Abstract
Fission yeast cells identify growing regions at the opposite ends of the cell, producing the rod-like shape. The positioning of the growth zone(s) and the polarized growth require CLIP170-like protein Tip1 and the Ndr kinase Orb6, respectively. Here, we show that the mor2/cps12 mutation disrupts the localization of F-actin at the cell ends, producing spherical cells and concomitantly inducing a G2 delay at 36°C. Mor2 is important for the localization of F-actin at the cell end(s) but not at the medial region, and is essential for the restriction of the growth zone(s) where Tip1 targets. Mor2 is homologous to the Drosophila Furry protein, which is required to maintain the integrity of cellular extensions, and is localized at both cell ends and the medial region of the cell in an actin-dependent fashion. Cellular localization of Mor2 and Orb6 was interdependent. The tyrosine kinase Wee1 is necessary for the G2 delay and maintenance of viability of the mor2 mutant. These results indicate that Mor2 plays an essential role in cell morphogenesis in concert with Orb6, and the mutation activates the mechanism coordinating morphogenesis with cell cycle progression.
Keywords: actin/checkpoint/CLIP170/Furry/growth polarity
Introduction
Cell morphogenesis and the cell cycle are coordinately regulated. The fission yeast Schizosaccharomyces pombe is an ideal system to study cell morphogenesis (Snell and Nurse, 1993; Verde et al., 1995; Hirata et al., 1998b; Mata and Nurse, 1998; Brunner and Nurse, 2000). The fission yeast cell has a cylindrical rod-like shape (8–14 µm in length and 3 µm in diameter), and cell growth occurs only at the tips (Mitchison, 1970). The growth polarity dynamically changes during three stages of the cell cycle: (i) the initiation of growth upon cell division; (ii) new end take off (NETO; Mitchison and Nurse, 1985); and (iii) septum formation after anaphase. After cell division, cortical F-actin moves from the new end that is newly produced by division to the old end that existed in the previous cell cycle. The cell growth is initiated from only the old cell end. Upon NETO, which occurs at 0.34 of the way through the cell cycle (Mitchison and Nurse, 1985), F-actin localization dynamically shifts from the old cell end to both cell ends. After NETO, the growth polarity changes from monopolar to bipolar. At the onset of mitosis, cell growth ceases, and the cortical F-actin patches at both ends then disappear. Prior to cytokinesis, actin reassembles into an F-actin ring in the middle region of the cell where septation will occur (Marks and Hyams, 1985; Marks et al., 1986).
In general, microtubules (MTs) mediate long-range transport of membranous organelles to the cell periphery, whereas actin mediates short-range transport and anchorage (Goode et al., 2000). Rod-shaped fission yeast cells grow in a polarized manner, and unlike in the case of the budding yeast, the correct positioning of the growth sites at cell ends requires interphase MTs. The Tea1 protein and interphase MTs are important for cell polarity in fission yeast (Mata and Nurse, 1997). Tea1 is localized on the plus ends of interphase MTs. Tea1 localization requires MTs, a kinesin-like protein Tea2, and the CLIP170-like protein Tip1 (Browning et al., 2000; Brunner and Nurse, 2000). The protein Tip1, which becomes localized at the distal tips of cytoplasmic MTs, enables the MTs to discriminate between the cell end(s) and other cortical regions, and regulates their dynamics accordingly (Brunner and Nurse, 2000). It has been suggested that there is a pre-existing marker at the cell ends recognized by Tip1 that promotes catastrophe by causing the dissociation of Tip1 from the MT tip (Brunner and Nurse, 2000). The mechanism targeting Tip1 to the cell ends remains to be clarified.
A series of checkpoints ensures that events proceed normally during the cell cycle. Several checkpoints, which monitor DNA replication, DNA damage, spindle assembly to the kinetochore and spindle orientation, have been identified. Cell morphogenesis and cell cycle progression are coordinately regulated, indicating that cells have a checkpoint(s) that monitors the cell morphogenesis at a specific stage(s) of the cell cycle. Indeed, the checkpoints that monitor the bud formation in budding yeast (Lew, 2000) and the completion of cytokinesis in fission yeast have been identified (Goff et al., 1999; Liu et al., 2000). In the ‘morphogenesis checkpoint’ monitoring the bud formation in budding yeast, the Cdc28 inhibitory kinase Swe1 is important for the G2 delay. In response to actin perturbations, the Swe1 protein is continuously accumulated during G2 delay by both increases in SWE1 transcription and inhibition of Swe1 degradation (Lew, 2000). The activation of the cytokinesis checkpoint in fission yeast requires the septation initiation network (SIN) pathway and the Cdc2 inhibitory kinase Wee1 (Goff et al., 1999; Liu et al., 2000).
The Drosophila protein Furry is important for maintaining the integrity of cellular extensions during morphogenesis (Cong et al., 2001). The furry mutation causes the formation of branched arista laterals, branched bristles and strong multiple hair cells. The Furry-like proteins have been evolutionarily conserved from yeast to humans. Genetic analysis has suggested that the furry gene functions in the same pathway as the tricornered gene that encodes Ndr (nuclear DBF2-related) kinase (Geng et al., 2000; Cong et al., 2001). The Ndr family is related to S.pombe Orb6 (Verde et al., 1998), Saccharomyces cerevisiae Cbk1 (Racki et al., 2000; Bidlingmaier et al., 2001; Colman-Lerner et al., 2001), Neurospora Cot1 (Yarden et al., 1992), Drosophila Warts/Lats (Justice et al., 1995; Xu et al., 1995), Caenorhabditis let-502 and Sax1 (Wissmann et al., 1997; Zallen et al., 2000), mammalian Rho-associated kinase (Leung et al., 1995; Ishizaki et al., 1996; Matsui et al., 1996) and human myotonic dystrophy kinase (DMPK) (Brook et al., 1992), and is required for the regulation of cell morphology and division. The Orb6 kinase of fission yeast is localized at the polarized growing sites, and its mutation results in a round morphology (Verde et al., 1995, 1998). Recently, it was shown that a budding yeast Furry-like protein, Pag1, is required for cell morphogenesis and forms a complex with the Ndr kinase Cbk1 (Du and Novick, 2002).
Here we show that the fission yeast Furry-like protein, Mor2, is required for the establishment of growth polarity and was localized to the cell ends and septation site in an actin-dependent fashion. The mor2 mutation disrupts the localization of F-actin, inducing a Wee1-dependent G2 delay. Furthermore, we demonstrate that the localization of Mor2 and Orb6 is interdependent for their function.
Results
Isolation of temperature-sensitive round mutants
To identify a mechanism coordinating morphogenesis with cell cycle progression in fission yeast, we visually screened for mutants that showed a round morphology and arrest of cell growth at 36°C. To distinguish the cell wall-defective mutants, we examined the sensitivity of the cells to the protein kinase inhibitor staurosporine (STS), because the PKC-related kinase Pck2 is important for the maintenance of the cell wall integrity and its mutation causes super-sensitivity to STS (Toda et al., 1993; Katayama et al., 1999). By this screening, four STS-insensitive mutants were isolated and were designated mor (morphological round). Complementation analyses between mor and other round mutants showed that the mor mutants could be classified into three loci, and that mor1 and mor2 were allelic to orb1 (Snell and Nurse, 1994) and cps12 (Ishiguro and Uhara, 1992), respectively. The orb1 mutant was isolated as one of the 12 orb loci that showed a round shape at 36°C, and the cps12 mutant as one of the 14 cps loci that showed super-sensitivity to the spindle poison chlorpropham (isopropyl N-3-chlorophenyl carbamate). In this paper, we focused on the novel mutant mor2/cps12. Hereafter, the originally isolated mor2 and cps12 mutants are referred to as mor2-786 and mor2-282, respectively.
Mor2 is a Drosophila Furry-like protein
The mor2+ gene was cloned by complementation of the phenotype of the mutant (temperature sensitivity and round morphology). Nucleotide sequencing of the cloned DNA fragment identified a novel gene that encoded 2196 amino acids (molecular mass 250 kDa) matching the genomic sequence of ORF SPBP19A11.04C in the S.pombe genome. Blast analysis indicated Mor2 to be a Drosophila Furry-like protein. The Furry-like proteins have been evolutionarily conserved from yeast to humans. The proteins exist in S.cerevisiae (Pag1/Tao3), Drosophila melanogaster (Furry), Caenorhabditis elegans (AAF 99910), Arabidopsis (T51397) and humans (HS85D21.1) (Figure 1A). The D.melanogaster, C.elegans and human proteins contain five similarity regions, but the S.pombe, S.cerevisiae and Arabidopsis proteins contain only the first three regions of similarity. Among these genes, the Drosophila essential gene furry was first to be identified and found to be important for maintaining the integrity of cellular extensions during morphogenesis (Cong et al., 2001). The S.cerevisiae tao3/pag1 mutant was originally identified as a mutant that caused altered transcription of the OCH1 gene (Saccharomyces Genome Database); however, recently it was shown that Pag1/Tao3 is important for cell morphogenesis (Du and Novick, 2002).
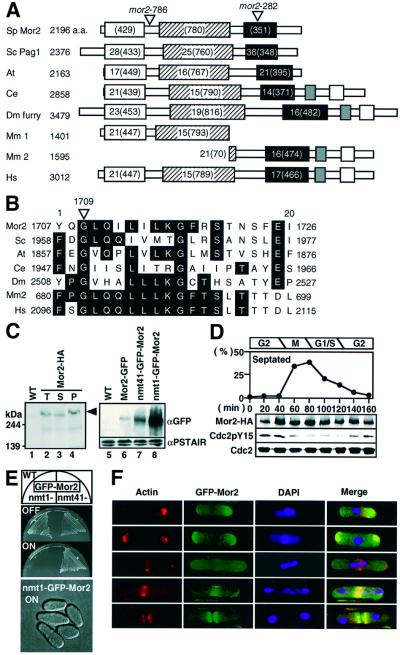
Fig. 1. Identification of Mor2. (A) A schematic presentation of the homology between Mor2 and Furry homologous proteins. Schizosaccharomyces pombe Mor2 is aligned with S.cerevisiae Pag1/Tao3 (Sc), Arabidopsis thaliana T51397 (At), C.elegans AAF99910 (Ce), D.melanogaster Furry (Dm), Mus musculus CAC42175.1 (Mm1), CAC42196.1 (Mm2) and Homo sapiens CAB42442 (Hs). Identities (%) and the number of amino acids are indicated. (B) The mutation site of the mor2-282 allele. (C) Identification of Mor2 protein. Lanes 1–4: total cell extract prepared from wild type (WT, 972), and the cells having mor2+:3HA:kanr (Mor2-HA, YS78-1) grown in YPD medium were electrophoresed on SDS–PAGE gels and immunoblotted with anti-HA (lanes 1 and 2). Total cell extracts (T, lane 2) prepared from the Mor2-HA strain were fractionated into soluble (S, lane 3) and insoluble fractions (P, lane 4) by centrifugation at 14 000 g. Lanes 5–8: wild type, and cells with mor2+:GFP:kanr (Mor2–GFP, YS77), kanr:nmt41-GFP:mor2+ (nmt41–GFP–Mor2, YS84-1) or kanr:nmt1-GFP:mor2+ (nmt1-GFP–Mor2, YS85) grown in Edinburgh minimal medium (EMM) containing 4 µM thiamine were transferred into EMM. After cultivation for 14 h, the cells were collected. Total cell extracts prepared from the cells were immunoblotted with anti-GFP or anti-PSTAIR antibodies. (D) Mor2 protein during the cell cycle. Early G2 cells of Mor2-HA strain were collected by centrifugal elutriation and cultured in YPD medium at 28°C. Samples of the culture were taken at the indicated times for calculating the percentage of septated cells. Total cell extracts prepared from the samples of the same cultures were immunoblotted with anti-HA (for Mor2-HA), anti-PSTAIR (for Cdc2) or anti-Cdc2 phosphorylated on tyrosine-15 (for Cdc2-P) antibodies. (E) Growth (upper) and morphology (lower) of the Mor2-over-expressing cells. Upper panel: the nmt1-GFP–Mor2 and nmt41–GFP–Mor2 strains were cultured on EMM (ON) or EMM plates containing 4 mM thiamine (OFF) at 25°C for 3 days. Lower panel: the nmt1-GFP–Mor2 strain grown in EMM containing 4 µM thiamine was transferred into EMM medium and cultured for 18 h at 25°C. (F) Localization of Mor2 and F-actin. The nmt41–GFP–Mor2 strain cells grown in EMM containing 4 µM thiamine were transferred into EMM medium. After cultivation for 14 h, cells were fixed and stained with rhodamine– phalloidin. Merged image (Merge): F-actin (red), GFP–Mor2 (green) and DAPI (blue).
The mutation sites of the two mor2 alleles were determined. The mor2-282 and mor2-786 mutants showed the substitution of an evolutionarily conserved glycine 1709 with aspartic acid [GGT to GAT (the mutated nucleotide is underlined)] and the substitution of tyrosine 558 (TAT) with cysteine (TGT), respectively (Figure 1A and B).
The mor2+ gene was disrupted by the one-step gene disruption method using the ura4+ gene as marker. Tetrad analysis indicated that two viable and two non-viable spores were obtained, and the viable spores produced Ura– colonies with an intact mor2+ gene. Microscopic observation of the non-viable spores showed that the putative Δmor2 spores failed to germinate or ceased growing immediately after germination. These results indicate that the mor2+ gene was essential for cell growth.
Mor2 is localized at cell ends and the septation site
To investigate the cellular localization of the Mor2 protein, we first introduced an epitope tag (3HA or 13Myc) or a green fluorescent protein (GFP) gene at the end of the chromosomal mor2+ gene in wild-type cells by using the polymerase chain reaction (PCR) tagging method (Bahler et al., 1998). The C-terminal tagging did not interfere with the function of the Mor2 protein, as the tagged strains grew as well as wild-type cells. We detected the proteins by western blotting using these tagged strains (Figure 1C), but we could not determine the specific cellular localization of Mor2. The Mor2-HA was found in both the soluble and insoluble fractions, suggesting that the protein was present in both cytoplasm and membrane regions (Figure 1C, lanes 2–4). The Mor2-HA protein level did not change drastically during the cell cycle (Figure 1D). To investigate the localization of the Mor2 protein, we next attempted to increase the expression level of the mor2+ gene by introducing the thiamine-repressible nmt41 or nmt1 promoter-driven GFP gene in front of the chromosomal mor2+ gene. The level of GFP–Mor2 protein expressed by the nmt41-GFP-mor2+ or nmt1-GFP-mor2+ strain grown under the inducible condition increased by 5- or 50-fold, respectively, compared with that expressed from the mor2+ promoter (Figure 1C, lanes 6–8). The excessive expression of GFP–Mor2 from the nmt1 promoter caused the defects in growth and morphology (Figure 1E), but the induced GFP–Mor2 protein from the nmt41 promoter did not affect growth and morphology. Therefore, we used an nmt41–GFP–Mor2 strain for the observation of the localization of Mor2 protein. In the monopolar growing cells in which the majority of F-actin localized to the old ends, GFP–Mor2 was localized predominantly at the growing end and faintly at the non-growing end. In the bipolar growing cells, GFP–Mor2 was localized at both ends of the cell. During mitosis, GFP–Mor2 was concentrated at the medial region where the septum formation would later occur (Figure 1F).
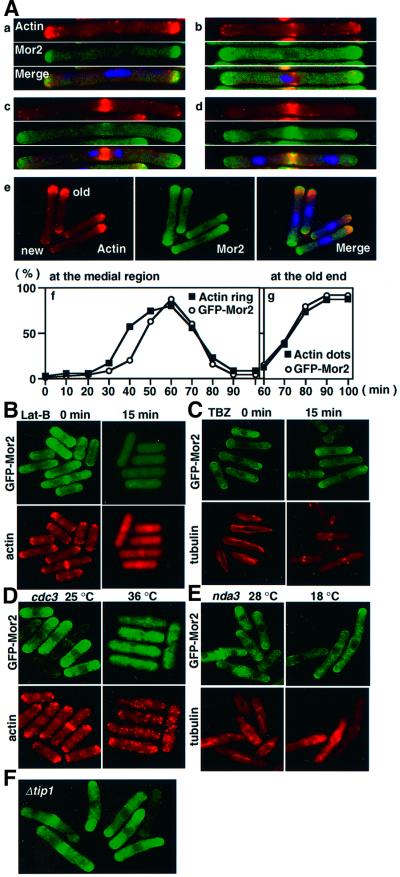
To investigate further the precise order of the localization of Mor2 and actin, we used synchronous cultures of cdc25-22 mutant cells (Figure 2A). The cells were arrested in late G2 phase at 36°C and were released at the permissive temperature of 25°C. After the release, we compared the appearance of the actin ring and the accumulation of GFP–Mor2 at the medial region in the time-course. In the G2-arrested cells, F-actin and GFP–Mor2 were localized at both ends of the cells (Figure 2A, a). At 40 min after the release, 60% of the cells formed an actin ring (Figure 2A, f). At this time, an actin ring formed in 40% of the cells, but the localization of GFP–Mor2 at the medial region was not observed (Figure 2A, b, c and f). The localization of GFP–Mor2 at the medial region followed the actin localization during cell division (Figure 2A, f). We next examined the appearance of actin dots and the accumulation of GFP–Mor2 at the old end of the cells. At the growth initiation after the cell division, the appearance of actin dots and the accumulation of GFP–Mor2 at the old end were observed simultaneously (Figure 2A, e and g). In the monopolar growing cells in which F-actin was predominantly localized at the old end, a small number of actin dots and the localization of GFP–Mor2 at the new end were observed. These results suggested that the appearance of actin dots but not the formation of an actin ring depends on Mor2.
Fig. 2. Role of cytoskeletons in Mor2 localization. (A) Mor2 localization in the cells released from G2 block. The cdc25-22 mutant cells having kanr:nmt41-GFP:mor2+ (DH294-4A) grown in EMM containing 8 µM thiamine were transferred into EMM medium. After cultivation for 11–12 h at 25°C, the cells were transferred to 36°C and kept there for 4 h. The medium was then was returned to 25°C, and the cells were fixed and stained with rhodamine–phalloidin (a–e). The appearance of the actin ring at the medial region (f) and actin dots at the old end (g) was examined with respect to Mor2 accumulation at their sites in the time course. (B and C) Effect of Lat-B and TBZ on Mor2 localization. The nmt41–GFP–Mor2 strain (YS84-1) cells grown in EMM containing 8 µM thiamine were transferred into EMM medium. After culti vation for 14–15 h at 25°C, the cells were incubated with 100 µM Lat-B (B) or 100 µM TBZ (C). Cells taken at the indicated times were fixed and stained with rhodamine–phalloidin (B) or anti-tubulin antibody (C). (D) Mor2 localization in the cdc3 mutant. The cdc3-6 (profilin) mutant cells with kanr:nmt41-GFP:mor2+ (DH299-2) grown in EMM containing 8 µM thiamine were transferred into EMM medium. After cultivation for 12 h at 25°C, the cells were transferred to 36°C for 3–4 h, fixed, and stained with rhodamine–phalloidin. (E) Mor2 localization in the nda3 mutant. The nda3-KM311 (β-tubulin) mutant cells with kanr:nmt41-GFP:mor2+ (DH297-1C) grown in EMM containing 8 µM thiamine were transferred into EMM medium. After cultivation for 11–12 h at 28°C, the cells were transferred to 18°C for 6 h, fixed, and immunostained with anti-tubulin antibodies. (F) Mor2 localization in tip1-deleted cells. The Δtip1 mutant cells with kanr:nmt41-GFP:mor2+ (DH421-2B) grown in EMM containing 8 µM thiamine were transferred into EMM medium and cultivated for 13 h at 25°C.
The actin cytoskeleton is important for the cellular localization of Mor2
To investigate whether the actin or MT cytoskeleton is important for the cellular localization of GFP–Mor2, we examined the localization of GFP–Mor2 in the cells treated with the F-actin inhibitor latrunculin-B (Lat-B; Spector et al., 1983) or the tubulin-binding drug thiabendazole (TBZ; Umesono et al., 1983). The normal localization of GFP–Mor2 was disrupted by Lat-B but not by TBZ (Figure 2B and C). Consistent with this result, the disruption of the actin cytoskeleton by the profilin mutation cdc3 (Balasubramanian et al., 1994), but not the disruption of MTs by the β-tubulin mutation nda3 (Hiraoka et al., 1984), prevented the normal localization of GFP–Mor2 (Figure 2D and E). Further, the deletion of the tip1+ gene, essential for the guidance of MT ends to their target region (Brunner and Nurse, 2000), did not affect the normal cellular localization of GFP–Mor2 (Figure 2F). These results thus indicated that the actin cytoskeleton, but not the MTs, was required to locate Mor2 at the cell ends and the septation site, and that the localization of Mor2 and F-actin at the cell end was interdependent.
Morphology of the mor2 mutant is reversed by temperature shift-down
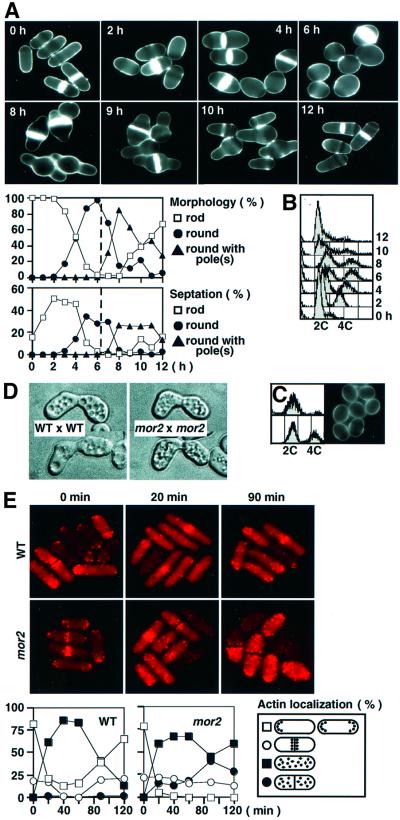
To investigate the morphological change in the mor2 mutant cells in detail, the mutant cells grown at the permissive temperature of 25°C were shifted up to the restrictive temperature of 36°C and kept there for 6 h, and then shifted down to 25°C and incubated for 6 h. The polarized growth ceased immediately after the shift-up, and the morphology completely changed to the round type after 6 h of the incubation at 36°C (Figure 3A). The cells finally showed two phenotypes: unseptated round with one nucleus (70%) and septated round with two nuclei (30%), indicating that Mor2 is important for the polarized growth during interphase and after mitosis. Fluorescence-activated cell sorting (FACS) analysis and the hemispherical nuclear structure indicated that the mor2 mutation induced a G2 delay at 36°C (Figure 3B). After the shift-down of the culture, the cell poles were re-established, and the morphology returned to a rod-shape through round with pole(s) (Figure 3A). The temperature-dependent reversible phenotype indicated that viability of the mutant was maintained at 36°C. To confirm G2 delay of the mor2 mutant, we performed FACS analysis of the unseptated round cells, selected by cell elutriation of the mutant culture at 36°C. The result indicated that the mutant was delayed in G2 (Figure 3C). Further, the double mutant cells (either mor2cdc10 or mor2cdc25) showed abnormal spherical morphology (data not shown), indicating that Mor2 is required for the maintenance of growth polarity in both G1 and G2 phases of the cell cycle.
Fig. 3. Phenotypes of the mor2 mutant. (A and B) Reversible phenotype of the mor2 mutant. mor2 mutant cells (DH107-4C) grown in YPD medium at 25°C were transferred to 36°C (time 0 h). After incubation for 6 h, the cells were shifted down to 28°C and incubated for 6 h (total 12 h). The cells at the indicated times were taken for observation of cell morphology after having been stained with Calcofluor (A) and for FACS analysis (B). (C) mor2 mutant cells (DH107-4C) grown in YPD medium at 25°C were transferred to 36°C and incubated for 4 h. The unseptated round cells in the culture were collected by centrifugal elutriation for FACS analysis (upper panels, the collected round cells; lower panels, the culture) and for observation of morphology (isolated round cells). (D) Shmoo morphology of the mor2 mutant. Wild-type (h–) or mor2 mutant cells (h– mor2-786) were crossed with wild-type (h+) or mor2 mutant cells (h+ mor2-786), respectively. After incubation for 24 h on malt extract (MEA) plate, the shmoo morphology of the cells was observed. (E) F-actin localization in the mor2 mutant. The wild-type (972) and mor2 mutant cells (DH192-1A) grown in YPD medium at 25°C were transferred to 36°C (time 0 h). The cells were taken at the indicated times for observation of F-actin localization after being stained with rhodamine–phalloidin.
The mor2 mutant cells could mate normally. To investigate whether Mor2 is important for the mechanism of cell polarization during conjugation, we examined the shmoo morphology of the mutant. As shown in Figure 3D, the shmoo morphology of the mutant was indistinguishable from that of wild-type cells.
We evaluated the cell wall integrity of the mutant by examining the kinetics of cell lysis upon β-glucanase treatment (Toda et al., 1996). The sensitivity of the mor2 mutant to the treatment was indistinguishable from that of wild-type cells (data not shown). We further examined whether an osmotic stabilizer, sorbitol, would rescue the mor2 mutant, because the phenotype of the cell wall-defective mutant is often suppressed by sorbitol (Ribas et al., 1991; Shiozaki and Russell, 1995). Neither the ts-growth nor round morphology of the mutant was restored by 1 M sorbitol (data not shown). These results indicated that the cell wall integrity of the mor2 mutant was not compromised.
Mor2 is required for re-localization of F-actin to the cell end(s)
To investigate the role of Mor2 in locating F-actin at the polarized growing sites, we examined the distribution of F-actin in the mor2 mutant at 36°C. Cortical F-actin in the wild-type cells is localized as patches at the growing end(s) of the cell and the septation site at 25°C (Marks and Hyams, 1985; Marks et al., 1986) (Figure 3E). In wild-type cells, F-actin at the cell end(s) but not at the septation site was dispersed throughout the cells within 20 min after the shift, presumably due to heat shock stress. By 90 min after the shift, however, F-actin had re-localized at the cell end(s). In the mor2 mutant cells, on the other hand, F-actin at the cell end(s) dispersed as in wild-type cells, but the re-localization of F-actin at the cell ends was not observed (Figure 3E). These results indicated that Mor2 was required for the re-localization of F-actin at the cell end(s).
Mor2 is important for the restriction of the growth zone(s)
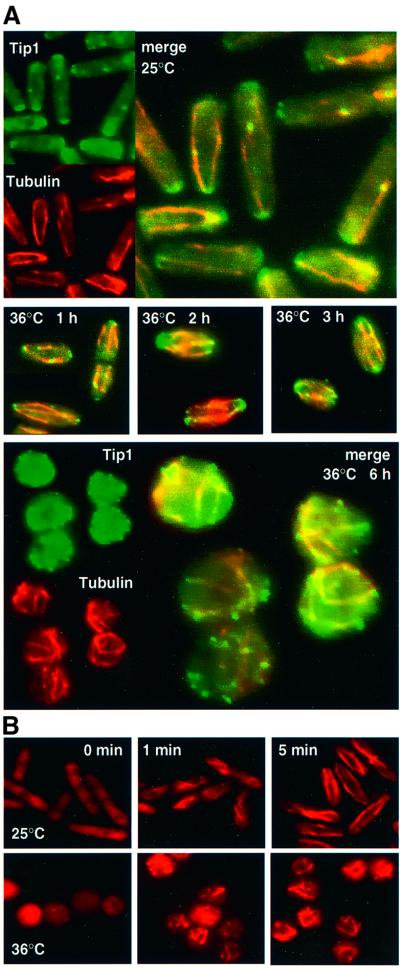
To investigate the role of Mor2 in the organization of the MT cytoskeleton, we examined the MT cytoskeleton and the MT end factor Tip1, which is required for the guidance of the MT ends to their target region at the cell end(s), in the mor2 mutant cells (Figure 4A). In the mutant cells at 25°C, cytoplasmic MTs and mitotic spindles were observed (Marks et al., 1986; Hagan and Hyams, 1988) (Figure 4A), and Tip1 was localized to both MT end(s) and cell tips, as in wild-type cells (Brunner and Nurse, 2000) (Figure 4A). At 36°C, with up to 3 h incubation, the MT-targeting region and normal cellular localization of Tip1 was maintained, but after 6 h the MT-targeting regions became dispersed, which resulted in Tip1 localizing throughout the cell. Despite this, Tip1 was still associated with MT end(s) (Figure 4A), suggesting that the Tip1 dispersion results from deposition by MT at random locations in the cell, but not from random dissociation from its localization sites. These results indicated that Mor2 is important for the restriction of the growth zone(s) to which Tip1 specifically targets.
Fig. 4. Role of Mor2 in the organization of the MT cytoskeleton. (A) MT structure and Tip1 localization in the mor2 mutant. The mor2 mutant cells (DH107-4C) grown in YPD medium at 25°C were transferred to 36°C. The cells were fixed and immunostained with anti-tubulin and anti-Tip1 antibodies at the times indicated. (B) MT nucleation in the mor2 mutant. Upper panels: the mor2 mutant cells having tip1+:GFP:kanr (DH360-2B) grown in YPD medium at 25°C were incubated on ice for 30 min, re-warmed to 25°C, and collected at the indicated times for immunostaining with anti-tubulin antibodies. Lower panels: the mor2 mutant cells grown in YPD medium at 25°C were transferred to 36°C and kept there for 6 h, incubated on ice for 30 min, and then re-warmed to 36°C and collected at the indicated times.
To investigate further whether Mor2 is involved in polymerization of MTs, we examined the nucleation of MTs disrupted by cold shock treatment. MTs were depolymerized by cold shock treatment, and then the cells were transferred to warm medium (Mata and Nurse, 1997). We performed two experiments. In the first experiment, the mor2 mutant cells grown at 25°C were incubated on ice for 30 min and then returned to 25°C (Figure 4B, upper panels). In the second experiment, the cells grown at 25°C were shifted up to 36°C for 6 h, incubated on ice for 30 min, and then returned to 36°C (Figure 4B, lower panels). Upon cold shock, no MTs were observed (Figure 4B, 0 min). At 25°C, the disrupted MTs were nucleated within 5 min after the return to 25°C (Figure 4B, upper panels). At 36°C, nucleation of the MTs was observed as it was at 25°C (Figure 4B, lower panels). These results indicated that Mor2 is not important for the nucleation of MTs.
The mor2 mutant protein is localized as dots at 36°C
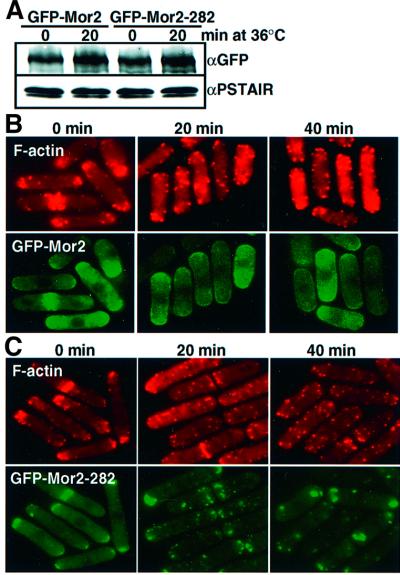
To compare the localization of the wild-type and mutant Mor2 proteins, we introduced an nmt41 promoter-driven GFP gene in front of the chromosomal mor2-282 mutant gene by the PCR-tagging method. The molecular weight of the Mor2 mutant protein was the same as that of the wild-type GFP–Mor2 protein, as judged by immunoblot analysis (Figure 5A). Using the nmt41–GFP–Mor2-282 cells, we examined the localization of the GFP–Mor2-282 mutant protein. At 25°C, the GFP–Mor2-282 mutant protein was localized at both ends and the septation site of the cell, as in the case of the wild-type protein (Figure 5B and C). After the shift to 36°C, a portion of the wild-type GFP–Mor2 protein was localized at the cell ends (Figure 5B), whereas the majority of GFP–Mor2-282 mutant protein was found as discrete punctate dots close to cell ends and the septation site (Figure 5C). After 6 h incubation, the GFP–Mor2-282 mutant protein was localized in cytoplasm as single large dot (data not shown). The expression levels of the wild-type and mutant Mor2 proteins were similar and no obvious change in the levels of these proteins was observed at 36°C (Figure 5A). This result indicates that the Mor2-282 protein is defective in proper cellular localization, namely at cell end(s) and the medial region of the cell, and supports the notion that Mor2 plays an essential role in the growth polarity control at the cell ends.
Fig. 5. Localization of the Mor2 mutant protein. (A–C) The nmt41–GFP–Mor2 [YS84-1 (B)] and nmt41–GFP–Mor2-282 [YY5-5 (C)] strains grown in EMM containing 8 µM thiamine were transferred into EMM medium. After cultivation for 14 h at 25°C, the cells were transferred to YPD medium at 36°C, fixed at the indicated times, and stained with rhodamine–phalloidin. The cells of the 36°C culture were taken at the indicated times for examination of the GFP–Mor2 and GFP–Mor2-282 proteins by immunoblotting (A).
Survival of the mor2 mutant at 36°C requires Wee1
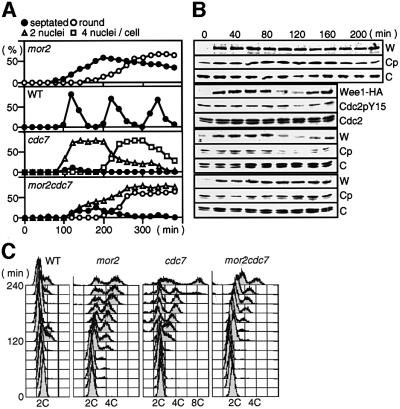
At present, five mitotic checkpoints, i.e. DNA structure (damage and replication), spindle assembly, spindle orientation and cytokinesis, have been identified in fission yeast. To investigate whether these checkpoint pathways are required for the survival and G2 delay of the mor2 mutant at the restrictive temperature, we examined the viability of the double mutants between Mor2 and Rad3 (essential for DNA structure checkpoint; Bentley et al., 1996), Mad2 (for spindlle assembly; He et al., 1997; Kim et al., 1998), Sty1 MAPK (for spindle orientation; Gachet et al., 2001) or Wee1 (inhibitory tyrosine kinase of Cdc2; Nurse, 1975; Lundgren et al., 1991). Among these mutants, only mor2wee1 substantially lost viability at 36°C (Figure 6A). Microscopic observation showed accumulation of the abnormally septated or anucleate cells (64% at 4 h; Figure 6B). We further examined the appearance of the abnormal cells in the mor2wee1 mutant by using the early G2 cells selected by centrifugal elutriation. After 3 h of the incubation at 36°C, 48% of the mutant cells had abnormal septa that were caused by undergoing a second mitosis before the completion of cytokinesis. These results indicated that the survival and G2 delay of the mor2 mutant required the function of Wee1, but not that of Rad3, Mad2 or Sty1.
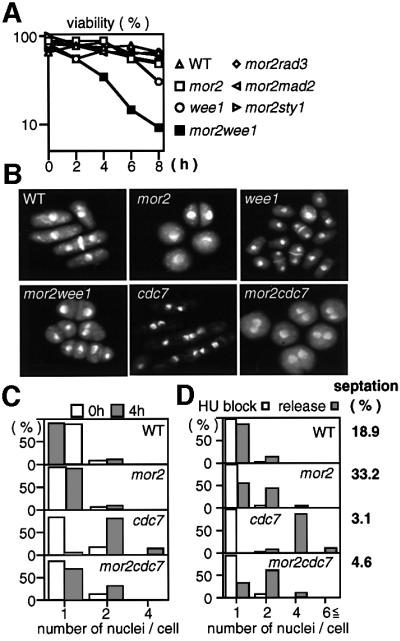
Fig. 6. Wee1-dependent G2 delay in the mor2 mutant. (A–C) Viability and nucleus number of the mutant. Wild-type (22), mor2 (DH107-4C), Δwee1 (IY1078), mor2Δwee1 (DH203-6A), mor2Δrad3 (DH205-5D), mor2Δmad2 (DH207-5A), mor2Δsty1 (DH214-3B), cdc7 and mor2cdc7 (DH295-2B) grown in YPD medium at 25°C were transferred to 36°C and collected at the indicated times. The cells were spread on YPD plates, incubated for 3–4 days at 28°C, and assessed for cell viability (A). The nuclear structures of the cells incubated at 36°C for 4 h were observed by DAPI staining (B), and the number of nuclei in the cells at the indicated times was examined (C). (D) Wild-type, mor2, cdc7 and mor2cdc7 mutant cells grown in YPD medium at 25°C were treated with 12 mM hydroxyurea (HU) for 3.5 h and shifted to 36°C for 1 h. HU-treated cells were washed with pre-warmed YPD medium, inoculated into fresh YPD medium, incubated at 36°C for a further 4 h, fixed, and stained with DAPI. HU-treated cells prior to release from HU are labelled ‘HU block’, and cells released from the HU block are labelled ‘release’. The septation index in ‘release’ is indicated.
As a portion of the mor2 mutant cells at 36°C (30% for 6 h) showed the septated round with two nuclei, we examined whether the G2 delay of the mor2 mutant was dependent on the cytokinesis checkpoint mechanism, which induces a Wee1-dependent G2 delay until the previous cytokinesis has been completed. The 1,3-β-glucan synthase-defective mutant cps1 arrests G2 at the restrictive temperature by this checkpoint (Goff et al., 1999; Liu et al., 2000). The double mutants between cps1 and the septation initiation mutant are unable to arrest the cell cycle and produce multi-nuclear cells at 36°C, indicating that the G2 delay induced by this checkpoint requires the SIN pathway (Goff et al., 1999; Liu et al., 2000). If the G2 delay of the mor2 mutant was caused by this checkpoint, the G2 delay would also require the SIN pathway, and the double mutant between mor2 and the SIN mutant cdc7 (Nurse et al., 1976) should have had multi-nuclei as in the case of cps1. However, the mor2cdc7 double mutant cells at 36°C had one or two nuclei, as in the mor2 mutant (Figure 6B and C), and the actin ring was not observed. These results indicated that the major mechanism inducing the G2 delay of the mor2 mutant was distinct from the cytokinesis checkpoint mechanism.
Fission yeast have a cell size control, such that the attainment of a critical cell size is required for the onset of mitosis (Nurse, 1975). Previously, Liu et al. (2000) showed that the cps1 mutant arrests at a two-nuclei stage without undergoing cytokinesis, even in the hydroxyurea (HU)-treated cells that are long enough to divide. The authors then concluded that the delay of the mutant is not cell size-dependent, instead being dependent upon cytokinesis per se (Liu et al., 2000). To investigate whether the G2 delay in the mor2 mutant is cell size- or cytokinesis-dependent, we performed the HU experiment, and asked whether nuclear division occurred in the mor2 mutant cells (at 36°C) that had been released from previous S-phase block at 25°C. If the G2 delay of the mor2 mutant was dependent on cell size control, the elongated mutant cells released from S-phase block should have accumulated more than two nuclei. This, however, was not the case. The HU-treated mor2 mutant cells elongated and arrested with one or two nuclei (Figure 6D). In contrast, in cdc7 mutant cells, after the release from S-phase block, 84% of cells accumulated more than four nuclei. Note that 33.2% of mor2 mutant cells had a septum, raising the possibility that in this case, G2 delay in mor2 mutants is induced by the cytokinesis checkpoint (Figure 6D). Therefore, to confirm further that the G2 delay of the mor2 mutant is not dependent on either cell size or cytokinesis, we performed the same experiment using the mor2cdc7 double mutant. After the release from S-phase block, 90.4% of mor2cdc7 double mutant cells arrested at the one- or two-nucleus stage (Figure 6D). These results indicate that the major mechanism inducing the G2 delay of the mor2 mutant is distinct from cell size control. However, it remains possible that cell size control partially contributes to the G2 delay in the mutant.
Wee1 protein levels are not reduced in the mor2 mutant
The Wee1 protein is known to oscillate during the cell cycle, its level decreasing in M and G1 phases (Aligue et al., 1997). To investigate whether the Wee1 protein and the inhibitory tyrosine phosphorylation of Cdc2 on tyrosine-15 (Nurse, 1990) were maintained in the mor2 mutant, we examined the amount of the tyrosine-phosphorylated form of Cdc2 in the mutant. The early G2 cells grown at 25°C were selected by elutriation and incubated at 36°C (Figure 7). In wild-type cells, the level of the phosphorylated Cdc2 was decreased by 120 min upon mitotic entry. The Wee1 protein level in wild-type cells was decreased by 100 min, just before the decrease in the phosphorylated Cdc2. In contrast, in the mor2 mutant, the Wee1 protein and the phosphorylated Cdc2 were maintained at high levels at 36°C until at least 4 h, when the septation index had already been decreased (Figure 7A and B).
Fig. 7. Maintenance of Wee1 protein levels in the mor2 mutant. (A–C) Early G2 cells of wild type (RA1224), mor2 (DH274-10B), cdc7 (DH306-8A) and mor2cdc7 (DH306-12B) were collected by centrifugal elutriation. The cells were released into YPD medium at 36°C, and taken at the indicated times for observation of cell morphology (A), immunoblotting (B) and FACS analysis (C). The total proteins (100 µg) were electrophoresed on SDS–PAGE gels. Wee1-HA protein, tyrosine phosphorylation of Cdc2, and Cdc2 protein in the samples were analysed by immunoblotting with antibodies specific for the HA epitope (Wee1-HA, W), Cdc2 phosphorylated on tyrosine-15 (Cdc2pY15, Cp) and an anti-PSTAIR antibody (Cdc2, C), respectively.
To confirm that the G2 delay of the mor2 mutant was independent of the cytokinesis checkpoint, we examined the Wee1 protein and the phosphorylated Cdc2 in the mor2cdc7 double mutant. In the cdc7 mutant, the Wee1 protein and the phosphorylated Cdc2 oscillated as in the wild-type cells. In the mor2cdc7 double mutant, however, both the Wee1 protein and the phosphorylated Cdc2 were maintained, as in the case of the mor2 mutant (Figure 7A and B). The FACS profile of the double mutant was also the same as that of the mor2 mutant (Figure 7C). Furthermore, the double mutant did not form a septum and arrested at the two-nuclei stage (Figure 7A). These results establish that the major mechanism inducing G2 delay of the mor2 mutant is independent of the cytokinesis checkpoint.
Functional relationship of Mor2 and Orb6
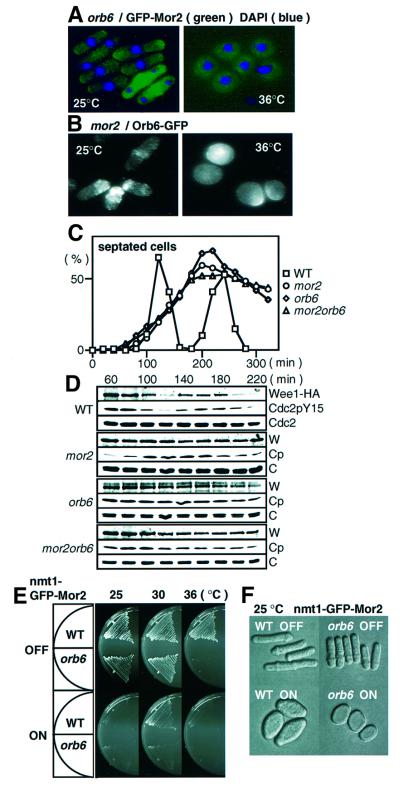
It has been suggested that a Furry-like protein functions in the same pathway as the Ndr kinase in flies and budding yeast (Cong et al., 2001; Du and Novick, 2002). The budding yeast Furry-like protein Pag1 forms a complex with the Ndr kinase Cbk1 (Du and Novick, 2002). To investigate the functional interaction of Mor2 with Orb6, we first examined by means of an immunoprecipitation experiment whether Mor2 could form a complex with Orb6. However, no interaction of Mor2 with Orb6 under our experimental conditions was found (data not shown). We next examined whether Mor2 and Orb6 could affect the cellular localization of each other. In the orb6 mutant cells at 36°C, the normal cellular localization of GFP–Mor2 was disrupted (Figure 8A). Similarly, in the mor2 mutant cells at 36°C, the Orb6–GFP protein was dispersed throughout the cell (Figure 8B). These results indicate that the localization of Mor2 and Orb6 is interdependent.
Fig. 8. Functional interaction between Mor2 and Orb6. (A) Localization of Mor2 in the orb6 mutant. The orb6 mutant cells with kanr:nmt41-GFP:mor2+ (DH451-3D) grown in EMM containing 8 µM thiamine were transferred into EMM medium. After cultivation for 12 h at 25°C, the cells were transferred to 36°C and kept there for 6 h. (B) Localization of Orb6 in the mor2 mutant. The mor2 mutant cells with orb6+:GFP:kanr (DH454–2B) and grown in YES5 medium at 25°C were transferred to 36°C and kept there for 6 h; they were then fixed and immunostained with anti-GFP antibodies. (C and D) Early G2 cells of wild type (RA1224), mor2 (DH274-10B), orb6 (DH463-2C) and mor2orb6 (DH436-12B) were collected by centrifugal elutriation. The cells were released into YPD medium at 36°C, and taken at the indicated times to determine frequency of the septated cells (C) and for immunoblotting (D). (E) Overexpression of Mor2 in the orb6 mutant. Wild-type and orb6 mutant cells with kanr:nmt1-GFP:mor2+ (WT, YS85; orb6, DH475-1C) were cultured on EMM (ON) or EMM plates containing 4 µM thiamine (OFF) at the indicated temperatures for 3 days. (F) The strains grown in EMM containing 4 µM thiamine were transferred into EMM medium and cultured for 18 h at 25°C.
As the overexpression of the orb6+ gene induces a Wee1-dependent G2 delay (Verde et al., 1998), we examined whether the maintenance of the Wee1 protein in the mor2 mutant required the function of Orb6. This was not the case. In the mor2orb6 double mutant, the G2 delay and the maintenance of Wee1 protein were observed as in the mor2 mutant (Figure 8C and D). In the orb6 mutant, Wee1 protein was maintained as in the mor2 mutant (Figure 8C and D). Further, the phenotype of the mor2orb6 double mutant was similar to that of the mor2 single mutant. These results suggested that Mor2 and Orb6 function in the same pathway for cell morphogenesis.
To investigate the functional relationship of Mor2 and Orb6 further, we examined the effect of mor2+ over-expression in the orb6 mutant. In the wild-type background, the over-expression of the mor2+ gene caused defects in growth (at 25 and 36°C) and morphology (round with poles) (Figure 8E and F). The over-expression of the mor2+ gene in the orb6 mutant background caused more severe defects in growth (no growth even at 30°C) and morphology (round) than it did in the wild-type background (Figure 8E and F). This result suggested that mor2+ over-expression prevented the polarity control in which Orb6 functions.
Discussion
Mor2 is essential for the establishment and maintenance of growth polarity
We have demonstrated that the essential protein Mor2 is required for the establishment and maintenance of growth polarity. In the absence of Mor2 function, cells lose polarity completely, which results in the failure of F-actin to localize at the cell ends. The Mor2-mediated polarity control is important for the restriction of the growth zone(s), where the MT end factor Tip1 targets. Mor2 is localized at both ends of the cell and the septation site. The maintenance of Mor2 localization is dependent on the actin cytoskeleton. Actin localization is also dependent on Mor2, except during cell division, when Mor2 follows actin localization. This result suggests that the actin ring, which forms first, does not depend on Mor2, but that the appearance of actin dots at the cell end(s) does. The localization of Mor2 and F-actin at the cell ends is interdependent. It is possible that Mor2 has a general role in organizing the growth machinery and the associated actin. The identification of the protein(s) interacting with Mor2 is important for the understanding of the role of Mor2 in cell morphogenesis.
The actin perturbation caused by Lat-B treatment induces the activation of the Sty1-Atf1-dependent mitotic checkpoint that ensures proper spindle orientation in fission yeast. It has been proposed that the formation of the cortical actin ring that encircles the early mitotic nucleus is monitored by this checkpoint (Gachet et al., 2001). The G2 delay in the mor2 mutant does not require the function of Sty1, suggesting that Mor2 is not important for the actin ring formation. Indeed, Mor2 follows actin localization during cell division. A mechanism that determines the growth polarity in septum formation would be different from that in interphase.
In fission yeast, Ras1, Cdc42 and Pak1/Shk1 participate in a pathway that regulates morphogenesis and mating (Chang et al., 1994; Miller and Johnson, 1994; Marcus et al., 1995; Ottilie et al., 1995). The mating and the shmoo morphology of the mor2 mutant seem to be normal, indicating that Mor2 is not important for the mating process. However, it remains a possibility that Mor2 participates in the Ras1–Cdc42–Pak1/Shk1 branch that is important for morphogenesis during vegetative growth. It is interesting to examine whether Mor2 interacts with this pathway.
Functional conservation and divergence of Furry-like proteins in yeasts
The Furry-like proteins in yeasts are involved in cell morphogenesis. However, we have found functional divergence between the fission yeast Mor2 and the budding yeast Pag1 proteins. Mor2 was important for the localization of actin at the cell ends during interphase, but not for the formation of shmoo during mating. In contrast, Pag1 was important for the organization of actin cytoskeleton during mating but not during vegetative growth (Du and Novick, 2002). These results indicate that the Furry-like proteins of the fission and budding yeasts are implicated, at least in part, in the different cellular processes, although both proteins are essential for cell morphogenesis. Furthermore, as the tao3/pag1 mutant was originally isolated as a mutant that caused the altered transcription of the OCH1 gene (J.Horecka and Y.Jigami, the Saccharomyces Genome Database), we examined the transcription level of the och1+ gene in the mor2 mutant. However, the mor2 mutation did not affect its transcription (N.Kishimoto and D.Hirata, unpublished results). This result also suggests that different proteins are functionally related to the Furry-like proteins in the budding and fission yeast.
Functional interaction between Furry-like Mor2 and Ndr kinase Orb6
It has been suggested that Furry-like proteins function in the same pathway as the Ndr kinase in D.melanogaster and budding yeast (Cong et al., 2001; Du and Novick, 2002). Although we have not found a physical interaction between Mor2 with Orb6, it is possible that these two proteins function in the same pathway. Indeed, the localization of Mor2 and Orb6 is interdependent, and phenotypes of the mor2orb6 double mutant were similar, if not identical, to those of single mutants. It remains possible that Mor2 interacts with Orb6 in the specific stage of the cell cycle. However, we also found a difference in the phenotype between the mor2 and orb6 mutants. The phenotype of the orb6 mutant but not that of the mor2 mutant was rescued by sorbitol (Y.Sogabe and D.Hirata, unpublished results), suggesting that Orb6 but not Mor2 is important for the maintenance of cell-wall integrity. Mor2 and Orb6 would therefore have somewhat different functions in cell morphogenesis.
The mor2 mutation induces a Wee1-dependent G2 delay
The mor2 mutation causes the loss of growth polarity, inducing G2 delay, suggesting that the defect in the establishment of growth polarity activates the mechanism coordinating morphogenesis with the cell cycle. A target of the mechanism is the tyrosine kinase Wee1. The mechanism inducing the G2 delay of the mor2 mutant might be similar to the morphogenesis checkpoint monitoring the establishment of polarity in bud formation in budding yeast (reviewed in Lew, 2000).
In the morphogenesis checkpoint, the Cdc28 inhibitory kinase Swe1 is continuously accumulated during the G2 delay by increases in both SWE1 transcription and inhibition of Swe1 degradation. Swe1 is targeted for rapid degradation in G2, but its degradation is inhibited by the checkpoint. Hsl1 (budding yeast homologue of the fission yeast Nim1 kinase) and Hsl7 play a direct role in targeting Swe1 for degradation. The downregulation of the Hsl1–Hsl7 pathway plays a role in the morphogenesis checkpoint response (McMillan et al., 1999; Shulewitz et al., 1999). In fission yeast, it is not known whether the Nim1 kinase targets Wee1 for degradation. The over-expression of the nim1+ gene did not affect the abundance of Wee1 (M.Suda and D.Hirata, unpublished results). Nim1 directly phosphorylates and inhibits Wee1 in fission yeast, but it is not known whether Hsl1 inhibits Swe1 activity in budding yeast. Further, Skb1 (fission yeast homologue of the budding yeast Hsl7) has a role as a mitotic inhibitor (Gilbreth et al., 1998). The putative role for Skb1 is opposite to that of Hsl7, as a negative regulator of Swe1. These results suggest that the regulatory mechanisms of the Cdc2/Cdc28 inhibitory kinase Wee1/Swe1 in the checkpoint response in both yeasts are different from each other.
Previously, Verde et al. (1998) showed that the over-expression of Orb6 induces Wee1-dependent G2 delay, and thereby concluded that the control of cell size required for division is linked via Orb6 to the growth machinery. We therefore examined whether Orb6 is important for the maintenance of Wee1 protein in the mor2 mutant; however, this was not the case. Furthermore, in the orb6 single mutant, the Wee1 protein was maintained as in the mor2 mutant. This result indicates that both loss and gain of Orb6 functions induce a Wee1-dependent G2 delay. It would be of interest to examine the Orb6 kinase activity in the mor2 mutant.
What defect(s) in the mor2 mutant produces a signal activating this mechanism and how Wee1 is upregulated by the signal are the next subjects to be addressed. To understand better the mechanism monitoring growth polarity in fission yeast, a G2-delay-defective mutant(s) in the mor2 mutant should be isolated.
Materials and methods
Media and general methods
All media and standard methods were followed as described previously (Moreno et al., 1991).
Isolation of conditional lethal mutants with polarity defects
Wild-type cells (HM123, h–leu1-32) were mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine as described previously (Uemura and Yanagida, 1984). In total, 200 960 colonies were screened for temperature sensitivity, and 2822 ts mutants were isolated. Mutants that showed altered cell shape were then selected visually by fluorescence microscopy after staining with Calcofluor White (Hirata et al., 1998a; Radcliffe et al., 1998). The isolated round mutants were examined further for sensitivity to the protein kinase inhibitor staurosporine (STS; 1.5 µg/ml).
Cloning and disruption of the mor2+ gene
An S.pombe genomic library constructed in the vector pAL-KS (a gift from Dr Nakamura) was used for the isolation of plasmids that complemented the ts mor2 mutant (DH107-4C). One out of 9600 colonies was capable of growing at 36°C and showed normal rod shape. The segregation analysis showed that the Ts+ phenotype was plasmid dependent. One plasmid (pMR301) containing 9.3 kb insert was isolated. The result of the transposon insertion indicated that an internal region of ∼6.5 kb was essential for the complementing activity of pMR301. The resulting plasmid (pMR310-9) lost the complementing activity, but Ts+ colonies appeared from transformants (DH107-4C) containing pMR310-9. We found that pMR310-9 containing the LEU2 marker was integrated into the genome via homologous recombination in Ts+ transformants. Tetrad analysis between the Ts+ integrant and a wild-type strain yielded 4 Ts+ spores in each tetrad, and further tetrad analysis between the integrant and the mor2 mutant yielded four viable spores, in which Ts+ Leu+ and Ts– Leu– segregated 2:2 in each tetrad. This demonstrated that pMR310-9 contained the mor2+ gene itself. For disruption of the mor2+ gene, a 5.1 kb BamHI–SacI fragment of pMR301 was subcloned into pUC119, and a 3.0 kb HindIII–HindIII fragment of the insert was replaced by a ura4+ fragment, yielding pMR309 (pΔmor2::ura4+). A 3.9 kb BamHI–SacI fragment containing the deleted mor2 gene (Δmor2::ura4+) was used to disrupt the mor2+ gene. The disruption was verified by Southern hybridization.
Epitope and GFP tagging
C- or N-terminal tagging of proteins with 3HA or GFP was performed using PCR-generated fragments (Bahler et al., 1998).
Immunological assay
Total cell extracts were prepared as described in Matsusaka et al. (1995). Protein samples were electrophoresed on SDS–PAGE gels and electrophoretically transferred onto nitrocellulose filters. The primary antibodies, anti-GFP (8362-1; Clontech), anti-HA antibodies (HA.11; BabCO), anti-PSTAIRE (sc-53; Santa Cruz Biotechnology) and anti-phospho-Cdc2 (9111S; Cell Signaling Technology), were used for detection of GFP–Mor2, Mor2-HA, Cdc2 and Cdc2 phosphorylated on Tyr15, respectively. Horseradish peroxidase-conjugated sheep anti-rabbit IgG or anti-mouse IgG (Amersham) was used as the second antibody. Enhanced chemiluminescence (ECL; Amersham) was used to detect bound antibody.
Cytological techniques
Cytological techniques were performed according to Alfa et al. (1993) and Matsusaka et al. (1995). For F-actin staining, rhodamine–phalloidin was used. For tubulin staining, a monoclonal anti-tubulin antibody (TAT-1; a gift from Dr Keith Gull) was used as the primary antibody, and Cy3-conjugated sheep anti-mouse IgG (Sigma) as the second antibody. Calcofluor white (Sigma) was used to monitor cell wall growth, and 4′,6-diamidino-2-phenylindole (DAPI; Sigma) was used for observing chromatin regions.
Acknowledgments
Acknowledgements
We thank Paul Nurse, Damian Brunner, Fulvia Verde, Tomohiro Matsumoto, Paul Russell, Jonathan B.A.Millar, Keith Gull and Taro Nakamura for strains, antibodies and genomic library. We are grateful to Damian Brunner, Mohan K.Balasubramanian, Tadashi Uemura, Kazuma Tanaka, Yoshikazu Ohya, Hiroto Okayama and Yoshinori Jigami for discussions. D.H. was supported by PREST, JST and grants from the Ministry of Education, Science and Culture of Japan. T.T. was supported by Cancer Research UK and a Human Frontier Science Program Research Grant.
References
- Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Experiments with Fission Yeast: a Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Aligue R., Wu,L. and Russell,P. (1997) Regulation of Schizosaccharomyces pombe Wee1 tyrosine kinase. J. Biol. Chem., 272, 13320–13325. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu,J., Longtine,M.S., Shah,N.G., McKenzie,A.III, Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., Hirani,B.R., Burke,J.D. and Gould,K.L. (1994) The Shizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J. Cell Biol., 125, 1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinhauer J.D., Hagan,I.M., Hegemann,J.H. and Fleig,U. (1997) Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol., 139, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley N.J., Holtzman,D.A., Flaggs,G., Keegan,K.S., DeMaggio,A., Ford,J.C., Hoekstra,M. and Carr,A.M. (1996) The Schizosaccharommyces pombe rad3 checkpoint gene. EMBO J., 15, 6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Bidlingmaier S., Weiss,E.L., Seidel,C., Drubin,D.G. and Snyder,M. (2001) The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook J.D. et al. (1992) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell, 68, 799–808. [DOI] [PubMed] [Google Scholar]
- Browning H., Hayles,J., Mata,J., Avelinne,L. and Nurse,P. (2000) Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J. Cell Biol., 151, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D. and Nurse,P. (2000) CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell, 102, 695–704. [DOI] [PubMed] [Google Scholar]
- Chang E.C., Barr,M., Wang,Y., Jung,V., Xu,H.-P. and Wigler,M.H. (1994) Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell, 79, 131–141. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A., Chin,T.E. and Brent,R. (2001) Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell, 107, 739–750. [DOI] [PubMed] [Google Scholar]
- Cong J., Geng,W., He,B., Liu,J., Charlton,J. and Adler,P.N. (2001) The furry gene of Drosophila is important for maintaining the integrity of cellular extensions during morphogenesis. Development, 128, 2793–2802. [DOI] [PubMed] [Google Scholar]
- Du L.-L. and Novick,P. (2002) Pag1p, a novel protein associated with protein kinase Cpk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell, 13, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y., Tournier,S., Millar,J.B.A. and Hyams,J.S. (2001) A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature, 412, 352–355. [DOI] [PubMed] [Google Scholar]
- Geng W., He,B., Wang,M. and Adler,P.N. (2000) The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics, 156, 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbreth M., Yang,P., Bartholomeusz,G., Pimental,R.A., Kansra,S., Gadiraju,R. and Marcus,S. (1998) Negative regulation of mitosis in fission yeast by the Shk1-interacting protein Skb1 and its human homolog, Skb1Hs. Proc. Natl Acad. Sci. USA, 95, 14781–14786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff X.L., Woollard,A. and Simanis,V. (1999) Analysis of the cps1 gene provides evidence for a septation checkpoint in Schizosaccharomyces pombe. Mol. Gen. Genet., 262, 163–172. [DOI] [PubMed] [Google Scholar]
- Goode B.L., Drubin,D.G. and Barnes,G. (2000) Functional cooperation between the microtubule and actin cytoskeletons. Curr. Opin. Cell Biol., 12, 63–71. [DOI] [PubMed] [Google Scholar]
- Hagan I.M. and Hyams,J.S. (1988) The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci., 89, 343–357. [DOI] [PubMed] [Google Scholar]
- He X., Patterson,T.E. and Sazer,S. (1997) The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl Acad. Sci. USA, 94, 7965–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Toda,T. and Yanagida,M. (1984) The NDA3 gene of fission yeast encodes β-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell, 39, 349–358. [DOI] [PubMed] [Google Scholar]
- Hirata D., Masuda,H., Eddison,M. and Toda,T. (1998a) Essential role of tubulin-folding cofactor D in microtubules assembly and its association with microtubules in fission yeast. EMBO J., 17, 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D., Nakano,K., Fukui,M., Takenaka,H., Miyakawa,T. and Mabuchi,I. (1998b) Genes that cause aberrant cell morphology by overexpression in fission yeast: a role of a small GTP-binding protein Rho2 in cell morphogenesis. J. Cell Sci., 111, 149–159. [DOI] [PubMed] [Google Scholar]
- Ishiguro J. and Uhara,Y. (1992) Isolation and characterization of mutants supersensitive to the spindle poison, Isopropyl N-3-chlorophenyl carbamate (CIPC) in the fission yeast Schizosaccharomyces pombe. Jpn J. Genet., 67, 97–109. [DOI] [PubMed] [Google Scholar]
- Ishizaki T. et al. (1996) The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J., 15, 1885–1893. [PMC free article] [PubMed] [Google Scholar]
- Jin H. and Amberg,D.C. (2001) Fission yeast Aip3p (spAip3p) is required for an alternative actin-directed polarity program. Mol. Biol. Cell, 12, 1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice R.W., Zilian,O., Woods,D.F., Noll,M. and Bryant,P.J. (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev., 9, 534–546. [DOI] [PubMed] [Google Scholar]
- Katayama S., Hirata,D., Arellano,M., Perez,P. and Toda,T. (1999) Fission yeast α-glucan synthase Mok1 requires the actin cytoskeleton to localize the sites of growth and plays an essential role in cell morphogenesis downstream of protein kinase C function. J. Cell Biol., 144, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin,D.P., Matsumoto,S., Kitazono,A. and Matsumoto,T. (1998) Fission yeast Slp1: an effector of the mad2-dependent spindle checkpoint. Science, 279, 1045–1047. [DOI] [PubMed] [Google Scholar]
- Korinek W.S., Copeland,M.J., Chaudhuri,A. and Chat,J. (2000) Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science, 287, 2257–2259. [DOI] [PubMed] [Google Scholar]
- Lee L., Tirnauer,J.S., Li,J., Schuyler,S.C., Liu,J.Y. and Pellman,D. (2000) Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science, 287, 2260–2262. [DOI] [PubMed] [Google Scholar]
- Leung T., Manser,E., Tan,L. and Lim,L. (1995) A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J. Biol. Chem., 270, 29051–29054. [DOI] [PubMed] [Google Scholar]
- Lew D.J. (2000) Cell-cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces rerevisiae. Curr. Opin. Genet. Dev., 10, 47–53. [DOI] [PubMed] [Google Scholar]
- Liu J., Wang,H. and Balasubramanian,M.K. (2000) A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J. Cell Sci., 113, 1223–1230. [DOI] [PubMed] [Google Scholar]
- Lundgren K., Walworth,N., Booher,R., Dembski,M., Kirschner,M. and Beach,D. (1991) mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell, 64, 1111–1122. [DOI] [PubMed] [Google Scholar]
- Marcus S., Polverino,A., Chang,E., Robbins,D., Cobb,M.H. and Wigler,M.H. (1995) Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA, 92, 6180–6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks J. and Hyams,J.S. (1985) Localization of F-actin through the cell division cycle of Schizosaccharomyces pombe. Eur. J. Cell Biol., 39, 27–32. [Google Scholar]
- Marks J., Hagan,I. and Hyams,J.S. (1986) Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J. Cell Sci., 5, 229–241. [DOI] [PubMed] [Google Scholar]
- Mata J. and Nurse,P. (1997) tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell, 89, 939–949. [DOI] [PubMed] [Google Scholar]
- Mata J. and Nurse,P. (1998) Discovering the poles in yeast. Trends Cell Biol., 8, 163–167. [DOI] [PubMed] [Google Scholar]
- Matsui T. et al. (1996) Rho-associated kinase, a novel serine threonine kinase, as a putative target for small GTP-binding protein kinase Rho. EMBO J., 15, 2208–2216. [PMC free article] [PubMed] [Google Scholar]
- Matsusaka T., Hirata,D., Yanagida,M. and Toda,T. (1995) A novel protein kinase ssp1+ is required for alteration of growth polarity and actin localization in fission yeast. EMBO J., 14, 3325–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J.N., Longtine,M.S., Sia,R.A.L., Theesfeld,C.L., Bardes,E.S.G., Pringle,J.R. and Lew,D.J. (1999) The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol., 19, 6929–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P. and Johnson,D.I. (1994) Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol. Cell. Biol., 14, 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison J.M. (1970) Physiological and cytological methods for Schizosaccharomyces pombe. In Prescott,D.M. (ed.), Methods in Cell Physiology, Vol. 4. Academic Press, New York, NY, pp. 131–165.
- Mitchison J.M. and Nurse,P. (1985) Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci., 75, 357–376. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nurse P. (1975) Genetic control of cell size at cell division in yeast. Nature, 256, 547–551. [DOI] [PubMed] [Google Scholar]
- Nurse P. (1990) Universal control mechanism regulating onset of M-phase. Nature, 344, 503–508. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux,P. and Nasmyth,K. (1976) Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146, 167–178. [DOI] [PubMed] [Google Scholar]
- Ottilie S., Miller,P.J., Johnson,D.I., Creasy,C.L., Sella,M.A., Bagrodia,S., Forsburg,S.L. and Chernoff,J. (1995) Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J., 14, 5908–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki W.J., Becam,A., Nasr,F. and Herbert,C.J. (2000) Cbk1p, a protein similar to the human mytonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J., 19, 4524–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe P., Hirata,D., Childs,D., Vardy,L. and Toda,T. (1998) Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell, 9, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas J.C., Diaz,M., Duran,A. and Perez,P. (1991) Isolation and characterization of Schizosaccharomyces pombe mutants defective in cell wall (1-3)β-d-glucan. J. Bacteriol., 173, 3456–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K. and Russell,P. (1995) Counteractive roles of protein phosphatase 2C (PP2C) and a MAP kinase kinase homolog in the osmoregulation of fission yeast. EMBO J., 14, 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulewitz M.J., Inouye,C.J. and Thorner,J. (1999) Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7123–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell V. and Nurse,P. (1993) Investigations into the control of cell form and polarity: the use of morphological mutants in fission yeast. Development, 289–299. [PubMed] [Google Scholar]
- Snell V. and Nurse,P. (1994) Genetic analysis of cell morphogenesis in fission yeast—a role for casein kinase II in the establishment of polarized growth. EMBO J., 13, 2066–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I., Shochet,N.R., Kashman,Y. and Groweiss,A. (1983) Latrunculins: novel marine toxins that disrupt microfilament organization in cultures cells. Science, 219, 493–495. [DOI] [PubMed] [Google Scholar]
- Toda T., Shimanuki,M. and Yanagida,M. (1993) Two novel protein kinse C-related genes of fission yeast are essential for cell viability and implicated in cell shape control. EMBO J., 12, 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Niwa,H., Nemoto,T., Dhut,S., Eddison,M., Matsusaka,T., Yanagida,M. and Hirata,D. (1996) The fission yeast sts5+ gene is required for maintenance of growth polarity and functionally interacts with protein kinase C and an osmosensing MAP-kinase pathway. J. Cell Sci., 109, 2331–2342. [DOI] [PubMed] [Google Scholar]
- Uemura T. and Yanagida,M. (1984) Isolation of type I and II topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J., 3, 1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K., Toda,T., Hayashi,S. and Yanagida,M. (1983) Cell division cycle genes nda2 and nda3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J. Mol. Biol., 168, 271–284. [DOI] [PubMed] [Google Scholar]
- Verde F., Mata,J. and Nurse,P. (1995) Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J. Cell Biol., 131, 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Wiley,D.J. and Nurse,P. (1998) Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl Acad. Sci. USA, 95, 7526–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A., Ingles,J., McGhee,J.D. and Mains,P.E. (1997) Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Dev., 11, 409–422. [DOI] [PubMed] [Google Scholar]
- Xu T., Wang,W., Zhang,S., Stewart,R.A. and Yu,W. (1995) Identifying tumor suppressors in genetic mosaic: the Drosophila lats gene eoncodes a putative protein kinase. Development, 121, 1053–1063. [DOI] [PubMed] [Google Scholar]
- Yarden O., Plamann,M., Ebbole,D.J. and Yanofsky,C. (1992) Cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J., 11, 2159–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J.A., Peckol,E.L., Tobin,D.M. and Bargmann,C.I. (2000) Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/Warts serine/threonine kinase family. Mol. Biol. Cell, 11, 3177–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]