Abstract
A novel polyphosphate kinase (PPK) was retrieved from an uncultivated organism in activated sludge carrying out enhanced biological phosphorus removal (EBPR). Acetate-fed laboratory-scale sequencing batch reactors were used to maintain sludge with a high phosphorus content (approximately 11% of the biomass). PCR-based clone libraries of small subunit rRNA genes and fluorescent in situ hybridization (FISH) were used to verify that the sludge was enriched in Rhodocyclus-like β-Proteobacteria known to be associated with sludges carrying out EBPR. These organisms comprised approximately 80% of total bacteria in the sludge, as assessed by FISH. Degenerate PCR primers were designed to retrieve fragments of putative ppk genes from a pure culture of Rhodocyclus tenuis and from organisms in the sludge. Four novel ppk homologs were found in the sludge, and two of these (types I and II) shared a high degree of amino acid similarity with R. tenuis PPK (86 and 87% similarity, respectively). Dot blot analysis of total RNA extracted from sludge demonstrated that the Type I ppk mRNA was present, indicating that this gene is expressed during EBPR. Inverse PCR was used to obtain the full Type I sequence from sludge DNA, and a full-length PPK was cloned, overexpressed, and purified to near homogeneity. The purified PPK has a specific activity comparable to that of other PPKs, has a requirement for Mg2+, and does not appear to operate in reverse. PPK activity was found mainly in the particulate fraction of lysed sludge microorganisms.
Activated sludge processes modified to achieve enhanced biological phosphorus removal (EBPR) are widely used to reduce the concentration of Pi in wastewater to acceptably low levels. During EBPR, excess Pi is taken up and stored as polyphosphate (polyP) inside microbial cells. This process appears to be induced by cycling the activated sludge from anaerobic to aerobic conditions. In the initial anaerobic zone, influent wastewater and sludge are mixed, volatile fatty acids are taken up and stored as polyhydroxyalkanoates, and Pi is released. Sludge then enters an aerobic zone, where stored carbon is oxidized for growth while Pi is taken up and polymerized to form polyP. This polyP is subsequently broken down when the sludge is recycled back to the anaerobic zone.
EBPR microbial ecology is poorly understood. Existing mechanistic models for the biochemical transformations during EBPR (19) are based on data from enrichment cultures composed of diverse assemblages of organisms with widely varying metabolic capabilities. A definitive metabolic model has eluded researchers, mainly because no pure culture is available that can carry out all of the characteristic carbon and phosphorus transformations. Until recently, Acinetobacter spp. were thought to be primarily responsible for EBPR, since these organisms are readily isolated from EBPR sludge and can accumulate polyP under certain conditions. Acinetobacter spp. were later shown not to be present in sludge in significant numbers (38).
Recent efforts using rRNA-based techniques have identified the dominant polyP-accumulating organism (PAO) in acetate-fed, laboratory-scale, sequencing batch reactors (SBRs). This PAO belongs to the phylogenetically defined Rhodocyclus group in the β subclass of the Proteobacteria (7, 9, 12), though it cannot grow in pure culture under conditions suitable for characterized Rhodocyclus spp. (12). Recent findings suggest that these organisms are responsible for a significant amount of polyP accumulation in some full-scale plants (42).
The polyP metabolism of PAOs is of particular interest, since polyP accumulation is the direct mechanism by which Pi is removed from wastewater. PolyP is present in numerous bacterial and archaeal cells, in microbial Eucarya, and in plant and animal tissue. PolyP degradation was the focus of many studies of Acinetobacter spp. metabolism (for examples see references 36 and 37). PolyP synthesis in model bacteria (Escherichia coli, Pseudomonas aeruginosa, Neisseria meningitidis, Acinetobacter spp.) has been studied extensively (15). In most of these organisms, polyphosphate kinase (PPK) is thought to be the enzyme primarily responsible for polyP synthesis. This enzyme catalyzes the transfer of the terminal phosphate of ATP to a growing chain of polyP—a reaction that, with E. coli PPK, is reversible in vitro (16). Apparent PPK homologs are found in many pathogens (34) and in most microbial genomes analyzed to date (15). The ubiquity of PPK is underscored by its role in fundamental physiological processes in bacteria that include motility, virulence, biofilm formation, quorum sensing, and RNA processing (for a review see reference 15). The high degree of PPK sequence conservation among so many bacteria should enable the retrieval of ppk genes from uncultivated organisms by using molecular techniques.
PCR has been used to retrieve fragments of highly conserved metabolic genes from groups of microorganisms. Genes retrieved from uncultivated microbes include those encoding enzymes involved in nitrogen fixation (nifH) (41), DNA repair (radA) (26), ammonia oxidation (amoA) (24), carbon fixation (cbbL, cbbM) (11), denitrification (nirK, nirS) (8), and sulfate respiration (dsr) (20). Degenerate primers were useful for characterizing functional gene diversity and distribution in natural populations. Such analyses have provided information on the metabolic capabilities of microbial communities beyond that obtained by ribosomal DNA (rDNA) gene-based analyses alone.
Although the taxonomic identity of a dominant PAO has been confirmed by using rDNA and rRNA-based methods, nothing is known of its genetics or biochemistry beyond that derived from bulk measurements of carbon and phosphorus storage products. The objective of this study was to retrieve a functional PPK from the principal PAO in a laboratory-scale SBR and to demonstrate that the gene is transcribed during EBPR.
MATERIALS AND METHODS
SBR operation and cultivation of pure strains.
Two reactors, one 1-liter (HP1) and one 4-liter (HP2), were operated to produce two similar sludges. HP1 was originally inoculated with mixed liquor from the Southeast Water Pollution Control Plant, San Francisco, Calif., and had been performing EBPR under various operating conditions for about two years (A. Schuler and D. Jenkins, unpublished data). HP2 was inoculated with HP1 sludge. Operating conditions are described in Table 1. The SBRs operated on a 6-h cycle, with 30-min draw and fill, 120-min anaerobic phase, 180-min aerobic phase, and 30-min settling. The hydraulic residence time was 12 h, and mean cell residence time was 4 days. The pH was maintained at 7.0 to 7.3, and the temperature was 23 ± 3°C. SBR feed was a mineral salts medium (39 and A. Schuler and D. Jenkins, unpublished) with Pi added in various amounts to achieve desired ratios of chemical oxygen demand to Pi (COD:Pi) (Table 1). A fixed loading of 115 mg of COD per liter of mixed liquor per cycle was maintained by using a separate carbon feed containing 92 mg of acetate per liter of total feed and 15 mg of Casamino Acids per liter of total feed. Mineral salts feed was added after the draw phase, and carbon feed was added after 40 min of N2 (gas) stripping to remove any oxygen remaining from the previous aerobic cycle. To aid in floc development, blended glass fiber filters (diameter, 47 mm; Gelman type A/E) in suspension were added daily to each SBR at 20 mg per liter of mixed liquor. Further details on SBR operation will be documented elsewhere (A. Schuler and D. Jenkins, unpublished). Rhodocyclus tenuis (ATCC 25093) was grown anaerobically under light at room temperature with acetate and bicarbonate as carbon sources, as recommended by Trueper and Imhoff (32). E. coli strains DH10B and BL21(DE3), Acinetobacter sp. strain ADP1, and P. aeruginosa strain PAO were grown in Luria-Bertani (LB) medium at 37°C.
TABLE 1.
Reactor operating conditions and sludge characteristics
| Operating parametera | Measurement for sludge:
|
|
|---|---|---|
| HP1 | HP2 | |
| Influent PO4-P (mg-P liter−1) | 12.6 | 7.0 |
| Influent COD:Pi (mg mg-P−1) | 16 | 14 |
| TSS (mg liter−1) | 961 | 938 |
| VSS (mg liter−1) | 606 | 576 |
| Phosphorus content of biomass [(mg-P) (mg VSS)−1] | 0.16 | 0.18 |
| Anaerobic Pi releasedb (mol C-mol−1) | 0.67 | 0.66 |
| Anaerobic PHA synthesized (C-mol C-mol−1) | 1.17 | NDc |
| Pi in effluent (mg-P liter−1) | 0.10 | 0.12 |
TSS, total suspended solids; VSS, volatile suspended solids; PHA, polyhydroxyalkanoate. All phosphate measurements are reported as P.
Normalized to the amount of acetate taken up during the anaerobic phase. Values are reported in terms of carbon-moles (C-mol), with acetate containing 2 C-mol per mol.
ND, not determined.
Analytical techniques.
Total suspended solids, volatile suspended solids, soluble Pi, and total phosphorus were measured by using Standard Methods 2540B, 2540E, 4500-P C, and 4500-P B.5, respectively (5). Acetate was measured by using gas chromatography (Model 3800; Varian, Palo Alto, Calif.) equipped with a flame ionization detector and a 0.53-mm-diameter, 15-m-long DB-FFAP capillary column (J&W Scientific, Folsom, Calif.). Polyhydroxyalkanoates were measured according to a modified propanolysis and esterification method, followed by gas chromatographic detection (22).
Nucleic acid extraction.
Bulk genomic DNA for use in PCR was extracted from centrifuged sludge and pure cultures by using a phenol:chloroform bead-beating method, precipitated with sodium acetate and isopropanol and further purified by passage through a Chromaspin-1000 size selection column (Clontech, Palo Alto, Calif.) to remove fragments less than 1 kb in length, as previously described (10). Standard precautions to eliminate RNase contamination, including treatment of reagents and glassware with diethylpyrocarbonate, were used while working with RNA (25). Total RNA was extracted from sludge and E. coli by using a low-pH, hot-phenol, bead-beating protocol (28) and was precipitated with sodium acetate and ethanol (25). After suspension in water, total RNA was further purified by using a Qiagen RNeasy column (Qiagen, Valencia, Calif.), treated with RNase-free DNase I (Pharmacia, Piscataway, N.J.), and purified again on a fresh RNeasy column. Nucleic acid concentrations were estimated with spectrophotometry at 260 nm. RNA integrity and concentration were verified by agarose gel electrophoresis.
PCR amplification of SSU rRNA genes.
Community small subunit (SSU) rDNAs were amplified from DNA extracted from sludge HP1 by using bacteria-specific primers (Table 2). Amplification of community rDNAs was carried out with 10 to 100 ng of bulk DNA in reaction mixtures containing 1× PCR buffer II (Perkin Elmer, Foster City, Calif.), 2.5 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, 400 nM concentrations of each forward and reverse primer, and 0.025 U of AmpliTaq Gold (Perkin Elmer)/μl on an MJ Research DNA Engine thermal cycler. The PCR was conducted with an initial denaturation step at 94°C for 12 min, followed by 25 cycles of 94°C for 1 min, 50°C for 45 s, and 72°C for 2 min and then a final extension at 72°C for 12 min.
TABLE 2.
Oligonucleotides used for PCR and FISH
| Oligonucleotide (abbreviation) | Sequence (5′-3′) | Target organism and/or gene | Reference |
|---|---|---|---|
| S-D-Bact-0008-a-S-20 (8F) | AGAGTTTGATCCTGGCTCAG | Virtually all bacteria, SSU rRNA | 18 |
| S-D-Bact-0338-a-A-18 (Bact338) | GCTGCCTCCCGTAGGAGT | Virtually all bacteria, SSU rRNA | 4 |
| S-*-Univ-1492-a-A-19 (1492R) | GGYTACCTTGTTACGACTT | Virtually all organisms, SSU rRNA | 18 |
| S-*-PAO-0846-a-A-21 (PAO846) | GTTAGCTACGGCACTAAAAGG | PAO cluster, SSU rRNA | 9 |
| NLDE-0199F (NLDE-F)a | CGTATGAATTTTCTTGGTATTTATTGTACTAATCTngaygarttytb | Most ppk homologs | This study |
| TGNY-1435R (TGNY-R)a | GTCGAGCAGTTTTTGCATGAwarttnccngtb | Most ppk homologs | This study |
| IPCRppk-0289Ra | GCGTTTCGTCGCAGACCAGGCG | Type I ppk | This study |
| IPCRppk-1375F | CACGCCAAGATGCTGATGATCG | Type I ppk | This study |
| NppkI-F | GGTGGTCATATGAACACCGCCGCCACAc | Type I ppk | This study |
| NppkI-R | GGTGGTTGCTCTTCCGCATCCTGCAGAGTCCTTCAGCAGTTCGATCAGGTc | Type I ppk | This study |
Primers are numbered according to the full-length Type I ppk sequence, with the number referring to the location of the base at the 3′ end of the primer.
Primers NLDE-F and TGNY-R were designed with CODEHOP software (23). The consensus clamp is shown in capital letters, while the degenerate core is shown in lowercase letters.
Restriction sites used for subcloning (NdeI and SapI) are underlined.
PCR amplification of putative ppk gene fragments.
Degenerate oligonucleotides targeting genes encoding PPK homologs were designed by using web-based CODEHOP software (23) and an amino acid alignment of 12 PPKs identified by using tools on the National Center for Biotechnology Information (NCBI) website, including BLAST (Basic Local Alignment Search Tool) (3) and Entrez Genomes. The alignment is available upon request. These primers, named NLDE-F and TGNY-R (Table 2), should amplify an internal ppk fragment approximately 1,300 bp in length. Amplification was carried out on 100 to 1,000 ng of bulk DNA from sludge HP1 by using the same reaction conditions described above but with a touchdown PCR program consisting of an initial 12-min denaturing step at 94°C followed by 10 cycles of 94°C for 1 min, 53°C for 45 s (decreasing 0.5°C per cycle), and 72°C for 2 min. An additional 25 cycles were carried out with the same denaturing and extension conditions but with 45°C annealing for 45 s followed by a final 12-min extension at 72°C. An internal ppk fragment was amplified from R. tenuis DNA by using the Advantage-GC cDNA PCR kit (Clontech) with 1 M GC-melt and touchdown PCR as described above, except with 45 s of denaturing and 53°C annealing, touching down to 45°C annealing.
Library construction, clone screening, and DNA sequencing.
PCR products were cloned by using a TOPO TA Cloning kit with vector pCR2.1-TOPO or pCR4-TOPO (Invitrogen, Carlsbad, Calif.). Plasmids were prepared with a 96-well alkaline lysis procedure (14). Approximately 90 clones from each library were screened by using restriction fragment length polymorphism (RFLP) analysis to identify unique sequence types (10). The rDNA library was screened with both HinP1 I and MspI (New England Biolabs, Beverly, Mass.) in the same digest. ppk libraries were screened with either MspI or HaeIII (Boehringer Mannheim, Mannheim, Germany). Representatives of unique types were chosen for partial or complete sequencing with vector primers and internal primers when necessary. DNA sequencing was carried out with both strands either by using the ABI Prism BigDye terminator sequencing kit (PE Applied Biosystems, Foster City, Calif.) and an ABI 373 Stretch DNA sequencer or by the DNA Sequencing Facility on the University of California, Berkeley, campus. Sequences were edited and assembled with the ABI Autoassembler software package.
Inverse PCR and cloning of the full-length PPK.
Genomic DNA was extracted from SBR sludge as described above, digested with the restriction enzyme AscI, and purified by standard phenol-chloroform extraction and ethanol precipitation methods (25). The digested DNA was ligated, purified by phenol-chloroform extraction and ethanol precipitation, and used as template in inverse PCR (25). Primers iPCRppk-0289R and iPCRppk-1375F were designed to target the Type I ppk on the basis of sequences obtained in the library (Table 2). The product of the inverse PCR was gel extracted, cloned with the TOPO TA Cloning kit with vector pCRXL-TOPO (Invitrogen), and fully sequenced. Primers NppkI-F and NppkI-R were subsequently designed to target the full-length Type I ppk (Table 2). The primers contain restriction sites for digestion with NdeI and SapI so that the PCR product could be cloned into vector pTYB1 (New England Biolabs). Amplification was carried out on 100 to 1,000 ng of bulk DNA from sludge HP1 in reaction mixtures containing 1× Pfu Turbo Buffer with 2 mM MgCl2 (Stratagene, La Jolla, Calif.), 200 μM concentrations of each deoxynucleoside triphosphate, 400 nM concentrations of each forward and reverse primer, and 0.05 U of Pfu Turbo (Stratagene)/μl for high fidelity. The touchdown PCR program consisted of an initial 1-min denaturing step at 94°C followed by 10 cycles of 94°C for 30 s, 70°C for 45 s (decreasing 0.5°C per cycle), and 72°C for 3 min. An additional 20 cycles were carried out with the same denaturing and extension conditions but with 67°C annealing for 45 s followed by a final 12-min extension at 72°C. The 2.1-kb product was gel extracted and TOPO TA cloned as described above. Subcloning into pTYB1 was carried out by standard methods (25) to produce pKDM12.
Phylogenetic analyses.
Sequences were compared against available databases with the BLAST network service on the NCBI website (3). rDNA sequences were aligned and analyzed by using the ARB software package. ppk fragments were translated, and the amino acid sequences were aligned against 32 known and putative PPK sequences by using the Seqlab program in the GCG software package, version 10.0 (Genetics Computer Group, Madison, Wis.). Preliminary phylogenetic analyses were carried out with amino acid alignment, masked manually to exclude positions with gaps in more than 15% of sequences, by using the software package PHYLIP, version 3.6a (J. Felsenstein, Department of Genetics, University of Washington, Seattle). DNA sequences were then aligned according to the respective aligned protein sequences. The alignment was masked manually to exclude positions with gaps in more than 15% of sequences, and 1,137 aligned nucleotide positions were exported for analysis with the PAUP∗ software package, version 4.0b8. The alignment and mask are available upon request. Trees were constructed by using the following phylogenetic inference methods available in the software: maximum parsimony, minimum evolution under the HKY85 substitution model, and maximum likelihood. Heuristic searches were unrooted and performed with random, step-wise addition of taxa incorporating the tree bisection-reconnection branch-swapping algorithm.
FISH.
Hybridizations to rRNA were carried out with whole cells essentially as described previously (27, 38). Oligonucleotide probes (Table 2) were synthesized, 5′ end labeled with fluorescein or Cy3, and high-performance liquid chromatography purified by Operon Technologies. Hybridized samples were mounted in Prolong antifade reagent (Molecular Probes, Eugene, Oreg.). For quantitative fluorescent in situ hybridization (FISH), samples were visualized on a Nikon Labophot-2 epifluorescent microscope with filters G-2A and B-2A for Cy3 and fluorescein detection, respectively. Hybridized cells were enumerated following image capture with a cooled charge-coupled device camera (Princeton Instruments, Princeton, N.J.) and IPLab software (Scanalytics, Fairfax, Va.). At least 20 fields were examined, and at least 2,000 cells were counted in each hybridization experiment.
Dot blot hybridizations.
Dot blot hybridizations were conducted as previously described for rRNA (21), with the following modifications. Sense and antisense reference RNA was generated by using the MAXIscript in vitro transcription kit (Ambion, Austin, Tex.) with either T3 or T7 RNA polymerase, depending on the ppk fragment orientation in the construct used for probe synthesis, according to the manufacturer's instructions. These RNAs were purified on BioSpin 30 columns (Bio-Rad, Richmond, Calif.) and were carefully quantified spectrophotometrically at 260 nm. In preparation for blotting, total (either extracted or in vitro transcribed) RNA was denatured with 50% formamide, 7% formaldehyde, and 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0) for 15 min at 68°C, followed by snap cooling on ice and dilution with 1× SSC to reach a final RNA concentration of 12.5 μg ml−1 (sludge and E. coli RNA) or 38 pg ml−1 (reference RNAs). A Schleicher & Schuell Minifold I dot blot vacuum manifold was used to apply 400 μl of diluted RNA per spot to Nytran SuPerCharge nylon membranes (Dassel, Germany). E. coli total RNA was included as a negative control. Membranes were baked at 80°C for 2 h and were prehybridized and hybridized at 65°C in buffer containing 50% formamide, 5× SSPE (0.9 M NaCl, 50 mM sodium phosphate buffer [pH 7.2], 5 mM EDTA), 4.3× Denhardt's solution, 0.5% sodium dodecyl sulfate (SDS), and 50 mg of poly(A)/liter (25). Total RNA was hybridized with radiolabeled polyribonucleotide probes synthesized from ppk fragments in clones HP1ppk-02, HP1ppk-57, HP1ppk-24, and HP1ppk-48. Since pCR4-TOPO has T3 and T7 promoters flanking the multiple cloning site, either sense or antisense RNA could be transcribed from each clone. Radiolabeled antisense transcripts were used to detect sense mRNA in the sludge, and a sense transcript from HP1ppk-02 was used to verify that RNA preparations from sludge were free of genomic DNA. Positive and negative controls included appropriate antisense and sense transcripts derived from the same clones listed above. Probes were generated by in vitro transcription with [α-32P]CTP according to the manufacturer's instructions with the MAXIscript in vitro transcription kit (Ambion). After 12 to 16 h of hybridization, membranes were washed twice at 65°C for 1 h in 1× SSPE-0.5% SDS and once in 0.1× SSPE-0.5% SDS for 30 min. Membranes were exposed to PhosphorImaging cassettes, scanned with a Typhoon 8600 Variable Mode Imager, and processed with Image Quant Software (Molecular Dynamics, Sunnyvale, Calif.). Because probe preparations of identical specific activity could not be obtained, different PhosphorImager screen exposure lengths were used to equalize the positive control signals. Because the same amount of each reference RNA (15 pg per spot) was blotted, signal equalization allowed comparison between membranes hybridized with different probes. The experiment was replicated three times.
Protein expression and purification.
Type I recombinant PPK (rPPK) was overexpressed from vector pKDM12 in E. coli BL21(DE3) (Novagen, Madison, Wis.). Exponentially growing cells were used to inoculate 1 liter of LB with 50 mg of carbenicillin/liter and 1% glucose. The culture was grown at 37°C to an optical density at 600 nm of 0.8 and was shifted to 16°C, and 0.4 mM isopropylthiogalactoside was added. Induction was overnight (14 h) at 16°C to a final optical density at 600 nm of 3.9. Cells were harvested by centrifugation, and the cell pellet was frozen at −80°C overnight. The cells were thawed on ice and suspended in 40 ml of lysis buffer (50 mM Tris, 50 mM NaCl, 10 mM MgCl2, pH 8.0). DNase I was added to a concentration of 20 mg liter−1, and the cells were lysed by French press. Solid KCl was added to a final concentration of 1 M and Na2CO3 was added to 25 mM, and the protein mixture was gently swirled on ice for 30 min. The lysate was sonicated on ice for three sets of five 10-s pulses and was centrifuged (30,000 × g, 4°C, 1 h). The clarified lysate was filtered through a 0.45-μm mixed cellulose ester filter, diluted to 100 ml with lysis buffer, and loaded onto a chitin bead column (New England Biolabs) equilibrated with Buffer 1 (20 mM Tris, 1 M NaCl, 10 mM MgCl2, pH 8.0) at a rate of 0.5 ml min−1. Flow was by gravity, at 1 ml min−1 unless otherwise noted. The column was washed with Buffer 1 (20 column volumes) and then was flushed with Buffer 2 (20 mM Tris, 500 mM NaCl, 10 mM MgCl2, 50 mM dithiothreitol, pH 8.0) at 3 ml min−1. The 50 mM dithiothreitol promotes self-cleavage of the intein tag, leaving the chitin-binding domain on the column. The column was incubated in cleavage buffer at 4°C for 36 h, and the rPPK was eluted from the column in 1-ml fractions. The protein concentration was estimated in each fraction, and the seven most concentrated fractions were pooled and concentrated in Centricon YM-10 centrifugal filter units (Millipore, Bedford, Mass.) to a final volume of 2 ml. The concentrated protein was loaded onto a size exclusion column (HiPrep 26/60 Sephacryl S200; Amersham-Pharmacia) on an AKTA Explorer chromatography system (Amersham-Pharmacia) at a flow rate of 1.3 ml min−1 after the column was equilibrated with Buffer 3 (20 mM Tris, 500 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, pH 8.0). Column eluent absorbance at 210 nm was monitored, and 5-ml fractions were collected for 1 column volume. The four fractions with highest absorbance were pooled and concentrated in Centriplus YM-10 centrifugal filter units (Millipore). This final preparation (0.2 mg of protein ml−1) was used to characterize the Type I rPPK. Protein concentrations were measured by using the Bradford assay with the Protein Assay Kit (Bio-Rad) and a bovine serum albumin standard diluted in the appropriate buffer.
PPK assays.
Three PPK activity assays were used.
(i) Measurement of metachromasy.
Forward rPPK activity was assayed by determining polyP production over a 10-min time period, as described previously, including the ATP regeneration system (30, 31). Standard assay conditions were room temperature in 40 mM glycine-glycine-KOH, 10 mM KH2PO4, 10 mM MgCl2, 15% glycerol, 1 mM ATP, 6 mM phosphoenolpyruvate, 20 U of pyruvate kinase (Sigma, St. Louis, Mo.) ml−1, pH 7.2. ATP and MgCl2 concentrations, pH, and temperature were changed in kinetic studies. PolyP was quantitated by measuring the metachromatic shift in maximum absorbance from 630 to 530 nm in a solution of 25 μM Toluidine Blue by using sodium polyP standards (chain length, 75; Sigma).
(ii) Production of ATP from polyP.
Reverse PPK activity was assayed under essentially the same conditions as those described above but with 1 mM ADP and 6 mM pyruvate instead of ATP and phosphoenolpyruvate; also, a 30-min reaction time was used. Sodium polyP (chain length, 75) was included in the reaction mixture at 200 μM. The amount of reverse PPK activity was determined by measuring the final ATP concentration with luciferase as described previously (6).
(iii) Incorporation of [32P]phosphate.
Forward PPK activity in sludge lysate was measured by monitoring incorporation of [32P]phosphate into polyP. Sludge samples from the end of the anaerobic phase were centrifuged (5,000 × g, 10 min) and resuspended in lysis buffer (50 mM Tris-HCl, 10% sucrose, 1 mM EDTA, 300 mM NaCl, 5 mg of lysozyme ml−1, pH 7.4). Following 30 min of incubation on ice, the sludge was sonicated at 30% power for 20 pulses, 10 s each with a Virsonic 600 Sonifier (Virtis, Gardiner, N.Y.). DNase I and RNase A were each added at concentrations of 10 mg liter−1, and MgCl2 was added to 10 mM. Lysed sludge was centrifuged (15,000 × g, 5 min), the supernatant was reserved, and the pellet was resuspended in the same volume of lysis buffer. Supernatant and pellet fractions were assayed individually. Assay conditions were the same as those for measurement of metachromasy assay described above, except that [γ-32P]ATP (7,000 Ci mmol−1; ICN, Irvine, Calif.) was added to give a specific activity in the assay of 100 mCi mmol−1 and the reaction was carried out for 30 min. Samples were taken at 0, 5, 10, 15, 20, and 30 min for polyP measurement. Synthesized polyP was bound to Glassmilk in the presence of guanidine isothiocyanate and ethanol and was eluted in 50 mM Tris-HCl (pH 8.0) as described previously (6). PolyP production was determined by liquid scintillation counting. The activity was calculated by using the initial rate of polyP accumulation. A separate aliquot of the particulate fraction was solubilized in lysis buffer containing 6 M guanidinium hydrochloride for protein concentration determination.
For both forward assay methods, 1 U of activity was defined as the activity that produced 1 pmol of polyP (in monomers) per min. All assays were conducted in triplicate.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequences determined in this study are AF502188 to AF502244.
RESULTS
SBR operation.
The SBRs were operated to produce two high-polyP-content sludges (Table 1). Both SBRs were sampled at steady state while exhibiting characteristic EBPR carbon and phosphorus transformations (complete anaerobic acetate uptake with concomitant Pi release and polyhydroxyalkanoate synthesis, followed by aerobic Pi uptake) (Table 1).
Microbial community structure. (i) rDNA clone library.
Sludge HP1 microbial community structure was assessed by analyzing 92 clones from a PCR-based SSU rDNA library. RFLP analysis identified 22 RFLP types in the library. SSU rDNA sequences were assigned tentative phylogenetic affiliations on the basis of the closest cultured relative of the best match obtained by using BLAST. These affiliations were then confirmed with the ARB software package. These sequences and other Rhodocyclus-like PAO rDNA sequences available in GenBank were aligned against an existing data set and masked as described elsewhere (10, 13). A neighbor-joining algorithm was used to insert the sequences into an existing evolutionary distance tree (data not shown). Dominant phylotypes in the library included members of the Rhodocyclus and Comamonadaceae groups (also known as the β-1 and β-2 subgroups, respectively) in the β subclass of the Proteobacteria (20 and 8 clones, respectively), members of the Rhodobacter and Rhizobiaceae groups in the α subclass of the Proteobacteria (20 and 14 clones, respectively), and the Bacteroidetes (15 clones). Other sequences that did not obviously associate with taxonomically defined groups included 9 from the α-Proteobacteria, 4 from γ-Proteobacteria, and 1 each from Actinobacteria and Verrucomicrobiales. One of the phylotypes (representing 20 clones) shared 99% sequence identity with the Rhodocyclus-like SSU rRNA genes previously correlated with high rates of EBPR (7, 9, 12). These clones appear to be derived from the same type of organisms that Hesselmann and coworkers refer to as the R6 type, tentatively named “Candidatus Accumulibacter phosphatis” (12).
(ii) rRNA-targeted FISH.
To confirm the community structure inferred by the nonquantitative PCR-based clone library technique, previously developed oligonucleotide probes targeting the uncultivated sequence type from the SBR were used to detect the Rhodocyclus-like organism in sludge HP2. All of the sequenced Rhodocyclus-like clones were perfect matches to probe PAO846. Organisms binding the probe PAO846 dominated the community, comprising 80% ± 5% of cells binding Bact338 (data not shown).
Retrieval of partial ppk homologs from R. tenuis and sludge. (i) Development and characterization of ppk-specific PCR primers.
With primers designed to amplify an internal ppk fragment (NLDE-F and TGNY-R; Table 2), PCR conditions were optimized to produce a single product approximately 1,300 bp in length from either cloned ppks or genomic DNA from pure cultures. PCR products of the appropriate sizes were obtained from plasmid constructs containing full-length ppks cloned from E. coli (pSPK1) (35) and Acinetobacter sp. strain ADP1 (pPLT8) (31) and from genomic DNA extracted from E. coli strain DH10B, Acinetobacter sp. strain ADP1, and P. aeruginosa strain PAO. It was found that larger than normal amounts of genomic DNA were required to amplify the ppk fragments, and it was usually necessary to titrate the template concentration to optimize product yield.
(ii) Partial ppk homolog from R. tenuis.
A 1,311-bp fragment of a putative ppk homolog was cloned from DNA extracted from a pure culture of R. tenuis by using the new ppk-specific degenerate PCR primers. The translated fragment was 61% identical to a probable PPK from Ralstonia solanacearum (NP_519657) across 412 amino acids, the closest match obtained from the alignment of the partial protein sequence to entries in GenBank by the TBLASTN program. It was also 60% identical to the well-characterized N. meningitidis PPK (accession no. U16262) (29).
(iii) Partial ppk homolog from sludge.
The new ppk-specific PCR primers were used to amplify partial ppk gene fragments of approximately 1,300 bp in length from DNA extracted from sludge HP1, and these PCR products were used to construct a clone library. Six distinct RFLP patterns were identified when 88 clones were screened by RFLP analysis. Representatives of each pattern (usually three) were sequenced, resulting in a total of 17 sequenced clones. These novel ppks were further arbitrarily classified into four types (I to IV) by using pairwise comparisons of both nucleic acid and deduced amino acid sequences. Two genes were classified as the same type if they shared ≥95% identity at the nucleotide sequence level. The putative ppk sequences were translated and aligned against other ppks, and representative sequences of each type were chosen for DNA-based phylogenetic analyses: HP1ppk-02 (Type I), HP1ppk-57 (Type II), HP1ppk-24 (Type III), and HP1ppk-48 (Type IV). The amino acid alignment was used to perform phylogenetic analyses with the entire data set (17 sequences) to confirm this classification (data not shown). Types I and II shared significant similarity to each other and to the ppk fragment from R. tenuis, as described below. Types I and II shared 91.6% amino acid similarity with each other, and 86.0 and 87.4% similarity, respectively, with R. tenuis ppk. These two types comprised 69 and 11% of the clones in the library, respectively. Types III and IV represented 3 and 5% of the library, respectively.
Phylogenetic analyses of novel ppk homologs.
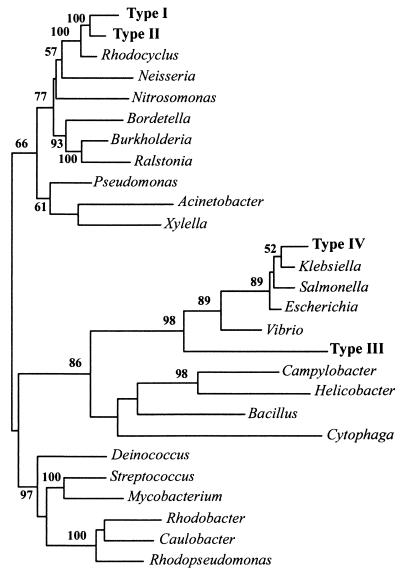
Comparative analysis of the four novel ppk sequence types, the R. tenuis ppk, nine confirmed ppk sequences from public databases, and 22 putative ppks identified by searching public databases and ongoing genome-sequencing projects further revealed the relatedness of these genes. An alignment consisting of 1,137 homologous nucleotides from 27 taxa was used to construct phylograms based on several different algorithms in the PAUP∗ software package. The phylogram in Fig. 1, based on nucleotide sequences, represents the best tree obtained by using maximum likelihood analysis with heuristic search.
FIG. 1.
Unrooted phylogram indicating inferred relatedness of ppk genes based on nucleotide sequence. Maximum likelihood analyses were carried out with PAUP∗ with 100 bootstrap resamplings. The bootstrap values for topologies supported in more than 50% of the trees are shown. Full organism names and the sources of DNA sequences are as follows, with the GenBank gene identification number or accession number given unless genome sequencing is unfinished, in which case the name of the sequencing facility is given. Acinetobacter sp. ADP1 (2462044), Bacillus halodurans C-125 (10173727), Bordetella pertussis (Sanger Center), Burkholderia pseudomallei (Sanger Center), Campylobacter coli (2239078), Caulobacter crescentus (13423119), Cytophaga hutchinsonii (Joint Genome Institute [JGI]), Deinococcus radiodurans (6459715), E. coli K12 (1788839), Helicobacter pylori J99 (4154939), Klebsiella aerogenes (391737), Mycobacterium tuberculosis (3261671), N. meningitidis (755166), Nitrosomonas europaea (JGI), P. aeruginosa PAO (2463578), Ralstonia metallidurans (JGI), Rhodobacter sphaeroides (JGI), R. tenuis (AF502199), Rhodopseudomonas palustris (JGI), Salmonella enterica serovar Typhimurium (3550416), Streptomyces coelicolor (7636006), Vibrio cholerae (3452464), and Xylella fastidiosa (9107809). Unfinished genome projects were searched with the TBLASTN program on the NCBI website with the Type I ppk amino acid sequence. Novel ppk homologs retrieved from reactor sludge are the following: Type I (AF502189), Type II (AF502196), Type III (AF502194), and Type IV (AF502195), all from this study.
Dot blot hybridizations.
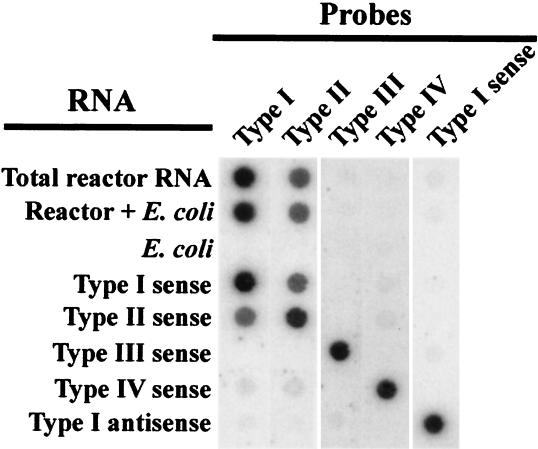
Dot blot hybridizations were carried out to detect ppk mRNAs in total RNA extracted from sludge 30 min after the addition of acetate in the anaerobic phase (Fig. 2). A strong signal was obtained with the Type I ppk antisense probe. A slightly fainter signal was observed with the Type II ppk antisense probe, and no significant signal resulted from hybridizations with Type III and Type IV antisense probes or with Type I sense probes. Hybridizations with probes for types III and IV indicate that these ppks were not transcribed in the sludge community at detectable levels. A lack of signal from E. coli RNA preparations with all probes demonstrated probe specificity. A 1:1 mixture of sludge and E. coli RNA was used to show that nontarget RNA did not interfere with detection of ppk mRNAs.
FIG. 2.
Dot blot hybridization of total RNA extracted from sludge HP2 during the anaerobic phase with five different probes. Single-stranded radiolabeled polyribonucleotide probes were designed to be either sense or antisense. Probes were antisense unless otherwise indicated. Positive and negative controls were in vitro-transcribed RNAs corresponding to each of the four types of ppk. All were sense strands unless otherwise indicated. E. coli total RNA served as a negative control to rule out nonspecific hybridization to rRNA. Total sludge RNA was mixed in equal proportions with E. coli total RNA to demonstrate reconstitution of the signal.
Inverse PCR and cloning of the full-length PPK.
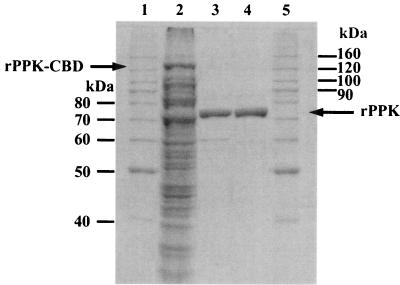
Inverse PCR was used to obtain the rest of the Type I gene sequence. As expected, the 8-kb amplified fragment contained the 5′ and 3′ flanking regions of the Type I fragment, and these were used to design PCR primers (NppkI-F and NppkI-R) to amplify the entire gene. When applied to HP2 sludge DNA, these primers produced a 2.1-kb product. Amplicons were TA cloned from three independent rounds of PCR carried out with Pfu polymerase, and they were subsequently sequenced. All three PCRs produced identical gene sequences that were 96% identical to the Type I ppk fragment obtained with degenerate primers NLDE-F and TGNY-R. One of these was subcloned into the pTYB1 vector for expression, creating pKDM12. This vector produced the Type I PPK with a C-terminal chitin-binding domain fusion to assist in purification by affinity chromatography with chitin beads.
Analysis of recombinant Type I PPK.
Type I rPPK was purified from a culture in which it was overexpressed from plasmid pKDM12 in E. coli. SDS-polyacrylamide gel electrophoresis was used to verify its purity following elution from a chitin affinity column and from a size exclusion column (Fig. 3). Following size exclusion, the specific activity was 33,700 U per μg of protein, and the preparation was greater than 99% pure (by densitometry of a Coomassie-stained gel). Based on size exclusion chromatography, the native rPPK appears to be a 75-kDa monomer (data not shown). The dependence of activity on ATP concentration, Mg2+, temperature, and pH were assessed by measuring polyP production (data not shown). The initial reaction rate was determined at ATP concentrations between 0 and 5 mM. Nonlinear regression analysis suggested a Vmax of 88,000 U per μg of protein and a Km of 1.2 mM. PPK activity depended strongly on Mg2+, with an optimum at 10 mM. Activity was measured over the pH range 5 to 9; optimal activity was at pH 7.4. The temperature optimum was 30°C. The rPPK had only minimal reverse activity under the assay conditions used, producing 267 ± 41 pmol of ATP per min per μg of protein.
FIG. 3.
SDS-polyacrylamide gel electrophoresis of protein purification. Lanes 1 and 5 contain molecular size markers. Lane 2 is clarified cell lysate (13 μg). The rPPK-chitin-binding domain (CBD) fusion protein, with an approximate predicted size of 130 kDa, is identified. Lane 3 is pooled fractions eluted from the chitin beads (0.8 μg). Lane 4 is pooled and concentrated fractions eluted from the size exclusion column (0.8 μg). Proteins were stained with Coomassie blue.
PPK activity in SBR sludge.
Forward PPK activity was assayed in HP2 sludge lysate, obtained at the beginning of the aerobic phase, by measuring the amount of [32P]phosphate incorporated into polyP. The lysate supernatant and pellet were assayed separately (see Materials and Methods). Supernatant PPK had an activity of 6.3 ± 0.7 U per μg of protein, and the cell debris pellet had an activity of 13.2 ± 0.5 U per μg of protein. The total activity in both the pellet and supernatant corresponded to 2,750 ± 120 U per mg of volatile suspended solids.
DISCUSSION
Elucidation of the link between EBPR microbiology and process performance requires assessment of both community structure and function. Although SSU rDNA-based community analysis is a useful way to track and enumerate a phylogenetically defined group of PAOs, it says nothing about the metabolism of these organisms. Because rDNA-defined taxonomic relatedness does not necessarily imply physiological similarity (1), the characterization and prediction of metabolic function requires the study of gene products directly involved in EBPR metabolism. PPK was used as a starting point because, in model organisms, it participates in the primary polyP synthesis pathway in bacteria (15).
The SBR enrichment cultures used in this study exhibited all known features of an acetate-fed EBPR community. At low influent COD:Pi, sludges aerobically accumulated large amounts of polyP and anaerobically took up acetate, stored polyhydroxyalkanoates, and released Pi (Table 1). Analysis of SSU rDNA clone libraries constructed from the sludges (verified by SSU rRNA-based FISH) indicated that the Rhodocyclus-like organism was present in high abundance. The discrepancy between the abundance of Rhodocyclus-like sequences in the clone library versus the cell abundance determined by FISH is not surprising given the nonquantitative nature of PCR-based techniques (40). Similar results have been noted by other researchers working with very similar sludges (9). Overall, these findings confirm previous reports (7, 9, 12) and provide evidence for the ubiquity of Rhodocyclus-like PAOs. Their predominance in SBRs on three continents (Europe, Australia, and North America) suggests that EBPR systems are exquisitely selective for these apparently highly specialized organisms.
To address the mechanism of polyP synthesis during EBPR, we investigated the diversity of putative ppk genes in a community highly enriched in PAOs. A library of ppk homologs was constructed by using PCR on genomic DNA extracted from sludge accumulating high amounts of polyP (HP1). Two sequence types (I and II) constituted the vast majority of clones in the library. Their phylogenetic relatedness to the ppk amplified from R. tenuis (Fig. 1) suggested that they were derived from the Rhodocyclus-like organism.
To determine whether Type I and/or Type II PPK is expressed during EBPR, ppk mRNA abundance in sludge was investigated. Semiquantitative dot blot hybridizations to extracted total RNA with a radiolabeled Type I ppk antisense probe resulted in a strong signal, indicating the presence of mRNA transcribed from this gene during the anaerobic phase (Fig. 3). The lack of signal from hybridization with the corresponding sense probe showed that genomic DNA was not present in significant enough quantities to bind either sense or antisense probes. The cross-hybridization of the Type I probe to the Type II positive controls and vice versa was not surprising given the high degree of identity shared between these two sequence types. The slightly fainter signal from hybridization of sludge RNA with Type II antisense probe could either be attributed to the presence of some Type II ppk mRNA or to cross-hybridization with Type I ppk mRNA. In either case, these results demonstrate that a ppk closely related to Type I is transcribed during EBPR.
The cloning of full-length Type I PPK from PAO-enriched sludge shows that established molecular techniques can be used for biochemical analysis of organisms that cannot be grown in isolation. The pKDM12 construct enabled production of large quantities of acceptably pure rPPK by chitin bead affinity chromatography. The rPPK produced polyP with a specific activity (33,700 U/μg of protein) comparable to that of purified Acinetobacter sp. ADP1 PPK (11,900 U/μg [31]) and E. coli PPK (29,000 U/μg [2]).
Demonstration of PPK activity in sludge would provide further evidence for expression of this enzyme during EBPR. Previous attempts to measure sludge PPK activity were unsuccessful (17, 33), but these experiments examined only the soluble fraction of lysate preparations. In some organisms, PPK is associated with the bacterial membrane and can be difficult to separate from particulate fractions of cell lysate (2, 30). Notably, N. meningitidis, the closest relative of the Rhodocyclus-like organism with a characterized PPK, requires extensive treatment with detergent to dissociate PPK from the cell membrane (30). We observed significant PPK activity in sludge HP2, but much of it was found in the insoluble cell debris. Initial attempts to disrupt the cells by routine sonication resulted in 20-fold less specific activity in the supernatant than in the pellet (data not shown). Additional experiments performed after excessive sonication resulted in improved cell lysis, and the insoluble pellet activity was only around twice as high as the supernatant activity. Therefore, it would appear that previous attempts to measure PPK activity in soluble sludge lysate fractions were unsuccessful because most activity was trapped in the discarded insoluble cell debris, as a result of insufficient cell lysis and dispersion.
These findings provide new evidence for the involvement of PPK in polyP synthesis during EBPR and constitute a new set of tools for the study of polyP metabolism in sludge. The primers can be used to retrieve ppk fragments from full-scale sludges and investigate the diversity of this gene in wastewater treatment plants with various configurations. The Type I ppk fragment can be used to probe the abundance of mRNA in samples taken from SBRs operating under different conditions. Polyclonal antibodies are presently being raised against the Type I rPPK and will be used in future studies to detect the enzyme in both laboratory-scale and full-scale EBPR systems. Ultimately, these reagents can be used to correlate the rate and extent of Pi removal with mRNA abundance and PPK enzyme levels, leading to an improved understanding of EBPR metabolism.
Acknowledgments
The authors wish to thank Vincent Martin and Jack Newman for helpful discussions and Heinz Kuo and Darren Cormack for technical assistance.
This research was supported in part by National Science Foundation Grant BES-9912472 to D.J. and J.D.K. and by an EPA STAR graduate fellowship to K.D.M.
Footnotes
This work is dedicated to the memory of our friend and colleague, Michael Anthony Dojka, Jr.
REFERENCES
- 1.Achenbach, L. A., and J. D. Coates. 2000. Disparity between bacterial phylogeny and physiology. ASM News 66:714-715. [Google Scholar]
- 2.Ahn, K. H., and A. Kornberg. 1990. Polyphosphate kinase from Escherichia coli - purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 265:11734-11739. [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association/American Water Works Association/Water Environment Federation, Washington, D.C.
- 6.Ault-Riche, D., C. D. Fraley, C.-M. Tzeng, and A. Kornberg. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bond, P. L., P. Hugenholtz, J. Keller, and L. L. Blackall. 1995. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl. Environ. Microbiol. 61:1910-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crocetti, G. R., P. Hugenholtz, P. L. Bond, A. Schuler, J. Keller, D. Jenkins, and L. L. Blackall. 2000. Identification of polyphosphate accumulating organisms and the design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 66:1175-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dojka, M. A., P. Hugenholtz, S. K. Haak, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon and chlorinated solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsaied, H., and T. Naganuma. 2001. Phylogenetic diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from deep-sea microorganisms. Appl. Environ. Microbiol. 67:1751-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hesselmann, R. P. X., C. Werlen, D. Hahn, J. R. van der Meer, and A. J. B. Zehnder. 1999. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 22:454-465. [DOI] [PubMed] [Google Scholar]
- 13.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 18:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 16.Kornberg, S. 1957. Adenosine triphosphate synthesis from polyphosphate by an enzyme from Escherichia coli. Biochim. Biophys. Acta 26:294-300. [DOI] [PubMed] [Google Scholar]
- 17.Kortstee, G. J. J., K. J. Appeldoorn, C. F. C. Bonting, E. W. J. van Niel, and H. W. van Veen. 2000. Recent developments in the biochemistry and ecology of enhanced biological phosphorus removal. Biochemistry (Moscow) 65:332-340. [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 19.Mino, T., M. C. M. Van Loosdrecht, and J. J. Heijnen. 1998. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32:3193-3207. [Google Scholar]
- 20.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittman, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riis, V., and W. Mai. 1988. Gas chromatographic determination of poly-β-hydroxybutyric acid in microbial biomass after hydrochloric acid propanolysis. J. Chromatogr. 445:285-298. [Google Scholar]
- 23.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sandler, S. J., P. Hugenholtz, C. Schleper, E. F. DeLong, N. R. Pace, and A. J. Clark. 1999. Diversity of radA genes from cultured and uncultured Archaea: comparative analysis of putative RadA proteins and their use as a phylogenetic marker. J. Bacteriol. 181:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K.-H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tinsley, C. R., and E. C. Gotschlich. 1995. Cloning and characterization of the meningococcal polyphosphate kinase gene: production of polyphosphate synthesis mutants. Infect. Immun. 63:1624-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tinsley, C. R., B. N. Manjula, and E. C. Gotschlich. 1993. Purification and characterization of polyphosphate kinase from Neisseria meningitidis. Infect. Immun. 61:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trelstad, P. L., P. Purdhani, W. Geissdorfer, W. Hillen, and J. D. Keasling. 1999. Polyphosphate kinase of Acinetobacter sp strain ADP1: purification and characterization of the enzyme and its role during changes in extracellular phosphate levels. Appl. Environ. Microbiol. 65:3780-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trueper, H. G., and J. F. Imhoff. 1992. The genera Rhodocyclus and Rubrivivax, p. 2556-2561. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed. Springer-Verlag, New York, N.Y.
- 33.T'Seyen, J., D. Malnou, J. C. Block, and G. Faup. 1985. Polyphosphate kinase activity during phosphate uptake by bacteria. Water Sci. Technol. 17:43-56. [Google Scholar]
- 34.Tzeng, C. M., and A. Kornberg. 1998. Polyphosphate kinase is highly conserved in many bacterial pathogens. Mol. Microbiol. 29:381-382. [DOI] [PubMed] [Google Scholar]
- 35.van Dien, S. J., S. Keyhani, C. Yang, and J. D. Keasling. 1997. Manipulation of independent synthesis and degradation of polyphosphate in Escherichia coli for investigation of phosphate secretion from the cell. Appl. Environ. Microbiol. 63:1689-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Groenestijn, J. W., M. M. Bentvelsen, M. H. Deinema, and A. J. Zehnder. 1989. Polyphosphate-degrading enzymes in Acinetobacter spp. and activated sludge. Appl. Environ. Microbiol. 55:219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Groenestijn, J. W., M. H. Deinema, and A. J. B. Zehnder. 1987. ATP production from polyphosphate in Acinetobacter strain 210A. Arch. Microbiol. 148:14-19. [Google Scholar]
- 38.Wagner, M., R. Erhart, W. Manz, R. Amann, H. Lemmer, D. Wedi, and K.-H. Schleifer. 1994. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl. Environ. Microbiol. 60:792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wentzel, M. C., R. E. Loewenthal, G. A. Ekama, and G. V. R. Marais. 1988. Enhanced polyphosphate organism cultures in activated sludge systems—part 1: enhanced culture development. Water SA 14:81-92. [Google Scholar]
- 40.Wintzingerode, F. V., U. B. Goebel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 41.Zehr, J. P., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zilles, J. L., J. Peccia, M.-W. Kim, C.-H. Hung, and D. R. Noguera. 2002. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl. Environ. Microbiol. 68:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]