Abstract
The Saccharomyces cerevisiae MOT3 gene encodes a nuclear protein implicated in both repression and activation of transcription. However, a mot3Δ mutation causes only mild phenotypes under normal growth conditions. To learn more about Mot3 function, we have performed a synthetic lethal screen. This screen identified PAN1, a gene required for normal endocytosis, and VPS41, a gene required for vacuolar fusion and protein targeting, suggesting a role for Mot3 in the regulation of membrane-related genes. Transcriptional analyses show that Mot3 represses transcription of ERG2, ERG6 and ERG9, genes required for ergosterol biosynthesis, during both aerobic and hypoxic growth. Chromatin immunoprecipitation experiments suggest that this repression is direct. Ergosterol has been shown to be required for endocytosis and homotypic vacuole fusion, providing a link between Mot3 and these processes. Consistent with these results, mot3Δ mutants have a number of related defects, including impaired homotypic vacuole fusion and increased sterol levels. Taken together, our data suggest that proper transcriptional regulation of ergosterol biosynthetic genes by Mot3 is important for normal vacuolar function and probably for the endocytic membrane transport system.
Keywords: ergosterol/Mot3/repression/transcription/vacuole
Introduction
MOT3 encodes a transcription factor that modulates the expression of a large number of genes in Saccharomyces cerevisiae. Mot3 is nuclear (Grishin et al., 1998), is rich in charged amino acids, has two Zn-fingers and binds DNA in a Zn-dependent manner (Madison et al., 1998). The Mot3 Zn-finger region is highly homologous to that of Msn2 and Msn4, two S.cerevisiae proteins involved in the stress-response pathway. MOT3 was originally identified in a genetic screen for high-copy-number suppressors of the spt3Δ mot1-24 double-mutant lethality (Madison et al., 1998) and in an unrelated screen for mutations that cause an altered response to α-factor (Grishin et al., 1998). More recently, Mot3 has been shown to repress the transcription of two genes during aerobic growth: ANB1, encoding a translation initiation factor; and DAN1, encoding a cell wall mannoprotein (Kastaniotis and Zitomer, 2000; Kastaniotis et al., 2000; Cohen et al., 2001). Mot3 has also been shown to activate transcription of CWP2, a gene encoding a cell wall protein (Abramova et al., 2001a). Thus, Mot3 can serve as both a repressor and an activator of transcription.
As described in this paper, Mot3 also plays a role key in the regulation of genes required for ergosterol bio synthesis. Ergosterol is the major sterol in S.cerevisiae membranes and serves a role similar to that of cholesterol in mammalian cells. Ergosterol is required for proper fluidity and function of cellular membranes (Sturley, 2000), and it plays a role in both endocytosis (Munn et al., 1999) and homotypic vacuole fusion (Kato and Wickner, 2001). Ergosterol biosynthesis is regulated by intracellular ergosterol and oxygen levels, and some regulation is known to occur at the transcriptional level for many ERG genes (Sturley, 2000). Both cis-acting promoter elements and trans-acting factors, including the heme activator proteins Hap1 and Hap2/3/4 as well as Rox1, have been identified that affect ERG gene transcription; these findings have suggested that transcription of ERG genes is sensitive to oxygen levels (Turi and Loper, 1992; Kennedy et al., 1999; Jensen-Pergakes et al., 2001; Kennedy and Bard, 2001; Leber et al., 2001; Vik and Rine, 2001). Most recently, Upc2 and Ecm22, two direct activators of ERG2 transcription, have been identified and shown to bind to the Sterol Regulatory Element (SRE) in the ERG2 promoter (Vik and Rine, 2001). However, the mechanisms that regulate transcription of ERG genes in response to changes in ergosterol and oxygen levels remain largely unknown.
The identification of Mot3 as a transcriptional regulator of ergosterol biosynthetic genes arose from efforts to understand the roles of Mot3. Mot3 is not essential for growth under several conditions tested, although mot3Δ strains exhibit many mild mutant phenotypes (Grishin et al., 1998; Madison et al., 1998). To learn more about Mot3, we conducted a screen for mutations that cause inviability in combination with a mot3Δ mutation. Mutations were identified in two genes: PAN1, involved in endocytosis; and VPS41, involved in vacuolar protein sorting and fusion (Radisky et al., 1997; Tang et al., 2000). Interestingly, we have found that mot3Δ mutants have several defects in vacuolar function as well. Although endocytosis and vacuolar protein-sorting and fusion are separate functions, they are all part of the general process known as the endocytic membrane transport system and, as such, they require many functions of the cell membranes (Munn, 2000). Therefore, we hypothesized a connection between Mot3 and genes required for membrane function. Transcription and chromatin immunoprecipitation experiments show that Mot3 is a repressor of at least three of the genes required for ergosterol biosynthesis, ERG2, ERG6 and ERG9. These results provide strong evidence that Mot3 plays an important role in proper function of the endocytic membrane transport system.
Results
Mutations in PAN1 and VPS41 cause synthetic lethality with a mot3Δ mutation
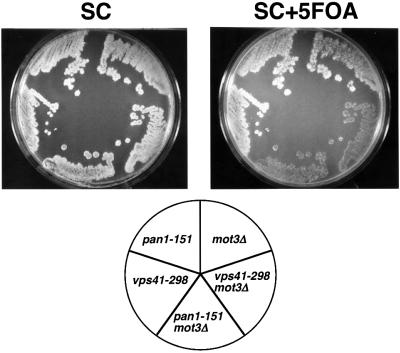
To identify genes that are functionally related to MOT3, we performed a synthetic lethal screen for mutations that cause inviability in a mot3Δ genetic background. We screened ∼87 000 colonies and identified two candidates. These mutants display additional mutant phenotypes even in a MOT3+ background, facilitating their genetic analysis: one of the synthetic lethal mutants is temperature sensitive for growth at 37°C and both mutants are sensitive to cadmium. Genetic analyses demonstrated that the synthetic lethality with mot3Δ and the additional phenotypes co-segregated 2:2 in each case (Materials and methods), demonstrating that all of the phenotypes are caused by a single mutation in each of the candidates. Additional genetic tests showed the two new mutations to be recessive and to complement each other, suggesting that they identify two separate genes. We confirmed that these mutations are synthetically lethal with mot3Δ by two tests. First, we performed plasmid-shuffle experiments to demonstrate that the viability of the double mutants depended upon a plasmid containing MOT3+. Secondly, we separated each mutation from mot3Δ by crosses to a MOT3+ strain and reconstructed the synthetic lethality by crossing each single mutant to a mot3Δ strain.
To clone the genes corresponding to the mot3Δ synthetic lethal mutations, plasmid library clones were isolated by complementation of the synthetic lethality or the temperature- and cadmium-sensitive phenotypes. The specific genes were identified by plasmid complementation with single genes and by linkage analysis. These experiments showed that the synthetic lethal mutation that causes temperature sensitivity is in PAN1 (designated pan1-151), and the other synthetic lethal mutation is in VPS41 (designated vps41-298) (Figure 1). PAN1 is an essential gene required for endocytosis (Tang and Cai, 1996; Tang et al., 1997, 2000; Wendland and Emr, 1998). VPS41 is a non-essential gene required for vacuolar protein targeting and homotypic vacuole fusion (Nakamura et al., 1997; Radisky et al., 1997; Darsow et al., 2001).
Fig. 1. A mot3Δ mutation is synthetically lethal with pan1-151 and vps41-298. mot3Δ (FY2070), mot3Δ vps41-298 (FY2073), mot3Δ pan1-151 (FY2072), vps41-298 (FY2075) and pan1-151 (FY2074), each carrying a URA3+MOT3+ plasmid, were grown on a YPD plate for 2 days at 30°C. The YPD plate was then replica plated onto an SC and an SC + 5-FOA plate and photographs were taken after incubation for 1 day at 30°C.
The results from this synthetic lethal screen show that mot3Δ is synthetically lethal with mutations in genes required for both endocytosis and vacuolar function. Previous studies have shown that mutants defective for endocytosis are often synthetically lethal with mutations that affect vacuolar function (Munn and Riezman, 1994; Riezman et al., 1996; Wesp et al., 1997; Munn, 2000, 2001). Thus, these data suggest that Mot3 could be important for both normal endocytosis and vacuolar functions, or generally required for proper function of the endocytic membrane transport system.
mot3Δ mutants have defective vacuoles
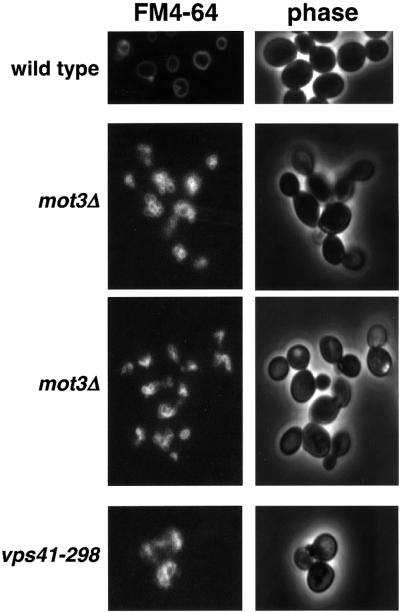
To test the hypothesis that Mot3 is required for proper function of the vacuole, we examined vacuolar morphology and function, comparing wild-type and mot3Δ strains. First, to test whether mot3Δ mutants have a morphologically abnormal vacuole, we stained a MOT3+ strain and two different mot3Δ mutant strains with FM4-64, a vacuole-specific vital dye (Vida and Emr, 1995). Mutations that impair homotypic vacuole fusion result in an increase in the number of vacuolar structures per cell. The criteria used for classifying vacuole morphology phenotypes have been described previously (Banta et al., 1988). Briefly, cells with a class A or wild-type vacuolar morphology have one to three vacuoles per cell; cells with a class B morphology have more than three vacuoles per cell; and cells with a class C morphology lack any visible vacuole structure. From this analysis, the MOT3+ strain exhibited a wild-type (class A) vacuolar morphology (Figure 2), with a single vacuole visible in each cell. In contrast, the mot3Δ mutants displayed a mutant class B vacuolar morphology phenotype, with greater than three vacuoles per cell. We also stained the vps41-298 mutant, which had no detectable vacuolar structures, suggesting a class C phenotype (Figure 2). We conclude from these experiments that mot3Δ mutants have abnormal vacuoles.
Fig. 2. A mot3Δ mutant has a defect in homotypic vacuole fusion. Saccharomyces cerevisiae strains were grown to 1–2 × 107 cells/ml in liquid YPD, stained with FM4-64 and visualized as described in Materials and methods. At least 100 cells were examined for each strain and the percentage that had the phenotype shown is as follows: wild type (FY2066), 95% class A; mot3Δ (FY2069), 78% class B; mot3Δ (FY2071), 82% class B; vps41-298 (FY2075), 100% class C.
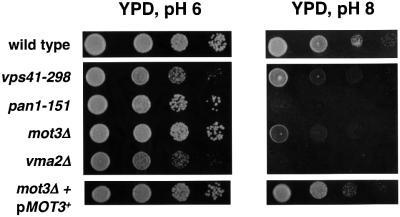
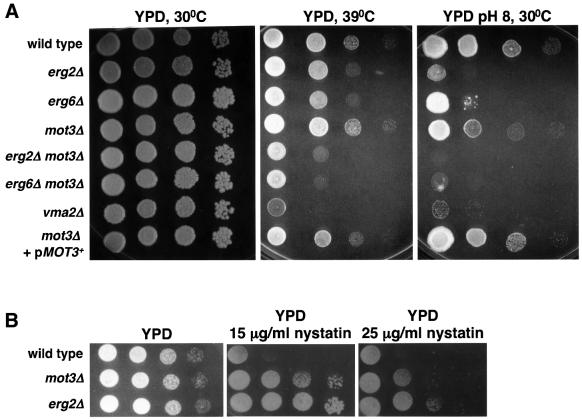
To test whether the aberrant vacuolar morphology exhibited by mot3Δ mutants correlated with a defect in vacuolar function, we assayed the ability of these mutants to grow in alkaline media. One important function of the yeast vacuole is to maintain a neutral intracellular pH (Klionsky et al., 1990). Therefore, mutants with a defective vacuole are unable to grow well in media buffered at either extreme acidic or alkaline pH (Klionsky et al., 1990). Indeed, the mot3Δ mutants grew poorly on solid alkaline buffered medium, similar to the defect we observed for the vps41-298 mutant (Figure 3) and vps41Δ (data not shown). Therefore, mot3Δ mutants are defective for vacuolar function. This phenotype of mot3Δ cells is fully complemented by providing MOT3+ on a plasmid. In addition, the pan1-151 mutant is unable to grow on alkaline buffered medium and its defect is as severe as that seen for a previously characterized vacuolar mutant, vma2Δ (Anraku et al., 1992). VMA2 encodes the regulatory subunit of the vacuolar ATPase (Anraku et al., 1992). These results, taken together with the aberrant vacuolar morphology of mot3Δ mutants, strongly suggest that Mot3 is important for homotypic vacuole fusion and proper vacuolar functions.
Fig. 3. A mot3Δ mutant is defective for growth at pH 8. Wild-type (FY2066), vps41-298 (FY2075), pan1-151 (FY2074), mot3Δ (FY2070), vma2Δ (FY2076) and mot3Δ (FY2070) with a URA3+ MOT3+ plasmid (pMOT3+) were tested for growth on YPD and YPD pH 8 plates by spot tests (see Materials and methods). To prevent growth of sensitive strains due to media acidification by growth from neighboring resistant strains, wild-type and mot3Δ+pMOT3+ were grown on a separate YPD pH 8 plate. YPD plates were photographed after 2 days of incubation at 30°C. YPD pH 8 plates were photographed after 6 days of incubation at 30°C.
Mot3 represses expression of ERG2, ERG6 and ERG9
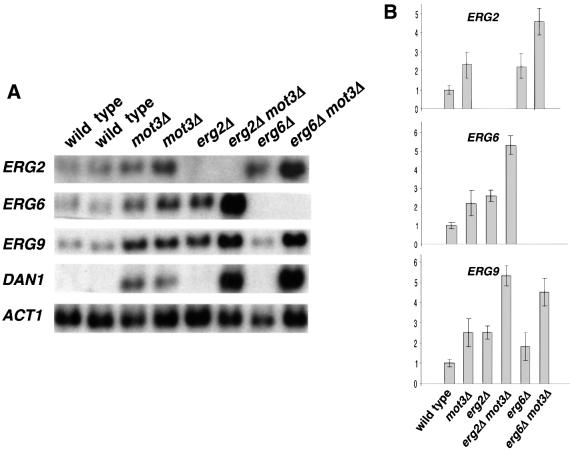
Mot3 is nuclear (Grishin et al., 1998) and has been shown to act as a repressor and an activator of transcription (Grishin et al., 1998; Madison et al., 1998; Kastaniotis et al., 2000; Abramova et al., 2001a; Cohen et al., 2001). Therefore, the mot3Δ synthetic lethal phenotypes and defects in vacuolar function are probably due to improper transcription of genes, such as those encoding proteins required for vacuolar function or vacuolar membrane fusion. To assess this possibility, we performed microarray analyses comparing mRNA from mot3Δ and wild-type strains. The results from these experiments showed minor (<1.5-fold) effects on the mRNA levels of many genes involved with vacuolar function (VPS, VAM and VMA genes; data not shown) and cell wall biosynthesis (PAU, DAN and TIR genes; data not shown). However, a more significant effect was observed on the mRNA levels of ERG2, ERG6 and ERG9, genes encoding ergosterol biosynthetic enzymes. The mRNA levels of these genes were increased in the mot3Δ mutant (data not shown). To confirm the microarray results, northern analyses were performed. These results show that ERG2, ERG6 and ERG9 mRNA levels were elevated ∼2.5-fold in mot3Δ mutants compared with wild-type strains (Figure 4), suggesting that Mot3 is a repressor of these ERG genes.
Fig. 4. ERG2, ERG6 and ERG9 mRNA levels are increased in mot3Δ mutants. (A) Representative northern analyses are shown of RNA prepared from wild-type (FY2066 and FY2068), mot3Δ (FY2069 and FY2071), erg2Δ (FY2077), erg2Δ mot3Δ (FY2078), erg6Δ (FY2079) and erg6Δ mot3Δ (FY2080) strains, probed for ERG2, ERG6, ERG9, DAN1 and ACT1 mRNAs. (B) Histograms showing the relative average mRNA levels for the genes indicated, normalized to the level of ACT1 mRNA. The values and standard errors are from seven independent experiments.
Previous studies suggest that ERG genes are tightly regulated at the transcriptional level, particularly in the later steps of the ergosterol biosynthetic pathway, through an uncharacterized feedback loop that ensures proper ergosterol levels (Sturley, 2000). Consistent with this model, it has been shown that mutations in some ERG genes affect the transcription of other ERG genes (Lees et al., 1995; Arthington-Skaggs et al., 1996; Smith et al., 1996; Kennedy et al., 1999). Therefore, we also tested for double-mutant phenotypes when mot3Δ was combined with erg2Δ and erg6Δ mutations (ERG9 is essential for aerobic growth, preventing similar tests with an erg9Δ mutation). As shown in Figure 4, ERG9 mRNA levels were increased in erg2Δ and erg6Δ mutants compared with wild-type levels. In the erg2Δ mot3Δ and erg6Δ mot3Δ double mutants, there is an additive effect as ERG9 mRNA levels are higher than in the single mutants. In addition, since ERG2 and ERG6 have been shown to be required for endocytosis (Munn et al., 1999; Munn, 2001), vacuolar fusion (Kato and Wickner, 2001) and growth in alkaline buffered media (Figure 5A), we tested for other phenotypes in the double mutants. Our results show that erg2Δ mot3Δ and erg6Δ mot3Δ double mutants had a more severe growth defect than the single mutants on YPD at 39°C and on alkaline buffered YPD (Figure 5A). In conclusion, there is a strong genetic interaction of MOT3 with ERG2 and ERG6, supporting a role for Mot3 in the normal transcriptional regulation of these genes required for proper ergosterol homeostasis.
Fig. 5. Genetic interactions of mot3Δ with erg2Δ and erg6Δ. (A) Wild-type (FY2066), erg2Δ (FY2077), erg6Δ (FY2079), mot3Δ (FY2069), erg2Δ mot3Δ (FY2078), erg6Δ mot3Δ (FY2080), vma2Δ (FY2076) and mot3Δ (FY2069) carrying a URA3+ MOT3+ plasmid (pMOT3+) strains were tested for growth on YPD and YPD pH 8 by spot tests (see Materials and methods). YPD plates were photographed after 2 days of incubation. YPD pH 8 plates were photographed after 6 days incubation. (B) Nystatin resistance of mot3Δ mutants. Wild-type (FY2066), mot3Δ (FY2069) and erg2Δ (FY2077) strains were tested for growth on YPD and YPD-nystatin plates. The YPD plate was photographed after 1 day of incubation at 30°C and the YPD-nystatin plates were photographed after 2 days of incubation at 30°C.
Ergosterol is not synthesized during anaerobic growth because oxygen is a cofactor for some of the ergosterol biosynthetic enzymes (Kennedy et al., 1999; Sturley, 2000). There is also evidence that oxygen levels control transcription of some of the genes required for ergosterol biosynthesis (Thorsness et al., 1989; Kennedy et al., 1999; Sturley, 2000; Kennedy and Bard, 2001). Therefore, we tested whether ERG2, ERG6 and ERG9 mRNA levels are altered during hypoxic growth and whether Mot3 plays a role under this growth condition. Our results show that, indeed, there was a >10-fold repression of ERG2 and ERG6 mRNA levels under hypoxic conditions (Figure 6). ERG9 mRNA levels were only decreased 2-fold during hypoxic growth (data not shown), in agreement with previous studies (M’Baya et al., 1989; Kennedy et al., 1999). In a mot3Δ background, the repression of ERG2 and ERG6 under hypoxic conditions was almost completely abolished (Figure 6). These results, then, show that Mot3 is required for the strong hypoxic repression of ERG2 and ERG6 transcription.
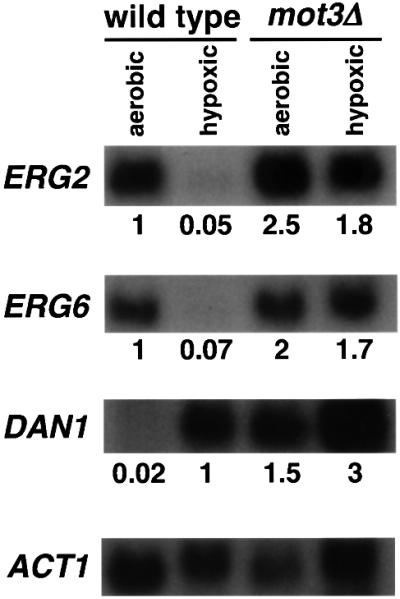
Fig. 6. Mot3 is required for hypoxic repression of ERG2 and ERG6. A representative northern analysis is shown of RNA prepared from aerobically and hypoxically grown wild-type (FY2066) and mot3Δ (FY2071) cells, probed for ERG2, ERG6, DAN1 and ACT1 transcripts. The numbers indicate the average values from three independent experiments, normalized to either the wild-type strain grown aerobic ally (for ERG2 and ERG6) or to the wild-type strain grown hypoxically (for DAN1). Standard deviations were <2%.
mot3Δ mutants have increased sterol levels
To test whether the increased levels of ERG2, ERG6 and ERG9 mRNAs in aerobically grown mot3Δ mutant cells affect sterol levels, we performed total sterol analyses on wild-type and mot3Δ cells. As shown in Table I, mot3Δ mutants have 21% more total sterols and 15% more ergosterol than wild-type cells. Despite this modest increase in total sterols and ergosterol, mot3Δ mutants are slightly resistant to nystatin (Figure 5B), a phenotype indicative of reduced ergosterol in plasma membrane (Lorenz and Parks, 1991). As nystatin primarily binds ergosterol, most erg mutants unable to synthesize ergosterol are resistant to nystatin (McLean-Bowen and Parks, 1982; Kovac et al., 1987). Taken together, these results suggest that there is a positive correlation between increased mRNA levels of these ERG genes and an increase in total sterol production in mot3Δ mutants; however, mot3Δ mutants probably have a reduced level of ergosterol in their plasma membranes (see Discussion).
Table I. Sterol levels in mot3Δ mutants.
| Relevant genotype | Total sterol | % increase | Total ergosterol | % increase |
|---|---|---|---|---|
| MOT3+ | 6.75 ± 0.26 | – | 4.27 ± 0.17 | – |
| mot3Δ | 8.16 ± 0.23 | 21 | 4.92 ± 0.24 | 15 |
The total sterol and ergosterol levels are reported as µg sterol/g yeast dry weight (see Materials and methods) of wild-type (FY2066) and mot3Δ (FY2069) cells. Values represent the average and standard deviations from three independent experiments.
Overexpression of UPC2 and ECM22 causes a defect in vacuolar morphology
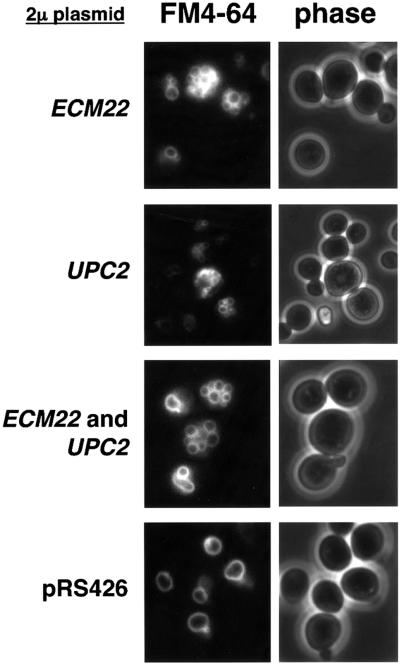
We observed a homotypic vacuole fusion defect in mot3Δ mutants, which have elevated ERG2, ERG6 and ERG9 mRNA levels compared with wild-type cells. To test whether the increased ERG gene transcription observed in mot3Δ mutants is responsible for the vacuolar defect, we increased ERG mRNA levels in a MOT3+ background. To do this we took advantage of the recent finding that overexpression of UPC2 and ECM22 causes increased ERG2 expression (Vik and Rine, 2001). Our results (Figure 7) show that a wild-type strain that contains UPC2, ECM22, or both genes on high-copy-number plasmids, had abnormal (class B) vacuolar morphology. In these experiments, ERG2 and ERG9 mRNA levels were increased ∼2-fold, while ERG6 mRNA levels were not significantly affected (data not shown). Thus, the increased expression of these ERG genes in mot3Δ mutants is likely to contribute to the mot3Δ vacuolar defect.
Fig. 7. Overexpression of UPC2, ECM22, or both, causes a vacuolar defect. High-copy-number plasmids carrying either UPC2, ECM22 or no insert were used to transform the MOT3+ strain FY2066 to either Ura+, Leu+ or Ura+ Leu+, respectively. Transformants were selectively grown and then stained with FM4-64 (see Materials and methods). The percentage of plasmid-containing cells that had the vacuolar phenotype shown above is as follows: ECM22, 71% class B; UPC2, 70% class B; EMC22 and UPC2, 75% class B; the plasmid control, 78% class A.
Mot3 binds to the promoter regions of ERG2 and ERG6 in vivo
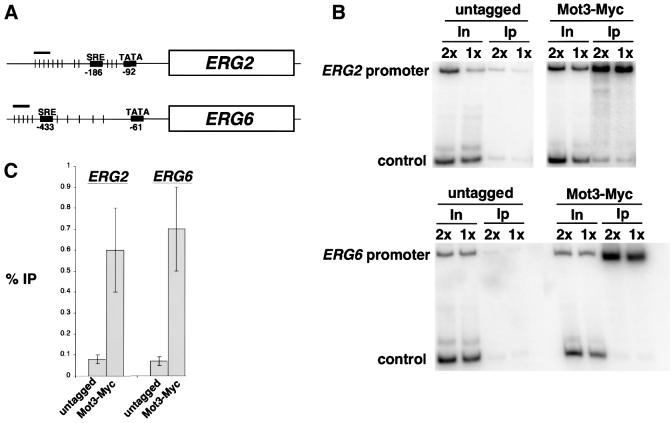
To test whether Mot3 acts directly in repression, we performed chromatin immunoprecipitation experiments. The strongest cases of Mot3 repression that we have observed are of ERG2 and ERG6 during hypoxic growth, and we found a cluster of putative Mot3 consensus binding sites in the regulatory regions of these genes (Figure 8A). Therefore, we examined Mot3 binding to the ERG2 and ERG6 regulatory regions in cells grown in hypoxic conditions. In these experiments, we used a version of Mot3 fused to the myc-epitope tag. Our results (Figure 8B and C) demonstrate specific chromatin immunoprecipi tation of the ERG2 and ERG6 promoter regions. These chromatin immunoprecipitation results, combined with the DNA binding properties of Mot3, the presence of Mot3 binding sites, and derepressed ERG2 and ERG6 transcription in mot3Δ mutants, strongly suggest that Mot3 directly represses ERG2 and ERG6 transcription.
Fig. 8. Mot3 binds to the promoter region of ERG2 and ERG6 in vivo. (A) Diagram of the ERG2 and ERG6 promoter regions. The short-thin vertical lines represent the positions of the Mot3 consensus binding site sequences [(C/T/A)AGG(T/A/C/G)(T/A)] and the short thick horizontal bar represents the position of the PCR product assayed in the chromatin immunoprecipitation experiments. (B) Chromatin immunoprecipitation was performed with extracts prepared from hypoxically grown Mot3-Myc18 (FY2081) and untagged (FY1339) cells. The abbreviations used indicate the input DNA (In) and immunoprecipitated DNA (Ip) used for PCR using primers that amplify the regions defined in (A). For input DNA, 2× and 1× indicate 1/500 and 1/1000 dilutions of the chromatin solution, respectively. For immunoprecipitated DNA, 2× and 1× indicate 1/10 and 1/20 dilutions of the immunoprecipitated chromatin solution, respectively. The non-specific control used in the reactions is described in Materials and methods. (C) A histogram showing the average values and their standard errors (% IP), calculated as previously described (Larschan and Winston, 2001), from three independent experiments.
Discussion
Our studies have demonstrated two new and related findings about the transcription factor Mot3: (i) Mot3 is required for normal vacuolar morphology and function; and (ii) Mot3 is a transcriptional repressor of the ERG2, ERG6 and ERG9 genes. These conclusions are based on several results. First, a genetic screen discovered synthetic lethality between mot3Δ and mutations in PAN1 and in VPS41, genes required for endocytosis (Tang and Cai, 1996; Wendland et al., 1996; Tang et al., 1997, 2000; Wendland and Emr, 1998) and for vacuolar protein targeting and fusion (Nakamura et al., 1997; Radisky et al., 1997), respectively. Previous studies have shown that mutations that affect vacuolar functions are frequently lethal in combination with mutations that impair endocytosis (Munn and Riezman, 1994; Munn, 2000, 2001). Secondly, mot3Δ mutants have defects in vacuolar function and morphology. Thirdly, mot3Δ mutants have increased levels of the ERG2, ERG6 and ERG9 mRNAs under both aerobic and hypoxic conditions. Fourthly, the repression of ERG2 and ERG6 by Mot3 appears to be direct, as chromatin immunoprecipitation experiments have shown Mot3 to be physically present at their promoters. Finally, mot3Δ mutants have increased sterol levels, consistent with the conclusion that Mot3 represses the transcription of these ERG genes. Taken together, these results suggest that Mot3 plays a critical role in a transcriptional regulatory system that modulates ergosterol levels to ensure a normal endocytic membrane transport system.
Although Mot3 has previously been implicated in the regulation of several different classes of genes (Grishin et al., 1998; Kastaniotis et al., 2000; Abramova et al., 2001a,b; Cohen et al., 2001), our results suggest that it is the defective repression of ERG genes that causes the vacuolar defects observed in mot3Δ mutants. This conclusion is based on our finding that when ERG genes are overexpressed in a MOT3+ genetic background, we observed vacuolar defects similar to those observed in mot3Δ mutants. Although we cannot rule out that the defect in homotypic vacuolar fusion caused by mot3Δ or by overexpression of UPC2 and ECM22 is due to other pleiotropic effects, we do observe similar effects on ERG2 and ERG9 mRNA levels in the two different situations. Recent studies have shown that ergosterol is required for homotypic vacuolar fusion (Kato and Wickner, 2001). Our work provides the first evidence that transcriptional regulation of genes required for ergosterol biosynthesis plays a significant role in vacuolar functions. The fact that ergosterol is also required for normal endocytosis (Munn et al., 1999) suggests that Mot3 is also required for this aspect of the endocytic membrane transport system.
Increased ERG gene expression in mot3Δ mutants and vacuolar defects
Our results have demonstrated that in mot3Δ mutants there is an increased level of ERG gene mRNAs and an increased level of sterols. However, mot3Δ mutants have a vacuolar defect similar to those observed in mutants defective in ergosterol synthesis (Kato and Wickner, 2001), an apparently opposite situation. The mild nystatin resistance of mot3Δ mutants suggests that the increased expression of ERG genes in mot3Δ mutants may actually result in a reduced level of ergosterol in the plasma membrane. There are several possibilities for how increased ERG gene expression might reduce the level of functional ergosterol and impair vacuolar morphology and function. For example, the increased level of ergosterol might accumulate in an aberrant form, thereby reducing the level of ergosterol available for membranes. This possibility is supported by the observation that mot3Δ mutants have an elevated level of lipid particles (M.Valachovic and M.Bard, unpublished results). Previous studies have shown that a common mechanism for disposing of excess intracellular sterol is sterol esterification and deposition into such lipid particles (Soustre et al., 1998; Zweytick et al., 2000). Therefore, the increased level of sterols in mot3Δ mutants might activate this mechanism of disposal, resulting in abnormal sterol trafficking, which has been correlated with an abnormal sterol composition of vacuole and plasma membranes (Tinkelenberg et al., 2000). Alternatively, increased ERG gene expression might lead to an accumulation of ergosterol precursors, resulting in an aberrant incorporation of sterols other than ergosterol into the membrane.
The role of Mot3 during hypoxic and aerobic growth
Our results have shown that Mot3 represses ERG2, ERG6 and ERG9 during both hypoxic and aerobic growth. The strongest effect that we observed on repression by Mot3 occurs at ERG2 and ERG6 during hypoxic growth. Previous studies showed that, under hypoxic growth conditions, S.cerevisiae is unable to synthesize ergosterol due to the unavailability of oxygen to function as a cofactor for ergosterol biosynthetic enzymes (Kwast et al., 1998). Our results now show that hypoxic control of ergosterol biosynthesis also occurs at the transcriptional level by Mot3 repression of ERG gene transcription. Results from two genome-wide expression studies that compared mRNA levels during aerobic and anaerobic growth differ from our results and from each other with respect to expression of ERG genes during anaerobic growth. One study did not detect a difference in mRNA levels for ERG2, ERG6 and ERG9 (ter Linde et al., 1999), and the other found a modest increase in ERG6 and ERG9 mRNA levels and no change for ERG2 (Kwast et al., 2002). Of note, the latter study also shows increased CWP1 and CWP2 mRNA levels during anaerobic growth, a condition in which these genes have been shown to be repressed (Abramova et al., 2001a). The reasons for the differences between our results and the microarray studies are not known, but could be due to different genetic backgrounds, growth conditions or the experimental techniques used. Regardless of the reason, our results have demonstrated that Mot3 is required for the hypoxic repression of ERG2 and ERG6 transcription.
During aerobic growth, Mot3 is also required for transcriptional repression of ERG2, ERG6 and ERG9, although the degree of repression is significantly less than during hypoxic growth. However, the modest repression of ERG genes by Mot3 during aerobic growth is clearly important, because under aerobic conditions mot3Δ mutations cause synthetic lethality with pan1 and vps41 mutations, defects in vacuolar morphology and function, and increased levels of total sterols and ergosterol. Furthermore, during aerobic growth, mot3Δ erg2Δ and mot3Δ erg6Δ double mutants have several synthetic phenotypes, suggesting that they impair a common pathway. Therefore, during aerobic growth, Mot3 may ensure that ERG gene transcription is modulated appropriately to achieve normal ergosterol levels.
Although there is previous evidence for regulation of ergosterol biosynthesis at the transcriptional level (Dimster-Denk et al., 1999), only a small number of regulatory factors have been identified. Some studies linking oxygen sensing and ERG gene transcription have identified Rox1 as a repressor and Hap1/2/3/4 as activators of ERG11 and HMG1 (Kwast et al., 1998). Recently, Vik and Rine (2001) identified two activators of ERG2, Upc2 and Emc22, which promote ERG2 transcription under conditions of sterol starvation. A recent study has identified Sut1 as a putative transcription factor involved in the regulation of sterol uptake during aerobiosis, suggesting a link between sterol uptake and ergosterol biosynthesis (Ness et al., 2001). Our studies have added to our understanding of ERG gene regulation by demonstrating that Mot3 is an important repressor of ERG2, ERG6 and ERG9 gene transcription. The role of Mot3 with respect to these other positive and negative regulators of ERG gene transcription has not yet been addressed.
Two important and related aspects of Mot3 repression of ERG2, ERG6 and ERG9 remain to be studied. First, the mechanism of repression by Mot3 at these genes is not understood. A previous study showed that, during aerobic growth, Mot3 may repress ANB1 and other genes by recruiting the Ssn6–Tup1 repressor complex (Kastaniotis et al., 2000). There is as yet no evidence that repression of ERG genes is dependent on Ssn6–Tup1, histone deacetylases or nucleosome remodeling complexes. Secondly, the difference in the degree of Mot3 repression of ERG gene transcription in cells grown hypoxically or aerobically is not yet understood. Since there are two clusters of putative Mot3 binding sites, 5′ and 3′ of the SRE elements in ERG2 and ERG6, it is possible that differential binding by Mot3 provides a means to control the level of repression in response to oxygen levels or sterol levels. Consistent with this idea, Mot3 binds to its four sites in the DAN1 promoter with different affinities (Cohen et al., 2001). Our future work will be aimed at elucidating the mechanism by which Mot3 represses expression of ERG genes during both hypoxic and aerobic growth.
Materials and methods
Yeast strains, genetic methods and media
The S.cerevisiae strains used in this study are listed in Table II. Strains designated FY are isogenic to S288C and are GAL2+ (Winston et al., 1995). Lower case letters indicate a recessive mutant allele and upper case indicates the wild-type allele. Strain construction and other genetic manipulations were carried out by standard methods (Guthrie and Fink, 1991). The mot3Δ2::HIS3 null allele, and the erg2Δ::kanMX and erg6Δ::kanMX null alleles have been described previously (Madison et al., 1998; Winzeler et al., 1999). The MOT3-Myc18::TRP1 allele was constructed as described previously (Knop et al., 1999). All yeast media, including YPD, synthetic complete (SC), omission media (SC–), sporulation media and media containing 5-FOA were prepared as described previously (Rose et al., 1990). For scoring vacuolar phenotypes, YPD was buffered at pH 8.0 by adding NaOH and a phosphate buffer consisting of 157 mM K2HPO4 and 4 mM KH2PO4 (final concentrations). For testing nystatin resistance, nystatin (Sigma) was dissolved in N,N-dimethylformamide and added to YPD to 15 or 25 µg/ml final concentration (YPD-nystatin). YPD-nystatin plates were kept at room temperature and used within 48 h after preparation. Cadmium sensitivity was tested on YPD supplemented with 100 µM cadmium. For growth under hypoxic conditions, in which S.cerevisiae requires ergosterol in the growth medium (Parks et al., 1985, 1995), liquid YPD medium was supplemented with a 1% final concentration of a stock solution containing 5 ml of Tween-80, 5 ml of ethanol and 20 mg of ergosterol (Sigma) (YPD-ergosterol), and cells were grown in 50 ml tightly capped polypropylene conical tubes, in a shaker at 30°C, to a density of 1–2 × 107 cells/ml. The strong induction of DAN1 expression (Figure 6) verifies that these conditions are hypoxic. Yeast transformants were selected on the appropriate SC media. All strains used for serial dilution plating were grown in the appropriate liquid media to a starting density of 1–2 × 107 cells/ml. Each row of spots represents a series of 10-fold dilutions in sterile water of the respective starting culture.
Table II. Saccharomyces cerevisiae strains.
| Strain | Genotype |
|---|---|
| FY84 | MATa ura3-52 his3Δ200 leu2Δ1 lys2-128δ |
| FY85 | MATα ura3-52 his3Δ200 leu2Δ1 lys2-128δ |
| FY1339 | MATa ura3Δ0 his3Δ200 trp1Δ63 |
| FY2066 | MATa ura3Δ0 his3Δ200 leu2Δ1 |
| FY2067 | MATa ura3-52 his3Δ200 leu2Δ1 lys2-128δ |
| FY2068 | MATα ura3-52 his3Δ200 leu2Δ1 lys2-128δ |
| FY2069 | MATα ura3-52 his3Δ200 lys2-128δ trp1Δ1 mot3Δ2::HIS3 |
| FY2070 | MATa ura3-52 his3Δ200 leu2Δ1 lys2-173R2 mot3Δ2::HIS3 |
| FY2071 | MATa ura3Δ0 his3Δ200 leu2Δ0 mot3Δ2::HIS3 |
| FY2072 | MATα ura3-52 his2Δ200 lys-128δ trp1Δ1 mot3Δ2::HIS3 pan1-151 <pCH2> |
| FY2073 | MATa ura3-52 his3Δ200 lys2-1732 leu2Δ1 mot3Δ2::HIS3 vps41-298 <pCH2> |
| FY2074 | MATα ura3-52 his3Δ200 leu2Δ1 lys2-173R2 pan1-151 |
| FY2075 | MATa ura3-52 his3Δ200 leu2Δ1 lys2-128δ vps41-298 |
| FY2076 | MATa ura3Δ0 his3Δ200 leu2Δ1 vma2Δ::kanMX |
| FY2077 | MATα ura3-52 his3Δ200 leu2Δ1 lys2-128δ erg2Δ::kanMX |
| FY2078 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ erg2Δ::kanMX mot3Δ2::HIS3 |
| FY2079 | MATα ura3Δ0 his3Δ200 leu2Δ1 lys2-128δ erg6Δ::kanMX |
| FY2080 | MATa ura3-52 his3Δ200 leu2Δ1 lys2-128δ met25Δ0 erg6Δ::kanMX mot3Δ2::HIS3 |
| FY2081 | MATa ura3Δ0 his3Δ200 trp1Δ63 MOT3-Myc18::TRP1 |
Bacterial strains and plasmids
Plasmids were constructed, amplified and isolated from Escherichia coli MH1 or DH5α, according to standard procedures (Sambrook et al., 1989). Plasmids were recovered from yeast as described previously (Robzyk and Kassir, 1992). The pRS series of vectors has been described previously (Sikorski and Heiter, 1989). Plasmid pCH2 was constructed by ligating a 4.3 kb BamHI–XhoI fragment containing MOT3 from pJM142 (Madison et al., 1998) into the BamHI–XhoI sites of pRS416. Plasmid pJM188 is a MOT3 LEU2 CEN plasmid (Madison et al., 1998). Plasmid pRL81 is the original PAN1 plasmid isolated from a yeast genomic URA3 CEN library (Rose et al., 1987). Plasmid pHL6 is the original VPS41 plasmid isolated from a yeast LEU2 CEN library (Spencer et al., 1988). Plasmid pAS249 is a PAN1 URA3 CEN plasmid (Sachs and Deardorff, 1992) (kindly provided by A.Sachs). Plasmid JK0024 is a VPS41 URA3 2µ plasmid (Radisky et al., 1997) (generously provided by J.Kaplan). Plasmid pCH7 is a 3.5 kb EcoRI fragment from pRL81 containing PAN1 subcloned into the EcoRI site of pRS416. Plasmid pCH9 is the 3.5 kb EcoRI fragment from pCH7 containing PAN1 subcloned into the EcoRI site of pRS305. Plasmid pCH11 is a 5.4 kb BamHI–NotI fragment from JK0024 containing VPS41 subcloned into the BamHI–NotI site of pRS315. Plasmid pCH12 is a 5.4 kb BamHI–NotI fragment from JK0024 containing VPS41 subcloned into the BamHI–NotI site of pRS305. Plasmid pJR2330 is an ECM22 LEU2 2µ plasmid (Vik and Rine, 2001) and plasmid pJR 2341 is a UPC2 URA3 2µ plasmid (both plasmids generously provided by B.Davies and J.Rine). Restriction enzymes were purchased from Boehringer Mannheim Biochemicals and used as recommended by the manufacturer.
Isolation of mot3Δ synthetic lethal mutations
Plasmid pCH2 (MOT3 CEN URA3) was used to transform two mot3Δ strains, FY2069 and FY2070 (Table II). The resulting strains were mutagenized with ethyl methanesulfonate by standard methods (Ausubel et al., 1988). Mutagenized cells were plated on YPD and incubated at 30°C. Approximately 42 000 colonies from FY2069 and 45 000 colonies from FY2070 were screened for the inability to grow without pCH2 by replica plating to 5-FOA plates. Those colonies that did not grow on 5-FOA but grew on SC-Ura and SC were chosen for further study. To demonstrate that the synthetic lethality was dependent on mot3Δ, plasmid shuffle experiments were conducted (Guthrie and Fink, 1991) using pJM188 (MOT3 LEU2 CEN) or the vector alone. Two mutants, FY2073 (from FY2069) and FY2072 (from FY2070), were able to lose pCH2 upon transformation with pJM188, but not when transformed with vector alone. To demonstrate that a single gene was responsible for the synthetic lethality with mot3Δ, FY2073 and FY2072 were crossed to FY2069 and FY2070, respectively. 5-FOA-sensitive double mutants were crossed to FY84 or FY85 to isolate the new mutations in a MOT3+ background. These single mutations were verified to cause synthetic lethality with mot3Δ by crossing each candidate to FY2069 or FY2070 carrying pCH2 and reconstructing the synthetic lethality by the inability to lose pCH2 on 5-FOA plates.
Cloning of synthetic lethal genes
To clone the synthetic lethal genes, a yeast genomic plasmid library in a URA3-marked vector (Rose et al., 1987) or a LEU2-marked vector (Spencer et al., 1988) was used to transform the mutant strains FY2074 and FY2073, respectively. Approximately 43 000 Ura+ transformants were screened by replica-plating for those that grew at 37°C and on YPD cadmium media. One transformant complemented both phenotypes. Approximately 30 000 Leu+ transformants were screened by replica-plating for those that grew on SC-Leu + 5-FOA. One transformant complemented the 5-FOA-sensitive phenotype. Library plasmids were isolated from these candidates and their insert DNAs were analyzed by sequence analysis of the ends of the inserts, followed by identification of the cloned sequence by a computer database search. Complementation of the mutation in FY2074 was achieved by transforming FY2074 with either pAS249 or pCH7, identifying PAN1 as the gene corresponding to the mutation, subsequently renamed pan1-151. Complementation of the mutation in FY2073 was achieved by transforming FY2075 with JK0024 or pCH11, identifying VPS41 as the gene corresponding to this mutation, subsequently renamed vps41-298. Linkage analysis using the integrating plasmids pCH9 and pCH12 showed that PAN1 and VPS41 were linked to the corresponding synthetic lethal mutations and were not unlinked suppressors.
RNA preparation and northern hybridization analyses
Total RNA was isolated by a hot-phenol method (Ausubel et al., 1988) and 20 µg of RNA were used in each sample. Northern transfer and hybridization were performed as described previously (Swanson et al., 1991). 32P-labeled probes were generated either with a Boehringer Mannheim Biochemical nick-translation kit or by random hexamer labeling (Ausubel et al., 1988). Northern results were quantitated using PhosphorImager screen and ImageQuant software (Molecular Dynamics).
Fluorescence microscopy
Yeast cells were grown in YPD or SC, supplemented with the appropriate amino acids for plasmid selection, to 1–2 × 107 cells/ml. Cells were harvested, resuspended in YPD and stained with FM4-64 (Molecular Probes), as described previously (Vida and Emr, 1995). Cells were viewed on a Zeiss AxiosKop2 microscope equipped with epifluorescence using a rhodamine filter, and images were obtained with a digital camera and IPlab software. The staining with FM4-64 is performed in YPD medium. Therefore, for strains carrying plasmids, the percentage of plasmid retention during the staining procedure was calculated by plating dilutions on YPD and SC lacking the appropriate amino acids after completion of the staining protocol.
Quantitative sterol extractions and sterol analysis
To determine the percentage of total cellular mass represented by sterol, quantitative sterol analysis was performed as described previously (Molzahn and Woods, 1972). To calculate the percentage of sterols per milligram of cell mass, 20 ml of the original culture, grown in SC medium, were harvested by vacuum filtration onto a pre-weighed 0.45 µm Millipore filter. The filters were pre-dried in a 105°C oven overnight and subsequently placed in a desiccator for 4 h before weighing. After vacuum fitration of harvested cells the filters were again placed into a 105°C oven overnight, followed by desiccation at room temperature for 4 h. The weight of the cells was determined immediately after opening the desiccator. The amount of individual sterols was calculated based on the area under each peak of the chromatograph relative to a known amount of ergosterol loaded onto the gas chromatograph using the Hewlett-Packard sterol quantitation program. Each sample was injected twice and the value reported for each sterol quantity was the average of two injections. Sterols were analyzed by gas chromatography using a Hewlett-Packard HP5890 series II chromatograph equipped with the Hewlett-Packard CHEMSTATION software package. The capillary column (DB-1) was 15 m × 0.25 mm × 0.25 µm (film thickness) (J&W Scientific, Folsom, CA) and was programmed from 195 to 280°C (1 min at 195°C and then an increase at 20°C/min to 240°C, followed by an increase at 2°C/min until the final temperature of 280°C was reached). The linear velocity was 30 cm/s, nitrogen was the carrier gas and all injections were run in the splitless mode.
Chromatin immunoprecipitation
For chromatin isolation, cells were grown in 50 ml of YPD-ergosterol medium in 50 ml tightly capped polypropylene conical tubes in a shaker at 30°C to a density of 1–2 × 107 cells/ml. All chromatin immunoprecipitations were performed as described previously (Dudley et al., 1999). Chromatin was sonicated to an average of 400 bp with a size range of 200–900 bp. A two-step immunoprecipitation was performed as described previously (Harlow and Lane, 1999). The rabbit polyclonal A14 anti-Myc antibody was used (Santa Cruz). Quantitative radioactive PCR was used to determine the percentage of ERG2 and ERG6 promoter DNA that co-immunoprecipitated with Mot3-Myc18, as described previously (Larschan and Winston, 2001). The linearity of all PCR reactions was assayed as described previously (Larschan and Winston, 2001). The relative enrichment for the specific ERG2 and ERG6 PCR products is reported after normalization to a control PCR product amplified within the same reaction. The control PCR product is a 150 bp region of chromosome V that is outside of any open reading frames (Komarnitsky et al., 2000). Quantitation was performed by PhosphorImager analysis (Molecular Dynamics). All calculations were carried out as described previously (Larschan and Winston, 2001). The primers used for the control PCR were described previously (Komarnitsky et al., 2000). All other primers used were created for this study and primer sequences are available upon request.
Acknowledgments
Acknowledgements
We thank Beverly Wendland, Dan Fraenkel, Brandon Davies and Jasper Rine for helpful discussions and advice. We also thank Martin Steffen, Aimée Dudley and George Church for help with microarray experiments. We are grateful to Beverly Wendland for providing FM4-64 and the staining protocol. We are also grateful to Alan Sachs, Jerry Kaplan, Brandon Davies and Jasper Rine for providing strains and plasmids. We thank Erica Larschan and Mary Bryk for critical reading of the manuscript. C.H. was supported by NIH Minority Predoctoral Fellowship 1F31GM19933-03. This work was supported by NIH grant GM45720 to F.W. and NIH grant GM62104 to M.B.
References
- Abramova N., Sertil,O., Mehta,S. and Lowry,C.V. (2001a) Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol., 183, 2881–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramova N.E., Cohen,B.D., Sertil,O., Kapoor,R., Davies,K.J. and Lowry,C.V. (2001b) Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics, 157, 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anraku Y., Hirata,R., Wada,Y. and Ohya,Y. (1992) Molecular genetics of the yeast vacuolar H+-ATPase. J. Exp. Biol., 172, 67–81. [DOI] [PubMed] [Google Scholar]
- Arthington-Skaggs B.A., Crowell,D.N., Yang,H., Sturley,S.L. and Bard,M. (1996) Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett., 392, 161–165. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1988) Current Protocols in Molecular Biology. Greene Publishing Associates/Wiley Interscience, New York, NY.
- Banta L.M., Robinson,J.S., Klionsky,D.J. and Emr,S.D. (1988) Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol., 107, 1369–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B.D., Sertil,O., Abramova,N.E., Davies,K.J. and Lowry,C.V. (2001) Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res., 29, 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T., Katzmann,D.J., Cowles,C.R. and Emr,S.D. (2001) Vps41p function in the alkaline phosphatase pathway requires homo-oligomerization and interaction with AP-3 through two distinct domains. Mol. Biol. Cell, 12, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimster-Denk D. et al. (1999) Comprehensive evaluation of isoprenoid biosynthesis regulation in Saccharomyces cerevisiae utilizing the Genome Reporter Matrix. J. Lipid Res., 40, 850–860. [PubMed] [Google Scholar]
- Dudley A.M., Rougeulle,C. and Winston,F. (1999) The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev., 13, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin A.V., Rothenberg,M., Downs,M.A. and Blumer,K.J. (1998) Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signaling in Saccharomyces cerevisiae. Genetics, 149, 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (eds) (1991) Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA.
- Harlow E. and Lane,D. (eds) (1999) Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Jensen-Pergakes K., Guo,Z., Giattina,M., Sturley,S.L. and Bard,M. (2001) Transcriptional regulation of the two sterol esterification genes in the yeast Saccharomyces cerevisiae. J. Bacteriol., 183, 4950–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastaniotis A.J. and Zitomer,R.S. (2000) Rox1 mediated repression. Oxygen dependent repression in yeast. Adv. Exp. Med. Biol., 475, 185–195. [PubMed] [Google Scholar]
- Kastaniotis A.J., Mennella,T.A., Konrad,C., Torres,A.M. and Zitomer,R.S. (2000) Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol. Cell. Biol., 20, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M. and Wickner,W. (2001) Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. EMBO J., 20, 4035–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M.A. and Bard,M. (2001) Positive and negative regulation of squalene synthase (ERG9), an ergosterol biosynthetic gene, in Saccharomyces cerevisiae. Biochim. Biophys. Acta, 1517, 177–189. [DOI] [PubMed] [Google Scholar]
- Kennedy M.A., Barbuch,R. and Bard,M. (1999) Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta, 1445, 110–122. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Herman,P.K. and Emr,S.D. (1990) The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev., 54, 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P., Cho,E.J. and Buratowski,S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev., 14, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac L., Bohmerova,E. and Necas,O. (1987) The plasma membrane of yeast protoplasts exposed to hypotonicity becomes porous but does not disintegrate in the presence of protons or polyvalent cations. Biochim. Biophys. Acta, 899, 265–275. [DOI] [PubMed] [Google Scholar]
- Kwast K.E., Burke,P.V. and Poyton,R.O. (1998) Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol., 201, 1177–1195. [DOI] [PubMed] [Google Scholar]
- Kwast K.E., Lai,L.C., Menda,N., James,D.T.,III, Aref,S. and Burke,P.V. (2002) Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol., 184, 250–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E. and Winston,F. (2001) The S.cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev., 15, 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber R., Zenz,R., Schrottner,K., Fuchsbichler,S., Puhringer,B. and Turnowsky,F. (2001) A novel sequence element is involved in the transcriptional regulation of expression of the ERG1 (squalene epoxidase) gene in Saccharomyces cerevisiae. Eur. J. Biochem., 268, 914–924. [DOI] [PubMed] [Google Scholar]
- Lees N.D., Skaggs,B., Kirsch,D.R. and Bard,M. (1995) Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae—a review. Lipids, 30, 221–226. [DOI] [PubMed] [Google Scholar]
- Lorenz R.T. and Parks,L.W. (1991) Physiological effects of fenpropimorph on wild-type Saccharomyces cerevisiae and fenpropimorph-resistant mutants. Antimicrob. Agents Chemother., 35, 1532–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M’Baya B., Fegueur,M., Servouse,M. and Karst,F. (1989) Regulation of squalene synthetase and squalene epoxidase activities in Saccharomyces cerevisiae. Lipids, 24, 1020–1023. [DOI] [PubMed] [Google Scholar]
- Madison J.M., Dudley,A.M. and Winston,F. (1998) Identification and analysis of Mot3, a zinc finger protein that binds to the retrotransposon Ty long terminal repeat (delta) in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 1879–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean-Bowen C.A. and Parks,L.W. (1982) Effect of altered sterol composition on the osmotic behavior of sphaeroplasts and mitochondria of Saccharomyces cerevisiae. Lipids, 17, 662–665. [DOI] [PubMed] [Google Scholar]
- Molzahn S.W. and Woods,R.A. (1972) Polyene resistance and the isolation of sterol mutants in Saccharomyces cerevisiae. J. Gen. Microbiol., 72, 339–348. [DOI] [PubMed] [Google Scholar]
- Munn A.L. (2000) The yeast endocytic membrane transport system. Microsc. Res. Tech., 51, 547–562. [DOI] [PubMed] [Google Scholar]
- Munn A.L. (2001) Molecular requirements for the internalisation step of endocytosis: insights from yeast. Biochim. Biophys. Acta, 1535, 236–257. [DOI] [PubMed] [Google Scholar]
- Munn A.L. and Riezman,H. (1994) Endocytosis is required for the growth of vacuolar H+-ATPase-defective yeast: identification of six new END genes. J. Cell Biol., 127, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn A.L., Heese-Peck,A., Stevenson,B.J., Pichler,H. and Riezman,H. (1999) Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell, 10, 3943–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Hirata,A., Ohsumi,Y. and Wada,Y. (1997) Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J. Biol. Chem., 272, 11344–11349. [DOI] [PubMed] [Google Scholar]
- Ness F., Bourot,S., Regnacq,M., Spagnoli,R., Berges,T. and Karst,F. (2001) SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. Eur. J. Biochem., 268, 1585–1595. [PubMed] [Google Scholar]
- Parks L.W., Bottema,C.D., Rodriguez,R.J. and Lewis,T.A. (1985) Yeast sterols: yeast mutants as tools for the study of sterol metabolism. Methods Enzymol., 111, 333–346. [DOI] [PubMed] [Google Scholar]
- Parks L.W., Smith,S.J. and Crowley,J.H. (1995) Biochemical and physiological effects of sterol alterations in yeast—a review. Lipids, 30, 227–230. [DOI] [PubMed] [Google Scholar]
- Radisky D.C., Snyder,W.B., Emr,S.D. and Kaplan,J. (1997) Characterization of VPS41, a gene required for vacuolar trafficking and high-affinity iron transport in yeast. Proc. Natl Acad. Sci. USA, 94, 5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H., Munn,A., Geli,M.I. and Hicke,L. (1996) Actin-, myosin- and ubiquitin-dependent endocytosis. Experientia, 52, 1033–1041. [DOI] [PubMed] [Google Scholar]
- Robzyk K. and Kassir,Y. (1992) A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res., 20, 3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Novick,P., Thomas,J.H., Botstein,D. and Fink,G.R. (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene, 60, 237–243. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Hieter,P. (eds) (1990) Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sachs A.B. and Deardorff,J.A. (1992) Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell, 70, 961–973. [DOI] [PubMed] [Google Scholar]
- Sambrook J.F., Fritsch,E.F. and Maniatis,T. (eds) (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast hosts strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.J., Crowley,J.H. and Parks,L.W. (1996) Transcriptional regulation by ergosterol in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustre I., Girard,P. and Karst,F. (1998) Biosynthesis and transport of sterols in the yeast Saccharomyces cerevisiae. C. R. Seances Soc. Biol. Fil., 192, 977–990. [PubMed] [Google Scholar]
- Spencer F., Connelly,S., Lee,S. and Hieter,P. (1988) Isolation and cloning of conditionally lethal chromosome transmission fidelity genes in Saccharomyces cerevisiae. In Kelly,T. and Stillman,B. (eds), Eukaryotic DNA Replication. Cold Spring Harbor Laboratory Press, New York, NY, pp. 441–452.
- Sturley S.L. (2000) Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim. Biophys. Acta, 1529, 155–163. [DOI] [PubMed] [Google Scholar]
- Swanson M.S., Malone,E.A. and Winston,F. (1991) SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol., 11, 4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.Y. and Cai,M. (1996) The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 4897–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.Y., Munn,A. and Cai,M. (1997) EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 4294–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.Y., Xu,J. and Cai,M. (2000) Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell. Biol., 20, 12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Linde J.J., Liang,H., Davis,R.W., Steensma,H.Y., van Dijken,J.P. and Pronk,J.T. (1999) Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol., 181, 7409–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness M., Schafer,W., D’Ari,L. and Rine,J. (1989) Positive and negative transcriptional control by heme of genes encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol. Cell. Biol., 9, 5702–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinkelenberg A.H., Liu,Y., Alcantara,F., Khan,S., Guo,Z., Bard,M. and Sturley,S.L. (2000) Mutations in yeast ARV1 alter intracellular sterol distribution and are complemented by human ARV1. J. Biol. Chem., 275, 40667–40670. [DOI] [PubMed] [Google Scholar]
- Turi T.G. and Loper,J.C. (1992) Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 α-demethylase (ERG11). J. Biol. Chem., 267, 2046–2056. [PubMed] [Google Scholar]
- Vida T.A. and Emr,S.D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol., 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vik A. and Rine,J. (2001) Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 6395–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B. and Emr,S.D. (1998) Pan1p, yeast eps15, functions as a multivalent adaptor that co-ordinates protein–protein interactions essential for endocytosis. J. Cell Biol., 141, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B., McCaffery,J.M., Xiao,Q. and Emr,S.D. (1996) A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol., 135, 1485–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesp A., Hicke,L., Palecek,J., Lombardi,R., Aust,T., Munn,A.L. and Riezman,H. (1997) End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell, 8, 2291–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard,C. and Ricupero-Hovasse,S.L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast, 11, 53–55. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A. et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Zweytick D., Athenstaedt,K. and Daum,G. (2000) Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta, 1469, 101–120. [DOI] [PubMed] [Google Scholar]