Abstract
Surface water and groundwater are continuously used as sources of drinking water in many metropolitan areas of the United States. The quality of water from these sources may be reduced due to increases in contaminants such as Escherichia coli from urban and agricultural runoffs. In this study, a multiplex fluorogenic PCR assay was used to quantify E. coli O157:H7 in soil, manure, cow and calf feces, and dairy wastewater in an artificial wetland. Primers and probes were designed to amplify and quantify the Shiga-like toxin 1 (stx1) and 2 (stx2) genes and the intimin (eae) gene of E. coli O157:H7 in a single reaction. Primer specificity was confirmed with DNA from 33 E. coli O157:H7 and related strains with and without the three genes. A direct correlation was determined between the fluorescence threshold cycle (CT) and the starting quantity of E. coli O157:H7 DNA. A similar correlation was observed between the CT and number of CFU per milliliter used in the PCR assay. A detection limit of 7.9 × 10−5 pg of E. coli O157:H7 DNA ml−1 equivalent to approximately 6.4 × 103 CFU of E. coli O157:H7 ml−1 based on plate counts was determined. Quantification of E. coli O157:H7 in soil, manure, feces, and wastewater was possible when cell numbers were ≥3.5 × 104 CFU g−1. E. coli O157:H7 levels detected in wetland samples decreased by about 2 logs between wetland influents and effluents. The detection limit of the assay in soil was improved to less than 10 CFU g−1 with a 16-h enrichment. These results indicate that the developed PCR assay is suitable for quantitative determination of E. coli O157:H7 in environmental samples and represents a considerable advancement in pathogen quantification in different ecosystems.
Constructed wetlands are commonly used for biotreatment of urban runoff and agricultural waste, such as dairy waste wash water from milk cows and on-farm waste from manure piles. These wastes harbor different bacterial species including human pathogens. Present management practices for dairy wash water on most commercial farms involve long-term storage in ponds where evaporation, percolation into groundwater, or spraying onto crops and/or disposal on land occurs. A constructed wetland treatment system maximizes the utility of existing storage ponds, reduces sediment loading in the wash water ponds through on-site treatment, increases pond capacity, and decreases the need to clean and scrape storage ponds. The goal is to have a final effluent water from the wetland that is suitable for on-site reuse and reduces the amount of contaminants entering groundwater supplies as a result of percolation of wash water stored in ponds and sprayed on disposal lands. This final product is expected to enhance water and air quality; reduce biological oxygen demand, nitrate-nitrogen, phosphorus, and ammonia emissions; and inactivate protozoan parasites and pathogens present in dairy wash water.
The wash waters from dairy and beef cattle operations harbor different bacterial species including human pathogens such as enterohemorrhagic Escherichia coli O157:H7. E. coli O157:H7 causes a wide spectrum of disease symptoms in humans, such as diarrhea ranging from mild to bloody, hemorrhagic colitis, and complications including hemolytic-uremic syndrome (HUS) and seizures that are particularly severe in children (14). E. coli O157:H7 strains are generally lysogenized with one or more phages carrying genes for Shiga-like toxins (7, 22). Shiga-like toxins play a major role in the pathogenesis of hemorrhagic colitis and HUS through cytotoxic effects on cells of the kidneys, intestines, central nervous system, and other organs (14, 23). E. coli O157:H7 also harbors a 43-kb pathogenicity island, termed the locus of enterocyte effacement, containing virulence attributes required for the formation of attaching and effacing lesions on the target host cells (17, 19, 26, 33). The formation of the attaching-effacing lesion phenotype requires interactions between intimin, a bacterial cell surface protein, and a translocated intimin receptor (Tir) (2, 17, 19, 20). Tir is secreted by bacterial cells into the host cells, where it is localized into the host cell membrane and serves as a docking point for bacterial intimin (15). Intimin and Tir are encoded by the genes eae and tir, respectively, which are present in the locus of enterocyte effacement (1). The intimin proteins of pathogenic E. coli strains are highly divergent in the amino acid sequence at the carboxy terminus, and based on this sequence variation, intimins have been classified into five subgroups with E. coli O157:H7 strains occurring in type Y (25).
Since dairy and beef cattle are considered important reservoirs for E. coli O157:H7 (6, 34), this pathogen could potentially enter the drinking water supply from cattle waste wash water. As wastewater is being increasingly used in the irrigation of crops, there is a danger of inadequate treatment to eliminate bacterial pathogens. Also, recent alterations in methods of food production and delivery to consumers have exacerbated problems with pathogenic bacteria such as E. coli O157:H7. The increasing popularity of organic food production has led to widespread composting of animal manure for use in crop production. Problems have often occurred as a consequence of temperatures during composting being insufficient to kill enteric bacteria.
Methods for the effective treatment of wastewater and manure are essential to ensure the safety of drinking water and crops. To ascertain the effectiveness of wastewater treatment, it is imperative to develop methods that would allow rapid and sensitive detection of bacterial pathogens in agricultural and urban water runoffs, soils, and cattle manure before and after treatment of these important recyclable resources. The availability of methods for effective treatment and detection of pathogens in environmental samples would reduce the incidence of human illnesses by preventing the transmission of bacterial pathogens to humans. This in turn would save millions of dollars in health-related medical costs and food recalls.
PCR has become an important method for the rapid, sensitive, and specific detection of bacterial pathogens. Recent advances in the synthesis of fluorogenic probes and the development of instrumentation for continuous monitoring of fluorescence have facilitated the development of real-time PCR assays for specific, automated detection and quantitation of amplified gene products (10). Real-time multiplex PCR has been used for the detection and quantification of E. coli O157:H7 in food and clinical samples (4, 21, 27, 28), but absolute quantification of this pathogen by this technology has not been tested vigorously with wastewater and environmental samples.
In this study, a real-time multiplex PCR assay with specific probes (Table 1) was used to detect and quantify E. coli O157:H7 in dairy wastewater prior to and following treatment by a subsurface constructed wetland. The main benefit of simultaneous multiplexing with quantification is cost efficiency and a higher throughput. The objective of the present study was to develop a real-time multiplex PCR assay to detect and quantify E. coli O157:H7 and to determine its suitability for monitoring the effectiveness of a subsurface constructed wetland system in reducing E. coli O157:H7 levels in dairy waste wash water. The real-time PCR method was also evaluated for the detection and quantification of E. coli O157:H7 in soil samples seeded with known numbers of the organism. An iCycle iQ detection system (Bio-Rad, Hercules, Calif.) consisting of a thermal cycler, an optical module, and detection software was used for the PCR assay validation. This instrument monitored incremental increases in fluorescence at successive PCR cycles in real time and used this data to establish a threshold cycle (CT) for the assay. During the early rounds of amplification, the changes in fluorescence are negligible and beyond the sensitivity of the system detector. At some point during cycling, a change will register with the detection system. This is called the threshold cycle (CT) and is proportional to the amount of the starting amount of nucleic acid (10). Since the threshold cycle is inversely proportional to the copy number of a target gene, this allowed for the construction of standard curves by using samples containing 10-fold dilutions of target genes.
TABLE 1.
Nucleotide sequences of primers and fluorogenic probes
| Primer or probea | Sequence (5′→3′) | Tm (°C) | Locationb within the target gene | Genec detected |
|---|---|---|---|---|
| stx1 forward | GAC TGC AAA GAC GTA TGT AGA TTC G | 60 | 90-114 | stx1 (150 bp) |
| stx1 reverse | ATC TAT CCC TCT GAC ATC AAC TGC | 59 | 240-217 | |
| stx1 probe | TGA ATG TCA TTC GCT CTG CAA TAG GTA CTC | 70 | 116-145 | |
| stx2 forward | ATT AAC CAC ACC CCA CCG | 59 | 184-201 | stx2 (200 bp) |
| stx2 reverse | GTC ATG GAA ACC GTT GTC AC | 58 | 392-373 | |
| stx2 probe | CAG TTA TTT TGC TGT GGA TAT ACG AGG GCT TG | 69 | 204-235 | |
| eae forward | GTA AGT TAC ACT ATA AAA GCA CCG TCG | 59 | 2494-2524 | O157:H7 eae (106 bp) |
| eae reverse | TCT GTG TGG ATG GTA ATA AAT TTT TG | 59 | 2599-2574 | |
| eae probe | AAA TGG ACA TAG CAT CAG CAT AAT AGG CTT GCT | 69 | 2572-2540 |
Primers and probes were designed by using the program Primer Express version 1.0 (PE Applied Biosystems). The melting temperature (Tm) of the primers ranged from 58 to 60°C, and that of the probes ranged from 68 to 70°C. The probes were conjugated with fluorescent reporter dyes FAM 490 (stx1), HEX 530 (stx2), and Texas red 575 (eaeO157:H7) at the 5′ ends and with the quencher dye BHQ 1 for FAM and HEX and BHQ 2 for Texas red at the 3′ ends.
The positions of the oligonucleotides are listed relative to the initiation codon (+1 adenine) of the respective gene.
MATERIALS AND METHODS
Bacterial strains and culture media.
The specificity of the multiplex PCR was determined with 33 different strains of E. coli O157:H7 and other related strains (Table 2), most of which have been previously characterized (28). From the 33 strains, 9 E. coli O157:H7 and non-O157:H7 strains were further evaluated to determine the sensitivity of the multiplex real-time PCR assay (Table 3). All strains were obtained from the National Animal Disease Center (Ames, Iowa) and were cultured on Luria-Bertani broth agar and sorbitol-MacConkey (SMAC) agar plates at 37°C.
TABLE 2.
Specificity of conventional multiplex PCR assay for detection of the stx1 and stx2 genes of STEC and the eae gene of E. coli O157:H7
| Bacterial strain type tested | Strain no. | Serotype | Genotypea
|
Origin/disease or status | ||
|---|---|---|---|---|---|---|
| stx1 | stx2 | eae | ||||
| Nontoxigenic E. coli | 63 | O78:H16 | − | − | − | Dog |
| 912 | OX3:H11 | − | − | − | Pig/normal | |
| EPEC | 1861 | O119 | − | − | − | Human/diarrhea |
| 1987 | O111:H2 | − | − | − | Human | |
| 1988 | O26:NM | − | − | − | Human | |
| 5722 | O55:NM | − | − | + | Human | |
| ETEC | 431 | O101 | − | − | − | Pig/diarrhea |
| 1477 | O149:H10 | − | − | − | Pig/diarrhea | |
| STEC | 5702 | O103:H2 | + | − | + | Human/HUS |
| 5354 | O157:H7 | + | − | + | Calf/normal | |
| 3048 | O157:NM | + | + | + | Calf | |
| 5264 | O157:NM | + | + | + | Calf/normal | |
| 4178 | O157:H7 | + | + | + | Human/HUS | |
| 5705 | O121:H19 | + | + | − | Human/HUS | |
| 3883 | O111:NH | + | − | + | Calf/normal | |
| 5710 | O26:H11 | + | + | + | Human/HUS | |
| 2871 | O157:H7 | − | + | + | Calf/normal | |
| 2725 | O157:H7 | + | + | + | HUS | |
| 5570 | O157:H7 | + | + | + | Human/diarrhea | |
| 3128 | O113 | + | + | − | Human/diarrhea | |
| 2799 | O157:NH | − | + | + | Calf/normal | |
| 5709 | O111:NM | + | + | + | Human/HUS | |
| 5981 | O157:NM | − | − | + | Calf | |
| 5701 | O126:H27 | + | − | − | Human/HUS | |
| 3861 | OX3:H2 | + | + | + | Calf/normal | |
| 2409 | O157:H7 | + | − | + | Food | |
| 1043 | O22 | + | − | + | Calf/diarrhea | |
| 2873 | O157:NM | + | + | + | Calf/normal | |
| 5708 | O45:H2 | + | − | − | Human/HUS | |
| 4700 | O157:H7 | + | + | + | Calf | |
| 3081 | O157:H7 | + | + | + | Calf | |
| 1524 | O132 | − | − | + | Rabbit/diarrhea | |
| 3244 | O119:H16 | − | + | − | Calf/normal | |
The presence of these genes had been determined by Sharma et al. (28) by fluorogenic PCR with DNA probes specific for these genes and confirmed in this study. +, gene detected by multiplex PCR; −, gene not detected by multiplex PCR.
TABLE 3.
Sensitivity of real-time multiplex PCR assay for detection and quantification of the stx1 and the stx2 genes of STEC, eae of E. coli O157:H7, and related strains
| Bacterial strain type | Strain no. | Serotype | Genotype |
CT of gene:
|
||
|---|---|---|---|---|---|---|
| stx1 | stx2 | eae | ||||
| Nontoxigenic E. coli | 63 | O78:H16 | 42.3 | −a | − | |
| Salmonella enterica serovar Typhimurium | 3244 | O119:H16 | 46.9 | − | − | |
| EPEC | 5722 | O55:NM | eae | 38.1 | 47.7 | 43.8 |
| ETEC | 431 | O101 | stap | 38.8 | − | − |
| STEC | 5701 | O116:H27 | stx1 | 24.6 | − | 47.8 |
| 4718 | O157:H7 | stx1 stx2 eae | 24.8 | 29.2 | 26.0 | |
| 4700 | O157:H7 | stx1 stx2 eae | 25.1 | 31.1 | 27.6 | |
| 3081 | O157:H7 | stx1 stx2 eae | 23.9 | 28.4 | 26.1 | |
| 5699 | O91:H21 | stx2 | 39.2 | 26.3 | − | |
−, CT not determined by real-time PCR.
Primer and probe design.
Primers and probes used for the detection and quantification of E. coli O157:H7 are shown in Table 1. The reporter dyes FAM (6-carboxyfluorescein), HEX (6-carboxyfluoroscein), and Texas red (sulforhodamine 101) were conjugated at the 5′ ends of the probes, and quencher dyes, Black Hole quencher (BHQ) dyes I and II (Biosearch Technologies, Novato, Calif.), were conjugated at the 3′ ends. The FAM-, HEX-, and Texas red-labeled probes were used for the detection of the stx1, stx2, and eae genes, respectively, and were synthesized by Biosearch Technologies. The BHQ dye was used as the quencher dye because of its broad quenching spectrum and a lower signal-to-noise ratio than that of other quenching dyes.
DNA isolation from pure culture and environmental samples.
Genomic DNA was isolated from a pure culture of E. coli O157:H7 strain 3081, grown for 12 h at 37°C, and extracted with the Qiagen tissue kit (QIAamp DNA Mini kit; Qiagen, Valencia, Calif.) according to the manufacturer's protocol. Total bacterial community DNA was extracted from environmental samples to determine the detection limits of E. coli O157:H7 strain 3081 by real-time PCR. The DNA was extracted from 500 mg of soil or fecal samples or 100 ml of water with UltraClean soil, fecal, and water DNA kits (MO BIO, Inc., Solana Beach, Calif.) according to the manufacturer's protocol. The 100 ml of water was filtered and concentrated into 0.25 ml for DNA extraction with the MO BIO water DNA kit.
Laboratory seeding experiments.
Arlington sandy loam soil was obtained from the University of California, Riverside, Agricultural Experiment Station. Soil was removed from the top 15 cm with a stainless steel shovel, passed through a 4-mm-pore-size stainless steel sieve, and treated with 500 mg of methyl bromide kg−1 to fumigate the soil. The soil was stored at 4°C and tested by bacteriological and PCR techniques to ensure that it was E. coli O157:H7 negative prior to use. An overnight culture of E. coli strain 3081 was serially diluted in tryptic soy broth, and the number of CFU of bacteria in each dilution was determined by plating on cefixime-tellurite (CT)-SMAC agar with BCIG (5-bromo-4-chloro-3-indoxyl-β-d-glucuronide) containing 0.05 mg of cefixime liter−1 and 2.5 mg of tellurite liter−1 (LAB M; IDG). The average titer (CFU milliliter−1) of three replicates was determined. For the spiked experiment, 10 g of soil was seeded with a 0- to 10-fold dilution series of E. coli O157:H7 strain 3081 in flasks containing 90 ml of modified Luria-Bertani broth consisting of 8 mg of vancomycin liter−1, 0.05 mg of cefixime liter−1, and 10 mg of cefsulodin liter−1. The inoculated flasks were incubated at 37°C with agitation at 160 rpm. Samples were vortexed for 30 s, and 2-ml aliquots were taken after 0, 2, 8, 16, and 24 h for DNA extraction. The DNA extracted from the enrichment broth was used as a template for real-time PCR. In experiments to compare real-time PCR with the conventional culture method, the enrichment broths of seeded soil samples were also plated onto CT-SMAC agar plates. Sorbitol-negative colonies were enumerated.
Layout of wetlands, sampling scheme, and microbiological analysis.
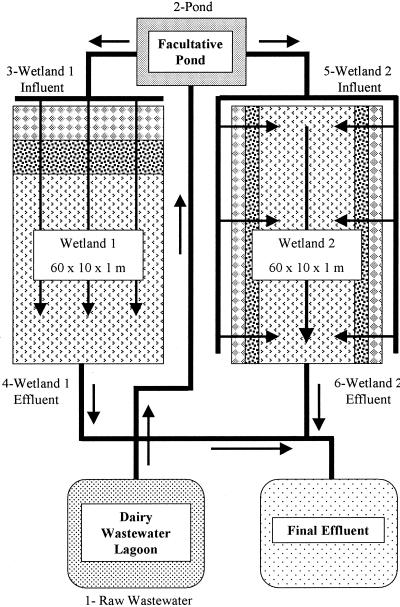
The project design included two wetlands operating in parallel, in addition to a raw and a facultative pond for central collection of wash water prior to treatment (Fig. 1). The constructed wetlands utilized a horizontal subsurface flow system where wash water treatment occurred beneath the surface of the gravel, which was planted with shallow but densely rooted herbaceous plants. Wetland 1 was an end-loading design, where the wash water entered through multiple inlets into coarse gravel, reeds (Phragmites communus), and bulrushes (Scirpus validus) and then drained into the collection box at the end of the basin. Wetland 2 was a side-loading basin, where wash water entered through multiple inlets along both sides of the wetland and passed through a narrow gravel bed, containing reeds and bulrushes, before being collected through a perforated pipe along the center of the wetlands which drained to the collection box.
FIG. 1.
Schematic of the dairy wash water wetlands used as a treatment facility for the removal of dairy waste and other compounds. All wastewater received in the wetlands was from the milking parlor. The wash water was drained into the raw water lagoon (1), moved through 3-in.-diameter polyvinyl chloride pipes by gravity to an aeration pond, and passed through two parallel wetlands. Water samples were collected from the raw dairy wastewater lagoon (1), facultative pond (2), wetland 1 influent (3), wetland 2 influent (5), wetland 1 effluent (4), and wetland 2 effluent (6).
Bacterial biofilms on the gravel supported the microbial degradation of nutrients in the wash water. The deep-rooted vegetation from bulrushes transported oxygen to the anaerobic zone, allowing for the nitrification of ammonium and the subsequent denitrification of nitrate in the wash water. Six sample locations were used as indicated in Fig. 1. Samples were collected and plated within 6 h. Fresh cow and calf fecal samples, as well as manure samples, were also collected for bacterial enumeration. Fresh fecal samples were collected within minutes of defecation with a stainless steel shovel, and the top portion was taken for laboratory analysis. Manure samples were taken from piles of fecal materials that had been deposited for about 2 weeks. DNA extraction of environmental samples was completed within 48 h of collection. All samples were collected from a commercial dairy farm in Chino, Calif., adjacent to the constructed wetland between December 2000 and September 2001.
One gram or 1 ml of environmental samples was added to 9 ml of tryptic soy broth, vortexed briefly, serially diluted, and plated for the enumeration of total heterotrophic bacteria, total E. coli, and E. coli O157 on tryptic soy agar, SMAC, and CT-SMAC agar, respectively. The plates were incubated at 25°C for heterotrophic bacterial counts and 37°C for total E. coli and E. coli O157 for 24 h. Six sorbitol-negative, translucent colonies per sample were tested by multiplex PCR to determine the presence of the three genes. Additionally, isolates that were sorbitol positive or β-glucuronidase positive (red-pink colonies with a purple center or green colonies) were enumerated as other E. coli strains.
Real-time multiplex PCR assay for quantification of E. coli O157:H7 strain 3081.
A single primer-probe reaction was compared with a multiplex primer-probe reaction to determine any significant differences in the CT values. E. coli O157:H7 strain 3081 genomic DNA (7.9 pg ml−1) was amplified in both single and multiplex reactions. To demonstrate the detection range of multiplex PCR, 10-fold serial dilutions containing 7.9 × 100 to 7.9 × 10−9 pg of genomic DNA ml−1 were assayed for the stx1, stx2, and eae genes. PCR was performed in a 50-μl volume containing 200 μM deoxynucleoside triphosphates (dNTPs), 2 μl of genomic DNA from each concentration, 2.5 U of AmpliTaq Gold polymerase, 5 μl of 10× TaqMan buffer (PE Applied Biosystems, Foster City, Calif.), 0.3 μM (each) primer, 0.1 μM (each) probe, and 3 mM MgCl2. Genomic DNA purified from E. coli O157:H7 strain 3081 was used as a template for the positive control. Reaction mixtures were dispensed into a 96-well, thin-wall PCR plate (Bio-Rad), covered with optically clear sealing film, and centrifuged briefly. PCR was performed with the iCycle iQ thermal cycler with the following cycle conditions: denaturation at 95°C for 10 min and 50 cycles of 94°C for 20 s, 55°C for 30 s, and 72°C for 40 s, followed by a 5-min extension at 72°C and holding at 4°C.
The iCycle iQ real-time PCR detection system and software were used for data analyses. This system used a thermal cycler, an optical module, and detection software to quantify PCR products in real time, as revealed by the increase of fluorescence signal by 5′-nuclease activity during the amplification process. The threshold cycle (CT) for each standard was plotted against the log of starting quantity to construct the standard curve used to quantify genes in the unknown samples.
Multiplex PCR quantification of environmental samples.
Template DNA (2 μl) was added to 48 μl of the reaction mixture as described above, except that the concentrations of MgCl2 and dNTPs were optimized to 5 mM and 450 μM, respectively. Amplification conditions were as described above for all environmental samples. E. coli O157:H7 concentrations in environmental samples were confirmed quantitatively by assaying serial dilutions (7.9 × 100 to 7.9 × 10−9 pg ml−1) of strain 3081 DNA in the same plate. Dilution series and environmental samples were tested in triplicate.
Standardization and amplification efficiency.
Standard curves generated from plotting CT versus log10 of starting DNA quantities (picograms) were used for determining the detection limit of the assay. The standard curve was constructed by using known quantities of genomic DNA (7.9 × 100 to 7.9 × 10−9 pg ml−1) extracted from samples containing 6.4 × 10−2 to 6.4 × 108 CFU of E. coli O157:H7 ml−1. The concentration of the extracted DNA was measured by an Ultrospec 4000 spectrophotometer with Swift II application software (Pharmacia Biotech, Cambridge, England), and numbers of CFU milliliter−1 were determined by plating culture dilutions on CT-SMAC. The titers (CFU milliliter−1) of E. coli O157:H7 present in unknown samples were determined from the standard curve. For a comparison of PCR amplification efficiencies and detection sensitivities among different experiments, slopes of the standard curves were calculated by performing a linear regression analysis with the iCycle iQ software. A mixture of all PCR reagents without any DNA was used as a negative control. Amplification efficiency (E) was estimated by using the slope of the standard curve and the formula E = (10−1/slope) − 1. A reaction with 100% efficiency will generate a slope of −3.32.
Standardization of DNA quantities between known and unknown samples was accomplished by dividing total numbers of CFU of E. coli O157:H7 strain 3081 milliliter−1 by the mean starting DNA concentration of that CFU milliliter−1 from the instrument analysis. This resulted in a CFU milliliter−1 index, which was used as a multiplier to calculate the CFU milliliter−1 for all unknown samples. The CFU milliliter−1 index was obtained from the highest DNA quantity to estimate CFU milliliter−1 from lower DNA quantities. This approach was used because the instrument can give reports in either concentrations or copy numbers, and for environmental samples, it is easier to understand results in actual numbers than in concentrations or copy numbers. For example, in the spiked soil calculated numbers of CFU milliliter−1 from plates for E. coli O157:H7 strain 3081 were 6.4 × 108 after 16 h of growth at 37°C. The mean starting quantity from the instrument was 2.8 × 101 CFU ml−1, resulting in a CFU milliliter−1 index of 2.29 × 107. This number was used as a factor for the determination of CFU milliliter−1 of unknown samples with starting mean DNA concentrations determined by the instrument analysis. This process allowed for the establishment of a consistent method for calculating DNA quantities and the corresponding CFU milliliter−1 in unknown samples. At the same time, DNA from serial dilutions with known CFU milliliter−1 was included in every reaction to cross check the accuracy of the calculations.
Statistical analysis.
Data analyses of environmental and laboratory samples were performed with SAS software (SAS/STAT User's Guide, Release 6.03; SAS Institute Inc., Cary, N.C.). Analyses of variance, means, and standard deviations were conducted to determine whether there were significant differences among mean quantities of pathogens for each sampling point.
RESULTS AND DISCUSSION
Specificity and sensitivity of multiplex real-time PCR assay.
The ability of the multiplex PCR assay to distinguish E. coli O157:H7 from other serotypes of E. coli was determined by analyzing 33 Shiga-toxigenic E. coli (STEC) and non-STEC E. coli strains (Table 2). These strains were selected from a variety of serotypes with various combinations of stx and eae genes. PCR analysis of the 33 strains yielded amplification results consistent with previously published features of these strains (28). The sensitivities of FAM-, HEX-, and Texas red-labeled probes in specifically detecting and quantifying stx1, stx2, and eae genes were determined by plotting the log DNA starting quantities of nine strains used for the test. Three different DNA concentrations with CT values of 23.9, 28.1, and 30.8 for stx1, stx2, and eae genes, respectively, from E. coli O157:H7 strain 3081 pure culture were tested in the real-time PCR assay to determine the sensitivities of the primers and probes in a multiplex reaction from different E. coli strains. Only CT values for the highest concentrations are shown (Table 3). Of the nine bacterial strains, E. coli O157:H7 strains 3081, 4700, and 4718 had CT mean values of 25 or less for the stx1 gene (150-bp amplicon size), 31 or less for the stx2 gene (200-bp amplicon size), and 27.6 or less for the eae gene (106-bp amplicon size) when 5 to 10 pg of DNA ml−1 was used as template. When E. coli O157:H7 strains that carry the stx1 gene, the stx2 gene, or the eae gene were tested, their CT mean values were also comparable to those in the multiple reaction. The non-STEC strains (nontoxigenic E. coli, enteropathogenic E. coli [EPEC], and enterotoxigenic E. coli [ETEC]) had mean CT values (38 to 47) outside the sensitivity limit of the assay for all three genes.
Standard curves, detection limits, and amplification efficiencies.
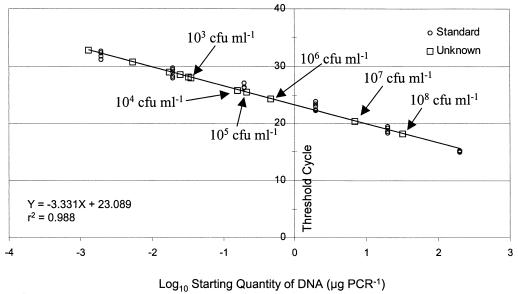
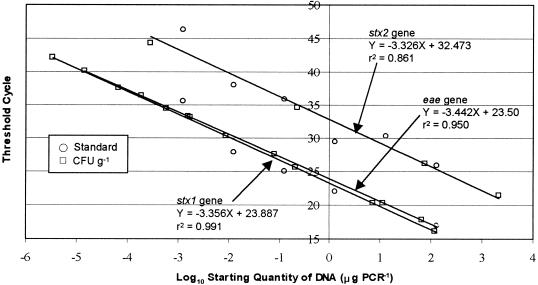
The detection sensitivity of the quantitative real-time PCR assay was determined by testing triplicate sets of genomic DNA prepared from serial dilutions (6.4 × 10−2 to 6.4 × 108 CFU ml−1) of E. coli strain 3081 (Fig. 2). The results were reported as threshold cycle numbers versus log starting quantities of DNA. Positive signals were found in all dilutions except those where the DNA concentrations were below 7.9 × 10−5 pg ml−1. The CT values were also plotted against CFU milliliter−1 (6.4 × 10−2 to 6.4 × 108 CFU ml−1) in 10-fold serial dilutions of the E. coli O157:H7 culture used for extracting genomic DNA. Based on this approach, a correlation was observed between the CT and the CFU milliliter−1 of the starting quantity of E. coli O157:H7 DNA. A detection limit of 7.9 × 10−5 pg of starting DNA ml−1, equivalent to 6.4 × 103 CFU ml−1, was determined. The efficiency of each assay was calculated from the slope of the standard curve. The standard curves generated from the PCR data resulted in reaction efficiencies of 93.6, 99.9, and 99.6% for the stx2, stx1, and eae genes, respectively, with correlation coefficients of 0.96, 0.99, and 0.98, respectively (Fig. 2 has data for the eae gene).
FIG. 2.
Standard curve for the real-time PCR analysis of E. coli O157:H7 strain 3081. The CT values are plotted against the corresponding E. coli O157:H7 DNA concentrations. Template DNA was extracted from E. coli O157:H7 DNA, diluted 10-fold, and used as a known DNA concentration in every experiment. DNA was extracted from E. coli O157:H7 strain 3081 in amounts between 6.4 × 10−2 and 6.4 × 108 CFU ml−1. The reaction efficiencies of the stx1 and eae genes were above 99%, while that of the stx2 gene was about 93%.
Recovery of E. coli O157:H7 from spiked soil.
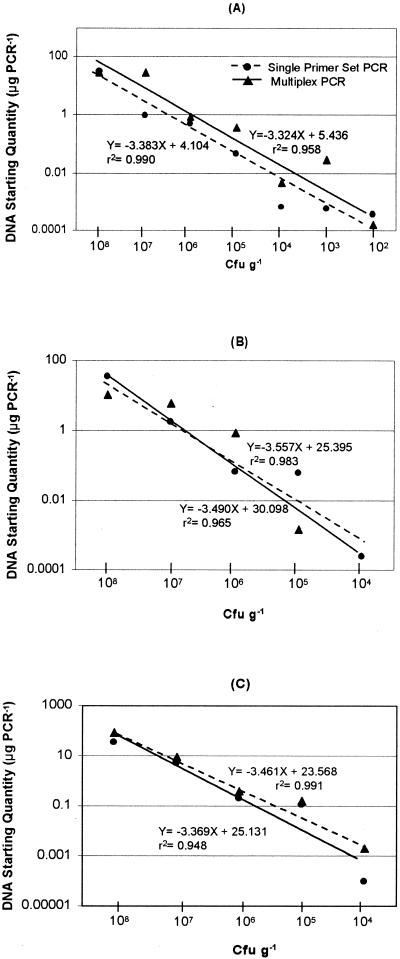
The ability to quantify E. coli O157:H7 in environmental samples was tested by spiking different dilutions of E. coli O157:H7 into methyl bromide-fumigated soil. Bacterial DNA was extracted from inoculated soil samples and used as the template in the multiplex PCR. Since the assay with spiked soil samples involved a multiplex real-time PCR assay, an effort was made to maximize and equalize the efficiency of amplification among each of the three primer-probe sets. Cross-reactivity between fluorophores was determined by comparing the mean CT values of wells containing one primer-probe set to those of wells containing primers and probes for all three genes in the spiked samples. By adjustment of the concentrations of dNTPs and MgCl2 to 450 μM and 5 mM, respectively, CT values of single primer-probe PCRs differed by ≤5% from the CT values generated in the multiplex PCR containing all three primer-probe sets (Fig. 3). Under these optimized conditions, the linearity between the CT values and the target concentrations was observed between 3.5 × 103 (stx1), 3.5 × 104 (stx2 and eae), and 3.5 × 108 CFU ml−1 dilutions of E. coli O157:H7. This demonstrates that quantification of target DNA was possible within this range. The detection sensitivity in soils spiked with 10-fold serial dilutions of E. coli O157:H7, therefore, ranged between 3.5 × 103 CFU ml−1 and 3.5 × 108 CFU ml−1 for the stx1 gene (Fig. 3A) and 3.5 × 104 CFU ml−1 and 3.5 × 108 CFU ml−1 for the stx2 and eae genes (Fig. 3B and C).
FIG. 3.
Determination of detection limits between the single reactions and the multiplex reactions in spiked soil. Standard curves for multiplex versus single-gene real-time PCR analysis of E. coli O157:H7 are shown for the three genes. A linear relationship was maintained between 6.4 × 103 and 6.4 × 108 CFU g−1 for the stx1 gene (A) and 6.4 × 104 and 6.4 × 108 CFU g−1 for the stx2 gene and the eae gene, respectively (B and C, respectively).
Effect of enrichment on detection sensitivity of multiplex PCR.
Most bacterial pathogens, including E. coli O157:H7, are present in very low numbers in soil, wastewater, and feces. Soil and feces also contain substances that are inhibitory to PCR and amplification efficiencies (31, 32). The amplification efficiency and the efficiency of the probe cleavages have direct effects on the CT measurements of the DNA starting quantities (11, 16, 24). Precise CT determination depends upon the efficient performance of the PCR amplification and detection of the reporter fluorophores. This problem is very critical when dealing with environmental samples (30). According to these authors, this problem was overcome by using probes designed to take advantage of quenching by fluorescence resonance energy transfer. Detection was improved in this study by the use of dark quenchers (BHQ dyes) with a very broad fluorescence spectrum within the context of fluorescence resonance energy transfer. This aided in the successful detection of equivalent numbers of E. coli O157:H7 in spiked soil.
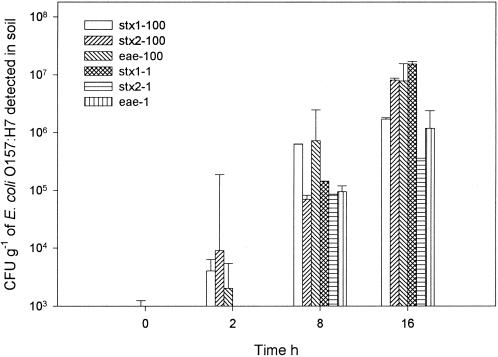
The detection of very low levels of bacterial contamination in soils and feces requires that the samples be cultured for a few hours in an appropriate enrichment broth. Enrichment dilutes out the inhibitory substances and provides conditions conducive to the growth and multiplication of bacterial pathogens to a detectable number. Plotting the mean CT values against the starting quantities of DNA generated the standard curve that was used to estimate E. coli O157:H7 concentrations in spiked soil samples (Fig. 4). The multiplex PCR assay allowed detection of 1.58 × 107, 7.52 × 106, and 4.96 × 106 CFU of E. coli O157:H7 g−1 in soil with the stx1-, stx2-, and eae-specific primer-probe sets, respectively, from soil spiked with 3.5 × 108 CFU of E. coli O157:H7 strain 3081 g−1 (data not shown). The detection limit of this PCR assay in soil samples without enrichment (0 h) was 3.5 × 104 CFU g−1. At this concentration, the assay efficiencies were within 5% of each other and were within the linear range of the standard curve. The concentrations were equivalent to 4.10 × 104, 4.88 × 104, and 4.85 × 104 CFU of E. coli O157:H7 g−1 based on detection of the stx1, stx2, and eae genes, respectively. After 2 h of enrichment, the assay detected 6.7 × 103 CFU g−1 on the average for the three genes when the soil was spiked with 100 CFU of E. coli O157:H7 ml−1 (Fig. 5). After 8 h, the soil samples spiked with 1 CFU ml−1 resulted in the detection of 1.60 × 105 CFU g−1 on the average for the three genes and 1.15 × 106 CFU g−1 when 100 CFU ml−1 was used as the inoculum density. By using a single enrichment process, the real-time PCR assay detected between 1 and 10 CFU of E. coli O157:H7 g−1 after 16 h of enrichment in artificially inoculated soils. The presence of heterotrophic bacterial flora at a level of 107 CFU g of soil−1 before enrichment had no effect on the detection sensitivity of this assay (data not shown). The detection sensitivity obtained in this assay was comparable to that of conventional multiplex PCR methods (13). The use of enrichment along with the real-time PCR approach described here offers the possibility for sensitive detection of E. coli O157:H7 in soil within 1 day. This approach has successfully been used for relative quantification of E. coli O157:H7 in other matrices (3, 21, 28).
FIG. 4.
Standard curves for multiplex real-time PCR analysis of E. coli O157:H7 in spiked soil. The reaction efficiencies for the stx1 and eae genes were higher than that for the stx2 gene as demonstrated by the closeness of the standard curves. These standard curves were used for the detection and quantification of E. coli O157:H7 from the enrichment experiment.
FIG. 5.
Detection and quantification of E. coli O157:H7 in soil spiked with different concentrations of pathogens and after 0 to 16 h of enrichment. The sensitivity of the multiplex PCR assay from the standard curve in Fig. 4 was used for the detection and quantification of the stx1 and stx2 genes of STEC and the eae gene of E. coli O157:H7 in spiked soil. Concentrations shown are for samples collected after incubation times of 0, 2, 8, and 16 h. Standard errors were derived from the means of triplicate data points.
Concentrations of total E. coli and E. coli O157 in feces, manure, and wetlands.
Total E. coli and E. coli O157 concentrations in different matrices from wetland sites 1 to 6 (Table 4) were determined between February 2001 and September 2001 by culture methods on SMAC and CT-SMAC agar. The numbers of total E. coli bacteria in the samples ranged from 1.1 × 102 (wetland 2 effluent) to 1.8 × 107 (cow and calf feces, manure, and raw wash water) CFU ml−1, and the numbers of E. coli O157 bacteria ranged from 48 (wetland 1 effluent) to 3.1 × 107 in manure samples. There was an overall reduction of 3 to 4 logs of total E. coli and E. coli O157 in this wetland demonstration project. The most efficient unit of the project in removing total E. coli and E. coli O157 was the facultative pond. To evaluate the effects of seasonal variations on the concentration of total E. coli and E. coli O157, samples were grouped and mean separation was carried out by the least significant difference test. The concentrations of total E. coli and E. coli O157 were not significantly different among the six sampling points in February, April, and September but were highly significant in July and August. On the average, the highest concentration of total E. coli and E. coli O157 was recorded in April and the lowest was recorded in September at all the sampling points in the wetlands.
TABLE 4.
Concentrations of total E. coli and E. coli O157 detected in environmental samples by culture methods
| Sitea | Value (CFU ml−1) for the month of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Februaryb | Aprilb | Julyb | Augustb | Septemberb | Aprilc | Julyc | Augustc | Septemberc | |
| Raw | 2.0 × 104 ± 0.4 × 103 | 3.9 × 107 ± 4.2 × 105 | 1.6 × 106 ± 1.7 × 105 | 1.1 × 107 ± 4.2 × 104 | 5.0 × 103 ± 1.7 × 103 | 3.8 × 106 ± 3.4 × 104 | 1.6 × 106 ± 1.5 × 104 | 1.1 × 106 ± 5.2 × 104 | 1.0 × 102 ± 1.8 × 101 |
| Pond | 2.2 × 104 ± 1.0 × 103 | 5.6 × 104 ± 7.4 × 103 | 1.2 × 103 ± 2.8 × 102 | 1.5 × 103 ± 2.5 × 102 | 1.2 × 103 ± 2.8 × 102 | 8.0 × 102 ± 1.2 × 102 | 8.2 × 102 ± 2.7 × 102 | 4.2 × 102 ± 1.0 × 102 | 8.9 × 102 ± 1.2 × 102 |
| W1 Inf | 1.1 × 104 ± 1.3 × 103 | 4.7 × 104 ± 3.9 × 103 | 2.4 × 103 ± 6.9 × 102 | 2.4 × 103 ± 6.9 × 102 | 1.1 × 104 ± 6.5 × 102 | 5.0 × 103 ± 2.3 × 103 | 2.1 × 103 ± 1.3 × 102 | 6.3 × 103 ± 1.5 × 102 | 5.1 × 102 ± 8.6 × 101 |
| W1 Eff | 2.4 × 102 ± 0.2 × 102 | 7.5 × 102 ± 1.4 × 102 | 1.3 × 102 ± 5.9 × 101 | 1.3 × 102 ± 5.9 × 101 | 1.0 × 103 ± 5.9 × 101 | 8.3 × 101 ± 1.3 × 100 | 4.8 × 101 ± 2.8 × 100 | 4.8 × 101 ± 3.2 × 100 | 7.0 × 101 ± 5.8 × 100 |
| W2 Inf | 1.7 × 106 ± 1.6 × 103 | 5.2 × 104 ± 3.8 × 103 | 2.5 × 103 ± 1.2 × 103 | 2.5 × 103 ± 1.3 × 103 | 1.2 × 104 ± 1.3 × 103 | 1.5 × 103 ± 1.1 × 103 | 1.8 × 103 ± 1.4 × 102 | 1.2 × 103 ± 2.5 × 102 | NDd |
| W2 Eff | 1.0 × 104 ± 0.9 × 102 | 6.6 × 102 ± 5.7 × 101 | 2.2 × 102 ± 8.0 × 100 | 2.2 × 102 ± 8.0 × 100 | 3.1 × 101 ± 1.0 × 100 | 3.2 × 102 ± 1.5 × 101 | 3.2 × 102 ± 1.8 × 102 | 1.2 × 102 ± 5.9 × 100 | 8.9 × 102 ± 2.8 × 101 |
| Calf F | NAe | 6.1 × 107 ± 6.2 × 104 | 3.7 × 107 ± 1.2 × 105 | 3.7 × 106 ± 1.1 × 105 | 1.9 × 107 ± 1.2 × 106 | 3.7 × 106 ± 2.4 × 103 | 3.2 × 106 ± 2.4 × 104 | 2.5 × 106 ± 3.8 × 104 | NA |
| Cow F | NA | 7.8 × 107 ± 4.1 × 104 | 9.6 × 105 ± 5.1 × 105 | 2.5 × 106 ± 5.2 × 105 | 3.6 × 106 ± 1.9 × 105 | 3.1 × 104 ± 1.9 × 103 | 6.7 × 104 ± 6.9 × 102 | 6.7 × 104 ± 8.2 × 102 | NA |
| Manure | NA | 6.5 × 107 ± 1.3 × 103 | 1.5 × 106 ± 7.8 × 105 | 9.5 × 105 ± 7.3 × 105 | 1.4 × 106 ± 4.9 × 104 | 2.4 × 105 ± 3.7 × 103 | 3.1 × 107 ± 5.8 × 105 | 1.7 × 107 ± 4.8 × 105 | NA |
All solid samples were from calf feces (Calf F), cow feces (Cow F), or manure, and the liquid waste samples were from the facultative pond (Pond), the raw pond (Raw), wetland 1 effluent (W1 Eff), wetland 1 influent (W1 Inf), wetland 2 effluent (W2 Eff), and wetland 2 influent (W2 Inf).
Total E. coli numbers from different samples determined on SMAC medium.
Total E. coli O157 numbers determined on CT-SMAC medium.
ND, not determined.
NA, samples not collected for analysis.
Quantification of E. coli O157:H7 from fecal and wetland samples by real-time PCR.
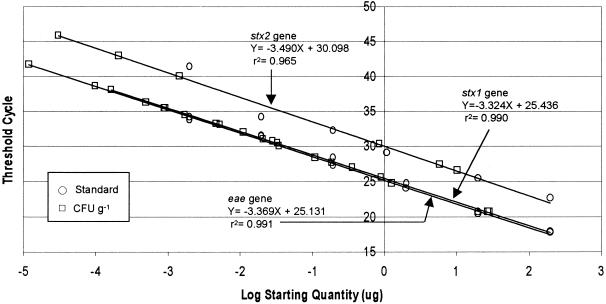
Successful quantification of E. coli O157:H7 in spiked soils led to the evaluation of wetland and fecal samples in this assay. The standard curves from August and September from a single experimental plate are shown as an example (Fig. 6). Quantitative real-time PCR analysis of the manure, fecal, and wetland samples for the three genes with the standard curves revealed linearity between the CT values and the starting quantities of DNA representing 104 to 108 CFU g−1. Amplification efficiencies for stx1 and eae genes were slightly higher than that for the stx2 gene in all the standard curves. These standard curves were used for estimating the numbers of E. coli O157:H7 in cattle or calf feces, manure, the facultative pond, the raw pond, and wetland influents and effluents over 1 year. Table 5 shows the number of E. coli O157:H7 bacteria detected over several months in feces, manure, wastewater, and wetland influents and effluents. Temporal fluctuation was noticed in the levels of E. coli O157:H7 in these samples. Most samples collected during February and September did not contain detectable levels of the three genes compared to samples collected in April. Samples from April showed significantly higher levels (P < 0.05) of E. coli O157:H7 than did those from the other months since the presence of the three genes was used for confirmation of the pathogen. The numbers quantified in these samples were in agreement with the work of Zhao et al. (34) and Shere et al. (29), who reported that concentrations of E. coli O157:H7 in cattle feces ranged from 102 to 105 CFU g of feces−1.
FIG. 6.
Standard curves for the multiplex real-time PCR analysis of E. coli O157:H7 from calf and cow feces, manure, and wetlands wastewater. E. coli O157:H7 concentrations were determined for the samples collected in August and September. Similar curves were done for December, February, April, and July.
TABLE 5.
Concentrations of E. coli O157:H7 detected in environmental samples by real-time PCR
| Sitea | Result (CFU ml−1) for gene and month
|
||||||
|---|---|---|---|---|---|---|---|
|
stx1
|
stx2
|
||||||
| February | April | July | August | September | February | April | |
| Raw | 1.3 × 105 ± 3.2 × 104 | 4.9 × 104 ± 1.1 × 103 | 1.5 × 104 ± 5.1 × 103 | 1.1 × 104 ± 4.4 × 103 | 8.1 × 103 ± 2.2 × 103 | NDb | 2.2 × 104 ± 3.2 × 103 |
| Pond | 2.3 × 103 ± 1.5 × 102 | 7.4 × 104 ± 2.1 × 103 | 1.1 × 103 ± 5.6 × 102 | 1.7 × 104 ± 7.1 × 103 | 1.9 × 104 ± 6.2 × 103 | ND | 2.2 × 104 ± 2.2 × 104 |
| W1 Inf | 4.1 × 103 ± 3.2 × 103 | 1.1 × 105 ± 3.4 × 104 | 3.1 × 103 ± 4.1 × 103 | 3.3 × 104 ± 5.2 × 103 | 3.4 × 104 ± 2.2 × 103 | 1.7 × 104 ± 1.1 × 102 | 4.4 × 103 ± 3.1 × 103 |
| W1 Eff | 3.2 × 103 ± 3.0 × 103 | 5.3 × 103 ± 4.2 × 103 | 3.4 × 102 ± 3.0 × 103 | 8.3 × 103 ± 2.2 × 103 | 1.0 × 104 ± 1.2 × 103 | ND | ND |
| W2 Inf | ND | 8.0 × 104 ± 1.5 × 104 | 2.3 × 103 ± 8.2 × 102 | 4.1 × 104 ± 6.2 × 103 | 5.0 × 103 ± 6.2 × 103 | ND | 3.2 × 104 ± 3.3 × 104 |
| W2 Eff | ND | 7.6 × 103 ± 2.3 × 103 | 1.8 × 103 ± 4.2 × 102 | 1.2 × 104 ± 9.1 × 104 | 4.3 × 103 ± 6.1 × 102 | ND | ND |
| Calf feces | 1.2 × 104 ± 2.2 × 103 | 5.9 × 103 ± 5.2 × 103 | 4.8 × 103 ± 3.8 × 103 | 2.9 × 104 ± 2.2 × 103 | 6.0 × 103 ± 8.3 × 103 | ND | 4.9 × 103 ± 3.5 × 103 |
| Cow feces | ND | 2.7 × 105 ± 1.1 × 105 | 3.2 × 103 ± 2.2 × 103 | 1.6 × 104 ± 1.1 × 103 | 4.2 × 104 ± 6.3 × 103 | ND | 6.5 × 104 ± 3.6 × 104 |
| Manure | 4.5 × 103 ± 4.2 × 102 | NAc | ND | 2.7 × 104 ± 9.1 × 103 | 9.5 × 104 ± 5.2 × 103 | 1.2 × 102 ± 2.2 × 101 | NA |
All solid samples were from calf feces (Calf F), cow feces (Cow F), or manure, and the liquid waste samples were from the facultative pond (Pond), the raw pond (Raw), wetland 1 effluent (W1 Eff), wetland 1 influent (W1 Inf), wetland 2 effluent (W2 Eff), and wetland 2 influent (W2 Inf).
ND, not determined.
NA, samples not collected for analysis.
The concentrations of E. coli O157:H7 obtained by real-time PCR (Table 5) were very close to the numbers obtained by the traditional culture methods on CT-SMAC (Table 4). The developed PCR assay, however, has the advantage of higher throughput, higher reproducibility, and less time required to screen many samples. While PCR may sometimes detect dead cells and degraded DNA, the data from this study showed only a small proportion of this artifact to be of concern for absolute quantification of pathogens in the environment with ribosomal DNA as a template for amplification. Therefore, the ability to quantify E. coli O157:H7 in the environment without using culture methods will be very helpful for developing models of pathogen transport in the environment and, subsequently, for risk assessment. Currently, most models and transport studies demonstrating the risk of E. coli O157:H7 in the environment depend on culture techniques (9). Other studies have looked at the detection of E. coli O157:H7 in soil, water, and feces (5, 8, 13). In this study, a fluorescent signal was converted into target cell densities and related directly to cell densities in soil, manure, feces, and wash water. This approach was made possible by relating the target DNA to the CFU milliliter−1 of a cultured E. coli strain. This procedure is in contrast to other studies of E. coli O157:H7, where presumptive detection by TaqMan PCR was used to estimate the population size of E. coli O157:H7 (4, 21). The quantification strategy used here was successful due to prior knowledge of E. coli O157:H7 DNA copy numbers and genome size. This strategy has also recently been applied to the detection and quantification of methyl tert-butyl ether-degrading strain PMI by real-time PCR (12) and to total E. coli and Pseudomonas fluorescens (18).
The automated PCR amplification and detection of target gene amplicons described in this study are conducive for screening large numbers of environmental samples in a single assay. This method is a significant tool for monitoring large numbers of environmental samples contaminated with cattle feces or manure that are subsequently transported either by horizontal flow to larger bodies of water or by vertical movement to groundwater. This problem is critical in the Chino-Santa Ana River Basin, Calif., where the surface water and groundwater are major sources of drinking water and where the water quality is progressively deteriorating due to intensive dairy operation and disposal of untreated wastewater into the Chino Basin. The use of constructed wetlands in treating wastewater from farm operations may improve the quality of water that drains into the Santa Ana River. The multiplex PCR assay in combination with culture methods can be a useful method for water districts in the area to monitor contamination by this pathogen in the major rivers and groundwater.
Table 5b.
| Result (CFU ml−1) for gene and month
| |||||||
|---|---|---|---|---|---|---|---|
|
stx2
|
eae
|
||||||
| July | August | September | February | April | July | August | September |
| 5.2 × 103 ± 6.6 × 103 | ND | 1.6 × 103 ± 2.2 × 102 | 5.2 × 103 ± 9.3 × 103 | 3.3 × 104 ± 5.2 × 103 | 2.1 × 104 ± 2.1 × 103 | 1.2 × 104 ± 7.2 × 103 | 1.6 × 103 ± 8.3 × 102 |
| 1.4 × 104 ± 4.2 × 104 | 8.7 × 103 ± 7.1 × 103 | ND | 1.4 × 104 ± 4.2 × 104 | 6.6 × 104 ± 2.2 × 104 | 4.1 × 103 ± 9.2 × 103 | 4.1 × 103 ± 3.2 × 103 | 6.3 × 103 ± 8.1 × 103 |
| 1.9 × 105 ± 2.1 × 103 | 2.1 × 104 ± 3.1 × 103 | ND | 1.9 × 105 ± 3.3 × 105 | 1.4 × 104 ± 7.2 × 104 | 3.1 × 104 ± 6.2 × 103 | 1.2 × 105 ± 3.2 × 103 | 2.0 × 103 ± 2.2 × 103 |
| 3.4 × 103 ± 3.1 × 102 | ND | ND | 4.3 × 103 ± 3.6 × 103 | ND | 3.5 × 103 ± 3.4 × 103 | 1.8 × 103 ± 3.2 × 104 | 1.0 × 103 ± 2.5 × 102 |
| ND | ND | ND | 6.8 × 104 ± 6.2 × 104 | 4.6 × 104 ± 4.2 × 104 | 1.2 × 104 ± 3.8 × 103 | 2.1 × 104 ± 5.1 × 103 | 8.7 × 104 ± 9.1 × 103 |
| ND | ND | ND | ND | 8.6 × 103 ± 4.4 × 103 | 6.2 × 103 ± 1.1 × 103 | 6.9 × 104 ± 1.1 × 103 | 9.7 × 104 ± 8.2 × 102 |
| ND | ND | ND | ND | 9.8 × 104 ± 2.0 × 103 | 9.6 × 104 ± 8.2 × 103 | 6.7 × 104 ± 6.2 × 103 | ND |
| ND | 8.9 × 103 ± 2.4 × 102 | ND | ND | 2.7 × 104 ± 3.0 × 104 | 5.8 × 104 ± 4.1 × 103 | 1.2 × 104 ± 7.2 × 103 | |
| 1.1 × 103 ± 7.2 × 102 | ND | ND | 4.0 × 102 ± 2.3 × 102 | NA | ND | 4.2 × 104 ± 4.2 × 103 | 7.6 × 102 ± 2.1 × 102 |
Acknowledgments
We thank Katherine O'Connor, project director of the Chino dairy wetlands, and the members of the staff of the Orange County Water District who were instrumental in the construction of the wetlands, especially the Technical Advisory Committee.
This research was supported by the 206 Manure and By-Product Utilization Project of the USDA-ARS.
Mention of trademark or proprietary products in this work does not constitute a guarantee or warranty of the property by the USDA and does not imply its approval to the exclusion of other products that may also be suitable.
REFERENCES
- 1.Abe, A., and H. Nagano. 2000. Functional analysis of the type III secretion system in enteropathogenic Escherichia coli O157:H45. Microbiol. Immunol. 44:857-861. [DOI] [PubMed] [Google Scholar]
- 2.Agin, T. S., J. R. Cantey, E. C. Boedeker, and M. K. Wolf. 1996. Characterization of the eaeA gene from rabbit enteropathogenic Escherichia coli strain RDEC-1 and comparison to other eae genes from bacteria that cause attaching-effacing lesions. FEMS Microbiol. Lett. 144:249-258. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, H. A., S. J. Flood, K. J. Livak, J. Marmaro, R. Knorr, and C. A. Batt. 1995. Use of fluorogenic probes in a PCR-based assay for the detection of Listeria monocytogenes. Appl. Environ. Microbiol. 61:3724-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellin, T., M. Pulz, A. Matusset, H.-G. Hempen, and F. Gunzer. 2001. Rapid detection of enterohemorrhagic Escherichia coli by real-time PCR with fluorescent hybridization probes. J. Clin. Microbiol. 36:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, G. R., J. Prosser, A. Glover, and K. Killham. 2001. Detection of Escherichia coli O157:H7 in soil and water using multiplex PCR. J. Appl. Microbiol. 91:1004-1010. [DOI] [PubMed] [Google Scholar]
- 6.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laaegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franke, S., D. Harmsen, A. Caprioli, D. Pierard, L. H. Wieler, and H. Karch. 1995. Clonal relatedness of Shiga-like toxin-producing Escherichia coli O101 of human and porcine origin. J. Clin. Microbiol. 33:3174-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fratamico, P. M., L. K. Bagi, and T. Pepe. 2000. A multiplex polymerase chain reaction assay for rapid detection and identification of Escherichia coli O157:H7 in foods and bovine feces. J. Food Prot. 63:1032-1037. [DOI] [PubMed] [Google Scholar]
- 9.Gagliardi, J. V., and J. S. Karns. 2000. Leaching of Escherichia coli O157:H7 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 11.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hristova, K. R., C. M. Luteneggerand, and K. M. Scow. 2001. Detection and quantification of methyl tert-butyl ether-degrading strain PM1 by real-time TaqMan PCR. Appl. Environ. Microbiol. 67:5154-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, Y., Q. Zhang, and J. C. Meitzler. 1999. Rapid and sensitive detection of Escherichia coli O157:H7 in bovine feces by a multiplex PCR. J. Appl. Microbiol. 87:867-876. [DOI] [PubMed] [Google Scholar]
- 14.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenny, B. 1999. Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol. 31:1229-1241. [DOI] [PubMed] [Google Scholar]
- 16.Lie, Y. S., and C. J. Petropoulos. 1998. Advances in quantitative PCR technology: 5′ nuclease assays. Curr. Opin. Biotechnol. 9:43-48. [DOI] [PubMed] [Google Scholar]
- 17.Louie, M., J. C. S. De Azavedo, M. Y. C. Handelsman, C. G. Clark, B. Ally, M. Dytoc, P. Sherman, and J. Brunton. 1993. Expression and characterization of the eaeA gene product of Escherichia coli serotype O157:H7. Infect. Immun. 61:4085-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig, W., and K.-H. Schleifer. 2000. How quantitative is quantitative PCR with respect to cell counts. Syst. Appl. Microbiol. 23:556-562. [DOI] [PubMed] [Google Scholar]
- 19.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKee, M. L., A. R. Melton-Celsa, R. A. Moxley, D. H. Francis, and A. D. O'Brien. 1995. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect. Immun. 63:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberst, R. D., M. P. Hays, L. K. Bohra, R. K. Phebus, C. T. Yamashiro, C. Paszko-Kolva, S. J. A. Flood, J. M. Sargeant, and J. R. Gellespie. 1998. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with internal fluorogenic probe and the 5′ nuclease (TaqMan) assay. Appl. Environ. Microbiol. 64:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Brien, A. D., and R. K. Holmes. 1987. Shiga and the Shiga-like toxins. Microbiol. Rev. 51:206-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obrig, T. G. 1992. Pathogenesis of shiga toxin (verotoxin)-induced endothelial cell injury, p. 405-419. In B. S. Kaplan, R. S. Trompeter, and J. L. Moake (ed.), Hemolytic uremic syndrome and thrombocytopenic purpura. Marcel Dekker, Inc., New York, N.Y.
- 24.Orlando, C., P. Pinzani, and M. Pazzagli. 1998. Developments in quantitative PCR. Clin. Chem. Lab. Med. 36:255-269. [DOI] [PubMed] [Google Scholar]
- 25.Oswald, E. H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt, H., C. Kernbach, and H. Karch. 1996. Analysis of the EHEC hly operon and its location in the physical map of the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:907-914. [DOI] [PubMed] [Google Scholar]
- 27.Sharma, V. K., and S. A. Carlson. 2000. Simultaneous detection of Salmonella strains and Escherichia coli O157:H7 with fluorogenic PCR and single-enrichment-broth culture. Appl. Environ. Microbiol. 66:5472-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma, V. K., E. A. Dean-Nystrom, and T. A. Casey. 1999. Semi-automated fluorogenic PCR assays (TaqMan) for rapid detection of Escherichia coli O157:H7 and other Shiga toxigenic E. coli. Mol. Cell. Probes 13:291-302. [DOI] [PubMed] [Google Scholar]
- 29.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stults, J. R., O. Snoeyenbos-West, B. Methe, D. R. Lovley, and D. P. Chandler. 2001. Application of the 5′ fluorogenic exonuclease assay (TaqMan) for quantitative ribosomal DNA and rRNA analysis in sediments. Appl. Environ. Microbiol. 67:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor, T. B., E. S. Winn-Deen, E. Picozza, T. M. Woudenberg, and M. Albin. 1997. Optimization of the performance of the polymerase chain reaction in silicon-based microstructures. Nucleic Acids Res. 25:3164-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tebbe, C. C., and W. Vahjen. 1993. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tesh, V. L., and A. D. O'Brien. 1992. Adherence and colonization mechanisms of enteropathogenic and enterohaemorrhagic Escherichia coli. Microb. Pathog. 12:245-254. [DOI] [PubMed] [Google Scholar]
- 34.Zhao, T., M. P. Doyle, J. Shere, and L. Garber. 1995. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 61:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]