Abstract
Xer site-specific recombination in Escherichia coli converts plasmid multimers to monomers, thereby ensuring their correct segregation at cell division. Xer recombination at the psi site of plasmid pSC101 is preferentially intramolecular, giving products of a single topology. This intramolecular selectivity is imposed by accessory proteins, which bind at psi accessory sequences and activate Xer recombination at the psi core. Strand exchange proceeds sequentially within the psi core; XerC first exchanges top strands to produce Holliday junctions, then XerD exchanges bottom strands to give final products. In this study, recombination was analysed at sites in which the psi core was inverted with respect to the accessory sequences. A plasmid containing two inverted-core psi sites recombined with a reversed order of strand exchange, but with unchanged product topology. Thus the architecture of the synapse, formed by accessory proteins binding to accessory sequences, determines the order of strand exchange at psi. This finding has important implications for the way in which accessory proteins interact with the recombinases.
Keywords: DNA-binding protein/PepA/site-specific recombination/Xer
Introduction
Site-specific recombination systems are implicated in a variety of DNA rearrangements in microorganisms (Hallet and Sherratt, 1997). Recombination is catalysed by recombinase proteins that carry out DNA strand cleavage and transfer reactions at short DNA sequences known as recombination core sites (Stark et al., 1992). Site-specific recombination often requires additional accessory proteins, which bind to accessory sequences adjacent to the recombination core. The recombinase and accessory proteins form a highly organized protein–DNA complex with the recombination site DNA, and together exert control over the efficiency and timing of the recombination reaction. The way in which accessory proteins bound at accessory sequences on a DNA molecule can regulate and control strand exchange reactions by recombinases is the subject of this study.
The Xer site-specific recombination system of Escherichia coli resolves multimeric forms of circular replicons to allow efficient segregation of monomers to daughter cells at cell division (Sherratt et al., 1995). Two related recombinases, XerC and XerD, act at the chromosomal site dif and at plasmid-borne sites such as cer of ColE1 and psi of pSC101 (Colloms et al., 1990; Blakely et al., 1993; Cornet et al., 1994). Xer recombination sites share an ∼30 bp core sequence, containing an 11 bp XerC-binding site and an 11 bp XerD-binding site separated by a 6–8 bp asymmetric central region. In addition, recombination at cer and psi requires accessory sequences and accessory proteins, which ensure that recombination is preferentially intramolecular, converting plasmid multimers to monomers and not vice versa (Stirling et al., 1989; Summers, 1989). PepA, an aminopeptidase with DNA-binding activity, and ArgR, the arginine repressor, bind to ∼180 bp of accessory sequences adjacent to the cer core and are both required for recombination at cer (Colloms et al., 1996; Alén et al., 1997). Similarly, PepA and ArcA, an anaerobic response regulator protein, act at ∼160 bp of accessory sequences adjacent to the psi core (Colloms et al., 1996, 1998).
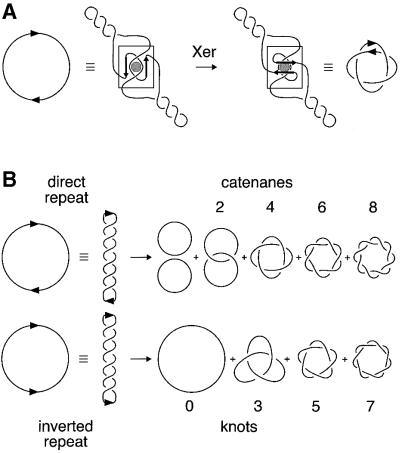
Xer site-specific recombination at psi has been reconstituted in vitro with purified PepA, XerC and XerD proteins. Although ArcA is essential for efficient recombination in vivo, it is not required in vitro (Colloms et al., 1998). Recombination on a substrate containing two psi sites in direct repeat generates two product rings, which are interlinked to form a specific four-noded catenane (Figure 1A; Colloms et al., 1997). Prior to strand exchange at psi, accessory proteins bind to two recombination sites and assemble a complex in which the DNA accessory sequences of both sites are wrapped a precise number of times around the accessory proteins (Colloms et al., 1997; Bath et al., 1999). This productive synaptic complex can only be formed easily between directly repeated sites on the same DNA molecule, and the recombinases only act efficiently at cer and psi cores within this complex. The requirement for this complex acts as a checkpoint (or topological filter) that favours resolution over inversion and intermolecular recombination. Because recombination occurs by a defined pathway, the products have a defined topology. Products of defined topology are produced by other systems that use accessory sequences and proteins to trap a fixed number of supercoils between recombining sites prior to recombination (e.g. Tn3/γδ resolvase and the DNA invertases; reviewed in Stark and Boocock, 1995). In other site-specific recombination reactions (such as those catalysed by λ integrase), recombination occurs after random collision of sites, trapping a variable number of supercoils between recombining sites. Such systems give mixtures of free circles and catenanes with even numbers of crossings from substrates with directly repeated sites, and knots with odd numbers of crossings from substrates with sites in inverted repeat (Figure 1B).
Fig. 1. Topology of site-specific recombination reactions. (A) Xer recombination at psi occurs only after the formation of a synaptic complex with a defined local structure (shown boxed). This productive synapse, formed by wrapping the accessory sequences of two recombination sites around the accessory protein PepA, traps a specific number of topological nodes. Strand exchange occurs by a defined mechanism and the product is a right-handed four-noded catenane with antiparallel psi sites. (B) Many site-specific recombination systems display no topological specificity. A random number of supercoils are trapped when the recombination sites come together, and recombination generates products of mixed topology. Sites in direct repeat yield two circles, which are either unlinked, or linked in catenanes with even numbers of topological crossings or nodes, as shown. Sites in an inverted repeat yield single circular products, which are either unknotted or knotted with an odd number of nodes. Knots and catenanes produced in these reactions are generally members of the torus family of knots and catenanes, as shown.
Xer recombination occurs by a pair of sequential single-strand exchanges, which first form and then resolve a Holliday junction (HJ) intermediate. At psi, these strand exchanges proceed with a strictly defined order: XerC first exchanges top strands and then XerD exchanges bottom strands. Xer cleavage and strand exchange at dif also proceed with a preferred order (Blakely et al., 1997; Neilson et al., 1999). The order of strand exchange at dif is determined by the sequence of the core site and its interactions with the recombinases (Hallet et al., 1999; Blakely et al., 2000). In this study, we show that the order of strand exchange at psi is dictated by the architecture of the synaptic complex formed by accessory proteins and sequences, so that on an appropriate DNA substrate the normal order of strand exchange is reversed. The cell division protein FtsK also alters the order of Xer strand exchange at dif (Aussel et al., 2002), so it seems likely that the results presented here will have implications for the mode of action of FtsK at dif.
Results
Efficient recombination at psi requires accessory sequences
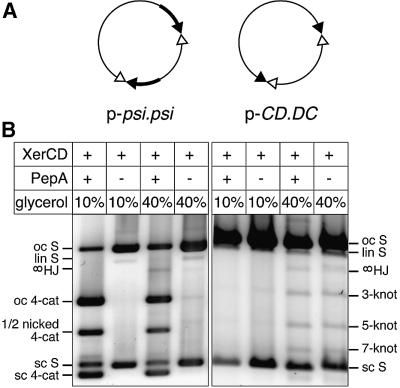
To investigate whether accessory sequences are absolutely required for recombination and topological selectivity at psi, we constructed a family of plasmids containing psi core sites with and without accessory sequences in different combinations. The ability of these plasmids to undergo recombination was tested in vitro using purified XerC and XerD in the presence and absence of PepA. Reactions were carried out under normal conditions (with 10% glycerol in the reaction buffer) and also in the presence of 40% glycerol, which has been shown previously to allow unusual Xer reactions to take place (Cornet et al., 1997; Hallet et al., 1999). Recombination between two full psi sites in the presence of PepA gave a four-noded catenane in both conditions, but 40% glycerol inhibited recombination and increased the yield of HJ intermediates (Figure 2). In the absence of PepA, no recombination was observed at 10% glycerol. However, a small amount of PepA-independent recombination was observed at 40% glycerol, giving products of mixed topology (four-, six- and eight-noded catenanes were observed).
Fig. 2. Recombination at psi sites with and without accessory sequences. (A) Maps of p-psi.psi, containing two psi sites in direct repeat, and p-CD.DC, containing two psi cores without accessory sequences in inverted repeat. Accessory sequences are shown as thick black lines; XerC- and XerD-binding sites in the psi core are shown as filled and open triangles, respectively. (B) Topological analysis of recombination products. Plasmids were reacted with XerC and XerD with or without PepA, with either 10 or 40% glycerol as indicated. Reactions were nicked and run on a 0.7% agarose gel. Bands are indicated as follows: oc S, open circle substrate; lin S, linear substrate; sc S, supercoiled substrate; oc 4-cat, four-noded catenated product nicked on both circles; 1/2 nicked 4-cat, four-noded catenated product nicked on the large circle but still supercoiled on the small circle; sc 4-cat, fully supercoiled four-noded catenane; 3-knot, 5-knot and 7-knot, nicked knotted inversion products with three, five and seven nodes, respectively; ∞HJ, HJ intermediate nicked at the HJ on the recombinant strand, with consequent loss of any knotting or catenation.
A plasmid with one full psi site in direct repeat with a psi core (p-psi.CD), as well as plasmids containing two psi cores in direct (p-CD.CD) or inverted (p-CD.DC) repeat, did not recombine efficiently in any of the conditions tested. The efficiency and topology of recombination on these substrates were not affected by the addition of PepA (Figure 2; data not shown). A low level of recombination was observed at 40% glycerol, yielding products of mixed topology (Figure 2; data not shown). These results demonstrate that two sets of accessory sequences together with PepA are required for efficient recombination at psi. In the absence of PepA and/or accessory sequences, very low levels of recombination occur only at 40% glycerol, giving products of mixed topology.
Recombination at the inverted-core psiDC site
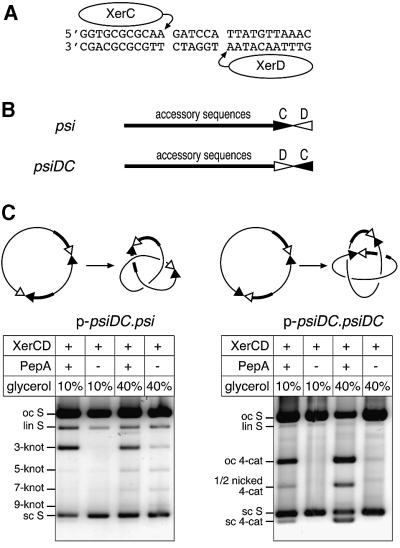
Wild-type psi consists of a 28 bp core, comprising 11 bp XerC- and XerD-binding sites separated by a 6 bp asymmetric central region (Figure 3A), with ∼160 bp of accessory sequences adjacent to the XerC-binding site. Xer cleavage and strand exchange take place within the psi core, on either side of the 6 bp central region. To investigate how accessory sequences affect alignment of recombination cores and subsequent activation of strand exchange, we constructed psiDC, in which the entire psi core has been inverted with respect to the accessory sequences (Figure 3B). Therefore, psiDC has an 11 bp XerD-binding site adjacent to the accessory sequences and an 11 bp XerC-binding site distal to the accessory sequences.
Fig. 3. Recombination at psiDC. (A) Sequence of the psi core showing XerC- and XerD-binding and cleavage sites. (B) Diagram of the psi and psiDC sites, showing accessory sequences, to which PepA binds, and the core site with XerC- and XerD-binding sites represented as filled and open triangles, respectively. (C) Diagrams of p-psiDC.psi and p-psiDC.psiDC and their major recombination products. Plasmids were reacted with XerC and XerD in the presence or absence of PepA, with 10 or 40% glycerol (as indicated). Reactions were nicked and run on a 0.7% agarose gel. Bands are indicated as in Figure 2.
The psiDC site was used to construct two recombination substrates (Figure 3C). p-psiDC.psi contains one wild-type psi site and one psiDC site. The accessory sequences of the two sites are directly repeated in this plasmid, but the core sites are in inverted repeat. Therefore, Xer recombination on p-psiDC.psi is expected to yield a single circular product, with the segment between the two cores inverted. The plasmid p-psiDC.psiDC contains two psiDC sites in direct repeat. The accessory sequences and the cores of the two sites in p-psiDC.psiDC are directly repeated. There fore, Xer recombination is expected to yield two circular resolution products, which may be catenated. Both plasmids were incubated with XerC and XerD in the presence and absence of PepA, at 10 and 40% glycerol. The DNA was nicked with DNase I or cleaved with XhoI, and products were separated on agarose gels (Figure 3C; data not shown).
p-psiDC.psiDC recombined relatively efficiently in the presence of PepA (Figure 3C). This PepA-dependent recombination was more efficient at 40% glycerol than at 10% glycerol, and yielded four-noded catenanes as the major product. There was no detectable recombination in the absence of PepA at 10% glycerol, but at 40% glycerol there was a small amount of recombination, yielding a mixture of four-, six- and eight-noded catenanes.
Restriction analysis confirmed that p-psiDC.psi gave the expected inversion product (data not shown). p-psiDC.psi recombined slightly less efficiently than p-psiDC.psiDC. In the presence of PepA, at 10 and 40% glycerol, the major product was three-noded knots, although traces of five- and seven-noded knots were detected at 40% glycerol. In the absence of PepA, low levels of recombination were observed at 40% glycerol; products were a mixture of three-, five- and seven-noded knots. This PepA-independent recombination on p-psiDC.psi was similar to the recombination observed between two psi cores in an inverted repeat on p-CD.DC (compare with Figure 2), supporting the hypothesis that there are two pathways of recombination: a PepA- and accessory sequence-independent pathway that is enhanced at 40% glycerol and gives products of mixed topology, and a more efficient pathway that requires PepA and two copies of the accessory sequences, giving products of a specific topology.
The order of strand exchange is reversed at psiDC
XerC cleaves and exchanges top strands of psi, adjacent to the XerC-binding site at the left end of the central region, whereas XerD cleaves and exchanges bottom strands at the right end of the central region (Figure 3A; Colloms et al., 1996; Blake et al., 1997). Xer recombination at psi proceeds with a defined order of strand exchanges (Colloms et al., 1996). XerC first exchanges top strands to generate a HJ intermediate. XerD then exchanges bottom strands to complete the reaction. If the order of strand exchange at psi is an intrinsic property of the way XerC and XerD bind to the psi core, then XerC will exchange the first pair of strands during recombination at both psi and psiDC. If, instead, the order of strand exchange at psi is determined solely by the architecture of the synapse formed by accessory proteins and sequences, then XerD will exchange the first pair of strands at psiDC.
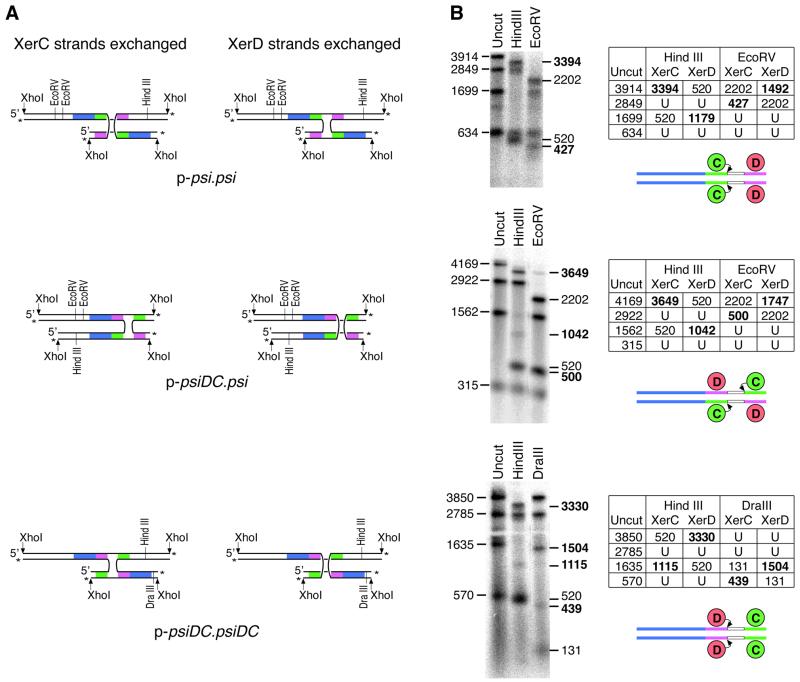
HJs made by XerC- and XerD-mediated strand exchange are not equivalent and can be distinguished by the recombinant strands they contain (Figure 4A). Strand-separating gel electrophoresis was used to reveal which strands had been exchanged in HJs formed during recombination between wild-type and inverted-core psi sites on p-psi.psi, p-psiDC.psi and p-psiDC.psiDC (Figure 4B). HJ yields were maximized by carrying out reactions at 40% glycerol, which leads to the accumulation of high levels of HJ intermediates in the presence of PepA (Hallet et al., 1999; this study). HJs were cleaved with XhoI to give χ-forms, and purified from an agarose gel. The χ-form HJs were 3′ end-labelled with 32P, and cleaved with restriction endonucleases that were predicted to give different sizes of single-stranded DNA fragments depending on which strands had been exchanged (Figure 4B). Samples were run on a denaturing agarose gel to analyse the separated single strands.
Fig. 4. Analysis of HJ intermediates formed by recombination at psi and psiDC. (A) HJ intermediates made by XerC or XerD strand exchange can be distinguished by the recombinant strands they contain. XhoI-cleaved χ-form HJs, 3′ end-labelled and then cleaved with restriction enzymes give different patterns of single-stranded DNA fragment depending on which strands have been exchanged. Accessory sequences are shown in blue, XerC- and XerD-binding sites are shown in green and pink, respectively, and 3′ end labels are indicated by asterisks. (B) XhoI-cleaved HJs from p-psi.psi, p-psiDC.psi and p-psiDC.psiDC were purified and 3′ end-labelled with 32P. Labelled χ DNA was run on a strand-separating agarose gel uncut or cleaved with HindIII, EcoRV or DraIII. Predicted sizes of labelled single-stranded DNA fragments for χ-forms produced by XerC or XerD strand exchange before or after cleavage with the appropriate restriction enzymes are tabulated. U indicates fragments unchanged by the restriction enzyme. Fragment sizes diagnostic for XerC or XerD strand exchanges are shown in bold. Sites taking part in each reaction are shown diagrammatically below each table. Curly arrows indicate recombinase partners initiating the first strand exchange reaction.
The results of this analysis confirmed that HJs produced by recombination between two wild-type psi sites are formed by exchange of XerC strands, as previously reported (Figure 4B; Colloms et al., 1996). HJs produced by recombination between psi and psiDC were also formed mainly by exchange of XerC strands (Figure 4B). In contrast, recombination between two psiDC sites yielded HJs formed by exchange of XerD strands (Figure 4B). Thus the order of strand exchange at psi is determined by the orientation of the core with respect to the accessory sequences. During recombination at identical sites, the recombinase that binds closest to the accessory sequences (XerC for psi, XerD for psiDC) exchanges the first pair of strands. However, when the two cores are in opposite orientations with respect to the accessory sequences in p-psiDC.psi, it appears that the natural preference for XerC to initiate recombination at psi cores dictates the order of strand exchange.
In addition to the major HJ with XerD strands exchanged in p-psiDC.psiDC, a small amount of HJ formed by XerC strand exchange was detected (Figure 4B). This probably comes from some PepA-independent recombination, which is known to occur in the high glycerol conditions used (see above). In this pathway, the location of the accessory sequences will have no effect and XerC strand exchange will probably occur first.
Recombination at psiDC with catalytic mutants of XerC and XerD
In the previous section, it was shown that XerD strands are exchanged in the HJ formed between two psiDC sites. However, it was not proven that XerD catalyses this strand exchange. It is possible that XerC binds to the XerD-binding site and exchanges XerD strands at psiDC. Catalytic mutants of XerC and XerD were therefore used to ascertain which recombinase carries out the first strand exchange at psiDC.
In vitro recombination of p-psi.psi and p-psiDC.psi was completely abolished when wild-type XerC was replaced by the catalytically inactive mutant XerCY275F. In contrast, when XerD was replaced with XerDY279F, HJs formed by XerC-mediated strand exchange accumulated on both of these plasmids (data not shown; see also Colloms et al., 1996). However, no HJs or recombination products were detected on p-psiDC.psiDC with either XerCY275F and XerD, or XerC and XerDY279F. Similarly, no HJs or products were obtained from p-psiDC.psiDC with other recombinase catalytic mutants, such as XerCR148K or XerDR148K, in combination with wild-type partners. The failure to produce HJs from p-psiDC.psiDC with catalytic mutants of XerC might be due to reduced activity or DNA binding affinity of these XerC mutants, or altered interactions with XerD (Arciszewska et al., 2000).
Therefore, to determine which recombinase exchanges the first pair of strands on p-psiDC.psiDC, recombination was examined in E.coli strains carrying chromosomal catalytic mutations in xerC or xerD (Table I). This approach is more sensitive because Xer recombination is more efficient in vivo than in vitro. HJs produced by Xer-mediated strand exchange in vivo can be processed, probably by replication or by HJ-resolving enzymes, to give fully recombinant product (Colloms et al., 1996). As expected, p-psi.psi recombined efficiently in the presence of XerC and XerDY279F, but not at all in the presence of XerCY275F and XerD. In contrast, p-psiDC.psiDC did not recombine in the presence of XerC and XerDY279F, but did recombine in the presence of XerCY275F and XerD. These results support the hypothesis that XerD exchanges the first pair of strands on p-psiDC.psiDC to give HJs that can be resolved in vivo to recombinant product.
Table I. In vivo recombination at psi and psiDC in E.coli strains carrying mutant xer genes.
| Plasmid | Genotype |
|||
|---|---|---|---|---|
| xerC+ xerD+ | xerC+ xerDYF | xerCYF xerD+ | xerCYF xerDYF | |
| p-psi.psi | +++ | +++ | – | – |
| p-psiDC.psiDC | ++ | – | + | – |
Plasmids were transformed into strains carrying the chromosomal copies of xer alleles indicated. The extent of recombination observed by agarose gel electrophoresis after overnight growth is indicated as: +++, 100% of substrate converted to product; ++, 50% of substrate converted to product; +, 15% of substrate converted to product; –, no detectable recombination.
Topology of HJ intermediates
The HJs produced in vitro at psiDC could be genuine intermediates in recombination, or they could be dead-end products from another pathway. If the HJs are genuine intermediates, their topology should be consistent with that of the fully recombinant products. HJs that are intermediates in the production of three-noded knots should have three trapped nodes and will co-migrate on agarose gels with a three-noded knot when singly nicked. Similarly, intermediates in the production of four-noded catenanes should have four entrapped catenation nodes and will co-migrate with a four-noded catenane (Colloms et al., 1996). The topology of HJs produced by XerC and XerD in the presence of PepA at 40% glycerol was determined by two-dimensional gel electrophoresis. Nicked products were separated according to their topology in the first dimension. Gel lanes were then excised and incubated with XhoI prior to electrophoresis in the second dimension, to ascertain the restriction patterns of the different products (Figure 5). This analysis showed that HJs produced in the reaction between psi and psiDC had three entrapped nodes, whereas HJs produced in the reaction between two psiDC sites had four entrapped nodes. Thus the HJs have the correct topology to be intermediates in the production of three-noded knots and four-noded catenanes, respectively.
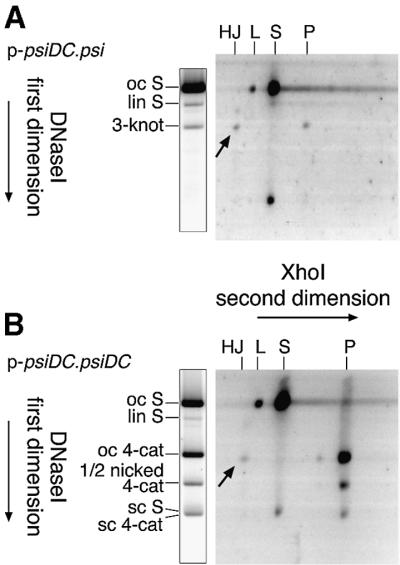
Fig. 5. Two-dimensional gel analysis of HJs formed by recombination between psi and psiDC sites. (A) Analysis of products formed by recombination on p-psiDC.psi. (B) Analysis of products formed by recombination on p-psiDC.psiDC. The first dimension separated substrates and products according to their topology, and the second dimension separated products according to their restriction pattern. Arrows indicate the positions of HJ intermediates on each gel. Migration of three-noded knots, four-noded catenanes and other species in the first dimension is indicated to the left of a duplicate first- dimension lane as in the legend to Figure 2. Second-dimension mobilities are shown above the gels as HJ, HJ intermediates; L, linear substrate; S, substrate restriction fragment; and P, product restriction fragment. Small product and substrate restriction fragments are not shown. Partial digestion with XhoI gave some linear substrate at the oc S position, and some uncut (open circle) large product circle at the four-node position in (B).
Recombination at psiDC with XerC[De]
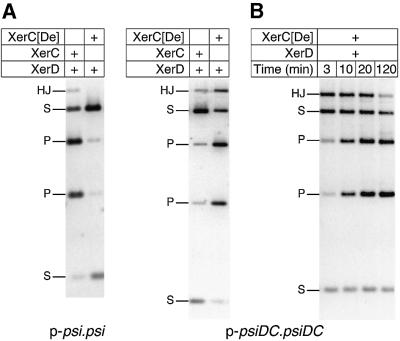
A derivative of XerC (XerC[De]), in which the C-terminal 21 residues of XerC have been replaced with the C-terminal 17 residues from XerD, has been shown to stimulate XerD strand exchange and impair XerC strand exchange, when tested on synthetic HJs (H.Ferreira and L.K.Arciszewska, unpublished results). Replacing XerC with XerC[De] inhibited in vitro recombination on p-psi.psi and p-psiDC.psi, but strongly stimulated recombination on p-psiDC.psiDC, giving increased yields of both HJ and recombinant product in a standard 1.5 h reaction (Figure 6A; data not shown). This result is consistent with the ‘stimulation of partner, impairment of self-catalysis’ phenotype of XerC[De]. Stimulation of XerD and impairment of XerC function gave higher amounts of HJs on p-psiDC.psiDC where XerD strand exchange occurs first. In contrast, the reduced activity of XerC[De] gave lower yields of HJs, and consequently lower levels of final recombination product on p-psi.psi and p-psiDC.psi where XerC strand exchange is first.
Fig. 6. Xer recombination at psi and psiDC in the presence of XerC[De]. (A) Recombination of p-psi.psi and p-psiDC.psiDC in the presence of XerD and either XerC[De] or XerC. (B) Time course of recombination on p-psiDC.psiDC with XerC[De] and XerD. Reactions contained 40% glycerol and were cleaved with XhoI. Bands are indicated as follows: HJ, HJ intermediates; S, substrate fragments; P, product fragments.
A time course of recombination on p-psiDC.psiDC using XerC[De] and wild-type XerD showed that HJs formed by XerC[De] and XerD are transient recombination intermediates which become resolved to the final product. Large amounts of HJs are observed at early time points, and gradually disappear at later time points (Figure 6B). HJs and products formed in this reaction have the same four-noded topology as those produced by wild-type XerC and XerD on this substrate (data not shown). Strand-separating gels confirmed that the HJs formed by XerC[De] and XerD on p-psiDC.psiDC were formed by XerD strand exchange (data not shown). These experiments therefore confirm that HJs in which XerD strands have been exchanged are intermediates in a complete recombination reaction between two psiDC sites.
Discussion
The results presented here demonstrate that two sets of accessory sequences are required for topological selectivity and efficient recombination at psi. The psi core appears to have been selected to be almost completely inactive in the absence of accessory sequences and accessory proteins. Recombination occurs only after the formation of the productive synapse, which acts as a checkpoint to ensure that sites are in a direct repeat in the same DNA molecule. This reflects the biological function of psi, which is to resolve plasmid multimers. A very small amount of accessory factor-independent recombination was observed at psi cores, consistent with the low level of recombination between psi cores previously seen in vivo (Blake et al., 1997). This recombination did not give products of a specific topology, showing that two full sets of accessory sequences are necessary for the formation of an interwrapped synapse. In contrast, Tn3 resolvase uses a similar mechanism to ensure that sites are only recombined if they are in a direct repeat on the same DNA molecule, but one set of res accessory sequences was enough to restrict the product topology to a single species (Bednarz et al., 1990).
Recombination between two psiDC sites occurred with the same topology as recombination between two psi sites. HJs formed by XerD strand exchange appeared with the correct topology and kinetics to be intermediates in the reaction between two psiDC sites. Therefore, XerD, rather than XerC, exchanged the first pair of strands in this reaction. As discussed below, our current model for recombination at psi predicts exactly this result; thus, the data presented here provide further support for our model of Xer recombination at psi (Figure 7).
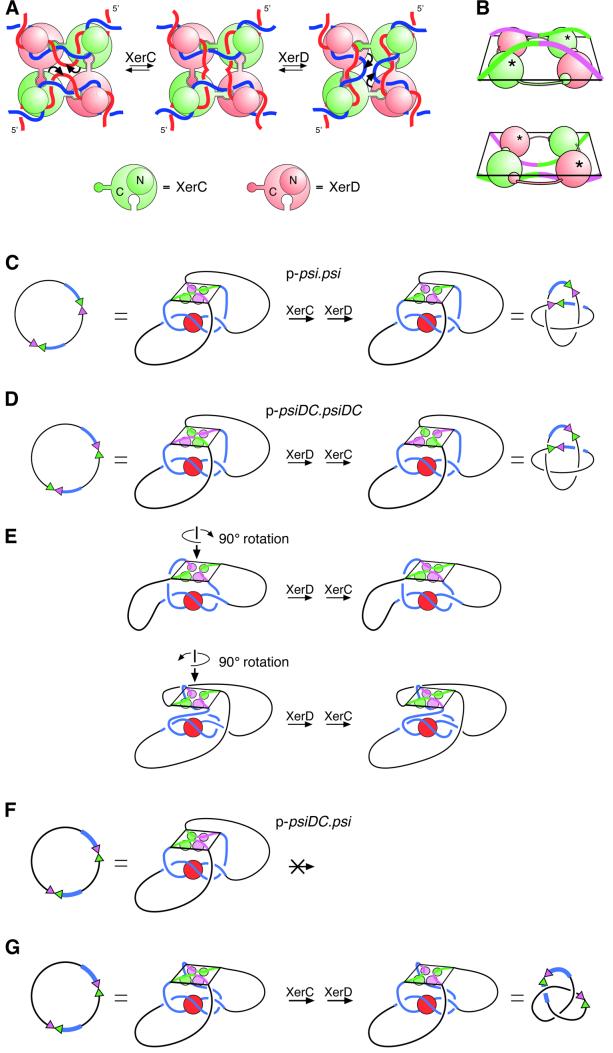
Fig. 7. Models for recombination at psi and psiDC. (A) The proposed mechanism of strand exchange by XerC and XerD at the psi core, based on X-ray crystal structures of the Cre recombinase bound to its recombination site. XerC (green) and XerD (pink) bind to a pair of psi core sites in an antiparallel conformation. A C-terminal extension on each monomer binds to the C-terminal domain of one of its neighbours in a cyclical arrangement. All four monomers bind to the DNA with their N-terminal domains (shown as small circles above the DNA) on one side of the HJ and their C-terminal domains (shown as large circles below the DNA) on the other side. XerC catalyses cleavage, strand exchange and re-ligation of one pair of strands (denoted by curly arrows) to form a HJ intermediate. XerD then exchanges the other pair of strands to form the recombinant product. Recombination can also occur by the reverse of this process (right to left) with XerD strand exchange preceding XerC strand exchange. (B) Perspective view of core–recombinase complexes in which XerC (top panel; equivalent to the left panel in A) or XerD (bottom panel; equivalent to the right panel in A) is about to catalyse strand exchange to form a HJ. The diagrams have been aligned so that DNA connectivity and the topological outcome of a complete strand exchange reaction will be the same in both cases. In this view, C-terminal domains have to be below the DNA for XerC strand exchange to proceed first, and above the DNA for XerD strand exchange to proceed first. XerC and XerD C-terminal domains are shown as green and pink spheres, respectively. N-terminal domains have been omitted for clarity. Active recombinase monomers are indicated with asterisks. (C) Synapsis of psi sites and interwrapping of accessory sequences around PepA define the order of strand exchange by XerC and XerD. During recombination between two psi sites, the accessory sequences (blue) of two psi sites wrap around PepA, with the psi cores brought together in an antiparallel fashion. XerC protein and binding sites are shown in green, and XerD protein and binding sites are shown in pink. This synapse is equivalent to that shown in Figure 1A. The recombinase–core complex is constrained so that the C-termini face towards PepA and the accessory sequences. By comparison with (A), it can be seen that the psi cores have been brought together such that XerC strand exchange will precede XerD strand exchange. The product is a four-noded catenane. (D) Two psiDC sites can form a synapse in which the DNA follows exactly the same path as in (C). In this complex, XerC occupies the position normally occupied by XerD, and vice versa. XerD strand exchange proceeds first in this synapse, and the product is a four-noded catenane. (E) The synapse shown in (D) can be rearranged into two geometrically different, but topologically equivalent synapses, by rotating the recombinase–core complex by ±90° about its 2-fold symmetry axis. XerC and XerD occupy the same positions in this synapse as in the synapse between wild-type psi sites, but the DNA path is altered to accommodate the psiDC sites. XerD strand exchange will proceed first and the product will be the observed four-noded catenane in both cases. (F) On p-psiDC.psi, interwrapping of accessory sequences as in (A) would bring the two cores together in a parallel conformation with forbidden XerC–XerD interactions. (G) To bring the two sites together in an antiparallel fashion, one of the cores must be rotated through 180° relative to its position in (F). If the core of psiDC is rotated, as depicted, XerC strand exchange will proceed first and the product will be a three-noded knot, as observed. However, an equivalent rotation of the psi core would give the same three-noded product but XerD strand exchange would proceed first. Alternatively, both cores might be rearranged to bring about the required antiparallel alignment.
Xer strand exchange almost certainly takes place by a mechanism analogous to that proposed for recombination by Cre, FLP and λ integrase (Figure 7A; Arciszewska et al., 1995, 1997; Nunes-Düby et al., 1995; Subramanya et al., 1997; Hallet et al., 1999). Recent crystal structures of Cre and FLP recombinases bound to their DNA substrates demonstrate that these proteins carry out recombination on core sites aligned in an antiparallel configuration, but bent so that the four arms exit the complex roughly at the corners of a square (Guo et al., 1997; Chen et al., 2000; Van Duyne, 2001). Recombin ation is catalysed by four recombinase monomers, each of which forms a C-shaped clamp around the DNA, with the N-terminal domains all on one side of the complex and the C-terminal domains on the other. In Cre, a C-terminal extension on each monomer interacts with the C-terminal domain of one of its neighbours in a cyclical arrangement (Figure 7A), whereas in FLP the cyclical exchanges involve other parts of the protein. The DNA is not planar in the complex; instead, all four arms are bent towards the recombinase C-terminal domains as they exit the complex (Figure 7B). The crystal structures suggest a model for strand exchange in which one pair of diagonally opposite recombinase monomers (e.g. XerC) exchanges one pair of strands to form a HJ intermediate. The other pair of recombinase monomers (e.g. XerD) then exchanges the second pair of strands to form recombinant product with the cores again aligned in an antiparallel configuration. For XerD strand exchange to precede XerC strand exchange, recombination must occur by the reverse of this pathway. The complex in which XerC is in an active configuration and is about to exchange XerC strands is distinct from the complex in which XerD is active (Figure 7A, compare left and right panels). The DNA has to bend one way within the complex to make a complex competent for XerC strand exchange, and the other way to perform XerD strand exchange first. Note also that the active monomer donates its C-terminal extension to the inactive monomer on the same duplex, whereas the inactive monomer donates its C-terminal extension to the active monomer on the other duplex. Therefore, the way in which the cores assemble into the complex defines which strands are exchanged first (Guo et al., 1999). Recombination reactions catalysed by Cre and λ integrase also occur with a preferred order of strand exchange. The order of strand exchange by Cre is dictated by asymmetry in the loxP core site, whereas the orientation of the attP core with respect to the P and P′ accessory sequences of attP defines the order of strand exchange by λ integrase (Kitts and Nash, 1988; Lee and Sadowski, 2001).
To account for the product topology of Xer recombination at wild-type psi sites on both circular and catenated substrates, recombination must occur in a synaptic complex equivalent to that shown in Figures 1A and 7C (Bath et al., 1999). This synapse has 2-fold rotational symmetry such that both recombination sites are equivalent, and the accessory sequences are intertwined so that the core sites are brought together in an antiparallel arrangement ready for strand exchange. The recombinases might be activated in the productive synapse simply by the correct juxtaposition of the core sites in a configuration that favours strand exchange. However, the recombinases might also be activated by direct contacts with PepA in the productive synapse.
The accessory sequences of two cer sites form an interwrapped synapse with PepA and ArgR, even in the absence of XerC and XerD (Alén et al., 1997), and it seems likely that the accessory sequences of psi can do the same thing. However, the synapse formed between two psiDC sites must differ from that formed between two psi sites, to accommodate the altered positions of the XerC- and XerD-binding sites. The path of the DNA could be unchanged, in which case XerC and XerD must change places (Figure 7D). Alternatively, PepA, XerC and XerD could occupy the same relative positions, in which case the DNA would have to follow a different path. In the former case, XerD occupies the position normally occupied by XerC, is in the active position in the recombinase–core complex and will carry out the first strand exchange (compare Figure 7D and A). The reaction will be completed by XerC strand exchange, giving products of the correct topology. However, if XerC and XerD proteins occupy their normal positions with respect to PepA, the path of the DNA could be altered in two different ways to give products of the correct topology (Figure 7E). These synapses are similar to that shown in Figure 7D except that the recombinase–core complex has been rotated relative to PepA, by plus or minus 90° around the 2-fold axis of the complex. XerD is again in the active position in both of these complexes and will exchange the first pair of strands.
Our results also demonstrate that the recombinase–core complex always forms with the recombinase C-terminal domains facing PepA and the accessory sequences (compare Figure 7A and C). If the recombinases assembled on wild-type psi with their N-terminal domains facing the accessory sequences, XerD would be in the active conformation and would catalyse the first strand exchange (compare Figure 7B and C). Similarly, if the N-terminal domains were facing the accessory sequences at psiDC, XerC strand exchange would proceed first. In both cases, the topology of the reactions would remain unchanged and the product would be the observed four-noded catenane. The direction in which the DNA arms exit the recombinase–core complex, pointing down towards the recombinase C-terminal domains, as well as the rotational phase of the DNA as it exits the accessory sequences complex, might fix the orientation of the recombinase complex within the synapse. Alternatively, interactions between the recombinases and PepA might fix this orientation. Although there is no evidence for interactions between PepA and the recombinases, it is interesting to note that any such interactions would be with the recombinase catalytic C-terminal domains. If XerC and XerD can exchange positions in the synapse to accommodate psiDC (as in Figure 7D), both recombinases would have to be capable of making equivalent interactions with PepA, although they are only 37% identical.
In recombination between psi and psiDC, the accessory sequences could assemble the same complex as that formed by the accessory sequences of two psi sites. However, this will bring the cores together in a parallel alignment in which they cannot recombine (Figure 7F). Recombination in this complex would lead to joining of two XerC-binding sites and two XerD-binding sites, would produce mismatches in the overlap region and would require forbidden XerC–XerD interactions. To carry out a legitimate XerC–XerD joining reaction, the cores have to be realigned in an antiparallel configuration, requiring a 180° rotation of one core with respect to the other. The recombination product will either be a (+) trefoil or a (+) five torus knot, depending on the direction of this rotation. The major product observed in the presence of PepA was trefoil (Figure 3C), so it appears that the cores are realigned preferentially by rotation in one direction (as in Figure 7G). It is interesting to note that PepA is still required for recombination on p-psiDC.psi, even though the synapse formed on this substrate must have quite a different structure from that formed on p-psi.psi.
Recombination between two psiDC sites is unusual amongst Xer reactions in that XerD strand exchange proceeds first. This has been seen previously only with combinations of proteins from different bacterial species (Blakely et al., 2000) or in Xer recombination reactions at dif in the presence of FtsK (Aussel et al., 2002). In the first case, it seems likely that altered interactions between partner recombinases that have not co-evolved to act together alter the preferred conformation of the recombinases bound at dif and thus the order of strand exchange. FtsK regulates Xer recombination to carry out its proper function at dif during chromosome partition. It is tempting to speculate that FtsK brings together two dif sites on a dimeric chromosome in a dividing cell such that XerD strand exchange occurs first, in a similar way to that proposed here for the action of accessory factors at psi and psiDC.
Materials and methods
Plasmids, DNA manipulations and proteins
The psi plasmids used in this work were derived from pMTL23 (Chambers et al., 1988). The wild-type psi recombination site was inserted into pMTL23 as a BamHI–EcoRI fragment of 381 bp from pSDC133 (Colloms et al., 1996). The CD core was inserted between the BamHI and SalI sites of pMTL23 as a double-stranded oligonucleotide with top strand sequence 5′-G′GATCCGGTGCGCGCAAGATCCA TTATGTTAAACG′TCGAC-3′. The DC core was inserted between the MluI and EcoRI sites of pMTL23 as a double-stranded oligonucleotide with the top strand sequence 5′-A′CGCGTTCGGGCGTTTAAC ATAATGGATCTTGCGCGCACCG′AATTC. The psiDC site was constructed by inserting the same oligonucleotide between the MluI and EcoRI sites of pLN5, which contains psi mutated to contain an MluI site between the accessory and core sequences (Colloms et al., 1996). The psiDC site was then moved to pMTL23 as a BamHI–EcoRI fragment. The 1.3 kb kanr gene from pUC4K (Pharmacia) was inserted into the EcoRI site of these pMTL23 derivatives as an EcoRI fragment. Substrate plasmids, with two recombination sites separated by kanr, were then constructed by ligating an ∼2.8 kb BsaI–BglII fragment from one of the kanr plasmids to an ∼1.7 kbp BsaI–BamHI fragment from the appropriate kans plasmid. Substrates were transformed into DS984 (Colloms et al., 1996), and supercoiled plasmid DNA was purified by caesium chloride ethidium bromide density gradient ultracentrifugation. DNA concentrations were estimated by measuring absorbance at 260 nm. PepA, ArcA, XerC and XerD were purified as described previously (Colloms et al., 1996, 1998; Subramanya et al., 1997).
Recombination assays
Recombination reactions contained 7.4 nM of supercoiled plasmid DNA in a volume of 20 µl. Reactions were set up in a buffer containing 50 mM Tris–HCl pH 8.0, 25 mM KCl, 5 mM spermidine, 1.25 mM EDTA, 25 µg/ml bovine serum albumin (BSA) with 10 or 40% glycerol. Reactions were started by adding PepA to 275 nM (hexamer), XerC to 147 nM and XerD to 140 nM. Reactions were normally carried out for 1.5 h at 37°C and stopped by the addition of phenol. After extraction with phenol, phenol–chloroform and then chloroform, DNA was recovered by ethanol precipitation, resuspended in TE, and treated with restriction enzymes as appropriate. DNase I nicking reactions were carried out for 1 h at 37°C in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 50 mM NaCl, 0.3 mg/ml ethidium bromide and 1 µg/ml DNase I. Ethidium bromide was removed from nicking reactions by extraction with phenol prior to gel electrophoresis. In vivo recombination was carried out by transforming plasmid DNA into AB1157 derivatives carrying chromosomal xerCY275F and xerDY279F mutations. Plasmid DNA was isolated from freshly transformed cultures using QIAprep kits (Qiagen) and analysed by electrophoresis on 1% agarose gels in TAE.
Gel electrophoresis
Recombination reactions treated with restriction enzymes were analysed on 1% agarose gels in TAE buffer. DNase I-treated reactions were analysed on 0.7% agarose gels in TAE. All gels were run at 3 V/cm for 16–24 h. For two-dimensional gels, recombination reactions were nicked with DNase I and run in duplicate lanes on 0.7% Seakem GTG agarose gels. One lane was stained with ethidium bromide and visualized by UV illumination. The other lane was excised from the gel and equilibrated for 2 h with two changes in XhoI restriction buffer (New England Biolabs NEBuffer 2 + BSA). The gel slice was then incubated in 5 ml of the same buffer at 37°C for 12 h with gentle shaking with 0.5 U/µl XhoI. The gel slice was cast into a new 1.2% agarose gel, and run in the second dimension. Gels were stained with Sybr Green I (Molecular Probes) and visualized on a Molecular Dynamics Fluoroimager 575.
Analysis of χ DNA by strand-separating gel electrophoresis
In vitro recombination reactions were carried out as above in 40% glycerol with PepA. After cleavage with XhoI, χ-form HJs were purified from an agarose gel using the QIAquick gel extraction kit (Qiagen). Purified χ-form DNA was 3′ end-labelled with [α-32P]dATP using Klenow polymerase in the presence of unlabelled dCTP, dGTP and dTTP. Labelled χ-form DNA, uncut and cut with appropriate restriction enzymes, was analysed by electrophoresis on a strand-separating 1% agarose gel in 50 mM NaOH, 1 mM EDTA, as described by Colloms et al. (1996). The DNA was transferred to Hybond-N membrane (Amersham) by capillary blotting and detected by autoradiography.
Acknowledgments
Acknowledgements
We wish to thank our colleages in Oxford and Glasgow (especially W.M.Stark) for stimulating conversation and useful comments on this manuscript. Thanks also to F.-X.Barre for providing xerC and xerD mutant strains, H.Ferreria for supplying XerC[De], and R.Baker for supplying other Xer proteins. The work was supported by a Wellcome Trust prize studentship to M.B., a Wellcome Trust Programme Grant to D.J.S., and Wellcome Trust Senior Fellowship number 57651 to S.D.C.
References
- Alén C., Sherratt,D.J. and Colloms,S.D. (1997) Direct interaction of aminopeptidase A with recombination site DNA in Xer site-specific recombination. EMBO J., 16, 5188–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska L., Grainge,I. and Sherratt,D. (1995) Effects of Holliday junction position on Xer-mediated recombination in vitro. EMBO J., 14, 2651–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska L.K., Grainge,I. and Sherratt,D.J. (1997) Action of site-specific recombinases XerC and XerD on tethered Holliday junctions. EMBO J., 16, 3731–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska L.K., Baker,R.A., Hallet,B. and Sherratt,D.J. (2000) Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J. Mol. Biol., 299, 391–403. [DOI] [PubMed] [Google Scholar]
- Aussel L., Barre,F.-X., Aroyo,M., Stasiak,A., Stasiak,A.Z. and Sherratt,D.J. (2002) FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of XerC and XerD recombinases. Cell, 108, 195–205. [DOI] [PubMed] [Google Scholar]
- Bath J., Sherratt,D.J. and Colloms,S.D. (1999) Topology of Xer recombination on catenanes produced by λ integrase. J. Mol. Biol., 289, 873–883. [DOI] [PubMed] [Google Scholar]
- Bednarz A.L., Boocock,M.R. and Sherratt,D.J. (1990) Determinants of correct res site alignment in site-specific recombination by Tn3 resolvase. Genes Dev., 4, 2366–2375. [DOI] [PubMed] [Google Scholar]
- Blake J.A., Ganguly,N. and Sherratt,D.J. (1997) DNA sequence of recombinase-binding sites can determine Xer site-specific recombination outcome. Mol. Microbiol., 23, 387–398. [DOI] [PubMed] [Google Scholar]
- Blakely G., May,G., McCulloch,R., Arciszewska,L.K., Burke,M., Lovett,S.T. and Sherratt,D.J. (1993) Two related recombinases are required for site-specific recombination at dif and cer in E.coli K12. Cell, 75, 351–361. [DOI] [PubMed] [Google Scholar]
- Blakely G.W., Davidson,A.O. and Sherratt,D.J. (1997) Binding and cleavage of nicked substrates by site-specific recombinases XerC and XerD. J. Mol. Biol., 265, 30–39. [DOI] [PubMed] [Google Scholar]
- Blakely G.W., Davidson,A.O. and Sherratt,D.J. (2000) Sequential strand exchange by XerC and XerD during site-specific recombination at dif. J. Biol. Chem., 275, 9930–9936. [DOI] [PubMed] [Google Scholar]
- Chambers S.P., Prior,S.E., Barstow,D.A. and Minton,N.P. (1988) The pMTL nic-cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene, 68, 139–149. [DOI] [PubMed] [Google Scholar]
- Chen Y., Narendra,U., Iype,L.E., Cox,M.M. and Rice,P.A. (2000) Crystal structure of a Flp recombinase–Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell, 6, 885–897. [PubMed] [Google Scholar]
- Colloms S.D., Sykora,P., Szatmari,G. and Sherratt,D.J. (1990) Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the λ integrase family of site-specific recombinases. J. Bacteriol., 172, 6973–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colloms S.D., McCulloch,R., Grant,K., Neilson,L. and Sherratt,D.J. (1996) Xer-mediated site-specific recombination in vitro. EMBO J., 15, 1172–1181. [PMC free article] [PubMed] [Google Scholar]
- Colloms S.D., Bath,J. and Sherratt,D.J. (1997) Topological selectivity in Xer site-specific recombination. Cell, 88, 855–864. [DOI] [PubMed] [Google Scholar]
- Colloms S.D., Alén,C. and Sherratt,D.J. (1998) The ArcA/ArcB two-component regulatory system of Escherichia coli is essential for Xer site-specific recombination at psi. Mol. Microbiol., 28, 521–530. [DOI] [PubMed] [Google Scholar]
- Cornet F., Mortier,I., Patte,J. and Louarn,J.M. (1994) Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J. Bacteriol., 176, 3188–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet F., Hallet,B. and Sherratt,D.J. (1997) Xer recombination in Escherichia coli. Site-specific DNA topoisomerase activity of the XerC and XerD recombinases. J. Biol. Chem., 272, 21927–21931. [DOI] [PubMed] [Google Scholar]
- Guo F., Gopaul,D.N. and Van Duyne,G.D. (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature, 389, 40–46. [DOI] [PubMed] [Google Scholar]
- Guo F., Gopaul,D.N. and Van Duyne,G.D. (1999) Asymmetric DNA bending in the Cre–loxP site-specific recombination synapse. Proc. Natl Acad. Sci. USA, 96, 7143–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallet B. and Sherratt,D.J. (1997) Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol. Rev., 21, 157–178. [DOI] [PubMed] [Google Scholar]
- Hallet B., Arciszewska,L.K. and Sherratt,D.J. (1999) Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol. Cell, 4, 949–959. [DOI] [PubMed] [Google Scholar]
- Kitts P.A. and Nash,H.A. (1988) An intermediate in the phage λ site-specific recombination reaction is revealed by phosphorothioate substitution in DNA. Nucleic Acids Res., 16, 6839–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. and Sadowski,P.D. (2001) Directional resolution of synthetic Holliday structures by the Cre recombinase. J. Biol. Chem., 276, 31092–31098. [DOI] [PubMed] [Google Scholar]
- Neilson L., Blakely,G. and Sherratt,D.J. (1999) Site-specific recombination at dif by Haemophilus influenzae XerC. Mol. Microbiol., 31, 915–926. [DOI] [PubMed] [Google Scholar]
- Nunes-Düby S.E., Azaro,M.A. and Landy,A. (1995) Swapping DNA strands and sensing homology without branch migration in λ site-specific recombination. Curr. Biol., 5, 139–148. [DOI] [PubMed] [Google Scholar]
- Sherratt D.J., Arciszewska,L.K., Blakely,G., Colloms,S., Grant,K., Leslie,N. and McCulloch,R. (1995) Site-specific recombination and circular chromosome segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci., 347, 37–42. [DOI] [PubMed] [Google Scholar]
- Stark W.M. and Boocock,M.R. (1995) Topological selectivity in site-specific recombination. In Sherratt,D.J. (ed.), Mobile Genetic Elements. IRL Press at Oxford University Press, Oxford, UK, pp. 101–129.
- Stark W.M., Boocock,M.R. and Sherratt,D.J. (1992) Catalysis by site-specific recombinases. Trends Genet., 8, 432–439. [PubMed] [Google Scholar]
- Stirling C.J., Colloms,S.D., Collins,J.F., Szatmari,G. and Sherratt,D.J. (1989) xerB, an Escherichia coli gene required for plasmid ColE1 site-specific recombination, is identical to pepA, encoding amino peptidase A, a protein with substantial similarity to bovine lens leucine aminopeptidase. EMBO J., 8, 1623–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanya H.S., Arciszewska,L.K., Baker,R.A., Bird,L.E., Sherratt,D.J. and Wigley,D.B. (1997) Crystal structure of the site-specific recombinase, XerD. EMBO J., 16, 5178–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D.K. (1989) Derivatives of ColE1 cer show altered topological specificity in site-specific recombination. EMBO J., 8, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne G. (2001) A structural view of Cre–loxP site-specific recombination. Annu. Rev. Biophys Biomol. Struct., 30, 87–104. [DOI] [PubMed] [Google Scholar]