Abstract
The particulate hepatitis core protein (HBcAg) represents an efficient carrier platform with many of the characteristics uniquely required for the delivery of weak immunogens to the immune system. Although the HBcAg is highly immunogenic, the existing HBcAg-based platform technology has a number of theoretical and practical limitations, most notably the “preexisting immunity” and “assembly” problems. To address the assembly problem, we have developed the core protein from the woodchuck hepadnavirus (WHcAg) as a new particulate carrier platform system. WHcAg appears to tolerate insertions of foreign epitopes at a greater number of positions than HBcAg. For example, both within the external loop region and outside the loop region a total of 17 insertion sites were identified on WHcAg. Importantly, the identification of an expanded number of insertion sites was dependent on additional modifications to the C terminus that appear to stabilize the various internal insertions. Indeed, 21 separate C-terminal modifications have been generated that can be used in combination with the 17 insertion sites to ensure efficient hybrid WHcAg particle assembly. This combinatorial technology is also dependent on the sequence of the heterologous insert. Therefore, the three variables of insert position, C terminus, and epitope sequence are relevant in the design of hybrid WHcAg particles for vaccine purposes.
The hepatitis B virus (HBV) core protein is a 21-kDa polypeptide, and 240 polypeptides spontaneously assemble into a particulate structure (HBcAg) (27 nm) in the course of virion assembly and during heterologous expression in both prokaryotic and eukaryotic systems. Recent crystallographic studies have elucidated the structure of HBcAg particles to a resolution of 3.3 Å (28). Dimers that align along the long vertical α-helical axis comprise the subunits of the particle. Dimer clustering of subunits produces spikes on the surface of the core shell which consist of radial bundles of four long α-helices. Previous studies have suggested that the immunodominant B-cell epitopes on HBcAg are localized around amino acids (aa) 76 to 82 (21, 24), which the structural studies showed formed an external loop that connected adjacent helices. Recent cryoelectron microscopy studies defined several B-cell epitopes localized within the external loop region as well as at least one B-cell epitope outside the loop region (1).
The immunogenicity of heterologous insertions in the loop region of HBcAg suggested that the inherent immunogenicity of the native HBcAg B-cell epitopes could be successfully “transferred” by placement of heterologous epitopes in the same position as the native dominant epitopes (i.e., at the tip of the spikes). It appears that the spacing of the spikes on the HBcAg shell is optimal for binding and cross-linking the naïve B-cell membrane immunoglobulin G (Ig) receptor (11, 14). Therefore, B cells appear to be the primary antigen-presenting cell (APC) for HBcAg (10, 14). A number of pathogen-specific B-cell epitopes (i.e., epitopes for HBV, human immunodeficiency virus type 1 [HIV-1], foot-and-mouth disease virus, human rhinovirus type 2, bovine leukemia virus, feline leukemia virus, HCV, Plasmodium berghei, P. yoelii, P. falciparum, murine cytomegalovirus, poliovirus type 1, and simian immunodeficiency virus) have been inserted by recombinant methods into HBcAg as a method to increase immunogenicity (16, 18). These studies, conducted in a number of independent laboratories, have yielded significant success, including complete protection against foot-and-mouth disease virus (6), P. berghei (26), and P. yoelii (25); these latter three studies prove that neutralizing B-cell epitopes presented in the context of HBcAg can elicit protective immunity.
Although HBcAg is a highly immunogenic particulate antigen, the existing HBcAg-based platform technology is plagued with a number of biochemical and immunological problems that may limit its full potential as a vaccine carrier for the human population. The two main limitations of the current HBcAg platform technology can best be described as the “preexisting immunity” problem and the “assembly” problem. Because HBcAg is derived from a human pathogen, preexisting anti-HBc antibodies are present in individuals previously exposed to HBV infection which are likely to immune complex with an HBcAg-based vaccine and may adversely affect immunogenicity. Further, the anti-HBc antibodies elicited by an HBcAg-based vaccine will compromise the usefulness of the anti-HBc assay currently employed as a diagnostic for current or recent HBV infection. Most importantly, T-cell immune tolerance towards HBcAg is present in individuals chronically infected with HBV (400 million globally). Because the rodent hepadnaviruses are not human pathogens, the “preexisting immunity” problems may be overcome by using, for example, the woodchuck hepatitis virus core antigen (WHcAg) or the ground squirrel hepatitis core antigen (GScAg) as a vaccine platform. In fact, recent studies have demonstrated that the rodent core proteins can serve as competent vaccine carrier platforms and, importantly, are not significantly cross-reactive with the HBcAg at the antibody level and only partially cross-reactive at the level of Th cell recognition (2). The lack of significant immune cross-reactivity between the rodent hepatitis core proteins and HBcAg suggests that their use as vaccine platforms will circumvent the “preexisting immunity” problems. Therefore, we have chosen the rodent hepadnavirus core proteins and, more specifically, WHcAg as a candidate vaccine platform to address the second “assembly” problem.
It is commonly acknowledged among investigators who use HBcAg as a vaccine platform that less than 50% of selected foreign sequences can be successfully inserted into HBcAg (8). This high failure rate is believed to be due to destabilization of particle assembly caused by inserting foreign sequences. Many parameters can affect the expression level and/or the correct assembly of hybrid core particles. Recent analysis of biochemical-biophysical properties of several proposed heterologous inserts revealed that parameters such as length, high hydrophobicity, high β-strand index, or large volume may impede the proper assembly-folding of chimeric core particles (9). The “assembly” problem is so severe that several groups working with HBcAg (8) or with other virus-like-particle (VLP) technologies (i.e., the L1 protein of the human papillomavirus) (3) have opted to chemically link the foreign epitopes to the wild-type (wt) VLPs as opposed to inserting the epitopes into the particles by recombinant methods. Chemical linkage of heterologous epitopes to VLPs may compromise a number of the advantages inherent in the recombinant hybrid particle technology. For example, chemical linkage will result in less than the 100% substitution achieved with the recombinant method, chemical linkage is not as reproducible, making manufacture difficult and more expensive, and in our experience, chemical linkage results in inferior immunogenicity compared to recombinant hybrid core particle results. Therefore, we have addressed the “assembly” problem by developing a so-called “combinatorial” technology using WHcAg as a new particulate carrier system. We demonstrate that (i) WHcAg appears to tolerate insertions of foreign epitopes at a greater number of positions (i.e., 17) than HBcAg both inside the external loop region (i.e., aa 76 to 82) and outside the loop region; (ii) the identification of an expanded number of insertion sites depended on developing 21 modifications to the C terminus that stabilize the various internal insertions and can be used in combination with the 17 insertion sites to ensure efficient hybrid WHcAg particle assembly; and (iii) the optimal combination of insertion site and stable C-terminal modification of WHcAg is also dependent on the sequence of the inserted epitope, and the charge of the insert sequence appears to be a very important variable.
MATERIALS AND METHODS
Construction and expression of recombinant hybrid WHcAg particles.
Full-length WHcAg (aa 1 to 188) (NCBI accession no. NC_004107) was expressed from the pUC-WHcAg vector under the control of the Lac operon promoter inserted between the NcoI-BamHI sites for further convenient subcloning. All the epitope sequences used in this study have been designed specifically to encompass unique restriction sites at both the 5′ and 3′ ends.
Creation of insertion sites.
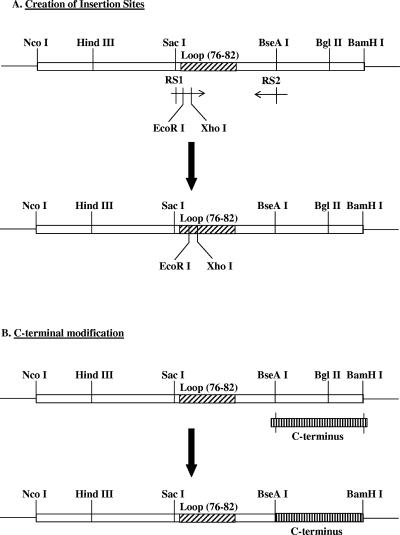
All insertions were accomplished by using the EcoRI-XhoI or SacI sites, with only the position of the insertion differing between constructs (Fig. 1A). Briefly, we have designed specific pairs of primers: one of the primers (forward or reverse) matches the exact sequence of the wt WHcAg gene and encompasses the restriction site RS1 (for example, SacI) and includes mismatches designed to encode two new restriction sites (EcoRI-XhoI); the other primer was homologous to the wt WHcAg gene and spanned the RS2 (for example, BseAI) restriction site. PCRs were performed using the forward-RS1-EcoRI-XhoI and the reverse RS2 as primers and the wt WHcAg plasmid as the template according to the procedure recommended by the manufacturer (La Roche). The resulting PCR product and the WHcAg wt plasmid were then digested by RS1-RS2 and ligated with T4 DNA ligase to create the new WHcAg with the corresponding insertion site. The EcoRI and XhoI restriction sites used in this study created the linkers Gly-Ile-Leu on the N terminus and Leu-Glu on the C terminus of the insert.
FIG. 1.
Schematic representation of the hybrid WHcAg constructs (see Materials and Methods).
Modifications of the C terminus.
We have created a library of C-terminal modifications to eliminate certain motifs (e.g., RNA/DNA binding motifs) and to accommodate the addition of other spacer sequences. The new C termini have been modified by designing oligonucleotides encoding the sequence of interest and flanked by 5′ BseAI and 3′ BamHI sites as a general pattern (Fig. 1B). Both the wt WHcAg gene and the pair of oligonucleotides were digested by the BseAI-BamHI sites and then ligated to replace the corresponding native fragment on the WHcAg wt plasmid. All the core constructs (insert sites and C termini, with or without foreign sequences) have been sequenced in both directions (Retrogen, Inc., San Diego, Calif.). Production and purification of the plasmids (MoBio Laboratories, Inc., San Diego, Calif.) and ligations (La Roche) were performed according to the manufacturers' procedures. Cloning and subcloning have been performed following protocols described by Sambrook et al. (23). The Top10 Escherichia coli strain was purchased from Invitrogen, and the PET11d plasmid and the BL21(DE3) E. coli strain were purchased from Stratagene (San Diego, Calif.). In some instances, the hybrid WHcAg construct was subcloned into another expression vector pET11d into NcoI-BamHI sites, which allowed expression of the corresponding protein under the control of an inducible T7/Lac operon promoter, and then transformed in the BL21(DE3) E. coli strain. Transformations of chemically competent Top10 and BL21(DE3) by heat shock were carried out according to the protocols of the manufacturers (Invitrogen and Stratagene).
Screening and characterization of hybrid WHcAg particles.
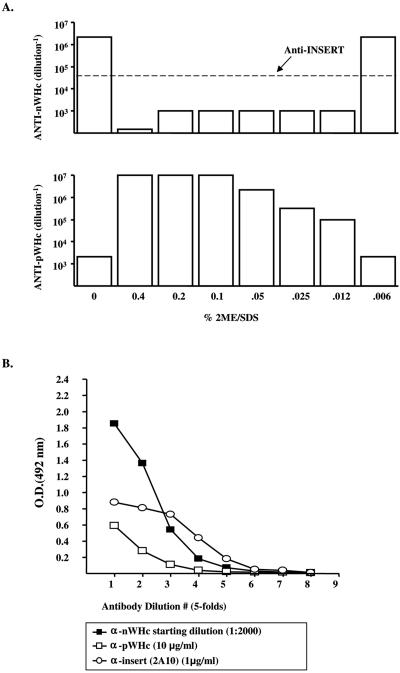
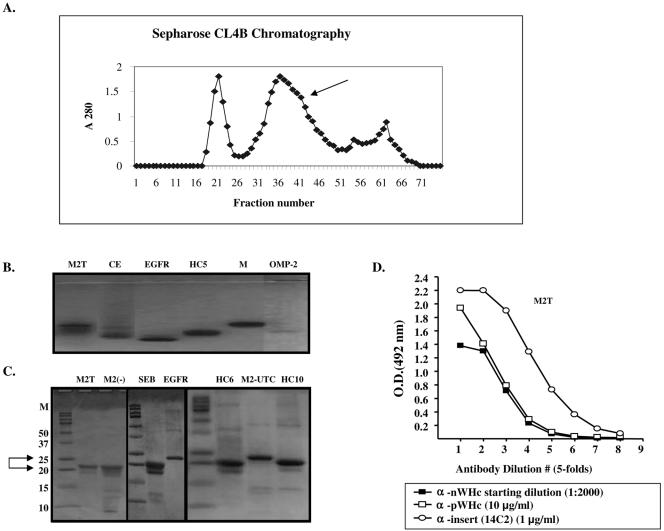
The transformed bacteria were grown overnight at low temperature (28°C) to avoid inclusion body formation before the expression of the protein was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mM for 4 h). The bacteria were lysed in a lysozyme-salt solution containing protease inhibitors. The resulting supernatant was precipitated overnight in the cold with 10 to 50% ammonium sulfate. Bacterial lysates were then screened for levels of protein expression, hybrid WHcAg particle assembly, and epitope antigenicity. Briefly, lysates were sequentially screened in capture enzyme-linked immunosorbent assays (ELISAs) designed either to detect the WHcAg polypeptide by use of a monoclonal antibody (MAb) (2221; Institute for Immunology, Tokyo University, Japan) specific for a peptide epitope (aa 129 to 140) on WHcAg (anti-pWHc) as a marker of protein expression or to detect the WHcAg particle by use of an antibody specific for a conformational epitope on WHcAg (anti-nWHc) as a marker for assembly. This polyclonal BALB/c anti-nWHc antibody only binds particulate WHcAg, and even mild denaturation of WHcAg destroys anti-nWHc antibody binding (Fig. 2A). Insert-specific antibodies (not sensitive to reduction-denaturation) were used to assess the expression level and antigenicity of the various inserts (Fig. 2A). The capture antibody was peptide specific and noncompetitive with the detecting antibodies. The levels of core protein expression and assembly competence detected in the capture ELISAs were assigned relative values (units) based on comparison with anti-pWHc and anti-nWHc antibody binding to wt WHcAg (see Fig. 6): for example, a value of 5 = wt binding (i.e., maximum binding level); 4 = 5× less than wt binding; 3 = 25× less than wt binding; 2 = 125× less than wt binding; 1 = 625× less than wt binding; 0 = no binding. “+,” “±,” and “−” correspond, respectively, to the relative scores of 5 to 3, 2 to 1, and 0. Based on relative expression level, assembly, and insert antigenicity detected in the lysates, optimal hybrid particle gene constructs were selected for further purification (Fig. 2B). The selected proteins were then purified through hydroxyapatite followed by gel filtration chromatography on Sepharose 4B columns as previously described (24). We have observed a 100% correlation between anti-nWHc antibody binding and the ability to purify over 50 hybrid WHcAg particles. A typical chromatograph profile is shown in Fig. 3A, demonstrating that the native or hybrid WHcAg particle is recovered from the second peak (arrow). Following this purification, each newly purified particle is then electrophoresed on a 1% agarose gel and on a 15% Tris-glycine gel under reducing (β-mercaptoethanol) and denaturing (sodium dodecyl sulfate) conditions to confirm the particulate structure (Fig. 3B) and the expected apparent molecular weight of its constitutive monomer (Fig. 3C), respectively. Finally, the purified particles are reanalyzed in the same capture ELISAs used to determine particulate structure and level of protein expression and the antigenicity of the insert in the lysate (Fig. 3D).
FIG. 2.
(A) Capture ELISAs were designed to detect either WHcAg polypeptide as a marker for expression or WHcAg particles as a marker for assembly in E. coli lysates. An MAb specific for a peptidic epitope on WHcAg is used as the detecting antibody to determine relative expression levels (anti-pWHc; bottom panel). A polyclonal antibody that recognizes only assembled particles (anti-nWHc) is used to determine relative assembly competence (top panel). The capture antibody is a noncompeting MAb specific for a peptidic epitope on WHcAg. Purified malaria-WHcAg hybrid particles were treated with the indicated concentrations of a reducing-denaturing buffer and analyzed in anti-pWHc and anti-nWHc capture ELISAs. A malaria M epitope-specific MAb (2A10) was used to detect the malaria repeat epitope, which was not sensitive to reduction-denaturation. (B) A bacterial lysate of a hybrid M-WHcAg particle was tested in capture ELISAs used to detect assembled particles (anti-nWHc) and levels of protein expression (anti-pWHc) and antigenicity (anti-insert; MAb 2A10). Antibody binding at the indicated dilution is expressed as optical density at 492 nm (OD492).
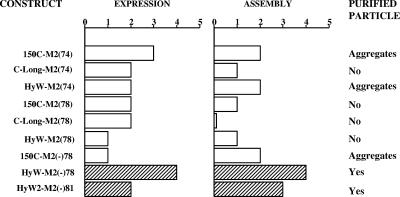
FIG. 6.
A list of M2-WHcAg or M2(−)-WHcAg hybrid constructs expressed in E. coli and analyzed by capture ELISAs for relative expression level and assembly competence. Lysates were sequentially screened with a MAb that preferentially recognizes denatured WHcAg and another antibody that only recognizes assembled WHcAg particles (see Fig. 2) and given scores (see Materials and Methods) relative to antibody binding to wt WHcAg.
FIG. 3.
(A) Typical gel filtration profile obtained after hydroxyapatite followed by Sepharose CL4B chromatography of hybrid WHcAg particles. The first peak represents the void volume, the hybrid WHcAg is recovered from the second peak (arrow), and the third peak corresponds to aggregates of dimers-monomers of hybrid WHcAg. (B) Migration of exemplar hybrid WHcAg particles through a 1% agarose gel. Note that the migration pattern differs depending on the C terminus, the insert position, and the epitope used. If particulate, the hybrid or wt WHcAg particles should migrate as a discrete band in native agarose (1%) gel electrophoresis. (C) Migration pattern through 15% Tris-glycine gel under denaturing (sodium dodecyl sulfate) and reducing (β-mercaptoethanol) conditions of exemplar hybrid WHcAg particles. The expected size of the monomer of each hybrid WHcAg particle corresponds to the band of strongest intensity and is generally between 20 and 25 kDa, depending on the length of the hybrid WHcAg polypeptide. For epitope abbreviations, see Table 1. (D) Hybrid WHcAg particles tested in capture ELISAs to detect assembled particles (anti-nWHc), level of protein (anti-pWHc), and antigenicity level (anti-insert; MAb14C2) after purification as described in Materials and Methods. The results obtained with an influenza A virus M2 hybrid WHcAg particle are shown.
Removal of endotoxins.
Endotoxin was removed from the core preparations by a modification of a phase separation method with Triton X-114 (13). A solution of the protein at a concentration of 5 mg/ml was made a 1% Triton X-114 and incubated at 4°C for 30 min with constant stirring. The solution was then incubated at 37°C for 10 min and then centrifuged at 20,000 × g for 10 min. The protein solution was recovered from above the detergent. This procedure was repeated four times. Finally, the protein was precipitated by lowering the pH to 5, whereas residual detergent remained in solution. The protein was recovered by centrifugation and dissolved in endotoxin-free buffer. Prior to Triton X-114 treatment the core preparations contain approximately 10 to 25 ng of endotoxin/μg HBcAg, and after phase separation with Triton X-114 the endotoxin content ranged from 0.1 ng/μg HBcAg to an undetectable amount, as determined by a QCL-1000 chromogenic Limulus amoebocyte lysate end point assay (Cambrex, East Rutherford, NJ).
Immunization of mice.
Groups of three to five female mice (either bred at the Vaccine Research Institute of San Diego, CA or obtained from Jackson Laboratories, Bar Harbor, ME) of various strains and approximately 6 to 8 weeks old were immunized intraperitoneally. Hybrid WHcAg particles (20 μg, primary; 10 μg, boost) were emulsified in incomplete Freund's adjuvant (IFA). Mice were bled preimmunization and at various times after primary and booster immunizations for anti-insert and anti-WHc antibody determinations. Anti-WHc or anti-insert antibodies were measured in pooled, murine sera by indirect solid-phase ELISA using solid-phase wt WHcAg (50 ng/well) or insert peptide (0.5 μg/well), and goat anti-mouse IgG (or IgG isotype-specific) antibodies were used as the secondary antibody. The data were expressed as the antibody titer representing the highest dilution yielding three times the optical density of the preimmunization sera. Synthetic peptides were synthesized by the simultaneous peptide synthesis method as previously described (22).
RESULTS
Production of a library of insertion sites on WHcAg.
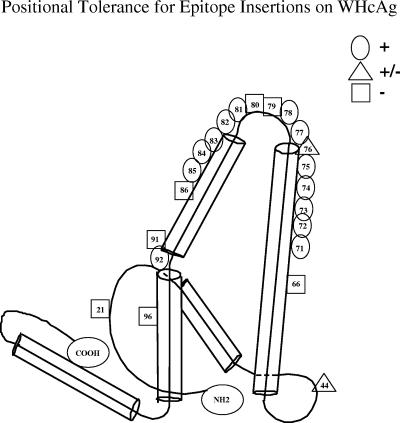
To determine the tolerance of WHcAg for insertion of foreign sequences we systematically inserted a malaria (M) (P. falciparum) circumsporozoite (CS) repeat sequence [NANPNVDP(NANP)3] (15, 29) into a variety of positions on the WHcAg platform by use of recombinant technology. In addition to positions 77, 78, 81, and 82 within the loop region and the NH2 and COOH termini, which are the sites previously used for HBcAg studies (19), we have identified a number of other internal insertion sites inside (position 76) and outside (including positions 44, 71, 72, 73, 74, 75, 83, 84, 85, and 92) the loop region on WHcAg which tolerated insertions of the M epitope used here as an exemplar sequence (Fig. 4). Tolerance for insertion of the M epitope was defined as the ability of the hybrid WHcAg particle to assemble and to detect the inserted epitope on the particle surface using an MAb specific for (NANP)3 (i.e., MAb 2A10). Note that positions 21, 66, 79, 80, 86, 91, and 96 on WHcAg did not tolerate insertions. Therefore, we have improved the core platform technology by producing a relatively large library of 17 competent insertion sites on the WHcAg platform. The capability of WHcAg to tolerate insertions in a variety of positions is not unique to the M epitope; a list of other insert sequences that have been tested is shown in Table 1. However, optimal insertion sites may differ depending on the insert sequence, as discussed below.
FIG. 4.
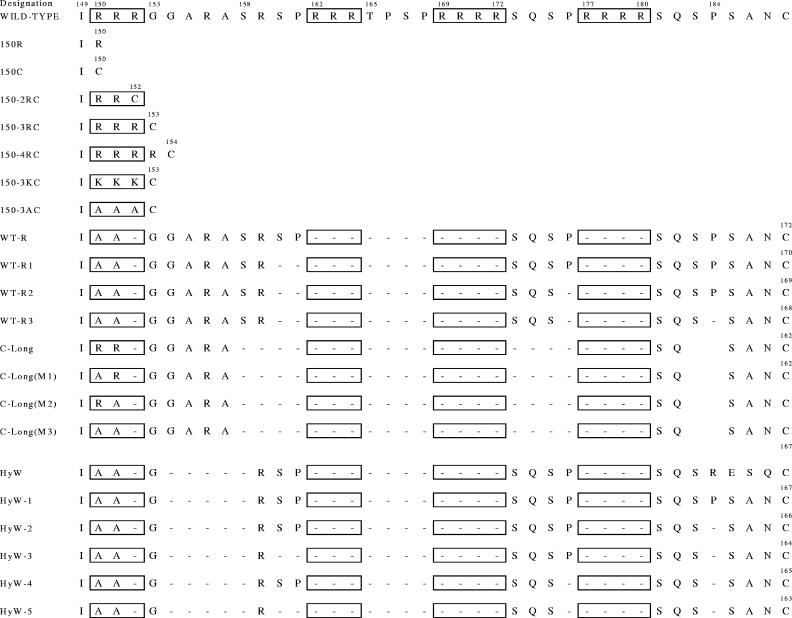
WHcAg accommodates the insertion of foreign epitopes at many positions. Insertion sites for foreign epitopes are indicated. Tolerant (+), intermediate (+/−), and nontolerant (−) insertion sites are indicated. Determinations of tolerance are based on relative scores (see Material and Methods). The C-terminus modifications tested were either 150C or HyW, HyW2, C-Long, 2RC, or 3RC.
TABLE 1.
Listing of model epitopes useda
| Epitope | Sequence | Length (aa) |
|---|---|---|
| AZ1 | DAEFRHDSGYEV | 12 |
| AZ2 | FRHDSGY | 7 |
| CE | FGFPEHLLVDFLQSL | 15 |
| EGFR | LEEKKGNYVVTDH | 13 |
| HC10 | Proprietary | 17 |
| HC17 | CFRKHPEA | 8 |
| HC18 | EATYSRCG | 8 |
| HC24 | HLHQNIVD | 8 |
| HC5 | Proprietary | 18 |
| HC6 | Proprietary | 8 |
| HC6L | Proprietary | 26 |
| HC8 | Proprietary | 8 |
| HIV4.1 | RIKQIGMPGGK | 11 |
| HIV5.1 | LLELDKWASL | 10 |
| HIV6.1 | EQELLELDKWASLW | 14 |
| HV1 | GEIKNCSFNISTSIRGKVQKEYAFI | 25 |
| HV2 | LTSCNTSVITQACPKVSFEPIPIHYC | 26 |
| HV3 | PKVSFEPIPIHYCAPAGEAILKCNN | 26 |
| HV4 | THGIRPVVSTQLLLNGSLAEEE | 22 |
| M | NANPNVDPNANPNANPNANP | 20 |
| MV | DRAAGQPAGDRADGQPAG | 18 |
| M2(−) | SLLTEVETPIRNEWGARANDSSD | 23 |
| M2T | SLLTEVETPIRNEWGSLLTEVETPIRNEWG | 30 |
| OMP-1 | RSDYKFYEDANGTRDHKKGRHTA | 23 |
| OMP-2 | EDANGTRDHKKGRHT | 15 |
| SEB | KKKVTAQELD | 10 |
We have successfully inserted 24 of the 26 listed epitopes into the WHcAg platform (92.5%). The two underlined entries represent epitopes have not been successfully inserted to date. Abbreviations: AZ, β-amyloid; CE, cholesteryl ester transfer protein; EGFR, epidermal growth factor receptor mutant VIII; HC, hepatitis C virus E2 protein; HIV and HV, HIV gp120 sequences; MV, malarial P. vivax CS repeat type I; M2(−), influenza A virus M2e extracellular domain mutant sequence lacking two cysteines; M2T, influenza A virus M2e extracellular domain tandem sequence; OMP-1 and OMP-2, outer membrane protein P5 of nontypable Haemophilus influenzae; SEB, staphylococcus enterotoxin B.
Generation of a library of C-terminal modifications on WHcAg.
Importantly, the expansion of the number of positions available for insertion of foreign epitopes was made possible by the generation of a library of C-terminal modifications to WHcAg which variably stabilize insertions at different positions. These modifications have been produced and comprise a very useful second library of 21 C-terminal modifications. Figure 5 lists the sequences of the various modified C termini. The C-terminal modifications were designed to eliminate RNA/DNA binding motifs (RRR/SPXX motifs), eliminate and/or replace prolines, replace the last five C-terminal amino acids, and eliminate or conserve nonhomologous regions between HBcAg and WHcAg. Wild-type or full-length WHcAg binds significant amounts of bacterial RNA/DNA, which is not desirable for a vaccine platform. We have been quite successful in eliminating RNA/DNA binding from these modified WHcAg particles, which bind little or no nucleic acid (data not shown).
FIG. 5.
List of amino acid sequences of the wt and the 21 modified C termini of WHcAg. (Note that “188,” as cited in the text, corresponds to the wild-type sequence).
Effect of C terminus on hybrid WHcAg assembly.
In addition to the insert position, a second variable that influences hybrid WHcAg particle assembly is the C terminus of WHcAg protein. For example, the M epitope inserted at position 74 results in hybrid WHcAg particle assembly in the context of 10 different C termini; however, 5 C termini are nonpermissive for assembly with the M epitope at position 74 (Table 2). The M epitope in position 78 appears less destabilizing, since most C termini, including all five of the C termini which were nonpermissive for this epitope inserted at position 74, are permissive. Therefore, nonpermissive C termini can be rescued by altering the insert position. Interestingly, the two nonpermissive C termini for the M epitope at position 78 are both permissive for the M epitope at position 74. This reciprocal relationship suggests that the mechanisms of destabilization caused by the M insert at positions 74 and 78 are different and can be stabilized by different C-terminal sequences. Other epitopes have been studied, although less extensively, and the results reveal that the HyW and HyW2 C termini appear to be significantly more permissive for a variety of inserted epitopes and positions than the 150C C terminus, i.e., the epitopes for cholesteryl ester transfer protein (CE), HV2, HV4, and the mutant M2 sequence lacking cysteine [M2(−)] (Table 2). There are also examples of situations in which a limited number of the C-terminal modifications permit assembly. For example, C-Long is the only permissive C terminus for the insert outer membrane protein 1 (OMP-1), and the four other C termini tested could not rescue this epitope (Table 2). Note also that the OMP-1 sequence permitted assembly only when inserted in position 81 (see Table 3). This example demonstrates that the insert position and the C terminus of WHcAg both affect the assembly of the hybrid particle. Further, successful production of hybrid OMP-1-WHcAg particles necessitated combining the libraries of insert positions and C-terminal modifications.
TABLE 2.
Effect of C-terminal modification on hybrid particle assembly
| Epitope | Insert Position | Assemblya | Low or nonassemblya |
|---|---|---|---|
| M | 74 | 188, 150R, 150-3RC, 150-4RC, 150-3KC, 150-3AC, C-Long(M3), HyW, HyW1, HyW2 | 150C, C-Long, C-Long(M1), C-Long(M2), WT-R |
| M | 78 | 150C, HyW, 150-2RC, 150-3RC, C-Long, C-Long(M1), C-Long(M2), C-Long(M3), WT-R | 150R, 188 |
| CE | 73 | HyW | 150C |
| HV2 | 75 | HyW2 | 150C |
| HV4 | 74 | HyW | 150C |
| M2(−) | 78 | HyW | 150C |
| OMP-1 | 81 | C-Long | 150C, 150-3KC, HyW2, WT-R |
Assembly and nonassembly were determined by ELISA using WHcAg assembly-dependent anti-nWHc antibody (see Materials and Methods). Boldface numbers depict C termini that can be “rescued” by altering the insert position.
TABLE 3.
Effect of insert position on hybrid particle assembly
| C terminus | Epitope | Assemblya | Low or nonassemblya |
|---|---|---|---|
| HyW/HyW2 | M | 44, 73, 74, 75, 78, 84, 85, 92, NH2, COOH | 21, 91, 96 |
| 150C | M | 75, 76, 77, 78, 81, 82, 83 | 66, 74, 79, 80, 86 |
| 188 | M | 74 | 78 |
| HyW/HyW2 | CE | 73 | 75, 78, 81, 84, 92 |
| C-Long | HC5 | 81 | 78 |
| HyW | HC6L | 73 | 78, 81, 84, 92 |
| HyW | HC10 | 73 | 78, 81, 84, 92 |
| HyW | HC8 | 84 | 73, 78, 81, 92 |
| HyW/HyW2 | HV4 | 74, 75 | |
| 150C | HV4 | 75, 78 | 74 |
| C-Long | OMP-1 | 81 | 75, 78, 92 |
Assembly and nonassembly were determined by ELISA using WHcAg assembly-dependent anti-nWHc antibody (see Materials and Methods). Boldface numbers depict insert positions that can be rescued by altering the C terminus. Numbers represent the amino acid position on WHcAg that precedes the inserted epitope, except for the NH2- and COOH-terminal inserts.
Effect of position of the insert on hybrid WHcAg assembly.
As noted above, the position of the inserted epitope within WHcAg can affect the ability of the hybrid WHcAg particle to assemble. As shown in Table 3, the M epitope in the context of either the HyW or the HyW2 C terminus permitted assembly in most positions tested, with the exception of positions 21, 91, and 96. Similarly, positions 75, 76, 77, 78, 81, 82, and 83 were permissive in the context of the 150C C terminus. Note that position 74 was not permissive in the context of the 150C C terminus, but this position is “rescued” in the context of HyW/HyW2 C termini. Similarly, position 78 is not permissive for assembly in the context of the 188C C terminus but is permissive in combination with HyW/HyW2 and 150C. Therefore, the position of the insert can affect assembly and nonpermissive insert positions can be rescued by combination with an alternate C terminus. This observation was not unique to the malaria insert, as other epitope sequences demonstrated even more dramatic positional effects, some of which could not to be rescued by altering the C terminus. For example, as shown in Table 3, positions 81, 73, and 84 are singularly permissive for four different epitopes derived from the HCV E2 protein: HC5, HC6L, HC10, and HC8. Furthermore, the CE epitope can only be accommodated in position 73, whereas five other insert positions do not permit hybrid CE-WHcAg particle assembly regardless of the nature of the C terminus. Similarly for the OMP-1 epitope, only position 81 is permissive for hybrid WHcAg assembly. Table 1 depicts 24 (92.3%) of the 26 different epitope sequences tested that have been successfully incorporated into hybrid WHcAg particles. Note that the sequences differ in length and amino acid content. In summary, no single insertion position or C-terminal modification will permit assembly of all epitope sequences into hybrid WHcAg particles; therefore, a combinatorial approach is required to accommodate the largest number of putative epitopes. For example, Table 4 lists eight of the model epitopes that have been successfully incorporated into hybrid WHcAg particles and the optimal insertion position and C-terminal modification for each are shown. Note that for each of these eight epitopes, a different combination of insert position and C-terminal modification was necessary to achieve the optimal construction as defined by particle assembly, protein expression, insert antigenicity, yield of purified hybrid WHcAg particles, and immunogenicity.
TABLE 4.
Optimal combination of insert position and C terminus
Rapid screening technology.
The approach of combining the optimal C terminus from a selection of 21 termini and the optimal insert position from a choice of 17 positions, in the context of a given epitope, requires a rapid screening technology that can be applied early in the manufacturing process. Therefore, an antibody-based method for detecting expression of the core polypeptide, for assembly of the polypeptide into core hybrid particles, and for assessing antigenicity of the inserted heterologous epitope was developed. This rapid screening technique was applied to lysates of transformed E. coli to assess the desirability of any given hybrid core to be produced. The influenza A virus M2 epitope serves as an example of this strategy. A series of M2-WHcAg hybrid constructs with four different C termini and three different insert sites was produced; the expression levels and assembly competence scores relative to wt WHcAg are shown in Fig. 6. Note that all constructs harboring the native M2 sequence either assembled poorly (score <2) or purified as aggregates instead of core particles. Presumably the two cysteines in the native M2 sequence result in inappropriate inter- or intraparticle disulfide bridges. Therefore, an M2-specific MAb (MAb 14C2, which inhibits influenza A virus growth of most strains) (30) was tested for binding to a M2 peptide analog panel including cysteine-substituted peptides (data not shown). Because replacement of either or both cysteine residues did not affect the binding of the 14C2 MAb, hybrid core constructs carrying M2(−) inserted at position 78 were produced. The M2(−) sequence expressed in the context of the 150C C terminus still resulted in aggregates during purification. However, the M2(−) sequence inserted at position 78 in the context of the HyW-C terminus allowed assembly and was easily purified (Fig. 6). Subsequently, other combinations of C termini and insert positions such as HyW2-M2(−)81 have been found to accommodate the M2(−) sequence.
Importance of insert charge on hybrid WHcAg assembly.
In addition to the presence of cysteines, other characteristics of the insert sequence may affect hybrid WHcAg assembly. For example, during the development of WHcAg combinatorial technology it became apparent that the presence of a number of highly basic amino acids (especially K, R, and H) in candidate insert epitopes correlated negatively with the assembly of hybrid WHcAg particles. Table 5 represents a list of epitope sequences with the indicated isoelectric points (pI) and the effect on assembly of insertion of the sequences into the loop region of WHcAg. It appears from this survey that positively charged inserts (high pI) may adversely affect assembly of hybrid WHcAg particles. The pI of the wild-type WHcAg loop region (positions 76 to 82) is approximately 6.14. Because the wild-type WHcAg loop region (positions 76 to 82) is acidic, it was reasonable to predict that epitope inserts more positively charged than the wild-type sequence may have adverse effects on dimer formation (i.e., the particle subunit) and/or particle assembly. To more directly test the importance of insert epitope charge on hybrid particle assembly, several epitopes with pI values of 7 or greater, which did not permit assembly of hybrid WHcAg particles, were modified to contain the acidic amino acid glutamic acid (E) or were bracketed by glutamic acid residues. The ability of glutamic acid residues added as substitutions or bracketed around the inserted epitope to rescue hybrid core particle assembly was tested. As shown in Table 6, in all cases the addition of glutamic acid residues rescued particle assembly on WHcAg, HBcAg, and GScAg platforms. To address the specificity of the added amino acid, various amino acids were used to “bracket” a single basic (i.e., pI = 8.74) epitope sequence (OMP-2). As shown in Table 7, only amino acids that significantly lowered the insert pI [i.e., glutamic acid (E) and aspartic acid (D)] allowed hybrid WHcAg particle assembly. Therefore, in the case of five different epitope inserts which were positively charged (pI ≥ 7) and did not permit hybrid particle assembly on the WHcAg, HBcAg, or GScAg platforms, particle assembly could be rescued by the addition of glutamic acid or aspartic acid around the insert.
TABLE 5.
Positively-charged inserts (high pI) can adversely effect assembly of hybrid WHcAg particles
| Epitope | Sequence | Insert pI | Assemblya |
|---|---|---|---|
| HIV4.1 | RIKQIGMPGGK | 11.3 | − |
| IgE 413-435 | GETYQSRVTHPHLPRALMRSTTK | 11.13 | − |
| HV1 | GEIKNCSFNISTSIRGKVQKEYAFF | 9.41 | − |
| OMP-1 | RSDYKFYEDANGTRDHKKGRHTA | 9.33 | ± |
| OMP-2 | EDANGTRDHKKGRHT | 8.74 | − |
| HV3 | PKVSFEPIPIHYCAPAGFAILKCNN | 8.68 | − |
| SEB | KKKVTAQELD | 8.63 | ± |
| HV2 | LTSCNTSVITQACPKVSFEPIPIHYC | 7 | − |
| AZ2 | FRHDSGY | 7 | − |
| HV4 | THGIRPVVSTQLLLNGSLAEEE | 4.55 | + |
| MV | DRAAGQPAGDRADGQPAG | 4.2 | + |
| CE | FGFPEHLLVDFLQSL | 4.11 | + |
| AZ1 | DAEFRHDSGYEV | 4.08 | + |
| M2(−) | SLLTEVETPIRNEWGARANDSSD | 3.95 | + |
| HC10 | Proprietary | 3.43 | + |
| M | NANPNVDPNANPNANPNANP | 3.43 | + |
| MB | DPPPPNPNDPPPPNPN | 3.22 | + |
For explanation of assembly scores (+, ±, −), see Materials and Methods.
TABLE 6.
Addition of acidic substitutions or linker sequences can neutralize the destabilizing effect of positively charged inserts (high pI) and rescue hybrid-core particle assembly on the WHcAg, HBcAg, and GScAg platformsa
| Platform and epitope | Sequence | Insert pI | Assembly result |
|---|---|---|---|
| WHcAg | |||
| SEB | KKKVTAQELD | 8.63 | ± |
| SEB2E | EEKKKVTAQELDEE | 4.2 | + |
| AZ2 | FRHDSGY | 7 | − |
| AZ2E | EEFRHDSGYEE | 4.02 | + |
| OMP-1 | RSDYKFYEDANGTRDHKKGRHTA | 9.33 | ± |
| OMP-1aE | ESDYEEYEDANGTRDHKKGRHTA | 4.83 | + |
| OMP-2 | EDANGTRDHKKGRHT | 8.74 | − |
| OMP-2E | EEEDANGTRDHKKGRHTEE | 4.88 | + |
| HIV4.1 | RIKQIGMPGGK | 11.3 | − |
| HIV4.1E | EERIKQIGMPGGKEE | 4.74 | + |
| HBcAg | |||
| AZ2 | FRHDSGY | 7 | − |
| AZ2E | EEFRHDSGYEE | 4.02 | + |
| OMP-2 | EDANGTRDHKKGRHT | 8.74 | − |
| OMP-2E | EEEDANGTRDHKKGRHTEE | 4.88 | + |
| HIV4.1 | RIKQIGMPGGK | 11.3 | − |
| HIV4.1E | EERIKQIGMPGGKEE | 4.74 | + |
| GScAg | |||
| AZ2 | FRHDSGY | 7 | + |
| AZ2E | EEFRHDSGYEE | 4.02 | ++ |
| HIV4.1 | RIKQIGMPGGK | 11.3 | − |
| HIV4.1E | EERIKQIGMPGGKEE | 4.74 | + |
The three platforms were tested with HyW2 C terminus. For explanation of assembly scores (+, ±, −), see Materials and Methods. Underlined letters indicate acidic substitutions or linker sequences.
TABLE 7.
Only acidic amino acids can rescue assembly of hybrid core particles containing a positively charged inserta
| OMP-2 epitopeb | Sequenceb | Linker type | Insert pI | Assembly resultc |
|---|---|---|---|---|
| L | L-EDANGTRDHKKGRHT-L | Small, hydrophobic, nonpolard | 8.74 | − |
| EE | EE-EDANGTRDHKKGRHT-EE | Acidic | 4.88 | + |
| DD | DD-EDANGTRDHKKGRHT-DD | Acidic | 4.67 | + |
| P | P-EDANGTRDHKKGRHT-P | Bulky, hydrophobic, nonpolar | 8.74 | − |
| QQ-EDANGTRDHKKGRHT-QQ | Amide, hydrophobic, uncharged, polar | 8.74 | − | |
| TT | TT-EDANGTRDHKKGRHT-TT | Nucleophilic, uncharged, polar | 8.74 | − |
| YY | YY-EDANGTRDHKKGRHT-YY | Aromatic, hydrophobic, uncharged, polar | 8.56 | − |
The C terminus used for OMP-2 hybrid core particles was HyW2.
E, glutamic acid; D, aspartic acid; P, proline; Q, glutamine; T, threonine; Y, tyrosine.
For explanation of assembly scores (+, −), see Materials and Methods.
Leucine linker due to cloning restriction site.
The position of the heterologous insert affects immunogenicity.
The evaluation of the combinatorial technology has included determination of the in vivo humoral immune responses to the inserted epitopes as well as to the WHcAg carrier. Although a hierarchy of immunogenicity has been observed, most hybrid WHcAg core particles are quite immunogenic in terms of both anti-insert and anti-WHc antibody production. However, the immunogenicity of hybrid core particles composed of the same HyW-modified C terminus and the same M repeat epitope can differ depending on where the epitope is positioned (Fig. 7). Particles with insertions in (aa 78) or near (aa 74) the loop were more immunogenic in terms of the anti-insert response than particles with fusion to the NH2 terminus, and placement at the C terminus was poorly immunogenic both in terms of end point serum titer and delayed onset of antibody production. This correlation was not true for anti-carrier antibody production, which for the NH2- and C-terminal locations of the M epitope was greater than or equal to the production seen with the internal insertions (data not shown). Therefore, the position of the epitope did not alter the overall immunogenicity of the particle and the positional effects most likely reflect greater surface exposure and/or optimal spacing of the heterologous epitopes in or near the loop region. The high anticarrier responses to the NH2- and C-terminally fused epitopes were also predictable, because the native loop structure and the endogenous WHcAg B-cell epitopes are intact on these hybrid core particles.
FIG. 7.
The effect of the position of the inserted epitope (NANP)n on the immunogenicity of the malaria-WHcAg hybrid particle. The hybrid particles are identical except for the position of the insert: the NH2- or COOH-terminal insertion or the internal insertion at aa 78 or 74. Groups of four mice were primed with 20 μg of the indicated particles in IFA. Sera were collected, pooled, and analyzed for the presence of IgG anti-NANP antibody by ELISA. Anti-NANP antibody titers are expressed as the reciprocal of serum dilution that yields three times the OD492 value of preimmunization sera.
The C terminus affects immunogenicity.
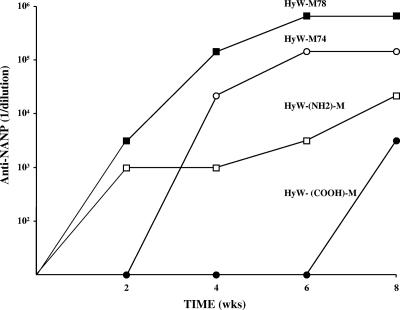
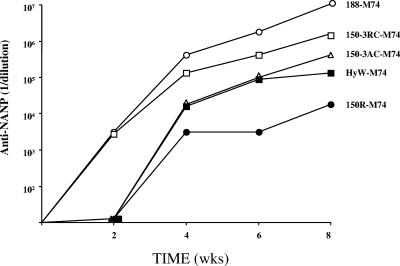
Hybrid WHcAg particles with the M epitope inserted at the same position (position 74) but with various C termini were also compared (Fig. 8). Particles with the native full-length (188-M74) or with the 150-3RC C terminus were more immunogenic in terms of serum titers of anti-NANP antibody as well as a quicker onset (week 2) compared to the 150 3AC and HyW C termini. The particle comprised of the 150R C terminus, which lacks a C-terminal cysteine, was weakly immunogenic. The 150R-M74 hybrid particle is the least stable in vitro and most likely in vivo, which probably explains the poorer immunogenicity results. However, factors other than hybrid particle stability may mediate the influence of the C-terminal sequence on antibody production with respect to epitopes inserted into the external loop. For example, it has been reported that mutation of aa 97 or the placement of a histidine tag on HBcAg affected antibody recognition of the loop region even though these regions are spatially distant (17). These data suggest that modifications to the C terminus of core proteins may induce conformational changes at a distance.
FIG. 8.
The effect on immunogenicity of modifying the C termini of hybrid M-WHcAg particles. Hybrid M-WHcAg particles containing the same M insert sequence inserted at the same position (position 74) but with various C termini were used to immunize mice. Groups of four mice were immunized with a single dose of 20 μg of the hybrid particles in IFA. Sera were collected, pooled, and analyzed by ELISA for the presence of IgG anti-NANP antibody.
DISCUSSION
We have attempted to increase the applicability of WHcAg as a VLP-based vaccine carrier by expanding the number of insertion sites (i.e., to 17) and C-terminal modifications (i.e., to 21) used. A number of self-assembling proteins have been proposed as vaccine carrier platforms. A universal and major problem has been the destabilizing effects on VLP assembly of adding or inserting foreign peptidic sequences (8). Hybrid VLP stability has represented such a serious problem that practitioners using HBcAg platform technology (8) and human papillomavirus (HPV) platform technology (3) have opted to chemically conjugate foreign epitopes to wild-type VLPs instead of attempting to produce hybrid particles by recombinant means. For example, it has been stated “that the ability of different HPV chimeric L1-self peptide chimeras to assemble into VLPs is highly unpredictable, limiting the general applicability of this technique” (3). Further, “the size and nature of epitopes that can be inserted into VLPs, in particular into their immunodominant regions, is restricted and VLPs containing peptides longer than 20 amino acids often fail to assemble” (8). Unfortunately, chemical linkage of heterologous epitopes to VLPs often compromises a number of the advantages inherent in the recombinant hybrid particle technology (i.e., 100% substitution, conservation of ordered structure, manufacturing reproducibility). Therefore, we have addressed and largely resolved this major problem by developing a “combinatorial” technology that combines the optimal insert position, C-terminal modification, and the epitope insert sequence that permits efficient self-assembly of the WHcAg platform.
The limited number of insertion sites and C-terminal modifications described for the HBcAg-based VLP technology may explain the fact that less than 50% of B-cell epitopes are successfully accommodated by the HBcAg platform (8). In contrast, the combined libraries of insertion sites and modified C termini we have accumulated for WHcAg have allowed us to be successful in attempts to insert 24 (92.3%) of 26 sequences. It is notable that the effect of the combination of insertion site modification and C-terminal modification on WHcAg is also dependent on the sequence of the inserted epitope. Therefore, three variables must be considered in designing a hybrid WHcAg particle: the insert position, the C-terminal sequence, and the epitope sequence. Because we have developed a rapid screening method to determine expression and assembly of hybrid core particles at the early bacterial lysate step, a combinatorial approach involving “shuffling” of the insert position and the C-terminal modification for each epitope of interest is feasible. But it is important to note that for eight model heterologous epitopes, eight different combinations of C terminus plus insert position were required (Table 4). This indicates that no one “universal” WHcAg platform will suffice for all heterologous epitopes and that a combinatorial approach is necessary for the widest possible application of this technology.
Because the inserted epitope sequence can affect hybrid core assembly or stability, it is useful to perform mutational analysis of the epitope to map the necessary antibody contact residues. Nonessential residues can be subsequently replaced with less-disruptive residues as needed. This strategy is also useful for identifying analogs with improved antibody binding. Karpenko et al. have examined the physical and chemical properties of epitope inserts that may affect the folding and assembly of chimeric proteins into HBcAg hybrid particles (9). This study defined three parameters of the epitope insert that prevented self-assembly of hybrid HBcAg particles, specifically, high epitope hydrophobicity, large epitope volume, and a high β-strand index for the epitope. We report herein that the epitope charge may be a more important factor to be considered when constructing hybrid core platforms. If possible, a negatively charged epitope should be selected; however, when this is not possible, a positively charged epitope can be modified to include acidic amino acid substitutions and/or be bracketed by acidic residues to permit efficient hybrid particle assembly. The addition of acidic amino acids can be integrated directly into the platform or added to the inserted epitope. This modification may be extrapolated beyond hepadnavirus core hybrid particles. Similarly to the hepadnavirus core protein results, preferred insertion sites on many VLPs are immunodominant exposed loop structures that are accessible for antibody recognition and that may be less likely to compromise the structural integrity of the particle, in contrast to insertions into α-helical or β-sheet regions (4, 5, 7, 27). The finding that insertion of positively charged epitopes into the exposed loop region of HBcAg, WHcAg, and GScAg disrupts assembly of hybrid particles may not to be unique to hepadnavirus core antigen VLPs. Interestingly, insertion of a highly acidic epitope (the HBcAg sequence at positions 78 to 83) into each of six distinct surface loop domains of HPV16-L1 VLPs did not disrupt hybrid particle assembly (20). The use of acidic amino acids to neutralize a “cloud” of positive charge around an inserted epitope may represent a generally useful strategy for a variety of VLP vaccine platforms. In a previous study, an adequate balance between positive and negative charge densities of the nucleic acid and protein complex in the interior of full-length HBcAg was suggested to contribute to a more stable capsid structure (12).
There are also practical considerations for implementing WHcAg-based combinatorial technology described in this report for real-world vaccine design. Because the combinatorial technology has the potential to produce 374 gene constructs or hybrid particles (i.e., 17 insertion sites × 22 C termini) for each designated B-cell epitope, it is essential to be able to select optimal constructs early in the production process. Our approach has been to stratify the production of constructs in groups of 10 (first tier, second tier, etc.). The 10 first-tier constructs include three insertion sites within the loop (i.e., positions 78, 81, and 82) and two insertion sites outside the loop (i.e., positions 74 and 83) combined with two different C termini (HyW2 and C-Long). The 10 constructs are transformed into E. coli and grown in low-volume fermentation culture (0.5 liter). Bacterial lysates are then screened for relative levels of core protein expression, hybrid WHcAg particle assembly, and insert epitope antigenicity by use of the capture ELISAs as described above. The capture ELISAs performed with bacterial lysates reliably and reproducibly predicted the constructs that resulted in high yields of high-quality hybrid WHcAg particles after the purification process. This is a crucial and useful selection step because it eliminates many suboptimal constructs very early in the development pathway. When optimal constructs are not identified, the process is repeated with the next 10 constructs and so forth until optimal constructs are identified.
In summary, the development of a combinatorial technology, together with the use of WHcAg as a platform, has significantly advanced the applicability of hepadnavirus core-based VLP vaccine design.
Acknowledgments
We thank Ori Raz and Rudy Garcia for technical assistance and M. P. Testa and T. R. Phillips for helpful discussions and Helen Pederson for editorial assistance.
This work was supported by National Institutes of Health grants 5 R01 AI 49730 and 5 R01 AI 20720 and grants from the Swedish Cancer Foundation and Swedish Science Council.
REFERENCES
- 1.Belnap, D. M., N. R. Watts, J. F. Conway, N. Cheng, S. J. Stahl, P. T. Wingfield, and A. C. Steven. 2003. Diversity of core antigen epitopes of hepatitis B virus. Proc. Natl. Acad. Sci. USA 100:10884-10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billaud, J.-N., D. Peterson, F. Schödel, A. Chen, M. Sallberg, F. Garduno, P. Goldstein, W. McDowell, J. Hughes, J. Jones, and D. Milich. 2005. Comparative antigenicity and immunogenicity of hepadnavirus core proteins. J. Virol. 79:13641-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chackerian, B., D. R. Lowy, and J. T. Schiller. 2001. Conjugation of a self-antigen to papillomavirus-like particles allows for efficient induction of protective autoantibodies. J. Clin. Investig. 108:415-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chackerian, B., D. R. Lowy, and J. T. Schiller. 1999. Induction of autoantibodies to mouse CCR5 with recombinant papillomavirus particles. Proc. Natl. Acad. Sci. USA 96:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, N. D., N. M. Cladel, C. A. Reed, L. R. Budgeon, M. E. Embers, D. M. Skulsky, W. L. McClements, S. W. Ludmerer, and K. U. Jansen. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324-334. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, B. E., S. E. Newton, A. R. Carroll, M. J. Francis, G. Appleyard, A. D. Syred, P. E. Highfield, D. J. Rowlands, and F. Brown. 1987. Improved immunogenicity of a peptide epitope after fusion to hepatitis B core protein. Nature 330:381-384. [DOI] [PubMed] [Google Scholar]
- 7.Gedvilaite, A., C. Frommel, K. Sasnauskas, B. Micheel, M. Ozel, O. Behrsing, J. Staniulis, B. Jandrig, S. Scherneck, and R. Ulrich. 2000. Formation of immunogenic virus-like particles by inserting epitopes into surface-exposed regions of hamster polyomavirus major capsid protein. Virology 273:21-35. [DOI] [PubMed] [Google Scholar]
- 8.Jegerlehner, A., A. Tissot, F. Lechner, P. Sebbel, I. Erdmann, T. Kundig, T. Bachi, T. Storni, G. Jennings, P. Pumpens, W. Renner, and M. Bachmann. 2002. A molecular assembly system that renders antigens of choice highly repetitive for induction of protective B cell responses. Vaccine 20:3104. [DOI] [PubMed] [Google Scholar]
- 9.Karpenko, L. I., V. A. Ivanisenko, I. A. Pika, N. A. Chikaev, A. M. Eroshkin, T. A. Veremeiko, and A. A. Ilyichev. 2000. Insertion of foreign epitopes in HBcAg: how to make the chimeric particle assemble. Amino Acids 18:329-337. [DOI] [PubMed] [Google Scholar]
- 10.Lazdina, U., M. Alheim, J. Nystrom, C. Hultgren, G. Borisova, I. Sominskaya, P. Pumpens, D. L. Peterson, D. R. Milich, and M. Sallberg. 2003. Priming of cytotoxic T cell responses to exogenous hepatitis B virus core antigen is B cell dependent. J. Gen. Virol. 84:139-146. [DOI] [PubMed] [Google Scholar]
- 11.Lazdina, U., T. Cao, J. Steinbergs, M. Alheim, P. Pumpens, D. L. Peterson, D. R. Milich, G. Leroux-Roels, and M. Sallberg. 2001. Molecular basis for the interaction of the hepatitis B virus core antigen with the surface immunoglobulin receptor on naive B cells. J. Virol. 75:6367-6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Pogam, S., P. K. Chua, M. Newman, and C. Shih. 2005. Exposure of RNA templates and encapsidation of spliced viral RNA are influenced by the arginine-rich domain of human hepatitis B virus core antigen (HBcAg 165-173). J. Virol. 79:1871-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, S., R. Tobias, S. McClure, G. Styba, Q. Shi, and G. Jackowski. 1997. Removal of endotoxin from recombinant protein preparations. Clin. Biochem. 30:455-463. [DOI] [PubMed] [Google Scholar]
- 14.Milich, D. R., M. Chen, F. Schodel, D. L. Peterson, J. E. Jones, and J. L. Hughes. 1997. Role of B cells in antigen presentation of the hepatitis B core. Proc. Natl. Acad. Sci. USA 94:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milich, D. R., J. Hughes, J. Jones, M. Sallberg, and T. R. Phillips. 2001. Conversion of poorly immunogenic malaria repeat sequences into a highly immunogenic vaccine candidate. Vaccine 20:771-788. [DOI] [PubMed] [Google Scholar]
- 16.Milich, D. R., D. L. Peterson, J. Zheng, J. L. Hughes, R. Wirtz, and F. Schodel. 1995. The hepatitis nucleocapsid as a vaccine carrier moiety. Ann. N. Y. Acad. Sci. 754:187-201. [DOI] [PubMed] [Google Scholar]
- 17.Ning, B., and C. Shih. 2004. Nucleolar localization of human hepatitis B virus capsid protein. J. Virol. 78:13653-13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pumpens, P., G. P. Borisova, R. A. Crowther, and E. Grens. 1995. Hepatitis B virus core particles as epitope carriers. Intervirology 38:63-74. [DOI] [PubMed] [Google Scholar]
- 19.Pumpens, P., and E. Grens. 2001. HBV core particles as a carrier for B cell/ T cell epitopes. Intervirology 44:98-114. [DOI] [PubMed] [Google Scholar]
- 20.Sadeyen, J. R., S. Tourne, M. Shkreli, P. Y. Sizaret, and P. Coursaget. 2003. Insertion of a foreign sequence on capsid surface loops of human papillomavirus type 16 virus-like particles reduces their capacity to induce neutralizing antibodies and delineates a conformational neutralizing epitope. Virology 309:32-40. [DOI] [PubMed] [Google Scholar]
- 21.Salfeld, J., E. Pfaff, M. Noah, and H. Schaller. 1989. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J. Virol. 63:798-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sallberg, M., U. Ruden, L. O. Magnius, E. Norrby, and B. Wahren. 1991. Rapid “tea-bag” peptide synthesis using 9-fluorenylmethoxycarbonyl (Fmoc) protected amino acids applied for antigenic mapping of viral proteins. Immunol. Lett. 30:59-68. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Schodel, F., A. M. Moriarty, D. L. Peterson, J. A. Zheng, J. L. Hughes, H. Will, D. J. Leturcq, J. S. McGee, and D. R. Milich. 1992. The position of heterologous epitopes inserted in hepatitis B virus core particles determines their immunogenicity. J. Virol. 66:106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schodel, F., D. Peterson, D. R. Milich, Y. Charoenvit, J. Sadoff, and R. Wirtz. 1997. Immunization with hybrid hepatitis B virus core particles carrying circumsporozoite antigen epitopes protects mice against Plasmodium yoelii challenge. Behring Inst. Mitt. 97:114-119. [PubMed] [Google Scholar]
- 26.Schodel, F., R. Wirtz, D. Peterson, J. Hughes, R. Warren, J. Sadoff, and D. Milich. 1994. Immunity to malaria elicited by hybrid hepatitis B virus core particles carrying circumsporozoite protein epitopes. J. Exp. Med. 180: 1037-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slupetzky, K., S. Shafti-Keramat, P. Lenz, S. Brandt, A. Grassauer, M. Sara, and R. Kirnbauer. 2001. Chimeric papillomavirus-like particles expressing a foreign epitope on capsid surface loops. J. Gen. Virol. 82:2799-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]
- 29.Zavala, F., J. P. Tam, P. J. Barr, P. J. Romero, V. Ley, R. S. Nussenzweig, and V. Nussenzweig. 1987. Synthetic peptide vaccine confers protection against murine malaria. J. Exp. Med. 166:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zebedee, S. L., and R. A. Lamb. 1989. Growth restriction of influenza A virus by M2 protein antibody is genetically linked to the M1 protein. Proc. Natl. Acad. Sci. USA 86:1061-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]