Abstract
Known eukaryotic selenocysteine (Sec)-containing proteins are animal proteins, whereas selenoproteins have not been found in yeast and plants. Surprisingly, we detected selenoproteins in a member of the plant kingdom, Chlamydomonas reinhardtii, and directly identified two of them as phospholipid hydroperoxide glutathione peroxidase and selenoprotein W homologs. Moreover, a selenocysteyl-tRNA was isolated that recognized specifically the Sec codon UGA. Subsequent gene cloning and bioinformatics analyses identified eight additional selenoproteins, including methionine-S-sulfoxide reductase, a selenoprotein specific to Chlamydomonas. Chlamydomonas selenoprotein genes contained selenocysteine insertion sequence (SECIS) elements that were similar, but not identical, to those of animals. These SECIS elements could direct selenoprotein synthesis in mammalian cells, indicating a common origin of plant and animal Sec insertion systems. We found that selenium is required for optimal growth of Chlamydomonas. Finally, evolutionary analyses suggested that selenoproteins present in Chlamydomonas and animals evolved early, and were independently lost in land plants, yeast and some animals.
Keywords: Chlamydomonas reinhardtii/evolutionary analysis/SECIS elements/selenoproteins
Introduction
Selenocysteine (Sec) is a rare amino acid found in several proteins from various domains of life (Low and Berry, 1996; Stadtman, 1996; Rother et al., 2001a; Hatfield and Gladyshev, 2002). It is inserted into protein co-translationally in response to the codon UGA and the specific Sec insertion machinery. The Sec insertion machinery includes a cis-acting mRNA structure, designated the Sec insertion sequence (SECIS) element, and the trans-acting factors Sec tRNA, selenophosphate synthetase, Sec synthase, Sec-specific elongation factor and a SECIS-binding protein.
Sec-containing proteins have been identified in bacteria, archaea and eukaryotes, and the universal use of UGA to designate Sec in these organisms suggests a common origin of the Sec insertion system (Gladyshev and Kryukov, 2001). However, besides selenophosphate synthetase, which is involved in Sec biosynthesis (Stadtman, 1996), there is no overlap between the sets of prokaryotic and eukaryotic selenoproteins. Moreover, selenoproteins in prokaryotes are typically involved in catabolic processes, whereas eukaryotes employ selenoproteins for biosynthetic and antioxidant processes.
Details of Sec evolution are scarce and largely unclear (Atkins et al., 1999; Gladyshev and Kryukov, 2001). For example, while all currently known eukaryotic selenoproteins are of animal origin, no selenoproteins have been described in non-animal eukaryotes. Moreover, when genomes of the plant Arabidopsis thaliana and the yeast Saccharomyces cerevisiae were sequenced, their analysis revealed neither selenoprotein genes nor any of the components of the Sec insertion pathway. This lack of Sec-containing proteins contrasts with the essential role of selenoproteins in animals and bacteria (reviewed in Low and Berry, 1996; Stadtman, 1996). For example, disruption of the Sec tRNA gene in mice results in an inability to synthesize selenoproteins and embryonic lethality (Bosl et al., 1997), and mutation in a fruit fly gene for selenophosphate synthetase is also lethal (Serras et al., 2001). Sec-containing proteins are essential for Escherichia coli when grown under anaerobic conditions (Bock et al., 1991).
Characterization of Sec-containing proteins and Sec insertion systems in non-animal eukaryotes may elucidate these seemingly contradictory observations in Sec evolution and its essential requirement for some organisms. One report described the presence of a possible Sec-containing glutathione peroxidase 1 (GPx1) in a model plant system, Chlamydomonas reinhardtii (Shigeoka et al., 1991). This protein was isolated directly from green algae, reacted with antibodies raised against mammalian GPx1 and found to contain a stoichiometric amount of selenium. It was not clear whether Sec was present in the protein as no protein or nucleotide sequences were reported for Chlamydomonas GPx1.
In this report, we identified and characterized selenoproteins in C.reinhardtii. Remarkably, this organism contains at least 10 natural selenoproteins, including one that is specific to green algae. We also report the identification of Sec tRNA and the finding that Chlamydomonas requires selenium for optimal growth. These data are discussed with respect to evolutionary events that led to the accumulation and loss of the Sec insertion system in eukaryotes.
Results
Chlamydomonas contains specific selenoproteins
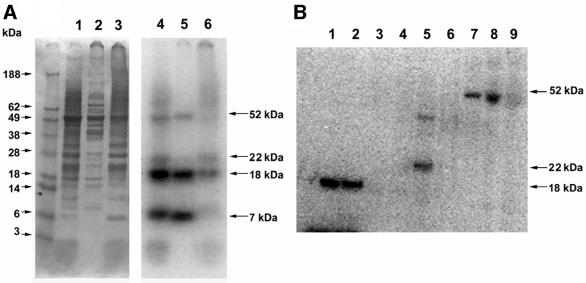
Chlamydomonas reinhardtii cells were labeled with 75Se and the cell extracts were analyzed by SDS–PAGE (Figure 1A, left panel) and detection of radioactivity on the gels (Figure 1A, right panel). This revealed four 75Se-labeled proteins that migrated as 52, 22, 18 and 7 kDa bands, with the 7 and 18 kDa proteins being the most abundant soluble selenium-containing proteins (Figure 1A, right panel, lane 5). This 75Se pattern did not match a Coomassie Blue staining profile (Figure 1A, left panel), suggesting the presence of specific Sec-containing proteins rather than labeling of proteins through non-specific selenium incorporation. Furthermore, the radioactive labeling pattern did not match that of animal cells, in which the major 75Se-labeled selenoproteins are a 25 kDa GPx1, a 20 kDa phospholipid hydroperoxide glutathione peroxidase, a 57 kDa thioredoxin reductase 1 and a 15 kDa Sep15 (Gladyshev et al., 1999a). To determine the identity of the Chlamydomonas selenoproteins, a large-scale purification of these proteins was conducted.
Fig. 1. Detection of Chlamydomonas selenoproteins. Chlamydomonas cells were grown in the presence of 75Se, collected by centrifugation, disrupted by sonication, and the resulting homogenate centrifuged at 18 000 g for 30 min. In (A), fractions were analyzed by SDS–PAGE. Lanes 1–3 (left panel) show homogenate, supernatant and pellet, respectively, stained with Coomassie Blue (protein markers are shown in the left-most lane and their sizes indicated on the left) and lanes 4–6 (right panel) show respective lanes in the same gel exposed to PhosphorImager detection of 75Se (sizes of detected selenoproteins are shown on the right). In (B), the soluble fraction [shown in (A), lane 2] was fractionated on a Q-Sepharose column. Lanes 1 and 2 show the flow-through fractions and contained the 7 and 18 kDa selenoproteins, and lanes 3–9 the fractions that eluted in the salt gradient (see Materials and methods). The 22 kDa selenoprotein shown in lane 5 eluted at ∼150 mM NaCl, and the 52 kDa selenoprotein shown in fractions 7–9 eluted at 400 mM NaCl in buffer B. Proteins were detected by PhosphorImager analysis of an SDS–PAGE gel.
Ammonium sulfate fractionation revealed that four proteins were present in a 20–80% ammonium sulfate fraction (not shown), which was subsequently chromatographed on an anion-exchange column (Figure 1B). This procedure separated the 22 and 52 kDa from the 7 and 18 kDa selenoproteins, with the latter two proteins being present in the flow-through fraction. Subsequent purification focused on the 7 and 18 kDa proteins due to their abundance in Chlamydomonas cells.
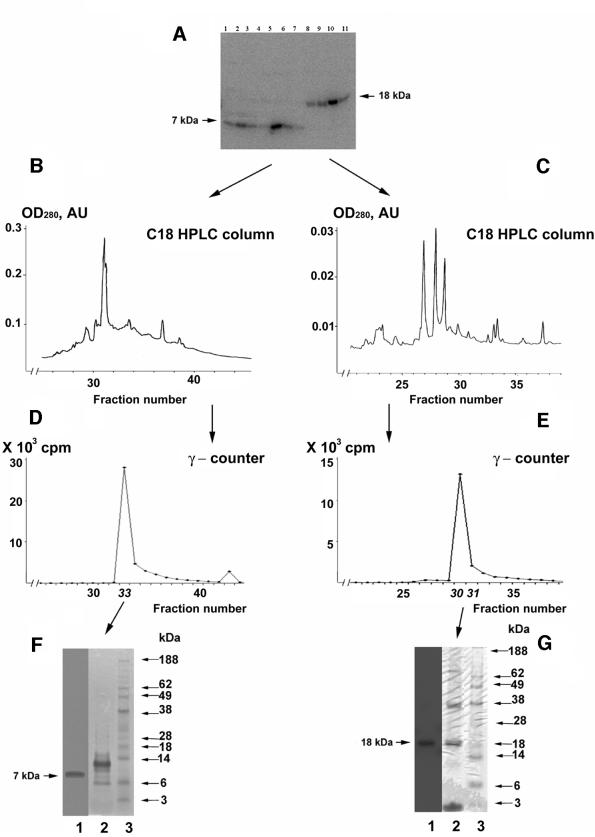
The use of HPLC hydrophobic interaction chromatography as the next purification step resulted in separation of the 7 and 18 kDa proteins (Figure 2A). Finally, the fractions containing these selenoproteins were fractionated separately on a C18 reversed-phase HPLC column (Figure 2B and C). Radioactive fractions corresponding to the 7 kDa selenoprotein eluted at 49% acetonitrile (Figure 2D) and the 18 kDa selenoprotein fraction at 47% acetonitrile (Figure 2E). These fractions were analyzed on SDS–PAGE gels (Figure 2F and G), and the protein bands were cut from the gels and analyzed by Edman degradation. Both selenoprotein sequences were found to be blocked, whereas protein contaminants in the selenoprotein bands were identified by N-terminal sequencing as ubiquitin (7 kDa band) and cyclophilin 1 (18 kDa band). To identify selenoproteins, the two selenoprotein bands cut from SDS–PAGE gels were digested with trypsin and analyzed by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (µLC/MS/MS). Twenty-one peptides derived from the 18 kDa selenoprotein band (Table I) and 33 peptides from the 7 kDa band (Table II) were sequenced by MS/MS.
Fig. 2. Purification of Chlamydomonas 7 and 18 kDa selenoproteins. The 7 and 18 kDa proteins were purified from the radioactive flow-through fractions from Q-Sepharose (see Figure 1B) by concentrating this material with ammonium sulfate, adjusting to a concentration of 0.4 M ammonium sulfate in buffer B, and fractionating on a phenyl–Sepharose HPLC column. Samples were analyzed as shown in (A): lanes 1–7 contain fractions eluted in a 0.4–0 M ammonium sulfate gradient in buffer B and lanes 8–11 show fractions eluted in a buffer B to water gradient as detected by PhosphorImager analysis; or further purified as shown in (B), (D) and (F): fractionation of the 7 kDa selenoprotein on a C18 reversed-phase HPLC column with a 0–70% acetonitrile gradient in 0.05% trifluoroacetic acid, γ-counter detection of 75Se in the eluted fraction, and SDS–PAGE analysis of fraction 33, respectively; and in (C), (E) and (G): fractionation of the 18 kDa selenoprotein on a C18 reversed-phase HPLC column with a 0–70% acetonitrile gradient in 0.05% trifluoroacetic acid, γ-counter detection of 75Se in the eluted fractions, and SDS–PAGE analysis of fraction 31, respectively. Lane 1 in (F) and (G) shows detection of 75Se by PhosphorImager analysis; lanes 2 and 3 show staining with Coomassie Blue of the column fractions and protein standards (masses are shown on the right).
Table I. Identification of Chlamydomonas PHGPx1 by tandem mass spectrometry.
Table II. Identification of Chlamydomonas SelW1 by tandem mass spectrometry.
| Protein | DDBJ/EMBL/GenBank accession No. | Nos of sequenced peptides |
|---|---|---|
| Selenoprotein W1 (7 kDa) | 3 | |
| Ubiquitin/ribosomal protein | UQKM | 5 |
| Enolase (2-phospho-d-glycerate hydrolyase) | P31683 | 1 |
| Oxygen-evolving enhancer protein 1 | P12853 | 2 |
| Probable acyl-coenzyme a binding | 15368290 | 4 |
| Probable RSZp22 splicing factor | 8286966,14242322 | 7 |
| Probable RNA-binding protein | 14237335 | 3 |
| Chitinase-like protein | 10775248 | 2 |
| DnaJ (Hsp40)-like protein | 15371690 | 2 |
| Unknown protein | 6548300 | 1 |
| Unknown protein | 8287459 | 1 |
| Chloroplast Cpn21-like protein | 6551572 | 1 |
| FtsH-like protease | 6549612 | 1 |
| Total: 13 | Total: 33 |
Selenoprotein W and glutathione peroxidase are major Chlamydomonas selenoproteins
Sequence analyses of MS/MS peptides revealed that the 7 kDa selenoprotein was a homolog of animal selenoprotein W (Whanger, 2000) [three peptides matched an open reading frame (ORF) predicted from Chlamydomonas expressed sequence tag (EST) sequences; Table II]. The 18 kDa protein was identified as a homolog of eukaryotic phospholipid hydroperoxide glutathione peroxidases by matches of five peptides to the ORF predicted from EST sequences (Ursini et al., 1997) (Table I). These two most abundant Chlamydomonas selenoproteins are designated as SelW1 and PHGPx1 in this paper.
In addition to the two selenoproteins, MS/MS analy sis identified 12 proteins that were either known Chlamydomonas proteins or Chlamydomonas homologs of proteins characterized in other species. Two additional proteins were also identified that have not been described previously in other organisms, but were represented by EST sequences (Tables I and II). A total of 16 Chlamydomonas proteins were identified by MS/MS analysis.
Sec is present in SelW1 and PHGPx1 sequences and is encoded by UGA
Based on the three SelW1 and five PHGPx1 peptide sequences, corresponding partial ORFs were constructed from EST sequences. At the time of analysis, SelW1 was not represented by EST sequences to generate a full cDNA sequence. Therefore, SelW1 cDNA was directly cloned from a Chlamydomonas cDNA library as described in Materials and methods. PHGPx1 and SelW1 ORFs contained in-frame UGA codons that corresponded to the Sec codons in their mammalian homologs (Figure 3A and B). Several PHGPx1 and SelW1 peptides that were encoded by nucleotide sequences located downstream of UGA codons were directly sequenced by MS/MS (Tables I and II), confirming that the UGA codon in these genes were read through and did not serve as terminator signals (Figure 3A and B). These observations, along with the fact that the isolated proteins contained selenium, indicated that UGA codons in SelW1 and PHGPx1 genes encoded Sec.
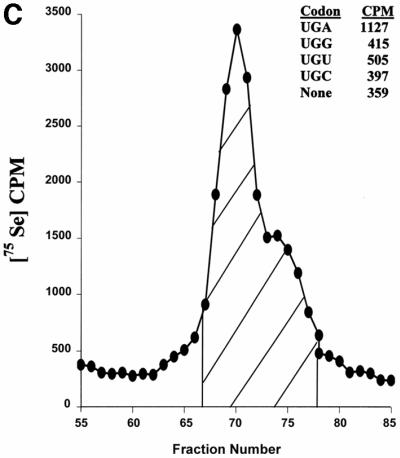
Fig. 3. Selenoproteins and Sec tRNA in Chlamydomonas. (A) Alignment of human, Arabidopsis, Clostridium, C.elegans, Schistosoma mansoni, yeast and Chlamydomonas PHGPx. The sequences of five tryptic peptides of Chlamydomonas PHGPx1, for which amino acid sequences were determined experimentally, are underlined. The DDBJ/EMBL/GenBank accession Nos are: NP_002076.1, human PHGPx; BF936124.1, Schistosoma PHGPx; AE007667.1, Clostridium PHGPx; AE007667.1, S.cerevisiae PHGPx; NP_497078.1, C.elegans PHGPx; AV623602, Chlamydomonas PHGPx1; BI721156 and AV623602, Chlamydomonas PHGPx2. Sec is present in human, Schistosoma and Chlamydomonas proteins, but is replaced by Cys in other PHGPx homologs. (B) Chlamydomonas cDNA sequence encoding selenoprotein W1. The amino acid U represents selenocysteine-14, which is encoded by TGA (underlined). The sequences of three tryptic peptides, for which amino acid sequences were determined experimentally, are also underlined. In the 3′-UTR, the position of the SECIS element is shown. (C) Chromatography of Chlamydomonas [75Se]selenocysteyl-tRNA. Chlamydomonas cells (2.5 g) were labeled with 75Se, and the labeled tRNA extracted and chromatographed on an RPC-5 column as described in Materials and methods. The large peak and trailing shoulder of 75Se-containing tRNA were pooled, as shown by the hatched area in the figure, prepared for ribosomal binding studies and the ribosomal binding studies carried out as described in Materials and methods. Binding to ribosomes in the absence of codon (designated None) or in the presence of codon is shown in the figure. Total CPM added to each reaction were 3200.
Chlamydomonas Sec tRNA decodes UGA
The finding of two specific Sec-containing proteins was surprising in that neither plants nor yeast are known to contain such proteins. Since Sec is inserted into all known natural selenoproteins by Sec tRNA, we isolated and characterized the Chlamydomonas Sec tRNA that recognizes UGA. Chlamydomonas cells were labeled with 75Se and the resulting labeled tRNA chromatographed on an RPC5 column (Figure 3C). The major peak and trailing shoulder were pooled as shown in the figure. The coding properties of this labeled tRNA were determined and it was found to recognize UGA, but not UGU, UGC (Cys codons) or UGG (Trp codon). These data strongly suggest that this 75Se-labeled tRNA recognizes specifically UGA. The amino acid attached to this tRNA was deacylated and identified as Sec as described in Materials and methods (data not shown). Chlamydomonas therefore contains Sec tRNA that decodes UGA.
Identification of seven additional selenoprotein genes
Chlamydomonas EST and genomic databases were analyzed for the presence of homologs of all known selenoproteins. Partial sequences derived from Chlamydomonas EST clones were identified that corresponded to several animal selenoproteins. Subsequently, whenever possible, EST clones were obtained and sequenced, and the remaining sequences were directly cloned from four Chlamydomonas cDNA libraries. These procedures resulted in the identification of seven additional Chlamydomonas selenoproteins. The ORFs of each of these selenoproteins were found to contain UGA at positions corresponding to Sec residues in animal proteins. The seven selenoprotein sequences were identified as: (i) a homolog of a recently identified mammalian selenoprotein M (further designated as SelM1) (Korotkov et al., 2002) (Supplementary figure A available at The EMBO Journal Online); (ii) a second selenoprotein M homolog (SelM2) (Supplementary figure A); (iii) a selenoprotein T homolog (SelT1) (Kryukov et al., 1999) (Supplementary figure B); (iv) a second selenoprotein W homolog (Supplementary figure C) (SelW2); (v) a second glutathione peroxidase homolog (PHGPx2) (Figure 3A); (vi) a homolog of a recently identified selenoprotein K (SelK1) (G.V.Kryukov and V.N.Gladyshev, unpublished data) (Supplementary figure D); and (vii) a homolog of animal thioredoxin reductase (TR1) (Sun et al., 1999) (Supplementary figure E).
Identification of Chlamydomonas SECIS elements
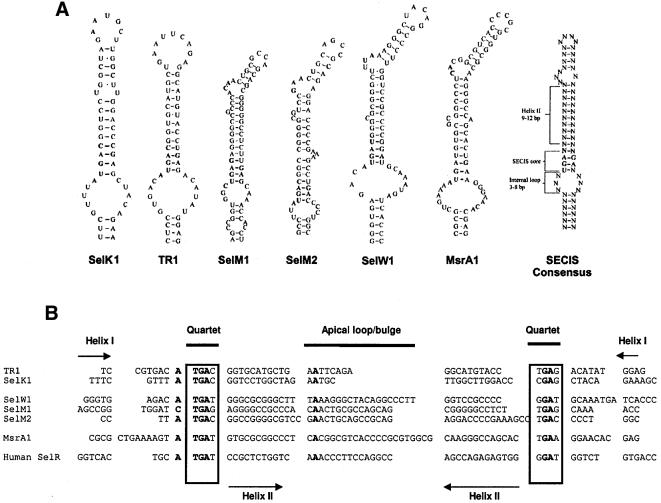
In animals, Sec is inserted in response to UGA when selenoprotein genes contain a stem–loop structure in 3′-UTRs, designated as the SECIS element (Low and Berry, 1996). Archaea also contain SECIS elements in their 3′-UTRs, but the secondary structure is different from that of animals (Rother et al., 2001b). In contrast, bacteria have a third type of SECIS element and it is located in the coding region of selenoprotein genes immediately downstream of Sec-encoding UGA codons (Bock, 2001). To determine which form of SECIS element is present in Chlamydomonas, we analyzed coding and 3′-UTR regions in the alga selenoprotein genes for which full cDNA sequences were available (SelK1, TR1, SelM1, SelM2 and SelW1) and identified SECIS elements in these genes (Figure 4A, five structures from the left) as well as a consensus RNA structural element (Figure 4A, right structure and B). The putative SECIS elements were found to reside in the 3′-UTR of selenoprotein genes. Overall structures of Chlamydomonas SECIS elements were found to be similar to those of animals. Both were composed of a conserved UGAN….NGAN sequence that formed a core (quartet) of non-Watson–Crick interacting nucleotides at the base of a strong helix (Figure 4B) (Low and Berry, 1996; Walczak et al., 1996). On top of the helix, unpaired nucleotides were present in either an apical loop or a bulge. Animal SECIS elements have been classified as Form 1 and Form 2 structures based on whether unpaired nucleotides are located in the loop or bulge (Grundner-Culemann et al., 1999), and both structures were interconvertible by mutations that remove or create a mini-helix in the apical loop. Interestingly, Chlamydomonas SECIS elements also occur as Form 1 (SelK1 and TR1 genes) and Form 2 (SelM1, SelM2 and SelW1 genes) structures (Figure 4A). These findings clearly indicate a common origin of Chlamydomonas and animal SECIS elements, and also distinguish the algal SECIS elements from those of bacterial and archaeal origin.

Fig. 4. SECIS elements in Chlamydomonas selenoprotein genes. (A) SECIS structures. SECIS element structures are shown for selenoproteins indicated below the structures. Proposed Chlamydomonas SECIS element consensus is shown on the right. (B) Alignment of the SECIS elements of Chlamydomonas selenoproteins genes. TR1 and SelK1 structures are Form 1 structures and the others are Form 2 structures. Human SelR SECIS element is aligned for comparison. Conserved nucleotides are shown in bold. Locations of helix 1, helix 2, quartet and apical loop are indicated. (C) Expression of Chlamydomonas SelK1 in mammalian cells. NIH 3T3 cells were transfected with a plasmid encoding GFP-chlamySelK (lane 1) or with GFP-C3 Vector (Clontech) as control (lane 2). Transfected cells were grown in the presence of [75Se]selenite and the resulting 75Se-labeled proteins were resolved by SDS–PAGE and visualized with a PhosphorImager. The location of GFP–SelK1 product is shown on the left. Locations and molecular weights of major selenoproteins, thioredoxin reductase 1 (TR1) and glutathione peroxidase 1 (GPx1), are also indicated. (D) Multiple alignment of Chlamydomonas MsrA1. The accession numbers are: NP_495540.1, C.elegans MsrA; P54149, bovine MsrA; NP_036463.1, human MsrA; NP_290851.1, E.coli MsrA; AAK83645, A.thaliana MsrA; 11342533, Fragaria ananassa MsrA; 7446683, Schizosaccharomyces pombe MsrA; AF494053, C.reinhardtii MsrA1.
In spite of an apparent common origin of animal and Chlamydomonas SECIS elements, the length of a helix that separated the core (quartet) and the unpaired nucleotides in the apical loop or bulge was 12–13 nucleotides in algal structures instead of 11–12 nucleotides as in the animal structures (Low and Berry, 1996; Walczak et al., 1996). Moreover, the identities of the nucleotide located immediately upstream of the quartet (A or G in animals) and the unpaired nucleotides in the loop/bulge (AA or CC in animals) were often different in that Chlamydomonas SECIS elements could tolerate additional nucleotides present in these positions (Figure 4A and B).
Expression of Chlamydomonas SelK1 in mammalian cells
To test whether the Chlamydomonas Sec insertion system was indeed similar to that of animals, we expressed Chlamydomonas SelK1 in mouse NIH 3T3 cells. This protein was selected because its SECIS element, which occurs as the Form 1 structure, was most closely related to animal SECIS elements (Figure 4A, left structure). We developed a construct encoding SelK1 fused in-frame downstream of GFP with the purpose of obtaining a selenoprotein form whose mass is different from those of natural mouse selenoproteins and could therefore be easily detected in transfected cells. When NIH 3T3 cells transfected with the GFP–SelK1 construct were labeled with 75Se, a band corresponding to a fusion protein was detected (Figure 4C). The data suggest that Sec was inserted into Chlamydomonas SelK1 by the mammalian Sec insertion system in response to a Chlamydomonas SECIS element. Since SECIS elements in other Chlamydomonas selenoprotein genes were more distantly related to animal SECIS elements, it is possible that Sec insertion into these algal selenoproteins in mammalian cells could be less efficient. Nevertheless, our data clearly demonstrate that the Chlamydomonas Sec insertion system functionally overlaps with that of animals.
Identification of a Chlamydomonas-specific selenoprotein
The large number of selenoprotein genes detected in Chlamydomonas cells was surprising as most organisms have only a few such genes. For example, the genome of Caenorhabditis elegans contains only a single selenoprotein gene (thioredoxin reductase) (Buettner et al., 1999; Gladyshev et al., 1999b), whereas the Drosophila genome encodes three selenoprotein genes (selenophosphate synthetase, G-rich and BthD) (Castellano et al., 2001; Martin-Romero et al., 2001). Eighteen selenoproteins were previously identified in mammals (Hatfield and Gladyshev, 2002), including homologs of all nine Chlamydomonas selenoproteins described above. Interestingly, in terms of a set of identified selenoproteins, Chlamydomonas appeared to be more similar to mammals than to any other organisms (for example, using BLAST analysis, the closest homolog of Chlamydomonas TR1 was human TR1).
We previously developed a program, called SECISearch, which allowed the identification of selenoprotein genes by searching for sequence, structural and thermodynamic characteristics of SECIS elements in animal nucleotide sequence databases (Kryukov et al., 1999; Martin-Romero et al., 2001). Since animal and Chlamydomonas SECIS elements were closely related, it was attractive to employ this program with the Chlamydomonas databases to search for additional algal selenoprotein genes. However, SECISearch could not be directly applied for identifying Chlamydomonas selenoprotein genes because of the differences in the length of helix 2, in the nucleotides in the apical loop and in the nucleotide preceding the core (quartet) between Chlamydomonas and animal SECIS elements. Therefore, we modified the program as described in Materials and methods to recognize SECIS elements in Chlamydomonas selenoprotein genes and applied the new program, Chlamy SECISearch, to the Chlamydomonas EST database.
A computational screen of 113 064 available sequences revealed only 84 unique sequences that satisfied primary sequence, secondary structure and energetic criteria of Chlamydomonas SECIS elements, including four sequences that corresponded to the already identified selenoprotein SelM1, SelK1, SelW1 and TR1 genes (Table III). The SECIS element in the SelM2 gene was not detected as it did not satisfy the algorithm employed.
Table III. Analysis of Chlamydomonas EST database with Chlamy SECISearch.
| Total No. of sequences | 113 064 (∼80 000 000 nucleotides) |
| Fit primary sequence and secondary structure consensuses | 1502 |
| Satisfy energy and fine structural criteria | 243 |
| Redundancy removal | 84 |
| Correspond to known selenoproteins | 4 |
| New selenoproteins | 1 (MsrA1) |
The remaining sequences were analyzed manually using Mammalian Selenoprotein Signature criteria (e.g. conservation of Sec and the presence of homologs containing cysteine in place of Sec) (Kryukov and Gladyshev, 2002) revealing at least one new SECIS element (Table III; Figure 4A, second structure from the right). The corresponding EST clone was obtained and its sequence determined, revealing a gene encoding methionine-S-sulfoxide reductase (MsrA1) (Figure 4D). The MsrA1 gene contained an in-frame UGA codon at the position corresponding to the active site cysteine present in all known MsrAs (Weissbach et al., 2002). No Sec-containing form of MsrA has been described previously, suggesting that we identified a new eukaryotic selenoprotein.
Selenium is required for optimal growth of Chlamydomonas cells
No requirement for selenium has previously been described for Chlamydomonas and, to our knowledge, no supplementation of the culture media for Chlamydomonas with selenium has been reported. Our initial experiments of supplementing Chlamydomonas culture with selenium as sodium selenite at levels of 0–1 mM revealed no differences in the growth characteristics of selenium-supplemented and -unsupplemented cells grown either photosynthetically or on an acetate-based medium (data not shown). However, selenium is known to be a common contaminant of sulfur-containing compounds. Thus, if the requirement for selenium is low for Chlamydomonas, the use of sulfur compounds of insufficient purity could potentially provide this organism with the required amount of selenium.
To test this possibility, we developed a defined Chlamydomonas medium, which was similar to a regular TAP medium except that it contained all ultra-pure grade chemicals. The growth of Chlamydomonas on this medium in the presence or absence of selenium as sodium selenite revealed that selenium-supplemented cells grew better than unsupplemented cells (Figure 5A). These data suggest that selenium is required for optimal growth of Chlamydomonas. It is very difficult to obtain a selenium-free medium, which may be necessary to determine whether selenium is essential for Chlamydomonas. Our experiments suggest that Chlamydomonas has a low requirement for selenium, possibly in the nanomolar or subnanomolar range.
Fig. 5. Growth of Chlamydomonas in selenium-deficient and -sufficient media. Cells were grown for 110 h in selenium-deficient TAP medium and in the presence of 50 nM selenite as described in Materials and methods. Initial cell counts were 103 cells/ml. Changes in cell numbers were determined with a hemacytometer, and each sample was analyzed three times. Lines were drawn according to a ‘sigmoidal fit’ functions of Origin, version 5.0 (OriginLab Corporation, Northampton, MA). Selenium-deficient (open circles) and selenium-suplemented (closed circles) cells were grown in the presence of 0.5 mM H2O2 (B), 0.75 mM methionine-S-sulfoxide (C) or 0.5 mM H2O2 and 0.75 mM methionine-S-sulfoxide (D) or in the absence of these compounds (A).
Since enzymes with known antioxidant function, including PHGPx, TR and MsrA, were among identified Chlamydomonas selenoproteins, we tested whether selenium-deficient algal cells were more sensitive to treatment with H2O2 and/or methionine-S-sulfoxide. We found no significant differences in sensitivities of selenium-supplemented and -deficient cells. For example, hydrogen peroxide with or without methionine-S-sulfoxide inhibited selenium-deficient and -supplemented cells to a similar extent (Figure 5B–D).
Evolution of Chlamydomonas selenoproteins
The close relationship between Chlamydomonas and mammalian selenoproteins, and the lack of all or almost all of these selenoproteins in insects, nematodes, plants and yeast, raised the possibility that selenoproteins and the Sec insertion system could have been transferred from vertebrates to Chlamydomonas by a horizontal gene transfer. Alternatively, these selenoproteins could have evolved prior to the separation of animal and plant kingdoms, but have been independently lost in invertebrates, land plants and yeast.
To distinguish between these two possibilities, we constructed phylogenetic trees for the 10 identified Chlamydomonas selenoproteins (Figure 6) and determined whether any of the algal selenoproteins were evolutionarily closer to animals than to plants. None of the tested Chlamydomonas sequences formed an alga/animal group, with the plant sequences being more distantly related. Indeed, Chlamydomonas PHGPx1, PHGPx2, SelK1, SelT1 and MsrA1 had close homologs in plants that had cysteine residues in place of Sec, whereas SelW1, SelW2, SelM1 and SelM2 and TR1 were not present in plants and yeast, and thus no evolutionary relationship could be obtained for these five proteins. The data are thus consistent with early evolution of eukaryotic Sec insertion machinery and selenoproteins present in mammals and Chlamydomonas, and their lineage-specific loss in other eukaryotes.
Fig. 6. Phylogenetic analyses of Chlamydomonas selenoproteins. Each selenoprotein is shown in an evolutionary tree as follows. (A) SelW tree. (B) SelT tree. (C) SelK tree. (D) MsrA tree. (E) PHGPx tree. (F) SelM tree. Accession numbers are given in the legends to Figures 3A, 4D and Supplementary figures. Selenium-containing proteins are indicated by asterisks.
Discussion
This study revealed an unexpected abundance of selenoprotein genes in a model plant cell system: the green alga C.reinhardtii. The number of selenoproteins found in Chlamydomonas is greater than in any other characterized non-vertebrate organism, a finding that contrasts sharply with the lack of selenoproteins and the Sec insertion system in land plants. Several identified Chlamydomonas selenoproteins were previously thought to be of animal origin, but our study places their origin much earlier in evolution. These observations have clear implications with regard to early accumulation of selenoproteins and evolution of Sec insertion in eukaryotes. Our study also increased the repertoire of known selenoproteins by adding MsrA to the list of 22 previously identified eukaryotic selenoproteins.
Five selenoproteins identified in Chlamydomonas, including two glutathione peroxidases, SelT1, SelK1 and MsrA1, had close cysteine-containing homologs in plants, suggesting that Sec/cysteine changes were relatively recent events. Interestingly, analysis of the Chlamydomonas EST database revealed that the alga had at least four additional glutathione peroxidases, in which cysteine was present in place of the catalytic Sec (data not shown). Genomic sequencing of Chlamydomonas is in the initial stages, so the number of glutathione peroxidases in this organism may be larger than the six enzymes identified in the present study.
In addition to the identification of cysteine homologs of PHGPx1 and PHGPx2, we detected three cysteine-containing homologs of selenoprotein MsrA1 (data not shown). MsrA proteins are present in all organisms except certain obligatory parasites and hyperthermophiles (Kryukov et al., 2002). For methionine sulfoxide reduction, these enzymes employ a strictly conserved cysteine that is located in the N-terminal portion of the enzymes and is the sulfoxide-attacking group (Lowther et al., 2000; Moskovitz et al., 2000). The presence of Sec in these enzymes has not been described previously. The replacement of the catalytic cysteine with Sec is consistent with the fact that, in all previously characterized selenoproteins, Sec residues are located at enzyme active sites (Low and Berry, 1996; Stadtman, 1996; Hatfield and Gladyshev, 2002). While the presence of Sec in methionine-S-sulfoxide reductases has been found thus far only in Chlamydomonas, methionine-R-sulfoxide reductases may also contain Sec (these proteins have previously been designated as selenoprotein R, and also as selenoprotein X). Selenoprotein forms of methionine-R-sulfoxide reductases appear to be specific for vertebrates (Kryukov et al., 2002). Indeed, we detected three methionine-R-sulfoxide reductases in Chlamydomonas, all containing cysteine in place of the catalytic Sec (data not shown).
The finding of multiple cysteine homologs of PHGPxs and MsrA1 in Chlamydomonas may explain the observation that low levels of these selenoproteins resulting from selenium deficiency did not translate into increased sensitivity of the green alga to either hydrogen peroxide or methionine-S-sulfoxide (Figure 5). Thus, it seems probable that PHGPx and MsrA1 selenoprotein deficiencies were compensated for by their cysteine homologs. On the other hand, we did observe decreased growth rates of selenium-deficient alga. Which selenoprotein(s) contributes to this phenotype is not known.
An additional important finding provided by our study was that Chlamydomonas cells contain a Sec tRNA that specifically recognizes the UGA codon. Identification of UGA-decoding tRNAs that contain Sec attached to them was previously proposed as a means of demonstrating the universal nature of Sec insertion, as such tRNAs were identified in several eukaryotes (Hatfield et al., 1991, 1992). However, the presence of these tRNAs does not always reflect their use in the Sec incorporation machinery. For example, S.cerevisiae was found to contain a minor tRNA with Sec attached to it, but the coding properties of this tRNA could not be determined (Hatfield et al., 1991). Since this tRNA contained Sec, it is not clear what its function is in protein synthesis. It is important in the present study that a Sec tRNA that decodes UGA specifically was identified in Chlamydomonas by the same criteria as those used in the earlier studies (Hatfield et al., 1991, 1992), but this organism clearly has the Sec insertion machinery. It is also of interest to note that our study provides evidence for the early occurrence of Sec tRNA in lower eukaryotes.
Recent genetic analyses reveal an important role of horizontal gene transfer between organisms (Koonin et al., 2001). The examples of gene transfer between bacteria and archaea are numerous, as also are the examples of gene transfer from various organisms, including humans to viruses. However, we found no examples of horizontal gene transfer involving Chlamydomonas selenoproteins (Figure 6). Our interpretation of these data is that most selenoproteins described in this paper probably evolved prior to the separation of animals and plants, and that these proteins were largely lost during evolution in invertebrate animals and non-animal eukaryotes. In support of this conclusion, we analyzed phylogenetic relationships of all (∼300) currently known Chlamydomonas proteins, and found no examples of horizontal gene transfer between animals and algae (S.V.Novoselov and V.N.Gladyshev, manuscript in preparation).
Our study also suggested the occurrence of a common eukaryotic SECIS element in Chlamydomonas and animals. However, the conservation profile of the algal structure appears to differ from that found in animal SECIS elements. It is possible that the primordial eukaryotic SECIS element that gave rise to Chlamydomonas and animal SECIS elements had little or no conservation at the nucleotide position preceding the quartet and in sequences of the apical loop, but increased its conservation profile along the evolutionary line to animals. Alternatively, evolution of Chlamydomonas apparently resulted in a greater flexibility in these nucleotides. It will be of interest to determine whether other lower eukaryotes contain a SECIS structure like that of animals or of Chlamydomonas.
Expression of animal selenoproteins has not been achieved in heterologous systems. The reasons are incompatibility of animal, bacterial and archaeal systems, the lack of Sec insertion systems in plants and yeast, and the low abundance of the Sec insertion system in insect cells. The only exception is animal TR, which contains a C-terminal penultimate Sec residue and therefore can tolerate sequences present downstream of Sec. Animal TRs were expressed in bacteria from constructs containing bacterial SECIS elements present downstream of the Sec codon in these enzymes (Arner et al., 1999; Gladyshev et al., 1999b). The ease of growing Chlamydomonas cells in large amounts and the abundance of selenoprotein genes in this organism suggest that Chlamydomonas may be employed as an important system for heterologous expression of selenoproteins.
Materials and methods
Cell growth and metabolic labeling
A strain of the alga C.reinhardtii, cwd/ARG, was grown in TAP medium supplemented with 50 µg/ml arginine and 1 nmol/l selenite at 25°C. For metabolic labeling, the cells were grown in TAP + arginine until mid-log phase and then labeled for 12 h with [75Se]selenious acid (190 Ci/mmol; Research Reactor Facility, University of Missouri, Columbia, MO), which had been freshly neutralized with KOH, at a concentration of 5 µCi/ml.
Protein purification
75Se-labeled cells were mixed with unlabeled cells (15 g wet weight), suspended in 50 mM Tris–HCI pH 8.3, 10% sucrose, 5 mM DTT, 1 mM EDTA, 1 mM AEBSF (buffer A) and disrupted by sonication. The homogenate was centrifuged at 18 000 g for 20 min and the supernatant fractionated with ammonium sulfate. The pellet between 20 and 80% saturation of ammonium sulfate was dissolved in 30 mM Tris–HCI pH 8.3, 5 mM DTT, 1 mM EDTA (buffer B) and dialyzed against buffer B. The solution was applied to a Q-Sepharose column (2.6 × 25 cm) that had been equilibrated with buffer B. The column was washed with a linear 0–600 mM gradient of NaCl in 500 ml of buffer B at 2.5 ml/min. Fractions containing 75Se were combined, concentrated and analyzed on SDS–PAGE gels. Selenium-containing proteins migrating as 7 and 18 kDa species were present in a flow-through fraction, whereas 22 and 52 kDa proteins were present in Q-Sepharose-bound fractions.
Subsequently, we concentrated our efforts on 7 and 18 kDa proteins, which were major Chlamydomonas selenoproteins. The flow-through fractions from the Q-Sepharose column were adjusted to 0.4 M ammonium sulfate in buffer B and applied to a phenyl–Sepharose HPLC column (TosoHaas; 21.5 mm × 15 cm) equilibrated with the same buffer. The radioactive fractions corresponding to 7 and 18 kDa proteins were eluted at 2 ml/min by application of a 60 ml linear gradient from 0.4 to 0 M ammonium sulfate in buffer B and a 30 ml linear gradient from buffer B to water, respectively. The radioactive fractions were pooled, concentrated, and analyzed by SDS–PAGE and PhosphorImager detection of 75Se on developed gels.
Radioactive fractions were further loaded onto a C18 reversed-phase HPLC column that had been equilibrated in 0.05% trifluoroacetic acid, a gradient of 0–70% acetonitrile in 0.05% trifluoroacetic acid applied, and 75Se-containing fractions corresponding to the 18 and 7 kDa proteins eluted at 43 and 49% acetonitrile, respectively. Fractions containing the 7 and 18 kDa selenoproteins from the C18 column were dried on a Speed-Vac SC110, dissolved in SDS–PAGE sample buffer and resolved on 10% SDS–NuPAGE gels (Novex). 75Se-labeled proteins were visualized with a Storm PhosphorImager system (Molecular Dynamics). Radioactive bands were cut and analyzed by Edman degradation at the UNL proteomics facility and by µLC/MS/MS on a Finnigan LCQ DECA quadrupole iontrap mass spectrometer at Harvard Microchemistry Facility.
Identification of C.reinhardtii selenoprotein ESTs
The TblastN program was used to search Chlamydomonas EST sequences for homologs of known eukaryotic selenoproteins. Identified Chlamydomonas sequences were further extended by similarity searches using BLAST programs and consensus sequence alignments of overlapping ESTs.
Screening of Chlamydomonas cDNA libraries and characterization of EST clones
Chlamydomonas EST clones containing full-size cDNA sequences for SelK1 (DDBJ/EMBL/GenBank accession No. AV632357) and TR1 (DDBJ/EMBL/GenBank accession No. AV626223) were obtained from Erika Asamizu, Kazusa DNA Research Institute. Their nucleotide sequences were determined at the UNL genomics facility. A set of four CC-1690 wild-type mt+ 21 gr individual libraries was purchased from the Chlamydomonas Genetics Center, Duke University. The libraries were generated from cells grown under the following growth conditions: (i) TAP medium in the light; (ii) TAP medium in the dark; (iii) HS medium plus 5% CO2, light; and (iv) HS medium, ambient CO2, light (Harris, 2001). Screening for several selenoprotein genes was performed by a PCR method. To amplify a SelW1 cDNA, forward 5′-GAGGCCAGCAACCATATCGCAACGATAAC and reverse 3′-TGG CAATAACAATGGGCCCCTGCGCAC primers were used. Forward 5′-TAGATATAAAGCCGCAGCGTTACGAGCCATGCGGGCGCTC-3′ and reverse 5′-CAGTAGACTGTCACTACAGAAGAGGGCATCTCGC CCCGAC-3′ primers were used to amplify SelM2 cDNA.
Expression of Chlamydomonas SelK1 in mammalian cells
Full-length cDNA of Chlamydomonas SelK1 was amplified with 5′-TAAACTCGAGATGCCTTACATCAGCCGAACAG and 3′-ATC GAAGCTTGCATGTACATCTCCACATTGCAAC primers, and cloned into XhoI–HindIII sites of pEGFP-C3 expression vector (Clontech). Escherichia coli strain NovaBlue (Novagen) was transformed with the resulting plasmid (GFP-chlamySelK) and the plasmid isolated using the Plasmid EndoFree Maxi Kit (Qiagen). NIH 3T3 cells were cultured in 60 mm culture dishes and transfected with 5 µg of GFP-chlamySelK plasmid in the presence of LipofectAMINE (Gibco-BRL) according to the manufacturer’s protocol. Metabolic labeling of transfected cells with 75Se was carried out as described previously (Kryukov et al., 1999), and the samples were analyzed on 10% SDS–NuPAGE gels (Novex). 75Se-labeled proteins were visualized with a Storm PhosphorImager system.
Identification of Chlamydomonas [75Se]selenocysteyl-tRNA
Chlamydomonas cells were grown to mid-log at 25°C in TAP medium supplemented with 50 µg/ml arginine and 5 × 10–7 M sodium selenite. Cells (2.5 g) were harvested, washed in fresh TAP media and resuspended in 100 ml of the same medium. [75Se]selenious acid (5.0 mCi) was then added to the cell culture and the cells labeled essentially as described previously (Hatfield et al., 1991, 1992). After 3 h incubation, cycloheximide (5 × 10–3 M; Sigma Chemical Co.) was added and the incubation period extended an additional hour. Cells were harvested, washed in fresh medium and the packed cells stored at –80°C until ready for use. Labeled aminoacyl-tRNAs were extracted from cells, chromatographed on an RPC-5 column, the resulting labeled tRNA fractions pooled, precipitated in ethanol, collected, and prepared for ribosomal binding studies (Nirenberg and Leder, 1964) or for deacylation and identification of Sec (Forchhammer et al., 1991) as described previously (Hatfield et al., 1991, 1992). The trinucleoside diphosphates UGU, UGC, UGA and UGG were the gift of M.W.Nirenberg.
Growth of Chlamydomonas in selenium-deficient and -sufficient media
All chemicals used as components of the TAP media that were employed to assess the requirement for selenium were of ultra-pure grade (99.9%+; Aldrich). The selenium content of a mineral mixture that was used for this medium was verified by inductively coupled plasma optical emission spectroscopy and flame atomic absorption (Galbraith Laboratories, Inc.). These assays revealed that the mineral mixture provides <10 pM selenium to the TAP medium. The medium that was prepared in this way was used as selenium-deficient medium. Selenium-sufficient medium was obtained by supplementing the selenium-deficient medium with 50 nM Na2SeO3. Cells were grown in selenium-deficient and -sufficient media with or without hydrogen peroxide and methionine-S-sulfoxide in parallel and in triplicates. Methionine-S-sulfoxide was kindly provided by Dr Ahmet Koc (Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, NE). Cell growth was measured by directly counting the number of Chlamydomonas cells in a hemacytometer. Each sample was analyzed three times (i.e. each point had nine independent measurements).
Phylogenetic analyses
Amino acid and nucleotide sequence alignments were generated using the PileUP program. The distances between sequences were calculated from these alignments using the DISTANCE program with the Kimura distance measuring methods and then the unrooted phylogenetic trees with unscaled distance branches were generated using the GrowTree program with the UPGMA method. All programs were part of the Wisconsin Package, Genetic Computer Group (GCG), Madison, WI.
Computational search for new selenoprotein genes in Chlamydomonas
All available Chlamydomonas EST sequences (113 064 at the time of our analysis) were extracted from DDBJ/EMBL/GenBank and analyzed with the Chlamy SECISearch, a program that recognizes Chlamydomonas SECIS elements. This program was developed by modifying SECISearch (Kryukov et al., 1999; G.V.Kryukov and V.N.Gladyshev, unpublished data) as follows. Primary sequence consensus was limited to NUGA/NGAN present in the SECIS element core. Helix II was ≥10 nucleotides with at most one mismatch and one single-nucleotide bulge on each side. Shoulders of the internal loop were set to be 2–8 nucleotides. Helix II was 11–12 nucleotides and contained less than four imperfections, including 1–2 mismatches and no more than one single-nucleotide bulge on each side of the helix. A single adenosine residue preceded by an unpaired nucleotide was required to be present in the 5′ branch of the apical loop or bulge. Type I and Type II SECIS element consensuses were incorporated separately in the searching pattern. For Type I SECIS elements, the length of the apical loop was restricted to 2–8 nucleotides. For Type II SECIS elements, the mini-stem was set at 3–8 nucleotides with one mismatch allowed and the apical loop was 3–6 unpaired nucleotides. The resulting pattern detected all Chlamydomonas SECIS elements except that of the SelM2 gene, whose predicted structure contained five bulged nucleotides. The free energy of a putative SECIS element including 10-nucleotide flanking regions was set at less than –20 kcal/mol. The free energy of the minimal SECIS element structure (e.g. quartet, helix II and apical loop) was set at less than –7.5 kcal/mol. Additional fine structural criteria (G.V.Kryukov and V.N.Gladyshev, unpublished data) were automatically applied. A set of Chlamy SECISearch-selected sequences was further automatically cleaned from redundancy caused by the presence of several ESTs corresponding to the same mRNA region. Finally, automatic BLAST analysis was employed to analyze the Chlamy SECISearch-selected sequences, and the candidate SECIS elements with ≥95% identity were replaced by the first representative hit.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Drs Gautam Sarath and Bill Lane for peptide sequencing, Dr Erica Asamizu for providing cDNA clones, Dr Marshall Nirenberg for providing trinucleotide codons and Dr Ahmet Koc for providing methionine-S-sulfoxide. This study was supported by the NIH GM61603 (to V.N.G.) and by the NSF 0115626 (to D.P.W.). cDNA sequences for Chlamydomonas SelM1, SelM2, SelW1, TR1, SelK1 and MsrA1 have been submitted to DDBJ/EMBL/GenBank.
References
- Arner E.S., Sarioglu,H., Lottspeich,F., Holmgren,A. and Bock,A. (1999) High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J. Mol. Biol., 292, 1003–1016. [DOI] [PubMed] [Google Scholar]
- Atkins J.F., Böck,A., Matsufuji,S. and Gesteland,R.F. (1999) Dynamics of the genetic code. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 637–673.
- Bock A. (2001) Selenium metabolism in bacteria. In Hatfield,D.L. (ed.), Selenium: Its Molecular Biology and Role in Human Health. Kluwer Academic, Norwell, MA, pp. 7–22.
- Bock A., Forchhammer,K., Heider,J. and Baron,C. (1991) Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem. Sci., 16, 463–467. [DOI] [PubMed] [Google Scholar]
- Bosl M.R., Takadu,K., Oshima,M., Nishimura,S. and Taketo,M.M. (1997) Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc. Natl Acad. Sci. USA, 94, 5531–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner C., Harney,J.W. and Berry,M.J. (1999) The Caenorhabditis elegans homologue of thioredoxin reductase contains a selenocysteine insertion sequence (SECIS) element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J. Biol. Chem., 274, 21598–21602. [DOI] [PubMed] [Google Scholar]
- Castellano S., Morozova,N., Morey,M., Berry,M.J., Serras,F., Corominas,M. and Guigo,R. (2001) In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO rep., 2, 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K., Leinfelder,W., Boesmiller,D., Veprek,B. and Bock,A. (1991) Selenocysteine synthase from Escherichia coli. Nucleotide sequence of the gene (selA) and purification of the protein. J. Biol. Chem., 266, 6318–6323. [PubMed] [Google Scholar]
- Gladyshev V.N. and Kryukov,G.V. (2001) Evolution of selenocysteine-containing proteins: significance of identification and functional characterization of selenoproteins. Biofactors, 14, 87–92. [DOI] [PubMed] [Google Scholar]
- Gladyshev V.N., Stadtman,T.C., Hatfield,D.L. and Jeang,K.-T. (1999a) Levels of major selenoproteins in T cells decrease during HIV infection and low molecular mass selenium compounds increase. Proc. Natl Acad. Sci. USA, 96, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev V.N., Krause,M., Xu,X.M., Korotkov,K.V., Kryukov,G.V., Sun,Q.A., Lee,B.J., Wootton,J.C. and Hatfield,D.L. (1999b) Selenocysteine-containing thioredoxin reductase in Caenorhabditis elegans. Biochem. Biophys. Res. Commun., 259, 244–249. [DOI] [PubMed] [Google Scholar]
- Grundner-Culemann E., Martin,G.W.,III, Harney,W. and Berry,M.J. (1999) Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA, 5, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (2001) Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol., 52, 363–406. [DOI] [PubMed] [Google Scholar]
- Hatfield D.L. and Gladyshev,V.N. (2002) How selenium has altered our understanding of the genetic code. Mol. Cell. Biol., 22, 3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D.L., Lee,B.J., Price,N.M. and Stadtman,T.C. (1991) Selenocysteyl-tRNA occurs in the diatom Thalassiosira and in the ciliate Tetrahymena.Mol. Microbiol., 5, 1183–1186. [DOI] [PubMed] [Google Scholar]
- Hatfield D., Choi,I.S., Mischke,S. and Owens,L.D. (1992) Selenocysteyl-tRNAs recognize UGA in Beta vulgaris, a higher plant and in Gliocladium virens, a filamentous fungus. Biochem. Biophys. Res. Commun., 184, 254–259. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Makarova,K.S. and Aravind,L. (2001) Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol., 55, 709–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov K.V., Novoselov,S.V., Hatfield,D.L. and Gladyshev,V.N. (2002) Mammalian selenoprotein in which selenocysteine (Sec) incorporation is supported by a new form of Sec insertion sequence element. Mol. Cell. Biol., 22, 1402–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov G.V. and Gladyshev,V.N. (2002) Mammalian selenoprotein gene signature: identification and functional analysis of selenoprotein genes using bioinformatics methods. Methods Enzymol., 347, 84–100. [DOI] [PubMed] [Google Scholar]
- Kryukov G.V., Kryukov,V.M. and Gladyshev,V.N. (1999) New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J. Biol. Chem., 274, 33888–33897. [DOI] [PubMed] [Google Scholar]
- Kryukov G.V., Kumar,R.A., Koc,A., Sun,Z. and Gladyshev,V.N. (2002) Selenoprotein R is a zinc-containing stereospecific methionine sulfoxide reductase. Proc. Natl Acad. Sci. USA, 99, 4245–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low S.C. and Berry,M.J. (1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci., 21, 203–208. [PubMed] [Google Scholar]
- Lowther W.T., Brot,N., Weissbach,H., Honek,J.F. and Matthews,B.W. (2000) Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc. Natl Acad. Sci. USA, 97, 6463–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Romero F.J., Kryukov,G.V., Lobanov,A.V., Carlson,B.A., Lee,B.J., Gladyshev,V.N. and Hatfield,D.L. (2001) Selenium metabolism in Drosophila: selenoproteins, selenoprotein mRNA expression, fertility and mortality. J. Biol. Chem., 276, 29798–29804. [DOI] [PubMed] [Google Scholar]
- Moskovitz J., Poston,J.M., Berlett,B.S., Nosworthy,N.J., Szczepanowski,R. and Stadtman,E.R. (2000) Identification and characterization of a putative active site for peptide methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. J. Biol. Chem., 275, 14167–14172. [DOI] [PubMed] [Google Scholar]
- Nirenberg M. and Leder,P. (1964) RNA codewords and protein synthesis. Science, 145, 1399–1407. [DOI] [PubMed] [Google Scholar]
- Rother M., Resch,A., Wilting,R. and Bock,A. (2001a) Selenoprotein synthesis in archaea. Biofactors, 14, 75–83. [DOI] [PubMed] [Google Scholar]
- Rother M., Resch,A., Gardner,W.L., Whitman,W.B. and Bock,A. (2001b) Heterologous expression of archaeal selenoprotein genes directed by the SECIS element located in the 3′ non-translated region. Mol. Microbiol., 40, 900–908. [DOI] [PubMed] [Google Scholar]
- Serras F., Morey,M., Alsina,B., Baguna,J. and Corominas,M. (2001) The Drosophila selenophosphate synthetase (selD) gene is required for development and cell proliferation. Biofactors, 14, 143–149. [DOI] [PubMed] [Google Scholar]
- Shigeoka S., Takeda,T. and Hanaoka,T. (1991) Characterization and immunological properties of selenium-containing glutathione peroxidase induced by selenite in Chlamydomonas reinhardtii. Biochem. J., 275, 623–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman T.C. (1996) Selenocysteine. Annu. Rev. Biochem., 65, 83–100. [DOI] [PubMed] [Google Scholar]
- Sun Q.A., Wu,Y., Zappacosta,F., Jeang,K.T., Lee,B.J., Hatfield,D.L. and Gladyshev,V.N. (1999) Redox regulation of cell signaling by selenocysteine in mammalian thioredoxin reductases. J. Biol. Chem., 274, 24522–24530. [DOI] [PubMed] [Google Scholar]
- Ursini F., Maiorino,M. and Roveri,A. (1997) Phospholipid hydroperoxide glutathione peroxidase (PHGPx): more than an antioxidant enzyme? Biomed. Environ. Sci., 10, 327–332. [PubMed] [Google Scholar]
- Walczak R., Westhof,E., Carbon,P. and Krol,A. (1996) A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA, 2, 367–379. [PMC free article] [PubMed] [Google Scholar]
- Weissbach H., Etienne,F., Hoshi,T., Heinemann,S.H., Lowther,W.T., Matthews,B., St John,G., Nathan,C. and Brot,N. (2002) Peptide methionine sulfoxide reductase: structure, mechanism of action and biological function. Arch. Biochem. Biophys., 397, 172–178. [DOI] [PubMed] [Google Scholar]
- Whanger P.D. (2000) Selenoprotein W: a review. Cell. Mol. Life Sci., 57, 1846–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]