Abstract
Planktonic crenarchaeotes are present in high abundance in Antarctic winter surface waters, and they also make up a large proportion of total cell numbers throughout deep ocean waters. To better characterize these uncultivated marine crenarchaeotes, we analyzed large genome fragments from individuals recovered from a single Antarctic picoplankton population and compared them to those from a representative obtained from deeper waters of the temperate North Pacific. Sequencing and analysis of the entire DNA insert from one Antarctic marine archaeon (fosmid 74A4) revealed differences in genome structure and content between Antarctic surface water and temperate deepwater archaea. Analysis of the predicted gene products encoded by the 74A4 sequence and those derived from a temperate, deepwater planktonic crenarchaeote (fosmid 4B7) revealed many typical archaeal proteins but also several proteins that so far have not been detected in archaea. The unique fraction of marine archaeal genes included, among others, those for a predicted RNA-binding protein of the bacterial cold shock family and a eukaryote-type Zn finger protein. Comparison of closely related archaea originating from a single population revealed significant genomic divergence that was not evident from 16S rRNA sequence variation. The data suggest that considerable functional diversity may exist within single populations of coexisting microbial strains, even those with identical 16S rRNA sequences. Our results also demonstrate that genomic approaches can provide high-resolution information relevant to microbial population genetics, ecology, and evolution, even for microbes that have not yet been cultivated.
Molecular phylogenetic surveys of rRNA genes have altered the perspective on naturally occurring microbial diversity and distribution (for a review, see references 11, 19, and 32). Despite a greater understanding of microbial identity, distribution, and abundance, rRNA-based gene surveys have provided little information about the biological properties of many planktonic microbes, especially groups for which there are no cultivated representatives. In addition, although rRNA microheterogeneity (variation within highly related rRNA sequence clusters) has been observed in virtually all taxa and environments sampled via rRNA gene surveys, the significance of this phenomenon remains uncertain. Recent advances in genomic sequencing techniques and the development of methods for cloning large genome fragments into fosmid (35, 41, 42, 45) or bacterial artificial chromosome (5, 6, 38) vectors now provide the means to characterize the gene content (41), metabolic potential (5), and population genetics (41) of uncultivated microorganisms, otherwise known solely by an rRNA sequence. The utility of such genomic approaches and their applicability to diverse questions in microbial ecology have only begun to be explored and exploited.
Members of the Archaea (48) are much more diverse and widespread than previously suspected. Representatives have now been detected in terrestrial environments, marine and lake sediments, and temperate ocean waters and polar seas (for a review, see reference 10). Marine planktonic archaea have been shown to occur in high relative abundance in the oceanic subsurface (13, 26, 27) and to dominate the prokaryotic fraction in the mesopelagic zone of the Pacific Ocean (20). Planktonic archaea also reach a relative seasonal maximum in winter Antarctic waters, approaching 10 to 30% of the total planktonic microbial population (12, 14, 28, 29). To gain additional information on yet-uncultivated Antarctic archaea, we constructed, by use of a fosmid vector (42), a recombinant DNA library that contained inserts of ∼40 kb from surface water picoplankton collected near Palmer Station, Antarctica, in late winter. Planktonic crenarchaeotal genome fragments that contained rRNA genes and originated from the same population were isolated and compared. These within-population genome comparisons yielded high-resolution information on genomic variations of uncultivated, sympatric archaeal cells. The entire sequences of one Antarctic crenarchaeotal clone (fosmid 74A4) and one temperate water subsurface crenarchaeotal clone (fosmid 4B7) (42) were also determined to compare the genomes of related, archaea inhabiting different oceanic provinces. This analysis provided comparative information on more distantly related crenarchaeotes derived from two different oceanic provinces. Our results demonstrate that microbial population structure can be determined at high resolution by examining genome divergence among highly related but genetically distinct cohorts coexisting in the same population. Insights into genomic variation, as it relates to rRNA sequence variation, also can be derived from comparisons of more distantly related microbial species sampled from different geographic locales.
MATERIALS AND METHODS
Sample collection, DNA extraction, and fosmid library preparation.
Coastal waters were collected near Palmer Station, Anvers Island, Antarctic Peninsula, in August 1996 during a period of high archaeal abundance (12). The samples were filtered by tangential flow filtration with an Amicon (Beverly, Mass.) DC-10 unit equipped with a 30,000-Da-cutoff polysulfone hollow-fiber cartridge. A total of 1,500 liters were concentrated to a volume of ∼900 ml. The cells were collected by centrifugation (4°C, 38,900 × g, 1 h) as previously described (12). The bacterioplankton pellet was embedded in agarose plugs as previously described (42). DNA extraction, preparation of the fosmid library, and multiplex PCR screening by using archaeon-biased 16S rRNA oligonucleotide primers were carried out as previously described (42). PCR primers used for screening the library were the 16S rRNA oligonucleotide primers Ar20-F (TTC CGG TTG ATC CYG CCR G) (13) and Arch958R (TCC GGC GTT GAM TCC AAT T) (9) and the 23S rRNA oligonucleotide primer LS2445a-R (CCC YGG GGT ARC TTT TCT ST) (13).
Subclone libraries.
A subclone library was constructed from fosmid 74A4 with DNA partially digested with RsaI (42). In this library, the content of the fosmid was not randomly represented; therefore, a second library was constructed. This library was prepared with randomly sheared DNA as described by Kawata et al. (21), except that the DNA was sheared by passage through a microemulsifying 25-gauge needle. DNA was cloned by using vector pCR 2.1 and the original TA cloning kit (Invitrogen Corporation, Carlsbad, Calif.). Fosmid subclone plasmids were purified by using a Mini-Prep 24 machine (MacConnell Research Corporation, San Diego, Calif.) according to the manufacturer’s instructions. Nucleotide sequences (800 bp, on average) were determined by the dideoxy termination reaction with fluorescence-labeled M13 forward and reverse primers, a SequiTherm EXCel II sequencing kit (Epicentre, Madison, Wis.), and a model 4200 automated DNA sequencer (LI-COR, Lincoln, Nebr.). The strategy for the construction of the subclone library and for the determination of the sequence of fosmid 4B7 was as previously described (41). Contiguous sequences were assembled by using SEQUENCHER 3.1.1 software (Gene Codes Co., Ann Arbor, Mich.).
Proteins, RNA genes, and motif search.
In-depth sequence analysis was based primarily on the use of the PSI-BLAST program (1) essentially as previously described (6). tRNAs were searched by using the tRNAscan-SE program (24). The BLAST-derived “e-values” (1) reported here take into account the statistics of database and local alignment size for the similarity scores obtained from local alignments. Signal peptides were predicted by using the SignalP program (30), and transmembrane segments were predicted by using the PHDhtm program (39, 40). Comparisons among 4B7, 74A4, and Cenarchaeum symbiosum (41) fosmids were performed with the BLAST 2 sequences program (43).
Phylogenetic analyses.
For distance and parsimony analyses of the inferred amino acid sequence of translation elongation factor 1-α (EF1-α), the program PaupSearch of the Wisconsin Package, version 10.0 (Genetics Computer Group, Madison, Wis.), was used.
Nucleotide sequence accession numbers.
Sequences reported in this study have been submitted to GenBank under the following accession numbers: AF393466, U40238, and AF393304 to AF393307.
RESULTS
Isolation of archaeal fosmid clones from Antarctic surface water.
An environmental fosmid library was constructed from microorganisms collected from late austral winter surface waters (−1.8°C) near Palmer Station, Antarctica, in 1996 (29). A total of 7,200 recombinants, each harboring 40 kb of Antarctic microbial DNA, were screened by using multiplex PCR (42) and archaeal-specific 16S rRNA primers. Six 16S rRNA gene-containing crenarchaeotal genome fragments were recovered in the library.
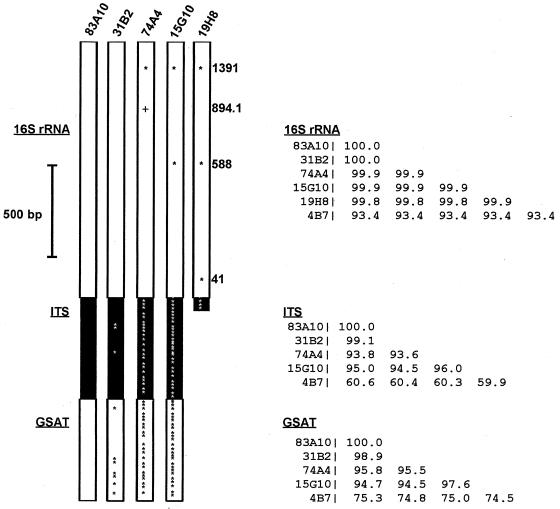
Five unique archaeal 16S rRNA-containing fosmids (15G10, 19H8, 31B2, 74A4, and 83A10) were identified by restriction fragment length polymorphism analysis (data not shown). Sequence analyses of the archaeal 16S rRNAs showed that all belonged to group I marine planktonic Crenarchaeota (9). The 16S rRNA gene sequences from fosmid clones 83A10 and 31B2 were identical to one another. One clone (15G10) contained an rRNA gene identical in sequence to a PCR-amplified 16S rRNA gene (ANTARCTIC 12) (14) that was recovered at the same site 3 years prior to the sampling reported here. The 16S rRNA gene sequence variation observed among the other Antarctic archaeal fosmids was limited, occurring at a total of four nucleotide residues within the 16S rRNA gene (Fig. 1 and 2). Relative to those in 83A10 and 31B2, 16S rRNAs in the other clones contained only two (74A4 and 15G10) or three (19H8) nucleotide sequence differences (Fig. 1).
FIG. 1.
Genetic variability in the 16S rRNA gene, intergenic spacer region (ITS), and GSAT gene in sympatric Antarctic archaea. Asterisks represent a base substitution at residues where sequence variation was observed, relative to the 83A10 sequence. The plus sign indicates an insertion. Numbers at the right indicate the sequence positions (Escherichia coli numbering system) where variation was observed. Clone 19H8 is missing the GSAT gene because the recombinant DNA insert terminates in the ITS. Similarity tables for the 16S rRNA gene, ITS, and GSAT gene are shown at the far right. Fosmid 4B7 from deep temperate Pacific waters is included as an outgroup.
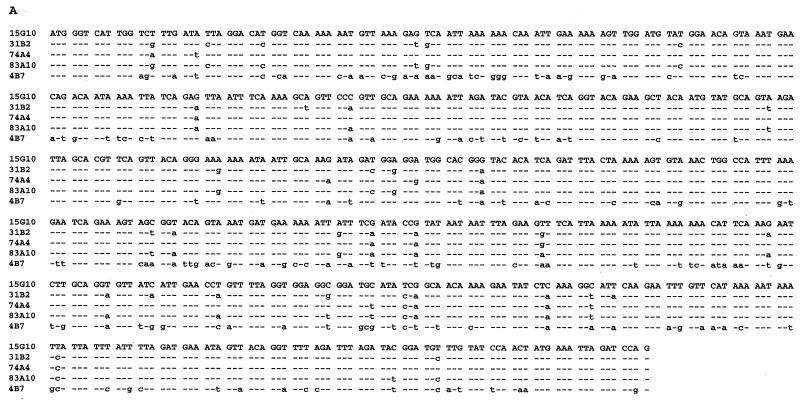
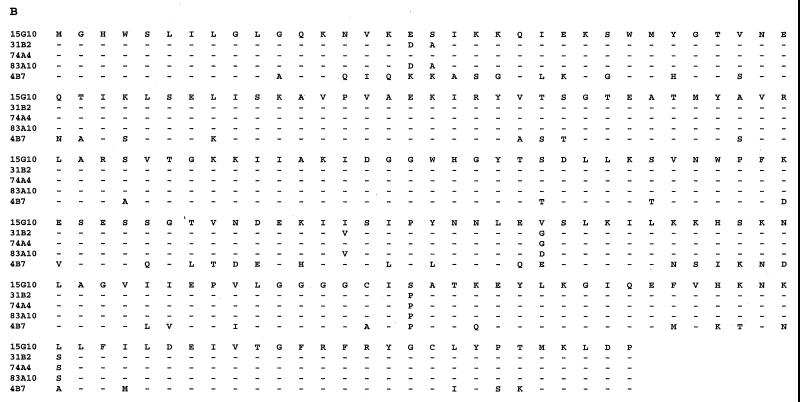
FIG. 2.
Nucleotide (A) and amino acid (B) sequence comparisons of the N terminus of the GSAT gene. The origin of the sequence is shown at the left. Triplets corresponding to amino acids are separated by spaces. Dashes indicate nucleotide residues identical to those of 15G10.
To gain more insight into the variation within the coexisting archaeal phylotypes, we PCR amplified and sequenced a region containing an intergenic spacer (593 bp) and a portion (590 bp) of the glutamate semialdehyde aminotransferase (GSAT) gene found adjacent to the 16S rRNA gene in planktonic marine group I crenarchaeotes (41, 42). The data revealed that the GSAT gene and the spacer between the GSAT gene and the 16S RNA gene were variable between the different sympatric archaeal genomes (Fig. 1). Most of the variation in the GSAT coding region occurred in the third coding position, not changing the amino acid composition of the coded protein (Fig. 2). Nevertheless, variation in the GSAT amino acid sequence was observed among sympatric archaea that differed by only two or three nucleotides in their 16S rRNA genes. Surprisingly, microvariation in the GSAT gene was also found between clones 31B2 and 83A10, clones that had identical 16S RNA gene sequences (Fig. 1 and 2).
Identification of rRNA and protein coding genes on fosmid 74A4.
The entire sequence of the 43.6-kb genome fragment contained on fosmid 74A4 was determined. Forty-nine predicted protein coding frames were identified on fosmid 74A4 by using the National Center for Biotechnology Information BLAST 2.0 program (1), allowing the prediction of protein structural features such as transmembrane segments and signal peptides. Of these predicted proteins, 28 showed significant similarity to the products of genes with known functions, allowing a clear functional prediction, and 7 proteins were homologs of other, uncharacterized proteins. The remaining 14 predicted proteins had no detectable homologs, but some of them were predicted to be either membrane or secreted proteins on the basis of the predicted corresponding structural features (Table 1). The majority of the encoded proteins showed the greatest sequence similarity to homologs from other archaea. No specific affinity was noted with homologs from the only completely sequenced crenarchaeotal genome, that of Aeropyrum pernix, but several proteins showed greatest similarity to homologs from the partially sequenced genome of another crenarchaeote, Sulfolobus (Table 1). The protein sequence of EF1-α that showed the highest similarity to EF1-α proteins from Crenarchaeota was used for phylogenetic analysis. Marine crenarchaeotal EF1-α contained a modified 11-amino-acid insert that is shared between eukaryotes and Crenarchaeota but is not found in Euryarchaeota (36). However, both distance and parsimony analyses of the amino acid sequence of EF1-α could not resolve the placement of marine crenarchaeotal EF1-α (data not shown). Affiliation with either euryarchaeotes or crenarchaeotes was not well supported by either method.
TABLE 1.
Predicted rRNA and protein coding genes in Antarctic crenarchaeotal fosmid 74A4a
| 74A4# | Nucleotide range (protein size, in no. of amino acids) | Dirb | Predicted function | Most similar homolog (e-value)c | Apparent phylo-genetic affinity | Comments |
|---|---|---|---|---|---|---|
| 1 | 372–962 (197) | + | NAD synthase | 5105656_AERPE (3e-46) | Crenarchaeotal | |
| 2 | 970–2355 (462) | + | Cysteinyl-tRNA synthetase | 7437720_PYRAB (e-103) | Euryarchaeotal or bacterial | |
| 3 | 2579–2758 (60) | + | Uncharacterized protein | None | NA | Paralog of 4B7#29 |
| 4 | 2761–3582 (274) | − | Conserved membrane protein | 6136692_ARCFU (2e-45) | Euryarchaeotal | |
| 5 | 3585–4346 (254) | − | Uncharacterized, conserved protein | 2649028_ARCFU (2e-11) | Euryarchaeotal | |
| 6 | 4674–5593 (274) | − | Periplasmic solute-binding protein | 3258088_PYRHO (6e-14) | Euryarchaeotal | Paralog of 4B7#4 |
| 7 | 5695–6849 (385) | + | Uncharacterized conserved protein | 3258294_PYRHO (2e-64) | Euryarchaeotal | Protein family so far detected only in archaea; highly conserved paralog of 4B7#3 |
| 8 | 6912–7496 (195) | − | Multitransmembrane protein | None | NA | |
| 9 | 7625–7957 (111) | − | Uncharacterized protein | None | NA | |
| 10 | 7990–8400 (137) | − | Predicted CoA-binding protein | 3257525_PYRHO (3e-23) | Euryarchaeotal or bacterial | Protein family highly conserved in many archaea and bacteria; Rossmann fold structure and CoA binding are readily predictable, but the function remains unknown |
| 11 | 9053–9409 (119) | + | Uncharacterized protein homologous to sugar epimerases | 10640618_THEAC (0.001) | Euryarchaeotal or bacterial | This predicted protein corresponds to the C-terminal portion of a highly conserved sugar epimerase family |
| 12 | 9455–9892 (146) | + | Uncharacterized protein | None | NA | Paralog of 4B7#22 |
| 13 | 10336–11022 (229) | + | Biotin (acetyl-CoA carboxylase) ligase | 12018159_METBA (9e-40) | Euryarchaeotal | Typical archaeal domain architecture—no DNA-binding HTH domain present in bacterial homologs |
| 14 | 11044–11430 (129) | − | Uncharacterized protein | None | NA | Lysine-rich repeats at the C terminus |
| 15 | 11553–13079 (509) | + | Extracellular (or periplasmic) Zn-dependent metalloprotease containing TPR repeats | 4454026_ARATH (3e-07) (TPR repeats), 5921827_RATNO (4e-05) (protease) | Eukaryotic | No protease with significant similarity detected in other archaea; paralog of 4B7#24 |
| 16 | 13091–13498 (136) | − | Xanthine-guanine phosphoribosyltransferase (crenarchaeotal euryarchaeotal) | 5105769_AERPE (1e-06) | Crenarchaeotal or euryarchaeotal | |
| 17 | 13741–14169 | + | NTP-binding protein of the UspA superfamily, potentially involved in stress response | 12313327_SULSO (1e-10) | Crenarchaeotal | |
| 18 | 14181–14648 | − | Peptide methionine sulfoxide reductase | 12230335_METTH (7e-47) | Eukaryotic or bacterial | This enzyme is nearly ubiquitous in bacteria and eukaryotes, but among archaea so far has been detected only in M. thermoautotrophicum |
| 19 | 14688–15065 (126) | − | Uncharacterized membrane protein | None | NA | Paralog of 4B7#36 |
| 20 | 15149–15913 (255) | − | SAM-dependent methyltransferase | 228451_SACER(1e-10) | Bacterial | |
| 21 | 16207–16587 (127) | + | Uncharacterized membrane protein | 10175820_BACHA (0.001) | Bacterial | |
| 22 | 23S RNA | |||||
| 23 | 16S RNA | |||||
| 24 | 21938–23224 (429) | + | Glutamate-1-semialdehyde aminotransferase | 3599375_CENSY (e-153) | Euryarchaeotal | Other than in Cenarchaeum, the most similar homologs are euryarchaeotal; highly conserved paralog of 4B7#34 |
| 25 | 23473–23961 (163) | + | TPR repeat protein | 2621120_METTH (4e-09) | Euryarchaeotal | |
| 26 | 24579–24974 (132) | + | Type I membrane protein | None | NA | |
| 27 | 25430–26248 (273) | + | TPR repeat protein | 3599376_CENSY (e-60) | Euryarchaeotal | Other than in Cenarchaeum, highly conserved homologs are from euryarchaeotes |
| 28 | 26249–26977 (243) | − | Double-stranded beta-helix fold enzyme | 11287381_CENSY (e-106) | Crenarchaeotal | The only two highly conserved homologs are from Cenarchaeum |
| 29 | 27091–27543 (151) | + | Double-stranded beta-helix fold enzyme, homolog of auxin-binding proteins | 4981537_THEMA (2e-08) | Bacterial | The only highly conserved ortholog is seen in Thermotoga |
| 30 | 27552–27914 (121) | + | Double-stranded beta-helix fold enzyme, homolog of mannose-6-phosphate isomerase and auxin-binding proteins | 2621410_METTH (2e-16) | Euryarchaeotal | Only distantly related to ORF29; highly conserved orthologs are in Methanococcus, Methanobacterium, and Mycobacterium |
| 31 | 27967–28635 (223) | + | Molecular chaperone of the DnaJ class, contains a C-terminal 3Fe-4S ferredoxin domain | 3915678_DROME (1e-09) (DnaJ), 232091_THELI (1e-04) (ferredoxin) | DnaJ portion—eukaryotic or bacterial, ferredoxin—euryarchaeotal | A unique combination of domains |
| 32 | 28638–29042 (135) | − | HIT superfamily hydrolase | 10581606_HALSP (1e-22) | Euryarchaeotal | |
| 33 | 29083–29415 (111) | − | Small membrane protein | None | NA | |
| 34 | 29533–30714 (394) | + | 3-Hydroxyacyl-CoA dehydrogenase | 2650351_ARCFU (8e-72) | Euryarchaeotal or crenarchaeotal | |
| 35 | 30710–31075 (122) | − | Rossmann fold nucleotide-binding protein | 10639670_THEAC (5e-13) | Euryarchaeotal or bacterial | |
| 36 | 31523–31939 (139) | + | Multiple Zn finger protein | None | NA | Proteins containing multiple Zn fingers are generally typical of eukaryotes |
| 37 | 31939–32139 (67) | + | C2H2 Zn finger protein | 2501713_RATNO (8e-08) | Eukaryotic | The first identification of this type of protein in prokaryotes |
| 38 | 32146–32358 (71) | − | Zn ribbon protein | 3258341_PYRHO (0.045) | Euryarchaeotal | |
| 39 | 32392–32697 (102) | − | Ribosomal protein S10 | 1350915_HALHA (9e-18) | Euryarchaeotal | |
| 40 | 32709–34004 (432) | − | EF-1α | 1361925_DESMO (e-128) | Crenarchaeotal | |
| 41 | 34134–35270 (379) | + | Uncharacterized conserved protein | 2984088_AQUAE (4e-79) | Euryarchaeotal or crenarchaeotal | Highly conserved in most archaea and Aquifex |
| 42 | 35338–35745 (136) | + | Uncharacterized secreted (periplasmic) protein | None | NA | |
| 43 | 35786–37552 (589) | + | ATP-dependent DNA ligase | 4099066_PYRAE (e-120) | Crenarchaeotal or euryarchaeotal | |
| 44 | 37622–38272 (217) | + | Coiled-coil protein | 2622819_METH (3e-08) | Euryarchaeotal | The only detectable ortholog is in Methanobacterium |
| 45 | 38368–38901 (178) | + | tRNA intron endonuclease | 5105334_AERPE (4e-22) | Crenarchaeotal | |
| 46 | 38904–39524 (207) | − | Uncharacterized protein | None | NA | |
| 47 | 39614–41383 (590) | + | Rossmann fold oxidoreductase, possibly glucose-1-dehydrogenase | 3599392_CENSY (0.0) | Bacterial | Other than in Cenarchaeum, all other strongly similar homologs are from bacteria |
| 48 | 41389–41835 (149) | + | Predicted acyl dehydratase | 7592627_STAAU (0.82) | Bacterial | |
| 49 | 41957–42727 (257) | − | Predicted permease | 2635735_BACSU (1e-29) | Bacterial or euryarchaeotal | |
| 50 | 42771–43397 (209) | − | Uncharacterized extracellular (periplasmic) protein | None | NA | |
| 51 | 43565–44038 (>158) | − | Uncharacterized conserved protein | 4982243_THEMA (8e-11) | Bacterial | Distant orthologs are detected in euryarchaeotes |
NA, not applicable; HTH, helix-turn-helix, TPR, tetracopeptide repeat; NTP, nucleoside triphosphate; SAM, S-adenosylmethionine; HIT, histidine triad.
Dir, direction of transacription, namely, rightward (+) or leftward (−).
The proteins are designated by their gene identification numbers (GenBank) followed by the abbreviated species name: AERPE, Aeropyrum pernix; ARATH, Arabidopsis thaliana; AQUAE, Aquifex aeolicus; ARCFU, Archaeoglobus fulgidus; BACSU, Bacillus subtilis; CENSY, Cenarchaeum symbiosum; CHLTR, Chlamydia trachomatis; DESMO, Desulfurococcus mobilis; DROME, Drosophila melanogaster; HALHA, Halobacterium halobium; METJA, Methanococcus jannaschii; METTH, Methanobacterium thermoautotrophicum; PYRAB, Pyrococcus abissi; PYRHO, P. horikoshii; RATNO, Rattus norvegicus; RICPR, Rickettsia prowazekii; SULSH, Sulfolobus shibatae; SULSO, S. solfataricus; THELI, Thermococcus litoralis; THEMA, Thermotoga maritima.
A partial analysis of a genome fragment from a marine group I crenarchaeote recovered from a depth of 200 m near the coast of Oregon (e.g., fosmid 4B7) was previously reported (42). To better compare the genome fragment of this subsurface archaeon to that of Antarctic fosmid 74A4, the entire 42-kb sequence of fosmid 4B7 was determined. Protein and rRNA coding genes from fosmid 4B7 are shown in Table 2. The average G+C contents for fosmids 74A4 and 4B7 were 32 and 34%, respectively, significantly different from the 55.6 and 57.1% reported for C. symbiosum (41). Genes on fosmids 74A4 and 4B7 appeared to be densely packed, as reported for other archaeal genomes, and homologs of archaeal genes were dispersed evenly over each of the inserts, strongly suggesting that the 74A4 and 4B7 inserts are contiguous genomic fragments derived from a marine archaeon. Several genes present on each of the analyzed marine archaeal fosmids showed a clear bacterial affinity (Tables 1 and 2; see also discussion below). We discuss below the predicted genes classified by functional categories.
TABLE 2.
Predicted rRNA and protein coding genes in marine crenarchaeal fosmid 4B7a
| 74A4# | Nucleotide range (protein size, in no. of amino acids) | Dir | Predicted function | Most similar homolog (e-value) | Apparent phylo-genetic affinity | Comments |
|---|---|---|---|---|---|---|
| 1 | 371–1459 (363) | + | Damage-induced, low-processivity DNA polymerase IV (DinB family) | 1706953_SULSO (5e-48) | Crenarchaeotal (Sulfolobus) or bacterial | So far not found in other archaea |
| 2 | 1449–2384 (312) | − | Uncharacterized multitransmembrane protein | None | NA | |
| 3 | 2426–3580 (385) | − | Uncharacterized conserved protein | 5457859_PYRAB (4e-71) | Euryarchaeotal | Protein family so far detected only in archaea; highly conserved paralog of 74A4#7 |
| 4 | 3671–4588 (306) | + | Periplasmic solute-binding protein | 7472739_DEIRA (3e-38) | Bacterial or euryarchaeotal | So far found only in archaea; highly conserved paralog of 74A4#6 |
| 5 | 4765–5034 (90) | − | Small membrane protein | None | NA | |
| 6 | 5246–5914 (223) | − | Periplasmic disulfide bond isomerase | 2649220_ARCFU (2e-16) | Euryarchaeotal or bacterial | Among archaea, found only in Archaeoglobus |
| 7 | 6272–7624 (451) | + | DEAD family RNA helicase | 10175004_BACHA (3e-73) | Bacterial or euryarchaeotal | Among archaea, found only in Methanobacterium |
| 8 | 7673–7906 (78) | + | Transcriptional regulator of the AsnC family | 3257044_PYRHO (3e-08) | Euryarchaeotal | |
| 9 | 8294–8734 (147) | + | Uncharacterized protein | None | NA | |
| 10 | 8769–9530 (254) | + | Uncharacterized secreted (periplasmic) protein | None | NA | |
| 11 | 9566–9847 (94) | + | Uncharacterized membrane protein | None | NA | |
| 12 | 9885–10202 (106) | + | Uncharacterized protein | None | NA | |
| 13 | 10205–12226 (674) | − | Highly conserved protein containing a thioredoxin domain | 586842_BACSU (e-141) | Bacterial | More distant homologs are seen in archaea |
| 14 | 12502–13218 (239) | − | Uncharacterized multitransmembrane protein | None | NA | |
| 15 | 13459–15651 (731) | − | EF2 | 461998_SULSO (0.0) | Crenarchaeotal | |
| 16 | 16479–16907 (143) | − | Uncharacterized protein | None | NA | Distantly related to the N-terminal portion of 4B7#19 |
| 17 | 17253–17567 (105) | − | Rhodanese-related sulfurtransferase or oxidoreductase | 1723283_SCHPO (2e-06) | Uncertain | So far not found in crenarchaeotes |
| 18 | 17595–17975 (127) | + | Uncharacterized protein | None | ||
| 19 | 17976–18818 (281) | − | Type I membrane protein containing a C-terminal cytoplasmic J domain (DnaJ) | 544179_METMA (2e-12) | Euryarchaeotal or bacterial | Unique domain architecture |
| 20 | 19397–21106 (570) | − | SWI/SNF family helicase | 3329163_CHLTR (1e-43) | Bacterial | Distinct subfamily of SWI/SNF helicases so far not seen in archaea |
| 21 | 21169–21903 (240) | − | Uncharacterized protein homologous to the N-terminal domain of Lon protease | 11354694_VIBCH (2e-08) | Bacterial | Domain potentially involved in proteolysis regulation; so far found in bacteria and eukaryotes but not in archaea |
| 22 | 21970–22482 (173) | + | Uncharacterized coiled-coil protein | None | NA | |
| 23 | 22485–23483 (333) | + | Bifunctional protein: biotin repressor and biotin (acetyl-CoA carboxylase) ligase | 773349_BACSU (1e-45) | Bacterial or euryarchaeotal | Typical bacterial domain architecture, with N-terminal DNA-binding domain and C-terminal enzymatic domain |
| 24 | 23549–25069 (507) | + | Secreted (periplasmic) Zn-dependent protease containing TPR repeats | 2688100_BORBU (3e-07) (TPR repeats), 5921827_RATNO (0.002) (protease) | Uncertain | The TPR repeats most closely resembles those found in euryarchaeotes; no close homologs of the predicted protease; paralog of 74A4#15 |
| 25 | 25072–25647 (192) | − | Ferritin (nonheme iron-containing protein) | 1707687_SULSO (e-49) | Crenarchaeotal | |
| 26 | 26062–26694 (211) | + | Transcription factor IIB | 1729909_SULSH (9e-52) | Crenarchaeotal | |
| 27 | 26866–27213 (116) | + | Uncharacterized nonglobular protein | None | NA | |
| 28 | 27359–27661 (101) | + | Uncharacterized protein | None | NA | |
| 29 | 27735–27956 (74) | − | Small membrane protein | None | NA | Paralog of 74A4#3 |
| 30 | 28121–28312 (64) | + | RNA-binding cold shock protein | 3097243_MYCLE (2e-13) | Bacterial | So far not seen in other archaea |
| 31 | 23S RNA | |||||
| 32 | 16S RNA | |||||
| 33 | 28470–28673 (68) | + | Small Cys- and His-rich protein | None | NA | Potentially involved in metal-dependent nucleic acid binding |
| 34 | 33434–34744 (437) | + | Glutamate-1-semialdehyde aminotransferase | 3599375_CENSY (e-158) | Euryarchaeotal or crenarchaeotal | Highly conserved paralog of 74A4#24 |
| 35 | 34782–35432 (217) | + | NAD (P) H-flavin oxidoreductase | 3915352_ARCFU (4e-11) | Euryarchaeotal | |
| 36 | 35435–35830 (158) | − | Uncharacterized membrane protein | None | NA | Paralog of 74A4#19 |
| 37 | 35989–36483 (165) | − | Uncharacterized conserved protein | 6015921_SULSO (2e-21) | Crenarchaeotal | Found in archaea and eukaryotes |
| 38 | 36518–37099 (194) | − | Uncharacterized protein | None | NA | |
| 39 | 37092–39980 (963) | − | Very large secreted (periplasmic) protein | None | NA | |
| 40 | 40120–40311 (64) | − | Zn ribbon protein | None | NA | Potentially involved in metal-dependent nucleic acid binding |
| 41 | 40653–41327 (225) | + | Methionine aminopeptidase | 3257034_PYRHO (3e-40) | Euryarchaeotal |
See footnotes to Table 1.
Transcription and translation.
Homologs of well-characterized components of the transcription system were found only on fosmid 4B7. These include a predicted SWI/SNF family ATPase, transcription factor IIB, an RNA-binding cold shock protein, and a predicted AsnC family transcriptional regulator. Three genes from 74A4 encode different types of Zn finger or Zn ribbon proteins that also could function as transcriptional regulators. Of particular interest is a small C2H2 Zn finger protein that belongs to a family that so far has been identified only in eukaryotes. Both fosmids contain several genes involved in translation. These include a single tRNA gene on 4B7 and a typical crenarchaeotal rRNA operon (16S-spacer-23S) on both fosmids. Protein components of the translation apparatus encoded on these clones include a cysteinyl-tRNA synthetase which is highly similar to other archaeal cysteinyl-tRNA synthetases, EF1-α, and ribosomal protein S10 on 74A4 and elongation factor 2 (EF2) on 4B7. The genes for EF1-α and S10 form a cluster that so far has not been detected in any other genomes. Another protein potentially involved in translation is a predicted RNA helicase encoded on 4B7.
DNA replication and repair.
One DNA repair enzyme, DinB/UmuC, recently identified as a repair-associated DNA polymerase (46), was identified on 4B7 and is most similar to the Dbh protein found in Sulfolobus solfataricus (22). Two other typical archaeal enzymes involved in replication and repair, DNA ligase (ATP dependent) and tRNA intron endonuclease, were identified on 74A4.
Transport and energy metabolism.
Both marine archaeal fosmids encoded several proteins implicated in energy conversion, particularly fatty acid metabolism. These included 3-hydroxyacyl-coenzyme A (CoA) dehydrogenase, acyl dehydratase, a predicted CoA-binding protein, glucose-1-phosphate dehydrogenase, and some other, poorly characterized oxidoreductases. Only one protein, a periplasmic solute-binding protein homologous to those found in iron(III) ATP-binding cassette transporters, was clearly assigned as a protein involved in transport. Other membrane transporters were tentatively identified on the basis of transmembrane segment predictions.
Miscellaneous proteins.
It was noted previously that moderately thermophilic archaea, such as Methanobacterium thermoautotrophicum or Methanosarcina barkeri, encode classical molecular chaperones of the hsp70 (DnaK) and hsp40 (DnaJ) families, whereas archaeal hyperthermophiles do not have those proteins (25). In agreement with this trend, two predicted marine archaeal proteins, 74A4#31 and 4B7#19, contain the J domain of the hsp40 family of chaperones and parts of the heat shock chaperone DnaJ, which interacts with and stimulates the hydrolysis of ATP by the cognate DnaK proteins (47). Protein 74A4#31 also contains a ferredoxin domain that is predicted to bind iron or possibly other ions and might be functionally analogous to the Zn clusters that are present in bacterial and eukaryotic DnaJ proteins. A J domain-ferredoxin fusion has not been reported so far. In contrast, protein 4B7#19 is predicted to be a type I membrane protein in which the J domain is the C-terminal cytoplasmic portion. Another interesting pair of paralogous proteins are 74A4#15 and 4B7#24, which are predicted membrane-associated, collagenase-like, metal-dependent proteases. A cluster of three genes on 74A4 encodes three predicted enzymes of the double-stranded beta-helix fold that might possess a variety of enzymatic activities, for example, that of sugar-phosphate isomerase.
Comparative genomic analyses of marine group I archaea.
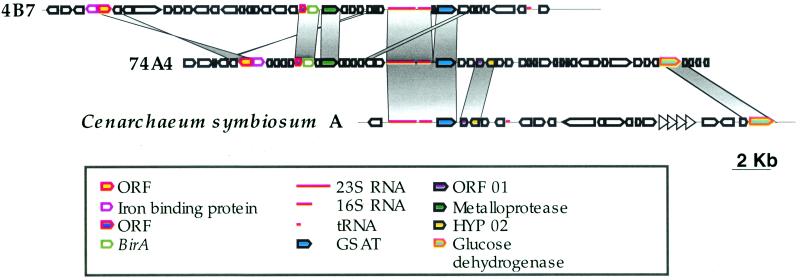
Protein and RNA gene organizations on fosmids 74A4 and 4B7 were compared to that of the C. symbiosum (variant A) fosmid (41) by using the BLAST 2 Sequences program (43). The 23S and 16S rDNA sequences were used as an alignment point for the sequences (Fig. 3). The typical crenarchaeotal rRNA operon (16S-23S) was shared by all three fosmids, and the operon was adjacent to the GSAT gene in all. Fosmids 74A4 and 4B7 shared two other regions in common. The first region includes an unknown open reading frame, the BirA protein gene, and a hypothetical metalloprotease gene. The second region represents an inversion between the two genomes and includes genes for a putative periplasmic-binding protein of an iron(III) ATP-binding cassette transporter and a neighboring hypothetical protein (Fig. 3). Fosmid 74A4 and the fosmid from C. symbiosum shared a region including hypothetical protein 02 and the product of ORF01, previously reported only for C. symbiosum (41), and a putative glucose dehydrogenase which was also reported only for C. symbiosum (GenBank accession number AAC62698).
FIG. 3.
Genomic organization in planktonic marine crenarchaeotes. Gene maps for fosmids 4B7 (42) and 74A4 and C. symbiosum A (35) were aligned based on ribosomal 16S and 23S sequences. Homologous regions are connected with lines.
DISCUSSION
This study was conducted to explore the use of large-scale genome sequence analysis to better describe the population genetics, genome content, and biological properties of naturally occurring, uncultivated microbial species. We focused on the description of an abundant but little understood component of marine plankton, the pelagic crenarchaeotes. Our approach facilitated (i) description of the structure and gene content of large genomic regions flanking the rRNA operon from uncultivated marine archaea, (ii) cross comparison of large genomic regions of allopatric marine crenarchaeotes (from Antarctica, deep temperate waters, and a marine sponge), and (iii) assessment of the genomic heterogeneity of sympatric crenarchaeotes that originate from the same population and that share identical or nearly identical rRNA sequences.
The 16S rRNA sequences of all five unique archaeal fosmids from the Antarctic picoplankton library were highly similar. One (15G10) was identical in sequence to a PCR-amplified 16S rRNA gene (ANTARCTIC 12) isolated from the same Antarctic waters in 1993 (3 years prior to the sampling of this report) (14). The sympatric archaeal 16S rRNA genes differed from one another by a maximum of three nucleotide substitutions over 1,418 nucleotide residues. However, the different restriction fragment length polymorphism patterns of the fosmids could not be explained solely by the distance between the rRNA operon and the cloning sites and suggested significant sequence divergence between these highly related variants. Since it was possible that the protein sequences and gene organization were less conserved than rRNA (as recently observed for natural variants of C. symbiosum) (41), we characterized the GSAT gene from the different clones. Microheterogeneity in the DNA sequence of the GSAT gene was observed for all clones, including 31B2 and 83A10, which were identical over the entire 16S rRNA gene. To our knowledge, this is one of the first studies that directly links 16S rRNA gene variation to heterogeneity in flanking protein coding genes in sympatric free-living microbes. Our study shows that within a single microbial population, considerable genomic variation exists, even among microbes with identical 16S rRNA gene sequences.
Sequence analysis of the 43- and 42-kb genome fragments derived from marine archaea from two different oceanic provinces showed many features typical of the domain Archaea. The majority of the genes identified, including those whose functions could not be predicted, most closely resembled archaeal protein genes (data not shown). Some features of these genome sequences, including the rRNA gene order, chromosomal organization, and nucleotide sequence of the genes for EF1-α and ribosomal protein S10, resembled those of other Crenarchaeota (7, 16, 17). The observation that the GSAT gene is located downstream of the ribosomal operon in all planktonic marine crenarchaeotes analyzed to date (41, 42; this study) also suggests some chromosomal organization common to marine group I crenarchaeotes. Specific gene sequences that we recovered might provide further insight into the relationship of marine crenarchaeotes to other cultivated species. For instance, EF1-α (EF-Tu in Bacteria) is a highly conserved protein that is found in all cellular organisms and that has proven extremely useful for global phylogenetic comparisons (2, 8, 37). Both distance and parsimony analyses of the EF1-α amino acid sequence derived from Antarctic fosmid 74A4, however, could not resolve its placement within the Crenarchaeota or Euryarchaeota (data not shown). Other important features of EF1-α are specific insertions and deletions among homologs that provide evidence for a specific evolutionary linkage between eukaryotes and crenarchaeotes. Antarctic crenarchaeotal EF1-α did contain an 11-amino-acid insertion (data not shown) that is characteristic of Eucarya and Crenarchaeota but not Euryarchaeota or bacteria (34). The sequence of the 11-amino-acid insertion of 74A4 most closely resembles the insertion of Pyrobaculum aerophilum, another crenarchaeote (four-amino-acid sequence difference; data not shown). This observation, together with the deep branching of planktonic archaeal 16S rRNA and of the EF2 amino acid sequence of fosmid 4B7 (42) in phylogenetic trees, could reflect a nonthermophilic origin of the crenarchaeotal subdivision. Alternatively, EF1-α homologs from as-yet-uncultured thermophilic relatives of low-temperature crenarchaeotes that have been detected in hot springs (3, 4) may branch more deeply, placing these thermophilic groups basal to cultivated crenarchaeotal lineages.
Small cold shock proteins were believed to be present only in bacteria and eukaryotes (18, 33). To date, no cold shock genes have been found in the archaeal genomes that have been entirely sequenced. It was therefore surprising to find a gene encoding a cold shock protein on fosmid 4B7. Based on amino acid similarity, this putative cold shock protein resembles those of bacteria. The observation that no other sequenced archaeal genomes encode members of the small cold shock protein family raises the possibility of lateral gene transfer of this gene into cold-adapted archaea from bacteria. Several other genes present on the two marine archaeal fosmids may have been acquired from bacteria, for example, the genes for double-stranded beta-helix fold proteins, the SWI/SNF helicase, and peptide methionine sulfoxide reductase. Even more unexpectedly, we identified a gene coding for a C2H2 Zn finger protein that so far has been found only in eukaryotes.
Analysis of the combined 80 kb of sequence data has identified several genes indicative of metabolic pathways (Tables 1 and 2). However, the lack of known transporters on the two fosmids makes it difficult to predict possible components being taken up by the archaeal cells. Additional data obtained for C. symbiosum and from other techniques, such as stable isotope and natural radiotracer analyses (34) and microautoradiography and fluorescence in situ hybridization (23, 31), should provide more insight into the potential metabolic traits of uncultivated marine archaea.
With regard to the marine planktonic crenarchaeotal clade in general, there exists considerable divergence and genome evolution. The 16S rRNA genes in fosmids 4B7 and 74A4 and C. symbiosum all share greater than 94% sequence similarity. However, the regions surrounding the rRNA operons vary substantially, indicating extensive genome rearrangements and various genome contents. These differences are also likely reflected in the phenotypic properties of the different crenarchaeotes that occupy the different oceanic regions.
Variation among homologous protein coding genes from microbes that share moderately similar (97%) 16S rRNA gene sequences has been reported for Prochlorococcus isolates derived from the same sample. Prochlorococcus isolates MED and SS120 shared 98% sequence similarity in their 16S rRNAs (44) but were only 76% identical based on their RNA polymerase C1 gene sequence (15). To our knowledge, however, genome variation among free-living, sympatric, uncultivated microbes has never been reported. Our data now provide a significant perspective on the extent of genome variation that can exist within a single population of free-living microbial cells that share identical (or nearly so) rRNA gene sequences. Our data suggest that the observed seasonal maximum of planktonic crenarchaeotes in Antarctic waters (29) is composed of (minimally) four highly related, yet nonidentical, co-occurring strains or variants. Of course, due to the labor and resource intensiveness of our procedures, our library screening procedure severely undersamples the actual population. Despite this undersampling, however, we did not recover any one dominant or identical genotype. Rather, we recovered identical or nearly identical rRNA phylotypes with significant differences in flanking genomic regions. Greater variation would be expected to be observed with larger sample size. These data strongly suggest that even within a single population, a very large amount of genomic heterogeneity exists that is undetectable by 16S rRNA sequence variation.
Presumably, genomic microheterogeneity can generate, eventually, physiological diversity. Even small variations among protein coding genes, such as those found here in sympatric archaeal cells that share identical or nearly identical rRNA gene sequences, could provide a selective advantage to the different genotypes under fluctuating environmental conditions. Such microvariations could confer greater fitness to the population as a whole under various environmental conditions, relative to any individual clonal phenotype. Our data strongly suggest that naturally occurring populations of bacteria and archaea can be viewed as nonclonal populations that harbor tremendous allelic variation.
Acknowledgments
We thank Chris Preston for advice and helpful discussions.
This work was supported by NSF grants OPP94-18442 and OCE0001619 and the David and Lucile Packard Foundation to E.F.D. O.B. was supported by a fellowship from the European Molecular Biology Organization. D.C.B., R.V.S., R.A.F., and J.L.S. were supported by Diversa Corporation.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldauf, S. L., J. D. Palmer, and W. F. Doolittle. 1996. The root of the universal tree and the origin of eukaryotes based on elongation factor phylogeny. Proc. Natl. Acad. Sci. USA 93:7749–7754. [DOI] [PMC free article] [PubMed]
- 3.Barns, S. M., C. F. Delwiche, J. D. Palmer, and N. R. Pace. 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA 93:9188–9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Béjà, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902–1906. [DOI] [PubMed] [Google Scholar]
- 6.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516–529. [DOI] [PubMed] [Google Scholar]
- 7.Ceccarelli, E., M. Bocchetta, R. Creti, A. M. Sanangelantoni, O. Tiboni, and P. Cammarano. 1995. Chromosomal organization and nucleotide sequence of the genes for elongation factors EF-1 alpha and EF-2 and ribosomal proteins S7 and S10 of the hyperthermophilic archaeum Desulfurococcus mobilis. Mol. Gen. Genet. 246:687–696. [DOI] [PubMed] [Google Scholar]
- 8.Creti, R., E. Ceccarelli, M. Bocchetta, A. M. Sanangelantoni, O. Tiboni, P. Palm, and P. Cammarano. 1994. Evolution of translational elongation factor (EF) sequences: reliability of global phylogenies inferred from EF-1 alpha(Tu) and EF-2(G) proteins. Proc. Natl. Acad. Sci. USA 91:3255–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong, E. F. 1998. Everything in moderation: archaea as ‘non-extremophiles.’ Curr. Opin. Genet. Dev. 8:649–654. [DOI] [PubMed] [Google Scholar]
- 11.DeLong, E. F. 2001. Microbial seascapes revisited. Curr. Opin. Microbiol. 4:290–295. [DOI] [PubMed] [Google Scholar]
- 12.DeLong, E. F., L. L. King, R. Massana, H. Cittone, A. Murray, C. Schleper, and S. G. Wakeham. 1998. Dibiphytanyl ether lipids in nonthermophilic crenarchaeotes. Appl. Environ. Microbiol. 64:1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLong, E. F., K. Y. Wu, B. B. Prezelin, and R. V. Jovine. 1994. High abundance of Archaea in Antarctic marine picoplankton. Nature 371:695–697. [DOI] [PubMed] [Google Scholar]
- 15.Ferris, M. J., and B. Palenik. 1998. Niche adaptation in ocean cyanobacteria. Nature 396:226–228. [Google Scholar]
- 16.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93–113. [DOI] [PubMed] [Google Scholar]
- 17.Garrett, R. A., J. Dalgaard, N. Larsen, J. Kjems, and A. S. Mankin. 1991. Archaeal rRNA operons. Trends Biochem. Sci. 16:22–26. [DOI] [PubMed] [Google Scholar]
- 18.Graumann, P. L., and M. A. Marahiel. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23:286–290. [DOI] [PubMed] [Google Scholar]
- 19.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510. [DOI] [PubMed] [Google Scholar]
- 21.Kawata, Y., S. Yano, and H. Kojima. 1998. Construction of a genomic DNA library by TA cloning. BioTechniques 24:564–565. [DOI] [PubMed] [Google Scholar]
- 22.Kulaeva, O. I., E. V. Koonin, J. P. McDonald, S. K. Randall, N. Rabinovich, J. F. Connaughton, A. S. Levine, and R. Woodgate. 1996. Identification of a DinB/UmuC homolog in the archaeon Sulfolobus solfataricus. Mutat. Res. 357:245–253. [DOI] [PubMed] [Google Scholar]
- 23.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K. H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macario, A. J., and E. Conway de Macario. 1999. The archaeal molecular chaperone machine. Peculiarities and paradoxes. Genetics 152:1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massana, R., E. F. DeLong, and C. Pedros-Alio. 2000. A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl. Environ. Microbiol. 66:1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massana, R., A. E. Murray, C. M. Preston, and E. D. DeLong. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara channel. Appl. Environ. Microbiol. 63:50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massana, R., L. T. Taylor, A. E. Murray, K. Y. Wu, W. H. Jeffrey, and E. F. DeLong. 1998. Vertical distribution and temporal variation of marine planktonic archaea in the Gerlache Strait, Antarctica, during early spring. Limnol. Oceanogr. 43:607–617. [Google Scholar]
- 29.Murray, A. E., C. M. Preston, R. Massana, L. T. Taylor, A. Blakis, K. Wu, and E. F. DeLong. 1998. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl. Environ. Microbiol. 64:2585–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6. [DOI] [PubMed] [Google Scholar]
- 31.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740. [DOI] [PubMed] [Google Scholar]
- 33.Phadtare, S., J. Alsina, and M. Inouye. 1999. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2:175–180. [DOI] [PubMed] [Google Scholar]
- 34.Preston, C. M. 1998. Prokaryotic diversity in marine sponges: a description of a specific association between the marine archaeon, Cenarchaeum symbiosum, and the marine sponge, Axinella mexicana. Ph.D. thesis. University of California, Santa Barbara.
- 35.Preston, C. M., K. Y. Wu, T. F. Molinski, and E. F. DeLong. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl. Acad. Sci. USA 93:6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivera, M. C., and J. A. Lake. 1992. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 257:74–76. [DOI] [PubMed] [Google Scholar]
- 37.Roger, A. J., O. Sandblom, W. F. Doolittle, and H. Philippe. 1999. An evaluation of elongation factor 1 alpha as a phylogenetic marker for eukaryotes. Mol. Biol. Evol. 16:218–233. [DOI] [PubMed] [Google Scholar]
- 38.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rost, B., R. Casadio, P. Fariselli, and C. Sander. 1995. Transmembrane helices predicted at 95% accuracy. Protein Sci. 4:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5:1704–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schleper, C., E. F. DeLong, C. M. Preston, R. A. Feldman, K. Y. Wu, and R. V. Swanson. 1998. Genomic analysis reveals chromosomal variation in natural populations of the uncultured psychrophilic archaeon Cenarchaeum symbiosum. J. Bacteriol. 180:5003–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein, J. L., T. L. Marsh, K. Y. Wu, H. Shizuya, and E. F. DeLong. 1996. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178:591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247–250. [DOI] [PubMed] [Google Scholar]
- 44.Urbach, E., D. J. Scanlan, D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 1998. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 46:188–201. [DOI] [PubMed] [Google Scholar]
- 45.Vergin, K. L., E. Urbach, J. L. Stein, E. F. DeLong, B. D. Lanoil, and S. J. Giovannoni. 1998. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64:3075–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, J., P. Gruz, S. R. Kim, M. Yamada, K. Matsui, R. P. Fuchs, and T. Nohmi. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4:281–286. [DOI] [PubMed] [Google Scholar]
- 47.Wall, D., M. Zylicz, and C. Georgopoulos. 1994. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J. Biol. Chem. 269:5446–5451. [PubMed] [Google Scholar]
- 48.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]