Abstract
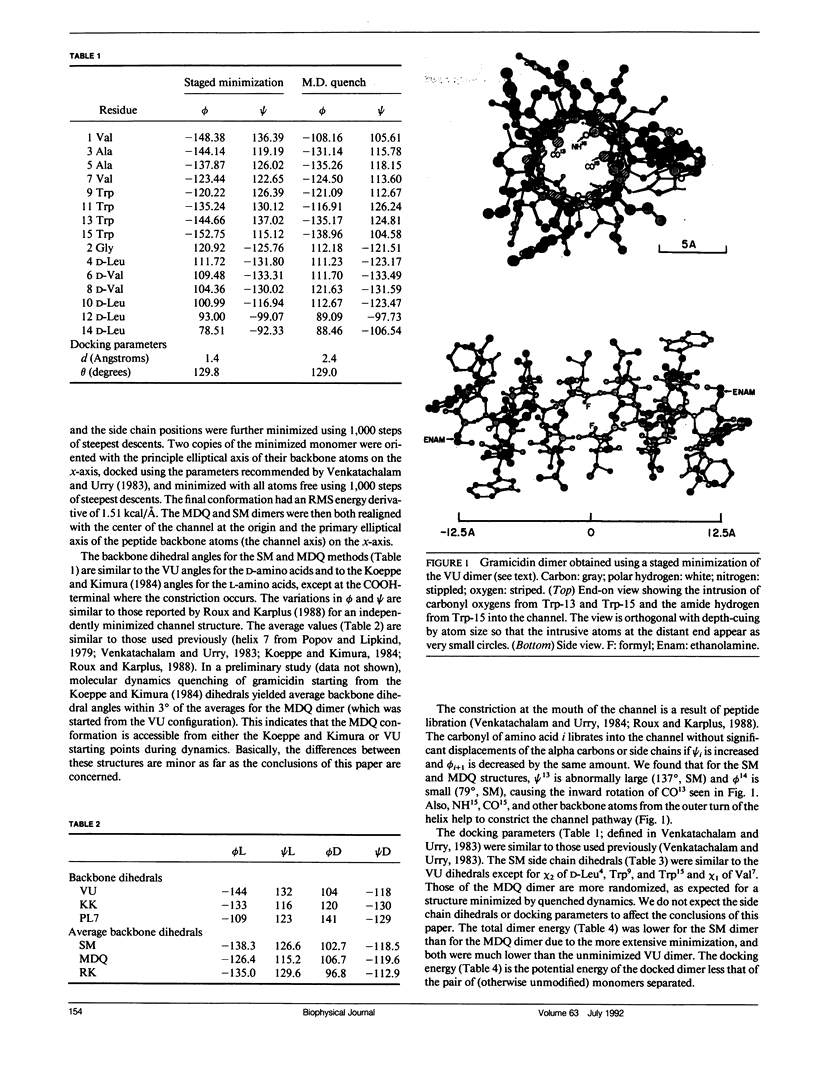
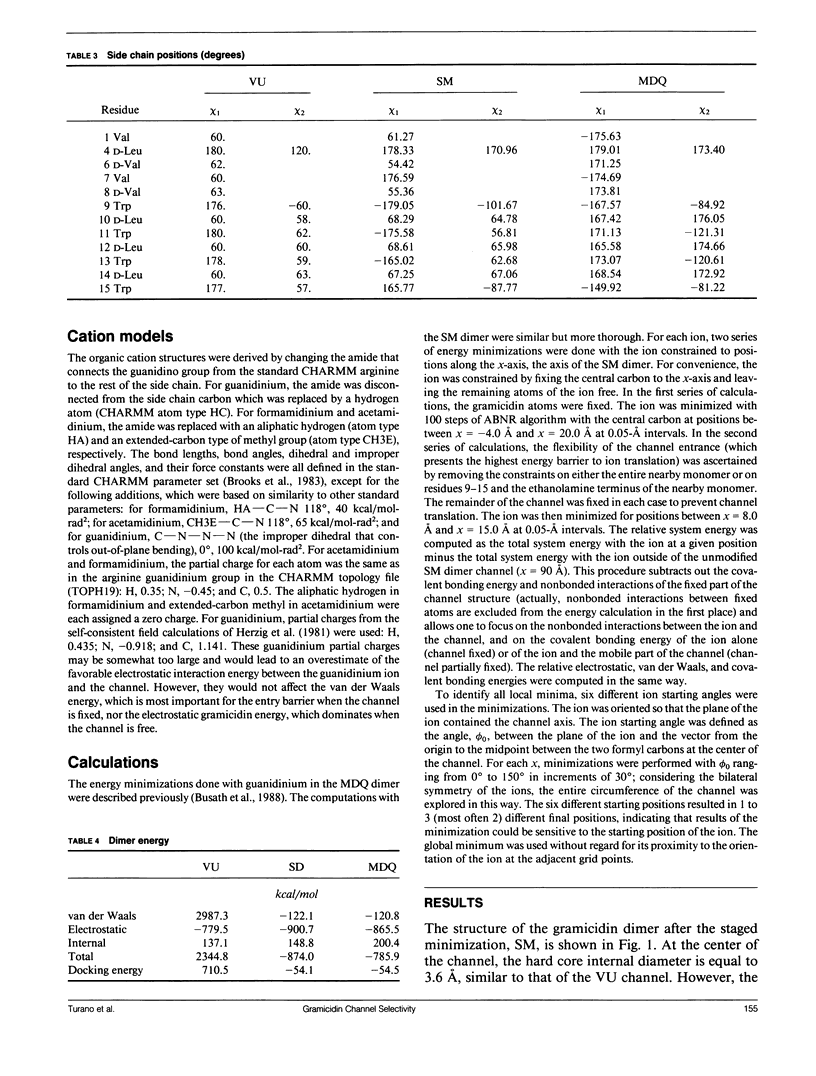
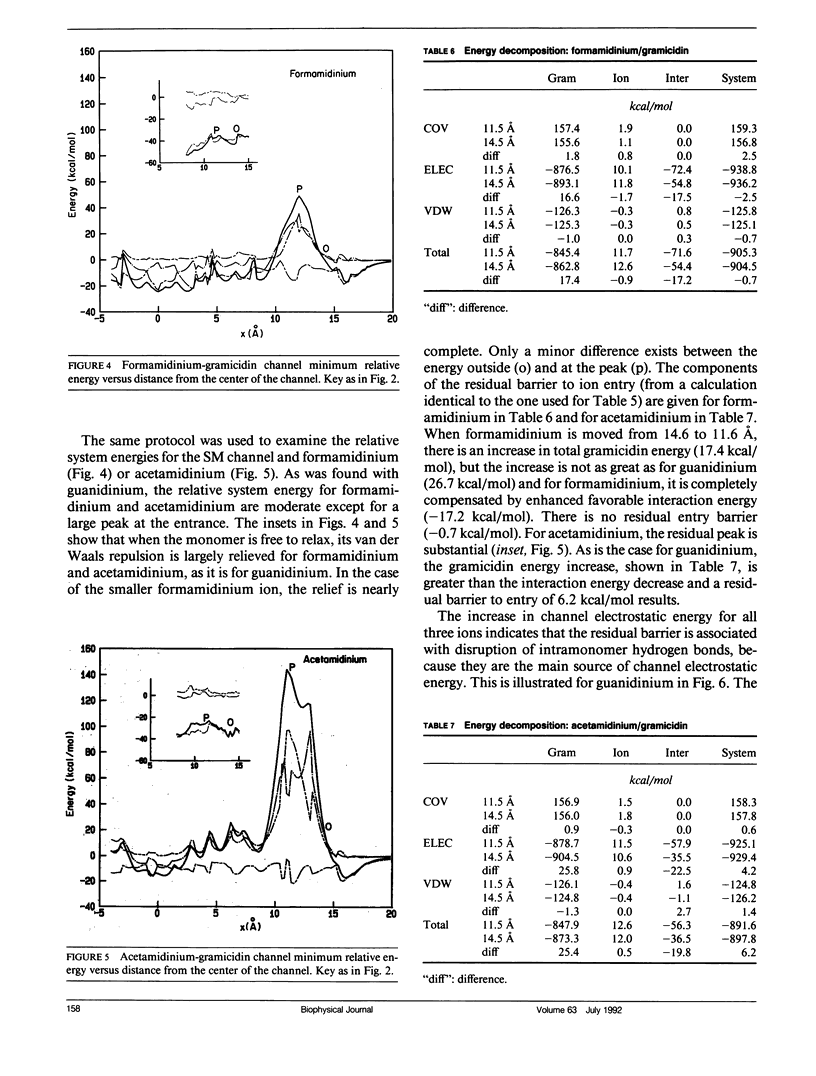
Empirical energy function calculations were used to evaluate the effects of minimization on the structure of a gramicidin A channel and to analyze the energies of interaction between three cations (guanidinium, acetamidinium, formamidinium) and the channel as a function of position along the channel axis. The energy minimized model of the gramicidin channel, which was based on the results of Venkatachalam and Urry (1983), has a constriction at the channel entrance. If the channel is not allowed to relax in the presence of the ions (rigid model), there is a large potential energy barrier for all three cations. The barrier varies with cation size and is due to high van der Waals and ion deformation energies. If the channel is minimized in the presence of the ions, the potential energy barrier to formamidinium entry is almost eliminated, but a residual barrier remains for guanidinium and acetamidinium. The residual barrier is primarily due, not to the expansion of the helix, but, to the disruption of hydrogen bonds between the terminal ethanoloamine and the next turn of the helix which occurs when the carbonyls of the outer turn of the helix librate inward toward the ion as it enters the channel. The residual potential energy barriers could be a possible explanation for the measured selectivity of gramicidin for formamidinium over guanidinium. The results of this full-atomic model address the applicability of the size-exclusion concept for the selectivity of the gramicidin channel.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen O. S. Gramicidin channels. Annu Rev Physiol. 1984;46:531–548. doi: 10.1146/annurev.ph.46.030184.002531. [DOI] [PubMed] [Google Scholar]
- Arseniev A. S., Barsukov I. L., Bystrov V. F., Lomize A. L., Ovchinnikov YuA 1H-NMR study of gramicidin A transmembrane ion channel. Head-to-head right-handed, single-stranded helices. FEBS Lett. 1985 Jul 8;186(2):168–174. doi: 10.1016/0014-5793(85)80702-x. [DOI] [PubMed] [Google Scholar]
- Bezanilla F., Armstrong C. M. Negative conductance caused by entry of sodium and cesium ions into the potassium channels of squid axons. J Gen Physiol. 1972 Nov;60(5):588–608. doi: 10.1085/jgp.60.5.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A., Karplus M. Dynamics of ligand binding to heme proteins. J Mol Biol. 1979 Aug 15;132(3):343–368. doi: 10.1016/0022-2836(79)90265-1. [DOI] [PubMed] [Google Scholar]
- Chiu S. W., Jakobsson E., Subramaniam S., McCammon J. A. Time-correlation analysis of simulated water motion in flexible and rigid gramicidin channels. Biophys J. 1991 Jul;60(1):273–285. doi: 10.1016/S0006-3495(91)82049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K. E., Gates P. Y., Eisenberg R. S. Diffusion theory and discrete rate constants in ion permeation. J Membr Biol. 1988 Dec;106(2):95–105. doi: 10.1007/BF01871391. [DOI] [PubMed] [Google Scholar]
- Cornell B. A., Separovic F., Baldassi A. J., Smith R. Conformation and orientation of gramicidin a in oriented phospholipid bilayers measured by solid state carbon-13 NMR. Biophys J. 1988 Jan;53(1):67–76. doi: 10.1016/S0006-3495(88)83066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell B. Gramicidin A--phospholipid model systems. J Bioenerg Biomembr. 1987 Dec;19(6):655–676. doi: 10.1007/BF00762301. [DOI] [PubMed] [Google Scholar]
- Etchebest C., Ranganathan S., Pullman A. The gramicidin A channel: comparison of the energy profiles of Na+, K+ and Cs+. Influence of the flexibility of the ethanolamine end chain on the profiles. FEBS Lett. 1984 Aug 6;173(2):301–306. doi: 10.1016/0014-5793(84)80795-4. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Wolynes P. G. Rate theories and puzzles of hemeprotein kinetics. Science. 1985 Jul 26;229(4711):337–345. doi: 10.1126/science.4012322. [DOI] [PubMed] [Google Scholar]
- Hemsley G., Busath D. Small iminium ions block gramicidin channels in lipid bilayers. Biophys J. 1991 Apr;59(4):901–907. doi: 10.1016/S0006-3495(91)82303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. The permeability of the sodium channel to organic cations in myelinated nerve. J Gen Physiol. 1971 Dec;58(6):599–619. doi: 10.1085/jgp.58.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik V. M., Krimm S. Vibrational analysis of the structure of gramicidin A. I. Normal mode analysis. Biophys J. 1986 Jun;49(6):1131–1145. doi: 10.1016/S0006-3495(86)83742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik V. M., Krimm S. Vibrational analysis of the structure of gramicidin A. II. Vibrational spectra. Biophys J. 1986 Jun;49(6):1147–1154. doi: 10.1016/S0006-3495(86)83743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson L. K., Cross T. A. Gramicidin cation channel: an experimental determination of the right-handed helix sense and verification of beta-type hydrogen bonding. Biochemistry. 1989 Nov 28;28(24):9379–9385. doi: 10.1021/bi00450a019. [DOI] [PubMed] [Google Scholar]
- Pullman A. Energy profiles in the gramicidin A channel. Q Rev Biophys. 1987 Nov;20(3-4):173–200. doi: 10.1017/s0033583500004170. [DOI] [PubMed] [Google Scholar]
- Roux B., Karplus M. The normal modes of the gramicidin-A dimer channel. Biophys J. 1988 Mar;53(3):297–309. doi: 10.1016/S0006-3495(88)83107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudelle Y., Daumas P., Heitz F., Etchebest C., Pullman A. Experimental and theoretical study of gramicidin P, an analog of gramicidin A with a methylamine C-terminal. FEBS Lett. 1987 May 25;216(1):11–16. doi: 10.1016/0014-5793(87)80747-0. [DOI] [PubMed] [Google Scholar]
- Urry D. W. The gramicidin A transmembrane channel: a proposed pi(L,D) helix. Proc Natl Acad Sci U S A. 1971 Mar;68(3):672–676. doi: 10.1073/pnas.68.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urry D. W., Walker J. T., Trapane T. L. Ion interactions in (1-13C)D-Val8 and D-Leu14 analogs of gramicidin A, the helix sense of the channel and location of ion binding sites. J Membr Biol. 1982;69(3):225–231. doi: 10.1007/BF01870401. [DOI] [PubMed] [Google Scholar]



