Abstract
Programmed telomere shortening in human somatic cells is thought to act as a tumor suppressor pathway, limiting the replicative potential of developing tumor cells. Critically short human telomeres induce senescence either by activating p53 or by inducing the p16/RB pathway, and suppression of both pathways is required to suppress senescence of aged human cells. Here we report that removal of TRF2 from human telomeres and the ensuing de-protection of chromosome ends induced immediate premature senescence. Although the telomeric tracts remained intact, the TRF2ΔBΔM-induced premature senescence was indistinguishable from replicative senescence and could be mediated by either the p53 or the p16/RB pathway. Telomere de-protection also induced a growth arrest and senescent morphology in mouse cells. However, in this setting the loss of p53 function was sufficient to completely abrogate the arrest, indicating that the p16/RB response to telomere dysfunction is not active in mouse cells. These findings reveal a fundamental difference in telomere damage signaling in human and mouse cells that bears on the use of mouse models for the telomere tumor suppressor pathway.
Keywords: p53/RB/telomere/TRF2/tumor suppressor
Introduction
In the absence of telomerase, human telomeres shorten by 50–200 bp per population doubling (PD) (Harley et al., 1990). After 50–100 PD, this process depletes the telomere reserve of primary human cells and results in replicative senescence. Forced expression of telomerase can avert senescence and immortalizes cells that are grown under optimal conditions (Bodnar et al., 1998). As senescence erects a proliferative barrier that can thwart the growth ambitions of pre-cancerous cells, programmed telomere shortening is viewed as a potential tumor suppressor system. Several observations argue in favor of this notion. First, in the majority of human tumors, telomere shortening is negated, usually through the upregulation of telomerase (reviewed in Shay and Bacchetti, 1997). Secondly, inhibition of telomerase in established tumor cell lines induces telomere shortening and eventually arrests culture growth (Hahn et al., 1999; Zhang et al., 1999; Damm et al., 2001). Thirdly, telomeres shorten much faster in telomerase-deficient human cells than in telomerase-negative yeast, flies or plants (Levis, 1989; Lundblad and Szostak, 1989; Fitzgerald et al., 1999), pointing to an active telomere attrition program in addition to passive sequence loss due to the ‘end-replication problem’.
Evidence in support of a telomere tumor suppressor pathway has been sought using mouse models. This approach is complicated by the fact that mice do not employ telomere attrition to limit their tumor burden. Mus musculus telomeres are much longer than human telomeres (Kipling and Cooke, 1990) and telomerase is not repressed in murine somatic cells (Prowse and Greider, 1995). However, the human telomere shortening program is recapitulated to some extent in mice lacking telomerase. The mTerc–/– mouse strain, which lacks telomerase due to a targeted deletion of the gene encoding the telomerase template RNA, has been used to explore the effect of telomere shortening on tumor formation (Blasco et al., 1997; Chin et al., 1999; Greenberg et al., 1999; Rudolph et al., 1999; Artandi et al., 2000). After 4–6 generations in the absence of telomerase, the telomeres of these mice are as short as those in human cells nearing senescence. Under these conditions, telomere shortening was demonstrated to have an inhibitory effect on tumorigenesis in Ink4a/Arf knockout (KO) mice (Greenberg et al., 1999), which develop tumors rapidly due to the absence of p19Arf and also lack a second tumor suppressor, p16 (Serrano et al., 1996; Krimpenfort et al., 2001; Sharpless et al., 2001; Sherr, 2001). The outcome of this study suggested that telomere shortening could suppress tumorigenesis, but the extent of the effect was rather small. At best, telomere shortening inhibited in vitro transformation and tumor outgrowth ∼2-fold. Furthermore, mice without a functional p53 gene did not show a reduction in tumor formation when their telomeres were shortened (Artandi et al., 2000).
Replicative senescence of human cells can be abrogated by suppression of both the p53 and the p16/RB pathways. For instance, suppression of these pathways by SV40 large T or the combined action of HPV16 E6 and E7 can fully suppress senescence, creating a cell population with an extended lifespan (Shay and Wright, 1989; Shay et al., 1991). In both instances, cells continue to grow and their telomere attrition proceeds unabated. The dwindling telomeres progressively fail to protect chromosome ends, and after an additional 20–30 PD such cultures usually succumb to karyotypic chaos (Counter et al., 1992). Oncoproteins that either repress p53 or inactivate RB proteins are unable to persuade cells to ignore their shortening telomeres (Shay et al., 1991). Although cells with diminished p53 activity gain a few additional cell divisions, the signal emanating from shortened telomeres eventually overcomes growth promoting forces, inhibits the phosphorylation of RB and results in senescence. Conversely, in the absence of a functional RB circuit, the p53 pathway can enforce a growth arrest when telomeres are shortened. Thus, in human cells there is redundancy in the signaling pathways that alert cells to dysfunctional telomeres and promote replicative senescence.
Recent evidence has implicated TRF2, a telomeric DNA binding protein, as a key player in the regulation of the senescence signal. Telomeres recruit a multimeric protein complex built up on two related sequence-specific DNA binding factors, TRF1 and TRF2, which both bind to the duplex array of TTAGGG repeats at chromosome ends (Broccoli et al., 1997). TRF2 is essential for protection of chromosome ends (van Steensel et al., 1998; reviewed in de Lange, 2002). Removal of TRF2 ‘uncaps’ telomeres, resulting in end-to-end chromosome fusions and rapid induction of a cell cycle arrest. In some settings, inhibition of TRF2 activates the ATM/p53 pathway and induces apoptosis (Karlseder et al., 1999). Conversely, overexpression of TRF2 can protect critically shortened telomeres and extend the replicative lifespan of primary fibroblasts (Karlseder et al., 2002). One interpretation of these findings is that replicative senescence is induced when short telomeres fail to recruit sufficient TRF2. It has been proposed that TRF2 is required to form t-loops, circular structures at chromosome ends in which the single-stranded telomeric terminus is tucked into the adjacent double-stranded telomeric repeat DNA (Griffith et al., 1999; Stansel et al., 2001). The formation of t-loops by TRF2 could represent a mechanism to sequester chromosome ends.
In this study, inhibition of TRF2 is used to examine how mammalian cells enter senescence in response to telomere dysfunction. Using a dominant-negative allele of TRF2, a premature senescence indistinguishable from replicative senescence could be induced in primary fibroblasts. The advantage of studying senescence using this system is that the response occurs within a few cell divisions, instead of requiring extensive cell growth, which could result in accumulation of other changes. The data emerging from the TRF2 inhibition studies reveal a crucial difference in how human and mouse cells respond to telomere dysfunction. While both the p53 and the p16/RB pathway can induce senescence in human cells, only the p53 pathway has this ability in mouse cells. When using mouse models to evaluate the role of telomere shortening in tumorigenesis, this important distinction must be taken into account.
Results
Induction of premature senescence through inhibition of TRF2
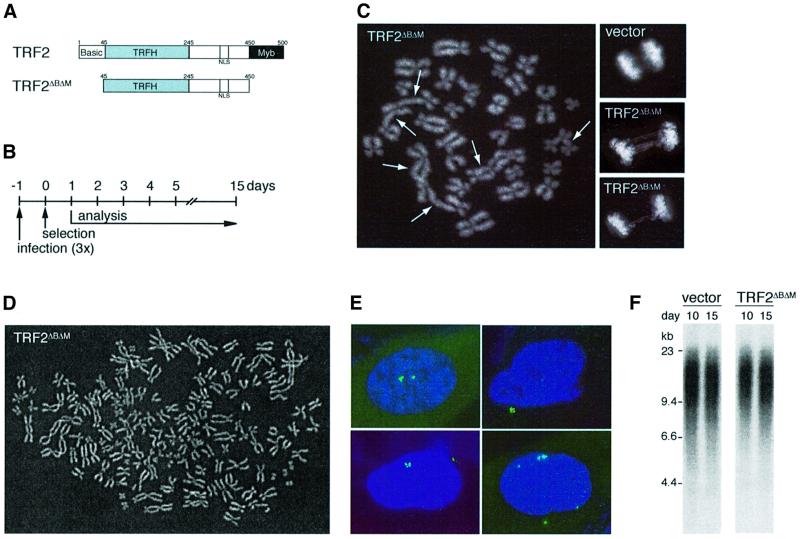
Retroviral gene delivery was used to express TRF2ΔBΔM (Figure 1A and B), a dominant-negative allele of TRF2 that removes the endogenous protein from telomeres (van Steensel et al., 1998). In tumor cell lines, TRF2ΔBΔM induces frequent end-to-end chromosome fusions (van Steensel et al., 1998; Karlseder et al., 1999). Similarly, primary human lung IMR90 fibroblasts expressing TRF2ΔBΔM showed chromosome end fusions detectable in metaphase spreads (Figure 1C, 219 fusions/100 metaphases) and anaphase cells often contained chromatin bridges (Figure 1C). The frequency of chromosome end fusions was not significantly increased in parallel control cultures infected with a virus expressing fully functional TRF2 or a virus lacking TRF2 (17 and 3 fusions per 100 metaphases, respectively). TRF2ΔBΔM also generated cells with a near tetraploid chromosome number (Figure 1D), a phenomenon not observed in control cultures. End-to-end chromosome fusions and tetraploid metaphases have previously been observed in senescent fibroblasts (Saksela and Moorhead, 1963; Wolman et al., 1964; Thompson and Holliday, 1975; Benn, 1976). The inhibition of TRF2 also caused a substantial fraction of the cells to accumulate more than two centrosomes (Figure 1E; Table I). Interestingly, centrosome amplification also occurred in fibroblasts undergoing replicative senescence (Table I), indicating that this is a new marker for replicative senescence. Thus, in terms of karyotypic and cytological characteristics, the phenotype created by inhibition of TRF2 is indistinguishable from replicative senescence. However, unlike cells undergoing replicative senescence, TRF2ΔBΔM-infected IMR90 cells did not show telomere shortening over the course of these experiments (15 days) (Figure 1F). Chromosome end fusions with retention of telomeric DNA were also observed in TRF2ΔBΔM-expressing primary HS68 and BJ fibroblasts, and in BJ fibroblasts immortalized with hTERT (data not shown). In previous studies, chromosome end fusions could be visualized on genomic blots as newly formed, larger telomeric restriction fragments (van Steensel et al., 1998). However, because the telomeres of the young fibroblasts used in this study are much longer, their fusion products could not be resolved by standard agarose gel electrophoresis.
Fig. 1. TRF2ΔBΔM induces telomere de-protection and karyotypic abnormalities in IMR90 cells without loss of telomeric DNA. (A) Schematic of TRF2 and TRF2ΔBΔM. (B) Experimental timeline. (C) Chromosome end fusions. Large panel, metaphase spread with multiple chromosome end fusions (arrows). Small panels, anaphase cells (top, control; bottom, TRF2ΔBΔM). (D) TRF2ΔBΔM-induced tetraploidy in IMR90. Diplochromosomes were observed in a subset of tetraploid metaphases. (E) TRF2ΔBΔM-induced centrosome amplification in IMR90. IF for γ-tubulin (FITC, green). (F) Persistence of telomeric DNA in TRF2ΔBΔM-expressing cells. Genomic blot of telomeric HinfI–RsaI fragments detected with a TTAGGG repeat probe. DNA was isolated from IMR90 cells on day 10 and 15 of selection, after infection with pWZLvector or pWZLTRF2ΔBΔM. (C–E) represent analysis on day 7 of selection and DAPI DNA staining.
Table I. Amplification of centrosomes in TRF2ΔBΔM-infected and senescent cells.
| Cells/passage | Retrovirus | Timepoint after selection | % cells with >2 centrosomesa |
|---|---|---|---|
| IMR90/p24 | Vector | Day 7 | 3 |
| Vector | Day 14 | 5 | |
| TRF2 | Day 7 | 4 | |
| TRF2 | Day 14 | 5 | |
| TRF2ΔBΔM | Day 7 | 15 | |
| TRF2ΔBΔM | Day 14 | 16 | |
| WI38/p21 | Vector | Day 5 | 9 |
| TRF2 | Day 5 | 12 | |
| TRF2ΔBΔM | Day 5 | 25 | |
| HS68/p18 | Vector | Day 5 | 3 |
| TRF2 | Day 5 | 4 | |
| TRF2ΔBΔM | Day 5 | 14 | |
| Senescent IMR90/p43 | None | 23 |
aCentrosome numbers were determined by staining with γ-tubulin antibody and the percentage of cells with more than two centrosomes was determined by microscopic inspection of ≥100 cells.
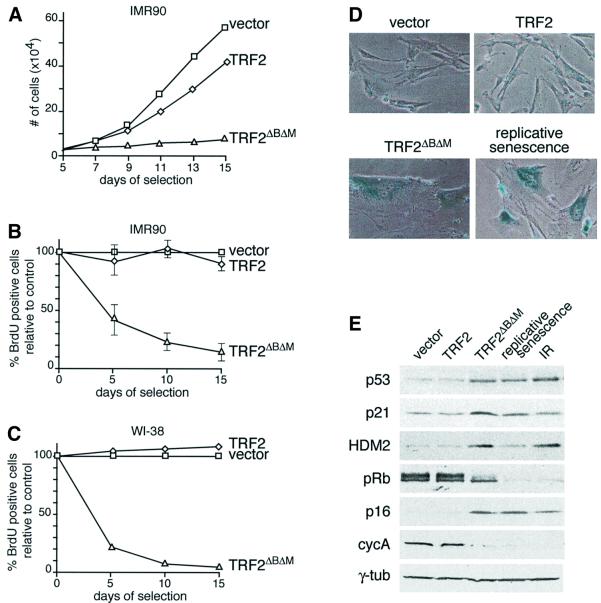
Consistent with the notion that inhibition of TRF2 induced premature senescence, TRF2ΔBΔM-expressing IMR90 cells ceased to proliferate while control cells continued to grow (Figure 2A). Moreover, TRF2ΔBΔM induced a rapid decline in the percentage of cells in S phase, as demonstrated by the reduced ability of the cultures to incorporate BrdU (Figure 2B). FACS analysis on day 15 revealed cells with either 2n or 4n DNA content, a profile matching that of cells undergoing replicative senescence (data not shown). It is possible that the population of cells with a 4n DNA content represents tetraploid cells rather than cells arrested in G2/M. A TRF2ΔBΔM-induced cell cycle arrest was also observed in primary human WI-38 fibroblasts (Figure 2C) and in human BJ fibroblasts that had been immortalized with hTERT (Bodnar et al., 1998) (data not shown). Concomitant with the arrest, the morphology of TRF2ΔBΔM-expressing fibroblasts changed progressively, with cells becoming enlarged and flat; their nuclei were often multi-lobed; and the cells stained positive for the senescence-associated β-galactosidase marker SA-β-gal (Dimri et al., 1995) (Figure 2D; data not shown). Based on these criteria, the cells were indistinguishable from fibroblasts at the end of their natural replicative lifespan.
Fig. 2. Premature senescence in primary human fibroblasts expressing TRF2ΔBΔM. (A) Growth curve of IMR90 cells expressing TRF2, or TRF2ΔBΔM and vector control cells. Cells were infected at passage 23 and analyzed according to the schematic in Figure 1B. After 3 weeks, TRF2-expressing cells proliferated at the same rate as vector control cells. (B) S-phase index in TRF2ΔBΔM-expressing IMR90 cells. The percentage of cells in S phase was measured using BrdU incorporation on day 5, 10 and 15 (see Materials and methods). Values represent the mean of 4–5 experiments and the SD is given. (C) S-phase index in TRF2ΔBΔM-expressing WI-38 cells. Method as in (B) but using WI-38 fibroblasts at passage 26. (D) TRF2ΔBΔM-induced senescent morphology and SA-β-gal expression. IMR90 cells (passage 22) infected with the indicated viruses as in (A) were stained for SA-β-gal on day 16. The bottom right panel shows IMR90 cells that have undergone replicative senescence (passage 42). (E) Immunoblot analysis of TRF2ΔBΔM-expressing fibroblasts, senescent fibroblasts and γ-irradiated fibroblasts. IMR90 cells (passage 22) infected with the indicated viruses and analyzed on day 15. Naturally senescent cells were collected at passage 42. Irradiated cells (10 Gray from a 137Cs source) were maintained for 15 days with daily feeding before preparation of the lysate. Lysates of 1.5 × 105 cell equivalents were loaded in each lane. Immunoblotting for the indicated proteins was performed as detailed in Materials and methods.
As previously reported, cells undergoing replicative senescence showed increased levels of p53, a concomitant increase in its downstream transcriptional target p21 and moderate induction of HDM2 (Figure 2E). Similarly, TRF2ΔBΔM-induced arrest was consistently accompanied by an induction of p53, p21 and HDM2 (Figure 2E). TRF2ΔBΔM-expressing cells also resembled cells undergoing replicative senescence in that p16 was upregulated and pRb was hypophosphorylated. Moreover, cyclin A levels were reduced in both the TRF2ΔBΔM-expressing cells and in senescent cells (Figure 2E). This profile also resembled that of irradiated IMR90 cells that have been arrested for 15 days (Figure 2E; Di Leonardo et al., 1994), consistent with the view that telomere de-protection may activate components of the DNA damage response pathway (Karlseder et al., 1999).
Independent induction of senescence by p53 and p16/RB
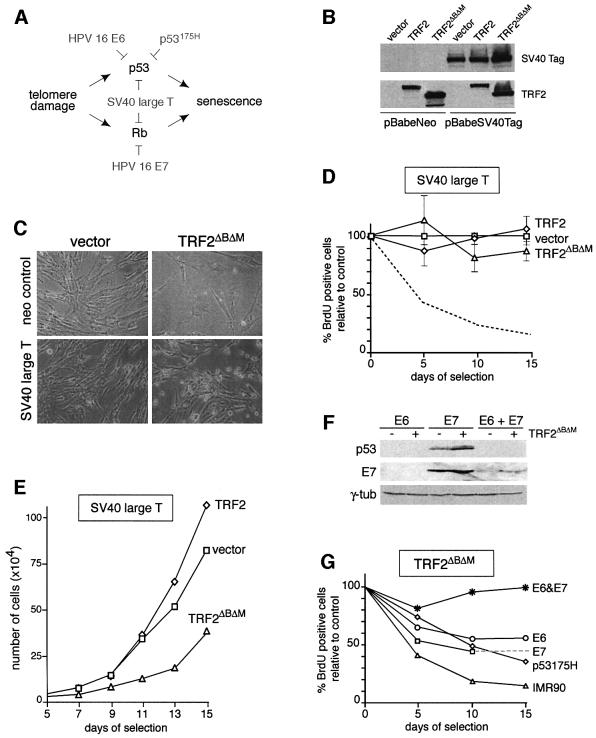
The role of the p53 and p16/RB pathways in TRF2ΔBΔM-induced senescence was examined using viral and cellular oncoproteins known to affect these tumor suppressors (see the schematic in Figure 3A). Young (passage 17) IMR90 cells were infected with a retrovirus expressing SV40 large T antigen and subsequently infected with a retrovirus expressing TRF2ΔBΔM. The expression of SV40 large T was monitored by immunoblotting (Figure 3B) and resulted in the expected high levels of inactive p53 (data not shown). In two independent experiments, the growth curves of these cultures indicated continued cell proliferation and their unaltered rate of BrdU incorporation showed that SV40 large T had eliminated the TRF2ΔBΔM-induced cell cycle arrest (Figure 3C–E). Furthermore, cells expressing SV40 large T did not adopt a senescent morphology in response to TRF2ΔBΔM, and they failed to express the SA-β-gal marker (Figure 3C; data not shown). In comparison, control fibroblasts infected with the pBabeNeo control vector responded to TRF2ΔBΔM as observed above, showing cell cycle arrest and adopting a senescent morphology (Figure 3C). Although the SV40 large T-transformed cells expressing TRF2ΔBΔM continued to enter S phase, the growth of these cultures was not as robust as SV40 large T-expressing cells infected with a TRF2 virus or the vector control (Figure 3E). Most likely this is due to cell death resulting from the extensive genome instability induced by TRF2ΔBΔM. Whether this diminished growth phenotype resembles the growth crisis seen in SV40-transformed human cells is not clear. In WI-38 cells, expression of SV40 large T similarly bypassed TRF2ΔBΔM-induced senescence. WI-38 cells expressing both SV40 large T and TRF2ΔBΔM continued to enter S phase (BrdU incorporation of 70–100% of the vector control), whereas the BrdU incorporation fell to 2–5% of the vector control when SV40 large T was absent (Figure 2C; data not shown).
Fig. 3. TRF2ΔBΔM-induced premature senescence in human cells mediated by p53 and p16/RB. (A) Oncoproteins used to inhibit the p53 and p16/RB pathways. (B) Immunoblot analysis of expression of SV40 large T antigen and retroviral TRF2 proteins. IMR90 cells infected with pBabeNeo or pBabeSV40Tag were superinfected with the indicated viruses and SV40 Tag and TRF2 (Ab 647) were detected as described in Materials and methods. (C) Morphology of SV40 Tag-expressing cells infected with the TRF2ΔBΔM virus. Cells were infected as in (B) and photographed on day 13 after selection. (D) S-phase index in cells expressing SV40 Tag and TRF2ΔBΔM. Procedure as in Figure 2B. Mean values and SDs were derived from three experiments. (E) Growth curve of IMR90 cells expressing SV40 Tag and TRF2ΔBΔM or control viruses (vector or TRF2). (F) Immunoblot analysis of IMR90 cells expressing HPV16 E6 and/or E7. IMR90 cells were infected with viruses for HPV16 E6, E7, or both, and superinfected with a control retrovirus (– lanes) or the TRF2ΔBΔM retrovirus (+ lanes). WCL (equal cell number equivalents) were analyzed on day 8 of selection for the expression of p53 and E7, as described in Materials and methods. Reduction of the basal level of p53 (– lanes) served as a marker for the activity of E6. γ-tubulin served as a loading control. (G) Effects of separate inhibition of the p53 and p16/RB pathways in TRF2ΔBΔM-induced premature senescence. Young (passage 19) IMR90 cells were first infected with viruses carrying HPV16 E6, HPV16 E7, HPV16 E6 and E7, p53175H, or with the pBabeNeo control virus (graph labeled IMR90). Subsequently, cultures were superinfected with the TRF2ΔBΔM virus or the vector control. The percentage BrdU-positive cells was determined in triplicate, as described in Materials and methods. The SDs were 1–5%. The dotted line extrapolates the data point for E7-expressing cells on day 10 to a day 15 data point from a different experiment.
To further analyze the requirements for the growth arrest in TRF2ΔBΔM-expressing cells, we inhibited the p53 and p16/RB pathways separately. Inhibition of p53 was achieved through the use of a dominant-negative allele (p53175H) or with HPV16 E6, and the RB family of proteins was inhibited using HPV16 E7 (Figure 3A). As expected, p53 was undetectable in cells expressing E6, even after infection with TRF2ΔBΔM (Figure 3F) or γ-irradiation, and E6-expressing cells failed to show a G1/S arrest after irradiation (data not shown). Expression of p53175H and E7 was confirmed using immunoblotting (Figure 3F; data not shown). Cells expressing HPV16 E6 or p53175H responded to expression of TRF2ΔBΔM with a strong decline in BrdU incorporation (Figure 3G), and their morphology resembled senescent cells (data not shown). Three independent experiments with E6 showed a consistent reduction of the BrdU incorporation to 40–50% of the controls. Similarly, in cells in which the p53 pathway was inhibited with the p53175H allele, TRF2 inhibition consistently resulted in a reduction of BrdU incorporation to 40% of the vector controls. Moreover, cultures expressing both p53175H and TRF2ΔBΔM hardly increased in cell number (doubling time >10 days compared with daily doubling of the controls), and the cultures did not grow faster than wild-type IMR90 cells infected with TRF2ΔBΔM (data not shown). The finding that p53 deficiency alone did not abrogate response to TRF2ΔBΔM is consistent with the ability of HT1080 fibrosarcoma cells, which lack a functional p53 pathway, to respond to telomere shortening or TRF2ΔBΔM (van Steensel et al., 1998; Damm et al., 2001).
Expression of HPV16 E7 alone was also insufficient to bypass the induction of senescence by TRF2ΔBΔM (Figure 3G), indicating that both the p53 pathway and the p16/RB pathway can independently enforce senescence in response to loss of TRF2. As expected, simultaneous expression of HPV16 E6 and E7 abrogated senescence in TRF2ΔBΔM-expressing fibroblasts, as evident from their continued growth, incorporation of BrdU and lack of senescent morphology (Figure 3G; data not shown). Thus, the genetics of the TRF2ΔBΔM-induced premature senescence pathway closely resembles that of replicative senescence in human cells.
TRF2ΔBΔM-induced arrest in mouse cells is abrogated by p53 deficiency
Since the TRFH dimerization domains of human TRF2 and mouse Trf2 are 95% identical (Broccoli et al., 1997), TRF2ΔBΔM removes mouse Trf2 from telomeres (Karlseder et al., 1999; A.Smogorzewska and T.de Lange, unpublished data). As is the case when human TRF2 is inhibited, diminished Trf2 function leads to de-protection of mouse telomeres, resulting in end-to-end fusions (Karlseder et al., 1999; A.Smogorzewska and T.de Lange, unpublished data) even though the telomeric DNA remains intact (R.Wang and T.de Lang, unpublished data). To test the response of mouse cells to this telomere dysfunction, wild-type mouse embryonic fibroblasts (MEFs) were infected with the TRF2ΔBΔM virus or a vector control. Like human cells, primary MEFs arrested rapidly after inhibition of Trf2, resulting in the cessation of culture growth (Figure 4A and B). The rate of BrdU incorporation reproducibly fell to 15–25% of the control cells after 2 weeks of expression of TRF2ΔBΔM. These cells adopted a senescent morphology, becoming flat and enlarged, and they stained positive for SA-β-gal (Figure 4C). Thus, mouse fibroblasts, like human cells, respond to TRF2ΔBΔM -induced telomere dysfunction with the induction of a growth arrest and show morphological changes resembling senescence in human cells.
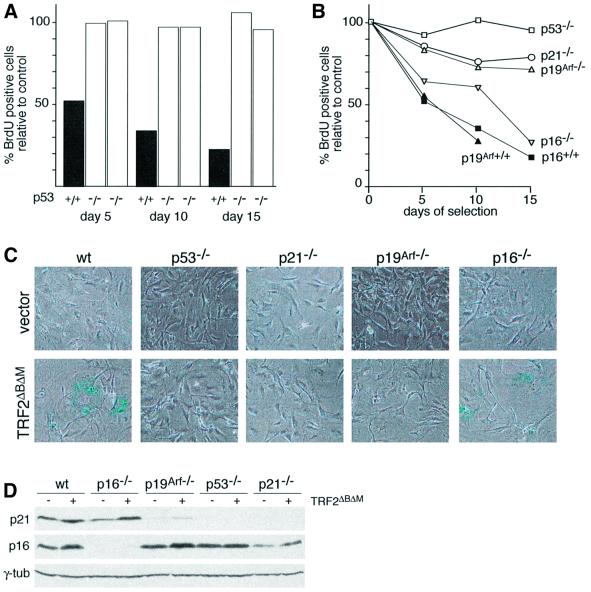
Fig. 4. TRF2ΔBΔM-induced growth arrest in murine cells depends on p53. (A) S-phase index of TRF2ΔBΔM-expressing p53+/+ and p53–/– MEFs. Cells of the indicated genotypes were infected with the TRF2ΔBΔM virus or the vector control, and the incorporation of BrdU in the parallel cultures was measured as described in Figure 2B. The bar graph shows the percentage of TRF2ΔBΔM-expressing cells in S phase compared with controls (control virus infection set at 100%) as measured using BrdU incorporation. Two experiments are shown for two independent batches of MEFs deficient in p53. (B) Effects of deficiency for p53, p21, p19Arf or p16 on the TRF2ΔBΔM-induced growth arrest in MEFs. Cells of the indicated genotypes were infected with TRF2ΔBΔM or a control virus. BrdU incorporation in TRF2ΔBΔM-expressing cells was normalized to control infected cells as described in Figure 2B. Values are derived from triplicate measurements. SDs were 1–5%. (C) Effects of p53, p21, p19Arf and p16 status on the TRF2ΔBΔM-induced morphology and SA-β-gal expression. Cells used in the experiment shown in (B) were plated at equal cell density on day 10 and assayed for β-gal activity 2 days later. (D) Levels of p21 and p16 in mouse cells expressing TRF2ΔBΔM. Cells of the indicated genotypes were infected with the TRF2ΔBΔM virus or the vector control. Lysates (104 cells/lane) were prepared on day 10.
The murine signaling pathway responsible for the TRF2ΔBΔM-induced arrest was examined using MEFs from genetically altered mice deficient for relevant cell cycle control genes. Strikingly, MEFs from p53–/– mice completely lacked the ability to respond to inhibition of Trf2 (Figure 4). In three independent experiments, in which two independent batches of p53–/– MEFs were infected with the TRF2ΔBΔM, the culture growth rate was unaltered and the incorporation of BrdU was the same as in control cells (Figure 4A and B). For instance, the fraction of p53–/– cells expressing TRF2ΔBΔM that incorporated BrdU was almost identical to the control (102 and 94% of the control in two independent experiments). As expected, the p53–/– MEFs lacked detectable p53 and p21 protein, but expressed high levels of p16 (Figure 4D). In addition to their continued proliferation, the p53–/– MEFs expressing TRF2ΔBΔM did not show a senescent morphology and did not contain elevated levels of SA-β-gal (Figure 4C). The p53–/– MEFs showed a very high rate of telomere fusions in response to TRF2ΔBΔM (51 fusions in 30 metaphases compared with 3 fusions in 30 metaphases in uninfected p53–/– MEFs), indicating that Trf2 was inhibited and that the telomeres had become de-protected. A parallel result was obtained with MEFs lacking p21 (Figure 4B–D). The p21–/– MEFs also continued to grow when infected with the TRF2ΔBΔM virus; the cells only showed a minor reduction in the rate of BrdU incorporation (78 and 67% of cells infected with the vector control in two independent experiments), did not adopt a senescent morphology and did not express SA-β-gal (Figure 4B and C). This result is consistent with p21 as the main conduit for the p53-mediated cell cycle arrest.
The p19Arf product of the Ink4a/Arf locus is thought to be a negative regulator of Mdm2 and cells lacking p19Arf have a diminished p53 response due to higher Mdm2 activity (Kamijo et al., 1997; Pomerantz et al., 1998; Zhang et al., 1998). In agreement with this view, the basal level of p21 expression in MEFs lacking p19Arf was considerably lower than in wild-type MEFs and the induction of p21 by telomere dysfunction is diminished in this setting (Figure 4D). These observations are also consistent with the previous demonstration that the DNA damage response is diminished in p19Arf null MEFs (Khan et al., 2000). The diminished activity of the p53 pathway in p19Arf null cells is reflected in the lack of cell cycle arrest in response to TRF2ΔBΔM. Consistent with the bypass of telomere-directed senescence in p53–/– MEFs, cells lacking p19Arf also did not respond normally to TRF2ΔBΔM, whereas MEFs derived from a wild-type littermate entered a senescent-like state (Figure 4B and C). Like p53–/– cells, the p19Arf–/– MEFs continued to incorporate BrdU (90 and 71% compared with cells infected with the control virus in two separate experiments), and they did not show a senescent morphology or detectable SA-β-gal activity. As expected, p19Arf–/– cells were slightly more responsive to TRF2ΔBΔM than p53–/– MEFs, probably reflecting their ability to induce modest levels of p53 upon a strong stimulus. Supporting this view, western analysis revealed a residual induction of p21 in response to TRF2ΔBΔM in p19Arf–/– MEFs, while induction of p21 could not be demonstrated in p53 null MEFs (Figure 4D). In sharp contrast to these findings, MEFs lacking the p16 component of the Ink4a/Arf locus (but retaining wild-type p19Arf; Sharpless et al., 2001) behaved like wild-type MEFs, arresting with a senescent morphology after expression of TRF2ΔBΔM (Figure 4B and C). The absence of p16 expression in the p16–/– MEFs was verified by immunoblot analysis (Figure 4D). Collectively, these data on the response of MEFs to Trf2 inhibition indicate that mouse cells glean information about the function of their telomeres primarily through the p53 pathway, with very little, or no independent contribution of the p16/RB pathway.
Discussion
This study utilized the human telomere protective factor, TRF2, to dissect the signaling pathway of telomere-directed growth arrest in human and mouse cells. Human cells expressing a dominant-negative allele of TRF2 arrest the cell cycle and adopt a phenotype that is indistinguishable from replicative senescence based on cell biological, cytogenetic and biochemical criteria. This finding, together with the demonstration that overexpression of TRF2 can delay senescence, suggests that the initiating signal for the induction of senescence emanates from chromosome ends lacking sufficient TRF2 protection. Using TRF2 inhibition as a tool to alter the status of the telomeres and mimic the events during replicative senescence, we discerned that senescence can be mediated by either the p53 pathway or by the p16/RB pathway, a redundancy that is consistent with previous studies on human cells undergoing replicative aging (Shay et al., 1991). Inhibition of mouse Trf2, which has the same protective function as the human protein, also resulted in a rapid growth arrest resembling the replicative senescence observed in human cells. However, the wiring of the murine response to telomere dysfunction differed significantly from the human system. MEFs lacking p53 continued to proliferate after Trf2 inhibition, indicating that in this setting the p16/RB pathway alone cannot respond adequately to telomere damage. This species-specific response to telomere dysfunction has important repercussions for the use of mouse models to study the human telomere tumor suppressor pathway.
Inhibition of TRF2 mimics replicative senescence
TRF2ΔBΔM-expressing cells and aged human fibroblasts are indistinguishable in terms of morphology, expression of SA-β-gal, cell cycle markers, DNA content, chromosomal aberrations and centrosome amplification. These striking parallels validate the use of TRF2 inhibition to dissect the signaling pathway of replicative senescence. Yet, inhibition of TRF2 does not result in the extensive loss of telomeric DNA associated with replicative senescence. This result further confirms the idea that telomeric DNA sequences, per se, are not sufficient to sustain chromosome end capping; the binding of TRF2 to these sequences appears to be an additional prerequisite for the capping function, including preventing the induction of senescence. Consistent with this idea, overexpression of TRF2 can protect critically shortened telomeres and delay the senescence response (Karlseder et al., 2002). Together, these findings support the view that replicative senescence in human cells is induced when telomeres have become too short to recruit the number of TRF2 molecules required for telomere protection. The resulting ‘uncapped’ state is the initiating event that signals to the p53 and p16/RB pathways in human cells. Although it is not excluded that the signal emanating from a single uncapped telomere is sufficient to induce senescence, the occurrence of multiple chromosome end fusions in senescent cells would suggest that several chromosome ends have lost their protection before the cells stop dividing.
Independent signaling of human telomere-directed senescence by p53 and p16/RB
The data obtained with TRF2ΔBΔM support the view that telomere-directed senescence in human cells can be mediated independently by the p53 and p16/RB pathways. The involvement of the p53 pathway in replicative senescence and telomere damage signaling had been established previously (Shay et al., 1991). However, the role of the p16/RB pathway had been confounded by induction of p16 under suboptimal culture conditions (Kiyono et al., 1998; Dickson et al., 2000; Munro et al., 2001). Because this induction of p16 is independent of the telomere dynamics, it was suggested that p16 upregulation is primarily a response to the growth conditions rather than telomere shortening. However, when the same cells are grown under optimal conditions, p16 is not induced and telomerase-mediated telomere maintenance is sufficient to endow cells with infinite replicative potential (Ramirez et al., 2001). These results suggested that p16 can be induced both by certain culture conditions and telomere shortening. Our observations of TRF2ΔBΔM-expressing cells further confirm that p16 levels increase in response to acute telomere dysfunction. The lack of p16 induction and continued growth of parallel control cultures indicated that the induction of p16 by TRF2ΔBΔM resulted from telomere dysfunction. Collectively, the data indicate that upregulation of p16 is part of the normal response to telomere dysfunction and that this pathway is likely to contribute to telomere-directed senescence. Although the strongest response to telomere dysfunction is observed in primary cells with intact p53 and p16/RB cells, cells with deficiencies in either the p53 or the p16/RB pathway retain the ability to execute a growth arrest when telomere protection is lost. Thus, human cells have two independent pathways that can respond to telomere status and induce senescence when telomere damage is extensive.
Mouse telomere damage signaling: p53 alone
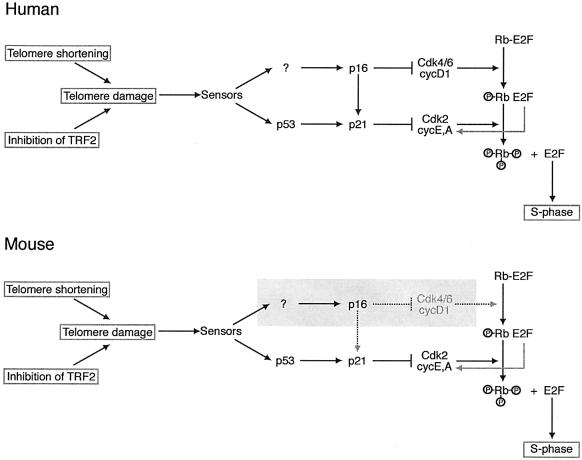
In striking contrast to p53-deficient human cells, mouse cells lacking p53 did not arrest when Trf2 was inhibited. Such p53–/– mouse cells continue to proliferate and have been maintained for several weeks without loss of TRF2ΔBΔM expression. Due to the lack of telomere protection, p53–/– cells lacking normal Trf2 function have extensive genome instability and accumulate end-to-end chromosome fusions (A.Smogorzewska, J.Karlseder, A.Jauch and T.de Lange, submitted). The importance of p53 status in the response to telomere dysfunction was confirmed with p19Arf–/– and p21–/– cells, which both have an impaired p53 pathway. Since each of these MEFs retained a wild-type p16/RB pathway, this pathway alone is not sufficient to respond to telomere damage. The activity of p16 also did not appear to be required for the arrest since p16–/– cells responded to Trf2 inhibition similarly to wild-type cells. Thus, these data argue that the response to telomere dysfunction in mouse cells is primarily dependent on the activation of p53 (Figure 5).
Fig. 5. Schematic illustrating the difference in the telomere damage signaling pathways in human and mouse cells. Telomere damage created by telomere shortening or removal of TRF2 is proposed to activate the same signaling pathway. The sensor(s) upstream of p16 and p53 have not been identified. In human cells, cell cycle arrest in response to telomere damage can be transduced through either the p53 or the p16/Rb pathways. In mouse cells, the telomere damage signal is transduced only through the p53 pathway. In the absence of p53, the p16/Rb pathway alone is insufficient to induce cell cycle arrest in response to telomere damage (shading). The arrow from p16 to p21 illustrates the ability of p16 to displace p21 from the CDK4/6 complex, resulting in a release of a latent pool of p21 that can inhibit cycE/A-dependent CDK2.
This conclusion is consistent with the finding that p53 deficiency can rescue many of the phenotypes of late generation mTerc–/– mice (Chin et al., 1999). While mTerc–/– mice fail to thrive and become infertile in the sixth generation, mTerc–/–p53–/– mice can be carried into the eighth generation. The fact that even the double KO lineage eventually dies out is most likely due to the extensive genome instability in this setting. The parallel between the observations on mTerc–/– mice experiencing telomere shortening and MEFs experiencing TRF2ΔBΔM-mediated telomere dysfunction argue that both systems employ the same basic signaling pathways. Therefore, we conclude that human and mouse fibroblasts are crucially different in their telomere damage response, with human cells using signals carried by p53 and p16/RB, and mouse cells using p53 alone.
Implications for mouse models of the telomere tumor suppressor pathway
The different telomere damage response pathways in human and mouse cells need to be taken into account when evaluating the effects of telomere shortening on tumorigenesis in the mouse. In order to measure the ability of telomere shortening to impede tumor outgrowth, it will be important to choose test systems in which the developing tumor cells still have the ability to respond to telomere dysfunction. In the mouse, this will necessitate a setting with an (initially) intact p53 pathway. If this prerequisite is not met, telomere shortening will not limit the proliferative potential of incipient tumor cells.
Indeed, when examined in the p53–/– tumor model, mice with mTerc–/– status and severely shortened telomeres develop tumors at rates at least as high as littermates with functional telomeres (Artandi et al., 2000). Furthermore, within the context of the Ink4a/Arf KO model, the short telomeres of G6 mTerc–/– mice only created a weak tumor suppressor effect (Greenberg et al., 1999). Since the Ink4a/Arf KO mice lack p19Arf, they have a defective p53 pathway, allowing the cells to continue to grow with telomere dysfunction and masking a potential tumor suppressor effect of shortening telomeres. These results with the p53 null mouse and the Ink4a/Arf model closely match our findings with Trf2-deficient MEFs. While p53–/– status was associated with a complete lack of response to Trf2 inhibition, p19Arf deficiency still allowed a minor response to telomere dysfunction in vitro. The central importance of the p53 pathway for the murine response to telomere damage also explains why critically shortened telomeres did not impair the immortalization of late generation mTerc–/– MEFs in a 3T3 protocol (Blasco et al., 1997), which involves the loss of either p19Arf or p53 (Harvey and Levine, 1991; Rittling and Denhardt, 1992; Kamijo et al., 1997). Furthermore, a test for in vitro transformation of mTerc–/– cells involved SV40 large T, an oncoprotein that inactivates p53 (Blasco et al., 1997). The result suggested that loss of telomere function did not interfere with tumorigenic transformation in vitro. However, it is now clear that the design of these assays was such that the effect of telomere shortening could not have been assessed. Thus, both the in vivo tumorigenesis assays as well as the in vitro assays for immortalization and transformation of mouse cells with critically shortened telomeres involved cells with an impaired telomere damage signaling pathway.
According to these considerations, it is pertinent to re-examine the tumor suppressor activity of critically short telomeres in mouse tumor models in which the p53 pathway is intact at the time tumorigenesis is initiated. Indeed, short telomeres appear to suppress the formation of DMBA/TPA-induced [7,12-dimethylbenz[a]anthracene (DMBA) and 12-O-tetradecanoylphorbol-13-acetate (TPA)] skin tumors (Gonzalez-Suarez et al., 2000) and intestinal carcinomas in the ApcMin mouse (Rudolph et al., 2001). Further studies of this type may reveal the true potential of telomere shortening as a tumor suppressor system and also illuminate the potential beneficial effects of telomerase inhibition in the oncology clinic.
Materials and methods
Cell culture
WI-38, HS68 and Phoenix cells were obtained from the ATCC. IMR90 cells expressing an ecotropic receptor (Figure 1C–E) were a gift from S.Lowe. Cells were grown in DMEM with 100 U of penicillin and 0.1 mg of streptomycin per milliliter, 2 mM l-glutamine, 0.1 mM non-essential amino acids and 15% FBS (or 10% FBS for Phoenix cells). One passage represents a 1:3 split. hTERT-BJ (Bodnar et al., 1998) (Clontech) were grown as suggested by the manufacturer. Primary p53–/– (Jacks et al., 1994), p21–/– (Deng et al., 1995), p19Arf–/– (Kamijo et al., 1997) and p16–/– (Sharpless et al., 2001) with the respective control (+/+) MEFs were grown in DMEM/15% FBS and supplements as above. All MEFs were at passage 5 or younger.
Retroviral gene delivery
pBabepuro, pLPCpuro and pWZLhygro (S.Lowe) were used to clone N-terminally myc-tagged (Figure 1C–E) or untagged (all other experiments) TRF2. pBabeSV40Tag was a gift from G.Hannon. PLXSNeoHPV16 E6, E7 and E6E7 were gifts from D.Galloway (Halbert et al., 1991). pWZLp53175H was provided by S.Lowe (Serrano et al., 1997). Retroviral gene delivery techniques have been described previously (Karlseder et al., 2002).
Chromosome analysis, centrosome analysis and genomic blotting
Chromosome analysis was performed as described previously (van Steensel et al., 1998). Centrosomes were detected on MeOH-fixed coverslips [10 min at –20°C, washed in PBS and processed for indirect immunofluorescence (IF) as described for TRF2, see below] with an anti γ-tubulin antibody (Sigma; GTU88). DNA was counterstained with DAPI. Genomic DNA was isolated as described elsewhere (van Steensel et al., 1998), digested with HinfI and RsaI, and fractionated on a 0.7% agarose gel. Blotting and hybridization with a telomeric probe was carried out as described previously (de Lange et al., 1990).
Growth curves, FACS analysis, BrdU labeling and SA-β-gal assay
After 3 days of selection in puromycin, only infected cells (infection rates ∼90% for mouse cells and 70–80% for human cells) remained in the cultures. On the fourth day after selection, cells were plated in duplicate at 3 × 104 cells per well in a 6-well plate and counted on subsequent days using a Coulter counter. Fluorescence-activated cell sorting (FACS) analysis was performed as described previously (Karlseder et al., 1999). For BrdU labeling, 1 day prior to labeling (days 4, 9 and 14), cells were plated on autoclaved coverslips at 1 × 105 cells per well in a 6-well plate. BrdU (10 µM; Roche) was added 24 h later and after 1 h coverslips were washed with PBS, fixed in 70% EtOH in 50 mM glycine–HCl buffer pH 2.0 for 45 min at room temperature, washed twice in PBS, denatured for 10 min in cold 4 N HCl, washed three times for 5 min in PBS and blocked in PBG [0.2% (w/v) cold water fish gelatin (Sigma), 0.5% (w/v) BSA (Sigma) in PBS]. Following a 2 h incubation with anti-TRF2 Ab 647, coverslips were washed in PBG (three times for 5 min) and incubated for 45 min with TRITC-conjugated donkey anti-rabbit antibody (Jackson) and FITC-conjugated BrdU antibody (Becton Dickinson and Co.). DNA was counterstained with DAPI and coverslips were mounted in 90% glycerol/10% PBS containing p-phenylene-diamine (1 mg/ml; Sigma). Using the dual labeling for BrdU and TRF2 overexpression (or overexpression of the dominant-negative allele), BrdU-positive cells that also expressed TRF2 or TRF2ΔBΔM were counted in triplicate on ∼700–1000 cells in total. Vector controls and cells expressing TRF2 alleles were always scored side by side. The fraction of TRF2 or TRF2ΔBΔM cells that also contained BrdU was determined and normalized to the vector control samples using the formula: (fraction of BrdU-positive cells in TRF2- or TRF2ΔBΔM -expressing cells/fraction of BrdU-positive cells in control vector cells) × 100%. The standard deviation (SD) in these measurements was always <5%. The SA-β-gal assay (Dimri et al., 1995) was performed 2 days after seeding 105 cells per well in a 6-well dish on day 14 (human cells) or on day 10 (mouse cells) (van Steensel et al., 1998).
Immunoblotting
For whole-cell lysates (WCL), cells were trypsinized, washed in cold PBS, counted and resuspended at 1 × 104 cells/µl in Laemmli buffer and the DNA was sheared through a 25 5/8 gauge needle. WCL (10–15 µl) were fractionated on SDS–PAGE (10% for TRF2, p53, SV40 Tag, cyclin A and γ-tubulin; 6% for pRb and HDM2; 12% for p16, p21 and E7) and transferred onto nitrocellulose or PVDF (for E7) membranes. Blocking (30 min at room temperature) and incubation with primary and secondary antibodies (overnight at 4°C) were performed in 5% milk/0.1% Tween-20 in PBS. Antibodies were as follows. TRF2: 647 (Zhu et al., 2000) or 05-521 (Upstate Biotechnology); human p53: 1801; mouse p53: PAB240 (A.Levine); HDM2: 3F3 (A.Levine); p21: F-5 (Santa Cruz Biotechnology); Rb: 14001A (PharMingen); human p16: NCLp16 (Novocastra Laboratories); mouse p16: M-156 (Santa Cruz Biotechnology); cyclin A: sc-239 (Santa Cruz Biotechnology); γ-tubulin: GTU88 (Sigma); SV40 Tag: PAB416 (A.Levine); and E7: Clone 8C9 (Zymed). After incubations with secondary antibodies (Amersham), all blots were developed using the Amersham ECL kit.
Acknowledgments
Acknowledgements
We are grateful to Scott Lowe for advice, discussion and gifts of plasmids and protocols for the retroviral expression system. D.Galloway, G.Hannon, T.Jacks, R.DePinho, M.Roussel, C.Sherr and A.Levine are thanked for reagents. J.Karlseder is thanked for helpful advice. F.Pearl is thanked for his contribution to this project. The members of the de Lange laboratory are thanked for advice and comments on this manuscript. A.S. is supported by NIH MSTP grant to the Cornell/RU/MSKCC Tri-Institutional MD/PhD program and by a training grant to RU. This work was supported by a grant from the NIH (NIA, AG16643) and the Ellison Medical Foundation.
References
- Artandi S.E., Chang,S., Lee,S.L., Alson,S., Gottlieb,G.J., Chin,L. and DePinho,R.A. (2000) Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature, 406, 641–645. [DOI] [PubMed] [Google Scholar]
- Benn P.A. (1976) Specific chromosome aberrations in senescent fibroblast cell lines derived from human embryos. Am. J. Hum. Genet., 28, 465–473. [PMC free article] [PubMed] [Google Scholar]
- Blasco M.A., Lee,H.W., Hande,M.P., Samper,E., Lansdorp,P.M., DePinho,R.A. and Greider,C.W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- Bodnar A.G. et al. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska,A., Chong,L. and de Lange,T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- Chin L., Artandi,S.E., Shen,Q., Tam,A., Lee,S.L., Gottlieb,G.J., Greider,C.W. and DePinho,R.A. (1999) p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell, 97, 527–538. [DOI] [PubMed] [Google Scholar]
- Counter C.M., Avilion,A.A., LeFeuvre,C.E., Stewart,N.G., Greider,C.W., Harley,C.B. and Bacchetti,S. (1992) Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J., 11, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm K. et al. (2001) A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J., 20, 6958–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. (2002) Protection of mammalian telomeres. Oncogene, 21, 532–540. [DOI] [PubMed] [Google Scholar]
- de Lange T., Shiue,L., Myers,R.M., Cox,D.R., Naylor,S.L., Killery,A.M. and Varmus,H.E. (1990) Structure and variability of human chromosome ends. Mol. Cell. Biol., 10, 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Zhang,P., Harper,J.W., Elledge,S.J. and Leder,P. (1995) Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell, 82, 675–684. [DOI] [PubMed] [Google Scholar]
- Di Leonardo A., Linke,S.P., Clarkin,K. and Wahl,G.M. (1994) DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev., 8, 2540–2551. [DOI] [PubMed] [Google Scholar]
- Dickson M.A., Hahn,W.C., Ino,Y., Ronfard,V., Wu,J.Y., Weinberg,R.A., Louis,D.N., Li,F.P. and Rheinwald,J.G. (2000) Human keratinocytes that express hTERT and also bypass a p16INK4a-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol., 20, 1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P. et al. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA, 92, 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M.S., Riha,K., Gao,F., Ren,S., McKnight,T.D. and Shippen,D.E. (1999) Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA, 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E., Samper,E., Flores,J.M. and Blasco,M.A. (2000) Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat. Genet., 26, 114–117. [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., Chin,L., Femino,A., Lee,K.H., Gottlieb,G.J., Singer,R.H., Greider,C.W. and DePinho,R.A. (1999) Short dysfunctional telomeres impair tumorigenesis in the INK4aΔ2/3 cancer-prone mouse. Cell, 97, 515–525. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- Hahn W.C. et al. (1999) Inhibition of telomerase limits the growth of human cancer cells. Nat. Med., 5, 1164–1170. [DOI] [PubMed] [Google Scholar]
- Halbert C.L., Demers,G.W. and Galloway,D.A. (1991) The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol., 65, 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- Harvey D.M. and Levine,A.J. (1991) p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev., 5, 2375–2385. [DOI] [PubMed] [Google Scholar]
- Jacks T., Remington,L., Williams,B.O., Schmitt,E.M., Halachmi,S., Bronson,R.T. and Weinberg,R.A. (1994) Tumor spectrum analysis in p53-mutant mice. Curr. Biol., 4, 1–7. [DOI] [PubMed] [Google Scholar]
- Kamijo T., Zindy,F., Roussel,M.F., Quelle,D.E., Downing,J.R., Ashmun,R.A., Grosveld,G. and Sherr,C.J. (1997) Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell, 91, 649–659. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Broccoli,D., Dai,Y., Hardy,S. and de Lange,T. (1999) p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science, 283, 1321–1325. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Smogorzewska,A. and de Lange,T. (2002) Senescence induced by altered telomere state, not telomere loss. Science, 295, 2446–2449. [DOI] [PubMed] [Google Scholar]
- Khan S.H., Moritsugu,J. and Wahl,G.M. (2000) Differential requirement for p19ARF in the p53-dependent arrest induced by DNA damage, microtubule disruption, and ribonucleotide depletion. Proc. Natl Acad. Sci. USA, 97, 3266–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D. and Cooke,H.J. (1990) Hypervariable ultra-long telomeres in mice. Nature, 347, 400–402. [DOI] [PubMed] [Google Scholar]
- Kiyono T., Foster,S.A., Koop,J.I., McDougall,J.K., Galloway,D.A. and Klingelhutz,A.J. (1998) Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature, 396, 84–88. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P., Quon,K.C., Mooi,W.J., Loonstra,A. and Berns,A. (2001) Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature, 413, 83–86. [DOI] [PubMed] [Google Scholar]
- Levis R.W. (1989) Viable deletions of a telomere from a Drosophila chromosome. Cell, 58, 791–801. [DOI] [PubMed] [Google Scholar]
- Lundblad V. and Szostak,J.W. (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell, 57, 633–643. [DOI] [PubMed] [Google Scholar]
- Munro J., Steeghs,K., Morrison,V., Ireland,H. and Parkinson,E.K. (2001) Human fibroblast replicative senescence can occur in the absence of extensive cell division and short telomeres. Oncogene, 20, 3541–3552. [DOI] [PubMed] [Google Scholar]
- Pomerantz J. et al. (1998) The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell, 92, 713–723. [DOI] [PubMed] [Google Scholar]
- Prowse K.R. and Greider,C.W. (1995) Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl Acad. Sci. USA, 92, 4818–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez R.D., Morales,C.P., Herbert,B.S., Rohde,J.M., Passons,C., Shay,J.W. and Wright,W.E. (2001) Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev., 15, 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittling S.R. and Denhardt,D.T. (1992) p53 mutations in spontaneously immortalized 3T12 but not 3T3 mouse embryo cells. Oncogene, 7, 935–942. [PubMed] [Google Scholar]
- Rudolph K.L., Chang,S., Lee,H.W., Blasco,M., Gottlieb,G.J., Greider,C. and DePinho,R.A. (1999) Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell, 96, 701–712. [DOI] [PubMed] [Google Scholar]
- Rudolph K.L., Millard,M., Bosenberg,M.W. and DePinho,R.A. (2001) Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet., 28, 155–159. [DOI] [PubMed] [Google Scholar]
- Saksela E. and Moorhead,P.S. (1963) Aneuploidy in the degenerative phase of serial cultivation of human cell strains. Proc. Natl Acad. Sci. USA, 50, 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lee,H., Chin,L., Cordon-Cardo,C., Beach,D. and DePinho,R.A. (1996) Role of the INK4a locus in tumor suppression and cell mortality. Cell, 85, 27–37. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin,A.W., McCurrach,M.E., Beach,D. and Lowe,S.W. (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell, 88, 593–602. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Bardeesy,N., Lee,K.H., Carrasco,D., Castrillon,D.H., Aguirre,A.J., Wu,E.A., Horner,J.W. and DePinho,R.A. (2001) Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature, 413, 86–91. [DOI] [PubMed] [Google Scholar]
- Shay J.W. and Bacchetti,S. (1997) A survey of telomerase activity in human cancer. Eur. J. Cancer, 33, 787–791. [DOI] [PubMed] [Google Scholar]
- Shay J.W. and Wright,W.E. (1989) Quantitation of the frequency of immortalization of normal human diploid fibroblasts by SV40 large T-antigen. Exp. Cell Res., 184, 109–118. [DOI] [PubMed] [Google Scholar]
- Shay J.W., Pereira-Smith,O.M. and Wright,W.E. (1991) A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res., 196, 33–39. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. (2001) The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell. Biol., 2, 731–737. [DOI] [PubMed] [Google Scholar]
- Stansel R.M., de Lange,T. and Griffith,J.D. (2001) T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J., 20, 5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson K.V. and Holliday,R. (1975) Chromosome changes during the in vitro ageing of MRC-5 human fibroblasts. Exp. Cell Res., 96, 1–6. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska,A. and de Lange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Wolman S.R., Hirschhorn,K. and Todaro,G.J. (1964) Early chromosomal changes in SV40-infected human fibroblast culture. Cytogenetics, 3, 45–61. [DOI] [PubMed] [Google Scholar]
- Zhang X., Mar,V., Zhou,W., Harrington,L. and Robinson,M.O. (1999) Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev., 13, 2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiong,Y. and Yarbrough,W.G. (1998) ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell, 92, 725–734. [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Kuster,B., Mann,M., Petrini,J.H. and Lange,T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet., 25, 347–352. [DOI] [PubMed] [Google Scholar]