Abstract
We explored the use of a cascade circuit for heterologous gene expression that consists of a regulatory module with a salicylate-inducible system that controls the expression of a second regulator, xylS2, whose product is activated by common inducers. Activation and increasing the concentration of the second regulator synergistically induced heterologous genes downstream of the Pm promoter in the expression module. This module can be placed in multicopy vectors or in the chromosome of a host strain by means of minitransposons. Using reporter genes, we evaluated gene regulation capacity and gross production of the system with different configurations. The highest yield was obtained when the expression module was in a multicopy plasmid after a 6-h induction. However, expression modules in plasmids showed low stability after induction even with selective pressure. The chromosomal configuration had the lowest basal levels and induced levels comparable to those of plasmid configurations, resulting in accumulation of more than 10% of the total protein. Unlike the configurations in plasmids, the yield was maintained for at least 3 days even without selective pressure. In conclusion, the cascade system in the chromosome configuration is more efficient for long-term fermentation because of the great stability of the overexpressing phenotype in spite of the high levels of expression.
Overexpression of cloned genes has been useful when purification of proteins is desired. Production of large amounts of cloned gene products has been traditionally achieved by combining gene amplification with strong promoters regulated by repressors. However, the usual strategy has the following disadvantages. (i) High levels of expression of recombinant proteins have been shown to reduce the host cell growth rate and, concomitantly, overall protein synthesis (2, 12). Gene amplification is achieved through a high-copy-number plasmid vector. Competition between host biochemical activity and the biochemical activity encoded by the plasmid, such as expression of plasmid-encoded genes and replication machinery, contributes to this detrimental effect. (ii) Maintenance of plasmid expression vectors requires the use of antibiotics, which results in an additional potential residual metabolic burden, additional costs, and potential contamination of products during large-scale industrial production (24). (iii) A low basal level of expression is most convenient for toxic protein production in order to prevent selection of expression-down mutations of the host cell transcription-translation machinery and to avoid accumulation of mutations in the recombinant protein products themselves (21). This is difficult to achieve when the cloned gene is in multicopy plasmid vectors. Furthermore, regulated systems based on a strong promoter and a plasmid-encoded repressor imply that there is transient expression of the toxic cloned gene before a high enough repressor level is obtained.
The nahR/Psal and xylS/Pm regulatory systems have been suggested to be alternative expression systems for the expression of heterologous genes on the basis of their tight regulation and the use of inexpensive inducers (18, 20, 28, 30). We previously designed a novel cascade control system that is able to amplify the gene expression capacity (4). The regulatory module consists of an nahR/Psal regulation system controlling expression of xylS2, a mutant of xylS (26), which is activated by salicylate. The expression module, comprising the Pm promoter and the gene of interest, is synergistically activated by induced synthesis of XylS2 by salicylate activation of the nahR/Psal system and the intrinsic activity of XylS2. The regulatory elements and the expressed gene are cloned in mini-Tn5 delivery vectors and thus can be inserted as single copies into bacterial chromosomes if desired. Although the number of gene copies of the expression system is reduced if the genes are delivered to the chromosome, the product yield may be increased by amplification of the gene expression capacity of the cascade regulatory circuit.
In this study, we investigated different parameters that influence the performance of the cascade system and tested different conditions to obtain the best results for different conceivable applications. The efficiency and stability of the system were tested by monitoring expression of lacZ. A chromosomal configuration of the whole system provided the best results for proteins that are going to be expressed for a long time in serial batch fermentations or continuous cultures. This configuration allows reduction of the basal expression levels and stabilization of production of heterologous proteins.
MATERIALS AND METHODS
Plasmids and strains.
All the bacteria and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacteria and plasmids
| Strain or plasmid | Description | Reference |
|---|---|---|
| E. coli strains | ||
| CC118 | phoA20 thi-1 rspE rpoB argE(Am) recA1 | 16 |
| CC118λpir | CC118 lysogenized with λpir phage | 16 |
| S17-1λpir | F−recA hsdR RP4-2 (Tc::Mu) (Km::Tn7) lysogenized with λpir phage | 9 |
| CC118FH26 | CC118 with mini-Tn5 xylS2/Pm::lacZ inserted into the chromosome, Kmr | 11 |
| CC118RSL9 | CC118 with mini-Tn5 nahR/Psal::lacZ inserted into the chromosome, Kmr | 11 |
| CC1184S2 | CC118 with mini-Tn5 nahR/Psal::xylS2 inserted into the chromosome, Kmr | This study |
| CC1184S2λpir | CC1184S2 lysogenized with λpir phage | This study |
| CC1184S2PT32 | CC1184S2 with mini-Tn5 Pm::trp′::′lacZ inserted into the chromosome, Kmr Sp/Smr | This study |
| CC1184S2PT97 | CC1184S2 with mini-Tn5 Pm::trp′::′lacZ inserted into the chromosome, Kmr Sp/Smr | This study |
| NCM631 | hsdS gal λDE3:lacI, lacUV5::gen1 (T7 RNA polymerase) Δlac linked to Tn10 | 15 |
| Plasmids | ||
| pUC18Not | Apr, identical to pUC18 (32) but with a NotI polylinker of pUC18-NotI as MCS | 16 |
| pUC18Sfi | Apr, identical to pUC18 but with an SfiI polylinker of pUC18-SfiI as MCS | 16 |
| pFH2 | Apr, similar to pBKT7-0 (18) but with NotI sites flanking the MCS | 14 |
| pFH28 | Apr, pUC18Sfi-KmR-xylS-Pm-Sfi | 11 |
| pCNB2-lacZ | Apr Kmr, R6Kori, pUT/mini-Tn5 xylS2/Pm::lacZ | 11 |
| pCNB4 | Apr Kmr, R6Kori, pUT/mini-Tn5 nahR/Psal | 11 |
| pCNB4-lacZ | Apr Kmr, R6Kori, pCNB4 with trp′::′lacZ reporter downstream of Psal | 11 |
| pCNB4-S2 | Apr Kmr, R6Kori, pCNB4 with nahR/Psal::xylS2 downstream of Psal | 4 |
| pTPm-lacZ | Apr Tcr, R6Kori, pUT/mini-Tn5 Pm::trp′::′lacZ | 4 |
| pTSPm | Apr Sm/Spr, R6Kori, pUT/mini-Tn5 Pm | This study |
| pTPm-lacZ | Apr Tcr, R6Kori, pUT/mini-Tn5 Pm::trp′::′lacZ | This study |
| pKK232-8 | Apr, ColE1ori, promoter probe plasmid, based in the CAT gene | 3 |
| pCCD5 | Apr, ColE1ori, NotI-rrnBT1-Pm→pBKT7-0 MCS-NotI | This study |
| pIZ1207 | Apr, ColE1ori, EcoRI-HindIII fragment with trp′::′lacZ cloned into pFH2 downstream of a T7 promoter | This study |
To compare the efficiencies of gene overexpression from plasmids and from the mini-Tn5 delivery expression vectors in the chromosome, a λpir lysogen of Escherichia coli CC1184S2 was made by using standard protocols (22). E. coli CC1184S2λpir lysogen can express the п protein that is required for plasmid R6K replication.
Construction of expression vectors and transconjugant strains for the cascade system.
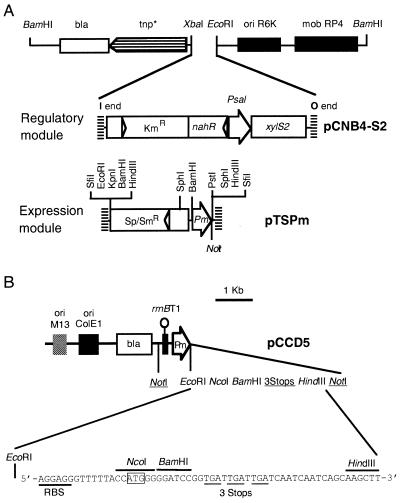
A delivery plasmid was constructed in which any gene cloned in mini-Tn5 auxiliary plasmids (8) could be inserted as a NotI fragment under the control of Pm. To construct this plasmid, the Sp/Smr interposon (13) was inserted into the BamHI vector fragment of pUC18Sfi-Kmr-xylS-Pm-Sfi (11). The resulting plasmid, having the structure SfiI-Sm/Spr-Pm-SfiI, was digested with SfiI, and the largest fragment was cloned into the SfiI-digested pUT backbone (16) to obtain the delivery plasmid pTSPm (Fig. 1).
FIG. 1.
Genetic elements used in the expression system. (A) The regulatory module contains the mini-Tn5 insertion sequences flanking a kanamycin resistance gene to select for transposition and a nahR-Psal regulatory system controlling the expression of xylS2. The expression modules constructed in this study are two plasmids, including pTSPm, a mini-Tn5 delivery vector that contains the Tn5 insertion sequences flanking a spectinomycin-streptomycin interposon, the Pm promoter, and a unique NotI site where a gene of interest may be cloned by using auxiliary plasmids (8). (B) pCCD5, a ColE1 vector containing a transcriptional terminator, Pm, and an MCS with an efficient translation initiation region, which are flanked by NotI sites. This plasmid allows subcloning of NotI fragments with Pm fusions to heterologous genes to the unique sites of mini-Tn5 delivery systems. RBS, ribosome binding site.
To avoid a requirement for the п protein for plasmid maintenance, we constructed a Pm-based expression plasmid with a fully independent replication origin (ColE1 derivative) (17). Plasmid pCCD5 was constructed based on pFH2 (14) and included its versatile multiple cloning site (MCS). To reduce readthrough from upstream promoters, an ApoI fragment (resulting in a fragment compatible with EcoRI termini) containing the rrnBT1 transcriptional terminator produced as a PCR fragment from pKK232-8 with the oligonucleotides 5′-GCAAATTTCCAGGCATCAAATAA and 5′-GGGAATTCCCTGGCAGTTTATGG was cloned into the EcoRI site of pFH2. After ligation, a unique EcoRI site resulted, and clones with the EcoRI site in equivalent MCS of pFH2 were selected. The Pm promoter was obtained by PCR by using the oligonucleotides A-Pm (5′-GTGTCAAATTTGATAGGGATAAGTCC-3′) and Pm-E (5′-GCCTGAATTCAGGCATTGACGAAGGCA-3′) as primers and pUC18Sfi-Kmr-xylS-Pm-Sfi as the template (11). Insertion of the amplified 0.4-kb Pm fragment digested with ApoI into the EcoRI-linearized intermediate plasmid resulted in vectors that contained rrnBT1 and the Pm promoter preceding the original MCS of pFH2, with all of the key elements flanked by NotI sites (Fig. 1). This last feature allowed transfer of the expression module to mini-Tn5 delivery vectors (8) and subsequent introduction of the regulatory module into the chromosome. A NcoI-HindIII lacZ fragment was introduced into pCCD5 to study lacZ expression from this plasmid, resulting in pCCD5-lacZ.
To compare stability results obtained with a commonly used expression system, a 3.2-kb EcoRI-HindIII fragment from pUJ9 (10), which contains the trp′::′lacZ hybrid gene, was cloned into pFH2 digested with the same enzymes. The trp′::′lacZ gene in the resulting plasmid was transcribed from the T7 promoter. The resulting plasmid (pIZ1207) was transformed into an E. coli NCM631 expression strain that contained a T7 RNA polymerase gene under the control of lacIq and Plac integrated into the chromosome (15).
Induction of bacterial cultures.
Cultures were grown at 37°C on Luria-Bertani (LB) medium with (strains with plasmids) or without ampicillin at a concentration of 100 mg/liter. Dilutions of the cultures (1:50 to 1:200, depending on the expected growth rate) were made in fresh medium and incubated for 2.5 h at 37°C. An inducer (2 mM) was added to the corresponding cultures, which were incubated at 30°C. NCM631(pIZ1207) was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
Protein analysis.
β-Galactosidase levels were measured enzymatically as described elsewhere (22). The relative amount of β-galactosidase was estimated by densitometry of Coomassie blue-stained protein gels (8% acrylamide sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] gels). SDS-PAGE was performed by the method of Laemmli (19). Densitometry studies of the SDS-PAGE gels were performed by using the ImageQuant program.
Characterization of the lacZ phenotype in the population.
The stability assay was performed by using cultures with or without antibiotic and in the presence or absence of 2 mM salicylate at 30°C. After at least 40 generations, cultures were diluted in 10 mM MgSO4 and plated on LB agar plates with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [50 mg/liter]). Blue and white colonies were counted to estimate the percentage of strains that maintained the ability to express lacZ. In some series, the blue colonies were tested for induction by 2 mM salicylate. Colonies were also studied for ampicillin resistance to check for the presence of plasmids. At least two independent experiments were performed for each strain and type of conditions.
Continuous culture experiments.
Continuous culture experiments were performed with a 2-liter microDCU fermentor (Braun Biotech) by using a final volume of 0.5 liter of LB medium or LB medium containing ampicillin (100 mg/liter) incubated at 30°C. We added 2.5 ml of a previously grown inoculum to the 0.5 liter (1:200). The cultures were incubated with a filtered air flux and stirred at 200 rpm. The culture medium was supplemented with Antifoam B (Sigma) according to the manufacturer's instructions. When the batch culture reached an optical density of 1.5, the culture was changed to the continuous mode at a dilution rate of 0.25 h−1. Salicylate (2 mM) was added to both the feed medium and the reaction vessel, and samples were removed for protein, enzymatic, and phenotype analyses for at least 48 h.
RESULTS
Evaluation of the capacity of the cascade expression system by using reporter genes.
To test the overproduction efficiency of the cascade system from the chromosome, the regulatory module in plasmid pCNB4-S2 was inserted into the CC118 genome by conjugation and subsequent transposition. The expression module was similarly inserted by using the pool of Rifr Smr transconjugant colonies as recipient strains in a subsequent mating with S17-1λpir(pTPm-lacZ) as the donor. Expression from 10 random colonies bearing both modules was analyzed. The basal β-galactosidase activity levels ranged from 150 to 800 Miller units, while the levels induced by salicylate (2 mM) ranged from 25,000 to 60,000 Miller units depending on the transconjugant, which corresponded to induction ratios of 100 to 400. The variations may have depended on the locations of the genetic constructs in relation to the chromosomal replication origin. It has been proposed that there are more copies of genes next to oriC per cell than of genes next to the replication termination region (23, 25, 27). The colony with the highest induction ratio (CC1184S2PT32) and the colony with the highest β-galactosidase activity (CC1184S2PT97) were selected for further study.
We compared lacZ expression from the cascade system to lacZ expression from other salicylate-responsive expression systems in which each regulator-promoter pair was used as a single scheme in a single copy (11). SDS-PAGE analysis of the total proteins revealed that the β-galactosidase hybrid protein accounted for more than 10% of the total protein of CC1184S2PT97 induced by salicylate, while expression from the single systems resulted in just 0.5% of the total protein. Both the induction ratios and the β-galactosidase activities for the single systems were at least 1 order of magnitude lower than the values for the cascade circuit (Fig. 2). These data suggested the potential utility of this system for overexpressing proteins with all the genetic elements inserted into the chromosome.
FIG. 2.
Overproduction of β-galactosidase by E. coli CC118 with the expression systems transposed into the chromosome. (A) Coomassie blue-stained SDS-PAGE gel containing total proteins from cells expressing lacZ and containing the transposable elements of pCNB4-lacZ, pCNB2-lacZ, and pCNB4-S2/pTPm-lacZ inserted into the chromosome. Induction and lack of induction by 2 mM salicylate are indicated by plus and minus signs, respectively. Accumulation of β-galactosidase is indicated by an arrow. (B) Quantification by densitometry. The bars indicate the relative amounts of β-galactosidase (β-gal).
The influence of the reporter gene on the regulatory capacity was also tested by using a reporter luciferase gene, lucOR (6, 31), encoding a less stable enzyme. The regulatory capacity when lucOR was used as the reporter, as defined by the induction ratio, was 3- to 10-fold higher than that obtained with the lacZ reporter gene (data not shown). These results indicate that use of the lacZ reporter may result in an underestimate of the regulatory capacity of the cascade system.
Inducer molecules of the cascade expression system.
The cascade system was designed for regulation in response to salicylate, a common inducer of XylS2 and NahR (4). We tested whether other inducers of the regulatory proteins could provide equivalent or greater activation of the system. Exponential cultures of CC1184S2PMT32 and cultures bearing simple circuits (CC118RSL9 and CC118FH26) were incubated for 5 h at 30°C with different benzoate derivatives. All the compounds that are common inducers of NahR and XylS2 resulted in amplification of the regulatory capacity of the cascade system compared to the capacity of the single systems (Table 2). Even with certain compounds that were inducers of NahR but not of XylS2 (4-chlorosalicylate and 3,5-dichlorosalicylate), the cascade still showed significant amplified regulatory capacity, indicating that the response to the inducers by NahR can be amplified by the residual constitutive activity of XylS2.
TABLE 2.
Ratios of induction of single expression and the cascade systems by different aromatic compoundsa
| Inducer | Induction ratios
|
||
|---|---|---|---|
| nahR/Psal | xylS2/Pm | nahR/Psal::xylS2/Pm | |
| Salicylate | 24 | 13 | 235 |
| Anthranilate | 22 | 11 | 285 |
| 2-Acetylsalicylate | 40 | 10 | 249 |
| 4-Chlorosalicylate | 33 | 1 | 191 |
| 5-Chlorosalicylate | 32 | 3 | 268 |
| 3,5-Dichlorosalicylate | 18 | 1 | 147 |
| 5-Methoxysalicylate | 3 | 46 | 183 |
| Benzoate | 1 | 44 | 82 |
| 3-Methylbenzoate | 11 | 63 | 240 |
| 2-Methoxybenzoate | 33 | 10 | 160 |
| 3-Methylsalicylate | 17 | 46 | 218 |
| 4-Methylsalicylate | 21 | 7 | 203 |
| 5-Methylsalicylate | 30 | 19 | 257 |
For most inducers induction was performed by using the inducer at a concentration of 2 mM; the only exception was 3,5-dichlorosalicylate, which was used at a concentration of 1 mM to avoid growth inhibition. The results are means from at least three independent experiments. The standard deviations were less than 28%.
Different configurations of the cascade system.
Since the cascade system allowed amplification of gene expression capacity, we tested the efficiency of the cascade system as a chromosomal integrated system and compared it to the efficiency of single expression schemes in plasmids. We also tested whether increasing the gene dose of the expression module of the cascade by placing the gene in a plasmid resulted in a further increase in the expression level. To do this, we used the delivery vectors described above established in CC118λpir, which allowed the R6K derivative plasmids to replicate. The single system showing the highest expression level in multicopy plasmids was that activated by xylS2 (pCNB2-lacZ) (Table 3). However, this system also showed very high basal levels of expression, which may be a major disadvantage for the production of many proteins. The induced expression levels of the strains bearing the whole cascade system in the chromosome were not as high as the level obtained with the xylS2 single system, but these strains had much lower basal expression levels. The highest protein yield was obtained with the strain containing the regulatory module in the chromosome and the expression module in plasmid pTPm-lacZ. In addition, this configuration showed much lower basal expression than the configuration with pCNB2-lacZ (Table 3). Therefore, the cascade system may provide either a higher regulatory capacity or greater protein production than the single expression systems, depending on the configuration.
TABLE 3.
Comparison of β-galactosidase activities and stabilities of the heterologous genes from the cascade and single expression systems in different configurations
| Strain | Plasmid | β-Galactosidase activity (Miller units)a
|
β-Galactosidase/total proteins (%)b
|
% of Lac+ coloniesc
|
|||
|---|---|---|---|---|---|---|---|
| −2OHB | +2OHB | −2OHB | +2OHB | −2OHB | +2OHB | ||
| CC118λpir | pTPm-lacZ | 1,211 | 1,358 | NDd | ND | 100 | 100 |
| CC118λpir | pCNB2-lacZ | 10,917 | 67,002 | 3.6 | 13 | 98, 81 | 20, 1.5 |
| CC118λpir | pCNB4-lacZ | 510 | 17,952 | ND | 3.9 | 100, 97 | 96, 85 |
| CC4S2PT32 | 171 | 30,383 | ND | 8.8 | 100 | 100 | |
| CC4S2PT97 | 408 | 39,143 | ND | 11 | 100 | 100 | |
| CC1184S2λpir | pTPm-lacZ | 1,487 | 78,257 | ND | 20 | 100, 99 | 5, 0 |
β-Galactosidase activity was assayed after 5 h of incubation without salicylate (−2OHB) or with 2 mM salicylate (+2OHB). The data are means from three independent experiments.
The relative amounts of β-galactosidase in cells were measured by densitometry of Coomassie blue-stained SDS-8% PAGE gels.
The stability assay was performed as described in Materials and Methods. Three independent experiments were performed for each strain and type of conditions. As expected, the values varied considerably for the experiments with the plasmid-containing strains, probably due to the stochastic appearance of the cured strains during culture. The maximum and minimum values are shown except for cases when identical results were obtained.
ND, not determined (0.5%).
The stability of the production phenotype of these systems in the absence of selective pressure was also tested. To do this, we measured the percentages of the Lac+ colonies plated on LB medium with X-Gal which came from batch cultures grown under different conditions. Under inducing conditions, the cascade expression systems in the chromosome could express lacZ for at least 40 generations without a loss of the production phenotype (Table 3). In contrast, the systems with plasmid configurations showed a major loss of the Lac+ phenotype under the inducing conditions, and this loss correlated to the expression levels (Table 3).
Impact of overexpression of lacZ on phenotype stability in serial batch cultures.
The experiments described above were equivalent to a fed batch fermentation that is widely used in industry to produce recombinant proteins. To evaluate the effect of the instability of plasmid-borne constructs on long-term protein production compared to that in strains with the chromosomal configuration, we studied the changes in β-galactosidase production in serial batch cultures. We monitored the lacZ expression of the two strains bearing the whole cascade system in the chromosome (CC1184S2PT32 and CC1184S2PT97) and the strains with the expression module of the cascade system in plasmids with different replication origins, CC1184S2λpir(pTSPm-lacZ) and CC1184S2λpir(pCCD5-lacZ).
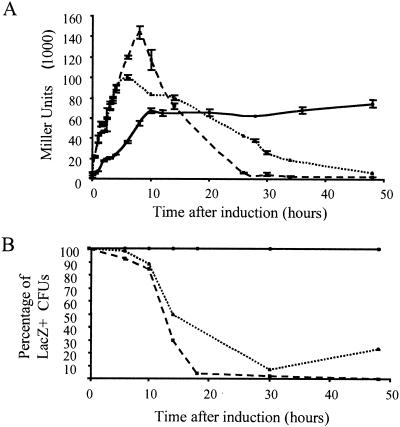
As shown in Fig. 3A, strain CC1184S2λpir(pTPm-lacZ) showed the highest β-galactosidase activity after 6 h of induction. However, the activities of the two strains bearing the expression module in a plasmid continuously declined with longer induction times until a basal level of activity was detected. In fact, in the absence of selective pressure (no ampicillin), the final activity of the induced cultures was even lower than the basal level of expression of these strains. This correlated with the domination (>99%) of bacteria cured of the plasmid in the cultures (see below). Therefore, overproduction was very unstable if the expression module was in a plasmid, regardless of the replication origin.
FIG. 3.
Stability of the overproduction phenotype. (A) Changes in β-galactosidase activities in batch cultures of organisms bearing the cascade system in different configurations 6, 12, 24, 36, 48, and 72 h after the addition of salicylate. Activity is represented by using a logarithmic scale. A 1:200 dilution of each culture in fresh medium was made every 12 h except for the last 24 h. The bars indicate the averages for at least two independent experiments in which less than 25% error was observed. (B) Total-protein samples of CC1184S2PT97 and CC1184S2λpir(pTPm-lacZ) cultures taken at different times after induction, resolved by SDS-PAGE. An arrowhead indicates the band corresponding to β-galactosidase. (C) Stability of the overproduction phenotype of the system based on the T7 RNA polymerase: changes in β-galactosidase activities in batch cultures of E. coli NCM631(pIZ1207) grown and diluted as described above for panel A. Symbols: ▵, uninduced, no ampicillin; ▴, induced, no ampicillin; ○, uninduced, ampicillin; •, induced, ampicillin. For comparison, the β-galactosidase activities of induced cultures of CC1184S2PT97 (⋄) and CC1184S2λpir(pTPm-lacZ) (▪) without selective antibiotic pressure are also shown.
In contrast, the strains with the cascade system in the chromosome maintained high expression levels during the whole experiment. This resulted in final levels of β-galactosidase activity that were at least 30-fold higher than those of the strains with the expression module in plasmids.
The level of β-galactosidase correlated with the relative amount of the recombinant polypeptide since SDS-PAGE of whole extracts from the batch cultures revealed that there was constant overproduction in the strains with the expression system in the chromosome, compared to the dramatic decrease in the level of the same polypeptide after 24 h of induction in strains bearing the expression module in a plasmid (Fig. 3B). Therefore, the observed loss of β-galactosidase activity was due to a loss of polypeptide production and not to production of inactive enzyme because of inappropriate folding of the protein.
To test the hypothesis that the instability of long-term overproduction by expression systems located in plasmids was not restricted to the cascade nahR/xylS2 system, we analyzed the production stability of the widely used T7 RNA polymerase expression system using strain NCM631(pIZ1207) (Fig. 3C). We observed a very high basal level of expression from the pT7 promoter (10,000 to 20,000 Miller units). The maximal level of activity (6 h after induction) was similar to that of the strains with the whole cascade system in the chromosome. With longer induction times, the activity rapidly decreased to values even lower than those obtained for the uninduced cultures after 24 h. Therefore, the high instability of the population overexpressing genes in plasmids compared to the stability of the population overexpressing genes in the chromosome seems to be a general feature and makes stable chromosomal integration of the gene of interest more convenient when a prolonged fermentation is desired.
Analysis of the terminal populations after long-term induction.
To explain the nature of the expression-down phenotypes of plasmids and the high stability of overexpression when the expression system was located in the chromosome, we analyzed the phenotypes of the bacterial populations of the different cultures grown after serial dilution by plating them on LB medium containing X-Gal. If organisms were grown under inducing conditions in the absence of selective pressure, the cultures of strains overexpressing from plasmids were dominated by Lac− bacteria (<99%) (Table 4). The white colonies obtained from these cultures were also sensitive to ampicillin, indicating that the plasmid was lost.
TABLE 4.
Percentages of Lac+ colonies in the final cell populations resulting from serially diluted batch cultures
| Strain | Presence of:
|
% of Lac+ CFUa | No. of CFU screened | |
|---|---|---|---|---|
| Ampicillin | Inducer | |||
| CC1184S2PT97 | − | − | 100 | 1,441 |
| − | + | 100 | 4,201 | |
| CC1184S2PT32 | − | − | 100 | 1,145 |
| − | + | 100 | 961 | |
| CC1184S2λpir (pTPm-lacZ) | − | − | 71.9 | 1,172 |
| − | + | 0 | 4,000 | |
| + | − | 97 | 2,100 | |
| + | + | 99.5 | 1,108 | |
| CC1184S2λpir (pCCD5-lacZ) | − | − | 96.5 | 733 |
| − | + | 0 | 2,185 | |
| + | − | 98.9 | 711 | |
| + | + | 95.4 | 1,637 | |
| NCM631 (pIZ1207) | − | − | 68.5 | 611 |
| − | + | 0 | 1,196 | |
| + | − | 97 | 1,146 | |
| + | + | 96 | 1,677 | |
The standard deviations were less than 28% except for NCM631(pIZ1207), for which values were obtained with duplicated series in the same experiment.
In contrast, most of the colonies from the cultures grown under inducing conditions in the presence of ampicillin maintained the Lac+ phenotype. A total of 40 colonies from cultures of strains overproducing from plasmids, which had been grown under these conditions, were analyzed to check induction of lacZ expression by salicylate. None of these colonies was able to respond to salicylate, and the colonies exhibited low levels of β-galactosidase activity in the presence of the inducer (data not shown). Therefore, most bacteria in the final population were mutants with an expression-down phenotype. To investigate if the mutations were located in the regulatory module (chromosome) or in the expression module (plasmid), we isolated plasmids from the mutants and retransformed them in CC1184S2 strains to see whether the mutation was linked. Approximately one-half of the mutations were located in the plasmids, since they could not be induced by salicylate in the new host, whereas the other half could be induced (putative mutation in the regulatory module). Lac+ plasmids unable to respond to salicylate had genetic rearrangements, as shown by the inability to amplify the Pm promoter with specific primers or by the restriction patterns of the plasmids (data not shown).
Analysis of stability of the overexpression phenotype in continuous cell cultures.
The stability of the overproducer phenotype was also tested in chemostat cultures in rich LB medium by using strains CC1184S2λpir(pTPm-lacZ) and CC1184S2PT97. We observed rapid induction by salicylate up to maxima of 140,000 and 70,000 Miller units, respectively. However, 12 h after salicylate addition, a decrease in β-galactosidase activity was observed in the culture overexpressing from the plasmid (Fig. 4A). After 48 h, the activity from CC1184S2λpir(pTPm-lacZ) was 33-fold lower than that from CC1184S2PT97 (2,300 versus 75,000 Miller units), which maintained equivalent high-level expression during the 2 days of culture.
FIG. 4.
Stability of the overproduction phenotype in continuous cultures. (A) Changes in β-galactosidase activities in chemostat cultures of CC1184S2PT97 without ampicillin (solid line), CC1184S2λpir(pTPm-lacZ) without ampicillin (dashed line), and CC1184S2λpir(pTPm-lacZ) with ampicillin (dotted line) after induction by 2 mM salicylate. (B) Percentages of the Lac+ phenotype in the cell populations during growth of the continuous cultures.
Analysis of the Lac phenotype in the culture of CC1184S2λpir(pTPm-lacZ) revealed the increasing dominance of the Lac− phenotype during growth of the culture even in the presence of ampicillin (Fig. 4B). This is not surprising since secretion of β-lactamase in the continuous culture at a high cell density could support growth of a non-β-lactamase-producing population. In contrast, all the colonies analyzed (≥4,000 colonies) from the CC1184S2PT97 culture were Lac+.
DISCUSSION
We tested the capacity of a previously described cascade circuit able to amplify gene expression as a system for overproducing recombinant proteins. The advantages of this system are positive regulation, in contrast to the regulation of lac-based expression systems, a modular structure, and activation by economic salicylate derivative inducers. The last advantage is of special interest for large-scale fermentations, in which the cost of other inducers, such as IPTG, may be significant. The modular structure of the system also allows insertion of the regulatory and expression modules into the chromosomes of gram-negative bacteria.
We tested the regulatory capacity, as estimated by induction ratios, and the maximal levels of expression of the cascade system in different configurations and compared the results to the results for single expression systems located in plasmids. The data show that the cascade system in which the expression module is located in a plasmid may improve both characteristics compared to the characteristics of the single systems in plasmids since it resulted in the highest expression level together with a significantly lower basal level of expression. The expression levels of a cascade system in which both the regulatory and expression modules were located in the chromosome were slightly lower than but comparable to those of the single systems in plasmids, but such a cascade system resulted in a significantly higher induction ratio because of its low basal level of expression (Fig. 2 and 3 and Table 3). Together, these data indicate that the cascade system may be more convenient for protein overproduction than single expression systems located in plasmids.
A major problem in large-scale fermentation is the ability of the overproducer strain to maintain a high level of expression since most of the expression systems are located in plasmids. Quick loss of plasmids with the heterologous gene under inducing conditions has been reported previously (1, 24). We found that this is also the case for different systems expressing genes from plasmids in the absence of selective pressure (Tables 3 and 4). Selecting for the presence of plasmids by adding an antibiotic to a growing culture did not solve the problem, since the production phenotype was also lost under these conditions when the culture was induced for more than 12 h (Fig. 4). Under these conditions, the overproduction phenotype was lost because the induced culture was rapidly taken over by a population of expression-down mutants. Analysis of such mutants from cultures of strains with the cascade system bearing the expression module in plasmids revealed that mutations were located either in the plasmid or in the chromosome, which was expected since both types of mutations may result in loss of the overproduction phenotype. These results indicate that high levels of expression from plasmids are not compatible with plasmid maintenance. A plausible explanation is that a high transcription rate interferes with replication of these small replicons. In addition, recombination hot spots due to high transcription rates could appear, which would increase the frequency of mutation due to rearrangements involving the regulatory sequences (29).
On the other hand, stability virtually identical to that of indigenous genes has been reported for expression systems inserted into the chromosome (5, 7, 28). However, the levels of expression from these systems in the chromosome are significantly lower than those of the expression systems in plasmids. In contrast, the cascade system when both modules were located in the chromosome allowed expression levels that were comparable to or even higher than those of expression systems located in plasmids (Table 3 and Fig. 3). In spite of the high levels of expression, stability analysis showed that the overproduction phenotype of the cascade system located in the chromosome is maintained by overproducing cultures for a long time after induction in both serially diluted batch cultures (Fig. 3) and continuous cultures (Fig. 4) without selective pressure. These results suggest that overproduction from the chromosomal configuration did not imply that there was a major loss of fitness of the overproducing strain, since otherwise expression-down mutants would have dominated the induced culture, and therefore that the chromosome is much more tolerant to high levels of expression than the standard episomal vectors.
In terms of biotechnological production, the cascade system in the chromosome configuration combines high levels of expression with great stability of the overproduction phenotype, which results in protein yields that are at least 1 order of magnitude higher than those of expression systems in plasmids when the time of fermentation is greater than 24 h.
Acknowledgments
This project was supported by the Spanish Ministerio de Ciencia y Tecnología (MCYT) with project FEDER: 1FD97-1788. A.C.R. was supported by a research postdoctoral grant from the Spanish Ministry of Science and Technology.
We thank Active Motif for the gift of pCCD5-lacZ.
REFERENCES
- 1.Bálbas, P., and F. Bolivar. 1990. Design and construction of expression plasmid vectors in Escherichia coli. Methods Enzymol. 185:14-37. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, W. E., N. Morkajalili, D. C. Anderson, R. H. Davis, and D. S. Kompala. 1990. Plasmid-encoded proteins: the principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnol. Bioeng. 35:668-681. [DOI] [PubMed] [Google Scholar]
- 3.Brosius, J. 1984. Toxicity of an overproduced foreign gene product in Escherichia coli and its use in plasmid vectors for the selection of transcription terminators. Gene 27:161-172. [DOI] [PubMed] [Google Scholar]
- 4.Cebolla, A., C. Sousa, and V. de Lorenzo. 2001. Rational design of a bacterial transcriptional cascade for amplifying gene expression capacity. Nucleic Acids Res. 29:759-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cebolla, A., F. Ruiz-Berraquero, and A. J. Palomares. 1993. Stable tagging of Rhizobium meliloti with the firefly luciferase gene for environmental monitoring. Appl. Environ. Microbiol. 59:2511-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cebolla, A., M. E. Vazquez, and A. J. Palomares. 1995. Expression vectors for the use of eukaryotic luciferases as bacterial markers with different colors of luminescence. Appl. Environ. Microbiol. 61:660-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lorenzo, V. 1994. Designing microbial systems for gene expression in the field. Trends Biotechnol. 12:365-371. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., L. Eltis, B. Kessler, and K. Timmis. 1993. Analysis of the Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., M. Herrero, V. Jakubzil, and K. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lorenzo, V., S. Fernández, M. Herrero, U. Jakubzik, and K. Timmis. 1993. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene 130:41-46. [DOI] [PubMed] [Google Scholar]
- 12.Dong, H., L. Nilsson, and C. G. Kurland. 1995. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J. Bacteriol. 177:1497-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez, S., V. de Lorenzo, and J. Perez-Martin. 1995. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol. Microbiol. 16:205-213. [DOI] [PubMed] [Google Scholar]
- 15.Govantes, F., J. A. Molina-Lopez, and E. Santero. 1996. Mechanism of coordinated synthesis of the antagonistic regulatory proteins NifL and NifA of Klebsiella pneumoniae. J. Bacteriol. 178:6817-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero, M., V. de Lorenzo, and K. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahn, M., R. Kolter, C. Thomas, D. Figurski, R. Meyer, E. Renaut, and D. R. Helinski. 1979. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 68:268-280. [DOI] [PubMed] [Google Scholar]
- 18.Kessler, B., K. Timmis, and V. de Lorenzo. 1994. The organization of the Pm promoter of the TOL plasmid reflects the structure of its cognate activator protein XylS. Mol. Gen. Genet. 244:596-605. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Mermod, N., J. L. Ramos, P. R. Lehrbach, and K. N. Timmis. 1986. Vector for regulated expression of cloned genes in a wide range of gram-negative bacteria. J. Bacteriol. 167:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mertens, N., E. Renaut, and W. Fiers. 1995. Tight transcriptional control mechanism ensures stable high level expression from T7 promoter based expression plasmids. Bio/Technology 13:175-179. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Miller, W. G., and R. W. Simons. 1993. Chromosomal supercoiling in Escherichia coli. Mol. Microbiol. 10:675-684. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson, J., and S. G. Skogman. 1986. Stabilization of Escherichia coli tryptophan-production vectors in continuous cultures: a comparison of three different systems. Bio/Technology 4:901-903. [Google Scholar]
- 25.Pavitt, G. D., and C. F. Higgins. 1993. Chromosomal domains of supercoiling in Salmonella typhimurium. Mol. Microbiol. 10:685-696. [DOI] [PubMed] [Google Scholar]
- 26.Ramos, J. L., A. Stolz, W. Reineke, and K. Timmis. 1986. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc. Natl. Acad. Sci. USA 83:8467-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sousa, C., V. de Lorenzo, and A. Cebolla. 1997. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology 143:2071-2078. [DOI] [PubMed] [Google Scholar]
- 28.Suarez, A., L. H. Staendner, M. Rohde, G. Piatti, K. Timmis, and C. Guzman. 1997. Stable expression of pertussis toxin in Bordetella bronchiseptica under the control of a tightly regulated promoter. Appl. Environ. Microbiol. 63:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilette, D., S. D. Ehrlich, and B. Michel. 1995. Transcription-induced deletions in Escherichia coli plasmids. Mol. Microbiol. 17:493-504. [DOI] [PubMed] [Google Scholar]
- 30.Winther-Larsen, H. C., K. D. Josefsen, T. Brautaset, and S. Valla. 2000. Parameters affecting gene expression from Pm promoter in Gram-negative bacteria. Metab. Eng. 2:79-91. [DOI] [PubMed] [Google Scholar]
- 31.Wood, K. V., Y. A. Lam, H. H. Seliger, and W. D. McElroy. 1989. Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science 244:700-702. [DOI] [PubMed] [Google Scholar]