Abstract
The mechanisms responsible for prokaryotic DNA segregation are largely unknown. The partitioning locus (par) encoded by the Escherichia coli plasmid R1 actively segregates its replicon to daughter cells. We show here that the ParM ATPase encoded by par forms dynamic actin-like filaments with properties expected for a force-generating protein. Filament formation depended on the other components encoded by par, ParR and the centromere-like parC region to which ParR binds. Mutants defective in ParM ATPase exhibited hyperfilamentation and did not support plasmid partitioning. ParM polymerization was ATP dependent, and depolymerization of ParM filaments required nucleotide hydrolysis. Our in vivo and in vitro results indicate that ParM polymerization generates the force required for directional movement of plasmids to opposite cell poles and that the ParR–parC complex functions as a nucleation point for ParM polymerization. Hence, we provide evidence for a simple prokaryotic analogue of the eukaryotic mitotic spindle apparatus.
Keywords: actin/DNA segregation/filaments/ParM/partitioning
Introduction
DNA segregation during mitosis is a highly dynamic process carried out by the mitotic spindle apparatus. The identification of a large number of proteins taking part in mitosis has provided a detailed understanding of the mechanisms underlying eukaryotic DNA segregation (Heald, 2000; Sharp et al., 2000). In contrast, the mechanism of prokaryotic DNA segregation has remained obscure. Early (Jacob et al., 1963) as well as contemporary (Nanninga, 2001) models invoke coupling of replicating DNA to the elongating cellular envelope as a means of bacterial DNA segregation. The recent discovery that DNA replication in Bacillus subtilis and Escherichia coli takes place at a stationary, centrally located replication factory has led to the proposal that bi-directional extru sion of newly replicated DNA from the replication factory followed by DNA condensation might constrain the motion of sister nucleoids to opposite sides of the division plane (Lemon and Grossman, 1998, 2000, 2001; Koppes et al., 1999; Gordon and Wright, 2000; Sawitzke and Austin, 2001; Onogi et al., 2002). Mounting evidence obtained by fluorescence microscopic techniques indicates, however, that replicated plasmids (Gordon et al., 1997; Niki and Hiraga, 1997; Jensen and Gerdes, 1999) and chromosomal origins of replication (Lewis and Errington, 1997; Webb et al., 1997, 1998; Niki and Hiraga, 1998; Jensen and Shapiro, 1999; Niki et al., 2000) move rapidly from mid-cell to fixed positions near the cell poles or at one-quarter and three-quarters of cell length, attesting to the presence of a prokaryotic mitotic-like machinery as well (Sharpe and Errington, 1999; Hiraga, 2000; Moller-Jensen et al., 2000).
Partitioning modules, found on many bacterial low copy number plasmids, confer genetic stability upon their replicons through specific subcellular positioning of the DNA (Gordon et al., 1997; Niki and Hiraga, 1997; Jensen and Gerdes, 1999). Generally, par systems are composed of three essential components: (i) an ATPase and (ii) a protein that binds to (iii) a cis-acting centromere-like DNA region (Gerdes et al., 2000). The genetic organization of the par locus of plasmid R1 is shown in Figure 1A. It encodes the ParM ATPase (M for motor), the DNA-binding protein ParR (R for repressor) and the cis-acting centromere-like region, parC, all of which are indispensable for plasmid partitioning. ParR binds to 10 direct repeats in parC, thereby forming a complex that mediates specific pairing of plasmid molecules in vitro (Jensen et al., 1998). ParM has been shown to interact with ParR in vivo and in vitro (Jensen and Gerdes, 1997). The specific role of ParM in the partitioning process has, however, remained unclear.
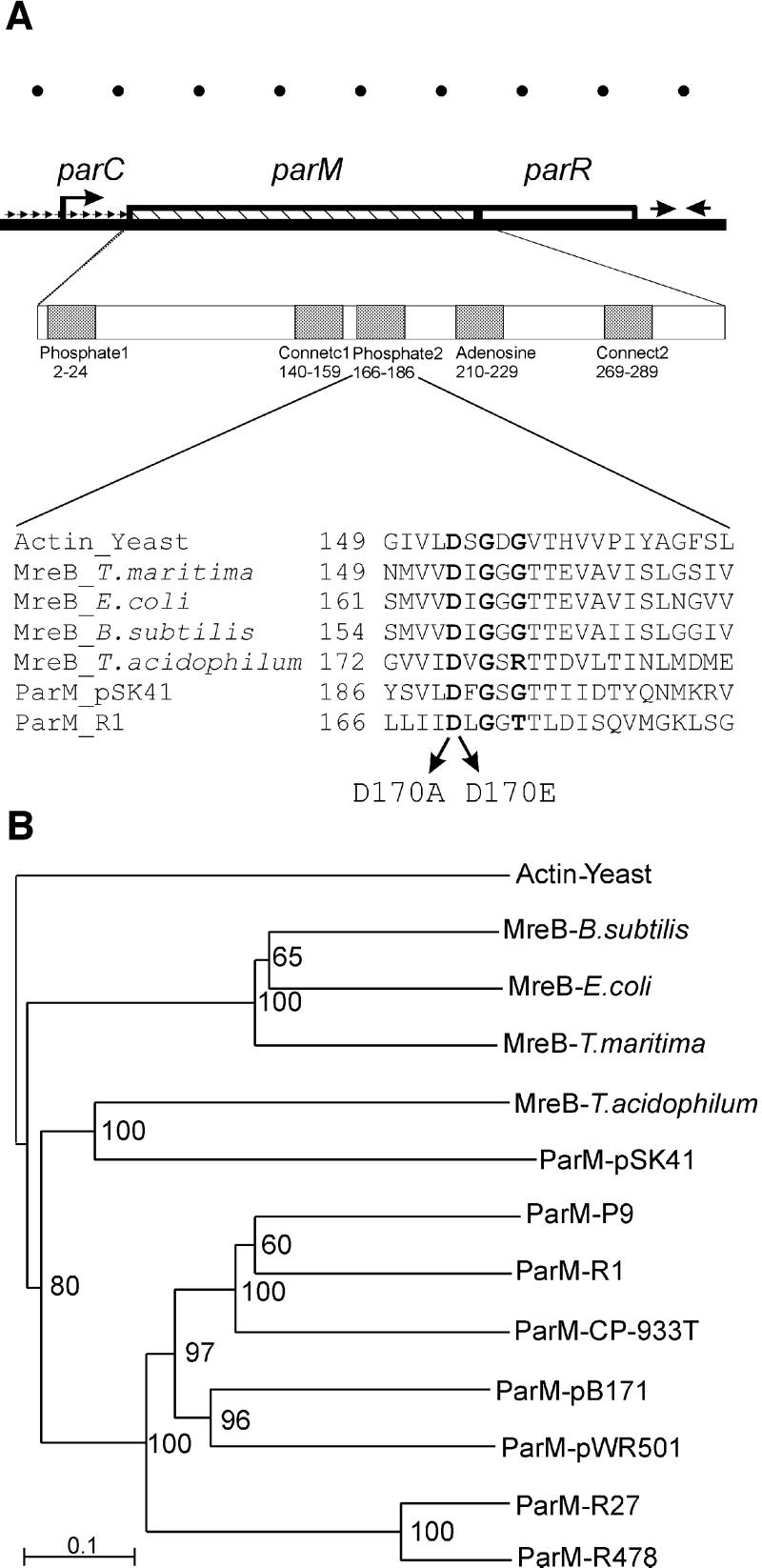
Fig. 1. (A) Genetic structure of the par locus from plasmid R1. parC is a centromere-like region that contains the par promoter and 10 direct repeats (indicated by small repeated arrows) to which ParR binds. ParR thereby autoregulates transcription of the par operon. The broken arrow indicates the par operon promoter. The parM (320 codons) and parR (117 codons) genes are shown as hatched and open boxes, respectively. Opposing arrows indicate a transcriptional terminator downstream of parR. The first enlargement shows the five regions in ParM that exhibit similarity to analogous regions present in all proteins with the actin fold (Bork et al., 1992). Numbers are amino acids in ParM. The second enlargement shows an alignment of the phosphate2 regions of actin (from yeast), MreBs from T.maritima, E.coli, B.subtilis and T.acidophilum, and ParMs from plasmids R1 and pSK41. Amino acid changes in the phosphate2 region of ParM of R1 analysed here are indicated with arrows. Conserved amino acids are shown in bold. Ruler units at the top of the figure represent 200 bp of DNA. (B) Phylogram showing the evolutionary relationships of actin, MreB and ParM proteins. The ParM proteins are located on plasmids, except for ParM of E.coli O157:H7 prophage CP-933T. The MreB proteins are encoded by prokaryotic chromosomes. Horizontal lines indicate relative evolutionary distances. The scale bar indicates arbitrary evolutionary distance. Bootstrap values are given at each branch point in the dendrogram. ParM and MreB form two distinct clades, whereas MreB from T.acidophilum and ParM from pSK41 group together. The phylogram was generated by ClustalX. Entrez accession Nos (GI): Actin-Yeast: 113309; MreB-B.subtilis: 7444226; MreB-E.coli: 1171017; MreB-T.maritima: 7444232; MreB-T.acidophilum: 10639751; ParM-pSK41: 3676423; ParR-pSK41: 3676424; ParM-ColIbP9: 9507465; ParR-ColIbP9: 9507466; ParM-R1: 78284; ParR-R1: 208645; ParM-CP-933T: 15802331; ParR&-CP-933T: 15802332; ParM-pB171: 10955418; ParR-pB171: 10955417; ParM-pWR501: 13449145; ParR&--pWR501: 13449144; ParM-R27: 10957204; ParR-R27: 10957203; ParM-R478: 1695865; ParR-R478: 1695866.
ParM belongs to a superfamily of ATPases that includes actin and its putative bacterial ancestor MreB (Bork et al., 1992; Jones et al., 2001; van den Ent et al., 2001). We therefore considered the possible role of ParM as a mechanochemical enzyme that couples ATP hydrolysis to the active movement of plasmid DNA. Here, we show that ParM forms filamentous structures extending along the longitudinal axis of E.coli cells. ParM filament dynamics were required for active segregation of the R1 plasmid. Filament formation and turnover were controlled by the ParR–parC complex and the ParM ATPase activity, respectively. In vitro, filament formation by purified ParM required magnesium and ATP, whereas hydrolysis of ATP was required for filament depolymerization. Consistently, the ATPase activity of ParM displayed cooperativity. Based on these results, a model for R1 plasmid segregation is proposed.
Results
Actin homologues are encoded by plasmids from Gram-negative and Gram-positive bacteria
The mreB gene encodes a prokaryotic actin homologue that is required to maintain the rod-shaped morphology of B.subtilis and E.coli cells (Wachi and Matsuhashi, 1989; Jones et al., 2001). MreB forms helical actin-like filaments in vivo (Jones et al., 2001) and in vitro (van den Ent et al., 2001). The structure of MreB from Thermotoga maritima was solved and showed that MreB contains the characteristic core of actin also found in Hsp70, sugar kinases and the cell cycle protein FtsA (van den Ent and Löwe, 2000). All the actin family members comply with the actin-fold characterized by two domains that form a cleft in which ATP binds (Kabsch and Holmes, 1995; van den Ent et al., 2001). Proteins belonging to the actin family contain five conserved sequence motifs that form the ATP-binding pocket and the interdomain hinge region (Bork et al., 1992). ParM (StbA) of plasmid R1 (R100) contains the five conserved motifs and thus belongs to the actin superfamily (see Figure 1A; Bork et al., 1992).
Using ParM from plasmid R1 as the query sequence in database searches (Gerdes et al., 2000), we found ParM homologues on a number of plasmids and a phage from enteric bacteria (Figure 1B). All the ParM homologues described in Figure 1B contain downstream putative ParR homologues (the accession numbers of the ParM and ParR homologues are given in the legend to Figure 1). We showed recently that the parMRC locus of pB171 constitutes an active partitioning system (Ebersbach and Gerdes, 2001).
We speculated that ParM might have evolved from an ancestral MreB gene. We therefore carried out BLAST searches with MreB homologues to identify new plasmid-encoded partitioning loci. Somewhat unexpectedly, we found that a plasmid from Staphylococcus aureus (pSK41) encodes a putative MreB homologue (here denoted ParM) that is most closely related to the MreBs from the archaeons Thermoplasma acidophilum and Archaeo globus fulgidus. A closely linked, putative parR gene is located downstream of parM of pSK41, and a series of direct repeats is located in the DNA upstream of parM (to be published elsewhere). We infer that pSK41 encodes a putative parMRC-like partitioning locus.
ParM of plasmid R1 forms axial filaments
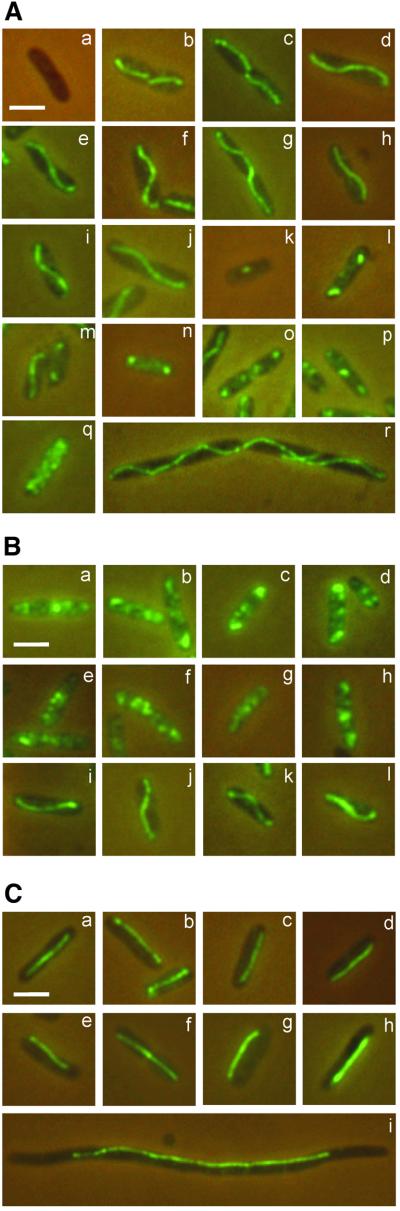
We performed immunofluorescence microscopy (IFM) on E.coli cells harbouring a plasmid R1 derivative carrying the par locus of R1. Figure 2 shows combined phase-contrast and fluorescence microscopy images of fixed cells stained with ParM-specific polyclonal antibodies. Cells in which the parM gene was absent displayed no fluorescence (Figure 2Aa). When expressed in its natural genetic context, ParM gave rise to an unexpected localization pattern (Figure 2Ab–q). Out of 1537 cells examined, 40% contained pole-to-pole axial filaments (Figure 2Ab–j), 20% contained fluorescent foci either at mid-cell or near the poles (Figure 2Ak–p) and 40% contained non-specifically located fluorescence (Figure 2Aq) or no signal. In some cells, two filaments appeared to extend from mid-cell to opposite poles (Figure 2Ab and c). The heterogeneous ParM localization pattern suggests that the filamentous structures are not static but rather undergo successive rounds of polymerization and degradation. ParM filaments often appeared as helical structures; this was especially evident in elongated cells obtained by inhibition of cell division by the addition of cephalexin (Figure 2Ar). In cells containing ParM filaments, most of the fluorescent signal appeared to be localized within the filamentous structures. To assess whether the fluorescent filament consisted of a single or multiple parallel arranged protofilaments, we determined the intracellular level of ParM molecules by quantitative immunoblotting (not shown). The amount of ParM protein was estimated to 15 000–18 000 molecules per cell. Assuming a longitudinal monomer spacing of 50 Å within ParM filaments (van den Ent et al., 2001), there is enough ParM to accommodate a filament of 15–20 times the cell length. Thus, the filaments shown in Figure 2A may well consist of several parallel protofilaments or even bundles of protofilaments (see Discussion).
Fig. 2. (A) ParM forms intracellular actin-like filaments. ParM localization was visualized by combined phase-contrast and immunofluorescence microscopy. Cells of E.coli MC1000 containing different plasmids were fixed and stained as described in Materials and methods. (a) Control cell containing the par– plasmid pOU82. The scale bar represents 2 µm. (b–r) Cells containing plasmid pKG491 carrying the wild-type par locus of R1. (r) Filamentous cell of MC1000/pKG491 obtained by addition of 10 µg/ml cephalexin to the growth medium 90 min prior to fixation. (B) ParM filamentation depends on the ParR–parC complex. ParM localization in the absence of other par components (a–d), in the presence of parR (e–h) or parR–parC on a second plasmid (i–l). In (a–d), ParM was expressed from pPH138 (ptac::parM) in the presence of pOU82 (par– control plasmid); in (e–h) in the presence of pMD1326 (parR+ plasmid); and in (i–l) in the presence of pDD1509 (parR+/parC+ plasmid). (C) Mutation of the ParM ATPase active site changes filament dynamics and morphology. (a–c) Cells containing pRBJ337 expressing ParM D170A in an otherwise par+ genetic context; (d–f) pRBJ338 expressing ParM D170E in a par+ context; (g and h) cells expressing ParM D170A and D170E in the absence of ParR and parC (from pRBJ212 and pRBJ213, respectively); (i) filamentous cell of MC1000/pRBJ338 obtained by addition of 10 µg/ml cephalexin to the growth medium for 90 min prior to fixation. ParM and mutant proteins were expressed to wild-type levels from an inducible ptac promoter construction by the addition of 20 µM IPTG to the growth medium.
ParM filamentation requires both ParR and parC
We examined the intracellular localization of ParM expressed from plasmid pPH138 (ptac::parM) with or without the presence of parR and parC on a compatible plasmid. ParM was expressed from ptac, an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter, at a level close to that expressed by the wild-type par locus (confirmed by western blotting; not shown). In the absence of the other par components, parR and parC, ParM did not form filamentous structures (Figure 2Ba–d). In all cells, a number of fluorescent foci were observed at different positions in the cytoplasm, indicating that ParM in the absence of the other par components is capable of some degree of aggregation, but not filamentation. Co-expression of ParR (without parC) from a co-resident plasmid (pPH138 + pMD1326) did not affect the localization pattern of ParM (Figure 2Be–h). However, when both parR and parC were present (pPH138 + pDD1509), ParM formed filaments indistinguishable from those observed in cells carrying the wild-type par system (Figure 2Bi–l). In the latter case, filamentation, fluorescent foci and non-specific localization were observed with frequencies similar to those of cells carrying wild-type par. This striking result strongly indicates that ParR bound to parC stimulates ParM filament assembly in vivo and is consistent with the proposal that the ParR–parC complex functions as a nucleation point for ParM filamentation. This inference is supported by results presented below.
ParM ATPase mutants exhibit changed filament morphology and dynamics
The phosphate2 sequence regions of actin, MreB and ParM are aligned in Figure 1B (Bork et al., 1992). The alignment shows that the aspartic acid residue at position 170 in ParM, which is involved in binding of Mg2+ (Kabsch and Holmes, 1995), is conserved in all proteins. We showed previously that amino acid changes D170A and D170E simultaneously abolished ParM ATPase activity and the capacity to support plasmid segregation (Jensen and Gerdes, 1997, 1999).
The localization patterns of ParM (D170A) and ParM (D170E) mutant proteins expressed in their natural genetic context were investigated by IFM. Both mutant proteins exhibited hyperfilamentation, forming straight filaments in >99% of all cells (Figure 2Ca–c and Cd–f). The filaments formed by the mutant ParM proteins appeared as less flexible, rod-like structures that extended along the longitudinal axis of the cells. In addition, they extended all the way from pole to pole less frequently than wild-type filaments. The changes in filament morphology were most evident in elongated cells obtained by inhibition of cell division (Figure 2Ci). These results show that the presence of filaments is not sufficient for plasmid segregation and that ATP hydrolysis is not required for ParM polymerization. Furthermore, the uniform appearance of filaments formed by the mutant ParM proteins confirms that the lack of filaments in ∼60% of cells carrying wild-type parM (Figure 2Ak–q) was not caused by artefacts, and thus validates the fixation and immunostaining procedures used here. The fact that mutant ParM filaments of uniform appearance were found in virtually every cell may indicate that these filaments are static rather than dynamic.
Mutant ParM proteins D170A and D170E were also expressed in the absence of parR/parC. Again, the two mutant ParM proteins formed filaments in >99% of the cells (Figure 2Cg–h). Thus, hyperfilamentation of the D170A and D170E mutant proteins was, in contrast to the case of the wild-type, not dependent on ParR and parC.
ParM filament formation requires ATP and Mg2+ ions
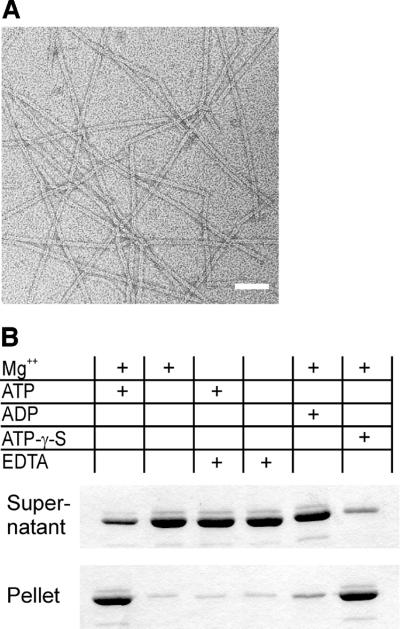
Using electron microscopy (EM), we investigated whether ParM might form polymers in vitro. In the presence of ATP or its non-hydrolysable analogue, ATP-γ-S, ParM formed straight protofilaments with a width of ∼7 nm (Figure 3A). In both cases, filament formation required the presence of Mg2+. ADP, however, did not induce filament formation detectable by EM. The filaments prepared by addition of ATP-γ-S were stable and could be diluted in polymerization buffer without nucleotide before application to the microscope grid (see Materials and methods). In contrast, filaments formed by addition of ATP were unstable and disintegrated rapidly when diluted.
Fig. 3. (A) ParM polymers visualized by electron microscopy. Typical ParM filaments formed by incubation in 30 mM Tris–HCl pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM DTT and 2 mM ATP-γ-S. Filament formation required Mg2+ and occurred in the presence of ATP or its non-hydrolysable analogue, ATP-γ-S, but not in the presence of ADP. Polymers have a cross-sectional diameter of ∼7 nm. Scale bar: 50 nm. (B) Nucleotide-dependent polmerization of ParM. Polymerization of 10 µM ParM was assayed in 30 mM Tris–HCl pH 7.5, 100 mM KCl and 1 mM DTT. MgCl2 (2 mM), EDTA (10 mM) and nucleotides (2 mM) were added as indicated. The protein was centrifuged immediately after mixing at 100 000 g for 15 min at ambient temperature. Supernatant and solubilized pellets were analysed by SDS–PAGE and Coomassie Blue staining.
By UV cross-linking, we found that purified ParM binds ATP and ADP (data not shown). To demonstrate directly the nucleotide requirements for ParM polymerization, we used an ultracentrifugation assay. ParM sedimented in the presence of Mg2+ and ATP (Figure 3B, lane 1), whereas omission of either nucleotide or Mg2+ led to impaired precipitation (lanes 2–4). Pelleting in the presence of Mg2+ could be achieved by addition of the non-hydrolysable nucleotide analogue, ATP-γ-S (lane 6), but not by ADP (lane 5).
ParM polymers are dynamic in vitro
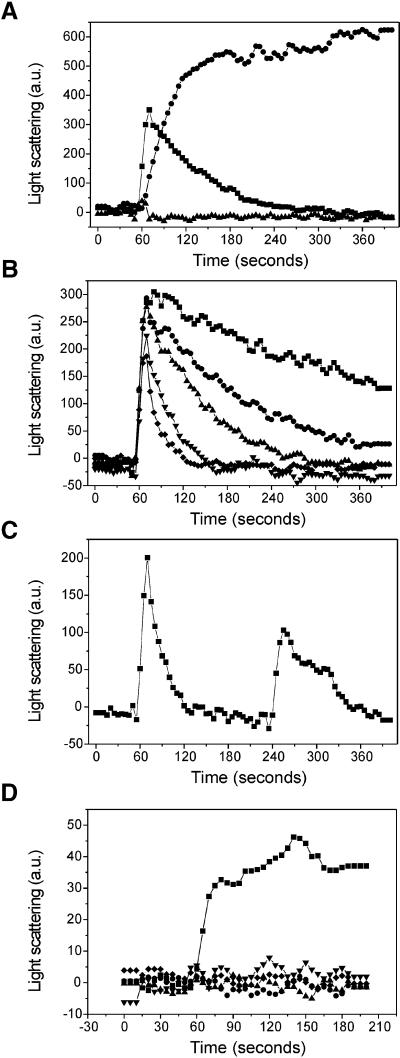
The heterogeneous ParM localization pattern observed with IFM suggested that the filamentous structures formed by ParM are transient. The finding that polymers prepared for EM could be diluted only if they were formed in the presence of non-hydrolysable ATP analogues further supported this notion. For actin, the intensity of the 90° angle light scattering has been found to be linearly proportional to the polymer weight concentration (Kang et al., 1999). To investigate the dynamics of ParM polymerization in vitro, we measured right angle light scattering after addition of different nucleotides in 100-fold molar excess (Figure 4A). The protein (5 µM) was allowed to equilibrate in polymerization buffer for 60 s before addition of nucleotide. ParM did not polymerize after addition of ADP, whereas addition of ATP resulted in a transient pattern indicative of very rapid polymerization followed by slower decay in overall polymerization state. ATP-γ-S addition resulted in a slower but stable polymerization of ParM. The slower rate of assembly indicates that the ParM affinity for ATP-γ-S is lower than that for ATP. However, in the presence of the non-hydrolysable nucleotide analogue, the light scattering signal reached a plateau level that was higher than the maximum signal produced by addition of ATP, indicating that a higher proportion, perhaps all, of the protein was polymerized stably. No depolymerization was observed after addition of ATP-γ-S, showing that this process is dependent on nucleotide hydrolysis. Compared with the ATP-γ-S curve, ATP addition only resulted in a maximum of ∼60% polymerization, indicating that depolymerization commences shortly after initiation of polymer formation.
Fig. 4. ParM polymerization is dynamic and is stimulated by the ParR–parC complex. (A) Polymerization of the ParM protein was assayed in the presence of different nucleotides: ATP (squares), ATP-γ-S (circles), ADP (triangles). The protein (5 µM) was allowed to equilibrate in the cuvette for 60 s before addition of 500 µM nucleotide. Light scattering at a 90° angle was measured as arbitrary units every 5 s. (B) Polymerization of ParM (5 µM) after addition of increasing amounts of ATP: 0.1 mM (diamonds), 0.2 mM (inverted triangles), 0.5 mM (upright triangles), 1 mM (circles) and 2 mM (squares). (C) Recycling of ParM protein. Light scattering signal measured after two successive additions of ATP: 100 µM ATP was added after 60 s and another 200 µM was added after 240 s. (D) Induction of ParM polymerization by ParR and parC DNA. Polymerization of 1 µM ParM was measured in the presence of ParR and/or parC DNA: ParR + pMD330 (pUC19-parC+) (squares), ParR (circles), pMD330 (pUC19-parC+) (upright triangles), ParR + pUC19 (control DNA) (inverted triangles), no addition (diamonds). ATP (500 µM) was added after 60 s of pre-equilibration.
The decay of the light scattering signal was dependent on the amount of ATP added to the reaction (Figure 4B). Saturating nucleotide concentrations ranging from 0.1 to 2 mM (20- to 400-fold molar excess) all gave rise to roughly the same maximum initial scattering signal. Increasing amounts of nucleotide led to a slower decay in scattering signal (i.e. overall polymeric state of the protein solution), showing that ATP is consumed during polymerization. These results indicate that ParM protein is recycled though polymeric states by a mechanism reminiscent of the ATP-dependent ‘treadmilling’ of actin.
To investigate whether depolymerized ParM retained the ability to re-polymerize, we measured right-angle light scattering after two successive ATP additions to the same protein sample (Figure 4C). After 60 s of equilibration, addition of 0.1 mM ATP resulted in a scattering signal, which decayed in ∼60 s. Then, after 240 s, another ATP addition, this time 0.2 mM, resulted in a second scattering signal. In spite of the double amount of ATP added, the second polymerization round gave rise to a weaker signal than the first round, indicating that fewer ParM molecules were polymerized. The decrease is probably due to a competitive inhibitory effect of the ADP resulting from the first round of polymerization. The fact that ParM could be mobilized for successive rounds of polymerization suggests that the protein is capable of nucleotide exchange in the absence of additional factors.
To investigate whether ParM polymerization in vitro was dependent on the presence of ParR and parC DNA, we assayed ParM light scattering in the presence of substoichiometric amounts of ParR and DNA (Figure 4D). At low concentration (1 µM), ParM did not respond to the addition of ATP. Addition of parC-containing DNA or ParR alone did not induce any light scattering; neither did simultaneous addition of ParR and non-specific DNA. However, addition of 20 nM ParR and 2 nM parC DNA induced a light scattering signal, showing that the partitioning complex is capable of inducing ParM polymerization in vitro.
ParM ATPase activity is cooperative
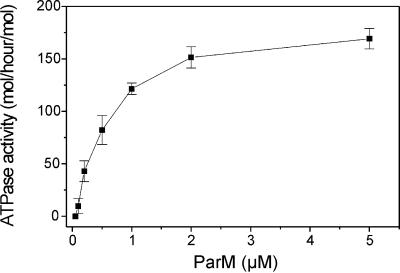
The light scattering analyses indicated that ATP hydrolysis was linked to the polymerization of ParM. We investigated the in vitro ATPase activity of ParM at varying concentrations of the protein (Figure 5). The specific activity increased dramatically with increasing ParM concentration. This result indicates that ParM polymerization stimulates its ATPase activity. Maximal activity occurred at 5 µM and, at concentrations below 0.1 µM, no ATPase activity could be measured. This cooperative nature of the ATPase activity indicates that nucleotide hydrolysis takes place within the filaments and is consistent with a role for ATP and ADP as allosteric effectors of filament turnover.
Fig. 5. ParM ATPase exhibits cooperativity. The ATPase activity of ParM (mol ATP hydrolysed/h/mol ParM) depicted as a function of the ParM concentration. Activities are averages of four independent determinations.
Discussion
Here we show that the actin homologue ParM of plasmid R1 forms dynamic filaments required for active plasmid DNA segregation. Filamentation of ParM was observed both in vivo and in vitro (Figures 2 and 3). In both cases, filament formation depended on the ATPase activity of ParM. In a culture carrying the wild-type par locus of R1, 40% of the cells displayed fluorescent filaments (Figure 2A). However, cells producing mutant ParM proteins unable to bind and hydrolyse ATP all contained filaments and did not segregate plasmid DNA correctly (Figure 2C). These results show that ParM filamentation in vivo is a dynamic process that is required for plasmid DNA segregation.
The intracellular localization of ParM visualized by IFM seemed to contrast with the previously reported localization pattern of a ParM–green fluorescent protein (GFP) fusion protein, which formed specific intracellular foci but not filaments (Jensen and Gerdes, 1999). As in the case of MreB of B.subtilis (Jones et al., 2001), the GFP moiety probably prevents polymerization of ParM–GFP, thus rendering the fusion protein non-functional in vivo.
In vitro polymerization studies show that ParM is closely related to actin. First, EM analyses showed that in the presence of ATP, ParM forms filaments with a width of 7 nm (Figure 3A). F-actin, consisting of two protofilaments, has a width of 6.5 nm, whereas MreB protofilaments have a width of 3.9 nm (Steinmetz et al., 1998; van den Ent et al., 2001). That filamentation of ParM depended on ATP and Mg2+ was supported further by results from the ultracentrifugation assay (Figure 3B). Thus, ParM filaments resemble those of F-actin with respect to both dimensions and ATP dependency.
Using light scattering, we showed that ParM polymers are dynamic (Figure 4). The transient signal observed after addition of ATP to purified ParM supports that the ATPase activity regulates filament formation (Figure 4A and B). Furthermore, addition of the non-hydrolysable ATP analogue ATP-γ-S to purified ParM led to formation of stable filaments (Figure 4A). Together, these results strongly support that ATP binding and hydrolysis are involved in regulation of filament dynamics.
Strikingly, ParM filament formation in vivo depended on the presence of both ParR and parC (Figure 2B). The strict dependency of filamentation on ParR–parC in vivo could in principle be due to filament stabilization by ParR via stoichiometric interaction between ParM and ParR along the filament. For several reasons, we do not favour this explanation. First, quantitative western blot analyses showed that ParM is present in 15 000–18 000 molecules per cell, whereas ParR is present in <1000 molecules per cell (data not shown). The ParM/ParR ratio argues against a direct filament-stabilizing activity of ParR. Secondly, ParR without parC did not induce ParM filamentation (Figure 2B). Thirdly, overproduction of ParR (without parC) also did not induce filamentation (not shown). Since the ParR–parC complex is present in low numbers (4–8 per cell), we favour a model in which the ParR–parC complex acts as a nucleation point that triggers ParM filamentation. This inference is supported by the finding that ParM polymerization in vitro was triggered by the presence of ParR and parC DNA in substoichiometric amounts (Figure 4D). In vivo, ParM may be present at a level such that polymerization depends on the ParR–parC complex.
The dynamic properties of ParM are fundamental to the plasmid partitioning process. We show that ParM filamentation is regulated by (at least) two mechanisms: by ATP hydrolysis and through interaction with the ParR–parC complex. Both mechanisms are also employed by actin to control its dynamic function. Actin changes its secondary and tertiary structure in response to the nucleotide state, thus providing a means of controlling its affinity for the polymeric state (Otterbein et al., 2001), and interactions between actin and a multitude of auxiliary proteins serve to regulate actin polymerization for its versatile functions. It remains to be determined whether ParM interacts with host-encoded factors during plasmid partitioning.
A new model for plasmid segregation
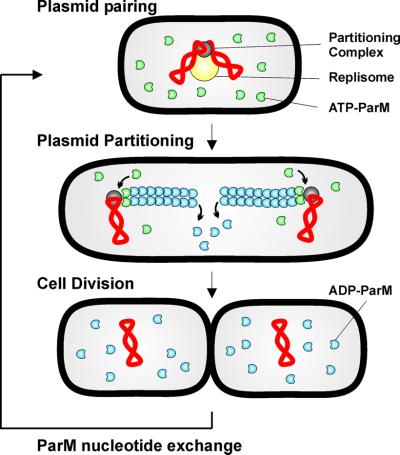
We previously showed that plasmids containing par of R1 move rapidly from mid-cell to positions near the cell poles in a symmetrical pattern (Jensen and Gerdes, 1999) and that ParR dimerizes parC-containing plasmids in vitro (Jensen et al., 1998). The results presented here allow us, for the first time, to present a model that invokes the function of ParM ATPase (Figure 6). The model assumes that plasmids are replicated at mid-cell by the replication factory. After replication, plasmids are paired rapidly via the ParR–parC complex (Jensen et al., 1998). Next, ParR molecules in the paired complex serve as nucleation points for ParM filamentation. Polymerization of ParM, perhaps arranged as multiple parallel fibres extending from the tandemly arranged ParR dimers bound at parC, generates the mechanical force that drives separation and subsequent movement of plasmid molecules to opposite cell poles. After cell division, the replication process relocates plasmid molecules to mid-cell and the cycle is repeated. The model would also function even if plasmid replication and/or separation do not occur at mid-cell. This is because the only demand on a physical separation system is to secure a certain minimal inter-plasmid distance, which, at cell division, should be longer than half the dividing cell. It should also be noted that the model described here also could work for clusters of plasmids or for chromosomal origin regions.
Fig. 6. Model for R1 par-mediated plasmid partitioning during the cell cycle. Plasmids (red) are replicated by the host cell replication machinery (yellow), which is located at mid-cell (Lemon and Grossman, 1998; Koppes et al., 1999). Replicated plasmid parC regions are paired through interactions with ParR protein (grey), thereby forming a partitioning complex. The partitioning complex forms a nucleation point for ParM filamentation. Continuous addition of ATP-ParM (green) to the filament poles provides the force for active movement of plasmid copies to opposite poles. Within the filaments, ATP is hydrolysed, leading to destabilization of the ParM polymer. Nucleotide exchange is required to recharge the ADP-ParM (blue) molecules for a subsequent round of partitioning.
The present data suggest that ParM polymerization per se is responsible for creating the force of plasmid movement. Invoking filament dynamics into the model, we propose that ParM filament formation and degradation is a cyclic process that is regulated to result in separation of paired plasmid replicates and, subsequently, movement of plasmid molecules to opposite cell poles. Initially in the process, ParM-ATP nucleates at ParR bound to parC. Additional ParM-ATP molecules interact with ParR bound to parC, thus providing a mechanism of recruitment of new ParM monomers into a growing ParM filament. Such an insertional polymerization mechanism provides the basis for actin-based motility observed in lamellipodium protrusion and intracellular propulsion of Listeria monocytogenes and Shigella flexneri (Pantaloni et al., 2001; Dickinson and Purich, 2002). The observed cooperativity of the ParM ATPase activity (Figure 5) indicates that nucleotide hydrolysis takes place within the filamentous structures. Conversion of ParM-ATP to ParM-ADP + Pi within the filament and subsequent diffusion of the inorganic phosphate may induce allosteric changes accounting for regulated destabilization of the ParM filament. Finally, rejuvenation of ParM by nucleotide exchange is required for another round of polymerization to commence. Light scattering measurements after two successive ATP additions (Figure 4C) did indicate that ParM rejuvenation could occur in the absence of specific nucleotide exchange factors. Whether plasmid partitioning is coupled to the host cell cycle remains to be determined, but absence or presence of ParM filaments was not correlated with cell size (data not shown).
Initial database analyses revealed that ParM homologues are confined to plasmids and phages of Gram-negative bacteria (Figure 1B). However, refined analyses using MreBs as query sequences revealed an actin homologue on a plasmid from a Gram-positive organism. This putative ParM orthologue from pSK41 is most closely related to MreB from the archaeon T.acidophilum. The finding that plasmids in both Gram-negative and Gram-positive bacteria encode MreB homologues, each more closely related to the MreB family of proteins than to each other, is consistent with the proposal that plasmids recruited MreB at least twice during evolution to perform the task of DNA segregation. It now seems reasonable to investigate whether MreB proteins are involved in segregation of prokaryotic chromosomes.
In conclusion, the R1 par system constitutes a unique example of a bacterial analogue of the eukaryotic spindle apparatus, including a centromeric protein-binding region (parC) at which spindle-like filaments nucleate.
Materials and methods
Strains and plasmids
ParM localization studies were carried out using the E.coli strain MC1000 [araD139 Δ(ara, leu)7697 ΔlacX74 galU galK atrA (Casadaban and Cohen, 1980)] harbouring plasmids carrying all or parts of the R1 par system. The plasmids used are described in Table I. Plasmid pPH138 was constructed by insertion of a BamHI–EcoRI restriction fragment containing the parM gene (bases 131–1199) into the pMD137 expression vector (Jensen and Gerdes, 1997).
Table I. Plasmids used and constructed.
| Plasmid | Relevant genotype | Replicon | Resistance | Reference |
|---|---|---|---|---|
| pOU82 | R1 par– | R1ts | bla | Jensen et al. (1994) |
| pKG491 | R1 par+ | R1ts | bla | Gerdes and Molin (1986) |
| pRBJ337 | parC+ parM170A parR+ | R1ts | bla | Jensen and Gerdes (1997) |
| pRBJ338 | parC+ parM170E parR+ | R1ts | bla | Jensen and Gerdes (1997) |
| pMD137 | ptac::parM (131–1360) lacIq | pUC | aphA | Jensen and Gerdes (1997) |
| pPH138 | ptac::parM (131–1199) lacIq | pUC | aphA | This work |
| pRBJ212 | ptac::parM170A lacIq | PUC | bla | Jensen and Gerdes (1997) |
| pRBJ213 | ptac::parM170E lacIq | PUC | bla | Jensen and Gerdes (1997) |
| pDD1509a | parC+ ΔparM parR+ | R1ts | bla | Jensen et al. (1994) |
| pMD330 | parC+ | pUC | bla | Dam and Gerdes (1994) |
| pMD1326 | plac::parR | pBR322 | bla | M.Dam (unpublished) |
| pUC19 | PUC | bla | Yanisch-Perron et al. (1985) |
apDD1509 is an R1 plasmid containing an in-frame deletion in parM and expresses a wild-type level of ParR (shown by quantitative western blot analysis).
IFM
For IFM, cells were grown in LB medium with antibiotics at 30°C with a generation time of 35 min. At mid-exponential growth phase, 200 µl of cell culture was transferred to 1 ml of cold methanol and stored at –20°C for at least 60 min. IFM was performed as described (Addinall et al., 1996) using affinity-purified rabbit anti-ParM antibodies at a 1:20 dilution and Alexa488-conjugated goat anti-rabbit IgG antibodies (Molecular Probes) at a 1:200 dilution. Cells were observed using a Leica DMRBE microscope with a PL APO 100×/1.40 objective. Combined phase-contrast and fluorescence microscopic images were obtained with a Leica DC200 camera and DC-viewer software.
EM
Solutions of purified ParM protein were centrifuged at 100 000 g for 15 min prior to nucleotide addition and subsequent microscopic examination. ParM (0.1 mg/ml) was incubated in 30 mM Tris–HCl pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM ATP at ambient temperature and applied onto a glow-discharged carbon-coated nickel grid for 1 min. The grid was blotted dry, washed with a drop of 1% uranyl acetate, stained with the same solution and blotted dry again. Samples were examined with a Philips EM208 electron microscope and images were acquired at ×50 000 magnification. ParM was purified as described previously (Jensen and Gerdes, 1997).
ATPase assay
ParM (0.02–10 µM) was incubated for 10 min in ATPase buffer (30 mM Tris–HCl pH 7.5, 200 mM KCl, 10 mM MgCl2, 1 mM DTT) at 37°C. Reactions were initiated by addition of 1 mM ATP containing 0.5 µCi of [γ-32P]ATP and allowed to proceed for 15 min. The amount of released 32PO43– was measured as described (Jensen and Gerdes, 1997).
Quantitative western analysis
Numbers of ParM and ParR proteins were determined by quantitative immunoblot analysis. A 1 ml cell culture grown in selective LB medium to an OD600 of 0.6 was harvested and resuspended in 50 µl of SDS–PAGE loading buffer (100 mM Tris–HCl pH 7.0, 1% SDS, 10% glycerol, 10 mM DTT, 0.05% bromophenol blue). Cells were disrupted by heating to 95°C for 3 min. Samples of 10 µl were run together with dilution series of purified ParM or ParR on 12.5 and 16% acrylamide gels, respectively, and transferred to PVDF membranes (Hybond-P, Amersham Pharmacia) by electroblotting. The membranes were subjected to immunoblotting using affinity-purified rabbit anti-ParM or anti-ParR and horseradish peroxidase (HRP)-conjugated secondary antibodies (Dako). Blots were developed using the ECF system (Amersham Pharmacia), essentially as described by the manufacturer, and scanned using the Storm system (Molecular Dynamics). The cell extract protein level was then determined using the ImageQuant software. The cell number per OD600 unit was determined by viable counting.
ParM polymerization assay
Solutions of purified ParM protein were cleared at 100 000 g for 15 min immediately before assaying to remove aggregates. Cleared ParM (10 µM) was mixed in a volume of 25 µl with 30 mM Tris–HCl pH 7.5, 100 mM KCl, 1 mM DTT. Mg2+ (2 mM) and nucleotides (2 mM) were added as indicated in Figure 3B and reactions were centrifuged immediately at 100 000 g in a Beckmann TA100 rotor for 15 min at ambient temperature. Supernatants were removed and adjusted to 50 µl in SDS–PAGE loading buffer and pellets were resuspended in 50 µl of SDS–PAGE loading buffer. Protein samples were analysed on 12.5% SDS–polyacrylamide gels and stained with Coomassie Blue.
Light scattering
Solutions of ParM were centrifuged at 100 000 g for 15 min prior to analysis. Light scattering at a 90° angle was measured using an LS-50B luminescence spectrometer (Perkin-Elmer). Excitation and emission wavelengths were set to 310 nm with slit widths of 2.5 nm. ParM (5 µM) was incubated in 400 µl of polymerization buffer (30 mM Tris–HCl pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM DTT) at 25°C in a quartz cuvette with a 1 cm path length. Polymerization was induced by addition of 10 µl of nucleotide solution at the concentrations indicated. Light scattering was measured every 5 s. ParR (20 nM) and/or DNA (2 nM) were added before induction of ParM polymerization, as indicated in the figure legends. Background scattering was determined prior to each experiment and subtracted for baseline normalization.
Database analyses
BLAST searches was accomplished using the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/). msf files were generated by the PILEUP program of the Wisconsin Package, Version 10.2, Genetics Computer Group (GCG) and transferred to ClustalX for a refined alignment and bootstrapping. The phylogenetic tree was drawn by NJPLOT associated with ClustalX.
Acknowledgments
Acknowledgements
We thank Pia Hovendal for excellent technical assistance, Fusinita van den Ent for discussions, Dr O.-G.Issinger for the use of his fluorescence microscope, Drs A.Maunsbach and P.Ottosen for providing EM facilities, and Dr M.Yarmolinsky for valuable suggestions on the manuscript. This work was supported by the Danish Biotechnology Program (CIS-FEM) and the Danish Biotech. Instrument Center (DABIC).
References
- Addinall S.G., Bi,E. and Lutkenhaus,J. (1996) FtsZ ring formation in fts mutants. J. Bacteriol., 178, 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Sander,C. and Valencia,A. (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin and hsp70 heat shock proteins. Proc. Natl Acad. Sci. USA, 89, 7290–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M.J. and Cohen,S.N. (1980) Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol., 138, 179–207. [DOI] [PubMed] [Google Scholar]
- Dam M. and Gerdes,K. (1994) Partitioning of plasmid R1. Ten direct repeats flanking the parA promoter constitute a centromere-like partition site parC, that expresses incompatibility. J. Mol. Biol., 236, 1289–1298. [DOI] [PubMed] [Google Scholar]
- Dickinson R.B. and Purich,D.L. (2002) Clamped-filament elongation model for actin-based motors. Biophys. J., 82, 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G. and Gerdes,K. (2001) The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc. Natl Acad. Sci. USA, 98, 15078–15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K. and Molin,S. (1986) Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J. Mol. Biol., 190, 269–279. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Moller-Jensen,J. and Bugge,J.R. (2000) Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol., 37, 455–466. [DOI] [PubMed] [Google Scholar]
- Gordon G.S. and Wright,A. (2000) DNA segregation in bacteria. Annu. Rev. Microbiol., 54, 681–708. [DOI] [PubMed] [Google Scholar]
- Gordon G.S., Sitnikov,D., Webb,C.D., Teleman,A., Straight,A., Losick,R., Murray,A.W. and Wright,A. (1997) Chromosome and low copy plasmid segregation in E.coli: visual evidence for distinct mechanisms. Cell, 90, 1113–1121. [DOI] [PubMed] [Google Scholar]
- Heald R. (2000) Motor function in the mitotic spindle. Cell, 102, 399–402. [DOI] [PubMed] [Google Scholar]
- Hiraga S. (2000) Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet., 34, 21–59. [DOI] [PubMed] [Google Scholar]
- Jacob F., Brenner,S. and Cuzin,F. (1963) On the regulation of DNA replication in bacteria. Cold Spring Harb. Symp. Quant. Biol., 23, 329–348. [Google Scholar]
- Jensen R.B. and Gerdes,K. (1997) Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR–parC complex. J. Mol. Biol., 269, 505–513. [DOI] [PubMed] [Google Scholar]
- Jensen R.B. and Gerdes,K. (1999) Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J., 18, 4076–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B. and Shapiro,L. (1999) The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl Acad. Sci. USA, 96, 10661–10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B., Dam,M. and Gerdes,K. (1994) Partitioning of plasmid R1. The parA operon is autoregulated by ParR and its transcription is highly stimulated by a downstream activating element. J. Mol. Biol., 236, 1299–1309. [DOI] [PubMed] [Google Scholar]
- Jensen R.B., Lurz,R. and Gerdes,K. (1998) Mechanism of DNA segregation in prokaryotes: replicon pairing by parC of plasmid R1. Proc. Natl Acad. Sci. USA, 95, 8550–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L.J., Carballido-Lopez,R. and Errington,J. (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell, 104, 913–922. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Holmes,K.C. (1995) The actin fold. FASEB J., 9, 167–174. [DOI] [PubMed] [Google Scholar]
- Kang F., Purich,D.L. and Southwick,F.S. (1999) Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem., 274, 36963–36972. [DOI] [PubMed] [Google Scholar]
- Koppes L.J., Woldringh,C.L. and Nanninga,N. (1999) Escherichia coli contains a DNA replication compartment in the cell center. Biochimie, 81, 803–810. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman,A.D. (1998) Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science, 282, 1516–1519. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman,A.D. (2000) Movement of replicating DNA through a stationary replisome. Mol. Cell, 6, 1321–1330. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman,A.D. (2001) The extrusion–capture model for chromosome partitioning in bacteria. Genes Dev., 15, 2031–2041. [DOI] [PubMed] [Google Scholar]
- Lewis P.J. and Errington,J. (1997) Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein. Mol. Microbiol., 25, 945–954. [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J., Jensen,R.B. and Gerdes,K. (2000) Plasmid and chromosome segregation in prokaryotes. Trends Microbiol., 8, 313–320. [DOI] [PubMed] [Google Scholar]
- Nanninga N. (2001) Cytokinesis in prokaryotes and eukaryotes: common principles and different solutions. Microbiol. Mol. Biol. Rev., 65, 319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H. and Hiraga,S. (1997) Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E.coli. Cell, 90, 951–957. [DOI] [PubMed] [Google Scholar]
- Niki H. and Hiraga,S. (1998) Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev., 12, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Yamaichi,Y. and Hiraga,S. (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev., 14, 212–223. [PMC free article] [PubMed] [Google Scholar]
- Onogi T., Ohsumi,K., Katayama,T. and Hiraga,S. (2002) Replication-dependent recruitment of the β-subunit of DNA polymerase III from cytosolic spaces to replication forks in Escherichia coli. J. Bacteriol., 184, 867–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterbein L.R., Graceffa,P. and Dominguez,R. (2001) The crystal structure of uncomplexed actin in the ADP state. Science, 293, 708–711. [DOI] [PubMed] [Google Scholar]
- Pantaloni D., Le Clainche,C. and Carlier,M.F. (2001) Mechanism of actin-based motility. Science, 292, 1502–1506. [DOI] [PubMed] [Google Scholar]
- Sawitzke J. and Austin,S. (2001) An analysis of the factory model for chromosome replication and segregation in bacteria. Mol. Microbiol., 40, 786–794. [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Rogers,G.C. and Scholey,J.M. (2000) Microtubule motors in mitosis. Nature, 407, 41–47. [DOI] [PubMed] [Google Scholar]
- Sharpe M.E. and Errington,J. (1999) Upheaval in the bacterial nucleoid. An active chromosome segregation mechanism. Trends Genet., 15, 70–74. [DOI] [PubMed] [Google Scholar]
- Steinmetz M.O., Hoenger,A., Tittmann,P., Fuchs,K.H., Gross,H. and Aebi,U. (1998) An atomic model of crystalline actin tubes: combining electron microscopy with X-ray crystallography. J. Mol. Biol., 278, 703–711. [DOI] [PubMed] [Google Scholar]
- van den Ent F. and Löwe,J. (2000) Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J., 19, 5300–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F., Amos,L.A. and Löwe,J. (2001) Prokaryotic origin of the actin cytoskeleton. Nature, 413, 39–44. [DOI] [PubMed] [Google Scholar]
- Wachi M. and Matsuhashi,M. (1989) Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J. Bacteriol., 171, 3123–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C.D., Teleman,A., Gordon,S., Straight,A., Belmont,A., Lin,D.C., Grossman,A.D., Wright,A. and Losick,R. (1997) Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B.subtilis. Cell, 88, 667–674. [DOI] [PubMed] [Google Scholar]
- Webb C.D., Graumann,P.L., Kahana,J.A., Teleman,A.A., Silver,P.A. and Losick,R. (1998) Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol., 28, 883–892. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira,J. and Messing,J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene, 33, 103–119. [DOI] [PubMed] [Google Scholar]