Abstract
Three different families of DNA (cytosine-5) methyltransferases, DNMT1, DUMT2, DNMT3a and DNMT3b, participate in establishing and maintaining genomic methylation patterns during mammalian development. These enzymes have a large N-terminal domain fused to a catalytic domain. The catalytic domain is homologous to prokaryotic (cytosine-5) methyltransferases and contains the catalytic PC dipeptide, while the N-terminus acts as a transcriptional repressor by recruiting several chromatin remodeling proteins. Here, we show that the human de novo enzymes hDNMT3a and hDNMT3b form complexes with the major maintenance enzyme hDNMT1. Antibodies against hDNMT1 pull down both the de novo enzymes. Furthermore, the N-termini of the enzymes are involved in protein–protein interactions. Immunocytochemical staining revealed mostly nuclear co-localization of the fusion proteins, with the exception of hDNMT3a, which is found either exclusively in cytoplasm or in both nucleus and cytoplasm. Pre-methylated substrate DNAs exhibited differential methylation by de novo and maintenance enzymes. In vivo co-expression of hDNMT1 and hDNMT3a or hDNMT3b leads to methylation spreading in the genome, suggesting co-operation between de novo and maintenance enzymes during DNA methylation.
Keywords: co-operation/de novo enzymes/DNA (cytosine-5) methyltransferases/human/maintenance enzymes
Introduction
Vertebrate DNA is modified by the addition of a methyl group at position 5 of certain cytosine residues. These modified bases, 5-methylcytosines, are found predominantly in symmetrical CpG dinucleotides. An estimated 80% of the CpGs are modified in the human genome (Cooper and Krawczak, 1989). DNA methylation is inversely correlated with the transcriptional activity of a gene (Holliday et al., 1996), either via disruption of transcription factor binding to methylated genes or through an alternative mechanism in which transacting factors, such as histone deactylase (HDAC), methyl DNA binding proteins, or DNMT1 as a transcriptional repressor (Nan et al., 1998; Fuks et al., 2000), can inhibit gene expression. HDAC modifies histones and creates a chromatin structure unfavorable for transcriptional initiation (Fuks et al., 2000). Apart from gene expression, DNA methylation is also believed to be involved in genomic imprinting (Sleutels et al., 2000), X-chromosome inactivation (Csankovszki et al., 2001), development (Li et al., 1992), mutation (Schmutte and Jones, 1998) and cancer (Baylin et al., 1998).
In mammals, three families of DNA (cytosine-5) methyltransferases, DNMT1, DNMT2, DNMT3a and DNMT3b, have been identified. DNMT1 is the largest methyltransferase with a molecular mass of 184 kDa (Smith et al., 1992; Yoder et al., 1996; Pradhan et al., 1997). The N-terminal two-thirds of the protein is considered to be the regulatory domain, and the C-terminal region contains the catalytic domain (Yen et al., 1992). The catalytic domain shares sequence homologies with all DNMTs (Kumar et al., 1994). Both N- and C-termini are required for catalysis (Pradhan and Roberts, 2000). DNMT1b, a splice variant of DNMT1 (Bonfils et al., 2000), and an oocyte-specific isoform lacking the first 118 amino acids have been identified (Mertineit et al., 1998). In proliferating cells, DNMT1 is found to be associated with replication foci (Leonhardt et al., 1992), ensuring methylation of the daughter strand during DNA replication. Knockout of Dnmt1 in the mouse genome resulted in global demethylation and embryonic lethality (Li et al., 1992). Homozygous DNMT1-knockout embryonic stem cells were viable and maintained de novo methyltransferase activity (Lei et al., 1996). In comparison with DNMT1, DNMT2 is much smaller, with a predicted molecular mass of 45 kDa, lacking the large N-terminus, but containing all the conserved methyltransferase motifs (Dong et al., 2001). No biological activity of this protein has been demonstrated. Knockout of this gene in the mouse had no phenotypic effect (Okano et al., 1998a). Thus, the role and function of this protein is unknown. The discovery of a third family of methyltransferases, DNMT3a and DNMT3b, confirmed the presence of de novo methyltransferases in mammals (Okano et al., 1998b). Both of these enzymes are crucial for embryonic development and are responsible for the de novo methylation during embryogenesis that establishes the somatic methylation pattern of the organism (Okano et al., 1999). DNMT3a and DNMT3b are intermediate in size (100–130 kDa) in comparison with DNMT1 (∼184 kDa) and DNMT2 (∼50 kDa), and possess a smaller N-terminal region. Mutation of human Dnmt3b is believed to be associated with ICF (immunodeficiency, centromeric instability and facial anomalies) syndrome, characterized by hypomethylated centromeric satellite sequences and genomic instability (Xu et al., 1999). Several alternatively spliced forms of DNMT3b were also found in specific tissues (Robertson et al., 1999). Transcripts of all three classes of methyltransferase are observed throughout development, but are highest in fetal tissue (Okano et al., 1999).
Murine DNMT3a and DNMT3b have no preference for hemimethylated versus unmethylated oligonucleotides in vitro (Okano et al., 1998b). Overexpression of DNMT3b in 293/EBNA cells resulted in the establishment of a distinct methylation pattern in an episome as compared with DNMT3a, suggesting different preferred targets or requirements for de novo methylation in vivo (Hsieh, 1999). In Drosophila cells, overexpression of DNMT3a resulted in de novo methylation of the genome that impaired viability (Lyko et al., 1999). It has been speculated that DNMT3a could mediate the prevalent non-CpG methylation in embryonic stem cells. In Drosophila, where the genome has a small amount of methylated DNA (Gowher et al., 2000), overexpression of DNMT3a resulted in CpA and CpT methylation (Ramsahoye et al., 2000). In human embryonic fibroblasts, CpA and CpT methylations were observed (Woodcock et al., 1997), suggesting the presence of catalytically active hDNMT3s. Recently, using a gene knockout approach, functional co-operation between hDNMT3b and hDNMT1 has been proposed in a human colorectal carcinoma line (Rhee et al., 2002), although biochemical information on DNA methylation and physical interaction between the enzymes is lacking.
Here, we have studied the interactions between various human DNMTs (hDNMT1, hDNMT3a and hDNMT3b) both in vivo and in vitro. Identical pre-methylated substrates were used to study target specificity and methylation stimulation. Overexpression of de novo and maintenance methyltransferases led to de novo methylation. Data presented here show that interaction between mammalian DNMTs is crucial for the spreading and establishment of methylation patterns in vivo.
Results
In vivo association of hDNMTs
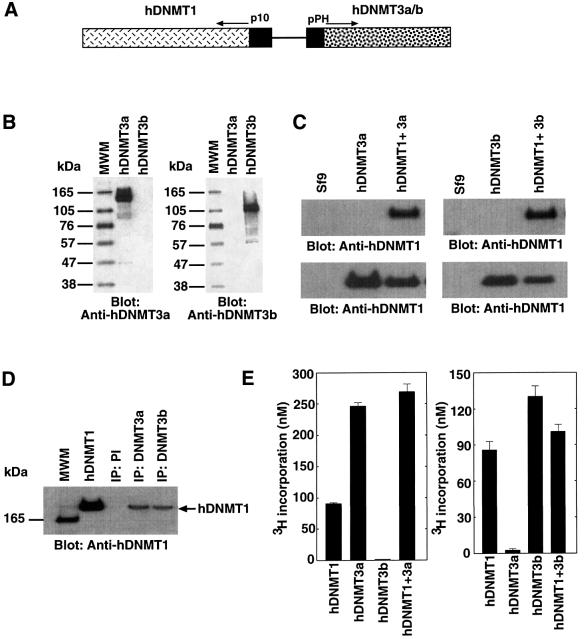
Establishment of the DNA methylation pattern of a mammalian genome is mediated by at least three enzymes working in concert. To identify the nature of co-operation between these different enzyme species, we constructed baculovirus transfection vectors able to express either hDNMT1 and hDNMT3a, or hDNMT1 and hDNMT3b (Figure 1A). Individual enzymes were also expressed separately. Expression was monitored using western blot analysis with antibody raised against hDNMT3a or hDNMT3b. These antibodies were specific for the respective antigen. Anti-hDNMT3a and -hDNMT3b were able to detect an ∼130 and ∼110 kDa antigen, respectively (Figure 1B). Upon transfection, the dual transfer vectors were integrated into the viral genome and expressed the respective enzymes in Sf9 insect cells (Figure 1C). Extracts from these cells were subjected to immunoprecipitation using antibodies specific for either hDNMT3a or hDNMT3b. Western blot analysis of the antibody- captured complexes revealed the presence of hDNMT1 (Figure 1D), suggesting that hDNMT1 was able to bind either hDNMT3a or hDNMT3b. Pre-immune serum could not capture hDNMT1 from either cell extract. To investigate whether these complexes are catalytically competent, Sf9 cell extracts expressing hDNMT1 and hDNMT3a, or hDNMT1 and hDNMT3b, were immunoprecipitated with saturating amounts of anti-DNMT3a or anti-DNMT3b antibodies, and assayed for methyl transfer activity using poly(dI–dC)·poly(dI–dC) substrate DNA. Significant methyltransferase activity was pulled down by all but the anti-hDNMT3b antibody in Sf9 cells expressing hDNMT1 and hDNMT3a (Figure 1E, left). A similar result was obtained with Sf9 cells expressing hDNMT1 and hDNMT3b, where the anti-hDNMT3a antibody immunoprecipitate had little methyltransferase activity (Figure 1E, right). Anti-hDNMT3a or -hDNMT3b antibodies were able to pull down slightly more, but not significantly higher methyltransferase activity than anti-hDNMT1. This might be due to competition for the epitope binding site of the hDNMT1 antibody by its interaction with hDNMT3a or hDNMT3b. Nevertheless, this result confirmed the antibody reactivity and the presence of catalytically active complexes.
Fig. 1. Recombinant hDNMT expression and binding analysis in vivo. (A) Construct used for the expression of DNMTs in Sf9 cells. hDNMT1 expression was driven by the baculovirus p10 promoter. In the same construct, the polyhedrin (pPH) promoter expressed hDNMT3a or hDNMT3b, as indicated by the arrow. (B) Specificity of hDNMT3 antibodies. Proteins, hDNMT3a or hDNMT3b are indicated at the top, and the antibody used at the bottom, of each panel. Each lane had 0.4 µg of purified antigen. Molecular weight markers are shown on the left. (C) Western blot analysis of the expression of the hDNMTs in insect cell extracts. Extracts from Sf9 cells expressing hDNMT3a alone, hDNMT3b alone, or DNMT1 plus hDNMT3a or hDNMT3b, are indicated at the top, and the antibody used at the bottom, of each panel. The Sf9 lane is a control cell extract. (D) Co-immunoprecipitation with anti-DNMTs in Sf9 cells expressing recombinant enzyme(s). Antibodies used for immunoprecipitation are indicated at the top. Monoclonal anti-GFP was used as a control. Insect cell extracts expressing hDNMT1 and hDNMT3a were immunoprecipitated with anti-hDNMT3a, and extracts from cells expressing hDNMT1 and DNMT3b were immunoprecipitated with anti-hDNMT3b. Pre-immune serum is marked as PI. The arrow shows the relative position of hDNMT1, along with purified hDNMT1 as a positive control. (E) Maintenance methyltransferase activity of co-immunoprecipitates with de novo enzymes. Extracts from Sf9 cells expressing hDNMT1 and hDNMT3a (left), or hDNMT1 and hDNMT3b (right), were immunoprecipitated with anti-DNMTs antibodies, as indicated. The immunoprecipitated product was assayed for hDNMT1 activity using poly (dI–dC)·poly (dI–dC) substrate DNA and tritiated AdoMet.
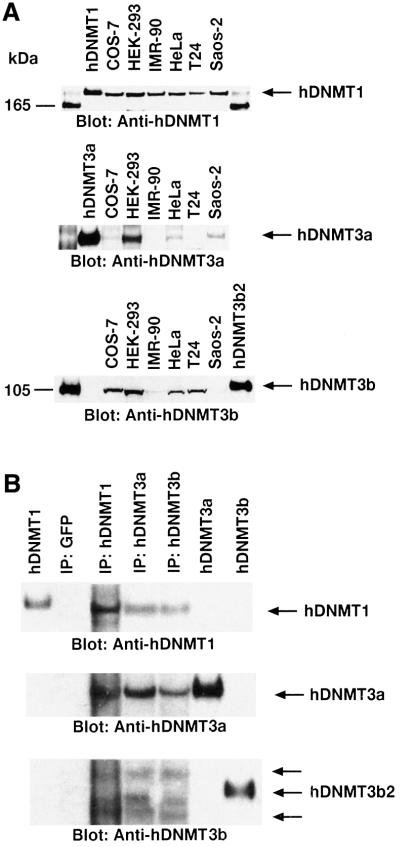
To find out whether similar complexes are formed in the mammalian nucleus, nuclear extracts from several mammalian cell lines were examined for the expression of hDNMTs. Western blot analysis of equal amounts of nuclear extract from one monkey and five human cell lines showed the presence of comparable amounts of DNMT1 (Figure 2A, top). However, expression of DNMT3a varied significantly between different cell lines (Figure 2A, middle). The highest expression of DNMT3a protein was present in the HEK-293 nuclear extract. The expression of DNMT3b was relatively uniform among the cell lines, except for Saos-2 cells (Figure 2A, bottom). Only one cell line, HEK-293, showed comparable levels of expression of all three DNMTs. Thus, HEK-293 nuclear extracts were subjected to immunoprecipitation using antibody specific to either one of the hDNMTs. The captured protein complexes were assayed for total DNA (cytosine-5) methyltransferase activity in the presence of substrate DNA and the cofactor tritiated AdoMet, and were found to be catalytically competent (data not shown). As expected, each anti-DNMT antibody was able to pull down DNA methyltransferase activity in comparison with a control GFP–monoclonal antibody. To determine the partners in the pull-down complexes, a western blot analysis was performed on the immunoprecipitated proteins using hDNMT1, hDNMT3a and hDNMT3b antibodies. Indeed, antibody against any one of the methyltransferases was able to pull down the other two, except the control GFP–monoclonal antibody (Figure 2B). This suggests that the human maintenance enzyme hDNMT1 is in close association with either one of the de novo methyltransferases, hDNMT3a, hDNMT3b or both, in vivo. Often, one higher and a slightly low molecular mass band appeared in the pull-down experiments with anti-hDNMT3b, indicating the association of more than one isoform of the enzyme. This confirms that hDNMTs form complexes in the nucleus.
Fig. 2. Expression profile and interactions between DNMTs in human cells. (A) Western blot analyses of DNMT1 (top), DNMT3a (middle) and DNMT3b (bottom) are shown. Nuclear extracts from different cell lines were used as indicated at the top, along with the positive control proteins hDNMT1, hDNMT3a and hDNMT3b. Molecular weight markers (in kDa) are in the extreme left and right lanes. Arrows indicate the relative locations of the endogenous DNMTs. Note that the same western blot gel was probed with three different antibodies for relative quantitation of the DNMTs. (B) Immunoprecipitation of hDNMTs in HEK-293 cell nuclear extracts. Antibodies used are indicated at the top. Control hDNMT1, hDNMT3a and hDNMT3b proteins were used as reference. Arrows on the right show the relative locations of the protein. Anti-hDNMT1, -hDNMT3a and -hDNMT3b antibodies were used for detection. Two additional arrows on the hDNMT3b western blot indicate the splice variants of hDNMT3b.
Association between hDNMTs involves N-terminal binding
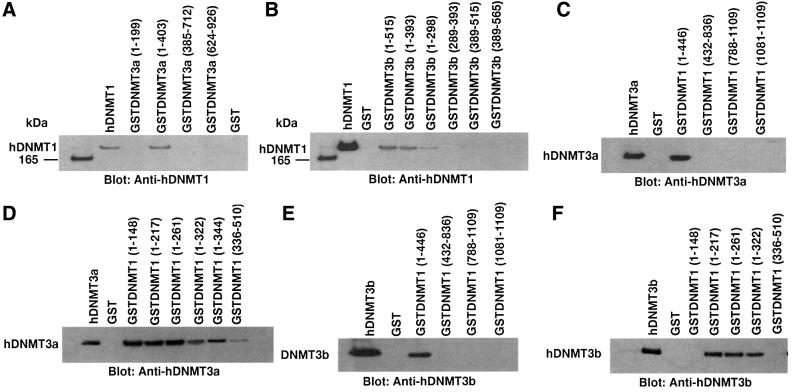
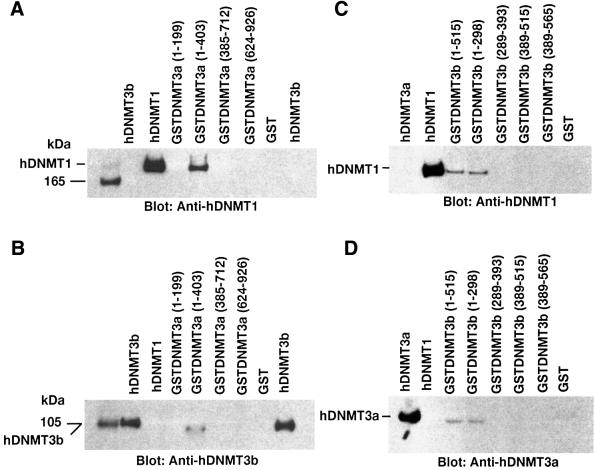
To identify the interacting amino acid residues, we first examined the ability of hDNMT3a to bind hDNMT1 in an in vitro GST pull-down assay. Fusion proteins consisting of GST and overlapping peptide fragments of hDNMT3a were bound to glutathione–Sepharose beads. These beads were incubated with full-length hDNMT1 to determine which region of hDNMT3a binds to hDNMT1. As shown in Figure 3A, amino acids 1–403 (GST–DNMT3a) of hDNMT3a bound to hDNMT1. In our assay conditions, GST protein alone or a GST fusion peptide containing 1–199 amino acids (GSTDNMT3a 1–199) was not able to pull down hDNMT1. These results suggest that amino acids between 199 and 403 of hDNMT3a are crucial for interaction with hDNMT1. A similar assay was performed to identify the interacting region of hDNMT3b with hDNMT1. The results are shown in Figure 3B. Amino acids 1–393 of hDNMT3b bound to hDNMT1 strongly. This association was reduced with GST fusion constructs containing amino acids 1–298 and abolished with amino acids 289–393 of hDNMT3b, suggesting amino acids 1–298 are essential for the interaction.
Fig. 3. Detailed mapping of the interacting regions of DNMTs. The GST–DNMT fusion peptides used for complex formation are indicated above each blot, with amino acid residue numbers shown in parentheses. Positive and negative controls are purified hDNMT1, hDNMT3a, hDNMT3b and GST protein, respectively. Molecular weight markers are in kDa. Antibodies used for the blots are indicated below each panel. (A) Immunoblot of bound hDNMT1 to various GST fusion fragments of hDNMT3a. (B) A similar immunoblot to that of (A) showing various GST fusion peptides of hDNMT3b bound to hDNMT1. (C) Identification of the hDNMT3a binding region of hDNMT1. (D) Finer mapping of the hDNMT1 binding region of hDNMT3a. (E) Immunoblot of bound hDNMT3b to various GST fusion fragments of hDNMT1. (F) Finer mapping of the hDNMT1 binding region of hDNMT3b. (G) hDNMT3a binding region of hDNMT3b. (H) hDNMT3b binding region of hDNMT3a. (I) Summary diagram of the various functional regions of DNMTs. Amino acid residues are indicated for various DNMTs. The relative locations of binding domains for other proteins are also indicated for reference. Proliferating cell nuclear antigen (PCNA), retinoblastoma gene product (Rb1), histone deacetylase (HDAC), DNA methyltransferase associated protein (DMAP) and methylated DNA-dependent allosteric activation domain (MDDAAD) are indicated. Another Rb binding region (Rb2) was recently discovered and mapped at the N-terminus of hDNMT1. Amino acid residues for protein–protein interaction are indicated for all hDNMTs.
To find the region of hDNMT1 that interacts with the de novo methyltransferases hDNMT3a and hDNMT3b, similar GST pull-down assays were performed. Overlapping GST–hDNMT1 fusion fragments were incubated with either full-length hDNMT3a or hDNMT3b. Surprisingly both of the de novo methyltransferases bound to the same region of hDNMT1 (amino acids 1–466, Figure 3C and E). Thus, a finer mapping was undertaken to characterize the binding regions of hDNMT1. Several smaller GST fusion peptides, GSTDNMT1 (1–148), GSTDNMT1 (1–217), GSTDNMT1 (1–261), GSTDNMT1 (1–322) and GSTDNMT1 (1–344), were constructed and GST pull-down assays were performed. Whereas hDNMT3a bound to the fusion protein containing amino acids 1–148 of hDNMT1 (Figure 3D), hDNMT3b did not. However, amino acids 1–217 but not amino acids 336–510 of hDNMT1 were able to bind to hDNMT3b, suggesting that the interacting region is between amino acids 149 and 217. Thus, the binding regions of hDNMT1 were distinct for both de novo methyltransferases.
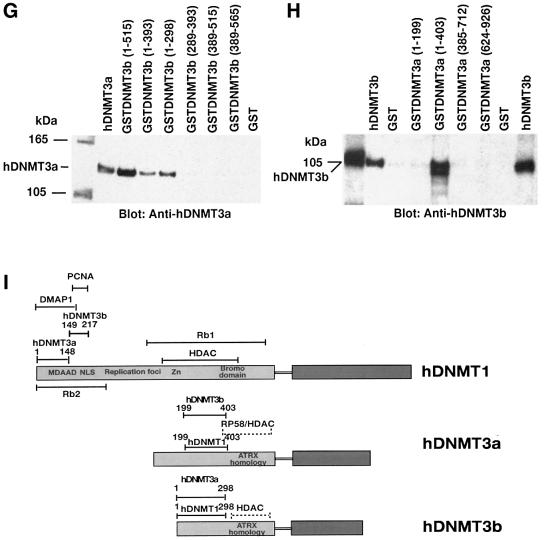
Increased expression of DNMT3a and DNMT3b enzymes during pre-implantation of the mammalian embryo has been proposed (Okano et al., 1999). It was suggested that, during this time, DNMT1 is not targeted to the nucleus, where DNMT3a and DNMT3b are located (Mertineit et al., 1998; Ramsahoye et al., 2000). This raises an interesting question about the mode of association between the two de novo methyltransferases. To study the interaction, two sets of GST pull-down assays were performed using GST-bound hDNMT3a or hDNMT3b. When GST–hDNMT3a was bound to beads, hDNMT3b was used as the binding partner, and vice versa. Indeed, hDNMT3a was able to bind hDNMT3b through amino acids 1–298 (Figure 3G). hDNMT3b was able to bind to hDNMT3a using amino acids 199–403 but not with 1–199 (Figure 3H). The interacting amino acids were located at the N-terminus of both enzymes, away from the HDAC binding region. These regions were identical to their binding motifs with hDNMT1, suggesting that both hDNMT3a and hDNMT33b can form complexes in the absence of the maintenance enzyme. In the presence of hDNMT1, both hDNMT3a and hDNMT3b can form complexes with hDNMT1, since they bind to different regions of hDNMT1. A summary of the binding regions for hDNMT1, hDNMT3a and hDNMT3b, along with other functionally identified regions, is presented in Figure 3I.
The order of enzyme binding and complex formation was studied using GST fusions of hDNMT3a or hDNMT3b immobilized on beads. The binding reaction was initiated by the addition of hDNMT1 to the beads, followed by washing off all the unbound hDNMT1. These binary protein complexes were challenged with purified hDNMT3a or hDNMT3b. The ternary complexes were washed to remove non-specific binding proteins and were analyzed by western blot analysis. Only GSTDNMT3a (1–403) was able to bind hDNMT1 (Figure 4A), which in turn bound to hDNMT3b (Figure 4B). Similarly, GSTDNMT3b (1–298/515) was able to bind hDNMT1 (Figure 4C), which further bound hDNMT3a (Figure 4D). hDNMT1 did not form a complex with pre-assembled hDNMT3a and hDNMT3b binary complex (data not shown), suggesting that enzyme recruitment might be an ordered phenomenon in vivo. Furthermore, the same binding regions of hDNMT3a and hDNMT3b are essential for hDNMT1 interaction (Figure 3I).
Fig. 4. Order of binding of hDNMTs. The GST–DNMT fusion peptides of hDNMT3a and hDNMT3b used for enzyme complex formation are indicated above each blot, with amino acid residue numbers shown in parentheses. Positive and negative controls are purified hDNMT1, hDNMT3a, hDNMT3b and GST protein, respectively. Molecular weight markers are in kDa. Antibodies used for the blots are indicated below each panel. (A) Immunoblot of bound hDNMT1 to various GST fusion fragments of hDNMT3a on GST beads. (B) A similar immunoblot showing various GST fusion peptides of hDNMT3b on GST beads bound to hDNMT1. (C) Identification of the full-length hDNMT3b binding to the hDNMT1–DNMT3a complex. (D) Identification of the full-length hDNMT3a binding to the hDNMT1–DNMT3b complex.
Localization of hDNMT1, hDNMT3a and hDNMT3b in COS-7 cells
In vitro binding studies led us to examine the intracellular distribution and co-localization of hDNMTs using immunocytochemistry. The interacting domain of hDNMT1 (amino acids 1–446) was cloned as a GFP fusion protein (pGFPDNMT446). The binding region of hDNMT3a (1–403) was cloned downstream of GST (pcDNAGST3A403). Thus, the GFPDNMT1 (1–446) fusion protein could be detected under UV light, whereas the GSTDNMT3a (1–403) protein could be detected with anti-GST monoclonal antibody. The other de novo methyltransferase, hDNMT3b (amino acids 1–515), was cloned with an N-terminal His6 tag (pcDNAHis3b515). Thus, the fusion protein could be identified in expressing cells using a monoclonal antibody to detect the His6 fusion peptide.
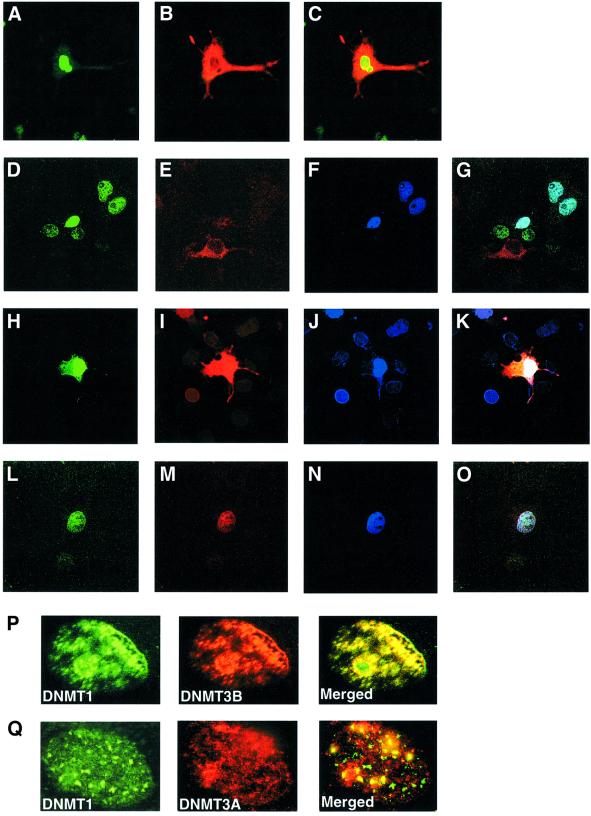
Log-phase COS-7 cells were transfected with each of the above constructs. Transfected DNA constructs expressed peptides of predicted size, as judged by a western blot assay (data not shown). Upon transfection of pGFPDNMT446, a GFP fusion protein was produced that could be visualized under UV as bright green spots. About 40–50% of the COS-7 cells showed distinct nuclear fluorescence in comparison with DAPI-stained nuclei (data not shown). Similar results were obtained for transfections with pcDNAGST3A403 or pcDNAHis3b515 using anti-GST or anti-His antibodies (data not shown), respectively. The fusion proteins appeared to be confined to the nucleus. Several sets of transfections were carried out to examine the localization profile of these proteins in different cell lines. Each transfection had either two or three constructs and the localization was analyzed using confocal microscopy. When pGFPDNMT446 and pcDNAGST3A403 were used in combination, the fusion peptide for hDNMT1 was expressed exclusively in the nucleus, as shown by the green fluorescence (Figure 5A), whereas GST–DNMT3a fusion proteins were expressed in both nucleus and cytoplasm (Figure 5B) with red fluorescence. A merged image of both GFP–DNMT1 and GST–DNMT3a showed a green–yellow nucleus and red cytoplasm (Figure 5C). When all three methyltransferases were expressed together, hDNMT1 and hDNMT3b were always present in the nucleus (Figure 5D, H and L for DNMT1 in green, and F, J and N for hDNMT3b in blue). A merge of the hDNMT1 and hDNMT3b images resulted in a whitish-blue nucleus (Figure 5G). hDNMT3a was present exclusively, either in the cytoplasm (Figure 5E) or nucleus (Figure 5M), or both nucleus and cytoplasm (Figure 5B and I). In some cells, all three DNMTs were co-localized in the nucleus (Figure 5L, M and N), suggesting a specific functional role for hDNMT3a in the nucleus. Merging of the fluorescence profiles for all three enzymes resulted in a white color in the nucleus (Figure 5K and O). Human DNMT1 in the nucleus appeared as bright green toroidal spots (Figure 5P and Q), as observed previously (Leonhardt et al., 1992). The His6-tagged DNMT3b fusion proteins (Figure 5F in blue and P in red, middle panel) behaved similarly. Both hDNMT1 and hDNMT3b merged to give either a light blue pattern (Figure 5G, upper right) or a greenish-yellow pattern (Figure 5P, merged), suggesting that amino acids 1–446 of hDNMT1 and 1–515 of hDNMT3b were capable of nuclear co-localization. Transfection of pGFPDNMT446 and pcDNAGST3A403 yielded a different localization pattern. Although GFP–hDNMT1 appeared as bright green toroidal spots in the nucleus (Figure 5Q), the red fluorescence pattern was diffuse for GST–hDNMT3a (Figure 5Q, middle panel). Nevertheless, some of the hDNMT3a co-localized with hDNMT1 (Figure 5Q, merged), suggesting that co-localization of human DNMTs takes place primarily in the nucleus.
Fig. 5. Representative confocal laser scanning microscopy images of COS-7 cells transfected with pGFPDNMT446 (hDNMT1), pcDNAGST3A403 (hDNMT3a) and pcDNAHis3b515 (hDNMT3b), or a combination of these clones expressing fusion/tagged hDNMT proteins. hDNMT1 appears in green, hDNMT3a in red and hDNMT3b in blue. Merged images of hDNMT1 and hDNMT3a are greenish-yellow in color; those of hDNMT1 and hDNMT3b are whitish-blue. Merged images of hDNMT1, hDNMT3a and hDNMT3b are white. Transfection and localization of hDNMT1 and hDNMT3a. (A) GFP–hDNMT1, (B) GST–hDNMT3a and (C) merge of hDNMT1 and hDNMT3a. Note that hDNMT3a is both nuclear as well as cytoplasmic. Transfection of all three hDNMTs: (D) GFP–hDNMT1, (E) GST–hDNMT3a, (F) His–hDNMT3b and (G) merge of all the hDNMTs. Note that hDNMT3a is only cytoplasmic (E and G). Another set of transfections showing all three hDNMTs: (H) GFP–hDNMT1, (I) GST–hDNMT3a, (J) His–hDNMT3b and (K) merge of all the hDNMTs. Note that in this set DNMT3a is localized both in the nucleus as well as cytoplasm (I and K). All three hDNMTs show nuclear co-localization. In another set of transfections, all three hDNMTs exclusively localized in the nucleus: (L) GFP–hDNMT1, (M) GST–hDNMT3a, (N) His–hDNMT3b and (O) merge of all the hDNMT1. (P and Q) Nuclear co-localization of hDNMTs. Expressions of GFP–hDNMT1 in green and GST–hDNMT3a or His–hDNMT3b in red are shown. The merged expression pattern is in yellow or greenish-yellow.
Pre-methylation stimulates DNA methylation
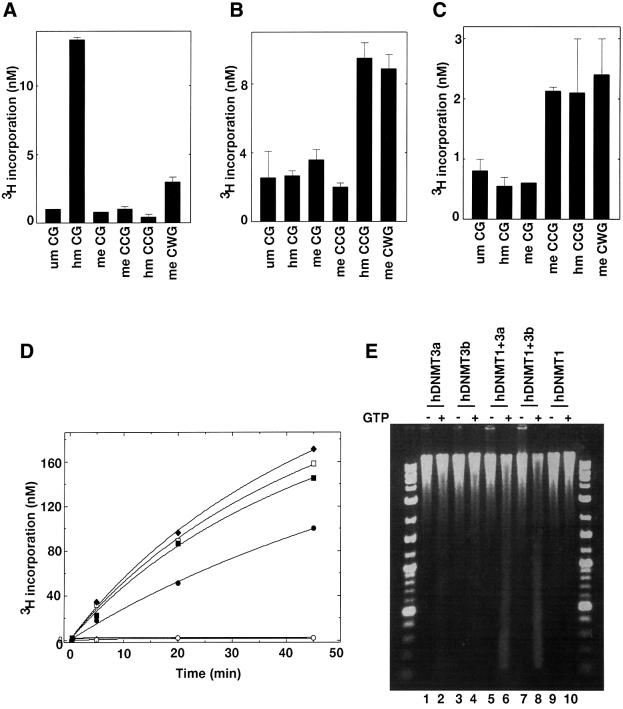
The murine male pronucleus is highly methylated. During fertilization, dynamic epigenetic modification occurs in which a wave of demethylation takes place and is followed by extensive de novo methylation (Santos et al., 2002). It is believed that DNMT3a and DNMT3b are the key players in this process. Immunohistochemical staining suggested the presence of small amounts of 5-methylcytosine during pre-implantation, when demethylase activity is highest, indicating that both hDNMT3a and hDNMT3b would have a pre-methylated substrate for de novo methylation. To examine the effect of pre-methylation on hDNMT3a and hDNMT3b activity, several duplex methylated and unmethylated oligonucleotides were synthesized and methylated in vitro in the presence of either one of the catalytically efficient enzymes. As expected, hDNMT1, the major maintenance methyltransferase, methylated hemimethylated CG (hm CG) duplex 12 times more efficiently (Figure 6A) than the unmethyl ated counterpart (um CG). This discrimination fits well with our previous observations (Pradhan et al., 1999). Pre-methylation in a non-CG context (me CWG) also stimulated DNA methylation (Figure 6A), suggesting de novo methylation of the available cytosine bases. Thus, non-CG methylation by hDNMT3a (Ramsahoye et al., 2000) might induce de novo methylation via hDNMT1 in vivo.
Fig. 6. Differential methylation of pre-methylated substrate DNA in vitro and methylation spreading in vivo. In vitro methylation using pre-methylated substrate with (A) hDNMT1, (B) hDNMT3a or (C) hDNMT3b. Equimolar concentrations of DNMTs were added to the reaction mix. Incorporation was measured after 30 min for hDNMT1 and 1 h for hDNMT3a or hDNMT3b. The experiment was conducted in duplicate and the average of two independent experiments is reported. (D) Co-operation for DNA methylation spreading in vitro. Reaction velocity curve of hDNMT1 and hDNMT3b (filled diamonds), hDNMT1, hDNMT3a and hDNMT3b (open squares), hDNMT1 and hDNMT3a (filled squares), hDNMT1 (filled circles), hDNMT3a (open circles), hDNMT3a and hDNMT3b (filled triangle), and hDNMT3b (open diamonds). Methylation by hDNMT3a, hDNMT3b and hDNMT3a, and hDNMT3b were similar and the data points are overlapping. The average values of two independent data points are plotted. The rates of methylation are shown in Table I. (E) Methylation spreading in vivo. Genomic DNAs from insect cells expressing different hDNMTs (indicated on top) were purified and digested with McrBC in the presence (+) or absence (–) of cofactor GTP. The digested DNA was analyzed on a 1.2% agarose gel. A 10 kbp to 100 bp ladder was used as a marker. um, unmethylated; hm, heminethylated; me, methylated.
To further elucidate the nature of pre-methylation versus de novo methylation by hDNMT3a and hDNMT3b, similar sets of methylase assays were carried out with the same substrates. The results are shown in Figure 6B and C. Neither DNMT3a nor DNMT3b preferred hemimethylated CGs, since incorporation in unmethylated and hemimethylated CG oligonucleotides was similar. However, hDNMT3a preferred hemimethylated CCG and methylated CWG substrates over unmethylated CG substrates, suggesting hDNMT3a can spread methylation in the presence of pre-methylated (hemimethylated CCG and methylated CWGs), but not with fully methylated CCGs. The same set of substrates exhibited a different methylation profile for hDNMT3b (Figure 6C). Hemimethylated and methylated CCG, and methylated CWG were good substrates for the de novo enzymes (Figure 6B and C). The data presented here show that de novo methylation in the mammalian genome might be a mixed mechanism, in which both processive and distributive mechanisms operate in concert with the help of maintenance and de novo DNMTs.
Co-operation between maintenance and de novo methyltransferases results in methylation spreading in vivo
The co-localization and association of hDNMTs shown above suggest that they might co-operate during genomic methylation. To test the above hypothesis in vitro, Micrococcus luteus genomic DNA was used as a substrate with various combinations of purified human de novo and maintenance methyltransferases. Equimolar amounts of enzymes were incubated with substrate DNA and tritiated AdoMet. The reaction progress curves were hyperbolic but showed linear product formation (methylation) in the first 15 min (Figure 6D). Although the M.luteus genome is 72% GC-rich, very little methylation was observed by hDNMT3a and hDNMT3b, either alone or in combination in vitro. The rate of methylation, as judged by nanomolar tritium incorporation into the substrate DNA for DNMT1 was 10- to 40-fold over the de novo enzymes hDNMT3b and hDNMT3a, respectively (Table I). However, combinations of de novo and maintenance enzymes (hDNMT1 + hDNMT3a, hDNMT1 + hDNMT3b and hDNMT1 + hDNMT3a + hDNMT3b) showed increased rates of methylation (Figure 6D). The rates of reaction of de novo and maintenance enzymes were higher than those of any of the individual enzymes (Table I). These results suggest that a small amount of methylation by either hDNMT3a or hDNMT3b can stimulate DNA methylation by hDNMT1 on genomic DNA (Bacolla et al., 1999, 2001).
Table I. Co-operation of human de novo and maintenance methyl transferases for DNA methylation.
| DNA methyltransferase | Methylation (nM CH3/h) |
|---|---|
| hDNMT1 | 15 ± 0.45 |
| hDNMT3a | 0.4 ± 0.04 |
| hDNMT3b | 1.25 ± 0.04 |
| hDNMT3a + hDNMT3b | 1.8 ± 0.03 |
| hDNMT1 + hDNMT3a | 18 ± 0.1 |
| hDNMT1 + hDNMT3b | 28 ± 0.8 |
| hDNMT1 + hDNMT3a + hDNMT3b | 26.1 ± 0.06 |
Each methylation reaction was performed twice in duplicate. Final methyl group incorporation was calculated after deduction of the background value.
DNAs from Sf9 cells expressing both de novo and maintenance methyltransferases (Figure 1A) were assayed to examine the enzyme co-operation in vivo. These cells do not normally contain a detectable level of methylated cytosine, thus allowing an examination of the methylation status after expression of various combinations of the three DNMTs. All the constructs expressed the respective enzymes, as judged by western blot and methyltransferase assays (Figure 1C). Genomic DNA was purified 48 h post-infection (peak enzyme activity), and the methylation status of the DNA was analyzed using an McrBC assay. In the presence of GTP and McrBC endonuclease, methylated DNA, but not unmethylated DNA, can be digested, thus providing a convenient assay. Extensive genomic DNA methylation was observed for cells expressing hDNMT1 and hDNMT3a (Figure 6E, lane 5 versus 6) or hDNMT1 and hDNMT3b (Figure 6E, lane 7 versus 8). Significantly smaller amounts of genomic DNA digests were obtained when either hDNMT3a or hDNMT3b was expressed individually. Cells expressing hDNMT1 showed little or no difference in their genomic DNA digestion pattern in the presence of GTP and McrBC, thus there was undetectable de novo methylation of the genome, as observed previously (Pradhan and Roberts, 2000). These results suggest that ectopically expressed de novo and maintenance methyltransferases can facilitate methylation spreading in the eukaryotic genome and both of the enzymes are required for such an event.
Discussion
We have demonstrated binding between human DNMTs involving the N-terminal regulatory regions. The biological significance of this association may be involvement in gene silencing via transcriptional repression or DNA methylation. Indeed, functional co-operation of human de novo methyltransferase DNMT3b and maintenance methyltransferase DNMT1 is essential for almost all of the methylation in the colorectal cancer cell line HCT116 (Rhee et al., 2002). Knockout of either DNMT3b or DNMT1 resulted in minimal effects on genomic methylation. However, a double knockout of DNMT1 and DNMT3b led to loss of most of the genomic methylation, despite the presence of the de novo methyltransferase DNMT3a. The loss extended to the repeated DNA sequences, such as satellite 2 and the Alu family repeats. Loss of insulin-like growth factor II (Igf2) imprinting and abrogation of silencing of the tumor suppressor gene p16INK4a were also observed in double-knockout cell lines. In mouse cells, functional co-operation is required between DNMT1 and DNMT3a and/or DNMT3b for methylation of a selected class of sequences, including abundant LINE-1 promoter sequences (Liang et al., 2002). This suggests that both de novo and maintenance enzymes must co-operate to carry out the maintenance of methylation and the silencing of the genes in the mammalian genome.
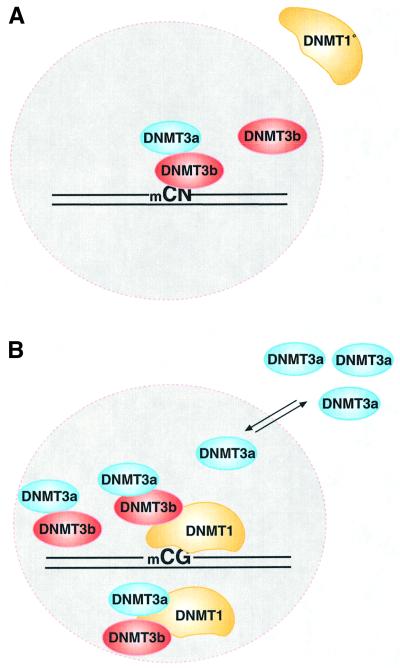
Methylation also plays an important role in establishing epigenetic patterns during mammalian development. In pre-implantation embryos, the mammalian genome is believed to be essentially unmethylated. At this stage, DNMT1 is transcribed and enzyme is made in the cytoplasm, but it is not targeted to the nucleus (Mertineit et al., 1998). However, 5-methylcytosine staining of early mouse embryos at different stages indicated the presence of methylated DNA (Santos et al., 2002). Transcripts of DNMT3a and DNMT3b could be detected as early as embryonic day 6.5, with the peak activity of DNMT3a at day 10.5 (Okano et al., 1999). These authors speculated that de novo enzymes are most active during the early developmental stages, where they establish the pattern of DNA methylation. In later developmental stages, DNMT1 activity is high and DNMT3a and DNMT3b levels go down. In fact, for DRM2 of mouse Igf2, an imprinted gene, all three enzymes were required for the correct establishment of methylation (Okano et al., 1999). Many human cell lines express hDNMT3a as well as hDNMT3b (Figure 2A), although it is not known whether this is merely an artifact of the establishment process. Although all the mammalian DNMTs recognize the same CG sites and modify them, the relationship between them, their mode of action and precise target recognition is still unknown. Recently, it has been demonstrated that DNMT3a and DNMT3b can modify non-CG sites such as CA, CT and to a lesser extent the 5′ C of the CC dinucleotide (Aoki et al., 2001; Gowher and Jeltsch, 2001; Yokochi and Robertson, 2002). Murine DNMT3a, the most studied de novo methyltransferase, shows a 3-fold preference for de novo methylation as compared with maintenance activity, and exhibits strand asymmetry (Lin et al., 2002; Yokochi and Robertson, 2002). One key question is how the unmethylated pre-implantation genome acquires its DNA methylation pattern during the short period of embryonic cell division. One possibility is that methylation spreading might be involved, with a small amount of de novo methylation by hDNMT3a and/or hDNMT3b followed by rapid allosteric activation of hDNMT1, a property encoded in the N-terminal region (Bacolla et al., 1999). Our binding results suggest that either hDNMT3a or DNMT3b could recruit DNMT1 and facilitate the rapid establishment of the de novo methylation pattern. Co-localization of hDNMT3b and hDNMT1 in the nucleus supports such a hypothesis. Since DNMT3a and DNMT3b are active during pre-implantation, both are likely to be involved in an initial wave of de novo methylation. With different target selectivity, both enzymes would be able to establish the initial blueprint of DNA methylation at CN (N is any nucleotide) sites. During this time, DNMT1 resides outside the nucleus, perhaps as a truncated version of DNMT1 (DNMT1°), as observed by Mertineit et al. (1998) (Figure 7A). After the pre-implantation stage, DNMT1 is transported into the nucleus. Nuclear DNMT1 co-localizes with DNMT3b and perhaps binds less strongly to DNMT3a. The presence of DNMT1 and DNMT3b in the nucleus brings out a wave of de novo and maintenance methylation. DNMT3a, which is capable of non-CG methylation, is perhaps escorted out of the nucleus, since further non-CG methylation is either not essential or may even be deleterious for later development (Figure 7B). Thus, co-operation between de novo and maintenance methyltransferases is crucial for the establishment of methylation. This co-operation might be lost during abnormal growth and development, such as in cancer.
Fig. 7. Model for DNA methylation. (A) mCpN methylation by DNMT3a and DNMT3b. DNMT3s can catalyze mCG, mCA, mCT and mCC methylation in vivo, either individually or together. This type of methylation is predominant in the pre-implantation stage of mammalian development. A shorter DNMT1, lacking 118 amino acids, stays in the cytoplasm, except at the 8-cell stage of the embryo (Mertineit et al., 1998). Lack of this N-terminal region might lead to an unfavorable interaction between de novo and maintenance methyltransferases. (B) DNA methylation by DNMTs. Both DNMT1 and DNMT3b co-localize in the nucleus. DNMT3a may interact with either DNMT3b or the full-length DNMT1. Excess DNMT3a is escorted out to the cytoplasm, or cytoplasmic DNMT3a is not allowed into the nucleus. DNMT1 ensures CG methylation in the cell. The nuclear membrane is shown as a dotted circle.
Thus, a model emerges in which DNMT3a and DNMT3b, in the absence of DNMT1, lay down an initial pattern of methylation that is spread once DNMT1 joins the methylation complex in the nucleus, perhaps via allosteric activation by methylated DNA. It is now crucial to determine what are the signals that cause DNMT3a and DNMT3b to set this initial pattern. It seems unlikely that DNA sequence alone is sufficient, and so chromatin structure in collaboration with various protein factors is probably responsible. Thus, accessibility of the target bases may be the key factor in establishing the methylation blueprint of the mammalian genome.
Materials and methods
Cloning of human DNMT3a, DNMT3b and expression constructs
The DNA sequence for the last ∼400 amino acids and 3′-UTR of hDNMT3a was obtained by sequencing expressed sequence tag (EST) clones (AA312013, W76111 and H04031) from the American Type Culture Collection (ATCC, Rockville, MD). To obtain 5′-UTR and N-terminal sequences, oligonucleotides N-029 (5′-TGCAGCAGCCA TTTTCCACTGCTC-3′) and N-031 (5′-CGTCTCCGAACCACATGA CCCAGC-3′) were designed according to a partial sequence deduced from EST AI023308, AI021920 and AA355824. These complement the sequence around amino acids 130 and 320, respectively. hDNMT3a cDNA was amplified using the 5′-RACE kit (Life Technologies Inc., Rockville, MD) with oligonucleotide N-021 (complementary to 3′-UTR) and human fetal liver poly(A)+ RNA (Clontech, Palo Alto, CA). Two rounds of PCR were run using this cDNA as a template [first-round primers N-031 and AAP (GGCCACGCGTCGACTAGTACGGGIIGG GIIGGGIIG); second-round primers N-029 and AUAP (5′-GGC CACGCGTCGACTAGTAC-3′)]. The Clontech Advantage-GC cDNA PCR kit was used in both PCRs. A final PCR product of ∼680 bp was obtained and directly sequenced to obtain the DNA sequence of the 5′-UTR and the first 100 amino acids. To clone the cDNA regions encoding the complete amino acid sequence, two rounds of PCR were performed using human liver cDNA (Clontech) and the same PCR kit as above [first-round primers N-078 (5′-TAACTCTCGCCTCCA AAGACCACGAT-3′, 5′-UTR) and N-021; second-round primers N-072 (5′-TCGGATATCAACATGCCTTCCAGCGGCCCCGGGG ACA-3′, N-terminal) and N-074 (5′-CCCAAGCTTCCCGGGACCCA CACACGCAAAATACTCCTTCAGCG-3′, C-terminal)]. An ∼2.7 kb DNA fragment was obtained, digested with EcoRV and HindIII, and cloned into the corresponding sites of pBR322. The final cDNA consensus sequence is 4258 nucleotides in total (DDBJ/EMBL/GenBank accession No. AF331856). It differs from the previously published DNMT3a sequence (accession No. AF067972) in several places. The biggest difference is in the 5′-UTR region, which is caused by different transcription start sites leading to different first exons. For all the other differences in the coding region and the 3′-UTR region, our sequence can be confirmed by a partially assembled genomic DNA sequence (DDBL/EMBL/GenBank accession No. NT_005204). Lastly, the hDNMT3a insert in pBR322 was moved, using a two-step ligation, into the baculovirus expression vector pVIC1. Again, the hDNMT3a insert in pVIC1 was confirmed by sequencing.
To clone the 5′-UTR and N-terminal regions of hDNMT3b, cDNA was synthesized similarly to DNMT3a, except that the primer used was N-083 (5′-CAACATGGGCTTCAGCTGGTCCTCCAATGA-3′, close to the 5′-end). Then two rounds of PCR were performed on the cDNA using the Advantage-GC cDNA PCR kit (Clontech); first, primers AAP and N-082 (5′-CATGGTACATGGCTTTTCGATAGGA-3′) with 35 cycles; second, primers AUAP and N-111 (5′-CGATAGGAGACGAGCTTATTGAA GGT-3′) with 30 cycles. The resulting 1.2 kb PCR fragment was directly sequenced using N-116 (5′-TCCTCTCCATTGAGATGCCTGGTG-3′) and N-117 (5′-CCGTTGACGAGGATCGAGTCTTCC-3′) to obtain the 5′-end sequences. Sequencing of several ESTs and PCR products (data not shown) gave the rest of the hDNMT3b sequence. The final assembled sequence (4335 nucleotides; accession no. AF331857) was confirmed by a genomic sequence entry (accession no. AL035071). This sequence differs from the previously published DNMT3b (accession no. AF156488) only in the 5′-UTR region, which appears to be the result of a different transcription start leading to different first exons.
For 5′-end cloning, a second round of PCR was carried out with DNA from the first PCR (see above sequencing step) as a template, using primers N-111 and N-115 (5′-CGGAATTCAACATGAAGGGAGA CACCAGGCATCTCAATGGAG-3′, translation start). The amplified ∼900 bp fragment was treated with Klenow enzyme, cut with EcoRI and cloned into the EcoRI and EcoRV sites of pBR322.
For 3′-end cloning, two rounds of PCR were carried out using human testis cDNA (Clontech, USA) as a template [PfuTurbo Polymerase; first-round primers N-120 (5′-GTCCTGGCTCTGCCACACACCCCAGT-3′) and N128 (5′-GCTATGTCTGGCATGCGGTGGGTC-3′); second-round primers N-065 (5′-TTCACATGCAAAGTAGTCCTTCAG-3′) and N-133 (5′-CAAGTTCTCCGAGGTCTCTGCAGACA-3′)]. A mixture of two bands of ∼1.5 and 1.7 kb, respectively, was obtained as the final product; both were cloned into the SmaI site of pTYB2. Sequencing of several clones revealed that the heterogeneity was the result of several splice variants. Nucleotides 1387–1446 are missing from all the clones, and the longest clone has all but this region. This clone is equivalent to the mouse DNMT3b2 gene. One clone has a 15 nucleotide insertion (5′-ACTCTG CCTTTGCAG-3′, corresponding to TLPLQ peptide sequence) between nucleotides 1242 and 1243; this insertion is absent from any mouse DNMT3b clones. The longest clone without this 15 nucleotide insertion was amplified using Vent DNA polymerase (New England Biolabs, Beverly, MA) and primers N-135 (5′-TGGGTCCAGTGGTT TGGCGATGGCAAGTTCTCCGAGGTCTCTGCAGAC-3′) and N-100 (vector region, 5′-GCGGACACTACGAGGTGTTCTAACA-3′), digested with BstXI and PshAI, and cloned into the corresponding sites from a pBR322-DNMT3b-5′ clone. The resulting clone was confirmed by sequencing to contain the complete coding region fused to part of the intein tag from the vector pTYB2. Lastly, the whole insert was excised by EcoRI and PshAI, and cloned into the corresponding sites in pVIC1. The final pVIC1-DNMT3b2 plasmid was confirmed by sequencing of the complete insert.
For GST fusion proteins, the indicated fragments from hDNMT1 (BamHI–EcoRI), hDNMT3a (EcoRI–SalI) and hDNMT3b (BamHI–SalI) were subcloned in-frame into pGEX-5X-1 (Amersham Pharmacia). For immunocytochemistry, hDNMT1 (amino acids 1–446) was ligated into the EcoRI and BamHI sites in pEGFP-C2 (Clontech). The resultant construct was pGFPDNMT446. For the hDNMT3a (amino acids 1–403) fusion construct, the vector pcDNA3.1(+) (Invitrogen Corp., Carlsbad, CA) was used. The final construct was pcDNAGST3a403. For the hDNMT3b (amino acids 1–515) fusion construct, a His6-based vector, pcDNA3.1, was used.
GST fusion protein and pull-down assay
GST fusion and GST control proteins were expressed in Escherichia coli BL21 or ER2502 cells. The recombinant proteins were induced with 0.1–0.3 mM IPTG at 30°C for 5 h. GST proteins were purified from bacterial crude cell lysates according to the manufacturer’s instructions. GST pull-down assays were performed as described previously (Pradhan and Kim, 2002).
Cell culture, transfection and immunocytochemistry
All of the cell lines were obtained from the ATCC: COS-7 (SV40 transformed, fibroblast; ATCC no. CRL-1651), HEK-293 human (embryonic kidney, transformed with adenovirus 5 DNA; ATCC no. CRL-1573), IMR-90 (normal human lung fibroblast; ATCC no. CCL-186), HeLa (human cervix, adenocarcinoma, epithelial; ATCC no: CCL-2), T24 (human, transitional cell carcinoma, urinary bladder, epithelial; ATCC no: HTB-4), Saos-2 (human osteosarcoma, bone; ATCC no: HTB-85). Cells were grown as per ATCC recommendations. For immunocytochemistry, epithelial COS-7 cells were cultured on glass cover slips in DMEM (Gibco-BRL) containing 10% FBS. Plasmid constructs were transfected into COS-7 cells using Fugene 6 Transfection Reagent (Roche, Indianapolis, IN), as directed in the product instructions. Post-transfection (24–48 h), cells were fixed with 100% methanol at –20°C for 10 min. Cover slips were blocked for 1 h with 10% goat serum, 1% BSA, 0.02% NaN3 in PBS at room temperature. The cover slips were incubated overnight at 4°C with primary antibodies, diluted in 1% BSA in 1× PBS. GST monoclonal antibody (Cell Signaling Technology, Beverly, MA) 1/800, GST antibody (Cell Signaling Technology) 1/500 and anti-HisG antibody (Invitrogen) 1/5000 were used. The following day, cover slips were first washed three times for 5 min with 1× PBS. Secondary antibodies against rabbit IgG or mouse IgG, conjugated with Alexa 488, Alexa 594 or Alexa 350 (Molecular Probes, Eugene, OR), were diluted at 1:1000 in 1% BSA in 1× PBS, and incubated individually and/or simultaneously on the cover slips for 30 min. Some samples were additionally stained with Hoechst dye (Molecular Probes) for nuclear staining, as per product instructions. Samples were washed again with PBS for 5 min, three times. Cover slips were mounted onto slides using Prolong Anti-Fade reagent (Molecular Probes), as per product instructions, and viewed by a Zeiss digital camera on a Nikon fluorescence microscope, or a confocal Carl Zeiss LSM510 microscope mounted on an Axioplan2 microscope. The objective lenses were 40× and 63×.
Insect cell culture, viral transfection, recombinant protein expression and purification
A pupal ovarian cell line (Sf9) from the fall armyworm Spodoptera frugiperda was used for co-transfection and expression of the human Dnmt3a and Dnmt3b, as described previously (Pradhan et al., 1999). Co-transfection of a monolayer of Sf9 insect cells was carried out using BaculoGold DNA, a modified, linearized Autographa california nuclear polyhedrosis virus (AcNPV) DNA (PharMingen) and the transfer vector pVIC1DNMT3a or pVIC1DNMT3b-2. The purified protein was stored at –20°C. Purity of the protein was checked by SDS–PAGE (4–20% Tris–glycine–SDS gradient gel). Purified protein was quantitated using the Bradford assay with BSA as standard.
DNA (cytosine-5) methyltransferase assay and data analysis
Methyltransferase assays were carried out at 37°C for 30–60 min in duplicate with a total volume of 25 µl of reaction mix. Reaction conditions were as described previously (Bacolla et al., 1999, 2001; Pradhan et al., 1999. Data obtained were plotted using the GraphPad PRISM program (GraphPad Software Inc., San Diego, CA).
Immunoprecipitations and western blot analysis
Sf9 insect cells were infected with recombinant baculovirus containing both hDNMT3 and hDNMT1 (Figure 1A). Post-infection (48 h), the cells were lysed in a binding buffer containing 50 mM Tris–HCl pH 7.5, 28 µM ZnCl2, 0.1% Triton X-100, 220 mM NaCl and 10% glycerol. Appropriate purified anti-DNMT antibodies (New England Biolabs) were used to capture the hDNMT complexes. The hDNMT–antibody complex was then trapped using protein A–Sepharose beads. After three washes (binding buffer containing 300 mM NaCl), the bound proteins were denatured and separated on a denaturing gel, blotted and probed with specific antibodies. For co-immunoprecipitation, HEK-293 cells were grown in MEM supplemented with 10% FCS. The nuclei were purified and co-immunoprecipitation was carried out in TNN buffer (40 mM Tris–HCl pH 8.0, 120 mM NaCl, 0.5% NP-40). After clarification of the extract with appropriate control IgGs, the protein complex was captured with antibodies conjugated to protein A or G magnetic beads (New England Biolabs) and analyzed by western blotting.
Cell extracts were mixed with SDS loading dye with 2 mM Tris–(2-carboxyethyl)–phosphine (TCEP) in place of DTT, boiled at 95°C for 5 min and loaded on a 4–20% Tris–glycine–SDS gradient gel. TCEP is a strong reducing agent and does not take part in the cleavage process. The proteins were blotted on an Immobilin-P membrane and probed with an anti-DNMT1, -DNMT3a or -DNMT3b antibody (New England Biolabs), followed by chemiluminescent detection of the human DNA cytosine methyltransferases.
Oligonucleotides and substrate DNA
Poly(dI–dC)·poly(dI–dC), average length 7000 bp, was obtained from Pharmacia. Micrococcus luteus DNA was obtained from Sigma. SNRPN exon 1 and FMR-1 locus oligonucleotide sequences were described in Pradhan et al. (1999). Oligonucleotides used for the methyltransferase stimulation assay, and their sequences, are listed in Table II.
Table II. Oligonucleotides used for the methyltransferase stimulation assay.
| Oligonucleotide | Sequence |
|---|---|
| um CG | AGACCGGTGCCAGCGCAGCTGAGCCGGATCTCTGGCCACGGTCGCGTCGACTCGGCCTAG |
| hm CG | AGACMGGTGCCAGMGCAGCTGAGCMGGATCTCTGGCCACGGTCGCGTCGACTCGGCCTAG |
| me CG | AGACMGGTGCCAGMGCAGCTGAGCMGGATCTCTGGMCACGGTCGMGTCGACTCGGMCTAG |
| hm CCG | AGAMCGGTGCCAGCGCAGCTGAGMCGGATCTCTGGCCACGGTCGCGTCGACTCGGCCTAG |
| me CCG | AGAMCGGTGCCAGCGCAGCTGAGMCGGATCTCTGGCMACGGTCGCGTCGACTCGGCMTAG |
| me CWG | AGACCGGTGCMAGCGMAGMTGAGCCGGATCTCTGGCCACGGTMGCGTMGAMTCGGCCTAG |
M represents 5-methylcytosine. um, unmethylated; hm, hemimethylated; me, methylated.
Acknowledgments
Acknowledgements
We thank Dr T.Evans for discussions and critical reviews. Technical assistance from Ms Nancy Badger for baculovirus expression of protein is highly appreciated. Support and encouragement from Dr D.G.Comb, New England Biolabs, is highly appreciated. J.N. was supported by SBIR R43 GM56535 to R.J.R.
References
- Aoki A., Suetake,I., Miyagawa,J., Fujio.,T. Chijiwa,T., Sasaki,H. and Tajima,S. (2001) Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 29, 3506–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A., Pradhan,S., Roberts,R.J. and Wells,R.D. (1999) Recombinant human DNA (cytosine-5) methyltransferase II. Steady-state kinetics reveal allosteric activation by methylated DNA. J. Biol. Chem., 274, 33011–33019. [DOI] [PubMed] [Google Scholar]
- Bacolla A., Pradhan,S., Larson,J.E., Roberts,R.J. and Wells,R.D. (2001) Recombinant human DNA (cytosine-5) methyltransferase II. Allosteric control, reaction order, and influence of plasmid topology and triplet repeat length on methylation of the fragile X CGG.CCG sequence. J. Biol. Chem., 276, 18605–18608. [DOI] [PubMed] [Google Scholar]
- Baylin S.B., Herman,J.G., Graff,J.R., Vertino,P.M. and Issa,J.P. (1998) Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res., 72, 141–196. [PubMed] [Google Scholar]
- Bonfils C., Beaulieu,N., Chan,E., Cotton-Montpetit,J. and MacLeod,A.R. (2000) Characterization of the human DNA methyltransferase splice variant Dnmt1b. J. Biol. Chem., 275, 10754–10760. [DOI] [PubMed] [Google Scholar]
- Cooper D.N. and Krawczak,M. (1989) Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum. Genet., 83, 181–188. [DOI] [PubMed] [Google Scholar]
- Csankovszki G., Nagy,A. and Jaenisch,R.(2001) Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol., 153, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A., Yoder,J.A., Zhang,X., Zhou,L., Bestor,T.H. and Cheng,X. (2001) Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res., 29, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F., Burgers,W.A., Brehm,A., Hughes-Davies,L. and Kouzarides,T. (2000) DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet., 24, 88–91. [DOI] [PubMed] [Google Scholar]
- Fuks F., Burgers,W.A., Godin,N., Kasai,M. and Kouzarides,T. (2001) Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J., 20, 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H. and Jeltsch,A. (2001) Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG sites. J. Mol. Biol., 309, 1201–1208. [DOI] [PubMed] [Google Scholar]
- Gowher H., Leismann,O. and Jeltsch,A. (2000) DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J., 19, 6918–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R., Ho,T. and Paulin,R. (1996) In Russo,V.E.A, Martienssen,R.A. and Riggs,A.D. (eds), Epigenetic Mechanism of Gene Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 47–61.
- Hsieh C.L (1999) In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol., 19, 8211–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Cheng,X.D., Klimasauskas,S., Sha,M., Posfai,J., Roberts,R.J. and Wilson,G.G. (1994) The DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 22, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H., Oh,S.P., Okano,M., Juttermann,R., Goss,K.A., Jaenisch,R. and Li,E.(1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development, 122, 3195–3205. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Page,A.W., Weier,H.U. and Bestor,T.H. (1992) A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell, 71, 865–873. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed] [Google Scholar]
- Liang G., Chan,M.F., Tomigahara,Y., Tsai,Y.C., Gonzales,F.A., Li,E., Laird,P.W. and Jones,P.A. (2002) Co-operativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol., 22, 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin I.G., Han,L., Taghva,A., O’Brien,L.E. and Hsieh,C.L. (2002) Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell. Biol., 22, 704–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F., Ramsahoye,B.H., Kashevsky,H., Tudor,M., Mastrangelo,M.A., Orr-Weaver,T.L. and Jaenisch,R. (1999) Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat. Genet., 23, 363–366. [DOI] [PubMed] [Google Scholar]
- Mertineit C., Yoder,J.A, Taketo,T., Laird,D.W., Trasler,J.M. and Bestor,T.H. (1998) Sex-specific exons control DNA methyltransferase in mammalian germ cells Development, 125, 889–897. [DOI] [PubMed] [Google Scholar]
- Nan X., Ng,H.-H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- Okano M., Xie,S. and Li,E. (1998a) Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res., 26, 2536–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Xie,S. and Li,E. (1998b) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet., 19, 219–220. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Pradhan S. and Kim,G.D. (2002) The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J., 21, 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S. and Roberts,R.J. (2000) Hybrid mouse-prokaryotic DNA (cytosine-5) methyltransferases retain the specificity of the parental C-terminal domain. EMBO J., 19, 2103–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S., Talbot,D., Sha,M., Benner,J., Hornstra,L., Li,E., Jaenisch,R. and Roberts,R.J. (1997) Baculovirus-mediated expression and characterization of the full-length murine DNA methyltransferase. Nucleic Acids Res., 25, 4666–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S., Bacolla,A., Wells,R.D. and Roberts,R.J. (1999) Recombinant human DNA (cytosine-5) methyltransferase I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem., 274, 33002–33010. [DOI] [PubMed] [Google Scholar]
- Ramsahoye B.H., Biniszkiewicz,D., Lyko,F., Clark,V., Bird,A.P. and Jaenisch,R. (2000) Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA, 97, 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I. et al. (2002) DNMT1 and DNMT3b co-operate to silence genes in human cancer cells. Nature, 416, 552–556. [DOI] [PubMed] [Google Scholar]
- Robertson K.D, Uzvolgyi,E., Liang,G., Talmadge,C., Sumegi,J., Gonzales,F.A. and Jones,P.A. (1999) The human DNA methyltransferases (DNMTs) 1, 3a and 3b: co-ordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res., 27, 2291–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F., Hendrich,B., Reik,W. and Dean,W. (2002) Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol., 241, 172–182. [DOI] [PubMed] [Google Scholar]
- Schmutte C. and Jones,P.A. (1998) Involvement of DNA methylation in human carcinogenesis. Biol. Chem., 379, 377–388. [DOI] [PubMed] [Google Scholar]
- Sleutels F., Barlow,D.P. and Lyle,R. (2000) The uniqueness of the imprinting mechanism. Curr. Opin. Genet. Dev., 10, 229–233. [DOI] [PubMed] [Google Scholar]
- Smith S.S., Kaplan,B.E., Sowers,L.C. and Newman,E.M. (1992) Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc. Natl Acad. Sci. USA, 89, 4744–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock D.M., Lawler,C.B., Linsenmeyer,M.E., Doherty,J.P. and Warren,W.D. (1997) Asymmetric methylation in the hyper methylated CpG promoter region of the human L1 retrotransposon. J. Biol. Chem., 272, 7810–7816. [DOI] [PubMed] [Google Scholar]
- Xu G.L. et al. (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature, 402, 187–191. [DOI] [PubMed] [Google Scholar]
- Yen R.W. et al. (1992) Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res., 20, 2287–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J.A., Yen,R.W., Vertino,P.M., Bestor,T.H. and Baylin,S.B., (1996) New 5′ regions of the murine and human genes for DNA (cytosine-5)-methyltransferase. J. Biol. Chem., 271, 30092–31007. [DOI] [PubMed] [Google Scholar]
- Yokochi T. and Robertson,K.D. (2002) Preferential methylation of unmethylated DNA by mammalian de novo DNA methyltransferase Dnmt3a. J. Biol. Chem., 277, 11735–11745. [DOI] [PubMed] [Google Scholar]