Abstract
STAT6 is a central mediator of IL-4-induced gene responses. STAT6-mediated transcription is depend ent on the C-terminal transcription activation domain (TAD), but the mechanisms by which STAT6 activates transcription are poorly understood. Here, we have identified the staphylococcal nuclease (SN)-like domain and tudor domain containing protein p100 as a STAT6 TAD interacting protein. p100 was originally characterized as a transcriptional coactivator for Epstein–Barr virus nuclear antigen 2. STAT6 interacted with p100 in vitro and in vivo. The interaction was mediated by the TAD domain of STAT6 and the SN-like domain of p100. p100 did not affect the immediate activation events of STAT6, but enhanced STAT6-mediated transcriptional activation and the IL-4-induced Igε gene transcription in human B-cell line. Finally, p100 associated with the large subunit of RNA polymerase II and was mediating interaction between STAT6 and RNA polymerase II. These findings identify p100 as a novel coactivator for STAT6 and suggest that p100 functions as a bridging factor between STAT6 and the basal transcription machinery.
Keywords: EBV/interleukin-4/p100/STAT6/transcription
Introduction
Cytokines are small soluble glycoproteins that play essential roles in regulation of the hematopoietic and immune systems. Interleukin-4 (IL-4) is a pleiotropic cytokine that functions on a variety of cells, including lymphoid, myeloid, stromal and endothelial cells. The biological responses to IL-4 stimulation are cell type-dependent, and activation of the IL-4 receptor (IL-4R) complex leads to proliferation, differentiation, cell survival or even apoptosis, depending on the cellular context (Chomarat and Banchereau, 1997; Nelms et al., 1999). In lymphoid cells, IL-4 mediates induction of Th2 differentiation as well as immunoglobulin (Ig) isotype switching and production of IgG1 and IgE, and Fc receptor for IgE (CD23) expression in B cells, and thereby orchestrates allergic responses (Kuhn et al., 1991). IL-4 stimulation results in activation of JAK1 and JAK3 tyrosine kinases (Johnston et al., 1994; Witthuhn et al., 1994) and subsequently in phosphorylation of the receptor recruited transcription factor signal transducer and activator of transcription 6 (STAT6) on C-terminal tyrosine 641 (Hou et al., 1994; Quelle et al., 1995). The tyrosine-phosphorylated STAT6 forms dimers that are translocated to the nucleus, where they bind to specific recognition sequences in the promoters of IL-4-responsive genes. STAT6 plays an indispensable role in IL-4-mediated transcriptional responses, as demonstrated by the similar phenotypes of IL-4–/– and Stat6–/– mice (Kuhn et al., 1991; Shimoda et al., 1996). Stat6-deficient mice do not develop T helper 2 cells, fail to produce significant levels of IgE and express only low levels of CD23, IL-4Rα and major histocompatibility complex class II molecules.
The immediate signaling events leading to DNA bind ing of STAT6 are well established, while relatively little is known about the molecular mechanisms of STAT6-stimulated transcription. Activation of gene expression involves the interplay between various sequence-specific transcription factors and transcriptional co-regulators with the basal transcriptional machinery of the cells. In IL-4-responsive promoters, STAT6 has been shown to cooperate with NF-κB, IRF-4, BSAP and C/EBPβ transcription factors (Mikita et al., 1998; Shen and Stavnezer, 1998; Gupta et al., 1999; Stutz and Woisetschläger, 1999), but the precise mechanisms by which these events are connected to activation of transcription are unknown. Sequence-specific transcription factors are considered to tether multisubunit co-regulator complexes through protein–protein interactions with the promoter. The functions of transcriptional coactivators can be broadly divided into two overlapping categories. First, coactivators can mediate chromatin modifications either through acetylation reactions mediated by histone acetyl transferases (HATs), such as CBP/p300, SRC, P/CAF or GCN5 (Xu et al., 1999), or through nucleosome remodeling complexes such as SWI/SNF (Muchardt and Yaniv, 1999). Alternatively, although not exclusively, some coactivators function as bridging factors mediating complex formation between sequence-specific transcription factors and RNA polymerase II or other components of the basal transcription machinery, such as general initiation factors TFIIA, -B, -D, -E, -F or -H (Orphanides et al., 1996; Green, 2000). The general coactivator CBP/p300 and NCoA-1 have been shown to cooperate with STAT6 (McDonald and Reich, 1999; Litterst and Pfitzner, 2001), as well as with most of the other STATs in transcriptional activation (reviewed in Shuai, 2000). However, the mode of interaction between STATs and the basal transcription machinery is currently unknown.
The transcription activation domain (TAD) of all STATs is localized in the C-terminus, which is the most divergent part of these proteins. Compared with other STATs, STAT6 has the longest TAD, which is rich in glutamine residues and functions as a potent independent transactivator (Mikita et al., 1996; Moriggl et al., 1997; Shen et al., 2001). This study was aimed at characterizing the molecular mechanism of STAT6-mediated transcription by analyzing STAT6-TAD interacting nuclear proteins. These studies led to the identification of human p100 protein as a STAT6 interacting protein. p100 was first identified as a transcriptional coactivator for Epstein–Barr virus nuclear antigen 2 (EBNA2). EBNA2 is essential for Epstein–Barr virus (EBV)-mediated B lymphocyte transformation and activates both viral genes such as latent membrane proteins LMP1 and LMP2, as well as several cellular genes (Wang et al., 1990). In addition, p100 has also been shown to interact with c-Myb, a transcription factor involved in the regulation of differentiation and proliferation of immature hematopoietic and lymphoid cells (Leverson et al., 1998). The predicted structure of p100 indicates that the protein is composed of four repeats homologous to the staphylococcal nuclease (SN)-fold, followed by a tudor domain (TD) in the C-terminus (Callebaut and Mornon, 1997), but the functions of these domains in p100 have not been characterized. We found that p100 interacted with STAT6 both in vivo and in vitro through its SN-like domains and enhanced STAT6-mediated transcriptional activation. Furthermore, p100, but not STAT6, interacted with the large subunit of RNA polymerase II, and p100 appears to function as a bridging factor between STAT6 and the basal transcription machinery.
Results
Identification of p100 as a STAT6-TAD interacting protein
To identify possible co-regulators of STAT6, the TAD of STAT6 was expressed as a glutathione S-transferase (GST) fusion protein (GST–St6TAD) and used to purify interacting nuclear proteins. Equal amounts of GST and GST–St6TAD fusion proteins were bound to glutathione-coupled beads and incubated with nuclear extracts from [35S]methionine-labeled or non-labeled Ramos B cells. As shown in Figure 1A and B, the GST–St6TAD fusion protein was found to interact specifically with several proteins.
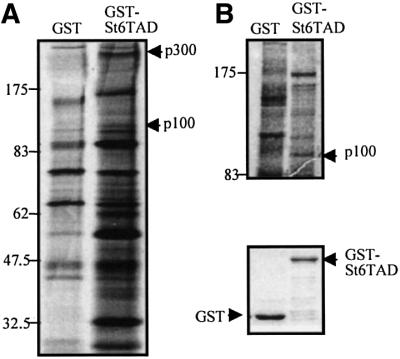
Fig. 1. Physical interaction between STAT6-TAD and p100. (A and B) STAT6-TAD interacts specifically with several nuclear proteins. Aliquots of nuclear lysates from [35S]methionine-labeled or unlabeled Ramos B lymphocytes were incubated with either GST alone or with GST–St6TAD. The bound proteins were subjected to SDS–PAGE and visualized by autoradiography (A) or silver staining (B). The band corresponding to the 100 kDa protein was cut out from silver-stained gel and subjected to trypsin digestion and MALDI-TOF mass spectrometry. The lower panel in (B) is the loading control of GST–St6TAD and GST proteins.
Initial analysis of the GST–St6TAD interacting proteins was carried out by immunoblotting using antibodies against some potential candidate proteins. Consistent with previous studies (Gingras et al., 1999; McDonald and Reich, 1999), western blot analysis revealed that the 300 kDa protein was CBP/p300 (data not shown). Other STAT6-TAD interacting proteins were identified by mass spectrometry or by N-terminal peptide sequencing. The band corresponding to a 100 kDa protein was recovered and subjected to in-gel trypsin digestion (Figure 1B). The molecular masses of the digested peptides were measured by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. The program ProFound was used to compare the mass maps obtained against theoretical tryptic peptide mass maps in the OWL protein sequence database. The comparison resulted in unequivocal identification of the 100 kDa protein as human p100 protein (Tong et al., 1995). Twenty-four peptides from the 100 kDa protein were found to match the calculated molecular masses of p100.
p100 interacts with STAT6 both in vivo and in vitro
To analyze whether STAT6–p100 complex formation is occuring in vivo, the protein interaction between STAT6 and p100 was investigated by coimmunoprecipitation assays. COS-7 cells were transfected with hemagglutinin (HA) epitope-tagged STAT6 (STAT6-HA) and Flag epitope-tagged p100 (p100-Flag), either separately or together. STAT6 was immunoprecipitated from the total cell lysates with anti-HA antibody, and the presence of p100 was detected by anti-Flag immunoblotting. As shown in Figure 2A, p100 was found to co-precipitate with STAT6. As a control, p100 was not detected in anti-HA immunoprecipitations from cells transfected with only STAT6-HA or p100-Flag. The middle and lower panels in Figure 2A are protein level controls for STAT6 and p100.
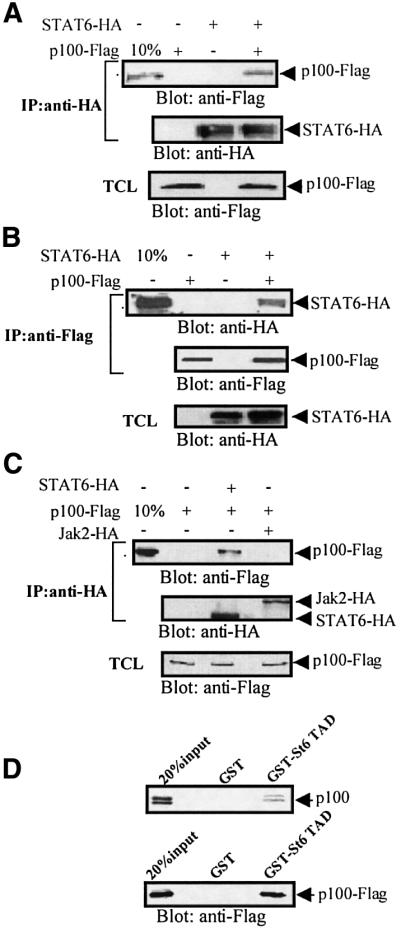
Fig. 2. p100 interacts with STAT6 in vivo and in vitro. (A) p100 co- immunoprecipitates with STAT6 in vivo. COS-7 cells were transfected with STAT6-HA and p100-Flag as indicated and immunoprecipitated with anti-HA antibody and blotted with anti-Flag antibody. Ten percent of total cell lysate (TCL) from p100-Flag-transfected cells was included as a control (upper panel). The same filter was stripped and re-blotted with anti-HA antibody for STAT6 (middle panel). The expression level of p100 from different lysates was detected by anti-Flag immunoblotting (lower panel). (B) STAT6 coimmunoprecipitates with p100. COS-7 cells were transfected as indicated and cell extracts were immunoprecipitated with anti-Flag and blotted with anti-HA antibody. Ten percent of TCL from STAT6-HA-transfected cells was included as a control (upper panel). The same filter was stripped and re-blotted with anti-Flag antibody (middle panel). The expression level of STAT6 from different lysates was detected by western blot analysis (lower panel). (C) Jak2 does not interact with p100. COS-7 cells were transfected with p100-Flag, HA-tagged Jak2 (Jak2-HA) or STAT6-HA expression plasmids. Cell extracts were immunoprecipitated with anti-HA and immunoblotted with anti-Flag antibody (upper panel). The same filter was stripped and re-blotted with anti-HA antibody (middle panel). The expression level of p100 in different lysates was detected by western blot analysis (lower panel). The positions of p100, STAT6 and Jak2 are indicated by arrows. (D) p100 interacts with STAT6-TAD in vitro. p100 was 35S-labeled by in vitro translation and incubated with beads loaded with GST–St6TAD or GST. The bound proteins were subjected to SDS–PAGE and visualized by autoradiography. Twenty percent of the in vitro-translated p100 was included as a control (upper panel). p100-Flag was expressed in COS-7 cells and the lysates were incubated with GST–St6TAD or GST. The bound proteins were blotted with anti-Flag antibody. Twenty percent of TCL from p100-Flag transfected cells was included as a control (lower panel).
The in vivo complex formation between STAT6 and p100 was also detected when the experiment was done reciprocally, by immunoprecipitating p100 with anti-Flag antibody and detecting STAT6 by blotting with anti-HA antibody (Figure 2B). The control blots showed similar amounts of p100 and STAT6 in different lysates. In order to confirm the specificity of p100 interactions, COS-7 cells were transfected with expression vectors encoding for either HA-tagged Jak2 or STAT6-HA and p100-Flag. As shown in Figure 2C, p100 was found to coimmunoprecipitate only with STAT6 but not with Jak2. Furthermore, the interaction between p100 and STAT6 was stable in stringent lysis conditions (1% NP-40, 250 mM NaCl).
The interaction between STAT6 and p100 was also analyzed in in vitro binding assays with GST–St6TAD or GST proteins and in vitro-translated p100 protein (Figure 2D, upper panel). The in vitro-translated p100 is detected as a doublet due to N-terminal cleavage of the protein (K.Carter and E.Kieff, manuscript in preparation). GST protein alone did not interact with p100, while GST–St6TAD interacted with both forms of the in vitro-translated p100. To substantiate the specificity of p100 and STAT6-TAD interaction, COS-7 cells were transfected with p100-Flag and the cell lysates were incubated with GST or GST–St6TAD fusion protein. Analysis of the bound proteins by anti-Flag immunoblotting showed that p100 interacted with GST–St6TAD, but not with GST protein (Figure 2D, lower panel). These results indicate that STAT6-TAD interacts with the p100 protein and suggest that the interaction is direct, although the possible presence of bridging protein(s) in reticulocyte lysate can not be excluded.
Cellular localization of STAT6 and p100 was also investigated in intact HeLa cells by dual immunostaining (data not shown). Without IL-4 stimulation, endogenous p100 and STAT6 were distributed in the cytoplasm as well as in the nucleus. In IL-4-stimulated cells, p100 and STAT6 colocalized uniformly in the nucleus. Taken together, these results indicate complex formation between p100 and STAT6, both in vitro and in vivo, most likely through a direct interaction.
p100 enhances STAT6-mediated transcription
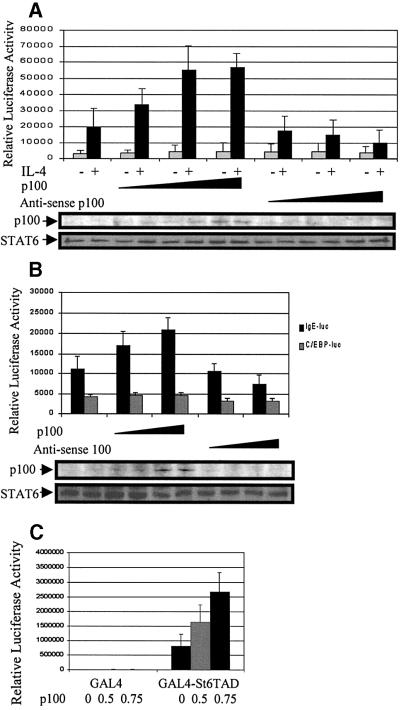
p100 functions as a transcriptional coactivator for EBNA2 (Tong et al, 1995), but in QT6 fibroblasts p100 has been reported to inhibit c-Myb-dependent promoter activation (Dash et al., 1996). To determine the functional consequences of the interaction between p100 and STAT6, Ramos B cells were transfected with STAT6-dependent Igε luciferase reporter gene, together with increasing amounts of p100-Flag or anti-sense p100. Ectopic expression of p100 did not affect the basal activity of the Igε reporter, but enhanced the IL-4-stimulated reporter gene activity in a dose-dependent manner (Figure 3A). OverB). Overexpression of p100 or anti-sense p100 constructs did not have any marked effect on the constitutively active C/EBP driven reporter gene (Figure 3B), and expression of anti-sense p100 affected only p100 protein levels but not STAT6 (Figure 3A and B), JAK1 or TBP protein levels (data not shown).
Fig. 3. p100 enhances STAT6-dependent transcriptional activation. (A) Overexpression of p100 enhances, while anti-sense p100 inhibits, transcriptional activation of STAT6. Ramos cells were transfected with Igε luciferase reporter construct (10 µg) and β-galactosidase vector (5 µg), together with increasing amounts (10, 20 and 30 µg) of p100 or anti-sense p100 expression plasmid, and treated with IL-4 as indicated. The normalized luciferase values are shown. The lower panels in (A) and (B) show p100 and STAT6 protein levels in the lysates. (B) p100 protein selectively enhances IL-4-induced transcriptional activation of STAT6. HeLa cells were transfected with STAT6 expression plasmid (1.0 µg), β-galactosidase vector (0.5 µg) and Igε luciferase reporter construct (1.0 µg) or C/EBP luciferase reporter constructs (1.0 µg), together with increasing amounts of p100 expression plasmid, and treated with IL-4. The normalized luciferase values are shown. (C) p100 enhances transcription of STAT6-TAD in a heterologous system. GAL4-St6TAD or GAL4-DBD alone (GAL4) expression plasmids (0.5 µg) were transfected into HeLa cells with reporter plasmid (GAL4)3TK-luciferase (0.5 µg), together with increasing amounts of the p100 expression vector. β-galactosidase and luciferase values were measured 48 h after transfection. Mean normalized luciferase values of three independent experiments with standard deviations are shown.
To characterize the mechanism of p100-mediated augmentation of STAT6 activity, we analyzed whether p100 would affect the immediate signaling events of STAT6. These experiments were carried out in COS-7 cells that do not express detectable levels of STAT6, but express a functional IL-4R (Moriggl et al., 1997; Pesu et al., 2000). In coexpression experiments, p100 did not have any effect on IL-4-induced tyrosine phosphorylation or DNA binding of STAT6 (data not shown); therefore, we considered the possibility that p100 would directly enhance the activity of STAT6-TAD. The TAD of STAT6 can stimulate reporter gene transcription autonomously when fused to the heterologous GAL4 DNA-binding domain (GAL4-DBD). GAL4–STAT6-TAD activated the reporter gene as expected (Figure 3C), and p100 increased the ability of STAT6-TAD to stimulate transcription in this heterologous system in a dose-dependent manner. Expression of p100 did not affect the basal activity of the promoter. In summary, these results indicate that p100 acts as a coactivator in the STAT6-mediated transcriptional activation and that the TAD of STAT6 is indispensable for functional interaction with p100.
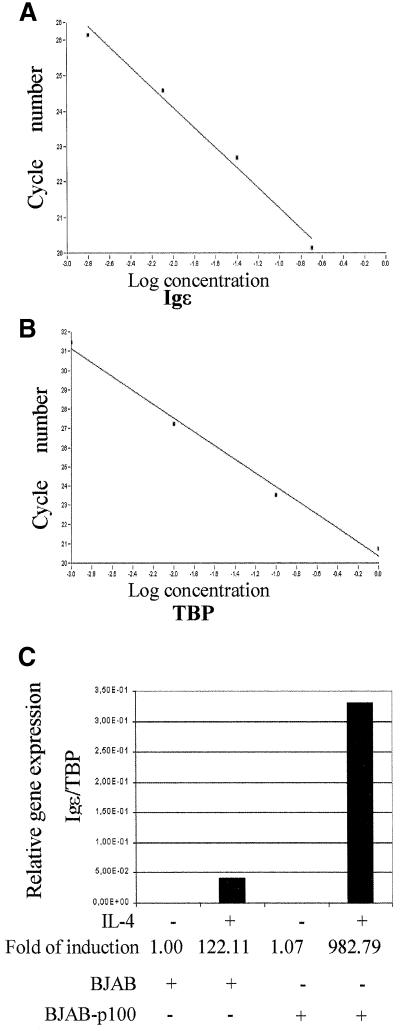
To confirm the p100-mediated enhancement of STAT6-dependent transcription in vivo, we analyzed the expression of Igε mRNA in BJAB B cells and in BJAB overexpressing p100 cells (Tong et al., 1995) by using real-time PCR analysis. The assay conditions were adjusted to allow quantitative comparison of Igε mRNA expression by normalizing the expression levels to the expression of the housekeeping TBP gene (Linja et al., 2001). As shown in Figure 4, stable overexpression of p100 did not affect the basal expression, but enhanced the IL-4-induced expression of Igε mRNA in human BJAB-p100 cells ∼8-fold when compared with parental cells.
Fig. 4. p100 enhances the transcription of Igε gene in BJAB cells. Real-time PCR analysis of Igε mRNA in BJAB and BJAB-p100 B-cell lines. Standard curves for Igε (A) and TBP (B) genes, plotting fractional cycle number at the fluorescent threshold on the y-axis and the logarithmic input cDNA concentration on the x-axis. (C) Relative expression of the Igε gene in untreated or IL-4-treated (100 ng/ml for 24 h) BJAB and BJAB-p100 B-cell lines. The results shown are representative of three experiments performed.
Mapping the interaction domain in p100 protein
To characterize further the interaction between p100 and STAT6, we delineated the domains of p100 engaged in their association. The structure prediction of p100 indicates that the N-terminus of the protein is composed of a repeat of four domains homologous to the SN-fold (Callebaut and Mornon, 1997), but their function is currently unknown. The C-terminus of p100 shows homology to the TD of Drosophila melanogaster tudor gene. The TD is found in proteins with putative functions in RNA-binding or protein-binding functions during RNA metabolism and transport (Ponting, 1997).
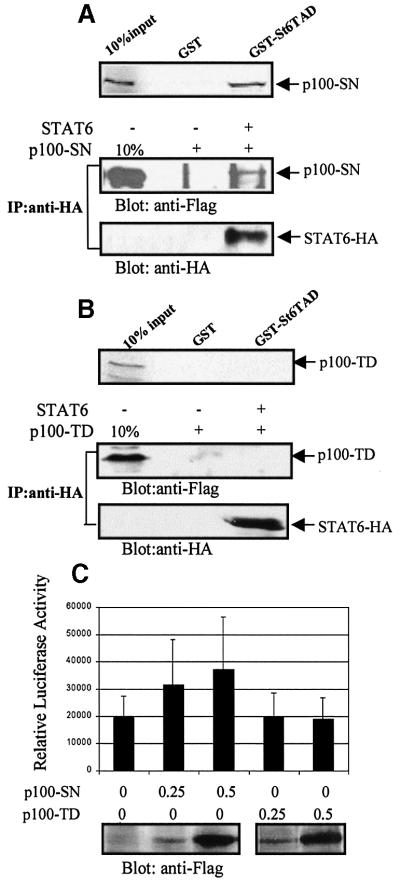
COS-7 cells were transfected with Flag-tagged expression constructs containing either the SN-like domains (p100-SN) or the TD of p100 (p100-TD), and GST pulldown assays were performed from the lysates. p100-SN was found to bind with GST–St6TAD fusion protein (Figure 5A, upper panel), while no interaction was detected with p100-TD (Figure 5B, upper panel). Consistent with the in vitro interaction results, in vivo association was detected only between STAT6 and p100-SN-like domain (Figure 5A and B, lower panels). Thus, the SN-like domain of p100 physically interacts with the TAD of STAT6. In reporter gene experiments, overexpression of the SN-like domain of p100 alone was sufficient to enhance STAT6-mediated reporter gene activity in response to IL-4 stimulation, although not as efficiently as the wild-type p100. Expression of the TD alone did not affect the activity of the reporter gene (Figure 5C).
Fig. 5. Mapping the STAT6 interaction domain of p100. (A) STAT6 associates with the SN-like domain of p100 (p100-SN) in vitro and in vivo. Upper panel: p100-SN interacts with STAT6-TAD in vitro. p100-SN was [35S]methionine labeled by in vitro translation and then incubated with beads loaded with GST or GST–St6TAD. After SDS–PAGE, the bound proteins were visualized by autoradiography. Lower panel: COS-7 cells were transfected with Flag-tagged p100-SN and STAT6-HA as indicated. Cell extracts were immunoprecipitated with anti-HA and blotted first with anti-Flag and then with anti-HA antibodies. (B) The TD of p100 (p100-TD) does not interact with STAT6. Upper panel: p100-TD was [35S]methionine-labeled by in vitro translation and then incubated with beads loaded with GST or GST–St6TAD, and the bound proteins were visualized by autoradiography. Lower panel: COS-7 cells were transfected with Flag-tagged p100-TD and STAT6-HA expression plasmids as indicated. Cell extracts were immunoprecipitated with anti-HA and blotted first with anti-Flag and then with anti-HA antibodies. (C) IL-4-induced transcriptional activation of STAT6 is enhanced by p100-SN but not by the TD of p100. HeLa cells were transfected with Igε luciferase reporter construct (0.5 µg), β-galactosidase vector (0.5 µg) and the indicated amounts of p100-SN or p100-TD expression plasmids, and treated with IL-4 for 6 h. The normalized luciferase values are shown. The expression levels of p100-SN and p100-TD are shown in the anti-Flag blot.
p100 interacts with RNA polymerase II in vitro and in vivo
Transcriptional coactivators usually function either through modifying chromatin organization or mediating scaffold functions. Since p100 has been shown to lack HAT activity (Wang et al., 2000), we considered the possibility that p100 might serve as a bridging factor between STAT6 and the basal transcription machinery. To investigate the potential interaction between p100 and RNA pol II, COS-7 cells were transfected with expression plasmids encoding for p100-Flag or STAT6-HA. Cell extracts were immunoprecipitated with antibody against the large subunit of RNA pol II or with the control antibody. As shown in Figure 6A, p100 coimmunoprecipitated with endogenous RNA pol II, but was not detected in the control immunoprecipitate. STAT6 was not detected in either anti-RNA pol II or control immunoprecipitates. The interaction between p100 with RNA pol II was also investigated by in vitro binding assays. Cell lysates were immunoprecipitated with antibodies against the large subunit of RNA pol II or unrelated control antibody under harsh conditions [1% NP-40, 0.1% sodium dodecyl sulfate (SDS), 300 mM NaCl], and the beads were washed with the lysis buffer to deplete any possibly associated proteins. The beads were then incubated in binding buffer with in vitro-translated p100 or STAT6, and after subsequent washes the RNA pol II, immunoprecipitate was subjected to SDS–PAGE and autoradiography. p100 associated with the immunopurified large subunit of RNA pol II, while no interaction could be detected between STAT6 and RNA pol II (Figure 6B). p100 and STAT6 were not detected in control immunoprecipitations. Next, we investigated whether p100 could function as a bridging factor between STAT6 and RNA pol II by adding exogenous p100 protein from p100-transfected 293T cells to the in vitro RNA pol II binding reactions. In vitro-translated STAT6 was detected in RNA pol II immunoprecipitates only in the presence of exogenous p100 protein (Figure 6C).
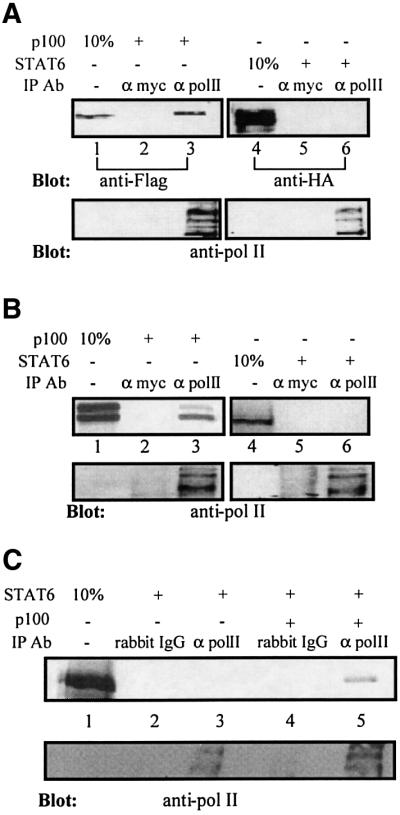
Fig. 6. p100 interacts with RNA polymerase II in vitro and in vivo. (A) p100 interacts with RNA polymerase II in vivo. COS-7 cells were transfected with p100-Flag or STAT6-HA expression plasmids. Upper panel: cell extracts were immunoprecipitated with anti-RNA pol II antibody (lanes 3 and 6) or unrelated antibody (anti-myc) as control (lanes 2 and 5) and immunoblotted with anti-Flag antibody for p100 or anti-HA antibody for STAT6. Anti-RNA pol II blot of the immunoprecipitates in the lower panel shows typical laddering due to differential phosphorylation and ubiquitylation. (B) p100 interacts with RNA polymerase II in vitro. Upper panel: HeLa cell lysates were immunoprecipitated with anti-RNA pol II antibody (lanes 3 and 6) or unrelated antibody (anti-myc) as control (lanes 2 and 5), and after extensive washes the immunoprecipitates were incubated with in vitro-translated and [35S]methionine-labeled p100 or STAT6. After washes, the bound proteins were subjected to SDS–PAGE and visualized by autoradiography. Lower panel: anti-RNA pol II blot of the immunoprecipitates. (C) HeLa cell lysates were immunoprecipitated with anti-RNA pol II antibody (lanes 3 and 5) or unrelated antibody (rabbit IgG) as control (lanes 2 and 4). After extensive washes, the immunocomplexes were incubated with in vitro-translated and [35S]methionine-labeled STAT6 together with control lysates lacking p100 (lanes 2 and 3) or with p100 protein lysate (lanes 4 and 5). After washes, the bound proteins were subjected to SDS–PAGE and visualized by autoradiography. Lower panel: anti-RNA pol II blot of the immunoprecipitates.
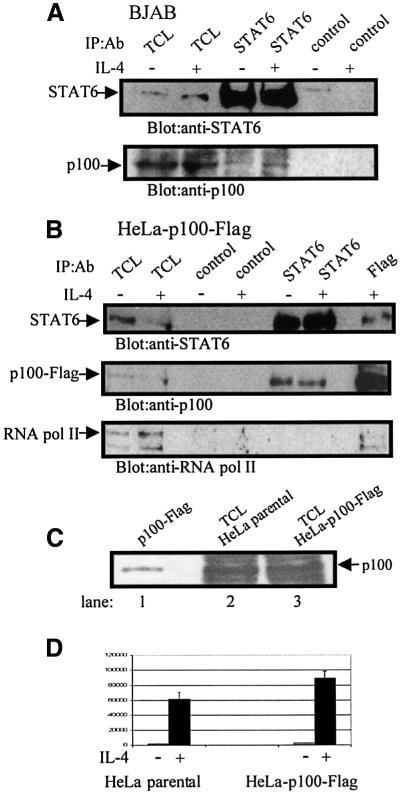
The interaction between endogenous p100 and STAT6 was analyzed by coimmunoprecipitation experiments in BJAB cells. p100 was consistently detected in anti-STAT6 immunoprecipitates, although at low level, and the interaction was IL-4-independent (Figure 7A). The polyclonal rabbit anti-p100 antibody does not work in immunoprecipitation, and subsequent experiments were performed in HeLa-p100-Flag stable cells, which allowed the use of better reagents for immunoprecipitation. This particular cell clone was chosen since it had only slightly increased p100 protein expression compared with the parental cells (Figure 7C). In functional assays, the HeLa-p100-Flag cells showed enhanced activation of Igε-luc in response to IL-4 stimulation (Figure 7D). As shown in Figure 7B, STAT6 and p100 readily coimmunoprecipitated in HeLa-p100-Flag cells. RNA pol II was detected in anti-Flag immunoprecipitates, but not in anti-STAT6 or control immunoprecipitates. Taken together, these results strongly suggest that p100 interacts with both STAT6 and RNA pol II, and also imply that p100 recruits RNA pol II to the STAT6-responsive promoter.
Fig. 7. p100 co-precipitates in vivo with STAT6 and RNA pol II. (A) p100 co-precipitates with STAT6 in intact BJAB cells. BJAB cells were treated with vehicle or IL-4 (30 min), and the lysates were immunoprecipitated with anti-STAT6 or anti-myc (control) antibodies. Upper panel shows blotting with anti-STAT6, and lower panel shows blotting with anti-p100. (B) p100 co-precipitates STAT6 and RNA pol II in HeLa-p100-Flag cells. Total cell lysates from vehicle or IL-4-treated (30 min) HeLa-p100-Flag cells were immunoprecipitated with anti-STAT6, anti-Flag and anti-myc (control) antibodies and subjected to anti-STAT6 (upper panel), anti-Flag (middle panel) or anti-RNA pol II (lower panel) immunoblotting. (C) Expression level of p100 in HeLa and HeLa-p100-Flag cells. Lane 1: immunoprecipitated p100-Flag from transfected COS-7 cells; lane 2: 25 µg of HeLa cell lysate; lane 3: 25 µg HeLa-p100-Flag cell lysate blotted with anti-p100 antiserum. (D) Activation of Igε-luc reporter gene in HeLa and HeLa-p100-Flag cells in response to IL-4 stimulation. HeLa and HeLa-p100-Flag cells were transfected with Igε luciferase reporter construct (0.5 µg) and β-galactosidase vector (0.5 µg) and treated with IL-4 as indicated. The normalized luciferase values are shown. Mean normalized luciferase values of three independent experiments with standard deviations are shown.
Discussion
A central, yet unresolved question related to cytokine-regulated gene transcription is the mechanism by which STATs are connected to activation of basal transcription machinery. Here, we show that the TAD of STAT6 is interacting with p100. p100 was found to enhance the STAT6-mediated transcription, and to bind to the large subunit of RNA pol II, thus providing a link between STAT6 and the general transcription apparatus.
p100 is a ubiquitously expressed protein that was initially identified as a protein interacting with the acidic TAD of EBNA2 (Tong et al., 1995). p100 enhanced transcriptional activity of EBNA2, but did not affect the function of another acidic TAD of VP16. Thus, p100 is not a general coactivator of transcription but its function requires specific protein interactions with transcription factors. Based on hydrophobic cluster analysis, the p100 protein is predicted to consist of four similar domains with homology to the SN domain, followed by a C-terminal TD (Callebaut and Mornon, 1997). The SN-like domains of p100 show homology to the oligonucleotide/oligosaccharide binding fold, suggesting that these domains may mediate interaction or binding functions. In line with this notion, our results demonstrate a stable interaction between the SN-like domain of p100 and STAT6-TAD both in vitro and in vivo. The STAT6-TAD is glutamine-rich, as opposed to the acidic TAD of EBNA2, thus suggesting that p100 is able to interact with different types of protein domains in transcription factors. The TD did not interact with STAT6 TAD, but the putative functions of TD in RNA metabolism warrant further study to address its function in STAT6-dependent transcription.
Transactivation domains recruit additional transcriptional co-regulators to the ligand-responsive promoters via physical association (Xu et al., 1999). The TADs of various STATs have been shown to associate and cooperate functionally with CBP/p300. Interestingly, the region of CBP that associates with STAT6 is different from the STAT1, STAT2 or STAT5 interaction domain (Gingras et al., 1999; Shuai, 2000). These findings indicate that the interaction mechanisms and composition of STAT–co-regulator complexes may vary between various STATs. The high degree of diversity among TADs of different STATs also supports this hypothesis. The mechanisms of STAT-mediated gene activation are currently characterized in most detail in relation to interferon-γ signaling, where STAT1-mediated gene activation involves CBP/p300 and P/CIP coactivators (Zhang et al., 1996; Torchia et al., 1997), and Ser727 phosphorylation regulated interaction with MCM5 proteins (Zhang et al., 1998). The TAD of STAT6 is rich in Pro, Ser, Thr, Leu and Glu residues, but lacks acidic residues commonly found in TADs. Serine phosphorylation of STAT6 (Pesu et al., 2000) may also increase its negative net charge and regulate the interaction with co-regulators. p100 has been linked to a signaling pathway from Pim-1 Ser/Thr kinase to c-Myb transcription factor (Leverson et al., 1998). The reported phosphorylation sites in p100 reside within the SN-like domain, therefore phosphorylation may potentially also regulate protein interactions between p100 and other transcription factors. Tyrosine phosphorylation of STAT6 per se was not required for the interaction with p100, but IL-4 stimulation induced nuclear entry of STAT6 and thereby nuclear colocalization of endogenous STAT6 and p100 proteins. p100 and STAT6 were uniformly distributed in the nucleus, which is consistent with a recent report demonstrating that activated STAT1 in living cells is not localized in defined structures, but diffuses freely in the nucleus (Lillemeier et al., 2001).
The mechanisms of p100-mediated transcriptional regulation have remained elusive. The SN-like domains of p100 lack residues required for the catalytic function, and p100 lacks HAT activity (Wang et al., 2000). We found that p100 enhanced transcriptional activation of STAT6-TAD without affecting tyrosine phosphorylation or DNA binding of STAT6. Our results suggested a direct interaction between p100 and the large subunit of RNA pol II, which plays an essential role in the regulation of initiation and elongation of transcription (Orphanides et al., 1996). In this interaction, however, we cannot completely exclude the possibility of an additional adapter with a high affinity for RNA pol II. The interactions between transcription factors and coactivators are generally rather weak, but p100 coimmunoprecipitated with STAT6 and RNA pol II under physiological conditions, and ternary complex formation between STAT6, p100 and RNA pol II was detected in vitro. Together, these results suggest that p100 functions as a bridging protein between STAT6 and the basal transcription machinery. Consistent with this notion, p100 has also been shown to be able to interact with TFIIE in vitro (Tong et al., 1995) and to be a component of the RNA pol II holoenzyme (K.Carter and E.Kieff, manuscript in preparation).
The interaction between STAT6 and p100 may also provide some interesting insight related to the mechanism of EBV-mediated gene activation and transformation. EBNA2 plays an important role in EBV-mediated transformation by inducing the expression of a viral gene encoding LMP1 and cellular genes such as CD23 (Wang et al., 1990). LMP1 shows functional similarity to CD40 and directly activates the JAK/STAT pathway (Gires et al., 1999). CD23, on the other hand, is a direct target gene for STAT6 (Tinnell et al., 1998) and the STAT6 response element of the CD23 promoter is localized within the EBNA2-responsive region (Wang et al., 1990). Interest ingly, EBNA2 and LMP1 cooperatively induce CD23 expression (Wang et al., 1990). EBNA2 has been shown to bind sequence-specific transcription factors such as PU.1, and by analogy a possible mechanism underlying the cooperation between EBNA2 and LMP1 could involve interaction between EBNA2 and STAT6, and formation of a higher-order complex of STAT6, EBNA2 and p100.
Transcriptional activation of cytokine-responsive genes requires coordinated cooperation between STATs and other sequence-specific transcription factors that recruit transcriptional co-regulators and components of the basal transcription machinery to the promoter. The results obtained from the estrogen receptor (ER)-mediated transcription propose a dynamic cofactor recruitment model, where ligand-activated ER first recruits HAT containing coactivators that modify chromatin structure, and thereby allows RNA pol II recruitment and its subsequent phosphorylation and progression of transcriptional elongation (Shang et al., 2000). Analogous sequence of events may also apply for STAT6-mediated transcription. Previously, CBP has been shown to associate and cooperate with STAT6, and the function of CBP appears to involve modulation of chromatin structure through histone acetylation (Shankaranarayanan et al., 2001). The interaction between STAT6, p100 and RNA pol II provide the first insight into the mechanisms underlying the connection between STATs and basal transcription machinery. It is, however, likely that the STAT6-transcriptional activation complex is a higher-order network involving additional co-regulators and cell-specific factors that contribute to the selective regulation of IL-4-responsive genes.
Materials and methods
Cell culture and transfection
Ramos 2G6 cells, 293T cells, HeLa cells, COS-7 cells, EBV-negative human Burkitt’s lymphoma B-cell line BJAB and p100-overexpressing BJAB cells were cultured as described previously (Tong et al., 1995; Pesu et al., 2000). Transfections of HeLa and 293T cells were done by the calcium-phosphate co-precipitation method (Paukku et al., 2000). Ramos cells were transfected by electroporation at 200 V/960 µF, and COS-7 cells were electroporated at 260 V/960 µF with a Bio-Rad gene pulser (Saharinen et al., 2000). The HeLa-p100-Flag stable cell line was cloned by cotransfecting pSG5-p100-Flag plasmid with hygromycin B DNA using the FUGENE 6 Transfection Reagent (Roche Diagnostics).
Plasmids
GST–St6TAD was constructed by cloning PCR products corresponding to amino acids (aa) 642–847 of human STAT6 into pGEX-4T-1 vector (Pharmacia). pCIneo-STAT6-HA was generated by PCR using primers containing the HA-epitope sequence inserted into the C-terminus of STAT6. GAL4–STAT6-TAD was kindly provided by Dr B.Groner (Moriggl et al., 1997). pSG5-p100-Flag was constructed by PCR amplification of full-length open reading frame from pBp100 and cloned into pSG5-Flag vector. The pSG5 vector expression plasmids containing SN-like domains of human p100 protein (aa 1–639) and the TD of p100 (aa 640–885) were generated by PCR using primers containing C-terminal Flag sequence. All PCR products were sequenced. The SN domain was cloned into pcDNA3 vector in reverse orientation to yield anti-sense-p100 plasmid. Igε-reporter and C/EBP reporter construct were as described previously (Pesu et al., 2000).
GST pulldown assays
For metabolic labeling, Ramos cells were incubated for 16 h at 37°C in the labeling medium (Dulbecco’s modified Eagle’s medium, 10 mM sodium pyruvate, 20 mM l-glutamine, 10% fetal bovine serum and 0.5 mCi/ml [35S]methionine–[35S]cysteine). GST pulldown experiments were performed essentially as described previously (Zhang et al., 1998). Briefly, 35S-labeled and unlabeled Ramos cells were washed once with ice-cold phosphate-buffered saline and then resuspended in ice-cold buffer A (20 mM HEPES pH 7.4, 1 mM EDTA, 10 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF, 50 mM NaF, 10 mM Na-pyrophos phate, 2 mM Na-molybdate, 2 µg/ml aprotinin). Nuclear fraction was collected by centrifugation for 5 min at 500 g and suspended in ice-cold buffer C (20 mM HEPES pH 7.4, 20% glycerol, 300 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 50 mM NaF, 10 mM Na-pyrophosphate, 2 mM Na-molybdate, 2 µg/ml aprotinin).
For binding assays, purified GST–St6TAD and GST proteins were bound on glutathione–Sepharose beads and then incubated with nuclear lysates or with in vitro-translated 35S-labeled p100 in binding buffer (12.5 mM HEPES pH 7.4, 0.1 mM EDTA, 0.05% NP-40, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 2 µg/ml aprotinin, 0.5% bovine serum albumin). After washing, the bound proteins were eluted with high-salt buffer, separated by SDS–PAGE and visualized by silver staining or autoradiography.
Mass spectrometry
The proteins separated by SDS–PAGE were excised and digested with trypsin as described previously (Nyman et al., 1998). The molecular masses of the peptide mixtures were determined by MALDI-TOF mass spectrometry. The molecular masses of the tryptic peptides of the 100 kDa protein were used to search the OWL protein sequence database for candidate proteins using the ProFound program.
Luciferase assay
Twenty-four hours after transfection, HeLa cells were starved overnight and stimulated with 10 ng/ml recombinant human IL-4 (PeproTech EC, London, UK) for 6 h. Ramos cells were stimulated with IL-4 without starvation. Forty-eight hours after transfection, cells were lysed and luciferase activity was measured as described previously (Saharinen et al., 1997). The luciferase values were normalized to β-galactosidase activity and are presented as the mean relative luciferase activity of three independent experiments. For all experiments, empty pSG5 vector DNA was used to balance the different amounts of DNA used in various transfections.
Immunoprecipitation and antibodies
Immunoprecipitations were carried out as described previously using 0.5 or 1% NP-40 lysis buffer (Saharinen et al., 2000) or RIPA lysis buffer (1% NP-40, 0.1% SDS, 300 mM NaCl, 50 mM Tris–HCl pH 8.0, 0.1 mM EDTA, 20% glycerol, 0.1 mM sodium orthovanadate). RIPA buffer was used for RNA pol II immunoprecipitation. Briefly, the lysates were immunoprecipitated with antibody against the large subunit of RNA pol II (Santa Cruz Biotechnology), and after extensive washes in RIPA buffer the beads were incubated with in vitro-translated 35S-labeled p100 protein or 35S-labeled STAT6 in binding buffer for 3 h at 4°C and then washed four times with binding buffer and separated by SDS–PAGE followed by autoradiography.
Immunodetection was performed using enhanced chemiluminescence (Saharinen et al., 2000). The following antibodies were used: anti-STAT6 antibody (ZYMED Laboratory, Inc.) anti-Flag M2 (Sigma), anti-HA (clone 16B12; BAbCO) and anti-myc (Boehringer Mannheim).
Quantification of Igε gene expression using real-time PCR
Total RNA was isolated from BJAB and BJAB-p100 cells using Trizol reagent (Life Technologies, Inc.) and used for the first-strand cDNA synthesis with M-MLV reverse transcriptase (Gibco-BRL) and random hexamers (Amersham Pharmacia) according to the manufacturer’s protocol. Total RNA from BJAB cells stimulated with IL-4 (100 ng/ml) for 48 h was used for preparing standard curve. After the first-strand cDNA synthesis, serial dilutions were made to correspond to cDNA transcribed from 750, 75, 7.5 and 0.75 ng of total RNA. Primers for the Igε and TBP have been described previously (Thienes et al., 1997; Linja et al., 2001).
The PCR reactions were performed in the LightCycler apparatus using the LightCycler-FastStart DNA Master SYBR Green I kit (Roche Diagnostics). The cycling conditions (45 cycles) consisted of denaturation at 95°C for 15 s (Igε) or 3 s (TBP), annealing at 55°C for 8 s (Igε) or 58°C for 13 s (TBP) and elongation at 72°C for 16 s (Igε) or 7 s (TBP). The LightCycler apparatus measured the fluorescence of each sample in every cycle at the end of the elongation step. After the PCR reaction, the logarithmic values of fluorescence were plotted against cycle number, and the relative Igε expression was calculated as described previously (Linja et al., 2001). PCR samples were also run in 1.2% agarose electrophoretic gels to ensure that a right-size product was amplified in the reaction.
Acknowledgments
Acknowledgements
We thank Paula Kosonen for excellent technical assistance, Dr B.Groner for reagents, Dr A.Tienhaara for FACS analysis and Drs T.Visakorpi, K.Savinainen and S.Kares for help with real-time PCR analysis. This work was supported by grants from the Academy of Finland, Medical Research Fund of Tampere University Hospital, CIMO, the Finnish Foundation for Cancer Research, the Sigrid Jusélius Foundation and the Tuberculosis Foundation of Tampere.
References
- Callebaut I. and Mornon,J.P. (1997) The human EBNA-2 coactivator p100: multidomain organization and relationship to the staphylococcal nuclease fold and to the tudor protein involved in Drosophila melanogaster development. Biochem. J., 321, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomarat P. and Banchereau,J. (1997) An update on interleukin-4 and its receptor. Eur. Cytokine Netw., 8, 333–344. [PubMed] [Google Scholar]
- Dash A.B., Orrico,F.C. and Ness,S.A. (1996) The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev., 10, 1858–1869. [DOI] [PubMed] [Google Scholar]
- Gingras S., Simard,J., Groner,B. and Pfitzner,E. (1999) p300/CBP is required for transcriptional induction by interleukin-4 and interacts with Stat6. Nucleic Acids Res., 27, 2722–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gires O. et al. (1999) Latent membrane protein 1 of Epstein–Barr virus interacts with JAK3 and activates STAT proteins. EMBO J., 18, 3064–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.R. (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci., 25, 59–63. [DOI] [PubMed] [Google Scholar]
- Gupta S., Jiang,M. and Pernis,A.B. (1999) IFN-α activates Stat6 and leads to the formation of Stat2:Stat6 complexes in B cells J. Immunol., 163, 3834–3841. [PubMed] [Google Scholar]
- Hou J., Schindler,U., Henzel,W.J., Ho,T.C., Brasseur,M. and McKnight,S.L. (1994) An interleukin-4-induced transcription factor: IL-4 Stat. Science, 265, 1701–1706. [DOI] [PubMed] [Google Scholar]
- Johnston J.A., Kawamura,M., Kirken,R.A., Chen,Y.Q., Blake,T.B., Shibuya,K., Ortaldo,J.R., McVicar,D.W. and O’Shea,J.J. (1994) Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature, 370, 151–153. [DOI] [PubMed] [Google Scholar]
- Kuhn R., Rajewsky,K. and Muller,W. (1991) Generation and analysis of interleukin-4 deficient mice. Science, 254, 707–710. [DOI] [PubMed] [Google Scholar]
- Leverson J.D., Koskinen,P.J., Orrico,F.C., Rainio,E.M., Jalkanen,K.J., Dash,A.B., Eisenman,R.N. and Ness,S.A. (1998) Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell, 2, 417–425. [DOI] [PubMed] [Google Scholar]
- Lillemeier B.F., Koster,M. and Kerr,I.M. (2001) STAT1 from the cell membrane to the DNA. EMBO J., 20, 2508–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linja M.J., Savinainen,K.J., Saramaki,O.R., Tammela,T.L., Vessella,R.L. and Visakorpi,T. (2001) Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res., 61, 3550–3555. [PubMed] [Google Scholar]
- Litterst C.M. and Pfitzner,E. (2001) Transcriptional activation by STAT6 requires the direct interaction with NCoA-1. J. Biol. Chem., 276, 45713–45721. [DOI] [PubMed] [Google Scholar]
- McDonald C. and Reich,N.C. (1999) Cooperation of the transcriptional coactivators CBP and p300 with Stat6. J. Interferon Cytokine Res., 19, 711–722. [DOI] [PubMed] [Google Scholar]
- Mikita T., Campbell,D., Wu,P., Williamson,K. and Schindler,U. (1996) Requirements for interleukin-4-induced gene expression and functional characterization of Stat6. Mol. Cell. Biol., 16, 5811–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikita T., Kurama,M. and Schindler,U. (1998) Synergistic activation of the germline epsilon promoter mediated by Stat6 and C/EBP β. J. Immunol., 161, 1822–1828. [PubMed] [Google Scholar]
- Moriggl R. et al. (1997) Comparison of the transactivation domains of Stat5 and Stat6 in lymphoid cells and mammary epithelial cells. Mol. Cell. Biol., 17, 3663–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C. and Yaniv,M. (1999) The mammalian SWI/SNF complex and the control of cell growth. Semin. Cell Dev. Biol., 10, 189–195. [DOI] [PubMed] [Google Scholar]
- Nelms K., Keegan,A.D., Zamorano,J., Ryan,J.J. and Paul,W.E. (1999) The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol., 17, 701–738. [DOI] [PubMed] [Google Scholar]
- Nyman T.A., Tolo,H., Parkkinen,J. and Kalkkinen,N. (1998) Identi fication of nine interferon-α subtypes produced by Sendai virus-induced human peripheral blood leucocytes. Biochem. J., 329, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- Paukku K., Valgeirsdottir,S., Saharinen,P., Bergman,M., Heldin,C.H. and Silvennoinen,O. (2000) Platelet-derived growth factor (PDGF)-induced activation of signal transducer and activator of transcription (Stat) 5 is mediated by PDGF β-receptor and is not dependent on c-src, fyn, jak1 or jak2 kinases. Biochem. J., 345, 759–766. [PMC free article] [PubMed] [Google Scholar]
- Pesu M., Takaluoma,K., Aittomaki,S., Lagerstedt,A., Saksela,K., Kovanen,P.E. and Silvennoinen,O. (2000) Interleukin-4-induced transcriptional activation by stat6 involves multiple serine/threonine kinase pathways and serine phosphorylation of stat6. Blood, 95, 494–502. [PubMed] [Google Scholar]
- Ponting C.P. (1997) Tudor domains in proteins that interact with RNA. Trends Biochem. Sci., 22, 51–52. [DOI] [PubMed] [Google Scholar]
- Quelle F.W. et al. (1995) Cloning of murine Stat6 and human Stat6, Stat proteins that are tyrosine phosphorylated in responses to IL-4 and IL-3 but are not required for mitogenesis. Mol. Cell. Biol., 15, 3336–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen P., Ekman,N., Sarvas,K., Parker,P., Alitalo,K. and Silvennoinen,O. (1997) The Bmx tyrosine kinase induces activation of the Stat signaling pathway, which is specifically inhibited by protein kinase Cdelta. Blood, 90, 4341–4353. [PubMed] [Google Scholar]
- Saharinen P., Takaluoma,K. and Silvennoinen,O. (2000) Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol. Cell. Biol., 20, 3387–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Shankaranarayanan P., Chaitidis,P., Kuhn,H. and Nigam,S. (2001) Acetylation by histone acetyltransferase CREB-binding protein/p300 of STAT6 is required for transcriptional activation of the 15-lipoxygenase-1 gene. J. Biol. Chem., 276, 42753–42760. [DOI] [PubMed] [Google Scholar]
- Shen C.H. and Stavnezer,J. (1998) Interaction of stat6 and NF-κB: direct association and synergistic activation of interleukin-4-induced transcription. Mol. Cell. Biol., 18, 3395–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. and Darnell,J.E.Jr (2001) Antiviral response in cells containing Stat1 with heterologous transactivation domains. J. Virol., 75, 2627–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda K. et al. (1996) Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature, 380, 630–633. [DOI] [PubMed] [Google Scholar]
- Shuai K. (2000) Modulation of STAT signaling by STAT-interacting proteins. Oncogene, 19, 2638–2644. [DOI] [PubMed] [Google Scholar]
- Stutz A.M. and Woisetschlager,M. (1999) Functional synergism of STAT6 with either NF-κB or PU.1 to mediate IL-4-induced activation of IgE germline gene transcription. J. Immunol., 163, 4383–4391. [PubMed] [Google Scholar]
- Thienes C.P., De Monte,L., Monticelli,S., Busslinger,M., Gould,H.J. and Vercelli,D. (1997) The transcription factor B cell-specific activator protein (BSAP) enhances both IL-4- and CD40-mediated activation of the human epsilon germline promoter. J. Immunol., 158, 5874–5882. [PubMed] [Google Scholar]
- Tinnell S.B., Jacobs-Helber,S.M., Sterneck,E., Sawyer,S.T. and Conrad,D.H. (1998) STAT6, NF-κB and C/EBP in CD23 expression and IgE production. Int. Immunol., 10, 1529–1538. [DOI] [PubMed] [Google Scholar]
- Tong X., Drapkin,R., Yalamanchili,R., Mosialos,G. and Kieff,E. (1995) The Epstein–Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol., 15, 4735–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia J., Rose,D.W., Inostroza,J., Kamei,Y., Westin,S., Glass,C.K. and Rosenfeld,M.G. (1997) The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature, 387, 677–684. [DOI] [PubMed] [Google Scholar]
- Wang F., Gregory,C., Sample,C., Rowe,M., Liebowitz,D., Murray,R., Rickinson,A. and Kieff,E. (1990) Epstein–Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol., 64, 2309–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Grossman,S.R. and Kieff,E. (2000) Epstein–Barr virus nuclear protein 2 interacts with p300, CBP and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl Acad. Sci. USA, 97, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witthuhn B.A., Silvennoinen,O., Miura,O., Lai,K.S., Cwik,C., Liu,E.T. and Ihle,J.N. (1994) Involvement of the Jak-3 Janus kinase in signalling by interleukins 2 and 4 in lymphoid and myeloid cells. Nature, 370, 153–157. [DOI] [PubMed] [Google Scholar]
- Xu L., Glass,C.K. and Rosenfeld,M.G. (1999) Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev., 9, 140–147. [DOI] [PubMed] [Google Scholar]
- Zhang J.J., Vinkemeier,U., Gu,W., Chakravarti,D., Horvath,C.M. and Darnell,J.E.,Jr (1996) Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc. Natl Acad. Sci. USA, 93, 15092–15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.J., Zhao,Y., Chait,B.T., Lathem,W.W., Ritzi,M., Knippers,R. and Darnell,J.E.,Jr (1998) Ser727-dependent recruitment of MCM5 by Stat1α in IFN-γ-induced transcriptional activation. EMBO J., 17, 6963–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]