When one adult meets another at a party, they often get to know each other by asking the question, “What do you do?” Translated, this question asks how you earn a living. When a microbiologist meets a newly isolated bacterium for the first time, the initial question is similar: What does it do? By this the scientist is really asking how that bacterium makes a living energetically; that is, what it will transform in its environment in order to generate more of itself. Bacteria do this a little more directly than humans, taking what they can from their environment without the intermediacy of money, but it amounts to the same thing.
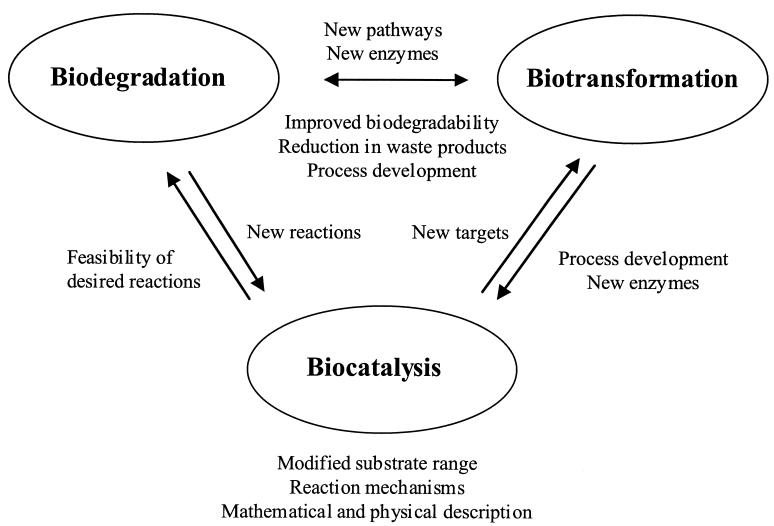
The American Society for Microbiology and the Society for General Microbiology cosponsored a Conference on Biodegradation, Biotransformation, and Biocatalysis (B3) that was held in San Juan, Puerto Rico, 2 to 6 October 2001. The conference was organized by Gary Sayler (University of Tennessee), David Gibson (University of Iowa), Hilary Lappin-Scott (University of Exeter), James Tiedje (Michigan State University), and Gary Toranzos (University of Puerto Rico). The conference dealt with how bacteria transform chemicals to make their living and how their activities can help humans make a living. In this context, the terms that comprise the title of the conference all deal with the same thing: microbial metabolism. Which term is used depends on the interest of the persons studying it. If their interest is in degrading environmental pollutants, they are said to study biodegradation. An industrialist using microbial metabolism is said to be conducting a biotransformation or to be using biocatalysis. In some cases, these interests can overlap (Fig. 1). In an illuminating talk by Mani Subramanian of The Dow Chemical Company, the process of biodegrading a waste compound is being integrated into manufacturing by biotransforming the unwanted side reaction product, 1,2,3-trichloropropane, into epichlorohydrin, a product of commercial value. In such an example, commerce and the environment benefit simultaneously.
FIG. 1.
Interdependence of the three main application areas of enzyme catalysis.
Biotransformations were observed by humans well before they were appreciated as having an underlying microbial cause. For example, food rot and the products of microbial fermentations have been dreaded and enjoyed for thousands of years, respectively. In 1858, Louis Pasteur provided evidence for the role of specific microorganisms conducting favorable and unfavorable fermentations of grape juice (72). The properties of enzymes, the principle biocatalysts, became generally appreciated from kinetic studies conducted in the early 1900s (62). An important industrial-scale fermentation to produce acetone to meet the wartime needs of Great Britain was developed in 1916 (35). In subsequent years, commodity chemicals largely came from petrochemical resources. But there is an increased realization that petroleum resources are limited. As a result, there is renewed interest in industrial biotransformations, with the input carbon derived from renewable sources such as corn starch. The B3 conference was exquisitely timed to bring together the common interests of engineers, chemists, industrialists, and basic biologists to help meet the needs of society for cleaner and more sustainable economic development.
The size of the B3 meeting (173 participants) allowed for excellent discussions during and outside the sessions. Ample time at the poster sessions also provided time for one-on-one and small-group interactions. Coming so soon after the September 11 tragedies, several registered participants were unable to attend, but time slots were graciously filled by volunteer speakers who arrived with presentations in hand. This minireview summarizes the results presented by the invited and replacement speakers as well as the six short presentations that were chosen from the poster abstracts.
In his keynote address, Jim Tiedje (Michigan State University) provided a perspective on the potential of microbes for practical application in the B3 fields. The application of our knowledge of biodegradative pathways may provide the most cost-effective technology for the remediation of pollutant-contaminated sites. This type of technology may also be the most acceptable to the public: it is a natural process that is essentially taking advantage of nature's recycling abilities. The most effective and acceptable systems strive for complete conversion of pollutants to inorganic products. Bacterial pathways for the degradation of many compounds that were previously thought to be nonbiodegradable are presently under study. For example, novel halorespiring microbes, such as Dehalococcoides spp., have the ability to catalyze the complete conversion of chlorinated ethenes to nontoxic metabolites (28, 60, 61). Pathways are being characterized from bacterial isolates that grow on and completely degrade aromatic hydrocarbons under anaerobic conditions (18, 42, 75, 91, 98). In addition, carbon tetrachloride can be metabolized by Pseudomonas stutzeri KC by a novel process (55-58). Four examples of bioremediation were discussed. In some cases biostimulation was required to allow indigenous bacteria to cometabolize a contaminant (e.g., addition of toluene or phenol to stimulate oxidation of trichloroethylene [TCE]; Moffett Federal Airfield, Mountain View, Calif.) (32, 46). At other sites, bioaugmentation with a chlorinated ethene-degrading coculture (Bachman Road, Oscoda, Mich.) or carbon tetrachloride-degrading P. stutzeri KC (Schoolcraft, Mich.) resulted in complete removal of chlorinated ethenes or carbon tetrachloride, respectively. In the former case, the bioaugmented plot completely degraded chlorinated ethenes to ethane within 6 weeks, while the parallel biostimulation plot took 3 to 4 months to achieve the same result. In the latter case, the strategy of niche adjustment was used to give the added microbes a competitive advantage as well as to stimulate production of the biodegradation catalyst (25). It is clear from these studies that every site is different, and site assessment is important in the development of an effective bioremediation strategy.
BIODEGRADATION OF HERBICIDES, PESTICIDES, AND ANTIMICROBIALS
Triclosan [5-chloro-2-(2,4-dichlorophenoxy) phenol] is an antimicrobial agent used in a wide range of consumer products, including soaps, toothpastes, hand creams, and plastics. Pseudomonas strains are typically resistant to triclosan, and this compound is the selective ingredient in Pseudomonas isolation agar (PIA; Difco Laboratories, Inc.). Anthony Hay (Cornell University) enriched for triclosan-degrading microorganisms by using samples from a municipal wastewater treatment plant. A stable bacterial consortium consisting of six microorganisms was capable of using triclosan as the sole source of carbon and energy, removing over 90% of the supplied triclosan with 35% mineralization of the compound (39).
Atrazine is a true xenobiotic compound that has been one of the most widely used herbicides for the control of broad leaf and grassy weeds. Michael Sadowsky (University of Minnesota) presented an up-to-date overview of atrazine degradation by Pseudomonas sp. ADP, a strain isolated from a herbicide spill site in Minnesota (95). Pseudomonas sp. ADP metabolizes atrazine to carbon dioxide and ammonia and uses atrazine as the sole nitrogen source for growth. The genes atzA, atzB, and atzC encode enzymes catalyzing the first three steps of atrazine degradation, converting it to cyanuric acid. These three genes are located separately on a 109-kb plasmid (pADP-1) in strain ADP and are highly conserved and globally distributed (21, 22). AtzA (atrazine chlorohydrolase) was purified, and its substrate range was recently determined (88). AtzA was capable of hydrolyzing an atrazine analog that carried a fluorine in place of the chlorine substituent, but not when azido, cyano, methoxy, thiomethyl, or amino groups were substituted. In addition, AtzA also catalyzed dechlorination of analogs with N-alkyl groups of various sizes. The recent sequencing of pADP-1 (59) revealed the presence of the atzDEF gene cluster, which is required for the conversion of cyanuric acid to carbon dioxide and ammonia. AtzD, a cyanuric acid amidohydrolase, converts cyanuric acid to biuret. AtzE and AtzF, members of an amidase family of proteins, convert biuret sequentially to allophanate and finally to CO2 and NH3. The plasmid sequence, originally identified in pR751, also revealed a mercury resistance gene cluster and numerous transposase genes.
Mike Manefield (Oxford) described the development of an RNA-based stable isotope labeling method to link function to phylogeny in complex microbial communities. In short, 13C-labeled substrate (in this case, phenol) is taken up by the organisms capable of its catabolism, and 13C is incorporated into cell biomass. By sequencing the 16S rRNA genes from the stable isotope-labeled (high density) RNA pool, the functional (phenol degrading) organisms were identified. With standard isolation techniques pseudomonads were typically isolated as the dominant phenol-degrading organisms in the aerobic reactor under investigation (97). A surprising result was observed with this innovative stable isotope labeling method. The dominant phenol degrader was a strain of Thauera aromatica, an organism that typically degrades aromatic compounds by reductive routes in the absence of molecular oxygen.
Christopher Smejkal (Hilary Lappin-Scott's group, University of Exeter) described the characterization of three strains of bacteria that degrade the herbicide mecoprop [2-(2-methyl-4-chlorophenoxy) propionic acid]. These strains were isolated by selective enrichments with mecoprop as the sole source of carbon and were identified as Alcaligenes denitrificans, Alcaligenes sp. CS1, and Ralstonia sp. CS2 (90). These strains were compared with 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria. All three mecoprop-degrading bacteria housed a large plasmid which contained the genes for mecoprop degradation. These genes showed high similarity to the tfd genes, which mediate 2,4-D degradation in bacteria (89).
Bernardo González (Universidad Católica de Chile) reported several interesting sets of experiments that demonstrated the versatility and plasticity of the 2,4-D degrading strain Ralstonia eutropha JMP134(pJP4). This strain grows on 3-chlorobenzoate (3-CB) as well as on 2,4-D, using pathways carried on plasmid pJP4. When maintained on 3-CB, a derivative was isolated that grew faster on 3-CB but lost the ability to grow on 2,4-D. Analysis of the plasmid present in this strain indicated that it had undergone an intermolecular rearrangement that resulted in a duplication of the tfdCIDIEIFI and tfdDIICIIEIIFII gene modules that are required for the conversion of 3-CB to β-ketoadipate. The rearrangement also resulted in the deletion of tfdA, which is required for the initial oxidation of 2,4-D (17). A strain that was transferred on 2-methylphenoxyacetate retained the ability to grow with several phenoxyacetates, including 2,4-D, but lost the ability to grow with 3-CB. The plasmid present in this strain had lost the tfd-I and tfd-II gene clusters but had a duplication of tfdA. By manipulating the copy number of the tfd-I and tfd-II gene clusters of cells growing on 3-CB and by monitoring enzyme activities, it became clear that different genes in each module are necessary for efficient growth on 3-CB (73). In particular, the contribution of tfdCI was critical to prevent the accumulation of the toxic metabolite chlorocatechol. Both JMP134(pJP4) and its cured derivative, JMP222, are able to grow on 2,4,6-trichlorophenol (TCP), suggesting that that pathway for TCP degradation is chromosomally encoded (16). However, while JMP222 can only grow with concentrations of TCP of up to 0.2 mM, JMP134(pJP4) can easily tolerate up to 0.5 mM TCP. Pathways for the degradation of 2,4-D and TCP converge at maleylacetate, and it was proposed that at higher TCP concentrations the plasmid-encoded maleylacetate reductase contributes to the ability of the strain to grow with higher levels of TCP (65).
B3 WITH OXYGENASES
The structure of the oxygenase component of naphthalene dioxygenase (52) has provided a wealth of information about multicomponent Rieske nonheme iron oxygenases. Rebecca Parales (University of Iowa) reported results of site-directed mutagenesis studies that were initiated on the basis of predictions made from the structure of the enzyme. In these studies, single or multiple amino acid substitutions were introduced into the α subunit of naphthalene dioxygenase. The aspartate residue at position 205 was shown to be essential for activity, and results support the hypothesis that Asp-205 is required for efficient electron transfer from the Rieske center in one α subunit to the mononuclear iron in an adjacent α subunit within the α3β3 hexamer (70). In addition, single amino acid substitutions near the active site resulted in enzymes with altered regio- and enantioselectivities. Enzymes with substitutions at position 352 within the α subunit had altered enantioselectivity with naphthalene, biphenyl, and phenanthrene and had major changes in regioselectivity with biphenyl and phenanthrene (69, 71, 100).
The application of microbial oxygenases for selective synthesis of mono- and dihydroxylated hydrocarbons and sulfur compounds has a long and successful tradition (8, 44, 47). Derek Boyd (The Queen's University of Belfast) pointed out the importance of isoenzyme selection and substrate geometry to achieve selective oxidations. Synthetic applications on a milligram-to-gram scale included the epoxidation of 2-(trifluoromethyl) methylbenzoate by the fungus Phellinus ribis (13) and selective tandem polyoxygenation of, e.g., indane derivatives and propylbenzene by toluene dioxygenases (6, 9) and larger polycyclic ring systems by biphenyl dioxygenase (10, 11). Coupling the biocatalytic formation of cis-diols to chemical oxidation and reduction allows selective synthesis of trans-diols (7, 13). The broad substrate range and reaction spectrum of bacterial dioxygenases (78) together with the availability of new enzyme derivatives from mutation experiments (12, 71) will make this class of enzymes even more interesting for future synthetic applications.
Peter Williams (University of Wales, Bangor) presented recent studies on the nag genes of Ralstonia sp. strain U2 which encode the pathway for naphthalene degradation (33, 101). This pathway is of particular interest because it differs from most of the previously characterized naphthalene degradation pathways in strains of Pseudomonas isolated from different geographical regions. The nah genes of Pseudomonas spp. mediate the metabolism of naphthalene to catechol and then to central metabolites via the meta-cleavage pathway. Peter Williams' group has now sequenced and characterized all of the nag genes encoding the enzymes involved in catabolism of naphthalene which are in a large cluster in strain U2. The Nag pathway degrades naphthalene to salicylate in a manner identical to that of the Nah pathway. In strain U2, however, salicylate is degraded via gentisate rather than catechol. The similarities between the nag and nah gene clusters for the conversion of naphthalene to salicylate suggest a common ancestry, but both appear to have acquired separate modules of genes for the terminal reactions from salicylate to central metabolites. The initial monooxygenase in the conversion of salicylate to gentisate, salicylate 5-hydroxylase (NagGH), is a member of the same family as the naphthalene dioxygenases, and in Ralstonia sp. strain U2 NagGH shares the same electron transport proteins (NagAa and NagAb) with its naphthalene dioxygenase (NagAcAd). Similarities in nucleotide sequence and gene organization suggest that the dioxygenase genes mediating nitrobenzene and nitrotoluene degradation have evolved from a nag-like rather than a nah-like naphthalene dioxygenase gene cluster (33).
The rhodococci are a remarkable group of microorganisms known for their ability to inhabit a diverse range of environments. They are able to metabolize a plethora of different carbon sources and have proven to be particularly adept at utilizing xenobiotic compounds as carbon and nitrogen sources for growth. The rhodococci are proving to be of significant interest to industries as biocatalysts. Michael Larkin (The Queen's University of Belfast) described how the biochemical and genetic characteristics of rhodococci from geographically distinct locations varied, using haloalkane metabolism as a specific example. Haloalkane dehalogenase genes from five species of rhodococci and one Pseudomonas strain isolated from contaminated sites in Europe, Japan, and the United States and the typed strain Rhodococcus sp. NCIMB13064 were compared (34, 76). Despite the fact that all five Rhodococcus strains were isolated independently from different continents, they were found to possess similar haloalkane catabolic gene clusters encoding the dehalogenase, an alcohol dehydrogenase, and an aldehyde dehydrogenase. The high degree of sequence similarity and the conservation of gene order suggests that these strains obtained the gene cluster as a preassembled unit from a common ancestral strain. These data also suggest that the distribution process occurred fairly recently and may be related to the widespread use of synthetic haloalkanes in industry and agriculture. In contrast, an interesting comparison was made between the pseudomonad and rhodococcal naphthalene dioxygenases. This enzyme catalyzes the oxidation of naphthalene to naphthalene cis-dihydrodiol. Of particular note is that the amino acid sequence of the catalytic component of the Rhodococcus naphthalene dioxygenase (ISPNAP α subunit) was greatly divergent from that of the analogous Pseudomonas subunit and raises a question as to the evolutionary origins of these enzymes (54). The crystal structures of the two enzymes, however, are highly conserved.
Nitrophenols are used as starting materials in the production of a variety of pharmaceuticals, pesticides, and explosives (102). Because of the use of nitrophenol derivatives in agricultural applications and their relatively high water solubilities, there have been concerns about the fate of nitrophenols in the environment. Gerben Zylstra (Rutgers University) reported the purification and characterization of the hydroxyquinol ring cleavage dioxygenase that is involved in p-nitrophenol degradation in Arthrobacter sp. strain JS443. This strain degrades p-nitrophenol by two sequential oxidations to hydroxyquinol (1,2,4-benzenetriol). Hydroxyquinol is then degraded through an ortho cleavage pathway (50, 102). PCR primers were designed on the basis of the N-terminal amino acid sequence of the purified protein, and Southern hybridizations were carried out to identify a cosmid carrying the hydroxyquinol dioxygenase gene (npdB). In the same gene cluster, genes encoding the reductase and oxygenase components of the p-nitrophenol monooxygenase (npdA1 and npdA2, respectively), which catalyzes the first two steps in p-nitrophenol degradation, maleylacetate reductase (npdC), which catalyzes the reaction following ring cleavage, and a regulatory protein (npdR) were identified. The substrate range of p-nitrophenol monooxygenase was investigated in mixtures containing crude lysate from an npdA2 expression clone, purified NpdA1, NAD, FAD, substrate, and an NADH regeneration system. The enzyme converted p-nitrophenol, p-chlorophenol, 4-nitrocatechol, 4-chlorocatechol, and 4-chlororesorcinol to hydroxyquinol at similar rates, converted phenol at a slower rate, and did not oxidize hydroquinone, catechol, or resorcinol at all. p-Nitrophenol monooxygenase catalyzed single oxidations on m-chlorophenol and nitrophenol to yield chloro- and nitrohydroquinols.
Chlorinated ethylenes are industrial solvents and common groundwater pollutants. TCE and tetrachloroethylene (PCE) can be degraded anaerobically, but in many cases degradation stops with the production of cis-dichloroethylene or vinyl chloride. The latter is a documented human carcinogen. Under aerobic conditions, PCE had not been degraded previously by any oxygenases. Tom Wood (University of Connecticut) reported that PCE is oxidized by toluene-o-monooxygenase from P. stutzeri OX1 (81) and that TCE, PCE, and toluene induce the production of the monooxygenase (80). Strain OX1 was shown to be chemotactic to PCE, and PCE also induced chemotaxis in this strain. This is the second example of a chemotactic response to PCE, the first having been reported for the toluene-degrading strain Pseudomonas putida F1 (68). In another study, Wood used DNA shuffling to improve the activity of toluene ortho-monooxygenase from Burkholderia cepacia G4. A single mutation in the α subunit of the hydroxylase (Val-106 to Ala) resulted in threefold higher activity with TCE and also yielded sixfold faster oxidation of naphthalene (15).
Ken Reardon (Colorado State University) presented the results of a collaborative project with Tom Wood's group (University of Connecticut) on the engineering of a strain of Escherichia coli capable of oxidizing PCE and TCE. Nonspecific oxygenases have been shown to degrade these solvents; however, the reaction rates are slow and the resulting chlorinated epoxides are toxic to the cell. To overcome the first of these limitations, an optimized toluene ortho-monooxygenase was produced by using methods of directed evolution. The effect of the expression of these recombinant proteins on the physiology of the host strain was investigated by proteomic analysis with two-dimensional polyacrylamide gel electrophoresis, and significant changes to the proteome were found.
Lindsay Eltis (University of British Columbia) discussed the mechanisms of inhibition of individual enzymes of the biphenyl/polychlorinated biphenyl (PCB) pathway. For example, biphenyl dioxygenase, which catalyzes the first step in the pathway, is partially uncoupled when provided with dichlorobiphenyls as substrates. The reaction results in the futile consumption of O2 and reducing equivalents and the production of hydrogen peroxide (49). The third enzyme in the pathway, 2,3-dihydroxybiphenyl 1,2-dioxygenase, is subject to competitive inhibition by 2′6′-dichloro-2,3-dihydroxybiphenyl. This compound binds tightly in the active site but is cleaved extremely slowly. The crystal structure of the enzyme-inhibitor complex demonstrated that one of the chloride substituents occupies the O2 binding site and prevents binding of O2 and catalysis. This enzyme also loses activity in the presence of the suicide inhibitor 3-chlorocatechol due to oxidation of the active-site ferrous iron (92). The serine hydrolase BphD, which catalyzes the hydrolysis of the ring cleavage product 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (HOPDA), is competitively inhibited by 3-chloro- and 4-chloro-HOPDAs (86, 87). Because many PCBs and their metabolites are successfully degraded by biphenyl pathway enzymes, it may be possible to engineer enzymes that will not be subject to inhibition by the small subset of toxic metabolites. If this is the case, then it should also be possible to construct efficient pathways for the degradation of PCB mixtures.
Methane monooxygenase (MMO) is one of the key enzymes in the global carbon cycle. Similar to naphthalene dioxygenase, this multiheteromeric enzyme complex contains a small electron transfer chain, but in contrast to naphthalene dioxygenase it contains a diiron cluster for oxygen activation. The particulate as well as the soluble isoforms of the enzyme have been used industrially for the preparation of various alcohols, ketones, and epoxides from cheap hydrocarbons. Methylococcus capsulatus (Bath) was, for example, shown to oxidize 220 different substrates. Howard Dalton (University of Warwick, Coventry, United Kingdom) reported on the interaction of the B subunit of soluble MMO with electron transport and effective substrate hydroxylation (96). The B subunit docks in a canyon of the oxygenase component as shown earlier. Activity tests of soluble MMO via electron flow analyses (cyclovoltammographic studies, performed together with Allen Hill, Oxford, United Kingdom) were done with a set of N-terminal deletion mutants of the B subunit and showed that proteins with deletions of more than 8 amino acids in length were inactive. These truncated B mutants also blocked the H2O2 shunt reaction cycle. In the oxygenase subunit, Tyr-213 was identified as being necessary for activity. These data may allow new engineering approaches for MMO in the direction of a catalytic cycle independent of electrons derived from NADH.
BIOCATALYSIS
Streptomyces griseus has kept Jack Rosazza's (University of Iowa) group busy for more than 25 years with identifying novel metabolites and transformations. It was the paradigm for the concept of microbial models for mammalian metabolism by catalyzing every type of phase I metabolic reaction with a wide variety of novel compounds. Of particular interest are the reactions this organism catalyzes with flavonoids. This is the most ubiquitous group of polyphenolic compounds in foods of plant origin. They are considered to produce a wide variety of beneficial effects, including protection against cardiovascular disease and certain forms of cancer. Flavonoids are thought to exert these biological effects through antioxidative activities whereby they scavenge free radicals. Microorganisms have been shown to carry out a diverse range of flavonoid transformations, including oxidation, reduction, and deglycosylation reactions. S. griseus was observed to hydroxylate and methylate a variety of flavonoid substrates. With S. griseus, methylation was only seen to occur with catechol moieties. A new catechol O-methyltransferase was purified from cells of S. griseus (23). The enzyme catalyzed the regiospecific transfer of the methyl group of S-adenosyl-l-methionine to the 6-hydroxyl group of 6,7-dihydroxycoumarin. An investigation of thiols in S. griseus revealed the presence of glutathione and the subsequent identification of the first glutathione S-transferase in a streptomycete.
There is considerable interest in reactions that can improve the yield of enantiomerically pure chiral compounds. Kinetic resolution of racemates is used successfully by industry; however, the theoretical yield can never exceed 50%. Nicholas Turner (University of Edinburgh) discussed three types of reactions that give yields greater than 50%: first, asymmetric synthesis using nonchiral compounds; second, dynamic kinetic resolution whereby the unreactive enantiomer is converted in situ to the reactive enantiomer; and third, a much less exploited approach by a process of deracemization. This can be achieved either by using two enzymes with opposite enantioselectivity (e.g., deracemization of secondary alcohols by alcohol dehydrogenases) or by using a single enzyme in combination with a chemical reagent (e.g., amino acid oxidase and a reducing agent for the deracemization of alpha amino acids). In 1971, Hafner and Wellner (37) reported the conversion of d-alanine to its respective l-enantiomer by using d-amino acid oxidase and sodium borohydride. They demonstrated that imino acids are intermediates in the reaction and that reduction of these intermediates was 36 times faster than that by hydrolysis. Subsequently, this method was extended to the deracemization of dl-proline (48). Turner's group has made considerable inroads in developing novel enzyme-based systems for the deracemization of racemic mixtures of chiral compounds by investigating different reducing agents and screening for improved oxidases. Earlier work used sodium borohydride as the reducing reagent, which is sensitive to water and easily decomposes. A more effective reducing agent was ammonium formate with palladium and/or charcoal.
The chemical synthesis of even simple carbohydrates is technically difficult and, thus, is prohibitively expensive on an industrial scale due to the need for complex protection group chemistry. Since complex carbohydrates play important roles in many aspects of biology, an inexpensive and convenient route to their synthesis is desirable. According to David Crout (University of Warwick), the use of glycosidases to catalyze the formation of carbohydrates (the reverse of their natural activity) presents a simple and cost-effective alternative (20, 85). Glycosidases are robust enzymes, and many are commercially available. However, incomplete regioselectivity is still a drawback in some cases. Synthesis examples comprise, e.g., the nodulation factors of rhizobia, chito-oligosaccharides, and adhesion molecules of Clostridium difficile (diarrhea) and Helicobacter pylori.
Epoxide hydrolases (EHs) are ubiquitous and are found in all mammalian cells as well as in microorganisms, where they function as detoxifying agents. The broad application of these readily available enzymes for the formation of chiral epoxides and diols from racemic mixtures of epoxides has been reported previously (3). EHs are highly interesting biocatalysts because they are cofactor independent and because various isoenzymes covering a broad substrate spectrum have been described, argued Roland Furstoss (Université de la Mediterranée, Marseille Cedex, France). The impressive presentation showed the applicability of EHs for production of epoxides and vicinal diols on a gram scale with excellent enantiomeric excess (ee) and enantioselectivity (E) values. Process development allowed the preparation of different enantiopure styrene oxide derivatives with Aspergillus niger EH at substrate concentrations of up to 2.5 M and productivities of up to 375 g liter−1 h−1 with 4 to 20 g of enzyme per liter. The new methodology compares very favorably with the presently known chemical approaches that require the use of heavy-metal-based catalysts. The A. niger EH is presently available from Fluka, thus opening the way to further exploration. A new spectroscopic assay for epoxide activity based on the oxidation of the aromatic diol products by NaIO4 to corresponding aldehydes (detection at 290 nm) will allow high-throughput screening of mutants and straightforward enzyme engineering to obtain better thermostability and higher E values for unnatural substrates.
Don Cowan (University of the Western Cape, Cape Town, South Africa) discussed the use of thermophilic nitrilases and nitrile hydratases for the convenient conversion of nitriles to the corresponding acids and amides. He reported the purification, characterization, and applications of two hydratases and a nitrilase from two thermophilic Bacillus strains (1, 2, 19, 36, 74). Each of the enzymes was more thermostable than corresponding enzymes isolated from mesophiles, but their temperature optima were marginally lower than the growth temperatures of the bacterial strains from which they were isolated (55 to 65°C). For the production of amides by nitrile hydratases, amidases that are typically present need to be mutated or inhibited or the nitrile hydratase genes need to be cloned and expressed in a background lacking amidase activity. In one case, the amidase(s), but not the hydratase, was inhibited by urea. Present studies are focused on the identification of nitrile-oxidizing enzymes from the metagenome. This strategy will circumvent the difficulties and limits of culturing thermophilic extremophiles in the search for highly thermostable enzymes.
B3 WITH NOVEL PATHWAYS AND ENZYMES; NEW USES FOR OLD PROTEINS
In his talk, Jim Spain (Air Force Research Laboratory, Tyndall Air Force Base, Fla.) emphasized that biodegradation pathways are continually evolving, especially pathways for the degradation of human-made compounds that have not been present in nature for more than a few decades. The study of pathways for the degradation of xenobiotic compounds often results in the identification of novel enzymatic reactions. In some cases the enzymes are recognizable as close relatives of enzymes present in pathways for the degradation of naturally occurring compounds, but in other cases it is not clear what the source of the genes encoding novel enzymes might have been. Thus, pathways for the degradation of human-made compounds may provide a source of novel biocatalysts. Novel enzymes involved in the reductive pathways of nitrobenzene degradation in Pseudomonas include nitrobenzene nitroreductase, hydroxylaminobenzene mutase, and 2-aminophenol dioxygenase (64). Together, nitrobenzene nitroreductase and hydroxylaminobenzene mutase catalyze the conversion of various nitroaromatic compounds to the corresponding o-aminophenols (40, 63). Substituted o-aminophenols can be used to produce polybenzoxazole polymers and starting materials for the production of compounds with electronic, medical, and pharmaceutical applications. Other enzymes from nitroarene-degrading pathways can convert aminodinitrotoluenes to aminonitrocatechols (51). Enzymes of the toluene and nitrobenzene degradation pathways catalyze biotransformations that result in the production of picolinic acids, compounds that can be used as starting materials for the production of pharmaceuticals, herbicides, dyes, and polymers (41). Many of the compounds that are produced by these biocatalytic routes are difficult to synthesize chemically, and some are novel.
Hans-Joachim Knackmuss (University of Stuttgart) discussed the underlying chemistry behind the recalcitrance of chloroethenes and nitroaromatics (79). Under aerobic conditions microorganisms degrade aromatic compounds by initial activation of the compound by oxygenation with the subsequent formation of catechols, followed by ring cleavage and subsequent dissimilation into pathways of central metabolism. Aromatic compounds become resistant to electrophilic attack by oxygenases with the presence of electron-withdrawing substituents, such as halo, azo, or nitro groups. Theoretically, these compounds are more likely to be reduced by enzymes rather than oxidized, particularly in anoxic microbial systems which are capable of catalyzing a wide range of reductive reactions. Reductive dechlorination, hydrogenolytic denitrations, and reduction of nitro groups decrease the redox potential of the xenobiotic compounds to such an extent that the metabolites generated are now susceptible to oxidative attack. Increased reactivity of the reduced products with molecular oxygen can facilitate a subsequent oxidative treatment process. An elegant example was presented, whereby groundwater contaminated with tetrachloroethene was treated with an integrated reductive/oxidative process by using a 30-liter packed bed reactor for the anaerobic dechlorination process and a 40-liter trickling filter for the subsequent aerobic oxidation step. Knackmuss also presented a detailed account of the reductive transformation of trinitrotoluene (TNT) under aerobic and anaerobic conditions. TNT is another example of a π-electron ring system with high electron deficiency. The nitro groups are readily reduced even under aerobic conditions. Importantly, nitro group reduction results in highly reactive species, which can form covalent bonds with the organic soil matrix. Mass balance studies indicated the enzyme-catalyzed formation of nonextractable TNT/soil complexes, opening the possibility of soil remediation through cometabolically induced immobilization. Perspectives on the mineralization of polynitroaromatic acids were presented, including the formation of hydride-Meisenheimer complexes of picric acid with reducing equivalents derived from coenzyme F420 (26, 43). Nitrite elimination results in the formation of 2,4-dinitrophenol, which can then be further degraded.
A large amount of land and ground water is now heavily contaminated with high explosives, such as TNT, hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), and nitroglycerin. The progressive accumulation of these toxic compounds in the soil and groundwater is deleterious to biological systems. Neil Bruce (University of Cambridge) described the isolation of bacteria from explosive-contaminated sites that can break down all of the major classes of explosives, in many cases to harmless products. Of particular interest is the evolutionary origin of the enzymes that have adapted to degrade explosives, since these compounds are xenobiotic in the strictest sense. Enterobacter cloacae strain PB2 was isolated from a munitions plant in the United Kingdom by its ability to utilize nitrate ester explosives as a sole nitrogen source for growth (5). The enzyme shown to catalyze the sequential liberation of two of the four nitro groups of pentaerythritol tetranitrate (PETN) and two of the three nitro groups of nitroglycerin was found to be an NADH-dependent reductase (PETN reductase). The enzyme forms part of a growing family of flavoproteins known to be homologous to Old Yellow Enzyme (30). Members of the Old Yellow Enzyme family have been identified as a consequence of their catalysis of a diverse range of transformations. The crystal structure of PETN reductase has now been elucidated, and it is a single domain protein with an Mr of 40,000 with a typical eight-stranded β/α barrel with a noncovalently bound flavin mononucleotide as the prosthetic group (4). Interestingly, PETN reductase confers on E. cloacae the ability to utilize TNT as a sole source of nitrogen for growth. The enzyme, like many flavoenzymes, is able to reduce the nitro groups of TNT, producing hydroxylamino- and amino-dinitrotoluenes. PETN reductase, however, also catalyzes the addition of hydride to the aromatic ring of TNT, yielding hydride and dihydride-Meisenheimer complexes, which appear to be further reduced, yielding nitrite and other as-yet uncharacterized end products (29). Expression of PETN reductase and a bacterial nitroreductase in plants provided an interesting example of how plants can be engineered to degrade and sequester explosives from the environment (31, 38).
Aromatic compounds are metabolized by microorganisms from a broad variety of different genera and species. Due to the wealth of information on its microbiology, biochemistry, and genetics, a very appealing example is E. coli. Its ability to degrade aromatic compounds was recently reviewed (24). Eduardo Diaz (Centro Investigaciones Biologicas-CSIC, Madrid, Spain) highlighted the metabolism of phenylacetic acid (PA) in E. coli W (14), its regulation, and the extension of this pathway to the complete degradation of styrene after heterologous expression of styrene upper pathway genes from Pseudomonas sp. strain Y2 (93). Styrene is transformed via styrene oxide and phenylacetaldehyde to PA. Subsequently PA is coupled to coenzyme A by PA-coenzyme A ligase (PaaK), a reaction strongly resembling the activation of hydrocarbons in anaerobic degradation pathways. In contrast, p-hydroxyphenylacetic acid is channeled into a meta-cleavage pathway via 3,4-dihydroxyphenylacetic acid by a monooxygenase with a very broad substrate range. These findings show that besides its use as a tool in molecular biology and as a host organism for heterologous production of interesting catalytic activities, the metabolic capabilities of E. coli itself may be exploited further for biodegradation and biotransformation.
Anaerobic biotransformations of hydrocarbons have mainly been reported and studied during the last 15 years. These reactions allow the anaerobic turnover and biodegradation of aromatic and nonaromatic hydrocarbons and their xenobiotic derivatives. Initial reactions are the activation of highly reduced compounds by, e.g., hydroxylation with oxygen derived from water, carboxylation, or addition to fumarate to form a succinyl derivative that is then thioesterified (75, 91, 98). Joseph Suflita (University of Oklahoma, Norman, Okla.) reported the anaerobic degradation of dodecane and hexane via fumarate addition reactions. Fumarate was added at position 2 of [1-13C]hexane to generate a succinylated product during anaerobic biotransformation experiments with strain ALDC4 (53). This finding is consistent with perdeuterated hexane metabolism by a denitrifying bacterium (77, 99). The transformation of toluene and xylenes by various anaerobic microorganisms resulted in the formation of methylbenzylsuccinic acid derivatives (27). 4-Methylbenzoic acid was converted to terephthalic acid, an interesting monomer used for new plastic materials in the chemical industry. Detailed characterizations of anaerobic metabolic reactions and their underlying microbial enzymes might well open completely new perspectives for industrial biocatalysis with respect to carbon-carbon bond formation, selective reductions, and oxidation reactions in the absence of oxygen. Finally, Suflita described studies in which anaerobic metabolites of alkanes and naphthalenes were detected at contaminated sites. The presence of these alkylsuccinic acids and naphthoic acids in environmental samples is indicative of actively occurring intrinsic biodegradation of petroleum hydrocarbon contaminants (34a).
B3: THE FUNCTIONAL BASIS FOR CHEMICAL BIOTECHNOLOGY
During the last century the research focus on applications of microbial metabolism and biology in general has gradually shifted. The range of topics goes from classical fermentations to environmental issues, including ever-important topics like waste treatment and bioremediation, to medical biotechnology. All areas are based on a thorough understanding of the underlying biochemical principles and enzymes catalyzing key chemical reactions (Fig. 1). In addition, our rapidly expanding knowledge about metabolic pathways (for example, see http://umbbd.ahc.umn.edu/), genomes (http://www.microbialgenome.org/), and enzymes (http://www.expasy.ch) together with advances in biochemical engineering has enabled the development of chemical biotechnology. In this fast-growing field, the combined research interests of microbiologists, chemists, and engineers have already allowed the development of mild and competitive production processes in the chemical industry (82).
From an industrial perspective, Sima Sariaslani (Dupont Central Research and Development, Wilmington, Del.) pointed out the key elements in the development of a bioroute to a chemical product, namely the correct choice of raw material and an in-depth knowledge of the microbiology, physiology, and biochemistry of the biocatalyst. Because glucose is one of the cheapest raw materials, metabolic engineering followed by fermentation and separation and purification with this starting material can result in an economically competitive process. This was illustrated by an industrial metabolic engineering project for the production of p-hydroxycinnamic acid from glucose by using Saccharomyces cerevisiae or, at a later stage, recombinant hosts like E. coli or Pseudomonas. Glucose is metabolized via shikimate to either phenylalanine or tyrosine. In an optimal route, tyrosine is converted directly to p-hydroxycinnamic acid by tyrosine ammonia lyase (TAL). Alternatively, in a phenylalanine-overproducing strain phenylalanine ammonia lyase (PAL) transforms phenylalanine to cinnamic acid, which is then hydroxylated to p-hydroxycinnamic acid by a cytochrome P450 monooxygenase. Ongoing catalyst development with molecular evolution includes engineering TAL by eliminating its PAL activity via point mutations, and it resulted in a mutant TAL with a 3.4-fold increase in the TAL/PAL activity ratio compared to that of wild-type TAL. Another strategy involved the application of phenylalanine para-hydroxylase in E. coli strain NST37, which allowed the preparation of p-hydroxycinnamic acid.
Alternatives to the conversion of glucose and other biofeed stocks to value-added products via fermentation are single- or multistep in vitro biotransformations (biocatalysis) to produce higher-value products from appropriate starting materials. In an interesting example, Mani Subramanian (The Dow Chemical Company, San Diego, Calif.) presented results on the application of haloalkane dehalogenase for industrial products. This enzyme catalyzes the hydrolytic dehalogenation of 1,2,3-trichloropropane (TCP) to dichlorohalohydrin, which can be reused in the process to make epichlorohydrin. TCP is a waste product in the production of epichlorohydrin from propylene. Up to 7% TCP is formed as waste in the process relative to 93% halohydrin. The first enzyme tested from Rhodococcus rhodochrous TDTM-003 showed a Km of 1.2 mM, a low stability at 55°C (half-life [t1/2] < 3 min), a product ee of 40%, product inhibition, and a kcat of only 0.17 s−1, features which had to be improved by at least 100-fold for a commercial application. Random mutagenesis via error-prone PCR and gene saturation mutagenesis were successfully used to obtain several thermostable mutants. The best mutant showed a t1/2 at 55°C of ≫29,000 min, with a kcat of 2.2 s−1 at room temperature. Gene reassembly of several mutant dehalogenase genes resulted in several improved mutants with a kcat of 1.6 s−1 and a Km of 1 mM. In parallel, a broad screening program to isolate better dehalogenase genes based on biopanning of DNA libraries prepared from 576 different environmental samples was done in collaboration with Diversa (San Diego, Calif.). This resulted in 25 full-length genes and enzymes with different properties compared to those of the wild type, such as improved catalytic activity, a broader substrate range, lower feedback inhibition, and different enantioselectivities. These results showed that the combination of environment-based gene discovery and directed evolution is a powerful tool for discovery and development of industrial biocatalysts.
Bacterial oxygenases often allow highly regio- and stereoselective incorporation of oxygen into hydrocarbons and are therefore interesting catalysts for organic synthesis. Biocatalyst and bioprocess development solutions for up to pilot-scale applications were reported by Andreas Schmid (Institute of Biotechnology ETH, Zurich, Switzerland). In most cases, oxygenase-catalyzed biotransformations use whole cells due to the necessity to recycle reduced cofactors and the instabilities of enzyme complexes. E. coli JM101 was used as the host organism to express styrene monooxygenase from Pseudomonas sp. VLB120 in whole-cell processes for the production of (S)-styrene oxide (ee > 99%) (67) and a range of different derivatives (83). Products were obtained in high yield with ee values exceeding 98% and productivities of up to 900 U/liter. The process was successfully scaled up to a 30-liter working volume for the preparation of 307 g of enantiopure (S)-styrene oxide (overall purity, >97%) by using distillation for product recovery and purification (66). In another example, E. coli JM101 expressing xylene monooxygenase from P. putida mt-2 was used for the two-step hydroxylation of pseudocumene to pure 3,4-dimethylbenzaldehyde in a two-phase liquid system on a similar scale. Alternatively, oxygenases can be used in isolated form, which allows uncoupling of catalyst preparation from its application, and in independent optimization of both processes. Cell-free preparations of 2-hydroxybiphenyl-3-monooxygenase (HbpA) were used as catalysts in organic aqueous emulsions for the preparation of pure 2,3-dihydroxybiphenyl from 2-hydroxybiphenyl with regeneration of NADH by formate dehydrogenase and productivities of up to 0.45 g liter−1 h−1 (84). Regeneration of NADH in biotransformations with HbpA can also be achieved by using electrochemical methods at productivities of up to 0.2 g liter−1 h−1 (45). Volumetric productivities thus achieved were in the range of those seen for optimized whole-cell processes based on the same oxygenases. Future work will aim at understanding and overcoming bottlenecks in stability and productivity of biocatalysts for oxidations based on whole cells and cell-free oxygenases.
In analogy to the scaling up of bioprocesses, the scaling down of bioprocesses is also of significant interest for the synthesis of functional materials. Jonathan Dordick (Rensselear Polytechnic Institute, Troy, N.Y.) reported on new developments in nanoscale biocatalysis as a tool in nanoscale and high-throughput discovery. This work is based on microfluidic technology with channels in the micrometer scale and electroosmotic solvent pumping controlled by voltages of 500 to 2,000 V. To this point, peroxidase (10 μg/ml) has been used for the coupling of phenols with H2O2 in a channel volume of 80 nl at 2,000 V and at a rate of 3.76 μM min−1 with a yield of 100%. Immobilization of peroxidase on microbeads prevents plugging of the channels with polyphenols. In Y-shaped microchannels, 10 mM phenol and 0.4 mM H2O2 are contacted and uniform mixing is obtained 0.35 μm after the mixing point. In the future the system will also be applied for detailed biochemical studies and metabolic pathway engineering of signal transduction, polyketide and isoprenoid synthesis, and central metabolism in order to examine the fundamental limits of these pathways.
SUMMARY
As John Archibald Wheeler said, “We live on an island of knowledge surrounded by a sea of ignorance. As our island of knowledge grows, so does the shore of our ignorance.” So as the B3 conference added to our collective store of knowledge, it also helped point out more of our areas of ignorance.
As we ponder the future, an important question to ask is the following: How broad is natural biocatalysis? This question is also important in the broader context of genomic biology, whereby genome annotators seek to match up sequences with functions. Since most microbial genes encode enzymes that catalyze reactions, we desperately need to know whether or not we are knowledgeable about most of the reaction types catalyzed by microbes. The answer to this question has important implications for genome annotation and biotechnological advancement.
There are some indicators suggesting that, regarding reaction types, our shore of ignorance is large. A significant proportion of the B3 conference was concerned with the action of oxygenases, which are critically important in aerobic biodegradation and for commercial biotransformations. However, we are still ignorant about the metabolism of several dozen chemical functional groups found in biologically produced compounds (94). This is despite the fact that many new reactions have been discovered recently; for example, biological equivalents of the following named organic reactions have been discovered: the Diels-Alder reaction, the Bamberger rearrangement, the Beckman rearrangement, and the Kolbe-Schmidt reaction. The B3 conference discussed novel biocatalysis and new ways of using well-known enzymes, but clearly more novel metabolism will be discovered before the next B3 conference convenes. Such discovery will increase the toolkit of enzymes that are usable in biotechnology. The expanded toolkit will provide new routes to synthesize a broad array of commodity chemicals from renewable resources. The use of renewable resources for clean and sustainable industry is clearly one of the major outcomes of B3.
Acknowledgments
We thank our gracious conference hosts from the various Puerto Rican universities. We also thank the B3 conference speakers for editing their individual sections for this review for accuracy and content.
Research in our laboratories is supported by the Strategic Environmental Research and Development Program (REP) and the Defense Science and Technology Laboratory, Ministry of Defense, and the Biotechnology and Biological Sciences Research Council (NCB). We also thank Pfizer, Inc., for financial support of the B3 conference.
REFERENCES
- 1.Almatawah, Q., and D. A. Cowan. 1999. Thermostable nitrilase catalysed production of nicitinic acid from 3-cyanopyridine. Enzyme Microbial Technol. 25:718-724. [Google Scholar]
- 2.Almatawah, Q. A., R. A. Cramp, and D. A. Cowan. 1999. Characterization of an inducible nitrilase from a thermophilic bacillus. Extremophiles 3:283-291. [DOI] [PubMed] [Google Scholar]
- 3.Archelas, A., and R. Furstoss. 2001. Synthetic applications of epoxide hydrolases. Curr. Opin. Chem. Biol. 5:112-119. [DOI] [PubMed] [Google Scholar]
- 4.Barna, T., H. Khan, N. C. Bruce, N. S. Scrutton, and P. C. E. Moody. 2001. Crystal structure of the explosive degrading flavoenzyme PETN reductase and complexes with steroid substrates and inhibitors. J. Mol. Biol. 310:433-447. [DOI] [PubMed] [Google Scholar]
- 5.Binks, P. R., C. E. French, S. Nicklin, and N. C. Bruce. 1996. Degradation of pentaerythritol tetranitrate by Enterobacter cloacae PB2. Appl. Environ. Microbiol. 62:1214-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers, N. I., D. R. Boyd, N. D. Sharma, P. A. Goodrich, M. R. Groocock, A. J. Blacker, P. Goode, and H. Dalton. 1999. Stereoselective benzylic hydroxylation of 2-substituted indanes using toluene dioxygenase as biocatalysts. J. Chem. Soc. Perkin Trans. 1 1999:1453-1461. [Google Scholar]
- 7.Boyd, D. R., and N. D. Sharma. Enzymatic and chemoenzymatic synthesis of arene trans-dihydrodiols. J. Mol. Catal. B Enzym., in press.
- 8.Boyd, D. R., N. D. Sharma, and C. C. R. Allen. 2001. Aromatic dioxygenases: molecular biocatalysis and applications. Curr. Opin. Biotechnol. 12:564-573. [DOI] [PubMed] [Google Scholar]
- 9.Boyd, D. R., N. D. Sharma, N. I. Bowers, J. Duffy, J. S. Harrison, and H. Dalton. 2000. Enantioselective toluene dioxygenase catalysed di- and tri-hydroxylations of monosubstituted benzenes. J. Chem. Soc. Perkin Trans. 1 2000:1345-1350. [Google Scholar]
- 10.Boyd, D. R., N. D. Sharma, J. S. Harrison, M. A. Kennedy, C. C. R. Allen, and D. T. Gibson. 2001. Regio- and stereo-selective dioxygenase-catalysed cis-dihydroxylation of fjord-region polycyclic arenes. J. Chem. Soc. Perkin Trans. 1 2001:1264-1269. [Google Scholar]
- 11.Boyd, D. R., N. D. Sharma, F. Hempenstall, M. A. Kennedy, J. F. Malone, C. C. R. Allen, S. M. Resnick, and D. T. Gibson. 1999. bis-cis-dihydrodiols: a new class of metabolites resulting from biphenyl dioxygenase-catalyzed sequential asymmetric cis-dihydroxylation of polycyclic arenes and heteroarenes. J. Org. Chem. 64:4005-4011. [Google Scholar]
- 12.Boyd, D. R., N. D. Sharma, L. V. Modyanova, J. G. Carroll, J. F. Malone, C. C. R. Allen, J. T. G. Hamilton, D. T. Gibson, R. E. Parales, and H. Dalton. 2002. Dioxygenase-catalyzed cis-dihydroxylation of pyridine-ring systems. Can. J. Chem. 80:589-600. [Google Scholar]
- 13.Boyd, D. R., N. D. Sharma, C. R. O'Dowd, and F. Hempenstall. 2000. Enantiopure arene dioxides: chemoenzymatic synthesis and application in the production of trans-3,4-dihydrodiols. Chem. Commun. 2151-2152.
- 14.Burlingame, R., and P. J. Chapman. 1983. Catabolism of phenylpropionic acid and its 3-hydroxy derivative by Escherichia coli. J. Bacteriol. 155:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canada, K. A., S. Iwashita, H. Shim, and T. K. Wood. 2002. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J. Bacteriol. 184:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clément, P., V. Matus, L. Cárdenas, and B. González. 1995. Degradation of trichlorophenols by Alcaligenes eutrophus JMP134. FEMS Microbiol. Lett. 127:51-55. [DOI] [PubMed] [Google Scholar]
- 17.Clément, P., D. H. Pieper, and B. González. 2001. Molecular characterization of a deletion/duplication rearrangement in tfd genes from Ralstonia eutropha JMP134(pJP4) that improves growth on 3-chlorobenzoic acid but abolishes growth on 2,4-dichlorophenoxyacetic acid. Microbiology 147:2141-2148. [DOI] [PubMed] [Google Scholar]
- 18.Coates, J. D., and R. T. Anderson. 2000. Emerging techniques for anaerobic bioremediation of contaminated environments. Trends Biotechnol. 18:408-412. [DOI] [PubMed] [Google Scholar]
- 19.Cramp, R. A., and D. A. Cowan. 1999. Molecular characterisation of a novel thermophilic nitrile hydratase. Biochim. Biophys. Acta 1431:249-260. [DOI] [PubMed] [Google Scholar]
- 20.Crout, D. H., and G. Vic. 1998. Glycosidases and glycosyl transferases in glycoside and oligosaccharide synthesis. Curr. Opin. Chem. Biol. 2:98-111. [DOI] [PubMed] [Google Scholar]
- 21.de Souza, M. L., J. Seffernick, B. Martinez, M. J. Sadowsky, and L. P. Wackett. 1998. The atrazine catabolism genes atzABC are widespread and highly conserved. J. Bacteriol. 180:1951-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Souza, M. L., L. P. Wackett, and M. J. Sadowsky. 1998. The atzABC genes encoding atrazine catabolism are located on a self-transmissible plasmid in Pseudomonas sp. strain ADP. Appl. Environ. Microbiol. 64:2323-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhar, K., and J. P. N. Rosazza. 2000. Purification and characterization of Streptomyces griseus catechol O-methyltransferase. Appl. Environ. Microbiol. 66:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Díaz, E., A. Ferrández, M. A. Prieto, and J. L. García. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65:523-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dybas, M. J., M. Barcelona, S. Bezborodnikov, S. Davies, L. Forney, H. Heuer, O. Kawka, T. Mayotte, L. Sepulveda-Torres, K. Smalla, M. Sneathen, J. Tiedje, T. Voice, D. C. Wiggert, M. E. Witt, and C. S. Criddle. 1998. Pilot-scale evaluation of bioaugmentation for in-situ remediation of a carbon tetrachloride-contaminated aquifer. Environ. Sci. Technol. 32:3598-3611. [Google Scholar]
- 26.Ebert, S., P.-G. Rieger, and H.-J. Knackmuss. 1999. Function of coenzyme F420 in aerobic catabolism of 2,4,6-trinitrophenol and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. J. Bacteriol. 181:2669-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elshahed, M. S., L. M. Gieg, M. J. McInerny, and J. M. Suflita. 2001. Signature metabolites attesting to the in situ attenuation of alkylbenzenes in anaerobic environments. Environ. Sci. Technol. 35:682-689. [DOI] [PubMed] [Google Scholar]
- 28.Fennell, D. E., A. B. Carroll, J. M. Gossett, and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 29.French, C. E., S. Nicklin, and N. C. Bruce. 1998. Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase. Appl. Environ. Microbiol. 64:2864-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.French, C. E., S. Nicklin, and N. C. Bruce. 1997. Sequence and properties of pentaerythritol tetranitrate reductase from Enterobacter cloacae PB2. J. Bacteriol. 178:6623-6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.French, C. E., S. J. Rosser, G. J. Davies, S. Nicklin, and N. C. Bruce. 1999. Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase. Nat. Biotechnol. 17:491-494. [DOI] [PubMed] [Google Scholar]
- 32.Fries, M. R., G. D. Hopkins, P. L. McCarty, L. J. Forney, and J. M. Tiedje. 1997. Microbial succession during a field evaluation of phenol and toluene as the primary substrates for trichloroethene cometabolism. Appl. Environ. Microbiol. 63:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuenmayor, S. L., M. Wild, A. L. Boyles, and P. A. Williams. 1998. A gene cluster encoding steps in the conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerrit, J., L. A. Kulakov, M. J. Larkin, J. E. T. Van Hylckama, and D. B. Janssen. 2000. Roles of horizontal gene transfer and gene integration in evolution of 1,3-dichloropropene and 1,2-dibromoethane-degradative pathways. J. Bacteriol. 182:2191-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Gieg, L. M., and J. M. Suflita. Detection of anaerobic metabolites of saturated and aromatic hydrocarbons in petroleum-contaminated aquifers. Environ. Sci. Technol., in press. [DOI] [PubMed]
- 35.Glazer, A. N., and H. Kikaido. 1995. Microbial bio/technology. W.H. Freeman and Co., New York, N.Y.
- 36.Graham, D., D. Barfield, R. A. Pereira, and D. A. Cowan. 2000. Nitrile transformation studies using free and immobilised cells of a themophilic Bacillus sp. Enzyme Microbial Technol. 26:368-373. [DOI] [PubMed] [Google Scholar]
- 37.Hafner, E. W., and D. Wellner. 1971. Demonstration of imino acids as products of the reactions catalyzed by D- and L-amino acid oxidases. Proc. Natl. Acad. Sci. USA 68:987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hannink, N., S. J. Rosser, C. E. French, A. Basran, S. Nicklin, J. A. H. Murray, and N. C. Bruce. 2001. Phytodetoxification of 2,4,6-trinitroluene by transgenic plants expressing bacterial nitroreductase. Nat. Biotechnol. 19:1168-1172. [DOI] [PubMed] [Google Scholar]
- 39.Hay, A. G., P. M. Dees, and G. S. Sayler. 2001. Growth of a bacterial consortium on triclosan. FEMS Microbiol. Ecol. 36:105-112. [DOI] [PubMed] [Google Scholar]
- 40.He, Z., L. J. Nadeau, and J. C. Spain. 2000. Characterization of hydroxylaminobenzene mutase from pNBZ139 cloned from Pseudomonas pseudoalcaligenes JS45. A highly associated SDS-stable enzyme catalyzing an intramolecular transfer of hydroxy groups. Eur. J. Biochem. 267:1110-1116. [DOI] [PubMed] [Google Scholar]
- 41.He, Z., and J. C. Spain. 2000. One-step production of picolinic acids from 2-aminophenols catalyzed by 2-aminophenol 1,6-dioxygenase. J. Ind. Microbiol. Biotechnol. 25:25-28. [Google Scholar]
- 42.Heider, J., A. M. Spormann, H. R. Beller, and F. Widdel. 1999. Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol. Rev. 22:459-473. [Google Scholar]
- 43.Heiss, G., and H.-J. Knackmuss. 2002. Bioelimination of trinitroaromatic compounds: immobilization vs. mineralization. Curr. Opin. Microbiol. 5:282-287. [DOI] [PubMed] [Google Scholar]
- 44.Holland, H. L. 1998. Microbial transformations. Curr. Opin. Chem. Biol. 2:77-84. [DOI] [PubMed] [Google Scholar]
- 45.Hollmann, F., A. Schmid, and E. Steckhan. 2001. First synthetic application of a monooxygenase employing indirect electrochemical NADH regeneration. Angew. Chem. Int. Ed. Engl. 40:169-171. [DOI] [PubMed] [Google Scholar]
- 46.Hopkins, G. D., and P. L. McCarty. 1995. Field evaluation of in-situ aerobic cometabolism of trichloroethene and three dichloroethene isomers using phenol and toluene as the primary substrates. Environ. Sci. Technol. 29:1628-1637. [DOI] [PubMed] [Google Scholar]
- 47.Hudlicky, T., D. Gonzalez, and D. T. Gibson. 1999. Enzymatic dihydroxylation of aromatics in enantioselective synthesis: expanding asymmetric methodology. Aldrichimica Acta 32:35-62. [Google Scholar]
- 48.Huh, J. W., K. Yokoiga, N. Esaki, and K. Soda. 1992. Synthesis of L-proline from the racemate by coupling on enzymatic enantiospecific oxidation and chemical nonenantiospecific oxidation and chemical nonenantiospecific reduction. J. Ferment. Bioeng. 74:189-190. [Google Scholar]
- 49.Imbeault, N. Y., J. B. Powlowski, C. L. Colbert, J. T. Bolin, and L. D. Eltis. 2000. Steady-state kinetic characterization and crystallization of a polychlorinated biphenyl-transforming dioxygenase. J. Biol. Chem. 275:12430-12437. [DOI] [PubMed] [Google Scholar]
- 50.Jain, R. K., J. H. Dreisbach, and J. C. Spain. 1994. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl. Environ. Microbiol. 60:3030-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson, G. R., B. F. Smets, and J. C. Spain. 2001. Oxidative transformation of aminodinitrotoluene isomers by multicomponent dioxygenases. Appl. Environ. Microbiol. 67:5460-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 53.Kropp, K. G., I. A. Davidova, and J. M. Suflita. 2000. Anaerobic oxidation of n-dodecane by an addition reaction in a sulfate-reducing bacterial enrichment culture. Appl. Environ. Microbiol. 66:5393-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larkin, M. J., C. C. R. Allen, L. A. Kulakov, and D. A. Lipscomb. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 181:6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee, C. H., T. A. Lewis, A. Paszczynski, and R. L. Crawford. 1999. Identification of an extracellular agent [correction of catalyst] of carbon tetrachloride dehalogenation from Pseudomonas stutzeri strain KC as pyridine-2,6-bis(thiocarboxylate). Biochem. Biophys. Res. Commun. 261:562-566. [DOI] [PubMed] [Google Scholar]
- 56.Lewis, T. A., M. S. Cortese, J. L. Sebat, T. L. Green, C. H. Lee, and R. L. Crawford. 2000. A Pseudomonas stutzeri gene cluster encoding the biosynthesis of the CCl4-dechlorination agent pyridine-2,6-bis(thiocarboxylic acid). Environ. Microbiol. 2:407-416. [DOI] [PubMed] [Google Scholar]
- 57.Lewis, T. A., and R. L. Crawford. 1995. Transformation of carbon tetrachloride via sulfur and oxygen substitution by Pseudomonas sp. strain KC. J. Bacteriol. 177:2204-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis, T. A., A. Paszczynski, S. W. Gordon-Wylie, S. Jeedigunta, C. H. Lee, and R. L. Crawford. 2001. Carbon tetrachloride dechlorination by the bacterial transition metal chelator pyridine-2,6-bis(thiocarboxylic acid). Environ. Sci. Technol. 35:552-559. [DOI] [PubMed] [Google Scholar]
- 59.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maymó-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maymó-Gatell, X., I. Nijenhuis, and S. H. Zinder. 2001. Reductive dechlorination of cis-1,2-dichloroethene and vinyl chloride by “Dehalococcoides ethenogenes.” Environ. Sci. Technol. 35:516-521. [DOI] [PubMed] [Google Scholar]
- 62.Michaelis, L., and M. L. Menten. 1913. Die kinetik der invertinwirkung. Biochem. Z. 49:333-369. [Google Scholar]
- 63.Nadeau, L. J., Z. He, and J. C. Spain. 2000. Production of 2-amino-5-phenoxyphenol from 4-nitrobiphenyl ether using nitrobenzene nitroreductase and hydroxyaminobenzene mutase from Pseudomonas pseudoalcaligenes JS45. J. Ind. Microbiol. Biotechnol. 24:301-305. [Google Scholar]
- 64.Nishino, S. F., J. C. Spain, and Z. He. 2000. Strategies for aerobic degradation of nitroaromatic compounds by bacteria: process discovery to field application, p. 7-61. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. CRC Press, Boca Raton, Fla.
- 65.Padilla, L., V. Matus, P. Zenteno, and B. González. 2000. Degradation of 2,4,6-trichlorophenol via chlorohydroxyquinol in Ralstonia eutropha JMP134 and JMP222. J. Basic Microbiol. 40:243-249. [DOI] [PubMed] [Google Scholar]
- 66.Panke, S., M. Held, M. Wubbolts, B. Witholt, and A. Schmid. 2002. Pilot scale production of (S) styrene oxide from styrene by recombinant Escherichia coli synthesizing styrene monooxygenase. Biotechnol. Bioeng. 80:33-41. [DOI] [PubMed]
- 67.Panke, S., M. G. Wubbolts, A. Schmid, and B. Witholt. 2000. Production of enantiopure styrene oxide by recombinant Escherichia coli synthesizing a two-component styrene monooxygenase. Biotechnol. Bioeng. 69:91-100. [DOI] [PubMed] [Google Scholar]
- 68.Parales, R. E., J. L. Ditty, and C. S. Harwood. 2000. Toluene-degrading bacteria are chemotactic to the environmental pollutants benzene, toluene, and trichoroethylene. Appl. Environ. Microbiol. 66:4098-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parales, R. E., K. Lee, S. M. Resnick, H. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parales, R. E., J. V. Parales, and D. T. Gibson. 1999. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J. Bacteriol. 181:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parales, R. E., S. M. Resnick, C. L. Yu, D. R. Boyd, N. D. Sharma, and D. T. Gibson. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the α subunit. J. Bacteriol. 182:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pasteur, L. 1858. Nouvelles recherches sur de fermentation alcoolique. C. R. Acad. Sci. (Paris) 47:224. [Google Scholar]
- 73.Pérez-Pantoja, D., L. Guzmán, M. Manzano, D. H. Pieper, and B. González. 2000. Role of tfdCIDIEIFI and tfdDIICIIEIIFII gene modules in catabolism of 3-chlorobenzoate by Ralstonia eutropha JMP134(pJP4). Appl. Environ. Microbiol. 66:1602-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pereira, R., D. Graham, and D. A. Cowan. 1998. A novel thermostable nitrile hydratase. Extremophiles 2:347-358. [DOI] [PubMed] [Google Scholar]
- 75.Phelps, C. D., and L. Y. Young. 2001. Biodegradation of BTEX under anaerobic conditions: a review. Adv. Agronomy 70:329-357. [Google Scholar]
- 76.Poelarends, G. J., M. Zandstra, T. Bosma, L. A. Kulakov, M. J. Larkin, J. R. Marchesi, A. J. Weightman, and D. Janssen. 2000. Haloalkane-utilizing Rhodococcus strains isolated from geographically distinct locations possess a highly conserved gene cluster encoding haloalkane catabolism. J. Bacteriol. 182:2725-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rabus, R., H. Wilkes, A. Behrends, A. Armstroff, T. Fischer, A. J. Pierik, and F. Widdel. 2001. Anaerobic initial reaction of n-alkanes in a denitrifying bacterium: evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical in n-hexane metabolism. J. Bacteriol. 183:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Resnick, S. M., K. Lee, and D. T. Gibson. 1996. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J. Ind. Microbiol. 17:438-457. [Google Scholar]
- 79.Rieger, P.-G., H.-M. Meier, M. Gerle, U. Vogt, T. Groth, and H.-J. Knackmuss. 2002. Xenobiotics in the environment: present and future strategies to obviate the problem of biological persistence. J. Biotechnol. 94:101-123. [DOI] [PubMed] [Google Scholar]
- 80.Ryoo, D., H. Shim, F. L. G. Arenghi, P. Barberi, and T. K. Wood. 2001. Tetrachloroethylene, trichloroethylene, and chlorinated phenols induce toluene-o-monooxygenase activity in Pseudomonas stutzeri OX1. Appl. Microbiol. Biotechnol. 56:545-549. [DOI] [PubMed] [Google Scholar]
- 81.Ryoo, D., H. Shim, K. Canada, P. Barberi, and T. K. Wood. 2000. Aerobic degradation of tetrachloroethylene by toluene-o-monooxygenase of Pseudomonas stutzeri OX1. Nat. Biotechnol. 18:775-778. [DOI] [PubMed] [Google Scholar]
- 82.Schmid, A., J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts, and B. Witholt. 2001. Industrial biocatalysis today and tomorrow. Nature 409:258-268. [DOI] [PubMed] [Google Scholar]
- 83.Schmid, A., K. Hofstetter, H.-J. Feiten, F. Hollmann, and B. Witholt. 2001. Integrated biocatalytic synthesis on gram scale: the highly enantioselective preparation of chiral oxiranes with styrene monooxygenase. Adv. Synth. Catal. 343:732-737. [Google Scholar]
- 84.Schmid, A., I. Vereyken, M. Held, and B. Witholt. 2001. Preparative regio- and chemoselective functionalization of hydrocarbons catalyzed by cell free preparations of 2-hydroxybiphenyl 3-monooxygenase. J. Mol. Catal. B Enzym. 11:455-462. [Google Scholar]
- 85.Scigelova, M., S. Singh, and D. H. G. Crout. 1999. Glycosidases-a great synthetic tool. J. Mol. Catal. B Enzym. 6:483-494. [Google Scholar]
- 86.Seah, S. Y. K., G. Labbé, S. R. Kaschabek, F. Reifenrath, W. Reineke, and L. D. Eltis. 2001. Comparative specificities of two evolutionarily divergent hydrolases involved in microbial degradation of polychlorinated biphenyls. J. Bacteriol. 183:1511-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seah, S. Y. K., G. Labbé, S. Nerdinger, M. R. Johnson, V. Snieckus, and L. D. Eltis. 2000. Identification of a serine hydrolase as a key determinant in the microbial degradation of poychlorinated biphenyls. J. Biol. Chem. 275:15701-15708. [DOI] [PubMed] [Google Scholar]
- 88.Seffernick, J. L., G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2000. Substrate specificity of atrazine chlorohydrolase and atrazine-catabolizing bacteria. Appl. Environ. Microbiol. 66:4247-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smejkal, C. W., T. Vallaeys, S. K. Burton, and H. M. Lappin-Scott. 2001. Substrate specificity of chlorophenoxyalkanoic acid-degrading bacteria is not dependent upon phylogenetically related tfdA gene types. Biol. Fertil. Soils 33:507-513. [Google Scholar]
- 90.Smejkal, C. W., T. Vallaeys, F. A. Seymour, S. K. Burton, and H. M. Lappin-Scott. 2001. Characterization of (R/S)-mecoprop [2-(2-methyl-4-chlorophenoxy) propionic acid]-degrading Alcaligenes sp. CS1 and Ralstonia sp. CS2 isolated from agricultural soils. Environ. Microbiol. 3:288-293. [DOI] [PubMed] [Google Scholar]
- 91.Spormann, A. M., and F. Widdel. 2000. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation 11:85-105. [DOI] [PubMed] [Google Scholar]
- 92.Vaillancourt, F. H., G. Labbé, N. M. Drouin, P. D. Fortin, and L. D. Eltis. 2002. The mechanism-based inactivation of 2,3-dihydroxybiphenyl 1,2-dioxygenase by catecholic substrates. J. Biol. Chem. 277:2019-2027. [DOI] [PubMed] [Google Scholar]
- 93.Velasco, A., S. Alonso, J. L. García, J. Perera, and E. Díaz. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J. Bacteriol. 180:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wackett, L. P., and C. D. Hershberger. 2001. Biocatalysis and biodegradation: microbial transformation of organic compounds. ASM Press, Washington, D.C.
- 95.Wackett, L. P., M. J. Sadowsky, B. Martinez, and N. Shapir. 2002. Biodegradation of atrazine and related s-triazine compounds: from enzymes to field studies. Appl. Microbiol. Biotechnol. 58:39-45. [DOI] [PubMed] [Google Scholar]
- 96.Walters, K. J., G. T. Gassner, S. J. Lippard, and G. Wagner. 1999. Structure of the soluble methane monooxygenase regulatory protein B. Proc. Natl. Acad. Sci. USA 96:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Whiteley, A. S., and M. J. Bailey. 2000. Bacterial community structure and physiological state within an industrial phenol bioremediation system. Appl. Environ. Microbiol. 66:2400-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259-276. [DOI] [PubMed] [Google Scholar]
- 99.Wilkes, H., R. Rabus, T. Fischer, A. Armstroff, A. Behrends, and F. Widdel. 2002. Anaerobic degradation of n-hexane in a denitrifying bacterium: further degradation of the initial intermediate (1-methylpentyl)succinate via skeleton rearrangement. Arch. Microbiol. 177:235-243. [DOI] [PubMed] [Google Scholar]
- 100.Yu, C.-L., R. E. Parales, and D. T. Gibson. 2001. Multiple mutations at the active site of naphthalene dioxygenase affect regioselectivity and enatioselectivity. J. Ind. Microbiol. Biotechnol. 27:94-103. [DOI] [PubMed] [Google Scholar]
- 101.Zhou, N.-Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zylstra, G. J., S.-W. Bang, L. M. Newman, and L. L. Perry. 2000. Microbial degradation of mononitrophenols and mononitrobenzoates, p. 145-184. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. CRC Press, Boca Raton, Fla.