Abstract
Nucleosome sliding is a frequent result of energy-dependent nucleosome remodelling in vitro. This review discusses the possible roles for nucleosome sliding in the assembly and maintenance of dynamic chromatin and for the regulation of diverse functions in eukaryotic nuclei.
Keywords: ATPase/chromatin assembly/chromosome structure/nucleosome positioning/remodelling
Introduction
All fundamental processes with chromatin substrate in eukaryotic nuclei, be it the replication of the genome, the transcription of genes, the sensing and repair of DNA damage, or the generation of genetic diversity through recombination, are initiated and regulated by DNA-binding proteins that scan the chromatin fibre in search of their preferred recognition sequences. Examples where active transcription factors are present in nuclei but are hindered from interacting with regulatory elements (Becker et al., 1987) highlight the discriminating role of the chromatin organization for factor access. Inaccessible chromatin is commonly characterized by DNA methylation and distinct patterns of histone modification. Rendering genes accessible involves unfolding of higher order structures through targeting of enzymes that alter these modifications (Jenuwein and Allis, 2001). At the level of the chromatin fibre, a second type of remodelling enzyme is needed; ATP-dependent nucleosome remodelling factors modulate chromatin structure in more subtle ways. They use the chemical energy freed by ATP hydrolysis to transiently disrupt the histone–DNA interactions that characterize canonical nucleosomes (Gregory et al., 2001; Becker and Hörz, 2002; Narlikar et al., 2002). Their action increases the accessibility of nucleosomal DNA and facilitates the relocation of histone octamers to adjacent DNA segments (‘sliding’), and may even lead to displacement of a histone octamer to a different DNA segment. Short-range nucleosome movements in cis seem to be a particularly attractive principle by which the overall packaging of DNA is maintained, yet rendered ‘transparent’ through stochastic exposure of individual DNA segments in the more accessible linkers between nucleosomes. Energy-dependent nucleosome remodelling not only serves to open chromatin structure, but is also involved in gene repression, chromatin assembly and the maintenance of higher order chromosome structure. Here, I will collect the available evidence for catalysed nucleosome sliding and speculate on the possible contributions of nucleosome sliding for chromatin dynamics in vivo.
Nucleosome sliding: early observations
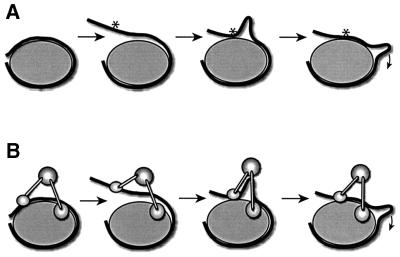
As the DNA double helix spools around the histone octamer to create a nucleosome core particle, it contacts the histone surface at 14 sites with clusters of hydrogen bonds and salt links (Luger and Richmond, 1998). Collectively, these weak interactions render the nucleosome a rather stable particle. Yet passive (non-catalysed) movement of histone octamers on DNA has been observed at moderately elevated temperature and ionic strength (Beard, 1978; Pennings et al., 1991; Meersseman et al., 1992). Dislocation of nucleosomal DNA requires that all interactions are broken and reformed. Disrupting all bonds at the same time would presumably lead to a prohibitive free energy penalty. Current models for passive histone octamer movement on DNA therefore assume that only segments of DNA (involving only few histone–DNA contacts) are peeled off the histone surface at any given time (Van Holde and Yager, 1985; Widom, 1999; Schiessel et al., 2001). The DNA at the ‘edge’ of the nucleosome comes off the histone surface most easily. Thermal twisting or other distortion of this DNA, such as bending it into a tight loop or ‘bulge’ will lead to the reformation of equivalent but non-identical histone–DNA interactions (Figure 1A), and the nucleosome would then contain a segment of distorted DNA (a ‘defect in stored length’; Schiessel et al., 2001) ‘looping’ from the nucleosome. Diffusion of this distortion over the particle until it emerges on the other side will lead to complete translocation of the DNA relative to hallmarks on the octamer (Figure 1A).
Fig. 1. Loop propagation model for passive (A) and active (B) nucleosome sliding. The nucleosome is represented by a grey ellipse around which the DNA (black string) winds. The initial steps of nucleosome mobilization are depicted. The asterisk represents a hallmark on the DNA. The remodelling machinery, of which no structure is known, is represented schematically by rod-connected spheres.
Nucleosome sliding catalysed by ATP-dependent nucleosome remodelling factors
Low levels of spontaneous nucleosome mobility may occur under physiological conditions, but it seems that the cell does not take chances. Rather, it invented nucleosome remodelling enzymes that lower the energy barrier, which limits spontaneous nucleosome movements, by coupling the disruption of histone–DNA contacts to ATP hydrolysis. The ATP-dependent (‘active’) sliding of nucleosomes over distances of up to 100 bp in arrays with physiological properties was first observed during the characterization of chromatin reconstituted in vitro in Drosophila embryo extracts (Varga-Weisz et al., 1995). During the following years, a zoo of ATP-consuming activities involved in chromatin metabolism were identified in these extracts, such as the nucleosome remodelling factor (NURF; Tsukiyama and Wu, 1995; Tsukiyama et al., 1995), the ATP-utilizing chromatin assembly and remodelling factor (ACF; Ito et al., 1997a) and the chromatin accessibility complex (CHRAC; Varga-Weisz et al., 1997). The shared subunit of these three complexes, the ATPase ISWI, endows these machineries with the ability to catalyse nucleosome sliding (for a review, see Längst and Becker, 2001b).
ISWI belongs to the large SWI2/SNF2 family of ATPases, members of which are involved in all prominent nuclear processes (Eisen et al., 1995). According to distinctive structural features, these ATPases define subfamilies, the prominent ones being represented by the bromodomain-containing SWI2/SNF2, by ISWI with C-terminal SANT-like modules and the chromodomain ATPase Mi-2 (Becker and Hörz, 2002; Narlikar et al., 2002). Despite the very different domain organization of the remodelling ATPases and their association with a variety of other subunits, all remodelling complexes are able to induce the movement of intact histone octamers on DNA in cis (Hamiche et al., 1999; Längst et al., 1999; Brehm et al., 2000; Guschin et al., 2000; Jaskelioff et al., 2000). However, whereas for ISWI-containing factors nucleosome sliding seems to be the predominant out come of nucleosome remodelling, ATPases related to yeast SWI2/SNF2 may also catalyse the disruption of histone–DNA interaction without relocation of the octamer or, at the other extreme, the complete dislocation of a histone octamer to free, competing DNA (Lorch et al., 1999; Phelan et al., 2000; Narlikar et al., 2001).
Whether, under physiological circumstances, nucleosomes will move on DNA in cis (sliding) or be shuffled to acceptor DNA in trans will undoubtedly depend on the nucleosome density and the availability of free DNA of suitable length close-by. My bias is that the former reaction is more likely to occur and that the inherent flexibility of the fibre (Woodcock and Dimitrov, 2001) will permit significant, short range nucleosome sliding.
Nucleosome sliding, chromatin assembly and nucleosome fibre folding
We envision global needs for nucleosome mobility, endowing the chromatin fibre with ‘fluidity’ and transparency. A first task may be posed by replication, when the nucleosomes of the parental strand are randomly distributed to the daughter strands and the resulting gaps in the fibre are filled by assembly of new octamers. Discon tinuities of the chromatin fibre may also arise throughout the cell cycle when DNA-binding regulators vacate their targets. It is unknown how the continuity of the nucleosomal fibre is assured in vivo, but in cell-free model systems, regular nucleosomal arrays are generated by an energy-dependent process that can be catalysed by some (but interestingly not all) nucleosome remodelling factors of the ISWI-type (Ito et al., 1997b; Varga-Weisz et al., 1997; Tsukiyama et al., 1999). Uniformity of inter-nucleosomal distances may define a low-energy state of the system since gaps in nucleosome arrays prevent the folding of the fibre (Fletcher and Hansen, 1996). Although counter-intuitive at first glance, it appears that the same factors that render DNA in chromatin accessible also have the potential to improve the folding of chromatin into higher order structures through their ‘spacing’ capability. A role for ISWI in the maintenance of chromosome integrity has recently been inferred from a particular phenotypic aspect of flies lacking ISWI. Analysing the polytene chromosomes of Drosophila larvae that die due to lack of ISWI, Tamkun and colleagues found the X chromosomes of male larvae massively deformed, consistent with a global impairment of chromatin folding (Deuring et al., 2000).
Once the nucleosomal fibre is properly assembled, ‘nucleosome spacing’ factors such as CHRAC and ACF may then continue to keep it in a ‘vibrant’ state, flexible to permit the interaction of DNA-binding regulators. Even movements of DNA relative to the histone surface over very short distances may have significant impact on factor binding. Some regulators are able to interact with DNA bent over the histone octamer, if their short recognition sequence faces outward from the histone surface (Beato and Eisfeld, 1997). Translocating a histone octamer by just 5 bp will expose any given sequence that was previously occluded by the histones. More profound movements will further change the accessibility; the closer a given DNA sequence to the nucleosome edge, the more accessible it will be. An important consequence of the existence of principles that assure global transparency of chromatin is that any DNA-binding regulator may profit, regardless whether its function is to activate or to repress, to open or to close chromatin.
Nucleosome sliding, nucleosome positioning and promoter architecture
Specific positioning of nucleosomes with respect to the underlying DNA sequence (as opposed to random localization of histone octamers in a population of genomes) is frequently observed close to regulatory elements (Wallrath et al., 1994). Are these positioned nucleosomes subject to the same kind of short distance motion or are they somehow exempt from energy-dependent mobilization? High resolution mapping after in vivo cross-linking revealed that even nucleosomes that appear tightly positioned at first glance, display a continuum of translational positions around preferred sites (Buttinelli et al., 1993; Fragoso et al., 1995; Tanaka et al., 1996). Presumably, these positions are in constant exchange in vivo due to nucleosome sliding and the mapping procedure captured a ‘snapshot’ of the equilibrium at the time of cross-linking.
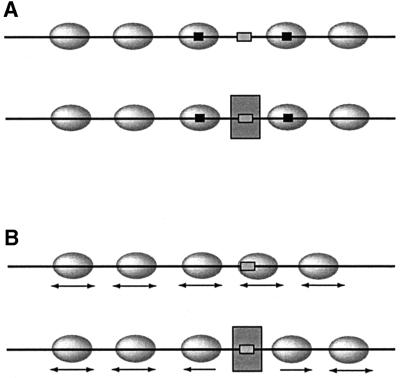
It is often assumed that the DNA structure around critical regulatory sites has a defining effect on nucleosome positions, such that, for example, binding sites for regulators are left nucleosome-free (Figure 2A). Since, at least in vitro, nucleosome remodelling ATPases are able to slide nucleosomes off even the strongest nucleosome positioning sequences (G.Längst, J.Widom and P.B.Becker, unpublished data), an alternative scenario must also be considered. Energy-dependent nucleosome mobilization may assure that histone octamers adopt all possible positions on a given DNA segment for a fraction of the time. A DNA-binding regulator may take advantage of the ‘window of opportunity’ to interact with its binding site when it is transiently exposed. Importantly, its binding will create a ‘boundary’, further constraining nucleosome movement. Nucleosome mobility will allow optimizing nucleosome positions according to the tendency of the system to avoid molecular clashes while maximizing the neutralization of DNA (Figure 2B). Adjustments of neighbouring nucleosomes in the array will lead to ‘nucleosome phasing’ with respect to a boundary. This principle, by which non-random nucleosome positions are brought about by an essentially stochastic mechanism, has already been envisioned early on by Kornberg and Stryer (1988), and supported by experiments in cell-free systems (Varga-Weisz et al., 1995; Kang et al., 2002) and is also consistent with in vivo observations (Fedor et al., 1988; Lomvardas and Thanos, 2001).
Fig. 2. Two models for the creation of promoter architecture. (A) Nucleosomes (ellipsoids) are positioned due to dedicated sequence elements (black boxes) such that the binding site (small grey box) for a regulator (large grey box) is left accessible. (B) Nucleosomes are not positioned but mobile. Due to statistical movement, the binding site for a regulator is transiently exposed. The interaction of a regulator brings about a positioning of adjacent nucleosomes.
Targeting nucleosome lubricants
While the proposed global functions of ATP-dependent nucleosome remodelling factors are still speculative to date, there is good evidence for the targeting of these enzymes to specific sites. Numerous interactions between sequence-specific transcription regulators and components of nucleosome remodelling complexes have been described. Direct or indirect recruitment by DNA-binding regulators along with effects on transcription suggests roles for ATP-dependent nucleosome remodelling factors in gene activation as well as repression (for reviews, see Peterson and Logie, 2000; Becker and Hörz, 2002; Narlikar et al., 2002). In some cases, targeting of remodelling factors to promoters has been directly correlated with changes in nucleosome positions in vivo (Goldmark et al., 2000; Kent et al., 2001; Lomvardas and Thanos, 2001). A particularly illustrative example is the targeting of the yeast Isw2 complex to the REC104 promoter via the DNA bound repressor Ume6p, leading to shifts in nucleosome positions towards the repressed state of the promoter (Goldmark et al., 2000). Other targeting principles may include interactions with components of constitutive heterochromatin (Bozhenok et al., 2002), and methylated DNA (Wade et al., 1999).
The workings of nucleosome sliding
The mechanisms by which nucleosome remodelling factors induce the sliding of histone octamers on DNA are still unknown. However, the ongoing analysis of different nucleosome remodelling factors in cell-free systems is providing a rich phenomenology from which general outlines can be derived. It is conceivable that catalysed nucleosome sliding is mechanistically related to passive nucleosome movements. For example, in the context of the model for passive nucleosome movement discussed above (Figure 1A), each of the following steps may be facilitated by enzymes: the peeling off of a segment of DNA from the histone octamer surface, the distortion of this DNA into a bulge, as well as the propagation of this DNA deformation over the histone octamer (Figure 1B).
Since nucleosome remodelling factors are very different in terms of subunit and domain composition, an interesting question is whether a unified mechanism for nucleosome sliding exists or whether individual machineries have their own approach to altering histone–DNA interactions. A common reaction catalysed by various ATPases is the distortion of DNA leading to torsional stress within a constrained domain (Havas et al., 2000; Gavin et al., 2001; Längst and Becker, 2001a; Liu et al., 2001). Notable differences in nucleosome remodelling phenomenology are particularly obvious if ISWI-type and SWI2/SNF2-type ATPases are compared (for a review, see Flaus and Owen-Hughes, 2001): (i) octamer transfer to competing DNA so far has only been observed in reactions driven by SWI2/SNF2-type ATPases; (ii) these ATPases cause obvious alterations of histone–DNA interactions within mononucleosomes, whereas such an ‘altered path’ of DNA has not been observed during ISWI-catalysed nucleosome sliding; (iii) a stable ‘remodelled state’ that can be best described as ‘nucleosome dimers with partially peeled off DNA’ (Schnitzler et al., 2001; and references therein) can be observed after remodelling with SWI/SNF, but not with ISWI; and (iv) ISWI requires the N-terminus of histone H4 for a productive ATPase cycle, whereas SWI/SNF-type enzymes can deal with nucleosomes lacking these domains. To date, all phenomenology related to reactions catalysed by ISWI can easily be explained by assuming that the sliding of intact histone octamers is the main outcome of the remodelling reaction. By contrast, some aspects of remodelling by SWI2/SNF2 (BRG1) points to changes in histone–DNA interactions without nucleosome movement.
Even though these distinctions are obvious, it may be premature to conclude about fundamentally different remodelling strategies. Conceivably, a overall similar in vitro remodelling reaction could lead to differential accumulation of intermediates, dead-end products or bona fide end products, depending on the reaction conditions. Intermediates of a nucleosome sliding reaction, such as looped-out DNA on the surface of a histone octamer may be stabilized under some circumstances and accumulate, leading to the observation of a stably altered path. Depending on the rate and directionality of propagation of these DNA distortions, nucleosome sliding or ‘remodelling’ without relocation may be observed (for a more detailed discussion, see Becker and Hörz, 2002).
Controlling nucleosome movements
It is likely that the activity of nucleosome remodelling factors will be regulated, and the first regulatory principles are emerging. While nucleosome remodelling ATPases alone can function in vitro, their activity is modulated by associated subunits in the physiological complexes (Ito et al., 1999; Phelan et al., 2000; Eberharter et al., 2001; Xiao et al., 2001). The modulation of stability and activity of Brg1 and hBrm by phosphorylation (Muchardt et al., 1996) is presumably only the first of many regulatory modifications to be discovered.
Not surprisingly, remodelling enzymes are also sensitive towards the modification status of exposed N-terminal ‘tails’ of the histones (Clapier et al., 2002; Corona et al., 2002). Interaction of the yeast SWI/SNF complex with chromatin is strengthened by histone acetylation (Hassan et al., 2001) and functional interaction between ATP-dependent nucleosome remodelling and histone acetylation is widely observed (for a review, see Becker and Hörz, 2002). Combinations of histone modifications are thought to determine the functional status of chromatin (Jenuwein and Allis, 2001), which obviously includes the degree of nucleosome mobility.
The ease with which nucleosomes can be moved on DNA also depends on the folding of the chromatin fibre. Association of histone H1 with nucleosomal linker DNA ‘seals’ the two DNA segments at the nucleosome edges into a ‘stem’ structure (Bednar et al., 1998) that restricts passive nucleosome movement (Pennings et al., 1994) as well as catalysed remodelling of nucleosomal arrays by enzymes of all major subclasses (Horn et al., 2002). Most interestingly, phosphorylation of histone H1 relieves these constraints, presumably due to weakened interactions with DNA (Horn et al., 2002). It is likely that the interplay between various linker-binding factors is a decisive determinant of chromatin fluidity. Remarkably, interaction of the abundant non-histone protein HMGB1 with linker DNA at the strategic nucleosomal edge, overlapping the ISWI interaction site not only is compatible with nucleosome sliding, but also facilitates mobilization of a nucleosome by ACF (T.Bonaldi, G.Längst, R.Strohner, P.B.Becker and M.E.Bianchi, submitted). In vivo, competition of HMGB1 with H1 for nucleosomal interaction may be facilitated by H1 phosphorylation.
The restrictive role of further levels of chromatin organization, such as the presence of heterochromatin components has not yet been defined. However, the recent observation of an inhibitory effect of a polycomb complex on nucleosome remodelling by Brg1 suggests that repressive chromatin structures are also characterized by reduced nucleosome mobility (Francis et al., 2001).
Conclusion
ATP-dependent, catalysed and regulated nucleosome sliding is likely to make important contributions to the plasticity and transparency of chromatin. Elucidation of the functional diversification of chromatin remodelling machines, their targeting, regulation and mechanism of action remains a worthwhile goal for the years to come.
Acknowledgments
Acknowledgements
I thank the members of my laboratory, past and present, for their commitment and dedication. I am grateful to the Deutsche Forschungsgemeinschaft for continuous support.
References
- Beard P. (1978) Mobility of histones on the chromosomes of simian virus 40. Cell, 15, 955–967. [DOI] [PubMed] [Google Scholar]
- Beato M. and Eisfeld,K. (1997) Transcription factor access to chromatin. Nucleic Acids Res., 25, 3559–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P.B. and Hörz,W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., 71, 247–273. [DOI] [PubMed] [Google Scholar]
- Becker P.B., Ruppert,S. and Schütz,G. (1987) Genomic footprinting reveals cell type-specific binding of ubiquitous transcription factors. Cell, 51, 435–443. [DOI] [PubMed] [Google Scholar]
- Bednar J., Horowitz,R.A., Grigoryev,S.A., Carruthers,L.M., Hansen,J.C., Koster,A.J. and Woodcock,C.L. (1998) Nucleosomes, linker DNA and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl Acad. Sci. USA, 95, 14173–14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhenok L., Wade,P.A. and Varga-Weisz,P. (2002) WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J., 21, 2231–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Langst,G., Kehle,J., Clapier,C.R., Imhof,A., Eberharter,A., Muller,J. and Becker,P.B. (2000) dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J., 19, 4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttinelli M., Di Mauro,E. and Negri,R. (1993) Multiple nucleosome positioning with unique rotational setting for the Saccharomyces cerevisiae 5S rRNA gene in vitro and in vivo. Proc. Natl Acad. Sci. USA, 90, 9315–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C.R., Nightingale,K.P. and Becker,P.B. (2002) A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res., 30, 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona D.F.V., Clapier,C.R., Becker,P.B. and Tamkun,J.W. (2002) Modulation of ISWI function by site-specific histone acetylation. EMBO rep., 3, 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R. et al. (2000) The ISWI chromatin remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Eberharter A., Ferrari,S., Langst,G., Straub,T., Imhof,A., Varga-Weisz,P., Wilm,M. and Becker,P.B. (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J., 20, 3781–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Sweder,K.S. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M.J., Lue,N.F. and Kornberg,R.D. (1988) Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J. Mol. Biol., 204, 109–127. [DOI] [PubMed] [Google Scholar]
- Flaus A. and Owen-Hughes,T. (2001) Mechanisms for ATP-dependent chromatin remodelling. Curr. Opin. Genet. Dev., 11, 148–154. [DOI] [PubMed] [Google Scholar]
- Fletcher T.M. and Hansen,J.C. (1996) The nucleosomal array: structure/function relationships. Crit. Rev. Eukaryot. Gene Expr., 6, 149–188. [DOI] [PubMed] [Google Scholar]
- Fragoso G., John,S., Roberts,M.S. and Hager,G.L. (1995) Nucleosome positioning on the MMTV LTR results from frequency-biased occupancy of multiple frames. Genes Dev., 9, 1933–1947. [DOI] [PubMed] [Google Scholar]
- Francis N.J., Saurin,A.J., Shao,Z. and Kingston,R.E. (2001) Reconsti tution of a functional core polycomb repressive complex. Mol. Cell, 8, 545–556. [DOI] [PubMed] [Google Scholar]
- Gavin I., Horn,P.J. and Peterson,C.L. (2001) SWI/SNF chromatin remodeling requires changes in DNA topology. Mol. Cell, 7, 97–104. [DOI] [PubMed] [Google Scholar]
- Goldmark J.P., Fazzio,T.G., Estep,P.W., Church,G.M. and Tsukiyama,T. (2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell, 103, 423–433. [DOI] [PubMed] [Google Scholar]
- Gregory P.D., Wagner,K. and Hörz,W. (2001) Histone acetylation and chromatin remodeling. Exp. Cell Res., 265, 195–202. [DOI] [PubMed] [Google Scholar]
- Guschin D., Wade,P.A., Kikyo,N. and Wolffe,A.P. (2000) ATP-Dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry, 39, 5238–5245. [DOI] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos,R., Gdula,D.A. and Wu,C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell, 97, 833–842. [DOI] [PubMed] [Google Scholar]
- Hassan A.H., Neely,K.E. and Workman,J.L. (2001) Histone acetyl transferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell, 104, 817–827. [DOI] [PubMed] [Google Scholar]
- Havas K., Flaus,A., Phelan,M., Kingston,R., Wade,P.A., Lilley,D.M. and Owen-Hughes,T. (2000) Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell, 103, 1133–1142. [DOI] [PubMed] [Google Scholar]
- Horn P.J., Carruthers,L.M., Logie,C., Hill,D.A., Solomon,M.J., Wade,P.A., Imbalzano,A.N., Hansen,J.C. and Peterson,C.L. (2002) Phos phorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol., 9, 263–267. [DOI] [PubMed] [Google Scholar]
- Ito T., Bulger,M., Pazin,M.J., Kobayashi,R. and Kadonaga,J.T. (1997a) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell, 90, 145–155. [DOI] [PubMed] [Google Scholar]
- Ito T., Tyler,J.K. and Kadonaga,J.T. (1997b) Chromatin assembly factors: a dual function in nucleosome formation and mobilization? Genes Cells, 2, 593–600. [DOI] [PubMed] [Google Scholar]
- Ito T., Levenstein,M.E., Fyodorov,D.V., Kutach,A.K., Kobayashi,R. and Kadonaga,J.T. (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev., 13, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskelioff M., Gavin,I.M., Peterson,C.L. and Logie,C. (2000) SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol., 20, 3058–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kang J.G., Hamiche,A. and Wu,C. (2002) GAL4 directs nucleosome sliding induced by NURF. EMBO J., 21, 1406–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N.A., Karabetsou,N., Politis,P.K. and Mellor,J. (2001) In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev., 15, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R.D. and Stryer,L. (1988) Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res., 16, 6677–6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G. and Becker,P.B. (2001a) ISWI induces nucleosome sliding on nicked DNA. Mol. Cell, 8, 1085–1092. [DOI] [PubMed] [Google Scholar]
- Längst G. and Becker,P.B. (2001b) Nucleosome mobilization and positioning by ISWI-containing chromatin remodeling factors. J. Cell Sci., 114, 2561–2568. [DOI] [PubMed] [Google Scholar]
- Längst G., Bonte,E.J., Corona,D.F.V. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- Liu R., Liu,H., Chen,X., Kirby,M., Brown,P.O. and Zhao,K. (2001) Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell, 106, 309–318. [DOI] [PubMed] [Google Scholar]
- Lomvardas S. and Thanos,D. (2001) Nucleosome sliding via TBP DNA binding in vivo. Cell, 106, 685–696. [DOI] [PubMed] [Google Scholar]
- Lorch Y., Zhang,M. and Kornberg,R.D. (1999) Histone octamer transfer by a chromatin-remodeling complex. Cell, 96, 389–392. [DOI] [PubMed] [Google Scholar]
- Luger K. and Richmond,T.J. (1998) DNA binding within the nucleosome core. Curr. Opin. Struct. Biol., 8, 33–40. [DOI] [PubMed] [Google Scholar]
- Meersseman G., Pennings,S. and Bradbury,E.M. (1992) Mobile nucleosomes—a general behaviour. EMBO J., 11, 2951–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C., Reyes,J.C., Bourachot,B., Leguoy,E. and Yaniv,M. (1996) The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J., 15, 3394–3402. [PMC free article] [PubMed] [Google Scholar]
- Narlikar G.J., Phelan,M.L. and Kingston,R.E. (2001) Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell, 8, 1219–1230. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Pennings S., Meersseman,G. and Bradbury,M.E. (1991) Mobility on nucleosomes on 5S rDNA. J. Mol. Biol., 220, 101–110. [DOI] [PubMed] [Google Scholar]
- Pennings S., Meersseman,G. and Bradbury,E.M. (1994) Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc. Natl Acad. Sci. USA, 91, 10275–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L. and Logie,C. (2000) Recruitment of chromatin remodeling machines. J. Cell. Biochem., 78, 179–185. [DOI] [PubMed] [Google Scholar]
- Phelan M.L., Schnitzler,G.R. and Kingston,R.E. (2000) Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell. Biol., 20, 6380–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessel H., Widom,J., Bruinsma,R.F. and Gelbart,W.M. (2001) Polymer reptation and nucleosome repositioning. Phys. Rev. Lett., 86, 4414–4417. [DOI] [PubMed] [Google Scholar]
- Schnitzler G.R., Cheung,C.L., Hafner,J.H., Saurin,A.J., Kingston,R.E. and Lieber,C.M. (2001) Direct imaging of human SWI/SNF-remodeled mono- and polynucleosomes by atomic force microscopy employing carbon nanotube tips. Mol. Cell. Biol., 21, 8504–8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Livingstone-Zatchej,M. and Thoma,F. (1996) Chromatin structure of the yeast URA3 gene at high resolution provides insight into structure and positioning of nucleosomes in the chromosomal context. J. Mol. Biol., 257, 919–934. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T. and Wu,C. (1995) Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell, 83, 1011–1020. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Daniel,C., Tamkun,J. and Wu,C. (1995) ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kD subunit of the nucleosome remodeling factor. Cell, 83, 1021–1026. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Palmer,J., Landel,C.C., Shiloach,J. and Wu,C. (1999) Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev., 13, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde K.E. and Yager,T.D. (1985) Nucleosome motion: evidence and models. In Nicolini,C. and Ts’o,P.O.P. (eds), Structure and Function of the Genetic Apparatus. Plenum Press, New York, NY, pp. 35–53.
- Varga-Weisz P.D., Blank,T.A. and Becker,P.B. (1995) Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J., 14, 2209–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Wilm,M., Bonte,E., Dumas,K., Mann,M. and Becker,P.B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature, 388, 598–602. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Gegonne,A., Jones,P.L., Ballestar,E., Aubry,F. and Wolffe,A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- Wallrath L.L., Quinn,L., Granok,H. and Elgin,S.C.R. (1994) Archi tectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. BioEssays, 16, 165–170. [DOI] [PubMed] [Google Scholar]
- Widom J. (1999) Equilibrium and dynamic nucleosome stability. Methods Mol. Biol., 119, 61–77. [DOI] [PubMed] [Google Scholar]
- Woodcock C.L. and Dimitrov,S. (2001) Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev., 11, 130–135. [DOI] [PubMed] [Google Scholar]
- Xiao H., Sandaltzopoulos,R., Wang,H., Hamiche,A., Ranallo,R., Lee,K., Fu,D. and Wu,C. (2001) Dual functions of the largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell, 8, 531–543. [DOI] [PubMed] [Google Scholar]