Abstract
The development of the lactic acid bacterial community in a commercial malt whisky fermentation occurred in three broad phases. Initially, bacteria were inhibited by strong yeast growth. Fluorescence microscopy and environmental scanning electron microscopy revealed, in this early stage, both cocci and rods that were at least partly derived from the wort and yeast but also stemmed from the distillery plant. Denaturing gradient gel electrophoresis (DGGE) of partial 16S rRNA genes and sequence analysis revealed cocci related to Streptococcus thermophilus or Saccharococcus thermophilus, Lactobacillus brevis, and Lactobacillus fermentum. The middle phase began 35 to 40 h after yeast inoculation and was characterized by exponential growth of lactobacilli and residual yeast metabolism. Lactobacillus casei or Lactobacillus paracasei, L. fermentum, and Lactobacillus ferintoshensis were detected in samples of fermenting wort examined by DGGE during this stage. Bacterial growth was accompanied by the accumulation of acetic and lactic acids and the metabolism of residual maltooligosaccharides. By 70 h, two new PCR bands were detected on DGGE gels, and the associated bacteria were largely responsible for the final phase of the fermentation. The bacteria were phylogenetically related to Lactobacillus acidophilus and Lactobacillus delbrueckii, and strains similar to the former had previously been recovered from malt whisky fermentations in Japan. These were probably obligately homofermentative bacteria, required malt wort for growth, and could not be cultured on normal laboratory media, such as MRS. Their metabolism during the last 20 to 30 h of fermentation was associated with yeast death and autolysis and further accumulation of lactate but no additional acetate.
Scotch malt whisky is distilled from the fermented hot-water extract of malted barley. The malted cereal is milled and infused with water (mashed) at about 63°C. After about 30 min, the first wort is removed, cooled, and pumped to the fermentation vessel. The second water, which is conducted at a higher temperature (about 75°C) to effect the maximal extraction of carbohydrate from the grist, is cooled and added to the first wort to fill the fermentation vessel. The wort is not boiled, as it is in a brewery in order to retain the activity of the soluble enzymes from the malt during the fermentation and to maximize alcohol yield. Consequently, bacteria from the malt that can survive mashing enter the fermentation, resulting in a mixed yeast-bacterial fermentation (11, 19). If large numbers of lactobacilli enter the fermentation (more than 106 cells/ml), they compete for nutrients with yeast cells and reduce the ethanol yield (9, 11, 20). In well-operated distilleries, however, the lactobacilli flourish after the yeast cells have reached stationary phase and grow on residual nutrients and autolysing yeast cells. This “late lactic fermentation” is encouraged by many distillers, since it is thought to have a beneficial effect on the flavor of the final spirit (13, 24).
Little is known about the composition of the bacterial community in malt whisky fermentation and its development as fermentation proceeds. Traditionally, these questions have been addressed for various natural fermentations by enumeration of bacteria on culture media followed by their identification (4, 17). Analysis of whisky fermentations in this way has revealed Lactobacillus brevis, Lactobacillus fermentum, and Lactobacillus paracasei as the most commonly isolated bacteria, with strains of Lactobacillus pentosus, Lactobacillus plantarum, and a new species, Lactobacillus ferintoshensis, being less commonly encountered (23). However, culture-dependent methods may underestimate the diversity of a bacterial community, particularly in such complex environments as fermented foods and beverages (4, 5).
In this study, we have adopted a polyphasic approach by using light and electron microscopy and denaturing gradient gel electrophoresis (DGGE) of PCR-amplified fragments of 16S ribosomal DNA (rDNA) to monitor the development of the lactic acid bacterial community throughout malt whisky fermentation. Our results reveal an underestimation of bacterial diversity by culture-dependent methods and the presence of novel lactobacilli and other taxa in malt whisky fermentation.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The reference strains of Lactobacillus used in this study were as follows: L. acidophilus LMG 7943 (Laboratorium voor Microbiologie, Universiteit Ghent, Ghent, Belgium), L. amylolyticus LA5 (Lehrstuhl für Technologie der Brauerei I, TU Munich, Germany), L. amylovorus NCIMB 13276 (National Collection of Industrial and Marine Bacteria, Aberdeen, United Kingdom), L. brevis R19-113 (23), L. buchneri CIP 103023 (Collection de l’Institut Pasteur, Paris, France), L. casei 541 (M. Collins, Agriculture and Food Science Centre, Belfast, United Kingdom), L. crispatus CIP 105003, L. ferintoshensis R7-9 (23), L. fermentum R22-70 (23), L. hilgardii CIP 103006, L. kefiri CIP 103007, L. paracasei R1-69 (23), L. pentosus R11-128 (23), L. plantarum R3-72 (23), and Lactobacillus sp. strain Y10 (S. van Beek and F. G. Priest, Abstr. 6th Symp. Lactic Acid Bacteria Genet. Metab. Appl., p. A34, 1999). All strains were maintained in MRS medium (10) containing 30% (wt/vol) glycerol at −70°C and were grown at 30°C statically in MRS broth at 30°C. Modified MRS agar was prepared by using sterile distiller’s wort rather than water. Distiller’s wort was prepared to an original gravity of 14.5°Plato in our pilot brewery, pasteurized at 90°C for 15 min, clarified by centrifugation, and stored at −20°C. Viable counts were determined in triplicate by plating appropriately diluted fermentation samples on modified MRS agar containing 50 mg of cycloheximide (Sigma)/liter and 10 mg of sodium azide (Sigma)/liter and incubating the samples at 30°C for 24 h.
Estimation of bacterial viability.
Fermentation samples were obtained from the Glenkinchie Distillery, a malt whisky distillery located in southern Scotland. They were stored at 4°C for 2 h to allow the bulk of the yeast to sediment, and 1 ml of the supernatant was clarified by centrifugation and washed twice with sterile distilled water. Viability was determined by using LIVE/DEAD BacLight bacterial viability kit L-7012 (Molecular Probes, Cambridge Bioscience, Cambridge, United Kingdom), which is based on mixtures of the green fluorescence nucleic acid stain, SYTO9, that labels all cells in a population and the red fluorescence nucleic acid stain, propidium iodine, that penetrates only bacteria with damaged membranes and quenches the green stain SYTO9. Thus, in an appropriate mixture of SYTO9 and propidium iodine, bacteria with intact cell membranes stain fluorescent green, whereas bacteria with damaged membranes stain fluorescent red. A bacterial suspension (1 ml) was stained with 3 μl of premixed dye according to the manufacturer’s instructions (http://www.molecularprobes.com), incubated at room temperature in the dark for 10 min, and immobilized on a 0.2-μm-pore-size Isopore polycarbonate filter membrane (Millipore, Watford, United Kingdom). Cells were viewed under an Axiophot microscope (Carl Zeiss, Welwyn Garden City, United Kingdom) equipped with a 50-W mercury arc lamp and Carl Zeiss filter set number 9 (excitation wavelength, 450 to 490 nm; emission wavelength, >520 nm). Photomicrographs were made with simultaneous light microscopy and epifluorescence microscopy on Fuji 1600-ASA color film. For each sample, 10 pictures were taken, each depicting 100 to 400 cells.
Environmental scanning electron microscopy (ESEM).
Fermentation samples were left at 4°C for 2 h, and 15 μl of appropriately diluted material was observed by using a Philips XL30 (LaB 6) environmental scanning electron microscope (FEI UK Ltd.). The microscope was operated at about 5 torr and 5°C, with a working distance of about 7.5 mm.
Preparation of DNA and RNA.
DNA from reference strains was isolated from 1 ml of late-exponential-phase culture (optical density at 600 nm, about 1.0) in MRS broth by using a PUREGENE DNA isolation kit (Philip Harris/Flowgen, Shenstone, United Kingdom) modified by the addition of 140 U of mutanolysin (Sigma)/ml to the lytic enzyme solution and incubating the cell suspension at 37°C for 45 min. Total DNA was isolated from 10-ml distillery fermentation samples which had been stored at 4°C for 2 h to allow the yeast to settle. Bacteria were collected from the supernatant by centrifugation, washed three times with distilled water, and resuspended in 1 ml of cell suspension solution from the PUREGENE DNA isolation kit. DNA was isolated as described above. The quality of the DNA was examined following electrophoresis on a 1% agarose gel in 40 mM Tris-acetate (pH 8.0)–0.1 mM disodium EDTA buffer. Nucleic acids were quantified by UV spectrometry (GeneQuant RNA/DNA calculator; Amersham Pharmacia Biotech, Buckingham, United Kingdom).
Total RNA was extracted from fermentation samples by using a PURESCRIPT RNA isolation kit (Philip Harris/Flowgen) with the same modifications as those used for DNA extraction. RNA was treated with DNase reagent and removal solution (Ambion/AMS Biotechnology Ltd., Abingdon, United Kingdom) to eliminate any genomic DNA contamination. The integrity of the RNA was checked by agarose gel electrophoresis under denaturing conditions. RNA (10 ng) was used as a template for reverse transcriptase (RT) PCR by using a RobusT RT-PCR kit (Philip Harris/Flowgen). First-strand cDNA synthesis was performed at 50°C for 45 min; inactivation of avian myeloblastosis virus RT and primer-RNA-cDNA denaturation were done at 94°C for 2 min; and second-strand cDNA synthesis and PCR amplification were accomplished during 40 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 40 s, and extension at 72°C for 1 min, followed by a final extension cycle at 72°C for 7 min. Simultaneously, a negative control reaction without RT was performed with each RNA template (results not shown).
PCR-DGGE analysis.
Purified DNA was amplified with primer pair 1 (HDA1-GC, 5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′ [the GC clamp is in boldface], and HDA2, 5′-GTA TTA CCG CGG CTG CTG GCA C-3′) (25), spanning the V2 region of the 16S rDNA gene (positions 339 to 539 in the Escherichia coli gene). PCRs were performed with a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer, Norwalk, Conn.). The reaction mixture (50 μl) consisted of 5 μl of reaction buffer, 1 μl of a 10 mM deoxynucleoside triphosphate mixture, 3 mM MgCl2, 0.1 pmol of each primer, 10 ng of genomic template DNA, and 0.5 U of Dynazyme EXT Taq DNA polymerase (Philip Harris/Flowgen). The amplification program was 96°C for 3 min; 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 60 s; and finally 72°C for 7 min. A second pair of primers targeted to an alternative region of the 16S gene was designed from an alignment of partial and complete Lactobacillus 16S rRNA sequences retrieved from GenBank: L. acidophilus (X61138), L. amylolyticus (Y17361), L. amylovorus (M58805), L. brevis (AF090328), L. buchneri (X61139), L. casei (D16552), L. crispatus (AF257097), L. fermentum (AF302116), L. ferintoshensis (AF275311), L. hilgardii (M58821), L. paracasei (D79212), L. pentosus (D79211), and L. plantarum (M58827). Primer pair 2 comprised HDA4C (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GGC GGT GGA GCA TGT GGT TTA-3′) and HDA5 (5′-CCT TCC TCC GGT TTG TCA CC-3′), covering positions 939 to 1163 in the E. coli gene. The PCR mixture was modified from that of primer pair 1 by reduction of the MgCl2 concentration to 1.5 mM and by use of 0.2 pmol of each primer. The amplification program was 96°C for 3 min; 30 cycles of 94°C for 30 s, 56°C for 40 s, and 72°C for 1 min; and an elongation step at 72°C for 7 min.
DGGE was performed with a DCode electrophoresis system (Bio-Rad, Hemel Hempstead, United Kingdom) and gels measuring 16 cm by 16 cm by 1 mm. Polyacrylamide gels (8%) were prepared and run in Tris-borate-EDTA buffer. The denaturing gradient was formed with 8% Acrylogel 2.6 (BDH, Poole, United Kingdom). PCR products were loaded onto a 40 to 55% gradient of urea and formamide and electrophoresed at a constant temperature of 60°C and a constant voltage of 50 or 60 V for 630 or 700 min, respectively. Gels were stained with ethidium bromide (0.5 mg/liter) in Tris-borate-EDTA buffer for 20 min, destained in sterile deionized water for 10 min, and viewed by UV transillumination.
DNA sequencing.
DGGE bands representing unknown organisms were excised from the gels, and a very small cut of each band, previously frozen at −70°C for 4 h, was used as a template for PCR with the primer pair used for DGGE. Subsequently, about 100 ng of PCR products was used as a template for sequencing amplification with one of the primers of each pair and 5% dimethyl sulfoxide in order to overcome difficulties due to the presence of secondary structures. The following cycling profile (25 cycles) was used: 96°C for 10 s, 55°C for 5 s, and 60°C for 4 min. Energy transfer dye terminator chemistry (Big Dye Terminator kit; Applied Biosystems, Warrington, United Kingdom) was used as described by the manufacturer for labeling the fragments. The excess of dye and buffer components was removed by isopropanol precipitation. The sequencing products were separated on an ABI 310 capillary sequencing system (Applied Biosystems). The partial 16S rDNA sequences obtained were compared to the sequences in the GenBank DNA database by using the BLASTN algorithm (2).
Analytical procedures.
Acetic acid and d- or l-lactic acid were assayed enzymatically by using commercially available kits (Boehringer Ingelheim Limited, Bracknell, United Kingdom). Residual sugars in fermentation samples were determined by high-pressure liquid chromatography (Waters 600E; Millipore, Bedford, Mass.). Samples were clarified by centrifugation and analyzed with an Aminex HPX-42A column (Bio-Rad) at 85°C and a flow rate of 0.5 ml of water/min. Ethanol and oligosaccharides up to maltoheptaose were detected by using a differential refractometer (Waters 410). Only late fermentation samples (>40 h) could be analyzed because the high concentrations of glucose and maltose in early samples obscured the analyses.
Nucleotide sequence accession numbers.
The GenBank accession number for the almost complete 16S rRNA sequence of Lactobacillus sp. strain Y10 is AY029223. See Table 2 for accession numbers for DGGE bands from uncultured strains.
TABLE 2.
Identities of bands obtained from DGGE analysis of the bacterial community
| Figure | Band | Closest relative | No. of nucleotides sequenced | % Identitya | Accession no. |
|---|---|---|---|---|---|
| 4B | a | ND | ND | ND | |
| bb | L. ferintoshensis | ||||
| 5B | a | Streptococcus thermophilus or Saccharococcus thermophilus | 206 | 98 | AF375001 |
| b | Streptococcus thermophilus or Saccharococcus thermophilus | 195 | 97 | AF375002 | |
| c | L. fermentum | 210 | 100 | ||
| d | L. fermentum | 208 | 100 | ||
| e | L. fermentum | 201 | 99 | AF375003 | |
| fb | L. ferintoshensis | ||||
| gb | L. casei, L. paracasei, or L. brevis | ||||
| hb | Lactobacilus sp. strain 19-2 | ||||
| ib | L. crispatus | ||||
| jb | L. plantarum | ||||
| k | Lactobacillus sp. strain Y10 | 223 | 100 | ||
| l | Lactobacillus sp. strain Y10 | 212 | 99 | AF375005 | |
| m | Lactobacillus sp. strain Y10 | 243 | 99 | AF375007 | |
| 6 | a | Streptococcus thermophilus or Saccharococcus thermophilus | 206 | 98 | Identical to AF375001 |
| b | ND | ND | ND | ND | |
| c | ND | ND | ND | ND | |
| d | L. fermentum | 200 | 99 | AF375004 | |
| e | L. fermentum | 220 | 100 | ||
| f | L. fermentum | 223 | 100 | ||
| g | L. fermentum | 201 | 99 | Identical to AF375003 | |
| hb | L. ferintoshensis | ||||
| ib | L. casei or L. paracasei | ||||
| jb | L. brevis | ||||
| kb | Lactobacillus sp. strain 19-2 | ||||
| lb | L. plantarum | ||||
| m | Lactobacillus sp. strain Y10 | 223 | 100 | ||
| n | Lactobacillus sp. strain Y10 | 212 | 99 | Identical to AF375005 |
Percentage of identical nucleotides in the sequence retrieved from the DGGE gels and the sequence of the closest relative found in GenBank. ND, not determined.
Band identified by comparison of migration distance with those of reference strains.
RESULTS
Growth of lactic acid bacteria during whisky fermentation.
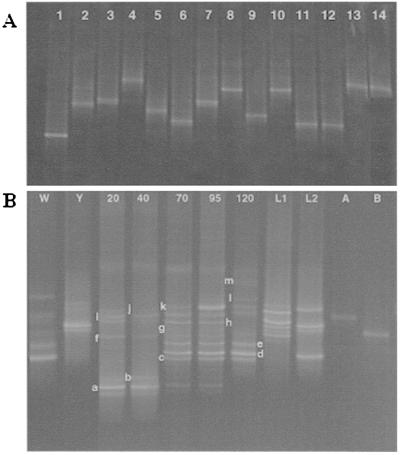
Initially, we amplified 16S rRNA genes from strains representing common distillery species by using primer pair 1 (positions 339 to 539 in the E. coli gene). DNA fragments from L. acidophilus and its phylogenetic close relatives L. amylolyticus, L. amylovorus, L. crispatus, and Lactobacillus sp. strain Y10 had similar migration distances (Fig. 4A, lanes 1, 2, 3, and 5, and Fig. 4B, lane E), as did products from L. paracasei and L. plantarum. Some strains, such as those of L. buchneri, were distinguishable from close relatives (L. kefiri) because they presented two bands, with one in common (Fig. 4A, lanes 10 and 11). When DNA amplified from distillery fermentation samples was examined by using these primers, two main groups of strains were revealed. Bacteria corresponding to L. fermentum were present from the early stage of the fermentation but were particularly pronounced at 46 h, reaching a maximum at 70 h and then decreasing slightly. A very faint band representing the L. acidophilus group (comigrating with Lactobacillus sp. strain Y10) was present before 70 h but increased from this point on, reaching a maximum at the very end of the fermentation. Such bacteria have been isolated from Japanese malt whisky fermentations but had not been detected previously in Scotch malt whisky fermentations (23). This band was accompanied by a second, slightly larger product, perhaps representing another species from this phylogenetic group. Other DNA fragments were less evident and corresponded to L. plantarum or L. paracasei in late fermentation samples (70 to 120 h) and to L. brevis from 30 to 70 h. The DNA fragment labeled “a” at 10 to 52 h could not be sequenced, and the band labeled “b” at 46 h was identified by migration distance as L. ferintoshensis (Table 2).
FIG. 4.
DNA-based DGGE gels of Lactobacillus reference strains (A) and fermentation samples (B) amplified by using primer pair 1 and separated in an 8% polyacrylamide gel containing a 40 to 55% denaturing gradient. (A) Lanes: 1, L. crispatus; 2, L. acidophilus; 3, L. amylolyticus; 4, L. pentosus; 5, L. amylovorus; 6, L. casei; 7, L. brevis; 8, L. fermentum; 9, L. ferintoshensis; 10, L. kefiri; 11, L. buchneri; 12, L. paracasei; and 13, L. plantarum. (B) Lanes 10 to 120, DNA profiles of samples taken after 10, 30, 46, 52, 70, 95, and 120 h of fermentation. Lanes A, B, C, D, and E, DNA fragments from pure cultures of strains of L. brevis, L. paracasei, L. plantarum, L. fermentum, and Lactobacillus sp. strain Y10, respectively. The bands labeled “a” and “b” are described in Table 2.
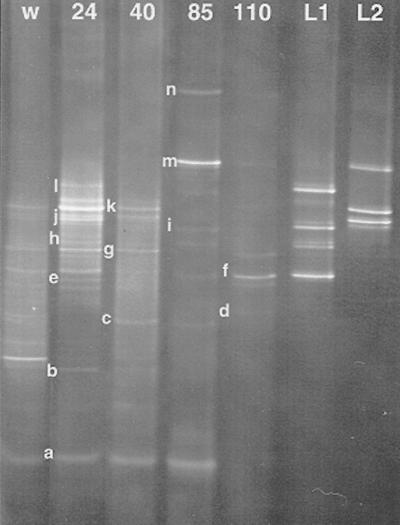
We next targeted a region of the 16S rDNA gene (nucleotides from positions 939 to 1163 of the E. coli gene) in which polymorphisms exist among species of the L. acidophilus group. Primer pair 2 gave improved discrimination of DNA fragments from species related to L. acidophilus, although strains of L. amylovorus and L. crispatus still migrated to the same position (Fig. 5A, lanes 7 to 10). Moreover, L. paracasei was now distinguished from L. plantarum, but strains of L. kefiri and L. buchneri (Fig. 5A, lanes 11 and 12), L. amylolyticus and L. casei (lanes 3 and 7), and L. pentosus and L. plantarum (lanes 13 and 14) were superimposed. Nevertheless, these data enabled us to analyze with different levels of discrimination the bacterial community in the fermentation (Fig. 5B). The development of the bacterial flora portrayed by these primers resembled that found earlier. Cocci were present in the early stage (labeled “a” and “b” in Fig. 5B) and declined as the middle stage began. L. fermentum occurred in the early stage but, on this occasion, continued into the end of the fermentation. With these primers, we recovered several bands corresponding to L. fermentum when DNA was isolated and sequenced (labeled “c,” “d,” and “e” in Fig. 5B). L. ferintoshensis (labeled “f” in Fig. 5B) was again present in the middle stage of the fermentation, and L. casei or L. paracasei was present in the middle to later stages. An L. acidophilus-like bacterium again proliferated in the final stage of the fermentation, but the single band seen with primer pair 1 was now resolved into several products of different compositions. A strain corresponding to Lactobacillus sp. strain Y10 was identified (labeled “k” in Fig. 5B), as was a new organism which we have now isolated in a pure culture and provisionally called Lactobacillus sp. strain 19-2 (labeled “h” in Fig. 5B). There were other bands (labeled “l” and “m” in Fig. 5B) which we have been unable to identify with confidence due to difficulties with sequencing, but they are probably members of the L. acidophilus group.
FIG. 5.
DNA-based DGGE gels of Lactobacillus reference strains (A) and fermentation samples (B) amplified by using primer pair 2 and separated in an 8% polyacrylamide gel containing a 40 to 55% denaturing gradient. (A) Lanes: 1, L. fermentum; 2, L. brevis; 3, L. casei; 4, L. paracasei; 5, L. hilgardii; 6, L. ferintoshensis; 7, L. amylolyticus; 8, L. amylovorus; 9, L. acidophilus; 10, L. crispatus; 11, L. kefiri; 12, L. buchneri; 13, L. pentosus; 14, L. plantarum. (B) Lanes W and Y, samples taken from the wort and yeast, respectively. Lanes 20 to 120, DNA profiles of samples taken after 20, 40, 70, 95, and 120 h of fermentation. Lanes L1 and L2, reference ladders, from top: L1, strain Y10, strain 19-2, L. paracasei, and L. ferintoshensis; L2, L. plantarum, L. brevis, and L. fermentum. Lanes A and B, DNA fragments from pure cultures of strains of L. crispatus and L. acidophilus, respectively. The bands labeled “a” to “m” are identified in Table 2.
On this occasion, we also examined DNA extracted from the cooled wort (prior to yeast inoculation) and from the yeast suspension before inoculation into the fermentation. The cooled wort provided a source of L. fermentum, Lactobacillus sp. strain Y10, and Lactobacillus sp. strain 19-2, and the major bands recovered from the yeast suspension represented L. brevis and L. plantarum.
Finally, we prepared a DGGE profile derived from total RNA extracted from the distillery fermentation and amplified by RT-PCR by using primer pair 2. Since active bacteria have higher numbers of ribosomes than dead or dormant cells, this procedure provided an indication of the relative activities of the major lactic acid bacteria present (Fig. 6). Cocci were evident in the wort and continued strongly until the later stage of the fermentation (85 h). The wort also appeared to be a source of Lactobacillus sp. strains 19-2 and Y10, L. brevis (labeled “j” in Fig. 6), and three different strains of L. fermentum. L. brevis and L. fermentum strains again flourished in the early stage, but one L. fermentum strain reappeared in the final sample. Lactobacillus sp. strain Y10 was growing particularly strongly at 85 h, and Lactobacillus sp. strain 19-2 was mostly represented in the earlier samples. Finally, L. plantarum (labeled “l” in Fig. 6) showed more activity in the early stage of the fermentation.
FIG. 6.
RNA-based DGGE detection of Lactobacillus strains during whisky fermentation (from wort to 110 h) by using primer pair 2 and the conditions described in the legend to Fig. 5. Lane W, sample taken from the wort. Lanes 24 to 110, RNA profiles from samples taken after 24, 40, 85, and 110 h of fermentation. Lanes L1 and L2, DNA fragment ladders from pure cultures of strains (from top to bottom): L1, L. plantarum, L. casei or L. paracasei, L. ferintoshensis, and L. fermentum; L2, Lactobacillus sp. strain Y10, Lactobacillus sp. strain 19-2, and L. brevis. The bands labeled “a” to “n” are identified in Table 2.
DISCUSSION
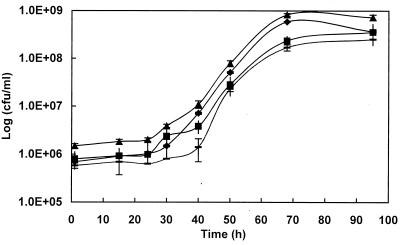
Many whisky distillers practice late lactic fermentation, during which lactic acid bacteria develop following yeast growth (22, 26). While gross bacterial numbers have been estimated in the past (11), this is the first time that the relative populations of the different types of bacteria present during a fermentation have been examined. Our results show that the fermentation can be divided into three phases: the period up to about 35 h from yeast inoculation, when yeast growth is rampant and bacterial growth is heavily suppressed; a second phase, from 35 h to about 70 h, when bacterial growth is exponential at the expense of exhausted yeast cells; and a final stationary or decline phase, when bacterial numbers no longer increase but lactic acid continues to accumulate (Fig. 1 and Table 1).
FIG. 1.
Bacterial growth during Scotch whisky fermentation. Symbols: triangles, total microscopic cell count; diamonds, viable microscopic cell count; squares, stressed/dead cell count; hyphens, viable culturable cells. Each point of the microscopic counts is the mean of three samplings, and for each sample, 10 microphotographs were taken. Each point of the plate counts is the result of triplicate counts. Standard deviations are shown (error bars).
TABLE 1.
Fermentation products and residual sugar concentrations during the late lactic fermentation
| Time (h) | g of the following product or sugar/litera
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lactic acid | Acetic acid | Ethanol | Residual sugarsb
|
||||||||
| G1 | G2 | G3 | G4 | G5 | G6 | G7 | >G10 | ||||
| 40 | 0.49 | 0.02 | 76.15 | 4.10 | 7.60 | 10.25 | 0.15 | 0.50 | 0.10 | 0 | 2.20 |
| 50 | 0.66 | 0.23 | 80.50 | 1.60 | 2.45 | 2.00 | 0.25 | 0.60 | 0.15 | 0 | 1.60 |
| 65 | 2.28 | 0.32 | 88.05 | 0.80 | 0.20 | 1.05 | 0.35 | 0.50 | 0.15 | 0 | 2.40 |
| 90 | 4.00 | 0.31 | 88.85 | 0 | 0 | 0.75 | 0.25 | 0.25 | 0.10 | 0 | 2.05 |
Values are means of triplicate determinations.
G1, glucose; G2, maltose; G3, maltotriose; G4, maltotetraose; G5, maltopentaose; G6, maltohexaose; G7, maltoheptaose.
Microscopic analysis of the wort revealed cocci and rods as potential inocula for the initial phase. However, it is difficult to assess the viability of this population given that the heat stress encountered may lead to a false interpretation of faintly red cells as dead, while these damaged cells may recover and grow (7). Nevertheless, the subsequent observation of cocci by ESEM (Fig. 3A and B) and the recovery of cocci related to Streptococcus thermophilus and Saccharococcus thermophilus in DGGE gels indicate that these bacteria do indeed subsequently grow. The exact nature of these bacteria is obscure because they were not previously identified in distillery fermentations. Partial 16S rRNA sequence analysis identified them as either Streptococcus thermophilus or Saccharococcus thermophilus. The former was previously associated only with milk, although streptococci related to Streptococcus bovis have been identified as a major component in the early stages of maize fermentations (3, 4, 5). Streptococcus thermophilus can survive a high temperature (60°C for 30 min) and can therefore survive a distillery mash, and both fluorescence microscopy and RNA-based DGGE suggested that viable cocci entered the fermentation from the wort. The alternative identification was Saccharococcus thermophilus (98% similarity over 224 nucleotides), a catalase-positive coccus that metabolizes sugars to lactic acid but is phylogenetically close to the thermophilic endospore-forming bacteria (21). This bacterium was originally isolated from sugar beet, and although it could have inhabited the distillery fermentation, its low tolerance of acid pH argues against this possibility. The 98% sequence similarity with known organisms covers a relatively conserved part of the 16S rRNA gene and thus does not allow for precise phylogenetic placement, and the identification of this coccus will require either isolation in a pure culture or cloning and analysis of complete rDNA sequences.
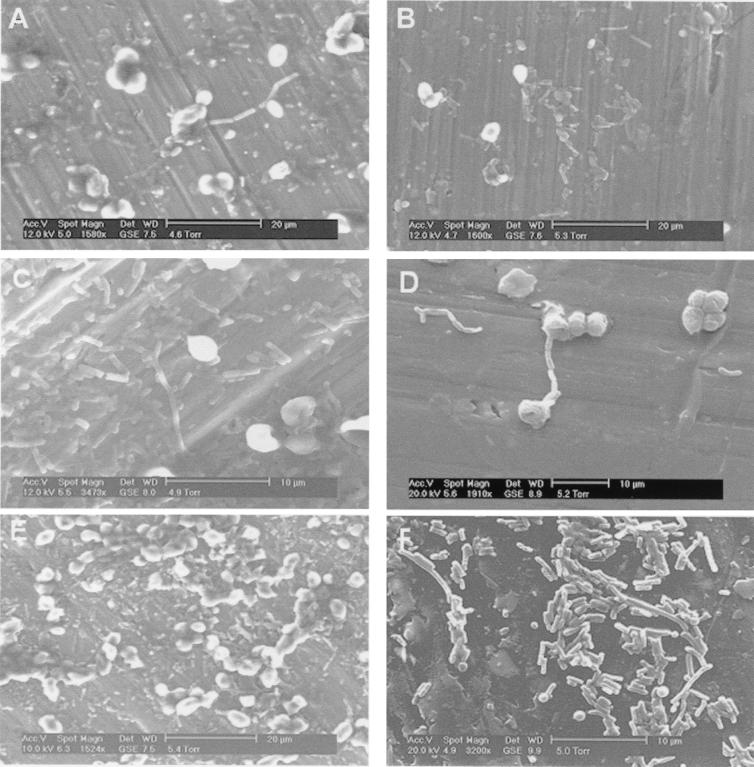
FIG. 3.
Environmental scanning electron micrographs of whisky fermentation samples 20 h (A), 40 h (B), 60 h (C), 70 h (D), 95 h (E), and 100 h (F) after yeast addition.
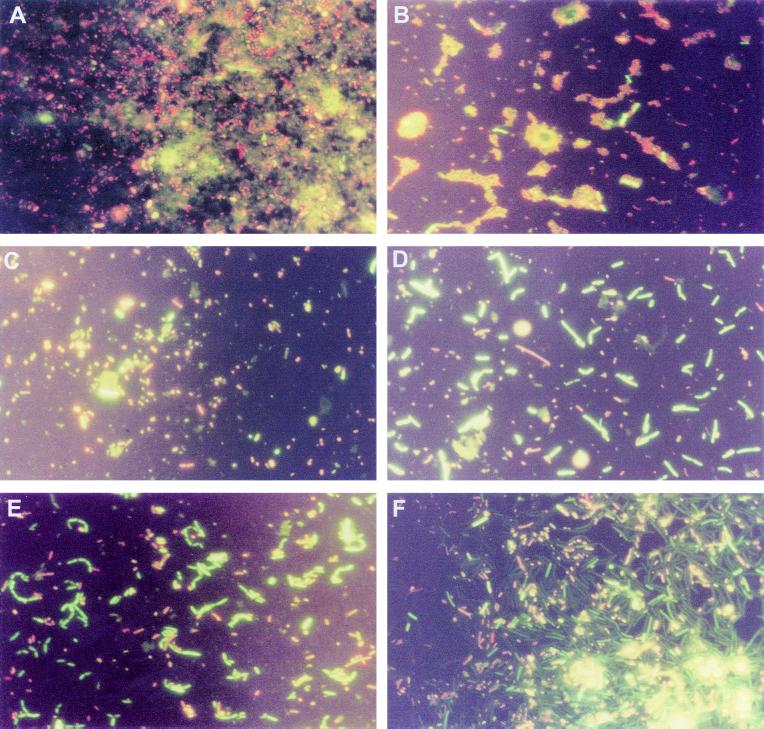
The first phase of the fermentation is also populated by rods from the wort and yeast cells which were identified by DGGE as mainly L. brevis, L. fermentum, and some bacteria, for example, L. ferintoshensis, that were not apparent in the incoming ingredients and were presumably colonizing the distillery plant. Previous studies examining the distributions of lactobacilli in geographically disperse distilleries concluded that individual distilleries may generate their own flora (23). During this first phase, the cocci and rods grew slowly and showed symptoms of stress under the fluorescence microscope, presumably due to the temperature shock of mashing and the massive competition provided by the yeast cells.
However, by 35 to 40 h, the second phase of the bacterial growth pattern was beginning. This middle phase was characterized by a reduction in yeast growth (ethanol production was 86% complete at 40 h), and the sugar content of the fermentation was about 4 g of glucose/liter, 7.6 g of maltose/liter, and some residual higher dextrins. Rod-shaped bacteria of various lengths (Fig. 2C and D and Fig. 3B and C) which stained strongly with the viability stain grew during this period alongside the cocci. During the second phase the difference between the microscopic viable counts and the plate counts was minimal, suggesting that most of the bacteria could be recovered on our modified MRS agar. L. fermentum dominated DGGE gels from this stage on, as it does in many cereal fermentations (1, 6, 14, 15, 17). Bands from neighboring locations on the gels were also identified as L. fermentum, reflecting the genomic diversity of this species, as previously indicated by ribotyping (23, 27) and randomly amplified polymorphic DNA analysis (16). L. fermentum was accompanied during the second phase by other heterofermentative species, such as L. ferintoshensis, and the homofermentative bacterium L casei or L. paracasei. The heterofermentative organisms were presumably responsible for the accumulation of acetic acid between 50 and 70 h as well as lactic acid (Table 1). High concentrations of acetic and lactic acids will have a detrimental effect on yeast viability and will contribute to the death of the yeast as the fermentation proceeds (20).
FIG. 2.
Fluorescence photomicrographs of whisky fermentation samples. (A) Wort after heat treatment; (B to F) samples at 15 h (B), 40 h (C), 55 h (D), 70 h (E), and 95 h (F) after yeast addition. Live cells stain green; stressed/dead cells stain orange/red.
The final phase of the fermentation began at about 70 h as the bacteria entered stationary phase and loss of viability ensued (Fig. 1A and Fig. 2D and E). There was a tendency for the cells to form long rods and chains, presumably as a result of stress (8). This characteristic accentuated the difference between microscopic viable counts and plate counts, since the latter would underestimate viability through growth of colonies from chains of cells. The yeast cells began to collapse and die during this period (Fig. 3E and F), and there was evidence for attachment of bacteria to yeast cells (Fig. 3D). It seems likely that the release of materials through leaking yeast membranes provided valuable nutrients for the bacteria as the sugar concentration of the fermentation dropped to almost zero. Bacterial growth in this later stage was largely represented by two unknown organisms that produced superimposed bands when DGGE was performed with primer pair 1 but could be distinguished with primer pair 2. One of these bacteria is the same as Lactobacillus sp. strain Y10, which was isolated from Japanese malt whisky fermentations and resembles the homofermentative bacterium L. acidophilus (unpublished results). This is the first time that such a bacterium has been detected in Scotch whisky fermentations, although it is relatively common in Japanese distilleries. Database comparisons of the almost complete 16S rDNA sequence of the second bacterium, which we have designated Lactobacillus sp. strain 19-2, indicated that its closest phylogenetic relative is Lactobacillus delbrueckii (data not shown). Both of these bacteria have complex growth requirements based on malt wort, and neither grows in or on standard laboratory media, such as MRS broth or agar. Full descriptions of these bacteria will be given elsewhere. The growth of these organisms in the final phase of the fermentation was accompanied by a large increase in lactic acid accumulation but no further acetic acid accumulation. These findings are consistent with homofermentative metabolism.
Other traditional fermentations have also been noted to follow three-stage community changes similar to those discovered here. For example, pozol, a Mexican fermented maize dough which has been studied in detail, is initiated by a high diversity of bacteria, including various streptococci, followed at the second stage by heterofermentative lactobacilli, notably L. fermentum, and finally homofermentative lactobacilli, in this case relatives of L. casei (5).
While the advantages of DGGE for the evaluation of complex community changes have been indicated in this study, the difficulty of developing a single set of primers for the differentiation of all species present has been highlighted. The close phylogenetic relationships of the lactobacilli made it impossible to differentiate all species in a single reaction. Members of the L. acidophilus cluster proved particularly difficult to differentiate by using primers HDA1-GC and HDA2 (25), targeted to the V2 region of the rRNA gene. The sequences of L. acidophilus, L. amylolyticus, L. amylovorus, L. crispatus, and Lactobacillus sp. strain Y10 are essentially identical over this region, with only seven polymorphic sites among all the strains, resulting in the failure of DGGE to resolve the PCR fragments. However, the area from positions 939 to 1163 (E. coli numbering) amplified by primers HDA4C and HDA5 was much smaller than the V2 region but contained 14 polymorphic sites for these bacteria and enabled discrimination. It is therefore important to substantiate DGGE gels of complex communities with multiple sets of primers directed to various parts of the rRNA gene in order to be confident that bands represent a single species.
A second drawback is that DGGE of DNA templates is not quantitative. Various ways to provide quantitative estimates involve the use of competitive quantitative PCR-based methods (12, 18) or 16S rRNA-targeted oligonucleotide probes (4). Here we simply used rRNA templates to provide a comparison with rDNA templates on the basis of the idea that actively growing cells will have large numbers of ribosomes compared with stationary-phase cells. The rRNA templates gave us a different sensitivity and, in particular, revealed the presence of L. brevis and L. casei or L. paracasei in the early stage of the fermentation, the continuation of the cocci well into the second stage, and the recurrence of L. fermentum in the final stage of the fermentation. Indeed, L. fermentum and L. paracasei were consistently recovered from late fermentation samples from various distilleries in a previous study (23).
In conclusion, we have shown that the Scotch whisky fermentation involves a changing community of bacteria starting with a diversity of cocci and rods and culminating in lactobacilli that fail to grow in or on standard laboratory media and are probably closely related to L. acidophilus or L. crispatus. The formation of lactic and acetic acids and other metabolites might have an effect on the flavor of the final spirit. Lactic acid reacts with ethanol during distillation to produce ethyl lactate, and spirits derived from long fermentations (greater than 55 h) in which lactic acid bacteria have flourished tend to have higher ester concentrations (13). It is possible that flavor could be modified by careful attention to the balance of the various bacteria present.
Acknowledgments
We are grateful to the owners and staff of the Glenkinchie Distillery for providing the fermentation samples and to Hisato Hikemoto and Takeshi Yonezawa (both of Suntory Ltd., Osaka, Japan) for help and valuable discussions. We also thank Jim Buckman for help with ESEM and Bertil Pettersson (Royal Institute of Technology, Stockholm, Sweden) for sequencing some of the strains used in this study.
This study was funded by Suntory Ltd.
REFERENCES
- 1.Agati, V., J.-P. Guyot, J. Morlon-Guyot, P. Talamond, and J. Hounhouigan. 1998. Isolation and characterization of new amylolytic strains of Lactobacillus fermentum from fermented maize doughs (mawè and ogi) from Benin. J. Appl. Microbiol. 85:512–520. [Google Scholar]
- 2.Altschul, S. F., W. Gish, E. Miller, W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 3.Ampe, F., N. ben Omar, and J. P. Guyot. 1999. Culture-independent quantification of physiologically-active microbial groups in fermented foods using rRNA-targeted oligonucleotide probes: application to pozol, a Mexican lactic acid fermented maize dough. J. Appl. Microbiol. 87:131–140. [DOI] [PubMed] [Google Scholar]
- 4.Ampe, F., N. ben Omar, C. Moizan, C. Wacher, and J. P. Guyot. 1999. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65:5464–5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ben Omar, N., and F. Ampe. 2000. Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl. Environ. Microbiol. 66:3664–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brauman, A., S. Léléké, M. Malonga, E. Miambi, and F. Ampe. 1996. Microbiological characterization of cassava retting, a traditional lactic acid fermentation for foo-foo (cassava flour) production. Appl. Environ. Microbiol. 62:2854–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunthof, C. J., S. van den Braak, P. Breeuwer, F. M. Rombouts, and T. Abee. 1999. Rapid fluorescence assessment of the viability of stressed Lactococcus lactis. Appl. Environ. Microbiol. 65:3681–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charpentier, E., R. Novak, and E. Tuomanen. 2000. Regulation of growth inhibition at high temperature, autolysis, transformation and adherence in Streptococcus pneumoniae by clpC. Mol. Microbiol. 37:717–726. [DOI] [PubMed] [Google Scholar]
- 9.Chin, P. M., and W. M. Ingledew. 1994. Effect of lactic acid bacteria on wheat mash fermentations prepared with laboratory backset. Enzyme Microb. Technol. 16:311–317. [Google Scholar]
- 10.De Man, P. J., M. Rogosa, and M. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130–135. [Google Scholar]
- 11.Dolan, T. C. S. 1976. Some aspects of the impact of brewing science on Scotch malt whisky production. J. Inst. Brew. 82:177–181. [Google Scholar]
- 12.Felske, A., A. D. Akkermans, and W. M. De Vos. 1998. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. Environ. Microbiol. 64:4581–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geddes, P. A., and H. L. Riffkin. 1989. Influence of lactic acid bacteria on aldehyde, ester and higher alcohol formation during Scotch whisky fermentations, p.193–199. In J. R. Piggot and A. Paterson (ed.), Distilled beverage flavour. Ellis Horwood, Chichester, United Kingdom.
- 14.Halm, M., A. Lillie, A. K. Sorensen, and M. Jakobsen. 1993. Microbiological and aromatic characteristics of fermented maize doughs for kenkey production in Ghana. Int. J. Food Microbiol. 19:135–143. [DOI] [PubMed] [Google Scholar]
- 15.Hamad, S. H., M. C. Dieng, M. A. Ehrmann, and R. F. Vogel. 1997. Characterization of the bacterial flora of Sudanese sorghum flour and sorghum sourdough. J. Appl. Microbiol. 83:764–770. [DOI] [PubMed] [Google Scholar]
- 16.Hayford, A. E., A. Petersen, F. K. Vogensen, and M. Jacobsen. 1999. Use of conserved randomly amplified polymorphic DNA (RAPD) fragments and RAPD pattern for characterization of Lactobacillus fermentum in Ghanaian fermented maize dough. Appl. Environ. Microbiol. 65:3213–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hounhouigan, D. J., M. J. Nout, C. M. Nago, J. H. Houben, and F. M. Rombouts. 1993. Characterization and frequency distribution of species of lactic acid bacteria involved in the processing of mawe, a fermented maize dough from Benin. Int. J. Food. Microbiol. 18:279–287. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S.-Y., J. Bollinger, D. Bezdicek, and A. Ogram. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62:3787–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makanjuola, D. B., A. Tymon, and D. G. Springham. 1992. Some effects of lactic acid bacteria on laboratory-scale fermentations. Enzyme Microb. Technol. 14:350–357. [Google Scholar]
- 20.Narendranath, N. V., S. H. Hynes, K. C. Thomas, and W. M. Ingledew. 1997. Effects of lactobacilli on yeast-catalyzed ethanol fermentations. Appl. Environ. Microbiol. 63:4158–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nystrand, R. 1984. Saccharococcus thermophilus gen. nov., sp. nov., isolated from beet sugar extraction. Syst. Appl. Microbiol. 5:204–219. [Google Scholar]
- 22.Simpson, A. C. 1968. Manufacture of Scotch malt whisky. Process Biochem. 33:60–65. [Google Scholar]
- 23.Simpson, K. L., B. Pettersson, and F. G. Priest. 2001. Characterization of lactobacilli from Scotch malt whisky distilleries and description of Lactobacillus ferintoshensis sp. nov., a new species isolated from malt whisky fermentations. Microbiology 147:1007–1016. [DOI] [PubMed] [Google Scholar]
- 24.van Beek, S., and F. G. Priest. 2000. Decarboxylation of substituted cinnamic acids by lactic acid bacteria isolated during malt whisky fermentation. Appl. Environ. Microbiol. 66:5322–5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson, D. C. 1983. Factors influencing the congener composition of malt whisky new spirit, p.79–92. In J. R. Piggot (ed.), Flavour of distilled beverages: origin and development. Ellis Horwood, Chichester, United Kingdom.
- 27.Zhong, W., K. Millsap, H. Bialkowska-Hobrzanska, and G. Reid. 1998. Differentiation of Lactobacillus species by molecular typing. Appl. Environ. Microbiol. 64:2418–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]