishes a general layout of chromosomes in which origin regions are localized towards cell poles throughout most of the cell cycle, while termini localize towards the middle of the cell. Regions between the origin and terminus localize between these sites, such that the chromosome appears to be condensed roughly according to its physical structure (Teleman et al., 1998). After the final separation of chromosomal termini, the division septum constricts at mid-cell, generating two equal daughter cells. Taken together, these data suggest that a mitotic-like apparatus might separate chromosomes in bacteria during ongoing DNA replication.
It is only poorly understood which components this apparatus consists of, and how it might work. Active DNA polymerase localizes to the middle of cells in Bacillus subtilis and Escherichia coli throughout most of the cell cycle (Lemon and Grossman, 1998; Koppes et al., 1999). Thus, DNA appears to move through a stationary replisome, rather than the polymerase moving along DNA. This finding has led to the proposal that DNA is moved towards cell poles through the threading of DNA through the replisome (Lemon and Grossman, 2001). However, DNA polymerase is not stationary in Caulobacter crescentus (Jensen et al., 2001). In addition, terminal regions of chromosomes cannot be separated towards both cell halves by pushing through the polymerase, so additional factors are required for complete segregation of chromosomes.
Key players in chromosome condensation, organization and segregation are SMC proteins. This group of proteins is found in prokaryotes and eukaryotes, and its members serve an essential role in a wide range of chromosome dynamics: chromosome condensation, cohesion, transcriptional repression of whole chromosomes and DNA recombination and repair (Hirano, 1999; Strunnikov and Jessberger, 1999; Graumann, 2001). SMC proteins consist of globular N- and C-terminal domains connected to a central hinge domain by two long coiled-coil regions. SMC proteins form antiparallel dimers (Melby et al., 1998) and, due to the flexible hinge domain, both ends of the symmetrical molecule can come together or stretch out to 180°. The globular N- and C-terminal domains within the dimer form one domain, called the head domain, which has a structure similar to ATP cassettes of ABC transporters (Hopfner et al., 2000; Lowe et al., 2001), and which has low ATPase activity in SMC proteins. Head domains appear to bind to DNA and undergo a conformational change upon ATP binding in Rad50, leading to dimerization of two head domains.
Eukaryotic SMC dimers act in several complexes, together with other proteins. The function(s) of condensin or cohesin complexes that mediate chromosome condensation or cohesion, respectively, require SMC heterodimers as well as additional proteins (Hirano et al., 1997; Michaelis et al., 1997). In vitro, the condensin complex can introduce positive writhe into DNA, which leads to negative supercoiling (Kimura et al., 1999). This activity is a means to condense DNA in vivo; however, its molecular mechanism is not well understood. The SMC-like protein Rad50 is involved in double-strand break repair by homologous recombination of DNA and acts only in the presence of its partner, the Mre11 endocuclease (Connelly et al., 1998; Hopfner et al., 2001). Purified SMC from B.subtilis displays ATP-dependent aggregation of single-stranded DNA (Hirano and Hirano, 1998). It is possible that SMC needs additional proteins for double-stranded DNA binding.
In contrast to eukaryotes that usually have four smc genes, archaeal and eubacterial genomes that have been sequenced contain only one or no smc gene (Soppa, 2001). MukB of E.coli, which belongs to the SMC protein family, and SMC of C.crescentus and of B.subtilis are essential for chromosome condensation, segregation and growth above 23°C (Britton et al., 1998; Graumann et al., 1998; Moriya et al., 1998; Jensen and Shapiro, 1999). Bacillus subtilis SMC localizes in foci (Britton et al., 1998) that are located close to the cell poles (Graumann et al., 1998), and is required for the complete separation of chromosomes, but not for the separation of chromosome origins (Graumann, 2000). SMC and MukB are also necessary for the proper arrangement of the chromosome (Graumann, 2000; Weitao et al., 2000). MukB acts in a complex with MukE and MukF (Yamazoe et al., 1999), which do not have homologues in species other than enteric bacteria. It is not known how SMC performs its function in the cells, and whether it also forms a complex with non-SMC proteins.
In this report, we characterize two novel genes found in a wide range of bacteria and archaea that are indispensable for chromosome segregation and condensation at temperatures above 23°C, similarly to smc. The products of these genes, ScpA and ScpB, interact with each other and with SMC in vivo, suggesting that these proteins form a complex. Fusions of ScpA and ScpB to yellow fluorescent protein (YFP) revealed a dynamic pattern of localization, indicating that a condensin-like complex operates in bacterial chromosome segregation to organize and separate chromosomes within both cell halves.
Results
ypuG (scpA) and ypuH (scpB) open reading frames encode proteins essential for chromosome condensation and segregation
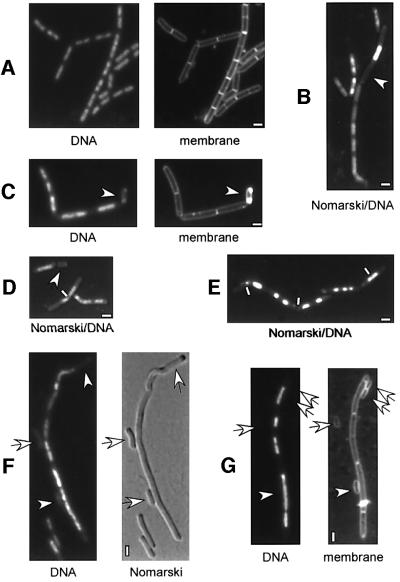
Using immunoprecipitation analysis, we have found that at least two 20–30 kDa B.subtilis proteins interact specifically with SMC in wild-type but not in smc mutant cells (see below). In our attempt to identify these proteins, we were intrigued by a new protein family whose genes are close to or overlap with smc genes in most archaeal genomes. A similar gene, ypuG, is located at 207.2° on the B.subtilis chromosome in a putative operon with ypuH and ypuI. It was shown previously that at 37°C, ypuI can be disrupted by single crossover integration of a plasmid, while ypuG and ypuH could not be disrupted (Vagner et al., 1998). To investigate whether disruption of ypuG and/or ypuH might have a similar phenotype to a deletion of smc (formation of slow growing colonies only below 23°C), we constructed strains in which 70–80% of ypuG, ypuH or ypuI were exchanged for the tetracycline resistance gene by double-crossover replacement. In contrast to the disruption of ypuI that resulted in wild-type-like colonies at 37°C, ypuG and ypuH mutant colonies were only obtained at 23°C, but not at 37 or 30°C. Investigation of nucleoid morphology in the mutant cells revealed decondensed and irregularly shaped nucleoids (Figure 1B and C) and the formation of 10–15% anucleate cells in the ypuG (JM11) and ypuH (PG32) strains, while ypuI mutant cells (strain PG36) had wild-type-like nucleoids and no detectable defect in chromosome segregation (Table II; data not shown). Occasionally, nucleoids appeared to be guillotined by septa (see Figure 1D, white line) in ypuG and ypuH mutant cells, similar to smc mutant strains. The genes were therefore renamed scpA and scpB (for segregation and condensation protein), respectively, and their products ScpA and ScpB, which is used hereafter. Similarly to smc mutant cells, scpA and scpB mutant cells were larger than wild-type cells (compare Figure 1A with C). To rule out the possibility that the phenotype of disruption of scpA is due to a polar effect on scpB, we constructed a strain in which a fusion of scpB to a variant of green fluorescent protein (gfp), cyan fluorescent protein (cfp), is expressed under the control of the xylose promoter at the amylase (amy) locus. In the presence of xylose, but not in its absence, scpB could be disrupted in strain JM10 (pxyl-scpB-cfp at amy) at 37°C. Growth at 37°C and nucleoid morphology were normal in the resulting strain PG40 (pxyl-scpB-cfp at amy, scpB::tet); therefore, for chromosome segregation, the scpB-cfp fusion complemented the scpB deletion in trans. Deletion of scpA in strain JM10 resulted in temperature-sensitive slow growth and formation of anucleate cells in the presence of xylose [similar to strain JM11 (scpA::tet), data not shown], so both scpA and scpB, are required for efficient chromosome segregation and condensation above 23°C in B.subtilis.
Fig. 1. Deletion of scpA and scpB leads to abnormal, decondensed nucleoid structure and formation of anucleate cells. Fluorescence microscopy of exponentially growing cells from strains (A) PY79 (wild type), (B) JM11 (scpA::tet), (C) PG32 (scpB::tet), (D) JM12 (scpA-B::tet), (E) PG37 (scpA::tet, spoIIIE::spec), (F) PG43 (scpB::tet, pspac-smc at amy locus, smc::kan) growing in the absence of IPTG (inducer) and (G) PG39 (scpB::tet, spo0J::spec). DNA was stained with DAPI, membranes with FM4-64 and outlines of cells by Nomarski digital interference contrast (DIC). Arrows indicate anucleate cells and white lines show chromosomes dissected (‘guilloutined’) by prematurely formed septa. (B), (D) and (E) are direct captures of DAPI fluorescence and Nomarski DIC. White rectangles indicate 2 µm.
Table II. Phenotypes of deletion strains at 23°C.
| Strain | Doubling time (min) | Anucleate cells (%) | Spore formation (%) |
|---|---|---|---|
| PY79 | 92 | <0.01 | 78 |
| PG31 (ypuI) | 98 | <0.01 | 77 |
| JM11 (scpA) | 196 | 11 | 8.5 |
| PG32 (scpB) | 224 | 12 | 39 |
| JM12 (scpAB) | 228 | 11 | n.d. |
| JM19 (scpA, smc) | 385 | 11 | n.d. |
| PG43 (scpB, smc) | 388 | 13 | n.d. |
| PG39 (scpB, spo0J) | 267 | 28 | n.d. |
| PG36 (ypuI, spoIIIE) | 96 | <0.01 | <0.001 |
| PG37 (scpA, spoIIIE) | 463 | <1 | <0.001 |
| PG38 (scpB, spoIIIE) | 448 | 1 | <0.001 |
| PGΔ388 (smc) | 386 | 12 | 0.4 |
The growth rate of scpA and scpB mutant cells was 2- and 2.5-fold lower than that of wild-type cells at 23°C, while growth of smc mutant cells (strain PGΔ388) is >4-fold lower (Table II). Sporulation efficiency was 8.5 and 39% in scpA and scpB mutant strains, respectively, while the smc mutant strain sporulated only with 0.4% efficiency compared with 78% in wild-type cells (Table II). Thus, although the formation of anucleate cells in scpA and scpB mutant cells is similar to that of the smc mutant strain, growth and sporulation efficiency are not as severely affected through the scpA or scpB deletion as through the smc deletion.
Genetic interactions of scpA and scpB
To test whether ScpA and ScpB act in the same genetic context, we constructed a double mutant by replacing both genes with a tetracycline resistance gene. The resultant strain, JM12 (scpA-B::tet) had a similar phenotype to that of the single mutants (Figure 1D; Table II). To investigate a possible genetic interaction with smc, we used a strain carrying a deletion of smc and an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible copy of smc at the amy locus (EP58; smc::kan, pspac-smc at amy) that grows at 37°C only in the presence, but not in the absence of IPTG. The scpA and scpB deletions were moved into strain EP58 at 23°C, generating strains JM19 (scpA::tet, smc::kan, pspac-smc at amy) and PG43 (scpB::tet, smc::kan, pspac-smc at amy). In the absence of IPTG, both strains grew with a doubling time similar to that of the smc mutant strain, accompanied by the formation of 10–15% anucleate cells (Figure 1F; Table II). This indicates that scpA and scpB act in the same pathway as SMC in chromosome dynamics.
It has been reported that the combination of a deletion of the spo0J gene, which leads to the formation of ∼1% anucleate cells but not to a reduction in growth rate (Ireton et al., 1994), with an smc deletion acerbates the phenotype of the smc deletion (Britton et al., 1998). To test whether this is also the case for the scpB deletion, we generated a spo0J scpB double mutant strain (PG39). We monitored the formation of 25–35% anucleate cells in PG39 (Figure 1G), as well as a reduction in growth rate compared with the single scpB deletion (Table II), suggesting that like SMC, ScpB acts on a different or only partially overlapping aspect of chromosome segregation than Spo0J.
In another report, Britton and Grossman (1999) found that a double mutant in smc and spoIIIE (involved in the rescue of chromosomes trapped within the division septum; Sharpe and Errington, 1995) is lethal. We therefore deleted scpA, scpB and ypuI in a spoIIIE mutant strain (which does not show a detectable phenotype at 37°C). In contrast to the ypuI spoIIIE double mutant strain that grew similarly to wild-type cells, the scpA spoIIIE and scpA spoIIIE double mutant strains were temperature sensitive and grew much more slowly than the single scpA or scpB mutant strains, and even more slowly than the smc deletion strain (Table II). DNA staining in the scp spoIIIE double mutant cells showed highly fragmented nucleoids that arise through trapping of non-segregated DNA by a prematurely formed septum (guilloutining, Figure 1E, indicated by white bars). Intriguingly, formation of anucleate cells was much lower in the scp spoIIIE double mutant strains (∼1%) than in the single scp mutants (>10%, Table II). This finding suggests that anucleate cells arise mainly through the rescue of guilloutined DNA into one daughter cell, rather than through pinching off of completely empty cell compartments. Taken together, these data suggest that ScpA and ScpB function in the same pathway as SMC in chromosome dynamics; however, loss of SMC is more dramatic for the cell than loss of ScpA and ScpB.
Cell cycle-dependent localization of ScpA and ScpB
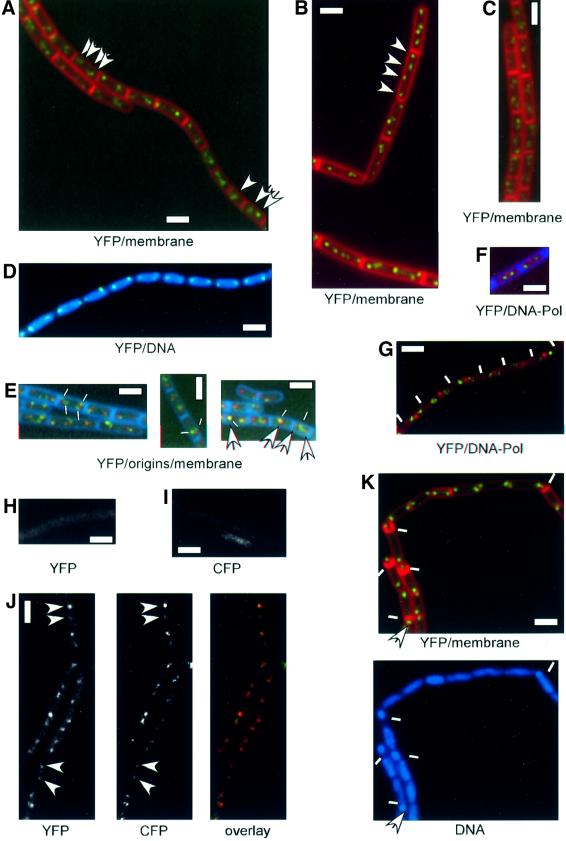
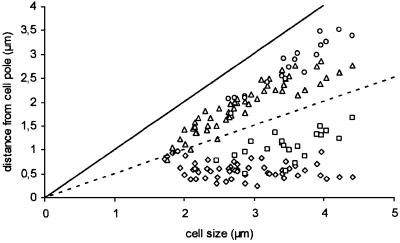
We integrated fusions of scpA and scpB to a variant of gfp, yfp, into the chromosome by single crossover, such that only the fusions are expressed. Both fusions, scpA-yfp and scpB-yfp, fully complemented growth at 37°C, and are therefore functional. Strain JM8 expressing ScpA–YFP showed delayed resumption of growth after re-inoculation from stationary phase, which might be due to slower folding of the fusion protein. Subcellular localization of ScpA–YFP and ScpB–YFP was investigated by fluorescent microscopy of live cells grown in minimal medium, which showed that both proteins are present in discrete fluorescent foci. In general, small cells contained one or two foci close to mid-cell (Figure 2A, arrow indicating the lower cell), while larger cells generally contained two bipolar foci (Figure 2A, two arrows indicating the lowest cell), or four foci, two of each close to but not at the cell poles (Figure 2A, four arrows indicating the upper cell). Statistical analysis of the position of ScpA–YFP foci (Figure 3) showed that the transition between central and bipolar foci occurred in the smallest cells and therefore early in the cell cycle. Four foci became apparent in cells >2.5 µm, which stayed in pairs that did not separate from each other before the cells reached ∼3.3–4 µm (Figure 3). Cell division of the largest cells would therefore give rise to daughter cells with bipolar ScpA foci. This pattern of localization was also seen for ScpB–YFP (Figure 2C). Bipolar ScpA–YFP foci were located mostly at the outer edges of the nucleoids (Figure 2D).
Fig. 2. Bipolar localization of ScpA and ScpB. Fluorescence microscopy of cells growing exponentially in minimal medium (A, C and D–J), in enriched minimal medium (B) or after entry into sporulation in sporulation medium (K). (A and B) JM8 (scpA-yfp), (C) JM9 (scpB-yfp), (D) JM8 (scpA-yfp), (E) PG27 (scpB-yfp, lacO-cassette at 359°, lacI-cfp at thr locus), (F) PG29 (scpA-yfp, dnaX-cfp), (G) PG30 (scpB-yfp, dnaX-cfp), (H) JM10 (pxyl-scpB-cfp at amy locus), (I) JM8 (scpA-yfp), (J) JM14 (scpA-yfp, pxyl-scpB-cfp at amy locus) and (K) JM8 (scpA-yfp). Arrows indicate the position of selected Scp–YFP signals and white lines indicate positions of septa in (G) and polar sporulation septa in (K). In (E), thin white lines indicate the position of ScpB–YFP (green) and arrows point to origin signals (red). In (F) and (G), Scps are green and DNA polymerase is red. DNA was stained with DAPI and membranes with FM4-64. White rectangles indicate 2 µm. Note that (B) is a composite image.
Fig. 3. Graphical representation of the position of ScpA foci in growing cells. The distance of foci to one cell pole (the one closest to a focus) dependent on cell size. Diamond, closest origin to cell pole; triangle, second origin in cells with two foci/third focus in cells with four foci; square, second closest focus in cells with four foci; circle, fourth focus in cells with four foci.
Cell growing in the minimal medium used in this study have one or two origin regions (Webb et al., 1998; and see below). At higher growth rate, cells contain two or more origin regions because of overlapping rounds of DNA replication (Sharpe et al., 1998). To test whether the number of Scp foci increases concomitantly with multiple origins, casamino acids were added to the medium, which led to a decrease in the number of cells with two foci in favour of cells with four foci. Most cells contained four foci that were well separated from each other, and up to eight foci could be observed within one cell (Figure 2B).
Entry into sporulation is characterized by the movement of origin regions to the extreme cell poles (Glaser et al., 1997; Lin et al., 1997; Webb et al., 1997; Graumann and Losick, 2001). Likewise, ScpA–YFP and ScpB–YFP were found at the extreme cell poles and within the forespore compartments 2 h after the entry into sporulation at 25°C (Figure 2K).
Dual labelling of ScpA, ScpB, origin regions on the chromosomes and DNA polymerase reveals a novel pattern of subcellular localization
The bipolar localization of ScpA and ScpB was very similar to the localization of origin regions on the chromosome (Glaser et al., 1997; Lin et al., 1997; Webb et al., 1997, 1998). To investigate whether ScpA and ScpB co-localize with origin regions, which is the case for Spo0J (Lewis and Errington, 1997; Teleman et al., 1998), we created strain PG27 expressing ScpB–YFP and possessing origin regions decorated with LacI–CFP. Cells from strain PG27 contained characteristic bipolar foci (Figure 2E); ∼12% of the cells had one focus (400 cells were analysed). Figure 2E shows that in general, ScpB–YFP foci did not co-localize with origin signals; however, 8% of origin signals were coincident with ScpB foci (Figure 2E, see right panel, left cell). Mostly, ScpB localized close to the origin regions and, frequently, two ScpB–YFP foci (indicated by thin white lines in Figure 2E) were observed to flank each bipolar origin region. ScpB–YFP was separate from origins even in small cells with one origin signal (Figure 2E, right panel, middle cell); however, we found 10 examples in which two bipolar origin signals flanked one central ScpB focus (Figure 2E, right panel, right cell). This finding suggests that origin regions move out towards both cell poles first, and that Scp foci separate shortly thereafter.
It is possible that ScpA and ScpB are associated, at least early in the cell cycle, with DNA polymerase that localizes to the mid-cell position throughout most of the cell cycle (Lemon and Grossman, 1998). Therefore, we simultaneously visualized ScpA–YFP or ScpB–YFP and DNA polymerase–CFP within the same cells. In all cells monitored, Scp foci were well separated from foci of DnaX–CFP, the τ-subunit (clamp) of the DNA polymerase complex (Figure 2F and G). Even in small cells in which Scp foci were close to mid-cell, DnaX and ScpA or ScpB foci were not coincident (data not shown). These data reveal that ScpA and ScpB localize in two and later in four bipolar clusters that are formed independently of origin regions or DNA polymerase, and which might locally condense DNA within both cell halves.
ScpA and ScpB co-localize
A fusion of ScpB to CFP integrated at the amy locus localizes in a similar manner to ScpB–YFP, and fully complements the function of ScpB in strain PG40 (scpB::tet, pxyl-scpB-cfp at amy) in the presence of xylose. We combined a fusion of ScpA–YFP with ScpB–CFP, to visualize both Scps simultaneously. At an exposure of 3 s (maximal exposure time taken in this work), no ScpB–CFP signal was detectable in the YFP filter (Figure 2H). Likewise, ScpA–YFP was not detectable in the CFP filter set (Figure 2I). The fluorescence of ScpB–CFP was very weak; however, we were able to collect a number of cells in which both signals were clearly visible. Figure 2J shows that ScpA and ScpB foci were coincident in all cells (53/53 cells), suggesting that they might form a complex in vivo.
Formation of ScpA and ScpB foci depends on DNA, SMC and both Scps
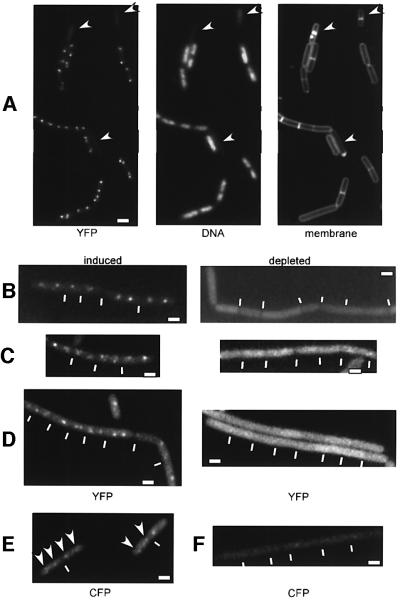
We wished to investigate whether ScpA and ScpB foci are associated with DNA. For this purpose, we moved the Scp–YFP fusions into spo0J mutant cells and analysed the localization of ScpA and ScpB in anucleate cells that are generated in this background at a frequency of 0.2–1.5% (Ireton et al., 1994). In contrast to cells containing DNA that showed bipolar localization of ScpA and ScpB, anucleate cells (12 of 12 and 52 of 52 analysed for ScpA and ScpB, respectively) did not show any foci (Figure 4A, indicated by arrowheads); Scp–YFP fluorescence in anucleate cells corresponded to background staining. We conclude that ScpA and ScpB are directly or indirectly associated with DNA in vivo.
Fig. 4. Fluorescence microscopy of exponentially growing cells from strains (A) PG35 (scpB-yfp, spo0J::spec), (B) PG33 (scpB-yfp, pspac-smc at amy locus, smc::kan), (C) JM17 (scpA-yfp, pspac-smc at amy locus, smc::kan), (D) PG41 (scpA-yfp, scpB::tet, pxyl-scpB-cfp at amy locus), (E) JM10 (pxyl-scpB-cfp at amy locus) and (F) JM18 (scpA::tet, pxyl-scpB-cfp at amy locus) in the presence of xylose. Depleted indicates that the inducer was removed during exponential growth 3–5 h prior to image acquisition. Arrows indicate anucleate cells in (A) and ScpB–CFP foci in (E), and white lines indicate septa between cells. Images ‘induced’ and ‘depleted’, and (E) and (F) are scaled. White rectangles indicate 2 µm.
Next, we monitored the formation of ScpA and ScpB foci in the presence or absence of SMC, which was achieved by moving the YFP fusions into strain EP58 (smc::kan, pspac-smc at amy), in which SMC can be depleted. At mid-exponential phase, cultures of the resultant strains, JM17 (scpA-yfp, smc::kan, pspac-smc at amy) and PG33 (scpB-yfp, smc::kan, pspac-smc at amy), were resuspended in medium containing or lacking inducer (IPTG). Fluorescent foci were observed in the presence of IPTG, but not in its absence (Figure 4B and C). In contrast to anucleate cells, SMC-depleted cells showed fluorescence that was substantially higher than background (Figure 4B and C), indicating that ScpA–YFP or ScpB–YFP were still present, but not able to form foci or associate with DNA. This was supported by western blot analysis, which showed that ScpA–YFP is not degraded in the absence of SMC (Figure 6, lane 4, middle panel).
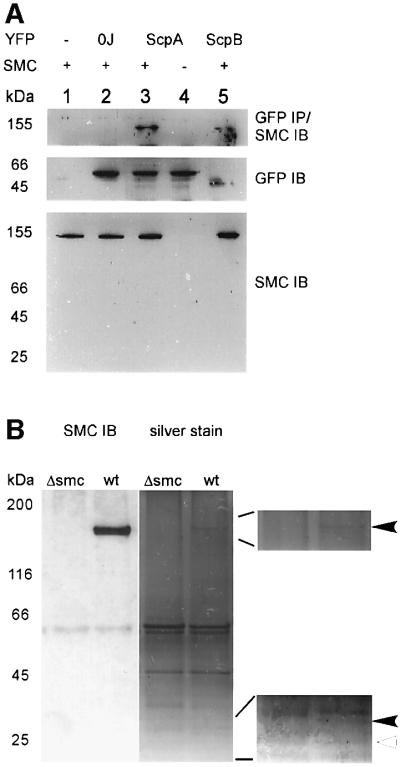
Fig. 6. Interaction of ScpA and ScpB with SMC. (A) Upper panel: immunoprecipitation (IP) of YFP fusions and blotting (IB) with anti-SMC antiserum. Middle panel: loading control, blotting with monoclonal GFP/YFP antibodies. Lower panel: blotting with SMC antiserum. Samples of cultures were taken during exponential growth. Lane 1, PY79 (wild type); lane 2, PG42 (spo0J-yfp); lane 3, JM8 (scpA-yfp); lane 4, JM17 (scpA-yfp, pspac-smc at amy locus (smc::kan) grown in the absence of inducer; lane 5, JM9 (scpB-yfp). The sizes of relevant protein markers are given. (B) Immunoprecipitation with SMC antiserum and western blot (SMC IB) or silver staining. Δsmc, PGΔ388 (smc::kan); wt, wild type. Right panels show enlargements of SMC (arrowhead, upper panel) and two interacting proteins (filled arrowhead, size of ScpA; open arrowhead, size of ScpB).
To investigate whether ScpA and ScpB form foci in the absence of the other Scp, we transformed the scpA-yfp fusion into strain PG40 (pxyl-scpB-cfp at amy, scpB::tet), in which ScpB–YFP can be depleted and which does not contain wild-type ScpB. At 3 h after removal of xylose (∼2 doubling times), ScpA foci were no longer detectable (Figure 4D). We also analysed strain JM18 (pxyl-scpB-cfp, scpA::tet) growing in the presence of xylose. In contrast to strain JM10 (pxyl-scpB-cfp at amy), which showed bipolar ScpB foci (Figure 4E), no such signal was observed in the scpA null background (Figure 4F). Therefore, the formation of Scp foci depends on the presence of both ScpA and ScpB.
ScpA and ScpB interact with each other and with SMC in vivo
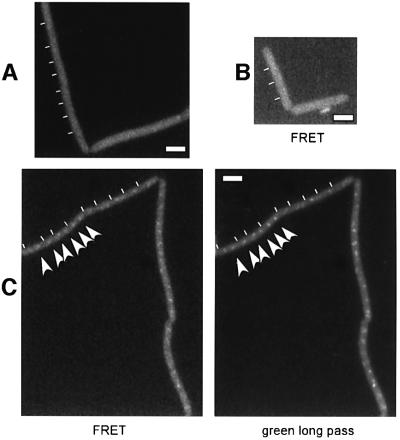
ScpA and ScpB might interact directly in vivo. We have used fluorescence resonance energy transfer (FRET) to test this possibility. In a modified filter set in which ScpB–CFP but not ScpA–YFP is excited, emission is monitored for YFP, but cut off for CFP. Figure 5A shows that emission from ScpB–CFP in strain PG40 (scpB::tet, pxyl-scpB-cfp at amy locus) is undetectable, and that ScpA–YFP is not excited in strain JM8 (scpA-yfp) (Figure 5B). However, foci are visible in strain PG41 (scpB::tet, pxyl-scpB-cfp at amy locus, scpA-yfp) that correspond to ScpA–YFP (Figure 5C, left panel), as seen in the long pass green filter (right panel). Foci were not seen in the absence of xylose (not shown). Fluorescence emission of ScpA–YFP is therefore due to excitation by fluorescence of ScpB–CFP, which is only possible if both protein fusions are in close proximity (10–100 Å) within the cell. These data show that ScpA and ScpB interact in vivo. To investigate whether ScpA and ScpB form a complex with SMC, analogous to that of MukB, MukE and MukF in E.coli (Yamazoe et al., 1999), we performed immunoprecipitation in strains expressing YFP fusions of ScpA and ScpB using monoclonal GFP antibodies that recognize all GFP variants. As controls, experiments were performed in wild-type cells, in a strain expressing Spo0J–YFP and in a strain expressing ScpA–YFP in which SMC was depleted. Figure 6A shows that SMC was co-precipitated with ScpA–YFP and with ScpB–YFP, but not in the control strains (upper panel). Possibly, SMC interacts more weakly with ScpB than with ScpA, or ScpB could be present in vivo in lower quantities than ScpA. The band does indeed correspond to SMC because no such signal is detected in a strain in which SMC was depleted for 5 h (lower panel). Under this condition, ScpA–YFP is stable, showing that depletion of SMC does not lead to degradation of ScpA–YFP. The middle panel in Figure 6A shows that the GFP antibody recognized all YFP fusions (Spo0J–YFP: 32 + 28 kDa, ScpA–YFP: 29.5 + 28 kDa, ScpB–YFP: 22 + 28 kDa). In support of protein interaction, we immunoprecipitated SMC, and monitored binding partners with silver stain. Figure 6B shows that clearly, a band corresponding to the size of ScpA (black arrowhead in lower magnification) co-precipitated with SMC (arrowhead, upper magnification) in wild-type but not in smc mutant (Δsmc) cells. A band corresponding to ScpB (open arrowhead) was extremely faint but detected reproducibly. The additional band in the Δsmc strain that runs slightly higher than the ScpA band in the wild-type lane corresponds well to the N-terminal fragment of SMC that is still synthesized in the deletion mutant. These data show that ScpA and ScpB interact with each other and with SMC in vivo, and suggest that SMC may form a condensin- or cohesin-like complex with ScpA and ScpB.
Fig. 5. FRET analysis of ScpB–CFP/ScpA–YFP. Fluorescence microscopy of cells growing in the presence of xylose from strains (A) PG40 (scpB::tet, pxyl-scpB-cfp at amy locus), (B) JM8 (scpA-yfp) and (C) PG41 (scpB::tet, pxyl-scpB-cfp at amy locus, scpA-yfp). FRET, excitation of CFP, emission of YFP; green long pass, excitation/emission of YFP. Arrows indicated ScpA–YFP foci. White rectangles indicate 2 µm.
ScpA and ScpB are two widespread protein families in eubacteria and archaea
ScpA and ScpB are small (29.5 and 22 kDa) acidic (pI 4.8 and 4.3) proteins. BLAST searches (see Supplementary figure 7A and B available at The EMBO Journal Online) revealed that both proteins are members of two widespread protein families, with homologues in many bacterial branches and in archaea. Sequence conservation ranges from 56/48% identity within bacteria to 27/28% between bacteria and archaea. In general, scpA and scpB form an operon in most bacterial genomes that were available for sequence analysis, while ypuI was only found in B.subtilis. Flanking genes of scpA and scpB were highly variable within bacteria, indicating that this operon has been movable throughout evolution. Conversely, in most archaea, scpA was found downstream of smc. Intriguingly, bacteria containing an smc gene also contained an scpA or an scpB gene but not necessarily both, while scpB is present in Deinococcus radiodurans, which does not possess smc or scpA. Enterobacteria that contain mukB, mukE and mukF, but no smc gene, do not contain scpA or scpB genes. It is likely that the muk operon is the enteric counterpart of smc, scpA and scpB of most other bacteria. We were not able to detect any known sequence motifs in ScpA and ScpB proteins; however, both proteins have a high content of leucine residues (Supplementary figure 7A and B). Deduced from their sequence, B.subtilis ScpA and ScpB are predicted to contain coiled-coil elements (Supplementary figure 8), suggesting that ScpA and ScpB may form homo- or heterodimers. Alternatively, the coiled-coil regions could be SMC-interacting domains. ScpA proteins contain some invariant residues, notably a lysine at position 71 that is framed by an invariant alanine and two leucines within a conserved motif (residues 59–78, Supplementary figure 7A). We suggest that this motif is essential for the function of ScpA-like proteins. Additionally, ScpA proteins have a conserved motif close to the C-terminus, containing an invariant leucine and a glutamine residue, and an unusual, highly acidic region (residues 79–96). ScpB proteins contain, in addition to several conserved leucine residues, an invariant aspartate residue close to the N-terminus, an arginine at position 119 and a TTXXF motif starting at position 154. The high degree of sequence conservation between prokaryotic ScpA and ScpB proteins suggests that these proteins perform similar functions in many bacteria and archaea.
Discussion
In this work, we identified two novel proteins involved in chromosome condensation and segregation in B.subtilis that are members of two widespread protein families in bacteria and archaea. Disruption of scpA or scpB leads to slow temperature-sensitive growth (below 24°C) in rich medium, aberrant chromosome structure and formation of >10% anucleate cells. This phenotype is strikingly similar to that of a deletion of the smc gene (Graumann, 2001). We have found that ScpA and ScpB interact with SMC protein in vivo and, in support of this, that SMC specifically interacts with at least one protein corresponding to the size of ScpA. An interaction of SMC with ScpA has also been found in an independent study (Soppa and Moriya, 2002). Because ScpA and ScpB are in close association in vivo, which we observed using the FRET technique, it is likely that SMC, ScpA and ScpB form a complex in vivo. The SMC–Scp complex appears to be similar to the MukB–MukE–MukF complex in E.coli (Yamazoe et al., 1999), where MukB is the counterpart of SMC (Niki et al., 1991). Although ScpA and ScpB do not have significant sequence homology with MukE or MukF, MukF and ScpA, and MukE and ScpB, respectively, are predicted to contain very similar predominantly α-helical secondary structures and coiled-coil regions (data not shown). Yamazoe et al. (1999) have shown that MukE and MukF form a complex even in the absence of MukB, and than MukF, but not MukE, binds to MukB in a direct manner. We suggest that SMC, ScpA and ScpB form a complex similar to MukB–MukE–MukF in most bacteria other than enterics.
The formation of an SMC–ScpA–ScpB complex in B.subtilis is consistent with genetic analyses. Depletion of SMC in scpA or scpB mutant strains led to a growth rate similar to the strain carrying an smc deletion, indicating that smc is epistatic to scpA and scpB and, therefore, ScpA and ScpB act in the same pathway as SMC for chromosome segregation and condensation. In contrast, deletion of spo0J (leading to 1% anucleate cells but not to a reduction in growth rate) in an smc mutant strain exacerbates the smc phenotype (Britton et al., 1998), which we also observed for an scpB spo0J double deletion. Interestingly, the scpB spo0J mutant strain produced a higher amount of anucleate cells than the scpB smc mutant strain, while its growth rate was higher than that of the latter strain. This shows that the slow growth rate of the smc mutant strain is not due simply to a high error rate in chromosome segregation, but that a checkpoint may reduce the formation of anucleate cells by delaying cell division. A segregation checkpoint has also been suggested to exist in C.crescentus (Jensen and Shapiro, 1999). It was shown that SpoIIIE performs a rescue function for chromosomes entrapped in septa in smc mutant cells, because a double smc spoIIIE mutation is lethal (Britton and Grossman, 1999), while the spoIIIE mutation has no detectable phenotype in growing cells (Wu and Errington, 1994). Likewise, disruption of spoIIIE in scpA or scpB mutant cells strongly increased the severity of the scp mutations. However, the fact that those mutant cells were viable indicates that SMC might perform a basic segregation function in the absence of ScpA and ScpB. This is supported by our finding that the growth rate of the scpA or scpB mutant strains was higher than that of the smc deletion strain. However, it is also possible that SMC confers a second ScpA- and ScpB-independent function. This function could be in chromosome cohesion, which has been shown to exist in E.coli (Sunako et al., 2001). Origin-proximal sites on the chromosome separated much earlier from each other in a mukB null strain than in wild-type cells, indicating that MukB and SMC could perform a dual function in chromosome condensation and cohesion, unlike in eukaryotes, where different SMC proteins are involved in both processes. In any event, deletion of SMC leads to chromosome decondensation, due to a decrease in negative supercoiling (Sawitzke and Austin, 2000), and its overproduction leads to pronounced chromosome condensation in vivo (C.Andrei-Selmer and P.L.Graumann, unpublished results), so SMC is a true condensation factor.
We analysed the subcellular localization of ScpA and ScpB in live cells using strains that exclusively express fusions with YFP or CFP, which revealed a pattern of localization that has not been observed previously for bacterial proteins. ScpA and ScpB co-localized to discrete foci that were present at the mid-cell position early in the cell cycle, and close to both cell poles in larger cells. Separation of Scp foci occurred within a short period of the cell cycle, similar to origin regions of the chromosomes (Webb et al., 1998). ScpA and ScpB co-localized with origin regions in a small percentage of cells; however, in most cells, two Scp foci were present close to and often flanking origin regions. ScpA and ScpB were associated exclusively with DNA, mostly close to the outer nucleoid borders. This suggests that ScpA and ScpB clusters do not bind in a sequence-specific manner, but might associate with different DNA regions during the cell cycle. Scp foci did not co-localize with but were well separated from DNA polymerase, indicating that the proteins are not involved in DNA replication. In agreement with its interaction with ScpA and with ScpB, an SMC–GFP fusion localizes in a similar, bipolar manner to ScpA and ScpB (J.Mascarenhas and P.L.Graumann, unpublished results), and thus similar to MukB–GFP (Ohsumi et al., 2001). Britton et al. (1998) also reported that SMC–GFP forms foci at the outer border of nucleoids. Immuno fluorescence microscopy has shown that SMC localizes in foci that are very close to the cell poles (Graumann et al., 1998). While these data are generally consistent, we believe that fixation of cells moves SMC foci closer to the cell poles than is true in live cells and, additionally, fixation condenses DNA.
Further support for complex formation of SMC and ScpA/ScpB was obtained from depletion studies, which showed that ScpA and ScpB distribute throughout the cells after depletion of SMC. In addition, Scp foci did not form in the absence of either ScpA or ScpB, showing that directly or indirectly, all three proteins are required for complex formation. The fact that fluorescence within the cells apart from the Scp foci is very close to background staining suggests that almost all ScpA and ScpB molecules are present within the foci and thus, probably, within the complex.
At the onset of sporulation, the chromosome assumes a special structure called the axial filament, in which the origin regions move out all the way to the cell poles to ensure that an origin is captured within the forespore compartment after asymmetric division (Webb et al., 1997; Graumann and Losick, 2001). Likewise, Scp foci were found at the cell poles during axial filament formation, which shows that this segregation machinery also switches its position. However, it does not appear to be needed after polar septation, because SMC is degraded at this point (Graumann et al., 1998). Interestingly, ScpA and ScpB were not epistatic for sporulation, because an scpB deletion had almost no effect on sporulation, while that of an scpA deletion was much more severe. This shows that at least for differentiation, ScpA and ScpB have a different function.
Taken together, our data provide strong predictions for the function of the SMC–Scp complex in B.subtilis. The localization of ScpA/ScpB close to the quarter sites of the cells following the bipolar movement of origin regions supports the model in which four ‘SMC-condensation factories’ within both cell halves locally condense DNA that is thereby organized and pulled towards the cell poles (Supplementary figure 9). Possibly, the energy of DNA translocation by DNA polymerase is harnessed in driving origin regions towards the cell poles (Lemon and Grossman, 2001). If both replication forks are in close proximity at mid-cell, as suggested by the findings of Lemon and Grossman (1998), four DNA strands would be expelled, two each towards both poles. We have found that two Scp foci frequently flank each origin region after origin regions have separated towards the poles, and we suggest that each focus constitutes a cluster of SMC–Scp complexes that condenses DNA as it extrudes from DNA polymerase (Supplementary figure 9). Thereby, DNA regions would be folded within future daughter cells in an ordered fashion, which could achieve the preferred arrangement of chromosomes in B.subtilis and E.coli, in which chromosomes are folded roughly according to their physical structure (Teleman et al., 1998; Niki et al., 2000). These data are consistent with the findings that SMC is not required for the movement of origin regions towards the cell poles, but for complete separation and proper folding of chromosomes (Graumann, 2000). Scp proteins could assist SMC head domains that bind to single-stranded DNA in vitro (Hirano and Hirano, 1998; Lowe et al., 2001) in binding to double-stranded DNA. SMC has the potential to bind to positions along the chromosome separated by up to 100 nm and to bring them together in an ATP-dependent manner (Melby et al., 1998; Hopfner et al., 2000), which would introduce a right-handed super helix that has been observed in vitro (Kimura et al., 1999). Investigations of the purified SMC–Scp complex will shed further light on its pivotal role in prokaryotic chromosome segregation.
Materials and methods
Bacterial strains and media
Escherichia coli XL1-Blue (Stratagene) was grown in Luria–Bertani (LB) rich medium supplemented with 50 µg/ml ampicillin where appropriate. Bacillus strains were grown in LB rich medium or in DSM sporulation medium. For microscopy, cells were grown in S750 defined medium (Jaacks et al., 1989), which was complemented with 1% casamino acids for studies of fast-growing cells. For induction of the xylose promoter, glucose in S750 medium was exchanged for 0.5% fructose, and xylose was added up to 0.5%. Threonine (0.5%) was added to strains PG25 and PG27 for growth in S750 medium.
Construction of deletion strains
The ypu genes were deleted by a method involving the direct transformation of a PCR fragment carrying a resistance gene with the flanking sequences of the original gene locus. Downstream regions were amplified using primers P1 and P2, and upstream regions with P3 and P4. P2 and P3 contain an ∼30 base homology to the tetracycline (tet) resistance gene, and the flanking sequences are used in a second round PCR to create a large PCR fragment that is boosted with primers P1 and P4. The flanking regions of the gene to be disrupted were amplified from the chromosomal DNA using the primers P1 (5′-ACGTGGTAC CGCTCATTTTCATAATAGATCGG-3′; for ypuI and ypuH/scpB) or P1n (5′-GCACCGTCAATAATGATCGCGC-3′; for ypuG/scpA and ypuGH/scpAB) and P2i (5′-GAACAACCTGCACCATTGCAAGAC GTTTACTCTCCCTTTTTCAGGG-3′; for ypuI) or P2h (5′-GAA CAACCTGCACAATTGCAAGAGAAAACTTTAACCAAACCTTCG AAG-3′; for ypuH /scpB) or P2g (5′-GAACAACCTGCACCATT GCAAGAGCTGATGAAAAATCAGCTGGTCC-3′). Upstream regions were amplified using P3i (5′-TTGATCCTTTTTTTATAACAGG AATTCGCTTCTTTCATGATAGCTTCTTCC-3′), P3h (5′-ATCGCC GCGGGGCTTCTTTCGTTTATGCCG-3′) or P3g (5′-TTGATCC TTTTTTTATAACAGGAATTCGTATCAATTTTCACTTGATATTC TTC-3′) and P4 5′-TTGATCCTTTTTTTATAACAGGAATTCGC CCTTCATCGCCTGCCGC-3′). For the double scpAB deletion, primers P2h and P3g were used. The tet resistance gene was PCR amplified from plasmid pDG1515 (BGSC, Wisconsin). Transformation of the resulting PCR fragments and selection for tet (10 µg/ml) resulted in strains JM11 (scpA::tet), JM12 (scpA-B::tet), PG31 (ypuI::tet) and PG32 (scpB::tet).
Construction of plasmids
To create a C-terminal fusion of scpA with yfp for single-crossover integration into the chromosome, the 3′ (∼500 bp) region of the scpA gene was amplified by PCR using primers 5′-CGGAATTCCCG AAGCAAGAGGAGG-3′ and 5′-CCGCTCGAGAGCCCCATGAATG GATTCAC-3′ and was cloned into pKL183 (Lemon and Grossman, 2000), resulting in pyG. The pspac promoter was excised from pSG1170 (Feucht and Lewis, 2001) and was cloned into the EcoRI site of pyG to ensure transcription of downstream genes in the operon, giving plasmid pyGs. A C-terminal fusion of scpB with yfp was created by cloning of the 3′ (∼500 bp) region of the scpB gene (amplified by PCR using primers 5′-CGGAGGTACCCTTCTTTATGCGGCAGG-3′ and 5′-CCGGAATTC GGTTTTTATATCTTCGAAGGTTTGG-3′) into KpnI and EcoRI sites of pSG1187 (Feucht and Lewis, 2001), resulting in pyH. A version of cfp C-terminally tagged to scpB was obtained by cloning scpB amplified by PCR using primers 5′-CCGGAATTCGGTTTTTATATCTTCGAAG GTTTGG-3′ and 5′-TAGGGTACCGGGGCTTGATATCGTGAATTG-3′ into the KpnI and EcoRI sites of pSG1192 (Feucht and Lewis, 2001), establishing pcHamy.
Immunoprecipitation
Cultures of cells (8–20 ml; adjusted for an OD of 1 at 600 nm) growing at mid-exponential phase were harvested and resuspended in 1 ml of lysis buffer (0.1 M Tris–HCl pH 7.5, 0.05 M EDTA). Cells were treated with lysozyme (2 mg/ml) and were disrupted with ultrasound (Branson Sonifier). After centrifugation, 1 ml of NET2 buffer (50 mM Tris–HCl, 0.05% NP-40, 150 mM NaCl) containing 2.5 mg of protein A–Sepharose and ∼1 µg of monoclonal GFP antibodies (Clontech) were added to cell extracts. After incubation overnight at 4°C, protein A–Sepharose was pelleted and washed eight times with NET2 buffer. The final pellet was resuspended in 15 µl of TE and 5 µl of SDS–PAGE loading buffer, and 10 µl were loaded onto gels. As loading control, 1 ml of cell culture was resuspended in 75 µl of TE buffer and was treated with lysozyme. After addition of 25 µl of loading buffer, 8–15 µl (adjusted for OD = 1) were loaded onto an SDS–polyacrylamide gel for blotting with anti-GFP serum or with anti-SMC serum. For immunoprecipitation of SMC, 0.3 l of culture was grown at 23°C for each strain to OD600 = 1. RNase and DNase were added and cells were disrupted by sonication. Extracts were clarified by two 15 min spins at 20 000 g. A 2 ml aliquot was taken for immunoprecipitation with the affinity-purified antibodies covalently coupled to CNBr-beads (8 h, 4°C, batch mode). After extensive washes, proteins were eluted with 2% SDS at 70°C.
Image acquisition
Fluorescence microscopy was performed on an Olympus AX70 microscope. Cells were mounted on agarose pads containing S750 growth medium on object slides. Images were acquired with a digital CCD camera; signal intensities and cell length were measured using the Metamorph 4.0 program. CFP and YFP filter sets were acquired from Olympus. For FRET analysis, exciter and dichroic filters for CFP were combined with emitter for YFP. The long pass green filter excites GFP and YFP, and visualizes green and red fluorescence (filter WIBA, Olympus). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; final concentration 0.2 ng/ml) and membranes were stained with FM4-64 (final concentration 1 nM).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Table I. Strains.
| Strain | Genotype |
|---|---|
| JM8 | scpA-yfp |
| JM9 | scpB-yfp |
| JM10 | pxyl-scpB-cfp at amy locus |
| JM11 | scpA::tet |
| JM12 | scpA-B::tet |
| JM14 | scpA-yfp, pxyl-scpB-cfp at amy locus |
| JM15 | scpB-yfp (cm::tet) |
| JM16 | smc::kan, pspac-smc at amy locus (cm::tet) |
| JM17 | scpA-yfp, smc::kan, pspac-smc at amy locus (cm::tet) |
| JM18 | scpA::tet, pxyl-scpB-cfp at amy locus |
| JM19 | scpA::tet, smc::kan, pspac-smc at amy locus |
| PG25 | lacI-cfp at thr locus |
| PG26 | lacO-cassette at 359°, lacI-cfp at thr locus |
| PG27 | scpB-yfp, lacO-cassette at 359°, lacI-cfp at thr locus |
| PG28 | dnaX-cfp |
| PG29 | scpA-yfp, dnaX-cfp |
| PG30 | scpB-yfp, dnaX-cfp |
| PG31 | ypuI::tet |
| PG32 | scpB::tet |
| PG33 | scpB-yfp, smc::kan, pspac-smc at amy locus |
| PG34 | scpA-yfp, spo0J::spec |
| PG35 | scpB-yfp, spo0J::spec |
| PG36 | ypuI::tet, spoIIIE::spec |
| PG37 | scpA::tet, spoIIIE::spec |
| PG38 | scpB::tet, spoIIIE::spec |
| PG39 | scpB::tet, spo0J::spec |
| PG40 | scpB::tet, pxyl-scpB-cfp at amy locus |
| PG41 | scpB::tet, pxyl-scpB-cfp at amy locus, scpA-yfp |
| PG43 | scpB::tet, smc::kan, pspac-smc at amy locus |
| EP58 | smc::kan, pspac-smc at amy locus |
| PGΔ388 | smc::kan (Graumann et al., 1998) |
Acknowledgments
Acknowledgements
We are indebted to Jose Eduardo Gonzales-Pastor for the gift of unpublished plasmids and strains and for the communication of the long flanking sequence PCR knockout method. We also thank Rob Britton, Katherine Lemon, Janet Lindow and Alan Grossman for the kind provision of strains and plasmids. This work was supported by the Deutsche Forschungsgemeinschaft (Emmy Noether Program).
References
- Britton R.A. and Grossman,A.D. (1999) Synthetic lethal phenotypes caused by mutations affecting chromosome partitioning in Bacillus subtilis. J. Bacteriol., 181, 5860–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton R.A., Lin,D.C.-H. and Grossman,A.D. (1998) Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev., 12, 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J.C., Kirkham,L.A. and Leach,D.R. (1998) The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl Acad. Sci. USA, 95, 7969–7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feucht A. and Lewis,P.J. (2001) Improved plasmid vectors for the production of multiple fluorescent protein fusions in Bacillus subtilis. Gene, 264, 289–297. [DOI] [PubMed] [Google Scholar]
- Glaser P., Sharpe,M.E., Raether,B., Perego,M., Ohlsen,K. and Errington,J. (1997) Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev., 11, 1160–1168. [DOI] [PubMed] [Google Scholar]
- Graumann P.L. (2000) Bacillus subtilis SMC is required for proper arrangement of the chromosome and for efficient segregation of replication termini but not for bipolar movement of newly duplicated origin regions. J. Bacteriol., 182, 6463–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P.L. (2001) SMC proteins in bacteria: condensation motors for chromosome condensation? Biochimie, 83, 53–59. [DOI] [PubMed] [Google Scholar]
- Graumann P.L. and Losick,R. (2001) Coupling of asymmetric division to polar placement of replication origin regions in Bacillus subtilis. J. Bacteriol., 183, 4052–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P.L., Losick,R. and Strunnikov,A.V. (1998) Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J. Bacteriol., 180, 5749–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M. and Hirano,T. (1998) ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J., 17, 7139–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. (1999) SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev., 13, 11–19. [DOI] [PubMed] [Google Scholar]
- Hirano T., Kobayashi,R. and Hirano,M. (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell, 89, 511–521. [DOI] [PubMed] [Google Scholar]
- Hopfner K.P., Karcher,A., Shin,D.S., Craig,L., Arthur,L.M., Carney,J.P. and Tainer,J.A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell, 101, 789–800. [DOI] [PubMed] [Google Scholar]
- Hopfner K.P., Karcher,A., Craig,L., Woo,T.T., Carney,J.P. and Tainer,J.A. (2001) Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell, 105, 473–485. [DOI] [PubMed] [Google Scholar]
- Ireton K., Gunther,A.D. and Grossman,A.D. (1994) spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol., 176, 5320–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaacks K.J., Healy,J., Losick,R. and Grossman,A.D. (1989) Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J. Bacteriol., 171, 4121–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B. and Shapiro,L. (1999) The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl Acad. Sci. USA, 96, 10661–10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B., Wang,S.C. and Shapiro,L. (2001) A moving DNA replication factory in Caulobacter crescentus. EMBO J., 20, 4952–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Rybenkov,V.V., Crisona,N.J., Hirano,T. and Cozzarelli,N.R. (1999) 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell, 98, 239–248. [DOI] [PubMed] [Google Scholar]
- Koppes L.J., Woldringh,C.L. and Nanninga,N. (1999) Escherichia coli contains a DNA replication compartment in the cell center. Biochimie, 81, 803–810. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman,A.D. (1998) Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science, 282, 1516–1519. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman,A.D. (2000) Movement of replicating DNA through a stationary replisome. Mol. Cell, 6, 1321–1330. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman,A.D. (2001) The extrusion–capture model for chromosome partitioning in bacteria. Genes Dev., 15, 2031–2041. [DOI] [PubMed] [Google Scholar]
- Lewis P.J. and Errington,J. (1997) Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol. Microbiol., 25, 945–954. [DOI] [PubMed] [Google Scholar]
- Lin D.C.-H., Levin,P.A. and Grossman,A.D. (1997) Bipolar localization of a chromosome partition protein to Bacillus subtilis. Proc. Natl Acad. Sci. USA, 94, 4721–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Cordell,S.C. and van den Ent,F. (2001) Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J. Mol. Biol., 306, 25–35. [DOI] [PubMed] [Google Scholar]
- Melby T.E., Ciampaglio,C.N., Briscoe,G. and Erickson,H.P. (1998) The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol., 142, 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk,R. and Nasmyth,K. (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell, 91, 35–45. [DOI] [PubMed] [Google Scholar]
- Moriya S., Tsujikawa,E., Hassan,A.K., Asai,K., Kodama,T. and Ogasawara,N. (1998) A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol., 29, 179–187. [DOI] [PubMed] [Google Scholar]
- Niki H. and Hiraga,S. (1998) Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev., 12, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Jaffe,A., Imamura,R., Ogura,T. and Hiraga,S. (1991) The new gene mukB codes for a 177 kDa protein with coiled-coil domains involved in chromosome partitioning of E.coli. EMBO J., 10, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Yamaichi,Y. and Hiraga,S. (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev., 14, 212–223. [PMC free article] [PubMed] [Google Scholar]
- Ohsumi K., Yamazoe,M. and Hiraga,S. (2001) Different localization of SeqA-bound nascent DNA clusters and MukF–MukE–MukB complex in Escherichia coli cells. Mol. Microbiol., 40, 835–845. [DOI] [PubMed] [Google Scholar]
- Sawitzke J.A. and Austin,S. (2000) Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl Acad. Sci. USA, 97, 1671–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe M.E. and Errington,J. (1995) Postseptational chromosome partitioning in bacteria. Proc. Natl Acad. Sci. USA, 92, 8630–8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe M.E., Hauser,P.M., Sharpe,R.G. and Errington,J. (1998) Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J. Bacteriol., 180, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J. (2001) Prokaryotic structural maintenance of chromosomes (SMC) proteins: distribution, phylogeny and comparison with MukBs and additional prokaryotic and eukaryotic coiled-coil proteins. Gene, 278, 253–264. [DOI] [PubMed] [Google Scholar]
- Soppa J., Kobayashi,K., Noirot-Gros,M-F., Oesterheld,D., Ehrlich,S.D., Dervyn,E., Ogasawara,N. and Moriya,S. (2002) Discovery of two new families of proteins, which are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members Ypu and YpuH. Mol. Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- Strunnikov A.V. and Jessberger,R. (1999) Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur. J. Biochem., 263, 6–13. [DOI] [PubMed] [Google Scholar]
- Sunako Y., Onogi,T. and Hiraga,S. (2001) Sister chromosome cohesion of Escherichia coli. Mol. Microbiol., 42, 1233–1242. [DOI] [PubMed] [Google Scholar]
- Teleman A.A., Graumann,P.L., Lin,D.C.H., Grossman,A.D. and Losick,R. (1998) Chromosome arrangement within a bacterium. Curr. Biol., 8, 1102–1109. [DOI] [PubMed] [Google Scholar]
- Vagner V., Dervyn,E. and Ehrlich,S.D. (1998) A vector for systematic gene inactivation in Bacillus subtilis. Microbiology, 144, 3097–3104. [DOI] [PubMed] [Google Scholar]
- Webb C.D., Teleman,A., Gordon,S., Straight,A., Belmont,A., Lin,D.C.-H., Grossman,A.D., Wright,A. and Losick,R. (1997) Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B.subtilis. Cell, 88, 667–674. [DOI] [PubMed] [Google Scholar]
- Webb C.D., Graumann,P.L., Kahana,J., Teleman,A.A., Silver,P. and Losick,R. (1998) Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol., 28, 883–892. [DOI] [PubMed] [Google Scholar]
- Weitao T., Dasgupta,S. and Nordstrom,K. (2000) Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol. Microbiol., 38, 392–400. [DOI] [PubMed] [Google Scholar]
- Wu L.J. and Errington,J. (1994) Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science, 264, 572–575. [DOI] [PubMed] [Google Scholar]
- Yamazoe M., Onogi,T., Sunako,Y., Niki,H., Yamanaka,K., Ichimura,T. and Hiraga,S. (1999) Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J., 18, 5873–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]