Abstract
In prokaryotes, Hfq regulates translation by modulating the structure of numerous RNA molecules by binding preferentially to A/U-rich sequences. To elucidate the mechanisms of target recognition and translation regulation by Hfq, we determined the crystal structures of the Staphylococcus aureus Hfq and an Hfq–RNA complex to 1.55 and 2.71 Å resolution, respectively. The structures reveal that Hfq possesses the Sm-fold previously observed only in eukaryotes and archaea. However, unlike these heptameric Sm proteins, Hfq forms a homo-hexameric ring. The Hfq–RNA structure reveals that the single-stranded hepta-oligoribonucleotide binds in a circular conformation around a central basic cleft, whereby Tyr42 residues from adjacent subunits stack with six of the bases, and Gln8, outside the Sm motif, provides key protein–base contacts. Such binding suggests a mechanism for Hfq function.
Keywords: Hfq/protein–RNA complex/RNA-binding protein/Sm/translation regulation
Introduction
The bacterial Hfq protein was first identified as a host factor required for the replication of Qβ RNA bacteriophage (Fernandez et al., 1968; Miranda et al., 1997; Schuppli et al., 1997; Su et al., 1997). Hfq proteins identified in several bacteria reveal that it is strikingly conserved and highly abundant; it has been estimated that there are 30 000–60 000 copies per Escherichia coli cell, localized primarily to the cytoplasm with ribosomes (Kajitani et al., 1994). Since its original discovery, it has been established that Hfq is a pleiotropically acting RNA-binding protein that is required for the degradation of some mRNA transcripts and the efficient translation of others (Kajitani et al., 1994; Azam et al., 2000). Hfq targets several mRNAs for degradation by binding to their poly(A) tails and stimulating poly(A) adenylation (Hajnsdorf and Régnier, 2000). It also represses mRNA translation by preventing ribosome binding, as observed for OmpA mRNA (Vytvytska et al., 2000). In addition, Hfq has been shown to interact with several small untranslated RNA regulatory molecules such as OxyS, DsrA, RprA and Spot42, and is required for RNA regulation of the σs gene by OxyS, DsrA and RprA (Zhang et al., 1998; Majdalani et al., 2001; Sledjeski et al., 2001; Wassarman et al., 2001). Recent data show that Hfq promotes contacts between the OxyS and Spot42 molecules and their target RNAs, suggesting that Hfq assists in bimolecular RNA–RNA interactions (Møller et al., 2002; Zhang et al., 2002). Moreover, studies carried out to identify additional small RNAs found that Hfq interacts with over half of these RNA molecules (∼9 RNA species) (Wassarman et al., 2001). The importance of Hfq is underscored further by the diverse pleiotropic effects caused by interruption of its gene, which include decreased growth rate, sensitivity to UV light and mutagens, and increased cell length (Tsui et al., 1994; Muffler et al., 1997).
Despite the important role that Hfq plays in translational processes, the molecular details of how it mediates such a range of functions through its interactions with RNA molecules is unknown. Because it appears to modulate RNA structures, it has been suggested that Hfq may function as an RNA chaperone (Muffler et al., 1997; Schuppli et al., 1997). Alternatively, because Hfq binding to mRNA poly(A) tails causes poly(A) polymerase to become processive, it has also been postulated that Hfq might be related to the eukaryotic poly(A)-binding protein II (Hajnsdorf and Régnier, 2000). New data have demonstrated clearly that Hfq forms a ring-like structure (Møller et al., 2002; Zhang et al., 2002). This finding and the fact that Hfq plays wide-ranging roles in RNA metabolism has led to the recent suggestion that Hfq may be similar structurally and functionally to eukaryotic Sm proteins. These proteins participate in many different RNA-processing reactions through their interactions with U-rich target RNAs (Branlant et al., 1982; Achsel et al., 1999; Salgado-Garrido et al., 1999; Bouveret et al., 2000; Tharun et al., 2000; Pillai et al., 2001).
Sm proteins contain two conserved regions termed the Sm1 and Sm2 motifs. These motifs are separated by a region, which is not conserved in its sequence or length, and named the variable region (Cooper et al., 1995; Hermann et al., 1995; Séraphin, 1995). Crystal structures of Sm proteins reveal that they contain an N-terminal α-helix followed by a twisted five-stranded β-sheet. This fold is remarkably conserved among eukaryotes and archaea (Kambach et al., 1999; Mura et al., 2001; Törö et al., 2001; Collins et al., 2002). In these structures, the Sm fold oligomerizes to form heptamers; the archaeal proteins AF-Sm1 and AF-Sm2 from Archaeoglobus fulgidus, SmAP from Pyrobaculum aerophilus and Lsmα from Methanobacterium thermoautotrophicum all form homo-heptameric rings (Mura et al., 2001; Törö et al., 2001; Collins et al., 2002), whilst the human Sm proteins B, D1, D2, D3, G, E and F form a hetero-heptamer (Walke et al., 2001). The details of how these protein complexes recognize and modulate RNA are not clear. However, the recent structure of AF-Sm1 with a small uridine oligonucleotide, although disordered, revealed some insight into RNA binding by Sm proteins (Törö et al., 2001).
The fact that Sm and LSm (Sm-like) proteins are found in all eukaryotic and archaeal cells (Cooper et al., 1995; Hermann et al., 1995; Séraphin, 1995; Salgado-Garrido et al., 1999) and are highly conserved between these kingdoms has led to the suggestion that the Sm family may have evolved from a early ancestor and, thus, Sm proteins may be present in bacteria. Here we report the crystal structures of the Staphylococcus aureus Hfq and an Hfq–RNA complex. These structures reveal that Hfq does indeed possess an Sm fold. However, unlike other described Sm proteins, Hfq forms a functional hexamer. Importantly, the Hfq–RNA structure demonstrates how it recognizes RNA and suggests how Hfq alters RNA structure, thus providing unifying insight into the multiple roles of Hfq in RNA metabolism.
Results and discussion
Overall structure of Hfq
The structure of the full-length 8.9 kDa (77 residue) S.aureus Hfq protein was determined by single isomorphous replacement (SIR) using a platinate derivative (K2PtCl4) (see Materials and methods, Table I). The current structure has an Rwork and Rfree of 23.7 and 25.9%, respectively, to 1.55 Å resolution. Hfq forms a symmetric hexameric ring with a diameter of ∼65 Å and width of ∼23 Å (Figure 1). The hexamer has a doughnut shape with a central hole ∼12 Å in diameter at the smallest constriction (Figure 1B). These dimensions are similar to, but smaller than, those obtained from recent electron microscopy studies on the E.coli Hfq protein (Zhang et al., 2002). Each subunit consists of an N-terminal α helix (α1, residues 7–18), followed by five β-strands (β1, residues 20–25; β2, residues 30–39; β3, residues 43–48; β4, residues 53–57; and β5, residues 60–66) (Figure 1A). Residues 1–6 are disordered in all but one subunit, and C-terminal residues 67–77 are missing in all subunits. There are two hexamers in the crystallographic asymmetric unit (ASU) (Figure 1C), and the 12 subunits are essentially identical with root mean square deviations (r.m.s.ds) between ∼0.22 and 0.60 Å (0.31, 0.34, 0.34, 0.31, 0.40, 0.26, 0.37, 0.40, 0.22, 0.47 and 0.60 Å) for all corresponding Cα atoms. The larger r.m.s.d. of 0.60 Å for one subunit is due to slight structural differences observed within its variable region turn (residues 49–52), which is the only region to display any structural variation among the subunits. The two hexamers in the ASU are also essentially identical, superimposing with an r.m.s.d. of 0.64 Å, and stack back to back (Figure 1C).
Table I. Selected crystallographic data for the apo Hfq structure determination.
| Crystal | Native 2 (SSRL) | Native 1 | K2PtCl4 |
|---|---|---|---|
| Space group | P21 | P21 | P21 |
| Cell constants (Å) | a = 67.0 | a = 68.5 | a = 68.8 |
| b = 90.0 | b = 90.7 | b = 91.2 | |
| c = 67.7 | c = 69.0 | c = 69.1 | |
| β = 98.0° | β = 96.8° | β = 97.0° | |
| Temperature (K) | 100 | 298 | 298 |
| Resolution (Å) | 22.4–1.55 | 68.1–2.78 | 68.1–2.72 |
| Total reflections (no.) | 460 147 | 47 080 | 39 936 |
| Unique reflections (no.) | 111 857 | 18 886 | 17 240 |
| Overall Rsym (%)a | 4.6 | 9.0 | 9.8 |
| Overall I/σ(I) | 7.6 | 10.9 | 9.5 |
| High resolution shell (Å) | 1.60–1.55 | 2.87–2.78 | 2.82–2.72 |
| Rsym (%) | 15.8 | 37.1 | 25.1 |
| I/σ(I) | 4.7 | 1.9 | 1.5 |
| Phasing powerb | 1.81 | ||
| Rcullisc | 0.51 | ||
| Riso (%)d | 25.7 | ||
| Heavy atom sites (no.) | 12 | ||
| Overall figure of merite | 0.23 | ||
| Refinement statistics: native 2 | |||
| Completeness (%) | 99.8 | ||
| Resolution (Å) | 22.40–1.55 | ||
| Rwork/Rfree (%)f | 23.7/25.9 | ||
| R.m.s.ds | |||
| Bond angles (°) | 1.57 | ||
| Bond lengths (Å) | 0.013 | ||
| B-values (Å2) | 3.3 | ||
| <B> (Å2) | 23.9 | ||
| Solvent/acetate molecules | 592/5 | ||
| Ramachandran analysis | |||
| Most favored (%/no.) | 94.4/636 | ||
| Additional allowed (%/no.) | 5.6/38 | ||
| Generously allowed (%/no.) | 0.0/0 | ||
| Disallowed (%/no.) | 0.0/0 |
aRsym = ΣΣ|Ihkl – Ihkl(j)|/ΣIhkl, where Ihkl(j) is the observed intensity and Ihkl is the final average value of intensity.
bPhasing power = r.m.s.(|Fh|/E), where |Fh| = the heavy atom structure factor amplitude and E = residual lack of closure error.
cRcullis = Σ||Fh(obs)| – |Fh(calc)||/Σ|Fh(obs)| for centric reflections, where |Fh(obs)| = observed heavy atom structure factor amplitudes and |Fh(calc)| = calculated heavy atom structure factor amplitude.
dRiso = Σ||Fph| – |Fp||/Σ|Fp|, where |Fp| is the protein structure factor amplitude and |Fph| is the heavy atom derivative structure factor amplitude.
eFigure of merit = <|ΣP(α)eiα/ΣP(α)|>, where α is the phase and P(α) is the phase probability distribution.
fRwork = Σ||Fobs| – |Fcalc||/Σ|Fobs| and Rfree = Σ||Fobs| – |Fcalc||/Σ|Fobs|, where all reflections belong to a test set of 5% of the data randomly selected and not used in the atomic refinement.
Fig. 1. The structure of Hfq. (A) Structure of the Hfq monomer shown as a ribbon diagram. Secondary structural elements are labeled, as are the first (N) and last (C) residues observed. Figures 1A–C, 2B–D, 3A, 4B and C and 6 were made with Swiss-PdbViewer and rendered with POVRAY (Guex and Peitsch, 1997; POVRAY, Persistence of Vision Raytracer version 3.1 http://www.povray.org). (B) Structure of the Hfq hexamer with each subunit colored differently. (C) The two Hfq hexamers in the crystallographic asymmetric unit. This view is rotated 90° to (B) along the vertical axis in the plane of the paper. Interactions between the two rings are made by residues from the hydrophobic surface of each hexamer (see Figure 5B).
Hfq contains an Sm fold
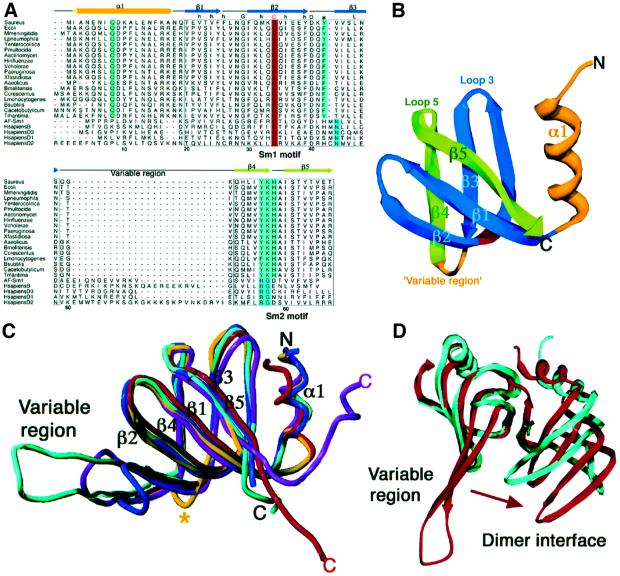
Sequence comparisons between Hfq and other Sm proteins reveal a region of homology within the Sm1 motif (Figure 2A). Specifically, Hfq and Sm proteins share a conserved pattern of hydrophobic residues as well as a highly conserved acidic residue (Asp40 in Hfq). A distinctive conservation between these proteins corresponds to a glycine, which is located in the middle of β2 and is critical to maintain the highly distorted Sm fold. However, a hallmark of the Sm1 motif, not shared by any Hfq protein, is a conserved asparagine residue near the N-terminus of β3. In contrast to the Sm1 motif, the C-terminal region of Hfq shows no strong sequence homology to the Sm2 motif of any Sm protein. For example, it is missing the RG motif at the end of β4 which, like the Sm1 asparagine, is a hallmark of the Sm2 motif. However, there are several potential sequence conservations between hydrophobic residues.
Fig. 2. Hfq is an Sm protein. (A) Structure-based sequence alignment of prokaryotic Hfq proteins (first 18 proteins) with the archaea AF-Sm1, and the human Sm proteins B, D3, D1 and D2. The alignment was based on an optimized superimposition (shown in C). The secondary structural elements of the Hfq protein are shown above the alignment and colored as in (B), where the non-Sm motifs, the N-terminal helix α1 and the variable region loop are colored yellow, the Sm1 motif region is colored blue and the ‘Hfq Sm2 motif’ is colored green. Both Sm motifs are boxed. Hfq residue Gly34, the sole conserved residue amongst Hfq and the Sm proteins, is blocked in red. Every 10th S.aureus Hfq residue is numbered. Highly conserved hydrophobic residues found in all Sm proteins within the Sm1 region are indicated by a lower case h, and the two highly conserved glycine and aspartic acid residues within the Sm1 motif are indicated by G and D, respectively. The absolutely conserved glutamine of helix α1 that is important for base recognition and the highly conserved tyrosine (or phenylalanine) are blocked in light green in the Hfq proteins, whilst the signature asparagine of the eukaryotic Sm proteins at the start of β3 is blocked in blue green. Within the Sm2 region, the ‘Hfq Sm2 motif YKH’ is colored light green, whilst the invariant RG dipeptide of the eukaryotic/archaea Sm2 motif is colored blue green. This figure was made with Alscript (Barton, 1993). (B) Ribbon diagram of an Hfq subunit colored to highlight the Sm1 and Hfq Sm2 motifs. The Sm1 motif is colored blue and the Sm2 motif is green to match the sequence alignment. Regions outside the two motifs, i.e. helix α1 and the variable region, are colored yellow. The conserved glycine, Gly34, is colored red. (C) Superimpositions of the structures of the AF-Sm1 of archaea (blue), the human SmB (cyan), and the D1 (magenta) and D2 (red) subunits onto Hfq (yellow). The resulting r.m.s.ds are 1.2 Å for 55 Cα atoms, 1.2 Å for 55 Cα atoms, 1.7 Å for 54 Cα atoms and 1.5 Å for 58 Cα atoms, respectively. These superimpositions clearly point out the remarkable structural similarity within the Sm1 and Sm2 motifs of these proteins and the much abbreviated variable region of Hfq (indicated by a yellow asterisk). (D) Comparison of an Hfq dimer (blue) with the human D3–B dimer (red) after superimposition of the human B subunit onto an Hfq monomer (where the Hfq dimer is rotated in the horizontal by ∼180° relative to the magenta/yellow dimer in Figure 1B). The less rotated (denoted by an arrow) dimer interface of Hfq might contribute to its hexameric oligomerization, in contrast to the heptameric oligomerization observed in the other Sm proteins, which contain large variable regions.
Though Hfq and LSm proteins exhibit faint sequence similarity to the Sm family of proteins, the crystal structure reveals that Hfq does indeed contain the distinctive Sm fold, which consists of a bent five-stranded antiparallel β-sheet capped by an N-terminal α-helix and separated by a variable region (Figure 2B). Structural superimpositions illustrate the strong similarity between Hfq and the archaeal AF-Sm1, M.thermoautotrophicum Lsmα, human B, human D3, human D1 and human D2 proteins, which result in r.m.s.ds of 1.2, 1.3, 1.2, 1.5, 1.7 and 1.5 Å for 55, 55, 55, 51, 54 and 58 corresponding Cα atoms, respectively (Figure 2C). Especially striking is that the structures of the Sm1 and ‘Sm2’ regions of these proteins are nearly identical.
Hfq is a hexameric Sm protein
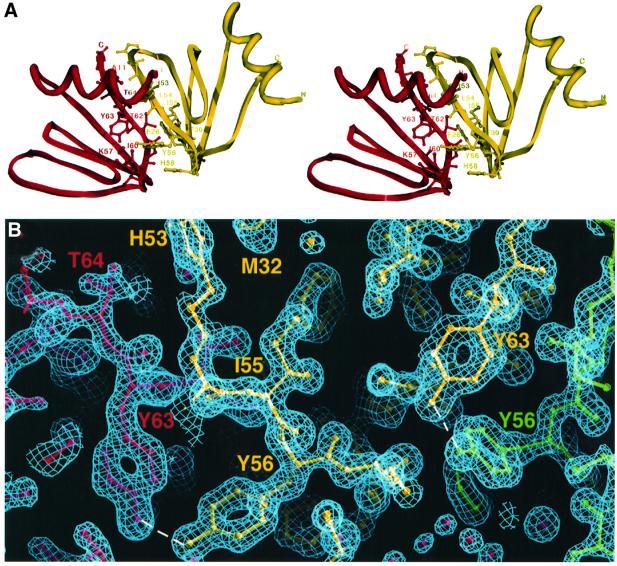
The dimer interface between subunits in Hfq buries 1333 Å2 of accessible protein surface. The Hfq dimer interface is formed, in part, by contacts between residues from α1 and the turn between β2′ and β3′ (where ′ indicates the other subunit) (Figure 3A). In addition, the side chain of Leu54′ from β4′ also contributes to this hydrophobic interface. β1 and β2 stack against the back of β5′ and β1′, with Phe26 from β1 and Phe39 from β2 anchoring this interaction. A key stabilizing dimer interaction is the intersubunit continuity of the β-sheet by the β4 and β5′ interface. This creates the continuous 30 stranded β-sheet of the hexamer (Figure 1B). A series of hydrogen bonds between Tyr56 (β4) and Tyr63′ (β5′) effectively latches this interface together (Figure 3).
Fig. 3. The Hfq dimer interface. (A) A cross-eyed stereo view of contacts within the Hfq dimer interface. One subunit is red and the other yellow. Interacting residues are shown as ball and sticks, and are labeled. (B) Simulated annealing 2Fo – Fc composite omit map (calculated with a starting temperature of 1500 K) contoured at 1.8σ showing the Tyr63 and Tyr56′ ‘kissing’ interactions that contribute to dimer and hexamer oligomerization. This figure and Figure 4A were made using O (Jones et al., 1991).
Two structural properties set Hfq apart from the other Sm proteins. First, Hfq contains only a very short turn between its Sm1 and ‘Sm2’ motifs (Figure 2B), whereas in other Sm proteins, not only are β-strands β3 and β4 extended to form a longer antiparallel sheet, but the region connecting these strands consists of a long turn or loop (Figure 2A and C). Secondly, Hfq oligomerizes to form a hexamer rather than a heptamer. The present study suggests that these two features may be correlated in the oligomerization of Hfq. Specifically, when an Hfq subunit is superimposed onto a subunit of any Sm protein, the absence of a large variable region within the Hfq subunit allows the neighboring subunit (i.e. of a dimer) to rotate closer. Such rotation is precluded in the other Sm proteins where the presence of a significant variable region structure impinges on the nearby dimer subunit (Figure 2D). Not surprisingly, residues from the variable regions of these Sm proteins contribute to the formation of the dimer interface. Though the abbreviated variable region in Hfq appears to be important in its oligomerization preference, more subtle contributions may also play a role in the oligomerization state of Sm and Sm-like proteins. Indeed, a complete understanding of the factors that govern oligomerization of Sm proteins will require further structural studies.
An internal, circular single-stranded RNA-binding site
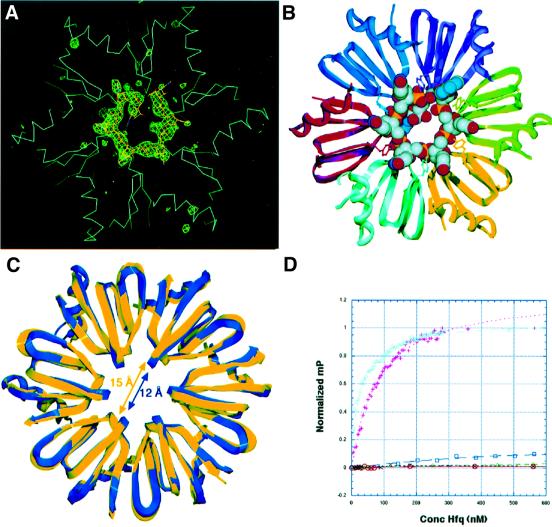
The doughnut-shaped structure and cavity dimensions of Sm proteins and the 10 Å resolution cryo-electron microscopy (EM) structure of the spliceosomal U1 small nuclear ribonucleoprotein particle have led to the idea that these proteins thread single-stranded RNA through their central hole (Kambach et al., 1999; Stark et al., 2001). To elucidate the mechanism of Hfq–RNA binding and, therefore, how Hfq functions to modulate RNA structure, we carried out crystallization trials of Hfq and several A/U-rich RNA sequences which bind Hfq (Figure 4). Data quality crystals of Hfq bound to the ribo-oligonucleotide 5′-AUUUUUG-3′ were obtained. This and similar RNA sequences were chosen for crystallization experiments because recent footprinting studies on Spot42 RNA demonstrated that Hfq binds to U-rich regions (usually stretches that contain four or five uridines) that are surrounded by a 5′ A and a 3′ G (Møller et al., 2002). Notably, 5′-AUUUUUG-3′ is also the canonical sequence recognized by Sm complexes (Kambach et al., 1999; Stark et al., 2001). The structure of the Hfq–RNA complex was solved by molecular replacement using the apo Hfq hexamer as a search model. Following initial refinement of the molecular replacement solution, clear Fo – Fc difference density was observed for the seven nucleotides of the single-stranded RNA molecule (Figure 4A). The current model has been refined to Rwork and Rfree values of 20.4 and 26.6%, respectively, to 2.71 Å resolution.
Fig. 4. Structure of an Hfq–RNA complex. (A) Initial Fo – Fc difference electron density map calculated following one round of SA of the Hfq–RNA complex before inclusion of the RNA. The trace of the Cα backbone of the Hfq hexamer is shown in light blue. The difference map (green mesh) is contoured at 3.4σ. (B) Ribbon diagram of the Hfq–RNA complex with the RNA represented as a CPK model. The oxygen, nitrogen, carbon and phosphorus atoms of the RNA are colored red, blue, gray and yellow, respectively. Also shown as balls and sticks are the Tyr42 residues from each subunit, which stack with the RNA bases. (C) A ribbon diagram of the overlay of the RNA-free (blue) and RNA-bound (yellow) Hfq hexamers. The shift of the loops within the Hfq pore upon RNA binding, which enlarges the pore from 12 to 15 Å, is highlighted by blue and yellow double-headed arrows. (D) Binding isotherms of Hfq to AUUUUUG (RNA-U) (plotted as pink plus signs), AAAAAAG (RNA-A) (cyan triangles), ACCCCCG (RNA-C) (green diamonds), AGGGGGG (RNA-G) (blue squares), dAdAdAdAdAdAdG (DNA-A) (red circles) and double-stranded DNA (black crosses).
In the Hfq–RNA structure, the RNA is bound in a circular, unwound, manner around the pore of the Hfq hexamer within a basic patch, which is observed on only one face of the hexamer (Figure 5A). This highly basic surface is circumscribed by an electronegatively charged region. The opposite side of the hexameric ring, used in crystal packing in both the apo and RNA-bound Hfq structures, is predominantly non-polar (Figure 5B). The AUUUUU nucleotides bind in separate, but linked binding pockets, which spiral around the pore (Figure 4B). Due to the symmetry of the interactions involving the uridines and adenine, it is possible that there is some disorder in which these nucleotides partially occupy all six sites. However, the electron density, in the initial Fo – Fc difference map and subsequent omit maps, indicates that the 3′-guanosine exits the back of the pore in a preferential position. In this location, there are no specific contacts to the guanine base, which instead is encircled by the side chains of His58 from three Hfq subunits and Leu27 from two subunits, whilst its O3′ hydroxyl contacts waters bound around the pore. Notably, the position of the guanine O3′ hydroxyl is not located ideally to allow extension of an RNA molecule through the pore, but rotation of the RNA chain would permit continued threading. Alternatively, the position of the guanosine might be the simple consequence of the small RNA used in this study. Thus, it is not possible to determine from this structure whether Hfq threads RNA through its pore. Clearly, additional structures of Hfq bound to longer RNA sites are needed.
Fig. 5. Electrostatic surface representation of Hfq in the Hfq–RNA complex. (A) Electrostatic surface representation of the RNA-binding side of the Hfq hexamer. Blue is electropositive and red is electronegative. The RNA is shown as a stick model, with oxygen, nitrogen, carbon and phosphorus atoms colored red, blue, white and yellow, respectively. The RNA-binding surface is clearly electropositive. Both (A) and (B) were made with GRASP (Nicholls et al., 1991). (B) Electrostatic surface representation of the opposite side of the Hfq hexamer, which highlights its non-polar character. This surface packs against itself in both RNA-bound and RNA-free Hfq crystal forms (see Figure 1C).
Interestingly, although very similar (the r.m.s.d. between all corresponding Cα atoms = 1.4 Å), the RNA-bound and apo Hfq structures display significant differences (Figure 4C). Namely, aside from small conformational differences in the variable region loops (which are also observed in the apo structure alone), loop 5 of the central binding ring at the mouth of the pore, which contains the conserved YKH motif, shifts appreciably in each subunit upon RNA binding. This leads to the expansion of the central pore from 12 Å in apo Hfq to 15 Å at the closest distance between loops (Figure 4C). Because this conformational change widens the pore at its smallest constriction, threading of the RNA through the pore could be facilitated.
The Hfq Sm1 and Sm2 motif: conserved residues mediate RNA interactions
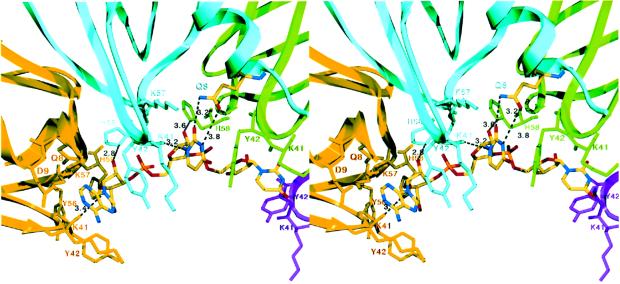
The Hfq–RNA structure reveals that Hfq utilizes residues from the Sm1 and Sm2 motifs from two adjacent subunits to build the six nucleotide-binding pockets (Figure 6). There are no intramolecular base stacking interactions within the RNA nucleotides, all of which adopt the C2′ endo conformation. The C2′ endo sugar pucker allows the bases to take a more extended conformation and fit into the individual binding pockets. A striking aspect of the Hfq–RNA interaction is the circularly permuted stacking of the extended RNA bases within the binding pore such that each base is sandwiched between two Tyr42 side chains, which are located on loop 3 (Figures 4B and 6). Residue Tyr42 is within the Sm1 motif, and examination of other Hfq protein sequences reveals that all Hfq proteins contain either a tyrosine or, more commonly, a phenylalanine at this position (Figure 2A). The presence or absence of the hydroxyl moiety appears to be of little functional consequence. In the AF-Sm1–RNA complex, the tyrosine (phenylalanine) is replaced by a histidine, which also stacks against the RNA bases (Törö et al., 2001).
Fig. 6. RNA recognition by Hfq. Cross-eyed stereo view of the 5′ adenine- and uracil-binding pockets. Hydrogen bonds are shown as black lines. All residues that contact the nucleotide are located within the Sm1 and Sm2 motifs (shown as sticks), with the exception of conserved residue Gln8, which is located on helix α1 and colored according to atom type (blue, red and yellow for nitrogen, oxygen and carbon).
One additional residue from the Hfq Sm1 motif, Lys41, also contributes to RNA binding via both its carbonyl oxygen and side chain amino group. Specifically, in each binding pocket, the Lys41 carbonyl oxygen interacts with the N1 atom of each uracil ring and the N9 of the adenine moiety, helping to anchor the bases into their binding pockets (Figure 6). In addition, the Lys41 side chain stacks with the base and contacts the O4 oxygen of some uracils. A possible role for Lys41 is to discriminate against cytidine by donor (N6)– donor (N4) clash. Unexpectedly, a key residue that dictates Hfq–RNA binding specificity is not found within its Sm1 or Sm2 motif. Instead, this residue, Gln8, is located near the N-terminus of α1, from which its Nε and Oε atoms contact the O2 and N3 atoms of the uracil base, respectively (Figure 6). Thus, it is not surprising that this residue is absolutely conserved among Hfq proteins and is, in fact, the only conserved residue within the α1 helix (Figure 2A). In contrast, the key specificity-determining residue of the AF-Sm1 protein appears to be the conserved asparagine within the Sm1 motif (Törö et al., 2001).
Residues within the ‘Hfq Sm2 motif’, namely the highly conserved KH motif, which is located on loop 5 and faces the pore, also contact the RNA. The lysine of the KH motif, Lys57, hydrogen-bonds with the uracil O2 atom, thus adding to the complement of base contacts in the complex and imparting additional discrimination against guanines. The imidazole side chain of conserved residue His58 makes contacts with the phosphate oxygens of one nucleotide as well as the ribose O2′ hydroxyl of the adjacent nucleotide. The latter contact would significantly disfavor DNA binding (Figure 6). Each phosphate is also anchored in the binding pocket by a water coordinated by the amide nitrogens of Lys57 and His58 and the carbonyl oxygen of Tyr42.
Hfq interacts with the 5′ adenine nucleotide in a similar manner but with some differences. In comparison with uracil, the larger adenine base better fills the Hfq base-binding pocket, suggesting a role for size, surface and shape complementarity. The Nε of Gln8 and the Nζ of Lys57 contact the adenine N3 in a manner analogous to the contacts to the O2 of uracil. The longer adenine base also stacks more optimally with the aliphatic atoms of the Lys41 side chain (Figure 6). The Hfq–RNA structure underscores the cardinal importance of protein–base contacts in protein interactions with single-stranded RNA.
Hfq–RNA as a model for Sm–RNA interactions
Recent structures of protein–RNA complexes have begun to reveal important details of these interactions, but the rules that govern specific complex formation are not yet clear. Emerging themes are the use of β-strands as RNA-binding scaffolds and the involvement of phenylalanine and/or tyrosine residues in base stacking with single-stranded RNA targets (Deo et al., 1999; Handa et al., 1999; Allain et al., 2000; Wang and Tanaka-Hall, 2001). Hfq shares these overall features; its structure is composed predominantly of β-strands, and Tyr42 plays a key role in RNA binding by stacking with the RNA bases. In gross overall terms, the Hfq–RNA complex shares some similarity with the TRAP–RNA complex (Antson et al., 1999) in that both oligomeric proteins adopt a symmetric ring and bind their RNA targets in a periodic circular pattern. Yet, in contrast to TRAP, which wraps its single-stranded RNA target like a belt around its outside surface, Hfq encases its RNA in a circularly permuted fashion within its central pore. As Hfq is clearly an Sm protein, it can be postulated that its mechanism of RNA recognition might be utilized by all members of the Sm protein superfamily. This is supported by the similarities in the structures of the AF-Sm1–RNA and our Hfq–RNA complex. Indeed, although the RNA in the AF-Sm1–RNA structure (Törö et al., 2001) was disordered (continuous density was not observed for more than three nucleotides), the nucleotides in that structure were bound similarly around the center of the pore, and were in the extended C2′ endo conformation (Törö et al., 2001).
Nucleotide-binding specificity of Hfq
The additive base contacts provided by Hfq residues Gln8, Lys41 and Lys57 discriminate against cytosine. In addition, the presence of the Gln8 side chain in the binding pocket suggests that guanine-containing nucleotides, in which the base is in the anti conformation, would bind poorly due to steric clash between the Gln8 side chain and the N2 guanine atom. On the other hand, as revealed by the structure, adenine is well accommodated by the Hfq base-binding pockets. Such preferences are consistent with accumulating data indicating that Hfq prefers A/U-rich RNA sequences (Zhang et al., 1998, 2002; Majdalani et al., 2001; Sledjeski et al., 2001; Wassarman et al., 2001; Møller et al., 2002). Moreover, the His58 contact to the O2′ hydroxy of the ribose rings as well as the shape complementarity between Hfq pocket residues and the O2′ hydroxyl group would disfavor DNA binding by Hfq (Takada et al., 1997). Indeed, only one study has suggested that Hfq may bind to DNA. However, the low affinity DNA binding in that study was reported to be non-specific, which sharply contrasts with results obtained from RNA-binding studies. Moreover, this study did not demonstrate that purified Hfq alone could bind DNA. More consistent with our Hfq–RNA structure are studies demonstrating that Hfq is localized mainly with ribosomes in the cytosol, which is consonant with its important RNA-binding translational regulatory roles (Kajitani et al., 1994; Azam et al., 2000).
To address directly the nucleic acid-binding preferences of Hfq, we carried out equilibrium dissociation binding studies using a fluorescence anisotropy/polarization (FA)-based binding assay (Lundblad et al., 1996). FA is a solution-based technique to determine protein–nucleotide and protein–protein affinities. This method permitted us to measure the affinities of several oligoribonucleotides including that crystallized in complex with Hfq, AUUUUUG (RNA-U), and oligoribonucleotides in which the five uracils were substituted by five adenines (RNA-A), five guanines (RNA-G) or five cytosines (RNA-C). In addition, the affinity of the oligodeoxyribonucleotide dAdAdAdAdAdAdG (DNA-A) was determined. The equilibrium dissociation constant, Kd, of Hfq for RNA-A and RNA-U was 30.5 ± 1.5 and 56 ± 2.4 nM, respectively (Figure 4D). In contrast, RNA-C, DNA-A and a double-stranded DNA control showed no binding. RNA-G displayed very weak binding that was not saturated at Hfq concentrations as high as 4000 nM (Figure 4D). These binding data demonstrate high affinity binding of A- or U-rich RNA sequences and no physiologically relevant DNA binding to Hfq, and are in agreement with our assertions derived from the Hfq–RNA crystal structure. Consistent with these binding data, only RNA-U and RNA-A crystallize with Hfq (see Materials and methods).
Implications for Hfq function
Hfq has been proposed to act as an RNA chaperone by its ability to modulate RNA structure. Such a role is supported by the finding that Hfq appears to denature the secondary structure of the 3′ end of the Qβ RNA positive strand (Fernandez et al., 1968; Miranda et al., 1997; Schuppli et al., 1997; Su et al., 1997). Hfq also affects the structure of the 5′-untranslated region of the rpoS gene, whereby it melts out the secondary structure of the translation inhibitor hairpin formed in this region at 37°C (Brown and Elliot, 1996; Muffler et al., 1996; Cunning et al., 1998). The structure of DsrA is also modulated by Hfq binding. Sledjeski and co-workers suggested that Hfq unfolds the first stem–loop of DsrA, thus aiding its binding to the RpoS mRNA leader sequence at low temperatures (Sledjeski et al., 2001). Thus, Hfq may promote RNA–RNA complex formation. Recent data have now demonstrated that, indeed, Hfq facilitates RNA–RNA interactions. Specifically, when Hfq binds to OxyS, a small untranslated RNA regulatory molecule, the interaction of OxyS with its target RNA, fhlA, is facilitated (Zhang et al., 2002). This interaction prevents ribosome binding to the fhlA mRNA, thus repressing its translation. Further, Hfq has been demonstrated to increase the interaction between the small regulatory antisense RNA, spot42 RNA and galK mRNA, thereby down-regulating the translation of the message (Møller et al., 2002). Thus, the regulation of translation by the mediation of RNA–RNA interactions is a critical function of Hfq.
One mechanism by which Hfq can promote RNA–RNA interactions as well as its other functions can be inferred from the Hfq–RNA structure. Simply, when Hfq binds single-stranded RNA, the target site RNA is unwound within its central pore. The structure indicates that Hfq could accommodate A/U-rich binding sites of up to six nucleotides. Such binding and unwinding would strongly destabilize surrounding RNA structures that are located several nucleotides on either side of a binding site, thereby permitting new RNA–RNA interactions, which were precluded previously by intramolecular secondary structure. This supposition is supported by the finding that Hfq binding to its target site within the OxyS RNA destabilizes a short stem–loop structure, which is located within a few nucleotides from its binding site (Zhang et al., 2002).
In conclusion, the structure of Hfq has revealed that it is an Sm protein, despite its lack of strong sequence homology to other Sm proteins and its homo-hexameric oligomerization. The Hfq–RNA structure reveals a striking circular binding mode of the RNA within the central basic pore of Hfq, suggesting a mechanism by which Hfq modulates RNA structure, thus providing insight into its diverse, pleiotropic functions.
Materials and methods
Crystallization and data collection: S.aureus Hfq
The full-length (77 residue) S.aureus Hfq protein was overexpressed in an E.coli Δhfq derivative of the ER2566 strain using the intein system (Impact-CN TM, New England Biolabs) and purified as described (Møller et al., 2002). Prior to crystallization, the protein was dialyzed extensively into a solution of 150 mM NaCl, 25 mM Tris pH 7.5 and 0.5 mM EDTA, to remove excess dithiothreitol (DTT). For crystallization, 20 mg/ml Hfq was mixed 1:1 with a reservoir of 2.5 M ammonium sulfate, 0.4 M acetic acid pH 4.6. The crystals take the space group P21. X-ray intensity data for the initial native and all derivatives were collected on an R-AXIS IV imaging plate system at 298 K and processed with BIOTEX. Cryo-protection conditions subsequently were established in which the crystals are suspended in 30% 2-methyl-2,4-pentanediol (MPD), 0.4 M acetic acid pH 4.6 for 2 min before placing in a liquid nitrogen stream. A 1.55 Å resolution intensity data set was collected at the Stanford Synchrotron Radiation Laboratory (SSRL) beamline BL 9-1 at 100 K and processed with MOSFLM.
Structure determination and refinement of S.aureus Hfq
The Hfq structure was solved by SIR using data from a crystal that was soaked in a 1 mM potassium tetrachloroplatinate solution for 2 days (Table I). The ASU contains two Hfq hexamers, and two sets of six heavy atoms sites (12 total sites) were obtained by difference Patterson and difference Fourier methods. Each set of six is arranged with a near perfect 6-fold symmetry. Heavy atom parameters were refined, multiple isomorphous replacement (MIR) phases calculated and density modification carried out using PHASES-95 (Furey and Swaminathan, 1997) (Table I). The handedness was determined by inspection of the electron density maps. The initial solvent-flattened SIR map revealed most of the β-strands of each hexamer, which were built into the map using O (Jones et al., 1991). Phase combination using this partial model greatly improved the density and permitted the rest of the model to be traced. The two hexamers were then subjected to simulated annealing (SA) using CNS (Brünger et al., 1998), followed by multiple cycles of SA and positional/thermal parameter refinement in CNS and rebuilding in O (Jones et al., 1991). The Rwork and Rfree converged to 21.0 and 28.9%, respectively, using all data to 2.75 Å resolution. This model was used as the starting model for refinement against the 1.55 Å resolution cryo data. Because of significant changes in the unit cell dimensions upon freezing, the structure was repositioned by molecular replacement using EPMR (Kissinger et al., 1999). This model was then subjected to rigid body refinement in CNS followed by SA/positional/thermal parameter refinement. Despite the fact that residues C-terminal to 66, which are not conserved in Hfq proteins, are disordered, the Rfree converged to 25.9%. The final model includes residues 6–66 of eight subunits, 6–65 of three subunits, 1–66 of one subunit, five acetate molecules and 592 water molecules; it has excellent stereochemistry (Table I) and no Ramachandran outliers (Laskowski et al., 1993).
Crystallization and data collection: Hfq–RNA complex
For co-crystallization trials, Hfq was dialyzed as described and mixed at various ratios with a variety of oligoribo- and oligodeoxyribonucleotides. Data quality crystals were obtained with 5′-AUUUUUG-3′ (RNA-U), by combining 0.5–1 mM Hfq and 1 mM RNA, followed by addition of an equal volume of 10% polyethylene glycol (PEG) monomethyl ether 550, 100 mM KCl, 15 mM magnesium chloride and 50 mM Tris pH 7.5. The crystals take the space group C2221. X-ray intensity data were collected at SSRL, beamline BL 9-1 at 298 K and processed with MOSFLM. Attempts were also made to crystallize Hfq with DNA-A, RNA-A, RNA-C and RNA-G. Thus far, only Hfq–(RNA-U) and small Hfq–(RNA-A) crystals have been obtained.
Structure determination and refinement of the Hfq–RNA structure
The Hfq–RNA structure was solved with the molecular replacement program MolRep in the CCP4 package (CCP4, 1994). The initial R-factor was 43.0%. The molecular replacement model was subjected to rigid body refinement followed by SA, after which an electron density map was calculated. The Fo – Fc map clearly revealed density for all phosphates and six bases of the RNA (Figure 4A). The RNA was built into the model and refined in CNS (Brünger et al., 1998). The ASU contains one Hfq hexamer and a 7mer RNA fragment. Following multiple cycles of SA/positional/thermal parameter refinement to 2.71 Å resolution, the model converged to an Rwork of 20.4% and an Rfree of 26.6%. The final model includes residues 6–65 of two subunits, residues 6–66 of four subunits, 7 nucleotides and 29 solvent molecules, and has excellent stereochemistry (Table II).
Table II. Selected crystallographic data for the Hfq–RNA structure determination.
| Space group | C2221 |
| Cell constants (Å) | a = 80.9, b = 115.6, c = 101.8 |
| Temperature (K) | 298 |
| Resolution (Å) | 2.71 |
| Overall Rsym (%)* | 6.4 |
| Overall I/σ(I) | 6.2 |
| Total reflections (no.) | 40 717 |
| Unique reflections (no.) | 11 950 |
| High resolution shell (Å) | 2.80–2.71 |
| Rsym(%)a | 36.7 |
| I/σ(I) | 2.0 |
| Refinement statistics | |
| Completeness (%) | 90.3 |
| Resolution range (Å) | 40.4–2.71 |
| Rwork (%)b/Rfree (%)c | 20.4/26.6 |
| R.m.s.ds | |
| Bond angles (°) | 1.20 |
| Bond lengths (Å) | 0.007 |
| B-values (Å2) | 2.7 |
| <B protein> (Å2) | 65.1 |
| <B RNA> (Å2) | 73.5 |
| Solvent (no.) | 29 |
| Ramachandran analysis | |
| Most favored (%/no.) | 86.9/291 |
| Additional allowed (%/no.) | 11.9/40 |
| Generously allowed (%/no.) | 1.2/4 |
| Disallowed (%/no.) | 0.0 |
aRsym = ΣΣ|Ihkl – Ihkl(j)|/ΣIhkl, where Ihkl(j) is the observed intensity and Ihkl is the final average value of intensity.
bRwork = Σ||Fobs| – |Fcalc||/Σ|Fobs|.
cRfree = Σ||Fobs| – |Fcalc||/Σ|Fobs|, where all reflections belong to a test set of 5% of the data randomly selected and not used in atomic refinement.
Fluorescence anisotropy/polarization
Fluorescence anisotropy/polarization measurements were collected with a PanVera Beacon Fluorescence Polarization System. Samples were excited at 490 nm and emission was measured at 530 nm. 5′-fluoresceinated oligonucleotides were purchased from Oligos Etc. (Wilsonville, OR). The binding buffer used for all measurements contained 20 mM sodium phosphate pH 7.0, 150 mM NaCl and 0.5 mM EDTA. Hfq (in 50 mM Tris 7.5, 150 mM NaCl) was serially titrated into the cuvette, which contained 1.5 nM 5′-fluoresceinated oligonucleotide. The measurements were performed at 298 K. Samples were incubated 15 s prior to each measurement, ensuring equilibrium binding. The data were plotted using Kaleidagraph, and the generated curves were fit by non-linear least squares regression assuming a bimolecular model such that the Kd values represent the protein concentration at half-maximal oligonucleotide binding. The fluoresceinated RNA-C, DNA-A and double-stranded DNA (top strand F-GAAAAAGAAAAGCTTTGC TTAGGG/plus a complementary strand without a 5′-fluorescein label) showed no binding even at Hfq concentrations >3000 nM. The fluoresceinated RNA-G oligonucleotide showed very weak and unsaturable binding up to 4000 nM Hfq. This latter binding isotherm was normalized to those obtained for the fluoresceinated RNA-U and RNA-A curves by assuming the same mPmax.
Coordinates
Coordinates and structure factors for the apo Hfq and the Hfq–RNA complex have been deposited with the Protein Data Bank under the accession codes 1QK1 and 1QK2.
Acknowledgments
Acknowledgements
Intensity data collection at the Stanford Synchrotron Radiation Laboratory (SSRL) was carried out under the auspices of the SSRL biotechnology program, which is supported by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program and by the Department of Energy, Office of Biological and Environmental Research. M.A.S. is a Burroughs Wellcome Career Development Awardee of Biomedical Science. This work was supported by grants GM 49244 from the National Institutes of Health to R.G.B and the Danish Natural Science Research Council to P.V-.H.
References
- Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of the U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain F.H., Bouvet,P., Dieckmann,T. and Feigon,J. (2000) Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J., 19, 6870–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antson A.A., Dodson,E.J., Dodson,G., Greaves,R.B., Chen, X-P. and Gollnick,P. (1999) Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature, 401, 235–424. [DOI] [PubMed] [Google Scholar]
- Azam T.A., Hiraga,S. and Ishihama,A. (2000) Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells, 5, 613–626. [DOI] [PubMed] [Google Scholar]
- Barton G.J. (1993) ALSCRIPT a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Bouveret E., Rigaut,G., Shevchenko,A., Wilm,M. and Séraphin,B. (2000) An Sm-like protein complex that participates in mRNA degradation. EMBO J., 19, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol,A., Ebel,J.P., Lazar,E., Haendler,B. and Jacob,M. (1982) U2 RNA shares a structural domain with U1, U4 and U5 RNAs. EMBO J., 1, 1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L. and Elliot,T. (1996) Efficient translation of RpoS σ factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol., 178, 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cooper M., Johnston,L.H. and Beggs,J.D. (1995) Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J., 14, 2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B.M., Harrop,S.J., Kornfeld,G.D., Dawes,I.W., Curmi,P.M.G. and Mabbutt,B.C. (2002) Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol., 309, 915–923. [DOI] [PubMed] [Google Scholar]
- Cunning C., Brown,L. and Elliot,T. (1998) Promoter substitution and deletion analysis of upstream region required for rpoS translation regulation. J. Bacteriol., 180, 4564–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo R.C., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Recognition of polyadenylate RNA by the poly(A) binding protein. Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- Fernandez F.M., Eoyang,L. and August,J.T. (1968) Factor fraction required for the synthesis of bacteriophage Qβ RNA. Nature, 219, 599–590. [DOI] [PubMed] [Google Scholar]
- Furey W.B. and Swaminathan,S. (1997) PHASE-95: a program package for the processing and analysis of diffraction data from macromolecules. Methods Enzymol., 277, 590–620. [DOI] [PubMed] [Google Scholar]
- Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hajnsdorf E. and Régnier,P. (2000) Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl Acad. Sci. USA, 97, 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H. and Shimura,Y. (1999) Structural basis for the recognition of the tra mRNA precursor by the sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- Hermann H., Fabrizio,P., Raker,V.A., Foulaki,K., Hornig,H., Brahms,H. and Lührmann,R. (1995) SnRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J., 14, 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.- Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kajitani M., Kato,A., Wada,A., Inokuchi,Y. and Ishihama,A. (1994) Regulation of the Escherichia coli hfq gene encoding the host factor for phage Qβ. J. Bacteriol., 176, 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C., Walke,S., Young,R., Avis,J.M., de la Fortelle,E., Raker, V.A., Lührmann, R, Li,J. and Nagai,K. (1999) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell, 96, 375–387. [DOI] [PubMed] [Google Scholar]
- Kissinger C.R., Gehlaer,D.K and Fogel,D.B. (1999) Rapid automated molecular replacement by evolutionary search. Acta Crystallogr., D55, 484–491. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W. and Thorton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lundblad J.R., Laurance,M. and Goodman,R.H. (1996) Fluorescence polarization analysis of protein–DNA and protein–protein interactions. Mol. Endocrinol., 10, 607–612. [DOI] [PubMed] [Google Scholar]
- Majdalani N., Chen,S., Murrow,J., St John,K. and Gottesman,S. (2001) Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol., 39, 1382–1394. [DOI] [PubMed] [Google Scholar]
- Miranda G., Schuppli,D., Barrera,I.C., Hausherr,C., Sogo,J.M. and Weber,H. (1997) Recognition of bacteriophage Qβ plus strand RNA as a template by Qβ replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol., 267, 1089–1103. [DOI] [PubMed] [Google Scholar]
- Møller T., Franch,T., Højrup,P., Keene,D.R., Bächinger,H.P., Brennan,R.G. and Valentin-Hansen,P. (2002) Hfq: a bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell, 9, 23–30. [DOI] [PubMed] [Google Scholar]
- Muffler A., Fischer,D. and Hengge-Aronis,R. (1996) The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev., 10, 1143–1151. [DOI] [PubMed] [Google Scholar]
- Muffler A., Traulsen,D.D., Fischer,D., Lange,R. and Hengge-Aronis,R. (1997) The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli.J. Bacteriol., 179, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura C., Cascio,D., Sawaya,M.R. and Eisenberg,D.S. (2001) The crystal structure of a heptameric archaeal Sm protein: implications for the eukaryotic snRNP core. Proc. Natl Acad. Sci. USA, 98, 5532–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B.H. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Will,C.L., Lührmann,R., Schümperli,D. and Müller,B. (2001) Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J., 20, 5470–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J., Bragado-Nilsson,E., Kandels-Lewis,S. and Séraphin,B. (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J., 18, 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B. (1995) Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J., 14, 2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppli D., Miranda,G., Tsui,H.C.T., Winkler,M.E. and Wever,H. (1997) Altered 3′-terminal RNA structure in phage Qβ adapted to host factor-less Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 10239–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D.D., Whitman,C. and Zhang,A. (2001) Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol., 183, 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H., Dube,P., Lührmann,R. and Kastner,B. (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature, 409, 539–541. [DOI] [PubMed] [Google Scholar]
- Su Q., Schuppli,D., Tsui,H.C.T., Winkler,M.E. and Weber,H. (1997) Strongly reduced phase Qβ replication, but normal phage MS2 replication in an Escherichia coli K12 mutant with inactivated Qβ host factor (hfq) gene. Virology, 25, 189–199. [DOI] [PubMed] [Google Scholar]
- Takada A., Wachi,M., Kaidow,A., Takamura,M. and Nagai,K. (1997) DNA binding properties of the hfq gene product of Escherichia coli.Biochem. Biophys. Res. Commun., 236, 576–579. [DOI] [PubMed] [Google Scholar]
- Tharun S., He,W., Mayes,A.E., Lennertz,P., Beggs,J.D. and Parker,R. (2000) Yeast Sm-like proteins functions in mRNA decapping and decay. Nature, 404, 515–518. [DOI] [PubMed] [Google Scholar]
- Törö I., Thore,S., Mayer,C., Basquin,J., Séraphin,B. and Suck,D. (2001) RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J., 20, 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H.C.T., Leung,H.C. and Winkler,M.E. (1994) Characterization of broadly pleotropic phenotypes caused by an hfq insertion mutation in an Escherichia coli K12. Mol. Microbiol., 13, 35–49. [DOI] [PubMed] [Google Scholar]
- Vytvytska O., Moll,I., Kaberdin,V.R., Gabain,A. and Bläsi,U. (2000) Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev., 14, 1109–1118. [PMC free article] [PubMed] [Google Scholar]
- Walke S., Bragado-Nilsson,E., Séraphin,B. and Nagai,K. (2001) Stoichiometry of the Sm proteins in yeast spliceosomal snRNPs supports the heptamer ring model of the core domain. J. Mol. Biol., 308, 49–58. [DOI] [PubMed] [Google Scholar]
- Wang X. and Tanaka-Hall,T.M. (2001) Structural basis for recognition of AU-rich element RNA by Hu proteins. Nat. Struct. Biol., 8, 141–145. [DOI] [PubMed] [Google Scholar]
- Wassarman K.M., Repoila,F., Rosenow,C., Storz,G. and Gottesman,S. (2001) Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev., 15, 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Altuvia,S., Tiwari,A., Argaman,L., Hengge-Aronis,R. and Storz,G. (1998) The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J., 17, 6061–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Wassarman,K.M., Ortega,J., Steven,A.C. and Storz,G. (2002) The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell, 9, 11–22. [DOI] [PubMed] [Google Scholar]