Abstract
PcrA, Rep and UvrD are three closely related bacterial helicases with a DExx signature. PcrA is encoded by Gram-positive bacteria and is essential for cell growth. Rep and UvrD are encoded by Gram-negative bacteria, and mutants lacking both helicases are also not viable. To understand the non-viability of the helicase mutants, we characterized spontaneous extragenic suppressors of a Bacillus subtilis pcrA null mutation. Here we report that one of these suppressors maps in recF and that previously isolated mutations in B.subtilis recF, recL, recO and recR, which belong to the same complementation group, all suppress the lethality of a pcrA mutation. Similarly, recF, recO or recR mutations suppress the lethality of the Escherichia coli rep uvrD double mutant. We conclude that RecFOR proteins are toxic in cells devoid of PcrA in Gram-positive bacteria, or Rep and UvrD in Gram-negative bacteria, and propose that the RecFOR proteins interfere with an essential cellular process, possibly replication, when DExx helicases PcrA, or Rep and UvrD are absent.
Keywords: Bacillus subtilis/PcrA/RecF/Rep/UvrD
Introduction
DNA helicases play key roles in many cellular processes by promoting the unwinding of the DNA double helix. In bacteria, attempts at making helicase null mutant strains have shown that two such helicases are essential: DnaB and PcrA. Whereas the reason for the essential character of DnaB has been known for a long time, as it is required for the progression of replication forks, the essential character of the pcrA gene remains a mystery.
The determination of the genome sequence of many bacterial species allows us to draw the landscape of their equipment in DNA helicases. Each species encodes a hexameric essential helicase equivalent to DnaB of Escherichia coli. In addition, each bacterial species encodes a set of helicases of the DExx family that are monomeric or dimeric, and that fulfill several, sometimes overlapping, functions. For example, E.coli possesses seven such helicases, UvrD, Rep, HelD and RecB classified in superfamily 1, and PriA, RecG and RecQ classified in superfamily 2 (for the classification see Gorbalenya and Koonin, 1993; for a recent review see Singleton and Wigley, 2002). None of them is essential. Their roles range from DNA repair for UvrD (DNA mismatches and UV lesions) to homologous recombination for RecB and RecG, and replication restart for PriA. The Rep and UvrD helicases share a high percentage of conserved amino acids (40%), and may share a common function, as each single mutant is viable, but the double rep uvrD mutant is not viable. Bacillus subtilis has eight such helicases, most of which are considered orthologs of the E.coli enzymes. They have been named according to their E.coli ortholog, except, for historical reasons, the AddA protein, which is the ortholog of RecB, and the RecS and YocI proteins, which are two RecQ orthologs (Fernandez et al., 2000; Carrasco et al., 2001). There is one noticeable exception, PcrA, which is highly similar to both UvrD and Rep helicases of E.coli (∼40% identity), and is essential. The same is true for the PcrA helicase of Staphylococcus aureus (Iordanescu, 1993) and, by extension, might be a hallmark of Gram-positive bacteria.
PcrA was the first DNA helicase for which the three-dimensional structure was solved (Subramanya et al., 1996; Velankar et al., 1999), and a set of in vitro experiments has led to the proposal that PcrA unwinds DNA using an active, inchworm mechanism (Bird et al., 1998; Soultanas et al., 1999, 2000). In vivo, the pcrA gene is required for rolling circle replication (Iordanescu, 1993; Petit et al., 1998), as are the Rep and UvrD helicases of E.coli (Scott et al., 1977; Yarranton and Gefter, 1979; Bruand and Ehrlich, 2000). An indication that the PcrA, UvrD and Rep helicases have, at least in part, overlapping functions comes from heterologous complementation studies: the expression of the pcrA gene product in E.coli allowed restoration of the viability of the uvrD rep double mutant, and it also conferred UV resistance to a uvrD mutant (Petit et al., 1998). In a previous attempt to elucidate the essential role of PcrA, a conditional mutant of pcrA was constructed, in which the gene was placed under the control of a repressible promoter. Analysis of DNA synthesis, by labeled thymidine incorporation into chromosomal DNA, of a strain depleted in PcrA revealed a modest but significant defect (Petit et al., 1998). This defect was weak compared with the profound defect observed in a strain depleted in DnaC, the replicative helicase of B.subtilis. These observations seemed to exclude a role for PcrA comparable with that of DnaC in the progression of the replication fork.
Homologous recombination in bacteria relies on the key role of RecA, a protein promoting the homology search between DNA molecules and the strand exchange reaction (for a review see Kuzminov, 1999). To facilitate the binding of RecA to the DNA in the first place, two main routes have been characterized in E.coli, one relying on RecBCD and the other on RecFOR proteins. The RecBCD complex enters DNA from a double-strand break and stimulates the further binding of RecA, whereas the RecFOR proteins are proposed to mediate the binding of RecA onto gapped DNA. The last stage of the recombination process, which allows the resolution of Holliday junction intermediates, depends on the RuvABC proteins or the RecG helicase. The same protein components are present in B.subtilis, with identical names except for the AddAB complex, which fulfills the role of RecBCD. Interestingly, B.subtilis possesses three additional routes promoting RecA-dependent recombination, apart from the AddAB and RecFOR pathways, which are less well characterized at present (Fernandez et al., 2000).
To gain further insights into the in vivo activities of PcrA, we isolated spontaneous mutations that render the pcrA1 strain viable. We found that mutations are extragenic suppressors of the pcrA1 mutation, and one of them was mapped in the recF gene. Mutations in recF, recL, recO and recR, which all belong to the same complementation group, were found to suppress pcrA1 lethality as well, albeit with a reduced efficiency compared with the spontaneous suppressor. In contrast, recA1 and addA5 mutations did not suppress pcrA, which suggests that only the initial step of homologous recombination mediated by RecFOR was toxic in helicase-deficient cells. The lethality of pcrA mutations appeared to be correlated with a recombination problem, as strains in which the amount of PcrA was reduced by a factor of 10 were hyper-recombinogenic, and this recombination was RecA and RecFOR dependent. In addition, we show that in E.coli, the presence of a recF, recO or recR mutation restores the viability of the rep uvrD mutant. Models are presented to account for these observations, in which the role of the DExx helicases PcrA, or Rep and UvrD, would be either to reverse blocked recombination intermediates or to facilitate replication by counteracting RecFOR.
Results
The PcrA helicase is abundant in B.subtilis cells
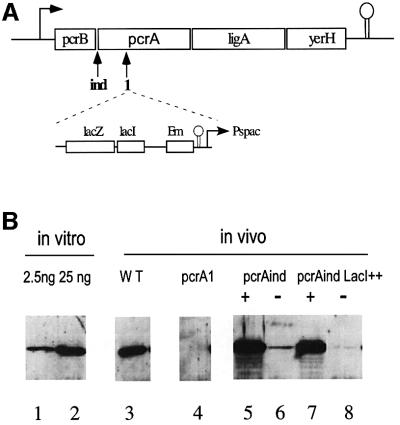
Two pcrA mutants were used in this study. One, pcrAind, corresponds to a transcriptional fusion of the pcrA gene to a LacI-repressible, isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible, Pspac promoter (Petit et al., 1998; Figure 1A). The other, pcrA1, is a pcrA gene disruption (Figure 1A; this strain is viable only in the presence of a suppressor mutation, see below). The amount of PcrA helicase present in wild-type B.subtilis cells, as well as in the pcrA mutants, was determined by western blot (Figure 1B). A strong signal, corresponding to ∼8000 molecules per cell, was obtained for the wild-type B.subtilis strain 168 (Figure 1B, lane 3). In the pcrAind strain, levels of PcrA ranged from 21 000 molecules per cell upon full derepression of the Pspac promoter (IPTG concentration >100 µM), down to 600 molecules upon repression by LacI (no IPTG; Figure 1B, lanes 5 and 6). This confirmed a previous observation that leaky transcription from the Pspac promoter allows the viability of the pcrAind strain in the absence of IPTG. Cell death was observed, however, when a plasmid overproducing LacI (pMAP65) was present in the strain. In this strain, 200 molecules of PcrA per cell were detected upon growth without IPTG for 12 generations (Figure 1B, lanes 7 and 8). This amount of PcrA is probably still the result of leaky transcription, since the simple dilution of 20 000 molecules over 12 generations should have resulted in some four molecules per cell. Finally, in a suppressed strain where the pcrA gene is disrupted (see below, strain MAS613), no PcrA signal was detected, as expected (Figure 1B, lane 4). From these data, we conclude that PcrA is abundant in B.subtilis, as is UvrD in E.coli (3000–5000 molecules per cell for the constitutive level, increasing 2.5-fold upon SOS induction; Arthur and Eastlake, 1983; Siegel, 1983). In contrast to UvrD, however, no SOS box was present at the pcrA promoter, and no induction of PcrA was observed upon UV irradiation of B.subtilis cells (not shown).
Fig. 1. (A) The operon encoding pcrA in B.subtilis. The two mutants used in this study carry the same cassette (shown below the operon), either in front of the pcrA ORF, resulting in a pcrA-inducible gene (ind mutation), or in the middle of the gene in the case of the pcrA1 null strain (mutation marked 1). (B) Western blot detection of PcrA in extracts of various B.subtilis strains. Lane 3, strain 168; lane 4, strain MAS613; lanes 5 and 6, strain HVS604; lanes 7 and 8, strain HVS604 with plasmid pMAP65. Cells were cultivated with 1 mM (lanes 5 and 7) or 0 mM (lanes 6 and 8) IPTG. Lanes 1 and 2 contained 2.5 and 25 ng, respectively, of purified PcrA.
The lethality of the pcrA1 disruption is suppressed by a mutation in recF
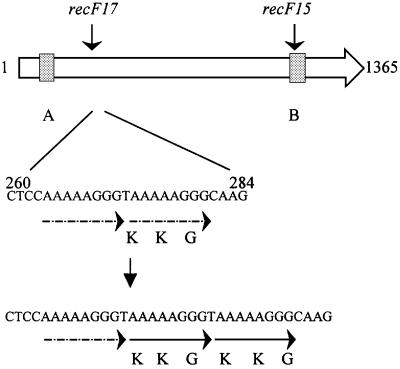
To investigate the causes of the lethality of a pcrA1 disruption, we analyzed spontaneous extragenic suppressors. For this purpose, a non-replicative plasmid containing an erythromycin resistance gene and an internal fragment of the pcrA gene, pMAP56 (Petit et al., 1998), was used to transform the wild-type B.subtilis strain, and transformants were selected on a synthetic medium containing erythromycin. Ghost colonies were obtained, unable to grow upon restreaking. Their presence was interpreted to mean that a sufficient amount of PcrA was present at the time of gene disruption to allow residual growth, but that viability was lost due to pcrA disruption. After a 48 h incubation at 37°C, fast growing papillae appeared at the surface of the ghost colonies. The papillae could be propagated and purified, and, in one strain, MAS613, the mutation was localized using a transposon mapping strategy (see Materials and methods). The mutation was found in the recF gene and was named recF17 (Figure 2). It consisted of a 9 bp addition at nucleotide 272 in the coding sequence, resulting in a KKG addition at the protein level. The wild-type sequence of recF harbors at this position an eight nucleotide duplication, which was probably expanded to generate this original and spontaneous mutation (Figure 2). The recF17 mutation was sufficient to confer the suppressor phenotype, because the pcrA gene was disrupted efficiently in the single recF17 mutant strain (see Materials and methods for the reconstruction strategy). Four other suppressor mutations were mapped to the region of the recF gene, but not studied further.
Fig. 2. The recF17 mutation. The recF ORF of B.subtilis is shown as an arrow. Gray squares indicate the ATP-binding motifs A and B. Below is shown an enlargement of the sequence of the wild-type and the recF17 mutation between nucleotides 260 and 284 after the start codon. The position of the recF15 mutation is indicated on top of the gene.
Mutations in recF, recL, recO and recR are suppressors of the pcrA1 lethality
The RecF protein, together with RecO and RecR, assists RecA binding to single-stranded DNA. In B.subtilis, a fourth gene, called recL, belongs to the recFOR complementation group, but its role and localization are unknown at present (Alonso et al., 1988). We tested whether the previously characterized mutations recF15, recL16, recR1 and recO1 also suppress the lethality of pcrA1. Mutations in recO1 and recR1 are gene disruptions, and recF15 maps in the Walker box B of RecF (Figure 2). These mutations can therefore be considered as null alleles. Competent cells of each rec mutant were transformed for pcrA1, using plasmid pMAP56 or chromosomal DNA of strain MAS613 as a donor. In all cases, pcrA1 rec double mutants were readily obtained, showing that recF, L, O and R mutations are all suppressors of pcrA1 lethality. The viability of the various pcrA1 suppressor strains was compared using three criteria: the generation time in synthetic medium (LM medium; see Materials and methods), the proportion of cells of a LM liquid culture able to form a colony on an LM plate (Table I, column c.f.u. per OD) and the sensitivity to rich medium. Results are presented in Table I. Interestingly, the viability of the pcrA1 recF17 strain was better than that of all other suppressed strains, including the pcrA1 recF15 strain. This suggests that recF17 is not a null allele, but rather a partial loss-of-function allele or even a gain of function (further evidence below suggests that recF17 is a partial loss-of-function allele).
Table I. Comparison of the viability of the pcrA1 recF, O or R derivatives.
| Genotype | Generation time (min) | c.f.u. per ODa (%) | Plating efficiency on rich mediumb (%) |
|---|---|---|---|
| Wild-type | 34 | 100 | 100 |
| recF17 | 40 | 100 | 67 |
| recF15 | 51 | 53 | 98 |
| recO1 | 50 | 46 | 91 |
| recR1 | 45 | 100 | 73 |
| pcrA1 recF17 | 46 | 63 | 55 |
| pcrA1 recF15 | 77 | 4 | 80 |
| pcrA1 recO1 | 78 | 4 | 63 |
| pcrA1 recR1 | 121 | 1 | 50 |
aNumber of colony-forming units (c.f.u.) per ml divided by the optical density of the culture, taking the wild-type strain as a reference (100% represents 1.5 × 108 c.f.u./ml in one unit of OD at 600 nm).
bRatio of the c.f.u. on LB and LM medium.
In conclusion, a mutation in any of the four genes belonging to the recF complementation group suppresses the lethality of the pcrA1 gene disruption in B.subtilis. This suggests that it is the combined activity of these four RecFLOR proteins that is toxic in a strain lacking PcrA.
Effects of mutations in addA and recA in strains defective for PcrA
Aside from RecFLOR, additional routes promoting RecA-dependent recombination are present in B.subtilis, among which the AddAB pathway is the more active (Alonso et al., 1988). We investigated whether an addA5 mutation suppresses pcrA1 lethality. Integration of genetic markers by homologous recombination is possible in a B.subtilis addA5 strain, due to the activity of the RecF pathway, and possibly three other pathways (Fernandez et al., 2000). No addA5 pcrA1 double mutant could be constructed, suggesting that addA5 is not a suppressor of pcrA1 lethality.
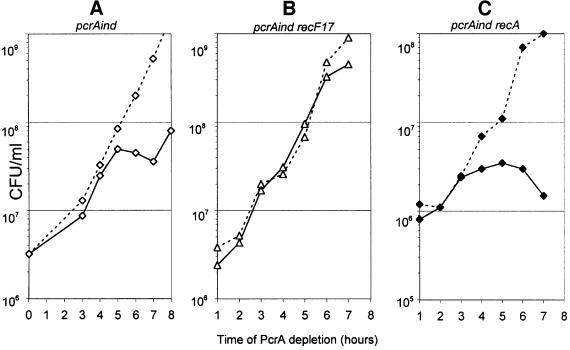
Finally, we tested whether the complete absence of homologous recombination in B.subtilis, mediated by a recA1 mutation, would permit the growth of a pcrA null strain. This could not be tested as above for the addA5 mutation, because the introduction of genetic markers into the chromosome by homologous recombination is prevented in a recA1 mutant. A different test was thus carried out, making use of the pcrAind strain. This strain stopped growing after 4 h without IPTG (Figure 3A), whereas the strain also carrying the recF17 mutation (MAS629; Table IV) grew to saturation (Figure 3B). The recA1 mutation did not allow growth of the pcrAind cells in the absence of IPTG (cells stopped growing after 3 h; Figure 3C).
Fig. 3. Inactivation of recA does not allow growth of B.subtilis cells depleted of PcrA. Cell growth of pcrAind derivatives in the absence (dark lines) or presence of 1 mM IPTG (dotted lines) was followed by plating appropriate dilutions.
Table IV. Bacillus subtilis strains.
| Strain | Genotype | Reference/source |
|---|---|---|
| 168 | trpC2 | C.Anagnostopoulos |
| HVS604 | trpC2 pcrAind | Petit et al. (1998) |
| MAS613 | trpC2 pcrA1::EmR recF17 | This work |
| MAS614 | trpC2 pcrA1::EmR recF17 rrnO::Tn10 (CmR) | This work |
| HVS16 | trpC threo5 recF15 | Laboratory collection |
| MAS305 | trpC threo5 recF17 | This work |
| BG128 | trpC2 metB5 amyE sigB xin-1 attSPβ recR1::CmR | Alonso et al. (1990) |
| BG107 | trpC2 metB5 amyE sigB xin-1 attSPβ recL16 | Alonso et al. (1987) |
| BG439 | trpC2 metB5 amyE sigB xin-1 attSPβ recO1::CmR | Fernandez et al. (1999) |
| MAS617 | trpC2 recR1::CmR | This work |
| MAS618 | trpC2 recO1::CmR | This work |
| MAS619 | trpC2 recR1::CmR pcrA1::EmR | This work |
| MAS620 | trpC2 recO1::CmR pcrA1::EmR | This work |
| MAS621 | trpC2 metB5 amyE sigB xin-1 attSPβ recL16 pcrA1::EmR | This work |
| GSY2258 | hisH2 metB5 addA5 | C.Anagnostopoulos |
| MAS622 | trpC2 pcrAind amyE::ligA (CmR) | This work |
| HVS567 | leuA8 metB5 recA::tet | Chedin et al. (1994) |
| MAS626 | trpC2 pcrAind amyE::ligA (PhleoR) | This work |
| MAS627 | trpC2 pcrAind amyE::ligA (PhleoR) pMAP65 | This work |
| MAS628 | trpC2 pcrAind amyE::ligA (PhleoR) pMAP65 recA::tet | This work |
| MAS629 | trpC2 pcrAind amyE::ligA (PhleoR) pMAP65 recF17 rrnO::Tn10 (CmR) | This work |
| CBB383 | trpC2 ΔilvA Kndup513 | Bruand et al. (2001) |
| MAS630 | trpC2 ΔilvA Kndup513 recF17 | This work |
| CBB529 | trpC2 ΔilvA Kndup513 recA::tet | C.Bruand |
| MAS631 | trpC2 thr5 recF15 rrnO::Tn10 (CmR) | This work |
| MAS632 | trpC2 ΔilvA Kndup513 pcrA1::EmR recF17 | This work |
| MAS633 | trpC2 ΔilvA Kndup513 pcrA1::EmR recF17 recA::tet | This work |
| MAS634 | trpC2 ΔilvA Kndup513 pcrAind amyE::ligA (CmR) | This work |
| MAS635 | trpC2 ΔilvA Kndup513 pcrAind amyE::ligA (CmR) recA::tet | This work |
| MAS636 | trpC2 ΔilvA Kndup513 pcrAind amyE::ligA (PhleoR) | This work |
| MAS637 | trpC2 ΔilvA Kndup513 pcrAind amyE::ligA (PhleoR)recF17 rrnO::Tn10 | This work |
| MAS638 | trpC2 ΔilvA Kndup513 pcrAind amyE::ligA (PhleoR) recF15 rrnO::Tn10 | This work |
| MAS639 | trpC2 ΔilvA Kndup513 pcrA1::EmR recF15 | This work |
| MAS640 | trpC2 ΔilvA Kndup513 pcrA1::EmR recF17 recR1::CmR | This work |
| TF8A | trpC2 attSPβΔPBSX ΔSkin | K.Devine |
| MAS641 | trpC2 attSPβΔPBSX ΔSkin amyE::dinR-lacZ (SpecR) | This work |
| MAS642 | trpC2 attSPβΔPBSX ΔSkin amyE::dinR-lacZ (SpecR) recF17 rrnO::Tn10 | This work |
| MAS643 | trpC2 attSPβΔPBSX ΔSkin amyE::dinR-lacZ (SpecR) recF15 rrnO::Tn10 | This work |
| MAS644 | hisH2 metB5 addA5 yhjP::EmR | This work |
| MAS645 | trpC2 attSPβΔPBSX ΔSkin amyE::dinR-lacZ (SpecR) addA5 yhjP::EmR | This work |
| MAS646 | trpC2 attSPβΔPBSX ΔSkin amyE::dinR-lacZ (SpecR) addA5 yhjP::EmR recF17 rrnO::Tn10 | This work |
| MAS647 | trpC2 attSPβΔPBSX ΔSkin amyE::dinR-lacZ (SpecR) addA5 yhjP::EmR recF15 rrnO::Tn10 | This work |
We conclude that the viability of a pcrA null strain cannot be restored by abolishing homologous recombination via addA or recA mutations, in contrast to recF, L, O or R mutations.
Not all defects of a pcrA null strain are corrected by the recF17 mutation
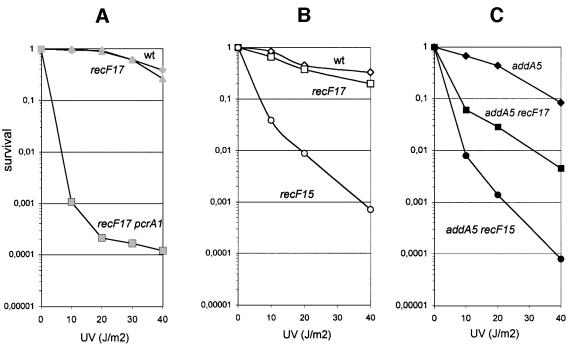
Two functions were reported previously for PcrA: it is required for the replication of rolling circle plasmids and it fully complements the UV sensitivity of a uvrD mutant of E.coli. We found that (i) a derivative of plasmid pC194, which replicates by a rolling circle mechanism, could not be established in the pcrA1 recF17 strain; and (ii) the pcrA1 recF17 strain was UV sensitive whereas the recF17 mutant was not (Figure 4A). Therefore, the presence of the recF17 mutation did not restore replication of a rolling circle plasmid or efficient UV repair in the absence of PcrA.
Fig. 4. UV sensitivity of various B.subtilis strains. The proportions of viable cells, relative to the unirradiated control, are shown as a function of UV dose (J/m2). The relevant genotype of each strain is reported on each curve.
The pcrA1 recF17 strain is hyper-recombinogenic
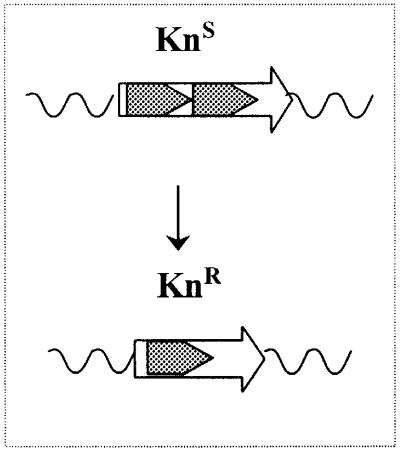
The fact that pcrA1 suppressor mutations map in genes involved in homologous recombination prompted us to examine recombination in the pcrA strains. Because recombination between tandem repeats had already been reported to increase in an E.coli rep mutant (Bierne et al., 1997b) and also in a uvrD mutant (Bierne et al., 1997a), a similar assay was used in B.subtilis. It consisted of measuring the frequency of recombination between two tandemly repeated sequences of 513 bp (Bruand et al., 2001). The repeats inactivate the kanamycin resistance (KnR) gene and, when one is deleted, the intact gene is restored (see Table III, scheme1). The cell thus becomes resistant to the antibiotic, and the recombination frequency can be estimated by measuring the proportion of KnR cells in independent cultures.
In a wild-type strain, the recombination frequency was 2 × 10–4. A defect in recA had no significant effect on the yield of recombinants (a 2-fold decrease; Table III). Such a modest role for recA in tandem repeat recombination has already been observed by others; it is due to the fact that replication slippage between the two repeats, which is recA independent, occurs at the same frequency of 10–4. RecA-dependent events become detectable only in strains where recombination is enhanced to frequencies >10–3 (Lovett et al., 1993; Bruand et al., 2001). The single recF17 mutation, similarly, did not modify the recombination frequency. In contrast, in the pcrAind strain, MAS634 propagated without IPTG (the Pspac promoter is leaky and cells contain ∼600 molecules of PcrA per cell under these conditions, see above) recombination was 15 times higher than in the wild-type strain (Table II). This recombination depended on RecA, as the recombination frequency decreased to the wild-type level in the recA1 derivative of the pcrAind strain. It also depended on RecF, as, in the recF17 or recF15 derivatives of strain MAS634, recombination frequency was at the wild-type level (Table II).
Table II. pcrA-inducible strains.
| Strain | Relevant genotype | Recombinant proportion (× 104) at 0 mM IPTG | Relative value | Recombinant proportion (× 104) at 1 mM IPTG | Relative value |
|---|---|---|---|---|---|
| MAS634 | pcrAind | 31.0 (± 11) | 15.5 | 4.7 (± 0.9) | 2.3 |
| MAS635 | pcrAind recA | 1.1 (± 0.6) | 0.5 | 4.3 (± 1.9) | 2.1 |
| MAS637 | pcrAind recF17 | 3.2 (± 1.3) | 1.6 | 4.0 (± 1.4) | 2 |
| MAS638 | pcrAind recF15 | 1.8 (± 0.9) | 0.9 | 3.2 (± 1.2) | 1.6 |
Recombination was studied next in pcrA1 derivatives. Due to its inviability, the single pcrA1 mutant could not be tested. In the pcrA1 recF17 strain MAS632, recombination was increased by a factor of 8.5, as compared with the wild-type strain (Table III). In contrast, no increase in recombination was observed in the pcrA1 recF15 strain. This difference between the recF17 and recF15 alleles suggests that the recF17 mutation only partially abolishes the recF function. Finally, inactivation of recA or recR in the pcrA1 recF17 strain (strains MAS633 and MAS640; Table III) restored a recombination frequency similar to the wild-type level. Altogether, these results show that recombination is increased in both the pcrAind and pcrA1 recF17 strains, and that it is dependent on RecA and on the RecFOR pathway.
Table III. pcrA-deleted strains and controls.
| Strain | Relevant genotype | Recombinant proportion (× 104) | Relative value |
|---|---|---|---|
| CBB383 | Wild-type | 2.0 (± 0.7) | 1 |
| MAS630 | recF17 | 3.2 (± 2.5) | 1.6 |
| CBB529 | recA1 | 1.1 (± 0.3) | 0.5 |
| MAS632 | pcrA1 recF17 | 16.7 (± 14.8) | 8.5 |
| MAS633 | pcrA1 recF17 recA1 | 4.7 (± 2.1) | 2.3 |
| MAS639 | pcrA1 recF15 | 3.2 (± 2.5) | 1.6 |
| MAS640 | pcrA1 recF17 recR1 | 1.5 (± 1.5) | 0.75 |
The recF17 mutation only partially inactivates the gene
Other than recombination, two phenotypes are associated with recF mutations, the sensitivity to UV and a defect in SOS induction. The recF17 allele was tested for these two phenotypes, and compared with the recF15 allele. The recF15 mutant was highly UV sensitive, whereas recF17 was not (Figure 4B). However, in the addA5 background where recombination relies mostly on RecFOR, the recF17 mutation conferred sensitivity to UV, although to a lesser extent than the recF15 mutation (Figure 4C). Similar to the UV results, the recF17 mutant was less sensitive to methylmethane sulfonate (MMS) than recF15 in an addA5 background (not shown). Altogether, these results indicate a partial loss of activity for the recF17 allele. The SOS response was next measured, using a dinR 2::lacZ fusion (Haijema et al., 1996). SOS induction upon UV irradiation was abolished by the recF15 mutation, as expected (Alonso and Stiege, 1991), whereas normal SOS induction was observed with the recF17 strain (data not shown).
In E.coli, inactivation of recF, recO or recR restores the viability of the uvrD rep double mutant
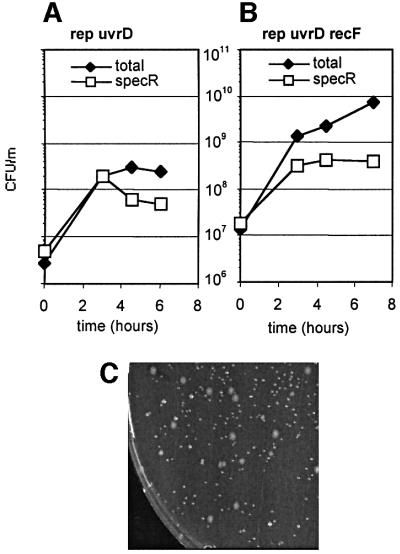
In order to generalize the observations made in B.subtilis, we investigated whether the lethality of the rep uvrD double mutant of E.coli is suppressed by mutations in recF, recO or recR. For this purpose, a plasmid shuffling experiment was carried out, using a thermosensitive pGB2ts plasmid derivative encoding the rep gene. This plasmid was introduced into the single rep::Cm strain, as well as the double mutant rep rec strains. Next, the uvrD::Tn5 mutation was introduced into all four strains by transduction. Cultures of the resulting strains were propagated in synthetic medium for 6 h at 42°C, a temperature at which the plasmid does not replicate. The total number of colony-forming units (c.f.u.) at 37°C and the number of SpecR colonies that still contain the Rep+ plasmid were determined as a function of time. Strains that tolerate the rep uvrD double mutation should grow in the absence of the plasmid, whereas those that do not should die after the loss of the plasmid. Results are reported in Figure 5.
Fig. 5. The E.coli rep uvrD recF, rep uvrD recO and rep uvrD recR strains are viable. Strains MAC556, MAC535, MAC569 and MAC574 were propagated for 6 h at 42°C so as to chase the thermosensitive pGB2TSrep plasmid, with a 100-fold dilution at 3 h. The number of total c.f.u. per milliliter, and the number of SpecR c.f.u. are reported as a function of time (multiplied by 100 after dilution). A representative experiment for the rep uvrD [pGB2TSrep] strain is shown in (A), and for the strain rep uvrD recF [pGB2TSrep] in (B). Plasmidless clones are smaller than plasmid-containing clones, as shown in (C) for the rep uvrD recR strain.
When the rep uvrD control strain was propagated at 42°C, the number of c.f.u. reached a plateau after 3 h of growth (Figure 5A). In contrast, the number of c.f.u. continued to increase after 3 h of growth at 42°C in the rep uvrD recF (Figure5B), rep uvrD recO and rep uvrD recR cultures (not shown, similar curves). At the 6 h time point, two kinds of colonies were found on plates (shown in Figure 5C for the rep uvrD recR strain): a majority (90%) of plasmidless small colonies and a minority of large colonies in which the Rep+ plasmid was still present. The small colonies could be propagated and were found to be rep uvrD recF, O or R triple mutants. Their viability was affected, as indicated by an increased doubling time in synthetic medium (65 min instead of 50 min for both parents, the rep recF and uvrD recF double mutants) and a plating efficiency defect (10% of the efficiency of uvrD recF or rep recF parents). However, uvrD rep recF (recO or recR) clones were not more sensitive to UV than the uvrD recF double mutant (not shown). The triple mutant strain rep uvrD recF was also constructed successfully by P1 transduction of the rep mutation in a recF uvrD strain (data not shown).
We conclude that in E.coli, as in B.subtilis, the lethality due to the absence of the two helicases Rep and UvrD, which are close orthologs of PcrA, can be overcome by eliminating the RecFOR activity. As in B.subtilis, the viability of the rep uvrD strain was not restored by a recA mutation (data not shown).
Discussion
In order to understand the causes of the lethality of a pcrA null mutant in B.subtilis, the characterization of extragenic suppressors was undertaken. We found that mutations in recF, recL, recO or recR allow the survival of a pcrA null mutant in B.subtilis. Furthermore, we report that in E.coli, mutations in recF, recO or recR allow the survival of a uvrD rep double mutant. This points to a common and essential role for the three closely related helicases PcrA, Rep and UvrD, which can be dispensed with in strains mutated for RecFOR activity. For the other essential bacterial helicase, the replicative helicase DnaB (DnaC in B.subtilis), no extragenic suppressor mutation has been reported. The lethality of the pcrA null mutant, rather than being the direct consequence of the absence of an essential function, appears to be indirect, as it can be overcome by inactivating another process, RecFOR-dependent recombination. The B.subtilis pcrA single mutant and the E.coli rep uvrD double mutant will be designated collectively as ‘helicase mutant’ strains below.
What function of RecFOR is toxic in helicase mutant strains?
Three roles, of varying importance, have been proposed for the RecFOR complex in bacteria. The main and best documented role for RecFOR is its participation in homologous recombination. In E.coli, where the proteins have been studied in more details, they are proposed to act by facilitating the formation of the RecA filament on gapped DNA (Hegde et al., 1996; Webb et al., 1997, 1999), but also more recently on 5′ ends of single-stranded DNA (Bork et al., 2001). The fact that helicase-deficient cells are rescued by inactivation of RecFOR, which reduces the level of homologous recombination, in both E.coli and B.subtilis, raises the possibility that the origin of the lethality in these helicase mutants is an excess of recombination. Indeed, the frequency of recombination in helicase mutants appears inordinately high. Bierne et al. have shown that recombination between tandem repeats increases by a factor of 10 in a uvrD mutant (Bierne et al., 1997a), and by a factor of 20 in a rep mutant of E.coli (Bierne et al., 1997b). Similarly, in B.subtilis cells where the concentration of the PcrA helicase is decreased 30-fold, recombination between tandem repeats increases 15-fold over the wild-type cells. This suggests that the absence of the PcrA, or UvrD and Rep, helicases leads to intense RecFOR-dependent recombination. The putative toxicity of recombination is unexpected, as recombination in bacteria is viewed mostly as a beneficial process, allowing the repair of broken chromosomes and arrested replication forks, and more largely promoting horizontal genetic exchanges (for a review see Michel, 2000). Nevertheless, there are precedents, as in E.coli, where the lethality of the triple rep ruv dif mutant was also suppressed by a mutation in recF or in recA (Michel et al., 2000). However, in the case of helicase mutants, the recA mutation does not restore viability, and the partial loss-of-function allele, recF17, promotes a better viability of the pcrA1 mutant than the recF15 allele (see Table I). This raises two non-exclusive possibilities: either a minimum level of homologous recombination is beneficial in the helicase mutants, or the accumulation of RecFOR–DNA complexes is toxic per se, regardless of its capacity to recombine, a hypothesis elaborated in the last part of the Discussion.
The RecF protein has also been found to play a role in the SOS response in E.coli and in B.subtilis. In its absence, the induction of SOS is delayed (Alonso and Stiege, 1991; Hegde et al., 1995; Whitby and Lloyd, 1995). We considered the possibility that recF, O or R mutations suppressed pcrA1 lethality by preventing SOS induction, indicative of a putative SOS toxicity. Two sets of data suggest that this is not the case. First, we found that a recA mutation that prevents SOS induction did not suppress the lethality of pcrA1. Secondly, we found that the recF17 allele, which does not affect the SOS response, restores viability of the pcrA1 strain better than the recF15 allele, which prevents SOS induction. Therefore, SOS induction cannot be the toxic process prevented in the pcrA1 recF17 strain.
Finally, there has been evidence pointing to a putative role for RecF in replication restart (Sandler, 1996; Courcelle et al., 1997). However, in the experiments reported by Courcelle et al. (1997), cells were treated with UV, whereas they were not in our study. Our observations are also different from those reported by Sandler (1996), because here the three mutations in recF, recO and recR all suppress the lethality of pcrA and of rep uvrD double mutants, whereas only recF but not recO nor recR is synthetic lethal with priA.
Are PcrA, Rep and UvrD involved in the editing of recombination intermediates?
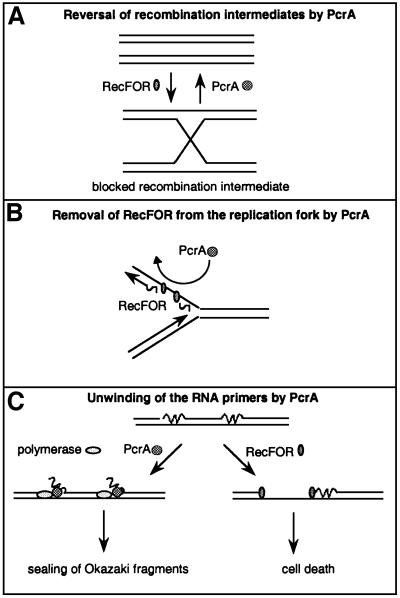
How would helicases prevent the toxicity of RecFOR? They may participate directly in the recombination process (Figure 6A). For instance, if recombination intermediates created via RecFLOR were unable to be processed to viable products, and thereby generate toxicity, the role of the helicases could be to reverse them to the un-recombined parental chromosome. Evidence for such an activity has been obtained in vitro for the UvrD helicase (Morel et al., 1993). Also, this is a current model proposed to account for observations in Saccharomyces cerevisiae with the Srs2 helicase, which is the closest ortholog to PcrA in this organism. The SRS2 gene is not essential in S.cerevisiae, and the combination of an srs2 deletion with mutations in genes involved in homologous recombination has contrasting effects, suggesting that the absence of Srs2 is better tolerated in a strain where no homologous recombination is taking place (Aboussekhra et al., 1992; Chanet et al., 1996). The Srs2 helicase is proposed to act as an editing helicase, intervening and removing recombination intermediates that are not processed correctly. A major difference between the two helicases is that PcrA is essential in B.subtilis, whereas Srs2 is not in yeast. However, the combination of the srs2 mutation with a mutation in SGS1, a gene encoding another helicase that is similar to the bacterial RecQ helicase, confers a severe growth defect. Interestingly, the viability of the double mutant srs2 sgs1 is greatly increased when homologous recombination is prevented, suggesting that some unprocessed recombination intermediates are toxic in yeast (Gangloff et al., 2000). However, the lethality of the single pcrA mutation (or rep uvrD double mutation) is probably not similar to the one observed for the srs2 sgs1 double mutation. To make a parallel, we would have to suppose that the PcrA helicase also has a RecQ-like activity, but B.subtilis already encodes two other recQ-like genes, recS and yocI. Furthermore, E.coli mutants in which recombination intermediates are expected to accumulate (ruv recG double mutant) are viable, which suggests that in bacteria at least, such an accumulation is not toxic.
Fig. 6. Three models illustrating the putative essential roles of the PcrA, Rep and UvrD helicases in the cell. (A) The helicases remove unprocessed recombination intermediates generated by RecFOR. This model makes no assumption about the DNA substrates used by RecFOR, which are therefore drawn as double-stranded molecules. (B) The helicases remove RecFOR bound to the lagging strand of the replication fork. (C) Left route: the helicases displace the RNA–DNA hybrid (RNA is shown as a wavy lane, DNA as a straight lane) and allow gap filling by DNA polymerase I. Right route: in the absence of helicases, gaps are created, and RecFOR enters into competition with the proper maturation of Okazaki fragments, which leads to cell death.
Do PcrA, Rep and UvrD compete with RecFOR during replication?
Rather than being toxic by promoting recombination, RecFOR may interfere with another essential process. Recent advances in the field of recombination have shown a close interplay of recombination and DNA replication (Michel et al., 2001 and references therein). Furthermore, we have shown that replication is slowed down in pcrA mutant cells (Petit et al., 1998). We therefore suggest that RecFOR might interfere with replication when the helicases are not present. Two possible entry points for RecFOR may be proposed (Figure 6B and C). RecFOR may bind to the single-stranded DNA on the lagging strand template (Figure 6B), which is generated during synthesis of the leading strand, and the function of the PcrA, UvrD or Rep helicases could be to remove the RecFOR–DNA complex and thus clear the way for the DNA polymerase on the lagging strand. Indeed, an in vitro study has shown the capacity of the Rep and UvrD helicases to displace the LacI–DNA complex (Yancey-Wrona and Matson, 1992). Such helicases appear to be at the replication fork, as it was reported that UvrD co-purifies with the DNA polymer ase III holoenzyme (Lahue et al., 1989). The antagonist function of RecFOR and the helicases would explain why the double mutant, pcrA recF, recovers viability.
An alternative is that due to the absence of the PcrA, UvrD or Rep helicases, the DNA substrate for RecFOR accumulates (Figure 6C). For instance, the function of the PcrA, Rep or UvrD helicases might be to facilitate the maturation of Okazaki fragments (OFs), by unwinding the RNA–DNA hybrid, a possibility supported by the unnoticed observation that the UvrD helicase is 10 times more active at unwinding a RNA–DNA hybrid than a DNA–DNA hybrid (Matson, 1989). Although DNA polymerase I (Pol I) is sufficient in vitro for the degradation of the RNA primer (5′ to 3′ exonuclease domain) and the gap-filling reaction (polymerase domain), the ΔpolA strain is viable both in E.coli (Joyce and Grindley, 1984) and in B.subtilis (E.Dervyn, unpublished observation), which suggests at least an alternative pathway for the removal of the RNA primer of OFs. In fact, the ΔpolA uvrD double mutant is not viable, as if UvrD was involved in this alternative pathway (Moolenaar et al., 2000). If the PcrA/UvrD/Rep helicases participate effectively in OF maturation, in a helicase mutant, OF may accumulate, and lead eventually to the creation of gaps on DNA, the substrate of RecFOR, which may become toxic due to its persistence on newly replicated DNA. In the absence of RecFOR, the way would be free for an alternative pathway of OF sealing, possibly relying on Pol I alone. This proposed role for the PcrA, Rep and UvrD helicases is reminiscent of that played by FEN1 and DNA2 in eukaryotic cells, except that these helicases are at the same time nucleases, whereas PcrA, Rep and UvrD are not. In bacteria, a separate nuclease would have to enter into play to remove the unwound RNA piece. Another similarity is the type of mutation generated in the recF gene of the pcrA1 recF17 suppressed strain, a 9 bp expansion, typical of rearrangements observed in FEN1 mutants (Tishkoff et al., 1997). At this stage, however, there is clearly no proof that the PcrA, Rep and UvrD helicases share a common function with the FEN1 and DNA2 enzymes in vivo.
In conclusion, we demonstrate that essential helicases act to prevent the toxicity of the recombination proteins. The toxicity could be due either to the accumulation of recombination intermediates or to the formation and persistence of nucleoprotein complexes that interfere with DNA replication. Future work should help in discriminating between these possibilities.
Materials and methods
Bacillus subtilis strain constructions
Bacillus subtilis strains are listed in Table IV. Strains were constructed and propagated in synthetic medium LM (Petit et al., 1998). Because the pcrA gene is part of an operon, and the next gene in this operon is essential (the ligA gene; see Figure 1A; Petit and Ehrlich, 2000), all strains used in this study were constructed so as to ensure a constitutive expression of ligA. In the case of the pcrA1 gene disruption, a Pspac promoter drives the transcription of ligA, and cells were always propagated in the presence of 0.5–1 mM IPTG (see Figure 1A). For the pcrAind derivatives, an additional copy of the ligA gene was introduced at the amyE locus as follows. The ligA gene was amplified by PCR so as to bring a BamHI site followed by an artificial promoter, consisting of a –10 extended sequence, before the ribosome-binding site of ligA, as underlined in the sequence of the relevant primer MAP191 (CGGGATCCTGCTATAATAAAGCAGTAATTGAAGAGGA). The reverse primer MAP192 included the end of the ligA open reading frame (ORF), as well as an MluI site. The DNA segment was next cloned into the pDH32 vector between the BamHI and MluI sites. The resulting plasmid, pMAP157, was then inserted into the B.subtilis chromosome by double crossing over, making use of the selectable chloramphenicol resistance (CmR) marker to replace the amyE gene. For some constructions, the CmR marker associated with ligA was replaced by a phleomycin resistance (PhleoR) gene. This was done in two steps. First, a pDH32 derivative, pMAP159, was constructed in which the CmR cassette was exchanged for a PhleoR cassette (a SphI–BanII fragment exchange with pUCphleo). Secondly, the ligA gene was cut out of pMAP157 by BamHI and BlpI and inserted into the BlpI site of pMAP159, resulting in plasmid pMAP163.
All other strain constructions in B.subtilis consisted of introducing alleles by transformation, either with a direct selection for an antibiotic resistance marker in the case of gene knock-outs, or selecting for an adjacent marker and screening for point mutations. In the case of recF alleles, the marker was a Tn10CmR inserted into rrnO16S (50% linkage); in the case of addA5, the marker was a pMUTIN2 EmR cassette inserted in yhjP (30% linkage). The screening for recF15 or addA5 mutations was done by a search for UV-sensitive clones. The sreening of the recF17 allele was done by PCR, using two primers flanking the 9 bp expansion mutation, and 170 bp apart (primers 100 and 110). The amplified products were then separated on a 12% acrylamide gel, and compared with control DNA, 170 and 179 bp long.
Escherichia coli strain constructions
The E.coli strains used are listed in Table V. All constructions and strain propagations were performed in M63 media supplemented with 0.2% casamino acids and 0.4% glucose, except for selection of kanamycin-resistant transductants, which was done on M9 supplemented with 0.2% casamino acids, 0.4% glucose, 1 mM MgSO4 and 0.1 mM CaCl2. Strains containing the pGB2TSrep plasmid were propagated at 30°C, unless otherwise stated. The introduction of the uvrD::Tn5 mutation in strains that were already resistant to kanamycin was done using the metE::Tn10 marker, which is 17 kb away from uvrD. Among the TetR clones, 10% were found to have a mutator phenotype, as judged by the proportion of rifampicin-resistant clones of an overnight culture, and were more UV sensitive than their parental strain. To cure strains from the pGB2TSrep plasmid, overnight cultures were diluted 100-fold, grown for 2 h at 30°C and then shifted for 6 h at 42°C. After 3 h at 42°C, cells were diluted 100-fold. The titers reported in Figure 6 are corrected for this 100-fold dilution.
Table V. Escherichia coli strains.
| Strain | Relevant genotypea | Reference/source |
|---|---|---|
| JJC451 | recF400::Tn5 | Michel laboratory |
| JJC404 | recO::Tn5 | Michel laboratory |
| JJC1521 | uvrD::Tn5 metE::Tn10 | Michel laboratory |
| JJC760 | Δrep::Cam[pGB2TSrep] | Michel et al. (2000) |
| JJC1183 | Δrep::Cam recF400::Tn5 [pGB2TSrep] | Michel et al. (2000) |
| JJC1193 | recR::Tn5 | Michel laboratory |
| MAC556 | Δrep::Cam uvrD::Tn5 [pGB2TSrep] | P1 JJC1521*JJC760 |
| MAC558 | Δrep::Cam recO::Tn5 [pGB2TSrep] | P1 JJC404*JJC760 |
| MAC564 | Δrep::Cam recR::Tn5 [pGB2TSrep] | P1 JJC1193*JJC760 |
| MAC535 | Δrep::Cam recF400::Tn5 uvrD::Tn5 metE::Tn10 [pGB2TSrep] | P1 JJC1521*JJC1183 |
| MAC569 | Δrep::Cam recO::Tn5 uvrD::Tn5 metE::Tn10 [pGB2TSrep] | P1 JJC1521*MAC558 |
| MAC574 | Δrep::Cam recR::Tn5 uvrD::Tn5 metE::Tn10 [pGB2TSrep] | P1 JJC1521*MAC564 |
| MAC546 | Δrep::Cam recF400::Tn5 uvrD::Tn5metE::Tn10 | MAC535 cured of pGB2TSrep |
| MAC575 | Δrep::Cam recO::Tn5 uvrD::Tn5 metE::Tn10 | MAC569 cured of pGB2TSrep |
| MAC584 | Δrep::Cam recR::Tn5 uvrD::Tn5 metE::Tn10 | MAC574 cured of pGB2TSrep |
aAll strains are JJC40 derivatives (AB1157 hsdR).
Mapping of the pcrA1 extragenic suppressor recF17
To map the pcrA1 suppressors, a tagging strategy based on Tn10 transposition was used. Transposition was allowed to proceed in the pcrA1-suppressed strain (‘pcrA1 sup 17’), and transposition events that occurred sufficiently close to the suppressing mutation to be recovered by genetic linkage were selected. To do so, a thermosensitive transposon delivery vector, derived from pHV1248 (Petit et al., 1990) and able to replicate in the pcrA1 sup strain, was first constructed (pMAP92). Cells harboring a transposon were selected on a Petri plate at 51°C, and total DNA was extracted from a pool of ∼75 000 transposition events. To recover transposons that were inserted next to the sup17 marker, a wild-type strain was transformed with the pooled DNA, selecting EmR CmR clones that would result from the simultaneous acquisition of the pcrA1 (EmR) allele, the sup17 allele and the Tn10CmR. Twenty-six clones were obtained, and DNA was extracted from each of them to screen for the clones in which Tn10 was linked effectively to either pcrA1 or sup17, as deduced from the percentage of CmR clones among pcrA1 sup17 transformants. Eleven clones were found to have a transposon linked (>3%), among which five were next to pcrA and six were grouped in the region of oriC in the following genes as ordered on the chromosome (the percentage linkage is given in parentheses): rocR (5.5%), yycA (3%), jag (9%), rrnO16S (15%), rrnO23S (13%), yabE (4%). This region is rich in essential genes or functions (oriC, dnaA, dnaN, gyrB and gyrA), and this may explain why no transposon with a high linkage was found. All genes between dnaA and gyrA were then sequenced in the pcrA1 sup17 strain, and a mutation in recF was found, which was designated recF17.
Reconstruction of a recF17 strain
To test whether the recF17 mutation was sufficient to restore the viability of a pcrA1 strain, it was necessary to construct a recF17 single mutant. A PCR was performed to amplify a 1.8 kb fragment encompassing the whole recF17 gene as well as 300–400 bp of flanking sequences. The product was phosphorylated and ligated, and subsequently used to transform the recF15 strain HVS16 of B.subtilis. This strain is sensitive to MMS, whereas the recF17 strain is resistant to MMS. Transformants resistant to 100 µg/ml of MMS were selected, and those having integrated the recF17 allele were screened by PCR with primers 100–110 (see above). Ten clones among 29 had received the recF17 allele. One of them was stored as strain MAS305.
Purification of PcrA and production of antibodies
Two oligonucleotides were designed to clone the pcrA ORF between a SapI and a NdeI site into the pCYB1 vector (Biolabs). As a result (plasmid pMAP112), the PcrA protein is produced as a C-terminal fusion to the intein–chitin domains. The plasmid was propagated in strain MC1061 containing plasmid pACmlacIq, which overproduces LacIq. To express the fusion protein, a 1 l culture was grown to an OD of 0.6 in LB at 37°C, and then shifted to 20°C with addition of 1 mM IPTG for 3 h. The cell pellet was resuspended in 20 ml of TNE buffer (50 mM Tris pH 7.5, 500 mM NaCl, 2 mM EDTA), and treated by sonication. The crude extract was then centrifuged for 30 min at 35 000 g. The supernatant was loaded onto a 2 ml chitin-binding column (Biolabs), washed with 170 ml of TNE, and then incubated overnight in TNE + 30 mM dithiothreitol. The reducing agent induced the intein cleavage, and allowed us to free the full size PcrA protein, which was eluted with TNE in a 4 ml peak containing between 0.8 and 2 mg of 80–90% pure PcrA, depending on the preparation. At this stage, 0.8 mg of protein was separated on a polyacrylamide gel, and the band corresponding to PcrA was collected and used to raise antibodies in rabbits (Eurogentec). Crude polyclonal antibodies were used for the westerns. To purify PcrA further, a protocol similar to that used for PcrA from Bacillus stearothermophilus (BstPcrA) was used (Bird et al., 1998). An ammonium sulfate cut at 50% was performed; PcrA was then dialyzed against buffer A200 (50 mM Tris pH 7.5, 2 mM EDTA, 200 mM NaCl, 1 mM DTT), ultracentrifuged at 100 000 g for 30 min and the supernatant was adjusted to A100 (the same as A200 but 100 mM NaCl) immediatly before loading onto a heparin–Sepharose column. PcrA was eluted with a salt gradient between 100 and 600 mM NaCl. The peak fraction was stored; its concentration was estimated as 0.125 mg/ml by UV absorption at 280 nm.
Detection of PcrA in B.subtilis extracts and estimate of cellular concentrations
Bacillus subtilis cells grown exponentially were collected, the OD was measured and dilutions were plated to estimate the cellular density. A 4 ml aliquot was pelleted and resuspended in a defined volume of Birnboïm solution I supplemented with 2 mg/ml of lysozyme. The volume was adjusted to 0.4 ml for an OD of 0.5. Cells were incubated for 10 min at 37°C and deep frozen–thawed three times. Equal amounts of total cellular proteins (20 µl, equivalent to 2 × 107 cells) were then separated by 10% SDS–PAGE and transferred to a Hybond PVDF membrane (Amersham) by immersion, at 2 A for 3 h, in ETB2 buffer (50 mM Tris, 380 mM glycine, 20% methanol, 0.01% SDS). PcrA immunodetection was carried out as described in the ECL+ kit (Amersham). PcrA antibodies were used at a 1:5000 dilution, and protein G–horseradish peroxidase (from Bio-Rad, 1:5000 dilution) was used as secondary antibody. PcrA was detected using the ECL+ reagent and several autoradiogram exposures. Signal intensities were compared with a range of purified PcrA dilutions present in control lanes.
Recombination assay
Strains were tested as described earlier (Bruand et al., 2001). Briefly, the proportion of recombinants in a 5 ml culture issued from a single colony picked from a Petri plate was estimated by titrating the number of kanamycin-resistant cells and the number of viable cells. Cells were propagated at 37°C in synthetic medium, at the same concentration of IPTG (either 0 or 1 mM) and with 0.5 µg/ml erythromycin, except for control strains (wild-type recA, and recF17) which were propagated in 60 µg/ml of spectinomycin. A minimum of five measurements per strain were made.
Acknowledgments
Acknowledgements
We are grateful to J.Alonso for providing strains. M.-A.P. is indebted to Claude Bruand and Bénédicte Michel for their decisive help in this work, to the students of the ‘DEA de génétique et physiologie des micro-organismes’ (year 2001), and to F.Fabre, P.Polard, S.Marsin and M.Velten for improving the manuscript.
References
- Aboussekhra A., Chanet,R., Adjiri,A. and Fabre,F. (1992) Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol., 12, 3224–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.C. and Stiege,A.C. (1991) Molecular analysis of the Bacillus subtilis recF function. Mol. Gen. Genet., 228, 393–400. [DOI] [PubMed] [Google Scholar]
- Alonso J.C., Viret,J.F. and Tailor,R.H. (1987) Plasmid maintenance in Bacillus subtilis recombination-deficient mutants. Mol. Gen. Genet., 208, 349–352. [DOI] [PubMed] [Google Scholar]
- Alonso J.C., Tailor,R.H. and Luder,G. (1988) Characterization of recombination-deficient mutants of Bacillus subtilis. J. Bacteriol., 170, 3001–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.C., Shirahige,K. and Ogasawara,N. (1990) Molecular cloning, genetic characterization and DNA sequence analysis of the recM region of Bacillus subtilis. Nucleic Acids Res., 18, 6771–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur H.M. and Eastlake,P.B. (1983) Transcriptional control of the uvrD gene of Escherichia coli. Gene, 25, 309–316. [DOI] [PubMed] [Google Scholar]
- Bierne H., Seigneur,M., Ehrlich,S.D. and Michel,B. (1997a) uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol. Microbiol., 26, 557–567. [DOI] [PubMed] [Google Scholar]
- Bierne H., Vilette,D., Ehrlich,S.D. and Michel,B. (1997b) Isolation of a dnaE mutation which enhances RecA-independent homologous recombination in the Escherichia coli chromosome. Mol. Microbiol., 24, 1225–1234. [DOI] [PubMed] [Google Scholar]
- Bird L.E., Brannigan,J.A., Subramanya,H.S. and Wigley,D.B. (1998) Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res., 26, 2686–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork J.M., Cox,M.M. and Inman,R.B. (2001) The RecOR proteins modulate recA protein function at 5′ ends of single-stranded DNA. EMBO J., 20, 7313–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C. and Ehrlich,S.D. (2000) UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol. Microbiol., 35, 204–210. [DOI] [PubMed] [Google Scholar]
- Bruand C., Bidnenko,V. and Ehrlich,S.D. (2001) Replication mutations differentially enhance RecA-dependent and RecA-independent recombination between tandem repeats in Bacillus subtilis. Mol. Microbiol., 39, 1248–1258. [DOI] [PubMed] [Google Scholar]
- Carrasco B., Fernandez,S., Petit,M.A. and Alonso,J.C. (2001) Genetic recombination in Bacillus subtilis 168: effect of ΔhelD on DNA repair and homologous recombination. J. Bacteriol., 183, 5772–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanet R., Heude,M., Adjiri,A., Maloisel,L. and Fabre,F. (1996) Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol., 16, 4782–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Carswell-Crumpton,C. and Hanawalt,P.C. (1997) recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 3714–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez S., Kobayashi,Y., Ogasawara,N. and Alonso,J.C. (1999) Analysis of the Bacillus subtilis recO gene: RecO forms part of the RecFLOR function. Mol. Gen. Genet., 261, 567–573. [DOI] [PubMed] [Google Scholar]
- Fernandez S., Ayora,S. and Alonso,J.C. (2000) Bacillus subtilis homologous recombination: genes and products. Res. Microbiol., 151, 481–486. [DOI] [PubMed] [Google Scholar]
- Gangloff S., Soustelle,C. and Fabre,F. (2000) Homologous recombin ation is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet., 25, 192–194. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino-acid sequence comparisons and structure–function relationships. Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- Haijema B.J., van Sinderen,D., Winterling,K., Kooistra,J., Venema,G. and Hamoen,L.W. (1996) Regulated expression of the dinR and recA genes during competence development and SOS induction in Bacillus subtilis. Mol. Microbiol., 22, 75–85. [DOI] [PubMed] [Google Scholar]
- Hegde S., Sandler,S.J., Clark,A.J. and Madiraju,M.V. (1995) recO and recR mutations delay induction of the SOS response in Escherichia coli. Mol. Gen. Genet., 246, 254–258. [DOI] [PubMed] [Google Scholar]
- Hegde S.P., Qin,M.H., Li,X.H., Atkinson,M.A., Clark,A.J., Rajagopalan,M. and Madiraju,M.V. (1996) Interactions of RecF protein with RecO, RecR and single-stranded DNA binding proteins reveal roles for the RecF–RecO–RecR complex in DNA repair and recombination. Proc. Natl Acad. Sci. USA, 93, 14468–14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanescu S. (1993) Characterization of the Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid pT181 replication. Mol. Gen. Genet., 241, 185–192. [DOI] [PubMed] [Google Scholar]
- Joyce C. and Grindley,N.D.F. (1984) Method for determining whether a gene of Escherchia coli is essential: application to the polA gene. J. Bacteriol., 158, 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev., 63, 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue R.S., Au,K.G. and Modrich,P. (1989) DNA mismatch correction in a defined system. Science, 245, 160–164. [DOI] [PubMed] [Google Scholar]
- Lovett S.T., Drapkin,P.T., Sutera,V.A.,Jr and Gluckman-Peskind,T.J. (1993) A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics, 135, 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson S.W. (1989) Escherichia coli DNA helicase II (uvrD gene product) catalyzes the unwinding of DNA·RNA hybrids in vitro. Proc. Natl Acad. Sci. USA, 86, 4430–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. (2000) Replication fork arrest and DNA recombination. Trends Biochem. Sci., 25, 173–178. [DOI] [PubMed] [Google Scholar]
- Michel B., Recchia,G.D., Penel-Colin,M., Ehrlich,S.D. and Sherratt,D.J. (2000) Resolution of Holliday junctions by RuvABC prevents dimer formation in rep mutants and UV-irradiated cells. Mol. Microbiol., 37, 180–191. [DOI] [PubMed] [Google Scholar]
- Michel B., Flores,M.J., Viguera,E., Grompone,G., Seigneur,M. and Bidnenko,V. (2001) Rescue of arrested replication forks by homologous recombination. Proc. Natl Acad. Sci. USA, 98, 8181–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenaar G.F., Moorman,C. and Goosen,N. (2000) Role of the Escherichia coli nucleotide excision repair proteins in DNA replication. J. Bacteriol., 182, 5706–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel P., Hejna,J.A., Ehrlich,S.D. and Cassuto,E. (1993) Antipairing and strand transferase activities of E.coli helicase II (UvrD). Nucleic Acids Res., 21, 3205–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M.-A. and Ehrlich,S.D. (2000) The NAD-dependent ligase encoded by yerG is an essential gene of Bacillus subtilis. Nucleic Acids Res., 28, 4642–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M.-A., Bruand,C., Janniere,L. and Ehrlich,S.D. (1990) Tn10-derived transposons active in Bacillus subtilis. J. Bacteriol., 172, 6736–6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M.-A., Dervyn,E., Rose,M., Entian,K.D., McGovern,S., Ehrlich,S.D. and Bruand,C. (1998) PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol., 29, 261–273. [DOI] [PubMed] [Google Scholar]
- Sandler S.J. (1996) Overlapping functions for recF and priA in cell viability and UV- inducible SOS expression are distinguished by dnaC809 in Escherichia coli K-12. Mol. Microbiol., 19, 871–880. [DOI] [PubMed] [Google Scholar]
- Scott J.F., Eisenberg,S., Bertsch,L.L. and Kornberg,A. (1977) A mechanism of duplex DNA replication revealed by enzymatic studies of phage φX174: catalytic strand separation in advance of replication. Proc. Natl Acad. Sci. USA, 74, 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel E.C. (1983) The Escherichia coli uvrD gene is inducible by DNA damage. Mol. Gen. Genet., 191, 397–400. [DOI] [PubMed] [Google Scholar]
- Singleton M.R. and Wigley,D.B. (2002) Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol., 184, 1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P., Dillingham,M.S., Velankar,S.S. and Wigley,D.B. (1999) DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol., 290, 137–148. [DOI] [PubMed] [Google Scholar]
- Soultanas P., Dillingham,M.S., Wiley,P., Webb,M.R. and Wigley,D.B. (2000) Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J., 19, 3799–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanya H.S., Bird,L.E., Brannigan,J.A. and Wigley,D.B. (1996) Crystal structure of a DExx box DNA helicase. Nature, 384, 379–383. [DOI] [PubMed] [Google Scholar]
- Tishkoff D.X., Filosi,N., Gaida,G.M. and Kolodner,R.D. (1997) A novel mutation avoidance mechanism dependent on S.cerevisiae RAD27 is distinct from DNA mismatch repair. Cell, 88, 253–263. [DOI] [PubMed] [Google Scholar]
- Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,H.S. and Wigley,D.B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- Webb B.L., Cox,M.M. and Inman,R.B. (1997) Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell, 91, 347–356. [DOI] [PubMed] [Google Scholar]
- Webb B.L., Cox,M.M. and Inman,R.B. (1999) ATP hydrolysis and DNA binding by the Escherichia coli RecF protein. J. Biol. Chem., 274, 15367–15374. [DOI] [PubMed] [Google Scholar]
- Whitby M.C. and Lloyd,R.G. (1995) Altered SOS induction associated with mutations in recF, recO and recR. Mol. Gen. Genet., 246, 174–179. [DOI] [PubMed] [Google Scholar]
- Yancey-Wrona J.E. and Matson,S.W. (1992) Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res., 20, 6713–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarranton G.T. and Gefter,M.L. (1979) Enzyme-catalyzed DNA unwinding: studies on Escherichia coli rep protein. Proc. Natl Acad. Sci. USA, 76, 1658–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]