Abstract
Inorganic polyphosphate (polyP) polymers are widely distributed in all kinds of organisms. Although the presence of polyP in members of the domain Archaea has been described, at present nothing is known about the enzymology of polyP metabolism or the genes involved in this domain. We have cloned, sequenced, and overexpressed an exopolyphosphatase (PPX) gene (ppx) from thermophilic Sulfolobus solfataricus. The gene codes for a functional PPX and possesses an open reading frame for 417 amino acids (calculated mass, 47.9 kDa). The purified recombinant PPX was highly active, degrading long-chain polyP (700 to 800 residues) in vitro at 50 to 60°C. The putative PPXs present in known archaeal genomes showed the highest similarity to yeast PPXs. In contrast, informatic analysis revealed that the deduced amino acid sequence of S. solfataricus PPX showed the highest similarity (25 to 45%) to sequences of members of the bacterial PPXs, possessing all of their conserved motifs. To our knowledge, this is the first report of an enzyme characterized to be involved in polyP metabolism in members of the Archaea.
Polyphosphate (polyP) is a linear polymer of hundreds of orthophosphate residues linked by high-energy phosphoanhydride bonds. These polymers are widely distributed in nature, having been found in every living organism, including those in the domain Archaea (14). polyP has a variety of physiological functions, such as serving as a reservoir of phosphate (Pi), substitution for ATP in kinase reactions, serving as a chelator of metals, and adaptation to nutrient starvation (4). While a direct role for polyP in the amino acid starvation response was recently established (18), other aspects of polyP, such as its likely prebiotic origin (11, 12, 36) and the enzymatic machinery related to its metabolism (16, 26), have remained elusive.
The best-known enzymes involved in the metabolism of polyP in bacteria are polyP kinase (PPK), which catalyzes the reversible conversion of the terminal phosphate of ATP into polyP, and exopolyphosphatase (PPX), which processively hydrolyzes the terminal residues of polyP to liberate Pi. These enzymes have been purified from Escherichia coli, and their genes have been cloned (1, 2). In Saccharomyces cerevisiae, the gene coding for a cytosolic PPX (ScPPX1) has also been described (35), and four genes involved in polyP accumulation have been identified (PHM1, PHM2, PHM3, and PHM4) (20). However, neither the ScPPX1 gene nor the PHM genes from S. cerevisiae seem to be homologous to the functionally equivalent E. coli ppx and ppk genes, probably because they belong to separate families. Very recently, an essential endopolyphosphatase gene was reported for S. cerevisiae (28).
Although the presence of polyP in the genus Sulfolobus (8, 14) and in other members of the Archaea (23, 25) has been confirmed, at present nothing is known about the enzymology of polyP metabolism or the genes involved in the domain Archaea. A gene homologous to ppk or PHM has not been described so far for the finished or unfinished archaeal genomes. The previously reported enzyme identified as a glycogen-bound PPK from Sulfolobus acidocaldarius (32) was further investigated and found to be a glycogen synthase (7).
The public availability of the Sulfolobus solfataricus genome before its release to the GenBank database allowed us to search for the putative genes involved in polyP metabolism. While no ppk or PHM genes were found, the existence of a ppx-like gene fragment prompted us to elucidate its functionality and to analyze the distribution of the ppx gene among several genomes. We found that the S. solfataricus genome contains an entire functional ppx gene, which shares a high level of homology with most bacterial ppx genes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. solfataricus DSM 1637 was grown at 70°C in medium 182 (Deutsche Sammlung von Mikroorganismen und Zellkulturen) with 0.1% yeast extract and 0.1% Casamino Acids. E. coli strains JM109, BL21(DE3)pLysS, NR100, and NR129 were cultivated in Luria-Bertani medium at 37°C with the appropriate antibiotics.
In vitro preparation of [33P]polyP750 as a substrate for PPX.
Radioactively labeled polyP with a chain length of 750 residues was prepared as previously described by Ault-Riché et al. (4), with the following modifications (7): the 10-ml reaction mixture contained 50 mM Tris-HCl (pH 7.4), 40 mM (NH4)2SO4, 4 mM MgCl2, 40 mM creatine phosphate, 20 μg of creatine kinase/ml, 1 mM [γ-33P]ATP (0.1 μCi/nmol), and 90,000 U of purified recombinant PPK from E. coli strain NR100 (17). After 3 h of incubation at 37°C, the mixture was cooled on ice for 5 min, and the reaction was stopped by the addition of 1 ml of 0.5 M EDTA.
The polyP reaction mixture was loaded over a cushion of 55 ml of 2.5 M CsCl-50 mM Tris-HCl (pH 7.4)-10 mM EDTA. After centrifugation at 30,000 rpm for 16 h at 4°C in a 647.5 rotor (Sorvall), aliquots of 5 ml were taken, and each one was added to 3.5 ml of isopropanol. After incubation at room temperature for 30 min and centrifugation at 11,000 rpm for 30 min in an Aj-20 rotor (Beckman), the supernatants were removed and the pellets were washed twice with 3.5 ml of 70% ethanol, dried overnight in a vacuum desiccator, and resuspended in 600 μl of distilled water. The identity and purity of [33P]polyP750 were determined by its susceptibility to hydrolysis by ScPPX1 as described previously (4, 7).
Preparation of cell extracts from S. solfataricus.
Forty milliliters of a culture grown to an optical density at 600 nm (OD600) of 0.4 was harvested by centrifugation (6,000 × g for 20 min). The pellet was resuspended in 200 μl of 50 mM Tris-acetate (pH 7) buffer and sonicated six times for 30 s each time. The lysate was centrifuged (5,000 × g for 5 min) to eliminate cellular debris, and the supernatant was used to measure PPX activity.
Assay for PPX activity.
PPX activity was determined as described by Akiyama et al. (2). The 50-μl reaction mixture contained 50 mM Tris-acetate (pH 7), 1 mM MnCl2, 175 mM KCl, and 50 to 250 μM [33P]polyP750. After incubation of the mixture for 15 to 30 min at various temperatures, the reaction was stopped by cooling the tubes on ice for 5 min and by adding 50 μl of 7% HClO4 and 5 μl of 20-mg/ml bovine serum albumin. The acid-precipitated remnant of [33P]polyP750 was collected on Whatman GF/C glass fiber filters and washed with 0.1 M pyrophosphate, 1 M HCl, and then ethanol. Quantitation was done by liquid scintillation counting. One unit of enzyme was defined as the amount releasing 1 pmol of phosphate from polyP per min.
TLC analysis of the reaction products of the PPX assay.
For thin-layer chromatography (TLC), samples of 2 μl were taken periodically from the reaction mixtures and loaded on polyethyleneimine-cellulose plates (Merck), which were then developed in 0.75 M KH2PO4. Radioactive spots were visualized by autoradiography.
Protein analysis.
Protein concentrations were determined by the method of Bradford (CoomassiePlus protein assay reagent; Pierce) (6a). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and staining with Coomassie blue were performed as described before (19).
DNA manipulations.
Restriction enzyme digestions and T4 DNA ligase reactions were performed according to the manufacturer's recommendations (Promega). Recombinant DNA techniques were carried out according to standard laboratory procedures (24). The dideoxy chain termination method was used to sequence DNA with [γ-33P]ATP and the dsDNA cycle sequencing system from GIBCO BRL. The DNA sequences were compiled and analyzed with the University of Wisconsin GCG package (version 9.1; Genetics Computer Group, Madison, Wis.).
Primers and PCR conditions.
The oligonucleotide primers were purchased from Genset Corporation. Taq polymerase and Pwo polymerase were obtained from Promega and Roche, respectively, and were used according to the manufacturers' recommendations. The oligonucleotide primer sequences were deduced from the available genome sequence of S. solfataricus (contig sh03g1150 (S. solfataricus genome site). The primers C5004NNdeI and C5003CXhoIH were designed from the 5′ end and the 3′ end of open reading frames (ORFs) c50_004 and c50_003, respectively, with recognition sites for the NdeI and XhoI restriction enzymes added. The sequences of these primers were as follows: C5004NNdeI, 5′-TTCATATGATATCGGCAGTTATAG-3′, and C5003CXhoIH, 5′-GCCTCGAGTACTCTTACACCGACAACGTACT-3′.
Sixty picomoles of each nucleotide and 25 ng of S. solfataricus total DNA were used in 50-μl reaction mixtures. PCR amplification conditions were 3 min at 95°C; 20 cycles at 95°C for 30 s, 54°C for 30 s, and 72°C for 2 min; and finally 3 min at 72°C. The DNA fragment (c50_004-3) was recovered from a 1% agarose gel and purified with Wizard PCR Prep (Promega).
DNA cloning and expression.
For DNA cloning and expression, we used the pGEM-T vector (Promega) and the pET system (Novagen). An A-tail was added to the purified S. solfataricus DNA fragment, and the fragment was ligated to the pGEM-T vector. The ligation products were used to transform E. coli strain JM109. The positive clones were analyzed by using colony PCR. Pure plasmids with inserts were obtained by using the Wizard Plus Miniprep DNA purification system (Promega). The DNA fragments and the pET-21b(+) vector were digested with NdeI and XhoI and ligated (Novagen). The ligation product [pET-21b(+)c50_004-3] and undigested pET-21b(+) were used to transform E. coli strain BL21(DE3)pLysS. The recombinant clones were selected on Luria-Bertani solid medium supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). The induction and expression analysis was done in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), added when the cultures reached an OD600 of 0.5. The expression of the recombinant protein (rPPX) was analyzed by SDS-PAGE of total cell extracts. One of the clones, SC21 [BL21(DE3)pLysS/pET-21b(+)c50_004-3], and control strain SC3 [BL21(DE3)pLysS/pET-21b(+)] were stored in 15% glycerol at −70°C until further analysis.
Western immunoblotting.
The total protein fractions corresponding to uninduced and induced SC3 and SC21 cells were separated by SDS-PAGE and electrotransferred to a polyvinylidene difluoride membrane as previously described (9). For the antigen-antibody reaction, the membrane containing the transferred proteins was treated with an anti-His tag monoclonal antibody (Novagen) (1:5,000 dilution) as the primary antibody and monoclonal anti-mouse antibodies conjugated with peroxidase (Amersham) as the secondary antibodies (1:2,000 dilution). A colorimetric method was used to develop Western blots as recommended by Promega.
Purification of rPPX.
rPPX was purified under denaturing conditions as follows. A 500-ml culture of E. coli strain SC21 was grown to an OD600 of 0.5 and induced with 1 mM IPTG. Cells were harvested by centrifugation, and the pellet was resuspended in 5 ml of His-nickel-nitrilotriacetic acid resin (Ni-NTA) binding buffer, containing 10 mM imidazole, 300 mM NaCl, and 50 mM NaH2PO4 (pH 8). Cell disruption was performed by adding lysozyme to a concentration of 250 μg/ml and by sonication (three times for 30 s each time). After centrifugation (20,000 × g for 30 min), the pellet (mainly a membrane fraction) was resuspended in 5 ml of Ni-NTA binding buffer containing 8 M urea and stirred for 1 h at room temperature. The sample was centrifuged (10,000 × g for 30 min) at room temperature, and the supernatant was collected. One milliliter of His-Ni-NTA binding resin (Novagen) was added to this fraction. After being stirred for 1 h at room temperature, the suspension was loaded onto a column and washed twice with 4 ml of Ni-NTA wash buffer, containing 20 mM imidazole, 300 mM NaCl, 50 mM NaH2PO4, and 8 M urea (pH 8). rPPX was eluted with 4 ml of Ni-NTA elution buffer, containing 250 mM imidazole, 300 mM NaCl, 50 mM NaH2PO4, and 8 M urea (pH 8). The collected fractions (0.5 ml) were analyzed by SDS-PAGE. Finally, the rPPX-containing fractions, which were essentially free from other proteins, were pooled and renatured by dialyzing the urea away in three sequential steps with 50 mM Tris-HCl, 100 mM KCl, and 15% glycerol (pH 7) containing 2 M, 500 mM, 100 mM, and no urea.
Sequence analysis.
Identity and similarity searching of the S. solfataricus genome site (http://niji.imb.nrc.ca; recently moved to http://www.-archbac.u-psud.fr/projects/Sulfolobus/sulfolobus.html) and of unfinished and finished genomes at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) was done by using the BlastP program (3). Multiple alignments were performed with ClustalX 1.81 (33). The alignments were improved by increasing the gap penalties to minimize gaps. In addition, the conservation of blocks of aligned residues was visualized by using BOXSHADE 3.21 (http://www.isrec.isb-sib.ch:8080/software/BOX_form.html). Prediction of secondary structures was done with PSIPRED (http://bioinf.cs.ucl.ac.uk/cgi-bin/psipred).
Nucleotide sequence accession number.
The nucleotide sequence of the ppx gene from S. solfataricus was deposited in the EMBL database under accession no. CAC39441. Almost simultaneously, the complete genome of S. solfataricus was published (29), and a putative ppx gene, 100% identical to our sequence, was released to the GenBank database under accession no NP_342652.
RESULTS
PPX activity in crude extracts from S. solfataricus.
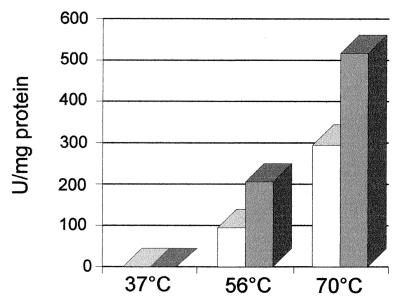
Contig sh03g1150 from the S. solfataricus unfinished genome contained two consecutive ORFs, c50_004 (133 amino acids) and c50_003 (279 amino acids), which were separated by a stop codon and a few nucleotides. The product of c50_004 was annotated as being similar to the N-terminal end of the PPX from E. coli (513 amino acids), and the product of c50_003 was described as a putative protein because of its unusual start codon, TTG. However, when we did a BlastP search, we found that the product of c50_003 was similar to the C-terminal end of the PPX from E. coli, suggesting the presence of a frameshift in the preliminary reported sequence. To find out whether a PPX was indeed present in S. solfataricus, we measured its activity in this archaeon. Crude extracts from a late-exponential-phase growing culture showed PPX activity (Fig. 1). Moreover, this activity was higher at 70°C, in agreement with the hyperthermophilic nature of S. solfataricus, and was twofold higher in the presence of Mn2+ than in the presence of Mg2+. The PPX activity detected strongly suggested that the frameshift previously reported was an error in the sequence of the genome, unless another gene was responsible for the PPX activity. To clarify this point, we decided to clone and sequence the entire region comprising ORFs c50_004 and c50_003 (c50_004-3) and to compare it with the equivalent region of the S. solfataricus genome.
FIG. 1.
PPX activity in crude extracts of S. solfataricus. Reaction mixtures (50 μl) containing cell extracts (50 μg of total protein), 50 μM [33P]polyP750, 50 mM Tris-acetate (pH 7), 175 mM KCl, and 1 mM MgCl2 (white bars) or 1 mM MnCl2 (gray bars) were incubated at the indicated temperatures for 30 min. PPX activity was determined by measuring polyP hydrolysis as described in Materials and Methods.
Cloning, sequencing, and identification of a putative ppx gene in the S. solfataricus genome.
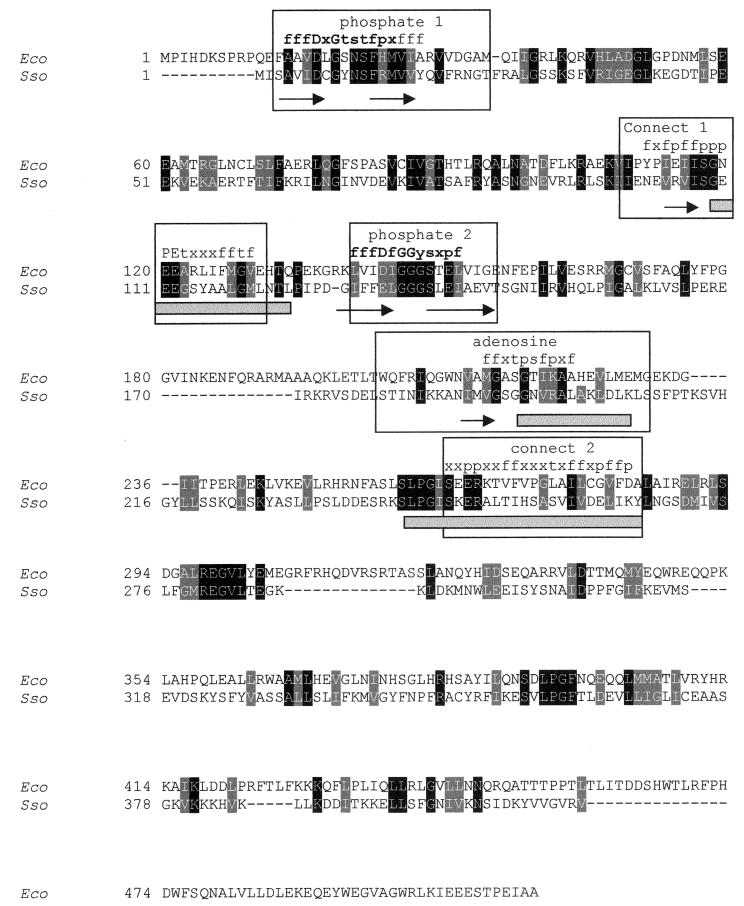
We amplified the fragment c50_004-3 from the genomic DNA of S. solfataricus, cloned it in the pGEM-T vector, and sequenced it. The fragment showed 100% identity with the corresponding sequence of the genome, except for the addition of one cytosine in the aforementioned frameshift, which resulted in its remotion to render a unique ORF (417 amino acids). The protein was 24% identical (44% similar) to the PPX from E. coli (513 amino acids). Two domains have been described for E. coli PPX based on its sensitivity to limited proteolysis (5), the N-terminal domain of approximately 300 amino acids and the C-terminal domain. The N-terminal domain contains the characteristic ATP binding motifs of the sugar kinase/actin/hsp70 superfamily: phosphate 1, connect 1, phosphate 2, adenosine, and connect 2 (6, 22). Conserved motifs have not been described for the C-terminal domain of E. coli PPX. However, equilibrium polyP binding experiments have suggested that this domain has a polyP binding site, although its exact position has not been established (5).
A pairwise alignment of the sequence of the product of the S. solfataricus c50_004-3 ORF with the amino acid sequence of E. coli PPX showed that the archaeal protein contained the described N-terminal and C-terminal domains, although the C-terminal domain was shorter than the E. coli counterpart and less conserved than the N-terminal domain (Fig. 2). The secondary structure prediction analysis revealed in both proteins the presence of the conserved β strands and α helices that cover the five motifs of the ATPase domain (6). A multiple-alignment analysis which included 45 homologous bacterial sequences (data not shown) revealed that the observed consensus sequences for the phosphate 1 and phosphate 2 motifs are in good agreement with those described for the superfamily (see consensus sequence in bold in Fig. 2). The conservation of the motifs connect 1, connect 2, and adenosine is less evident in this comparison (Fig. 2). However, these motifs have also been described for the PPX and the GppA proteins from E. coli (22).
FIG. 2.
Pairwise alignment of the amino acid sequences of E. coli PPX and the putative PPX from S. solfataricus. The deduced amino acid sequence from S. solfataricus (EMBL accession no. CAC39441) (Sso) was aligned with the E. coli PPX sequence (NCBI protein database accession no. NP_416997) (Eco). Identical residues (black shading) and similar residues (gray shading) are indicated. The ATPase conserved motifs (6) are boxed, and within these boxes β strands are indicated by arrows and α helices are indicated by gray rectangles. The consensus sequence motif among 45 aligned PPXs is indicated over the alignment in terms of the following amino acids groups: f, partly hydrophobic (VLIFWYMCGATKHR); t, tiny (GSAT); s, small (GSATNDVCP); p, tiny plus polar (GSATNDQEKHR); and x, any amino acid. The residues in bold correspond to the consensus sequences described previously (6).
From this set of analyses, we concluded that the DNA fragment present in contig sh03g1150 of the genome of S. solfataricus consists of a putative ppx gene.
Expression of the putative ppx gene from S. solfataricus and purification of archaeal rPPX.
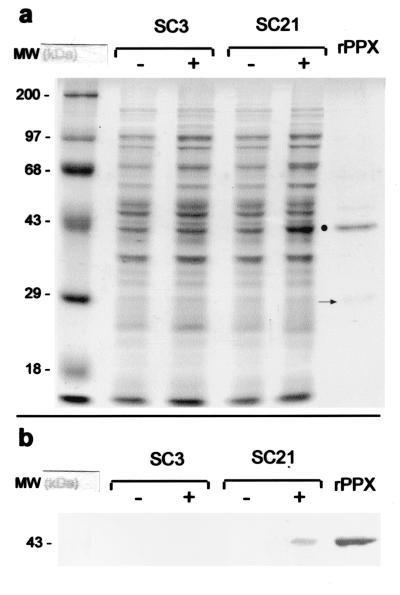
To gain insight into the functionality of the putative ppx gene from S. solfataricus, the fragment was subcloned into the expression vector pET-21b(+). The resulting plasmid, pET-21b(+)c50_004-3, contained the gene under the control of a T7 promoter, allowing the inducible overexpression of a recombinant protein (rPPX) with a C-terminal six-His-tagged sequence. This construct was used to transform E. coli strain BL21(DE3)pLysS to give the recombinant clone SC21. Crude extracts from uninduced and induced strain SC21 and control strain SC3 were analyzed by SDS-PAGE (Fig. 3). The overexpression of a protein of approximately 43 kDa was observed in the total protein fraction of induced SC21 (Fig. 3a). The molecular mass of the protein calculated from the amino acid sequence was 47.9 kDa. This slightly higher value, together with the rather poor levels of overexpression of rPPX observed, prompted us to investigate the presence of the entire recombinant protein by Western blotting with an anti-His tag monoclonal antibody (Fig. 3b). The His tag was detected only in the overexpressed protein and in purified rPPX (Fig. 3b), confirming the presence of the entire rPPX. Thus, the anomalous migration of this protein was assumed to be due to its association with membranes (data not shown), as has been observed for the PPK from E. coli (1).
FIG. 3.
Overexpression of rPPX from S. solfataricus in E. coli. (a) E. coli strain SC21 [BL21(DE3)/pLysS transformed with pET-21b(+) carrying the c50_004-3 insert] and E. coli strain SC3 [BL21(DE3)/pLysS transformed with pET-21b(+)] were grown for 2 h in the presence (+) or in the absence (−) of 1 mM IPTG. Membrane fractions were analyzed by SDS-PAGE and stained with Coomassie blue. The dot indicates the overexpressed rPPX protein. The arrow indicates the 30-kDa product possibly resulting from the degradation of rPPX (see the text). (b) Western immunoblot analysis of the fractions separated in panel a to detect the presence of His tag-containing polypeptides was done as described in Materials and Methods. MW, molecular weight (in thousands).
rPPX was purified under denaturing conditions by nickel affinity chromatography (Fig. 3). A barely detectable band of approximately 30 kDa was also present in the final fraction (Fig. 3a). Considering that the amount of this band diminished when 1 mM phenylmethylsulfonyl fluoride was present during purification, its molecular mass, and the absence of its detection by the anti-His tag antibody, we assumed that this protein corresponded to the N-terminal domain generated by proteolysis of rPPX. However, the C-terminal domain fragment was not observed either by Coomassie blue staining or by Western blotting with the anti-His tag antibody, as expected. This result was probably due to its complete degradation.
Functional analysis of PPX from S. solfataricus.
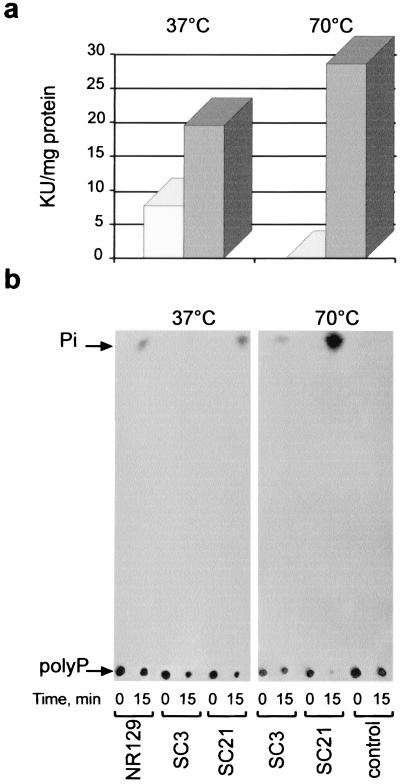
We measured PPX activity in crude extracts of induced SC3 and SC21 cells at different temperatures (Fig. 4). At 37°C, there was an increase in PPX activity in crude extracts obtained from induced SC21 cells compared with control cells (Fig. 4a). The PPX activity of rPPX was clearly evident at 70°C, which is very close to the optimal growth temperature of S. solfataricus; the PPX activity reached an increase of about 50% of its specific activity with the induced SC21 cell extracts, while the PPX activity of SC3 control cells was undetectable (Fig. 4a).
FIG. 4.
Functional analysis of rPPX. (a) The PPX activity present in cell extracts from E. coli strain BL21(DE3)/pLysS transformed with pET-21b(+) carrying the c50_004-3 insert (SC21 cells) (gray bars) or in SC3 control cells carrying only the vector (white bars) was measured at the indicated temperatures as the decrease in the radioactivity of polyP used as a substrate. Incubation was done for 15 min. KU, kilounits (103) of PPX activity. (b) TLC analysis of the reaction products obtained during the PPX assay. Samples (2 μl) from the reaction mixtures analyzed in panel a and a reaction mixture containing a cell extract from E. coli strain NR129 were taken at time zero and at 15 min of incubation and were applied to the origin of the plate. The arrows indicate the migration positions of [33P]polyP750 and H332PO4. Development of the TLC plates and autoradiography were done as described in Materials and Methods.
The release of Pi from polyP by rPPX over time was confirmed by using TLC analysis (Fig. 4b). For comparison, we used E. coli strain NR129, which overexpresses PPX from E. coli (Fig. 4b). To determine that the observed release of Pi at 70°C was not due to spontaneous hydrolysis of polyP, we ran in parallel a control tube without the protein fraction. The release of Pi from polyP was not observed (Fig. 4b). Taken together, these results demonstrate that the S. solfataricus cloned gene codes for a protein with PPX activity.
Effect of temperature and MnCl2 on the enzymatic activity of rPPX.
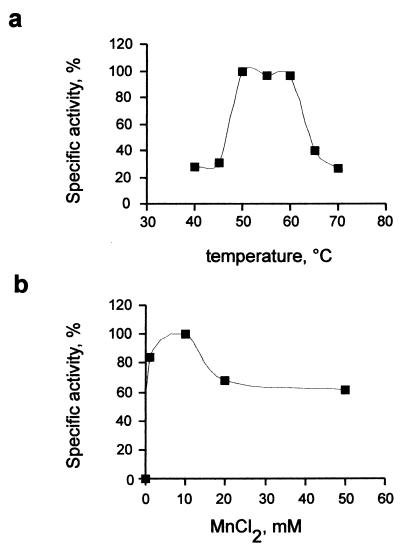
The activity of purified rPPX was assayed under different conditions. The optimal pH for the activity was found to be 7.0 (data not shown). Unexpectedly, the enzyme was optimally active at temperatures of between 50 and 60°C, while the specific activity was inhibited by 75% at 70°C (Fig. 5a). Enzymatic activity was influenced by the concentration of MnCl2. The activity increased up to 10 mM MnCl2 (Fig. 5b), a value much higher than the optimal concentration (1 mM) of bivalent cations for E. coli PPX (2). The highest specific activity that we obtained was 600,000 U/mg of protein, which is 10 times lower than the maximal specific activity reported for E. coli PPX (2). This difference is probably due to intrinsic properties of the archaeal PPX or a result of the perturbation of the native enzyme structure by the poly-His tag, as has been reported for recombinant PPK from E. coli (17).
FIG. 5.
Effects of temperature and MnCl2 on the activity of S. solfataricus rPPX. Reaction mixtures (50 μl) containing 350 ng of purified protein, 250 μM [33P]polyP750, 50 mM Tris-acetate (pH 7), 100 mM KCl, and 10 mM MnCl2 (a) or the indicated MnCl2 concentrations (b) were incubated at the indicated temperatures for 10 min (a) or for 10 min at 50°C (b). PPX activity was determined by measuring polyP hydrolysis as described in Materials and Methods.
Genomic analysis.
Prior to the release of the S. solfataricus genome, the only representatives of putative PPXs in Archaea were those genes of Archaeoglobus fulgidus and Methanococcus jannaschii. These ORFs were, respectively, 22 and 23% identical to ScPPX1. On the other hand, the predicted ScPPX1 protein did not show significant homology to any existing bacterial PPX (35). Unexpectedly, we found a ppx gene in the genome of S. solfataricus that was similar to most bacterial ppx genes. This finding prompted us to search for putative PPXs in the available finished and unfinished microbial genomes at the NCBI website. Given that an event of duplication of the ppx gene in E. coli has been described (10), we selected only completed genomes in order to search for the conservation of this duplication event. We also included in the analysis the eucaryal genomes available from the NCBI database.
A summary of the occurrence of ppx genes in all microbial genomes available is presented in Table 1. Among all the archaeal genomes, the ppx gene of S. solfataricus and that of its close relative Sulfolobus tokodaii turned out to be unique among 12 completed genomes in having a higher identity with most bacterial ppx genes. Both the A. fulgidus and the M. jannaschii ppx genes possessed higher identity with ScPPX1, while neither the crenarchaeote Aeropyrum pernix nor the euryarchaeotes Halobacterium sp. and Methanothermobacter thermoautotrophicus showed the presence of any ppx gene.
TABLE 1.
Distribution of PPX proteins encoded by 67 completed genomes
| Taxonomy | Organisma | Accession no. for PPX proteinb |
|---|---|---|
| Archaea (Crenarchaeota) | Aeropyrum pernix | |
| Archaea (Crenarchaeota) | Pyrobaculum aerophilum | |
| Archaea (Crenarchaeota) | Sulfolobus solfataricus | NP_342652 (417) |
| Archaea (Crenarchaeota) | Sulfolobus tokodaii | NP_377507 (417) |
| Archaea (Euryarchaeota) | Archaeoglobus fulgidus | NP_069590 (322) |
| Archaea (Euryarchaeota) | Halobacterium sp. strain NRC-1 | |
| Archaea (Euryarchaeota) | Methanococcus jannaschii | NP_247590 (307) |
| Archaea (Euryarchaeota) | Methanothermobacter thermoautotrophicus | |
| Archaea (Euryarchaeota) | Pyrococcus abysii | |
| Archaea (Euryarchaeota) | Pyrococcus horikoshii | |
| Archaea (Euryarchaeota) | Thermoplasma acidophilum | |
| Archaea (Euryarchaeota) | Thermoplasma volcanium | |
| Aquificales | Aquifex aeolicus | NP_213602.1 (312) |
| Cyanobacteria | Nostoc sp. strain PCC 7120 | NP_487592 (550) |
| Cyanobacteria | Synechocystis sp. strain PCC 6803 | NP_442969 (540) |
| Planctomyces or Chlamydia | Chlamydia trachomatis | |
| Firmicutes (low G+C) | Staphylococcus aureus | NP_372443 (309)c |
| Firmicutes (low G+C) | Bacillus subtilis | NP_391935 (309)d |
| Firmicutes (low G+C) | Bacilus halodurans | NP_242259 |
| Firmicutes (low G+C) | Clostridium acetobutylicum | NP_347258 (499), NP_348756 (310) |
| Firmicutes (low G+C) | Clostridium perfringens | NP_562578 (502) |
| Firmicutes (low G+C) | Lactococcus lactis | NP_267969 (314)d |
| Firmicutes (low G+C) | Listeria innocua | NP_470822 (308)c |
| Firmicutes (low G+C) | Listeria monocytogenes | NP_464973 (308)c |
| Firmicutes (low G+C) | Mycoplasma genitalium | |
| Firmicutes (low G+C) | Mycoplasma pneumoniae | |
| Firmicutes (low G+C) | Mycoplasma pulmonis | |
| Firmicutes (low G+C) | Streptococcus pneumoniae | NP_358982 (311)d |
| Firmicutes (low G+C) | Streptococcus pyogenes | NP_268701 (311)d |
| Firmicutes (low G+C) | Ureaplasma urealyticum | |
| Firmicutes (high G+C) | Mycobacterium leprae | NP_302578 (339), NP_301313 (317) |
| Firmicutes (high G+C) | Mycobacterium tuberculosis | NP_335491 (319), NP_334925 (344) |
| Spirochaetales | Borrelia burgdorferi | |
| Spirochaetales | Treponema pallidum | |
| Thermotogales | Thermotoga maritima | NP_228010 (288)d |
| Thermus or Deinococcus | Deinococcus radiodurans | NP_285509 (515), NP_296296 (314) |
| α Proteobacteria | Agrobacterium tumefaciens | NP_531842 (507), NP_531319 (341) |
| α Proteobacteria | Brucella melitensis | NP_541576 (473), NP_540121 (512) |
| α Proteobacteria | Caulobacter crescentus | NP_420516 (499), NP_420425 (391) |
| α Proteobacteria | Mesorhizobium loti | NP_108461 (372), NP_108317 (511) |
| α Proteobacteria | Rickettsia conorii | NP_36029 (473) |
| α Proteobacteria | Rickettsia prowazekii | NP_220678 (470) |
| α Proteobacteria | Sinorhizobium meliloti | NP_384861 (446), NP_385294 (507) |
| β Proteobacteria | Neisseria meningitidis | NP_274476 (502) |
| β Proteobacteria | Ralstonia solanacearum | NP_519658 (527) |
| ɛ Proteobacteria | Campylobacter jejuni | NP_281544 (486) |
| ɛ Proteobacteria | Helicobacter pylori 26695 | NP_207076 (484) |
| γ Proteobacteria | Escherichia coli K-12 | NP_416997 (513), NP_418226 (494) |
| γ Proteobacteria | Haemophilus influenzae | NP_438855 (323) |
| γ Proteobacteria | Pasteurella multocida | NP_246746 (516) |
| γ Proteobacteria | Pseudomonas aeruginosa | NP_253928 (506) |
| γ Proteobacteria | Salmonella enterica | NP_457037 (513), NP_457833 (493) |
| γ Proteobacteria | Salmonella enterica serovar Typhimurium | NP_461437 (513), NP_462804 (493) |
| γ Proteobacteria | Vibrio cholerae | NP_230371 (523), NP_229959 (497), NP_231323 (301) |
| γ Proteobacteria | Xylella fastidiosa | NP_299867 (508) |
| γ Proteobacteria | Yersinia pestis | NP_406345 (519) |
| Eucarya (Arthropoda) | Drosophila melanogaster | AAF45743 (405) |
| Eucarya (Ascomycota) | Saccharomyces cerevisiae | NP_0111262 (588), NP_012071 (397) |
| Eucarya (Ascomycota) | Schizosaccharomyces pombe | NP_594390 (384) |
| Eucarya (Nematoda) | Caenorhabditis elegans | |
| Eucarya (Microsporidia) | Encephalitozoon cuniculi | |
| Eucarya (Chordata) | Homo sapiens | |
| Eucarya (Chordata) | Mus musculus | |
| Eucarya (Chordata) | Rattus norvegicus | |
| Eucarya (Chordata) | Danio rerio (zebra fish) | |
| Eucarya (Chordata) | Fugu rubripes (torafugu fish) | |
| Eucarya (Embryophyta) | Arabidopsis thaliana | AAC24085 (568) |
Genomes are available at NCBI Genome Entrez (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = Genome). One constraint imposed was that the alignment must stretch over approximately the entire sequence length retrieved.
Most of the PPX proteins listed are putative. Accession numbers are from the NCBI database. The reciprocal best-hit relationship between each protein and the PPX families was confirmed for all sequences. The total lengths of the amino acid sequences are given in parentheses.
More divergent sequences were obtained from a BlastP search against the A. fulgidus PPX sequence for an amino acid expect (e) value of ≤10−50.
More divergent sequences were obtained from COG 0248 and COG1227 of the clusters of orthologous groups of proteins (COG) database at NCBI.
The ppx gene shown to be widespread in bacterial genomes and several other microorganisms was present in two copies (Table 1). Surprisingly, the low-G+C gram-positive group showed higher homology with ScPPX, with the exceptions of Bacillus halodurans and Clostridium perfringens. No putative ppx genes were present in the obligate parasites of the genus Mycoplasma, a phenomenon that was also observed in the order Spirochaetales.
DISCUSSION
In this study, we demonstrated the presence of a functional PPX gene in the archaeon S. solfataricus, characterized the gene product, and analyzed the distribution of the gene across the available microbial genomes. S. solfataricus PPX proved to be active as a hydrolase of long-chain polyP, like those synthesized in vitro by E. coli PPK. Although crude extracts of S. solfataricus showed the highest PPX activity at 70°C, this was not the case for pure rPPX, which was more active at 50 to 60°C. This result suggests that intracellular factors may contribute to its thermostability (34). Also, the requirement of a higher concentration of Mn2+ for this enzyme than for the mesophilic homologues could be related to the better temperature-stabilizing effect of Mn2+ than of Mg2+ for the same thermophilic enzyme (34).
It is likely that polyP is the only in vivo substrate for S. solfataricus PPX. E. coli GppA, a phosphohydrolase acting on guanosine pentaphosphate (pppGpp), has been shown to be active as a PPX in vitro (10). While S. solfataricus PPX could act in vitro as a GppA, we believe that its physiological function is PPX activity, given that the guanosine polyPs (pppGpp and ppGpp) do not seem to exist in Archaea (27) and the genes involved in their synthesis (relA and spoT) have not been found in archaeal genomes.
The ppx gene was shown to be widespread in known microbial genomes. One possible explanation for this type of occurrence is the involvement of polyP in the regulation of both enzyme activities and the expression of large groups of genes (14). These functions are the basis of survival for different microorganisms, including bacterial pathogens, during the stationary growth phase (14). Furthermore, Deinococcus radiodurans, Clostridium acetobutylicum, and Vibrio cholerae seem to be relicts of the coexistence of two putative ppx genes. If we take into account that polyP confers improved fitness under environmental stress (21), then the selective maintenance of ppx genes may be strongly associated with changing environments, while an immovable environment, such as a cell host, could cause gene loss, as suggested by the lack of ppx genes in Mycoplasma spp. (Table 1).
On the other hand, M. jannaschii and Bacillus subtilis have ppx genes that are homologous to ScPPX and whose products have experimentally confirmed pyrophosphatase activity (15, 30, 31). This alternative function for PPX could cause evolutionary constraints related to the conservation of the pyrophosphatase function. Nevertheless, the pyrophosphatase activity does not seem to be the more conserved function in this group of proteins, since ScPPX1 has been proven to lack this activity (35). Moreover, the widely spread family I of pyrophosphatases seems to provide this essential function across the genomes (31).
Taking into account that, prior to the release of the S. solfataricus genome, the only representatives of putative ppx genes in Archaea were highly homologous to ScPPX1, it was surprising for us to find in the genome of S. solfataricus an active gene coding for a PPX with a higher percent identity with the PPX from E. coli. The presence of this kind of ppx gene in the archaeal genomes of S. solfataricus and S. tokadaii might further facilitate the distribution of bacterial ppx genes across microbial genomes (13). Obviously, further research is needed to elucidate the possible evolutionary history of this gene as part of the disparity of polyP metabolic pathways. This aspect, together with the study of the antiquity of polyP, may in the future shed light on early cell evolution.
ADDENDUM IN PROOF
The complete genomic sequences of Methanosarcina mazei strain Goel (U. Deppenmeier et. al., J. Mol. Microbiol. Biotechnol. 4:453-461, 2002) and Methanosarcina acetivorans C2A (J. E. Galagan et al., Genome Res. 12:532-542, 2002) were recently published. Both of these archaeal genomes carry putative genes highly similar to bacterial exopolyphosphatases (ppx) and polyphosphate kinases (ppk).
Acknowledgments
This research was supported by FONDECYT projects 2990035 and 1000679, by Millennium Scientific Initiative project ICM P-99-031-F, and by ICGEB (project CRP/CHI00-04, contract 01/001). S.T.C. was the recipient of a DAAD Ph.D. scholarship.
We are very grateful to Arthur Kornberg for kindly providing us with E. coli strain NR100 in order to obtain [33P]polyP750 and NR129.
REFERENCES
- 1.Akiyama, M., E. Crooke, and A. Kornberg. 1992. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J. Biol. Chem. 267:22556-22561. [PubMed] [Google Scholar]
- 2.Akiyama, M., E. Crooke, and A. Kornberg. 1993. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 268:633-639. [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. L. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ault-Riché, D., C. D. Fraley, C.-M. Tzeng, and A. Kornberg. 1998. A novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolesch, D. G., and J. D. Keasling. 2000. Polyphosphate binding and chain length recognition of Escherichia coli exopolyphosphatase. J. Biol. Chem. 275:33814-33819. [DOI] [PubMed] [Google Scholar]
- 6.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye bindings. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Cardona, S., F. Remonsellez, N. Guiliani, and C. A. Jerez. 2001. The glycogen-bound polyphosphate kinase from Sulfolobus acidocaldarius is actually a glycogen synthase. Appl. Environ. Microbiol. 67:4773-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardona, S., F. Remonsellez, N. Guiliani, and C. A. Jerez. 2001. Polyphosphate metabolism in the archaeon Sulfolobus acidocaldarius, p. 345-354. In V. S. T. Ciminelli and O. Garcia, Jr. (ed.), Biohydrometallurgy: fundamentals, technology and sustainable development. Elsevier Science B. V., Amsterdam, The Netherlands.
- 9.Guiliani, N., and C. A. Jerez. 2000. Molecular cloning, sequencing, and expression of omp-40, the gene coding for the major outer membrane protein from the acidophilic bacterium Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 60:2318-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keasling, J. D., L. Bertsch, and A. Kornberg. 1993. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl. Acad. Sci. USA 90:7029-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keefe, A. D., and S. L. Miller. 1995. Are polyphosphates or phosphate esters prebiotic reagents? J. Mol. Evol. 41:693-702. [DOI] [PubMed] [Google Scholar]
- 12.Keefe, A. D., and S. L. Miller. 1996. Potentially prebiotic synthesis of condensed phosphates. Origins Life Evol. Biosph. 26:15-25. [DOI] [PubMed] [Google Scholar]
- 13.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornberg, A., N. N. Rao, and D. Ault-Riché. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn, N. J., A. Wadeson, S. Ward, and T. W. Young. 2000. Methanococcus jannaschii ORF mj0608 codes for a class C inorganic pyrophosphatase protected by Co(2+) or Mn(2+) ions against fluoride inhibition. Arch. Biochem. Biophys. 379:292-298. [DOI] [PubMed] [Google Scholar]
- 16.Kulaev, I. S., T. V. Kulakovskaya, N. A. Andreeva, and L. P. Lichko. 1999. Metabolism and function of polyphosphates in bacteria and yeast. Prog. Mol. Subcell. Biol. 23:27-43. [DOI] [PubMed] [Google Scholar]
- 17.Kumble, K. D., K. Ahn, and A. Kornberg. 1996. Phosphohistidyl active sites in polyphosphate kinase of Escherichia coli. Proc. Natl. Acad. Sci. USA 93:14391-14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda, A., K. Nomura, R. Ohtomo, J. Kato, T. Ikeda, N. Takiguchi, H. Ohtake, and A. Kornberg. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293:705-708. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa, N., J. DeRisi, and P. O. Brown. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11:4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reizer, J., A. Reizer, and M. H. Saier, Jr. 1993. Exopolyphosphatase and guanosine pentaphosphatase belong to the sugar kinase/actin/hsp70 superfamily. Trends Biochem. Sci. 18:247-248. [DOI] [PubMed] [Google Scholar]
- 23.Rudnick, H., S. Hendrich, U. Pilatus, and K.-H. Blotevogel. 1990. Phosphate accumulation and the occurrence of polyphosphates and cyclic 2,3-diphosphoglycerate in Methanosarcina frisia. Arch. Microbiol. 154:584-588. [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Scherer, P. A., and H. P. Bochem. 1983. Ultrastructural investigation of 12 Methanosarcinae and related species grown on methanol for occurrence of polyphosphatelike inclusions. Can. J. Microbiol. 29:1190-1199. [Google Scholar]
- 26.Schröder, H. C., B. Lorenz, L. Kurz, and W. E. G. Müller. 1999. Inorganic polyphosphate in eukaryotes: enzymes, metabolism and function, p. 45-74. In H. C. Schröder and W. E. G. Müller (ed.), Inorganic polyphosphates. Biochemistry, biology, biotechnology. Springer-Verlag KG, Berlin, Germany. [DOI] [PubMed]
- 27.Scoarughi, G. L., C. Cimmino, and P. Donini. 1995. Lack of production of (p)ppGpp in Halobacterium volcanii under conditions that are effective in the eubacteria. J. Bacteriol. 177:82-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sethuraman, A., N. N. Rao, and A. Kornberg. 2001. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shintani, T., T. Uchiumi, T. Yonezawa, A. Salminen, A. A. Baykov, R. Lahti, and A. Hachimori. 1998. Cloning and expression of a unique inorganic pyrophosphatase from Bacillus subtilis: evidence for a new family of enzymes. FEBS Lett. 439:263-266. [DOI] [PubMed] [Google Scholar]
- 31.Sivula, T., A. Salminen, A. N. Parfenyev, P. Pohjanjoki, A. Goldman, B. S. Cooperman, A. A. Baykov, and R. Lahti. 1999. Evolutionary aspects of inorganic pyrophosphatase. FEBS Lett. 454:75-80. [DOI] [PubMed] [Google Scholar]
- 32.Skórko, R., J. Osipiuk, and K. O. Stetter. 1989. Glycogen-bound polyphosphate kinase from the archaebacterium Sulfolobus acidocaldarius. J. Bacteriol. 171:5162-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vielle, C., and G. J. Zeikus. 2001. Hyperthermophilic enzymes: sources, uses and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wurst, H., T. Shiba, and A. Kornberg. 1995. The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J. Bacteriol. 177:898-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamagata, Y., H. Watanabe, M. Saitoh, and T. Namba. 1991. Volcanic production of polyphosphates and its relevance to prebiotic evolution. Nature 352:516-519. [DOI] [PubMed] [Google Scholar]