Abstract
The effects of single-base-pair near-terminal and terminal mismatches on the dissociation temperature (Td) and signal intensity of short DNA duplexes were determined by using oligonucleotide microarrays and neural network (NN) analyses. Two perfect-match probes and 29 probes having a single-base-pair mismatch at positions 1 to 5 from the 5′ terminus of the probe were designed to target one of two short sequences representing 16S rRNA. Nonequilibrium dissociation rates (i.e., melting profiles) of all probe-target duplexes were determined simultaneously. Analysis of variance revealed that position of the mismatch, type of mismatch, and formamide concentration significantly affected the Td and signal intensity. Increasing the concentration of formamide in the washing buffer decreased the Td and signal intensity, and it decreased the variability of the signal. Although Tds of probe-target duplexes with mismatches in the first or second position were not significantly different from one another, duplexes with mismatches in the third to fifth positions had significantly lower Tds than those with mismatches in the first or second position. The trained NNs predicted the Td with high accuracies (R2 = 0.93). However, the NNs predicted the signal intensity only moderately accurately (R2 = 0.67), presumably due to increased noise in the signal intensity at low formamide concentrations. Sensitivity analysis revealed that the concentration of formamide explained most (75%) of the variability in Tds, followed by position of the mismatch (19%) and type of mismatch (6%). The results suggest that position of the mismatch at or near the 5′ terminus plays a greater role in determining the Td and signal intensity of duplexes than the type of mismatch.
DNA microarray technology provides parallel nucleic acid hybridizations for hundreds to thousands of oligonucleotides or larger DNA fragments on a small surface area (35). In applied and environmental microbiology, this technology has been used for assessing gene expressions (31, 33, 40), characterizing whole genomes (8), and identifying bacteria (11). This technology assumes an adequate discrimination between probes and their targets, which is determined in part by the stability of probe-target duplexes and by hybridization and wash conditions.
The stability of any DNA duplex can be determined by monitoring the nonequilibrium dissociation rate (i.e., melting profile) and then calculating the dissociation temperature (Td) of the probe-target duplex. Acquiring information on how Td changes for a particular DNA duplex under different hybridization and washing conditions allows us to maximize discrimination by minimizing nonspecific hybridizations. However, complete discrimination is often difficult to achieve, particularly for short probe-target duplexes (∼18 to 25 nucleotides) with single-base-pair mismatches. Accurately predicting the Td of short probe-target duplexes for defined washing conditions, probe and target lengths, and nucleotide composition and order is a substantial challenge. Understanding the rules governing nucleic acid hybridizations of short probe-target duplexes is necessary and fundamental for the application of microarray technology to routine environmental microbiology because it would facilitate the design of good probes (i.e., those which have high specificity), minimize the chances of nonspecific hybridizations, and improve our ability to confidently interpret microarray data.
There is a paucity of data dealing with the Tds of short DNA duplexes, and even less is known about duplexes containing terminal mismatches (34). In general, a mismatch near or at the terminus of a short duplex is less destabilizing than an internal mismatch (38). However, experimental observations have shown that in at least some instances, the type of a mismatch (i.e., the potential loss of hydrogen bonds due to incorrect base pairing) can override the effects of position (39). Hence, there are no absolute rules for predicting the influence of mismatch position on duplex stability (38). The effects of the position of a single-base-pair mismatch and the type of mismatch on predicting Td of a probe-target duplex are nonlinear, with position being more important in some instances and type of mismatch being more important in others. Determining the rules controlling nonlinear factors is further hampered by conventional statistical methods (e.g., analysis of variance [ANOVA]) which use linear approaches to examine nonlinear data. An approach that provides the advantages of conventional statistics while accounting for the nonlinear nature of the experimental data and facilitating a more detailed examination of the data would be very advantageous.
Artificial neural networks (NNs) provide a useful tool for recognizing patterns in complex, nonlinear data sets such as those associated with predicting the Td of specific probe-target duplexes. NNs are particularly advantageous over conventional statistical methods because they can deal with the inherent variability associated with biological data. NNs are constructed by using computer software and consist of layers of neurons that make independent computations and pass on their outputs to other neurons (28). Each neuron in a layer is connected to neurons in the next layer, so that the output of each neuron affects the activation of all neurons to which it is connected. Neurons are adaptable and, through the process of learning from examples, store knowledge and make it available for use (1). In a training technique called back-propagation (32), a pattern is presented to an input layer of a network and the network produces output based on the sum of the weighted inputs. When the pattern of the output layer is compared to target values, the errors between them are computed. An error function is used to adjust the weights and biases of each neuron. The adjusted weights of a trained network can be used to recognize and predict patterns such as the Td of probe-target duplexes. The adjusted weights can also be used to provide information on functional relations between variables and an output. For example, sensitivity analysis of the adjusted weights can be used to determine the relative contribution of individual input neurons (i.e., position of the mismatch, type of mismatch, and formamide concentration) to an output neuron (e.g., Td).
The objective of this study was to investigate the effects of single-base-pair mismatches on Td and signal intensity of oligonucleotide duplexes using polyacrylamide gel-based DNA microarrays. Specifically, we examined the effects of mismatch type, position of the mismatch relative to the 5′ terminus of the probe, and formamide concentration in the washing buffer on Td and signal intensity using 31 oligonucleotide probes which target DNA sequences for 16S rRNA (rDNA). A second objective was to extract general rules establishing the relative contribution of each of these variables to duplex stability as needed for more informed probe design. To this end, we report on the use of a back-propagating NN for dealing with the high variability and nonlinearity of this data set. Sensitivity analysis of the trained NNs identified the factors that most contributed to training the NNs.
MATERIALS AND METHODS
Synthesized target DNA.
The 16S rDNA gene sequences of Staphylococcus epidermidis (accession numbers L37605 and X75943), which is known as an important opportunistic pathogen associated with infections of synthetic medical devices (14), and Nitrosomonas eutropha (accession number M96402, which is a well-studied lithoautotrophic nitrifying bacterium (15, 39), were obtained from GenBank at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/GenBank). A short section of these sequences was custom-synthesized as single-stranded DNA and fluorescently labeled with Cy3 at the 5′ terminus (Operon Technologies Inc., Alameda, Calif.). In addition, the length of the target DNA was extended by 10 nucleotides on both ends of the probe-binding site with flanking 16S rDNA sequence to promote increased stability of the probe-target duplex (41). The names, compositions, sizes, positions of the target using Escherichia coli numbering, and probe-binding sites (underlined) are as follows: for the S. epidermidis target, 5′-TCTGGTCTGTAACTGACGCTGATGTGCGAAAGCGTGGGG-3′ (39 nt, positions 737 to 775), and for the N. eutropha target, 5′-ACTACAAAGCTAGAGTGCAGCAGAGGGGAGTGGAATTC-3′ (38 nt, positions 643 to 680).
Oligonucleotide probe synthesis and oligonucleotide array fabrication.
A 19-base oligonucleotide probe (S-G-Staph-0747-a-A-19) targeting the genus Staphylococcus was designed by using the probe design function of ARB software (http://www.mikro.biologie.tu-muenchen.de). The specificity of the probe for the target was checked with the probe check function in the ARB software, the BLAST search (3) at the National Center for Biotechnology Information, and the Probe Match program in Ribosomal Database Project II (17). Self-complementarities were also examined by Ribosomal Database Project II. An 18-base oligonucleotide probe (S-✻-Nsom-0653-a-A-18) targeting halotolerant and obligately halophilic Nitrosomonas, which was designed and previously reported as NEU, was also used (43). Oligonucleotide probes were synthesized with an amino linker at the 3′ end at Argonne National Laboratory (5). Mismatch probes were designed to have various single-base-pair mismatches in the first to fifth positions from the 5′ terminus of the probe (Table 1). The microarray matrix, containing 100- by 100- by 20-μm polyacrylamide gel pads placed 100 μm from each other and fixed to a glass slide, was manufactured by photopolymerization (10) and activated as described previously (27). A total of 3 nl of 1 mM amino-oligonucleotide solutions was applied to each gel element containing aldehyde groups (42) which were designed and implemented by a robot arrayer (44). A total of 31 oligonucleotide probes were immobilized through reductive coupling of the 3′ amino group of the oligonucleotide with the aldehyde group of the activated gel pad on the microarrays (27).
TABLE 1.
Probes used in this study and effect of formamide concentration, position, and type of mismatch on Td
| Probea | Sequenceb |
Td with formamide concn (%) in washing bufferc
|
||||
|---|---|---|---|---|---|---|
| 0 | 10 | 20 | 30 | |||
| S. epidermidis | SPM | TCGCACATCAGCGTCAGTT | 45.4 ± 1.8 | 38.2 ± 1.7 | 34.8 ± 0.3 | 29.9 ± 1.0 |
| s1aa | aCGCACATCAGCGTCAGTT | 43.1 ± 0.8 | 37.3 ± 1.1 | 34.0 ± 0.9 | 29.0 ± 0.9 | |
| s1ga | gCGCACATCAGCGTCAGTT | 46.3 ± 1.8 | 38.8 ± 0.9 | 34.7 ± 0.7 | 29.8 ± 1.2 | |
| s1ca | cCGCACATCAGCGTCAGTT | 44.2 ± 1.1 | 37.4 ± 1.2 | 33.9 ± 0.3 | 29.2 ± 1.2 | |
| s2ag | TaGCACATCAGCGTCAGTT | 47.8 ± 1.4 | 39.5 ± 1.6 | 34.8 ± 0.7 | 29.7 ± 1.6 | |
| s2gg | TgGCACATCAGCGTCAGTT | 45.7 ± 1.2 | 37.7 ± 1.0 | 32.9 ± 0.9 | 28.4 ± 0.8 | |
| s2tg | TtGCACATCAGCGTCAGTT | 44.4 ± 1.1 | 36.6 ± 1.1 | 32.4 ± 0.8 | 27.9 ± 0.7 | |
| s3cc | TCcCACATCAGCGTCAGTT | 40.8 ± 0.4 | 33.7 ± 0.8 | 28.4 ± 1.8 | 24.3 ± 0.3 | |
| s3tc | TCtCACATCAGCGTCAGTT | 40.7 ± 0.5 | 33.9 ± 0.9 | 28.5 ± 2.0 | 25.0 ± 1.1 | |
| s3ac | TCaCACATCAGCGTCAGTT | 43.5 ± 0.4 | 36.1 ± 1.8 | 29.8 ± 1.1 | 25.4 ± 1.1 | |
| s4gg | TCGgACATCAGCGTCAGTT | 43.5 ± 0.4 | 35.8 ± 1.4 | 30.2 ± 1.1 | 25.6 ± 0.9 | |
| s4tg | TCGtACATCAGCGTCAGTT | 42.8 ± 0.5 | 35.1 ± 1.4 | 30.0 ± 1.3 | 25.4 ± 0.8 | |
| s4ag | TCGaACATCAGCGTCAGTT | 42.0 ± 0.6 | 34.4 ± 0.9 | 29.3 ± 1.9 | 24.5 ± 0.3 | |
| s5gt | TCGCgCATCAGCGTCAGTT | 44.5 ± 1.2 | 36.8 ± 1.2 | 31.5 ± 1.5 | 26.8 ± 0.8 | |
| s5ct | TCGCcCATCAGCGTCAGTT | 40.6 ± 0.6 | 33.0 ± 0.9 | 27.8 ± 2.1 | 24.2 ± 0.7 | |
| s5tt | TCGCtCATCAGCGTCAGTT | 43.2 ± 1.2 | 35.8 ± 1.9 | 29.8 ± 1.3 | 25.6 ± 1.3 | |
| N.eutropha | NPM | CCCCTCTGCTGCACTCTA | 43.0 ± 1.4 | 33.7 ± 1.3 | 30.8 ± 1.3 | 27.4 ± 0.8 |
| n1gg | gCCCTCTGCTGCACTCTA | 45.4 ± 1.4 | 37.4 ± 1.5 | 31.0 ± 0.4 | 26.9 ± 1.5 | |
| n1ag | aCCCTCTGCTGCACTCTA | 45.3 ± 1.8 | 36.2 ± 1.0 | 30.8 ± 0.8 | 27.2 ± 1.4 | |
| n1tg | tCCCTCTGCTGCACTCTA | 44.0 ± 1.8 | 35.2 ± 0.9 | 30.3 ± 0.7 | 26.6 ± 1.2 | |
| n2gg | CgCCTCTGCTGCACTCTA | 43.5 ± 1.5 | 35.8 ± 2.2 | 29.7 ± 1.1 | 25.3 ± 0.9 | |
| n2ag | CaCCTCTGCTGCACTCTA | 44.5 ± 0.8 | 36.1 ± 0.6 | 30.0 ± 0.9 | 25.7 ± 0.9 | |
| n2tg | CtCCTCTGCTGCACTCTA | 44.2 ± 0.9 | 36.6 ± 2.4 | 29.4 ± 0.9 | 25.0 ± 0.8 | |
| n3gg | CCgCTCTGCTGCACTCTA | 43.6 ± 0.9 | 36.0 ± 2.5 | 29.3 ± 1.3 | 24.8 ± 0.7 | |
| n3ag | CCaCTCTGCTGCACTCTA | 43.1 ± 1.3 | 35.3 ± 1.5 | 29.8 ± 1.3 | 25.4 ± 1.0 | |
| n3tg | CCtCTCTGCTGCACTCTA | 41.0 ± 1.3 | 33.2 ± 0.8 | 28.0 ± 1.6 | 24.1 ± 0.6 | |
| n4gg | CCCgTCTGCTGCACTCTA | 41.7 ± 0.5 | 33.5 ± 0.9 | 28.1 ± 1.4 | 22.9 ± 2.3 | |
| n4ag | CCCaTCTGCTGCACTCTA | 41.4 ± 0.8 | 33.4 ± 0.9 | 28.0 ± 1.4 | 24.3 ± 0.5 | |
| n4tg | CCCtTCTGCTGCACTCTA | 40.4 ± 1.0 | 32.7 ± 0.9 | 27.3 ± 1.6 | 23.5 ± 0.9 | |
| n5ga | CCCCgCTGCTGCACTCTA | 39.5 ± 0.8 | 32.4 ± 0.5 | 28.1 ± 1.5 | 24.9 ± 0.9 | |
| n5aa | CCCCaCTGCTGCACTCTA | 41.5 ± 0.9 | 33.3 ± 0.8 | 28.2 ± 1.4 | 24.6 ± 1.2 | |
Probe names incorporate the type of target (s, Staphylococcus; n, Nitrosomonas), position of the mismatch (positions 1 to 5 from 5′ end of probes), and the type of mismatch (probe/target). SPM and NPM, perfect match probes for Staphylococcus and Nitrosomonas targets, respectively.
Mismatches are in lower case.
Values are means ± standard deviations (n = 3).
Hybridization and washing protocols.
Hybridizations were carried out at room temperature (20°C) for 12 h in 40 μl of hybridization buffer containing 1 μg of each target DNA (final concentration, 50 ng/μl), 0.9 M NaCl, 20 mM Tris-HCl (pH 8.0), and 40% formamide. Following hybridization, the microarray was washed three times at room temperature with a washing buffer consisting of 20 mM Tris-HCl (pH 8.0), 5 mM EDTA, 4 mM NaCl, and 0, 10, 20, or 30% formamide. After the final wash, 100 μl of washing buffer was added to the washing chamber (Grace BioLabs, Bend, Oreg.) for image and melting profile analyses.
Image and melting profile analyses.
To generate melting profiles, the microarray was fixed on a thermotable mounted on the stage of a custom-designed epifluorescence microscope (State Optical Institute, St. Petersburg, Russia) and connected with a thermoelectric temperature controller (LFI-3751; Wavelength Electronics, Inc. Bozeman, Mont.) and a water bath (Cole Parmer Instruments Co., Chicago, Ill.). The microscope was equipped with appropriate fluorescence filters (Omega Optical, Brattleboro, Vt.) and a cooled charge-coupled device camera (Princeton Instruments, Trenton, N.J.) and manipulated with a software program which allows image acquisition, processing, and analysis (LabVIEW version 5.1; National Instruments Co, Austin, Tex.) (16). Melting profiles for all probes were monitored and recorded at 2°C intervals between 18 and 70°C by increasing the temperature at a rate of 1°C per min. The melting profile experiments were performed in triplicate and repeated on different days.
Td software.
The Td software was designed to automatically calculate the experimentally determined Td and signal intensity at the Td for each probe-target duplex by using data obtained from the image acquisition, processing, and analysis software. A Web-based interface for this software is available at http://stahl.ce.washington.edu. This interface contains a Td -cal.readme file, which describes how to use the software and what files and formatting are needed to make the Td calculator work (e.g., formatting of input files).
Normalization of the data was needed to compare various combinations of probe-target duplex profiles and to standardize the Td calculations. The data were normalized with the following equation: normalized value = (actual value − minimum value)/(maximum value − minimum value).
The temperature of each profile was normalized to a minimum value of 0, corresponding to 10°C, and a maximum value of 1, corresponding to 70°C. The intensities of every sample were normalized to a minimum of 0, corresponding to the lowest intensity in a profile, and a maximum of 1, corresponding to the highest intensity in a profile. If the difference between the maximum and minimum signal intensity was less than 0.1, the normalized value was set to 0.
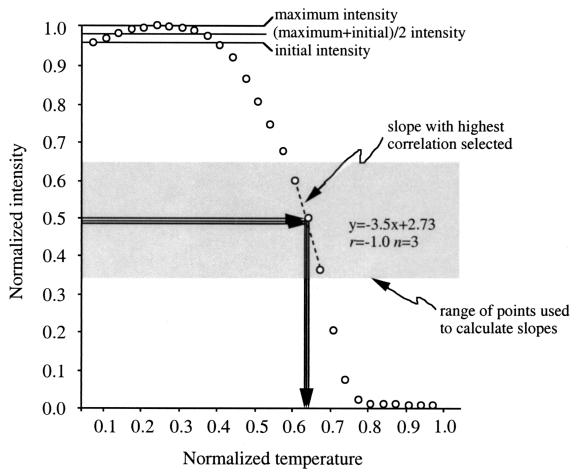
Preliminary experiments revealed difficulties in determining the intensity midpoint needed to calculate the Td because in some cases (approximately 30% of 369 samples), the maximum signal intensity was slightly greater than the intensity observed at the beginning of the experiment (Fig. 1), even when care was taken to avoid pixel saturation. In other cases, maximum intensity equaled the initial intensity (data not shown). For this reason, three Tds were calculated: one based on maximum intensity, another based on the initial intensity, and the mean of the two intensities.
FIG. 1.
Effect of maximum and initial signal intensities on the calculated Td. Three Tds were calculated based on the midpoints of initial, (maximum + initial)/2, and maximum intensities corresponding to the transition from DNA duplex to random coil. Shown is an intensity-temperature profile for the S. epidermidis target probe s2ag and the target sequence. The normalized Tds were calculated to be 0.63, 0.64, and 0.64, which correspond to Tds of 47.9, 48.3, and 48.1°C, respectively.
Normalized intensities in the range of 0.35 to 0.65 U were used to calculate slopes around the Td (Fig. 1). This range was chosen to maximize the number of points used to calculate the slope. The program was designed to consider all possible slopes within the specified range and to select the slope and intercept with the highest Pearson correlation coefficient. The selected slope and intercept were then used to calculate the normalized Tds. The normalized Tds were then converted to actual Tds by determining the equation defining the slope and intercept between normalized and actual temperatures and back-calculating the actual value. Similarly, the intensities at the Tds were calculated by determining the slope and intercept defining the equation between normalized and actual intensities and back-calculating the signal intensity at the Td. For Fig. 1, Td1 was calculated using the maximum normalized intensity in the profile, Td2 was calculated using the initial intensity (i.e., lowest temperature), and Td3 was calculated using the mean of the maximum normalized intensity and the initial intensity.
Data for the NN.
The complete input data set consisted of the following variables: the position of the single-base-pair mismatch, the type of mismatch, and the concentration of formamide used in the washing buffer. The position of the mismatch was given a value of 0 for a perfect match between probe and target and a value of 1 to 5 for mismatches occurring at one to five positions from the 5′ terminus of the probe, respectively. The type of mismatch refers to the number of hydrogen bonds which would be potentially lost from the target DNA due to incorrect base pairing. If the number of hydrogen bonds potentially lost from the target DNA due to incorrect base pairing was three (i.e., a G or C mismatch), the mismatch type was coded as 1. If the number of hydrogen bonds potentially lost from the target DNA was two (i.e., an A or T mismatch), the mismatch type was coded as 0. Perfect matches could not be coded as 0 or 1 for the type of mismatch. Since the NN required a value for type of mismatch, the number of perfect match records in the data set was duplicated, one-half of the records were coded as 0 for mismatch type, and the remainder were coded as 1. Formamide concentrations were coded as 0, 10, 20, and 30, corresponding to the percent formamide used in the washing buffer.
The output data set for determining the Td consisted of one variable: the experimentally determined Td. The output data set for determining the signal intensity consisted of one variable: the calculated signal intensity at the experimentally determined Td as determined by the Td software.
For NN and sensitivity analyses, the entire data set was normalized, with a minimum value of the data set being set at 0.1 and a maximum value set at 0.9 (18). At the end of the analyses, the NN predictions were converted back to their actual values by defining the equation between the normalized and actual data and back-calculating the value.
Statistical software.
The NN and sensitivity software was custom-designed by using C++ software and based on the recommendations of Bishop (6), Hagan et al. (12), Masters (18), and Basheer and Hajmeer (4). Stand-alone applications of the NN and sensitivity analysis used in this study are available on request (panoble@washington.edu), and a Web-based user interface is available at http://noble.ce.washington.edu. This Web-based interface contains neural.readme and sense.readme files, which describe how to use the software, and specifies the files needed to implement the back-propagating NN and sensitivity analysis (e.g., datax and datay).
For NN analysis, a logistic equation, f(x) = 1/(1+e−x), was used as the transfer function and error back-propagation was used to optimize the connection weights and biases. Full interconnection between the layers was used. The learning rate ranged from 0.1 to 0.5 U depending on the training run and was adjusted accordingly by the user. The input and output architecture of the NN consisted of three input neurons representing the position of the mismatch, the type of mismatch, and the concentration of the formamide and one output neuron representing the normalized Td or signal intensity at the Td.
For NN analysis, the order of the data records was randomized, and 90% of the data (n = 347) were selected for training the NN. The remaining 10% (n = 39) were used for testing the trained NN. In order to minimize the effective number of degrees of freedom in the network, training was stopped when the error measured with the independent test data started to increase (6, 24). This criterion was also used to select the optimal number of hidden neurons, which was determined to be three for all NNs.
Sensitivity analysis was used to determine which of the NN inputs (i.e., position of the mismatch, type of mismatch, and concentration of formamide) significantly contributed to predicting the Td or signal intensity at Td. A C++ program was custom-designed and is similar to the sensitivity programs described by Masters (18) and Noble et al. (24). Briefly, the relative sensitivity of the normalized Td was defined as the change in normalized Td relative to the change of an input. The sensitivity of each input was determined by increasing the minimum value of an input to its maximum value by using a step function for every possible combination and recording the change in normalized Td. In this study, four step intervals were used for each input value. Changes in Td for all variables in a sample were adjusted to a total value of 1, with each variable contributing to a portion of the total change in Td. Sensitivity analysis for this study involved a total of 4,632 combinations (3 inputs × 4 intervals × 386 samples). The overall relative sensitivity of the inputs was determined by calculating the mean change in normalized Td for each input (386 samples).
Conventional statistical analysis.
Pearson product-moment correlation was used to determine the degree of association between variables. Linear regressions were used to estimate the relationship of one variable to another (36). An ANOVA was used to determine the source of variability in the experimental data. The Student-Newman-Keuls (SNK) test was employed to determine whether the difference between any two means in a set of means was significant (20). ANOVA and SNK tests were conducted by using the GLM procedure in an SAS program (release 6.11; SAS Inc., Cary, N.C.). Student t tests were tabulated in MS Excel 98 (Microsoft, Inc., Redmond, Wash.) using a Macintosh 9.1 operating system.
RESULTS
Td software.
Two-tailed t tests of Td1 and Td2 using the entire data set (n = 386) revealed that there was no statistical difference in the mean Tds, suggesting that differences in the maximum and initial intensities had no affect on the calculated Td. However, it was necessary to consider the difference in Tds for samples not treated with formamide, since two-tailed t tests of Td1 and Td2 (n = 96) yielded significant differences (P < 0.04). Preliminary results suggested that this difference may have resulted from pixel saturation in samples containing low concentrations of formamide and decreased background signal originating from nonspecific binding. The results (presented below) support this, since high concentrations of formamide in the washing buffer decreased background signal intensity. For this reason, Td3 was used for all subsequent analyses.
Effect of formamide, position, and type of mismatch on Td.
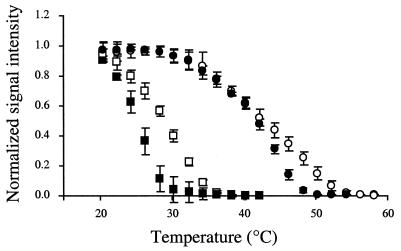
Preliminary studies using 0 to 70% formamide in the hybridization buffer revealed that 40% formamide was optimal because it yielded the highest signal intensities (data not shown). To further investigate how formamide affects signal intensity, we included formamide in the washing buffer. At room temperature (20°C), none of the probes was washed off the microarray at 0% formamide (Fig. 2). However, at 30% formamide, some of the probes with mismatches at positions 3 to 5 had reduced signal intensities. Figure 3 shows the effects of 0 and 30% formamide on melting profiles of a probe-target duplex with a single-base-pair mismatch at the fourth position from the 5′ terminus of the probe. Formamide shifted the melting profiles to the left, reducing the Td.
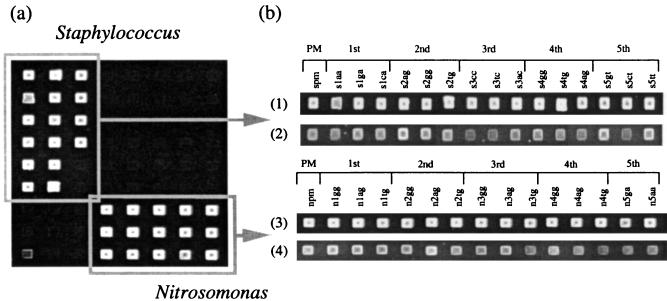
FIG. 2.
(a) Typical image of a DNA microarray; (b) fluorescence images of perfect match (PM) duplexes and duplexes with a single-base-pair mismatch. Single-base-pair mismatches are located at the first to fifth positions from the 5′ terminus of the probe. After hybridization, the microarrays were washed with a washing buffer containing 0 or 30% formamide (see Materials and Methods). 1, S. epidermidis, 0% formamide; 2, S. epidermidis, 30% formamide; 3, N. eutropha, 0% formamide; 4, N. eutropha, 30% formamide.
FIG. 3.
Effect of formamide concentration on the melting profiles of n4gg and N. eutropha target. The single-base-pair mismatch occurs at the fourth position from the 5′ terminus of the probe. Open circles, perfect-match Nitrosomonas probe in 0% formamide; closed circles, n4gg in 0% formamide; open squares, perfect-match Nitrosomonas probe in 30% formamide; closed squares, n4gg in 30% formamide.
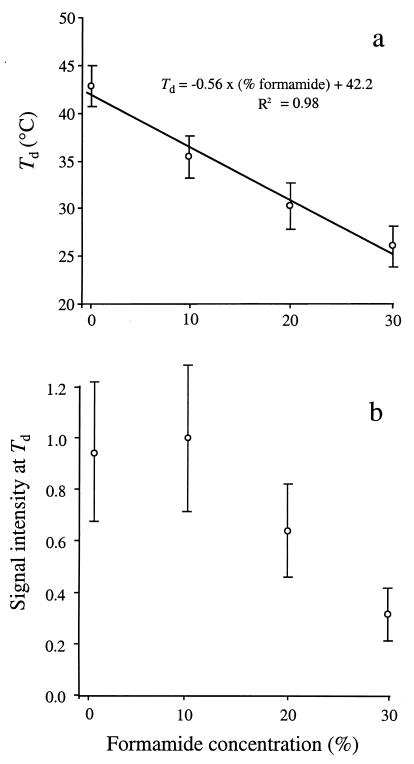
Table 1 lists the mean Tds as a function of the concentration of formamide in the washing buffer, the type of mismatch, and the position of the mismatch from the 5′ terminus of the probe. ANOVA revealed that the formamide concentration in the washing buffer, the type of mismatch, and the position of the mismatch significantly affected the Td (Table 2). In general, increasing the formamide concentration by 1% decreased the Td by approximately 0.6°C (Fig. 4a). The regression model indicated that there was a significant interaction between the concentration of formamide and the type of mismatch and Td (Table 2). At high formamide concentrations (e.g., 30%), the potential loss of three hydrogen bonds due to incorrect base pairing in a probe-target duplex (i.e., G or C mismatches) [Td = 42.1°C − (0.58 × % formamide); 250 samples; R2 = 0.90] had a greater effect on the Td than the potential loss of two hydrogen bonds (i.e., A or T mismatches) [Td = 42.1°C − (0.52 × % formamide); n = 119 samples; R2 = 0.83]. These results provide evidence for the nonlinear relationships between Td, formamide concentration, and mismatch type. SNK results (α = 0.05) indicated that the loss of three hydrogen bonds significantly decreased the Td by 0.9°C (mean ± standard deviation, 33.6 ± 6.9°C, n = 250) relative to the loss of two hydrogen bonds (34.4 ± 6.4°C, n = 119).
TABLE 2.
ANOVA of Td as a function of formamide concentration, position of the mismatch, and type of mismatch (369 samples)
| Source | df | Mean square | F | P |
|---|---|---|---|---|
| Formamide concentration (%) | 3 | 4,971.1 | 1,846.5 | <0.0001 |
| Position of the mismatch from 5′ end | 5 | 151.5 | 56.3 | <0.0001 |
| Type of mismatch (A/T or G/C) | 1 | 58.3 | 21.7 | <0.0001 |
| Formamide concentration (%) × type of mismatch (A/T or G/C) | 3 | 15.8 | 5.9 | 0.0006 |
FIG. 4.
Effect of formamide concentration on Td (a) and signal intensity at Td (b) of probe-target duplexes. Each circle is the mean of at least 92 Tds, and each error bar represents the standard deviation of the mean.
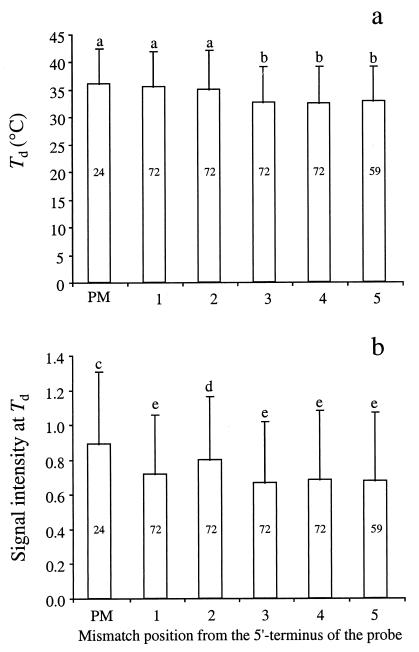
In general, moving the position of the mismatch away from the 5′ terminus to the center of the probe decreased the Td of the probe-target duplex (Fig. 5a). The Td of probes with a perfect match to the target and those with a single-base-pair mismatch at the first or second position from the 5′ terminus of the probe were not significantly different from one another (Fig. 5a) (SNK test). Similarly, the Td of probes having mismatches located at the third to fifth positions (Fig. 5a) (SNK test) were not significantly different from one another. Overall, probes containing a single-base-pair mismatch at positions 3 to 5 (32.6 ± 6.6°C, n = 212) decreased the Td by 2.8°C compared to probes with a single-base-pair mismatch at the first or second position (35.4 ± 6.6°C, n = 168).
FIG. 5.
Effect of position of the single-base-pair mismatch on Td (a) and signal intensity at Td (b). PM, Perfect-match duplex. Samples with the same letter are not significantly different (α = 0.05) as determined by the SNK test. Numbers in the center of the bars are the numbers of samples represented.
Effect of formamide, position, and type of mismatch on signal intensity.
ANOVA revealed that formamide concentration, type of mismatch, and position of the mismatch from the 5′ terminus of the probe significantly affected signal intensity of the probes (Table 3). Signal intensities at Td were not significantly different at 0 and 10% formamide. However, at 20 and 30% formamide, the signal intensity decreased linearly as well as the variability associated with the signal (Fig. 4b). The statistically insignificant increase in signal intensity at 10% formamide was presumably due to decreased nonspecific binding on the gel pads. Conditions favoring nonspecific binding (e.g., low formamide concentration) might have increased the background noise, decreasing the contrast and increasing the background correction of the image-processing software.
TABLE 3.
ANOVA of signal intensity at Td as a function of formamide concentration, position of the mismatch, and type of mismatch (369 samples)
| Source | df | Mean square | F | P |
|---|---|---|---|---|
| Formamide concentration (%) | 3 | 11.4 | 2.6 | <0.0001 |
| Position of the mismatch from 5′ end | 5 | 0.5 | 10.6 | <0.0001 |
| Type of mismatch (A/T or G/C) | 1 | 0.2 | 3.6 | 0.0011 |
In general, moving the position of a single-base-pair mismatch away from the 5′ terminus of the probe (i.e., to the center of the probe) decreased signal intensity (Fig. 5b). Probes with perfect matches to the target had significantly higher signal intensities than those with a single-base-pair mismatch at the first to fifth positions from the 5′ terminus of the probe. Single-base-pair mismatches at the second position had significantly higher signal intensities than mismatches at the first, third, fourth, or fifth position. Presumably, the low signal intensity at the first position was an artifact of the data set, because mismatches in the first position included AT and GC mismatches while the second to fourth positions included only GC mismatches. That is, two-hydrogen-bond mismatches were not adequately represented at the second to fourth positions in the targets. Clearly, a variety of targets is needed to elaborate and verify these findings.
NN and sensitivity analysis.
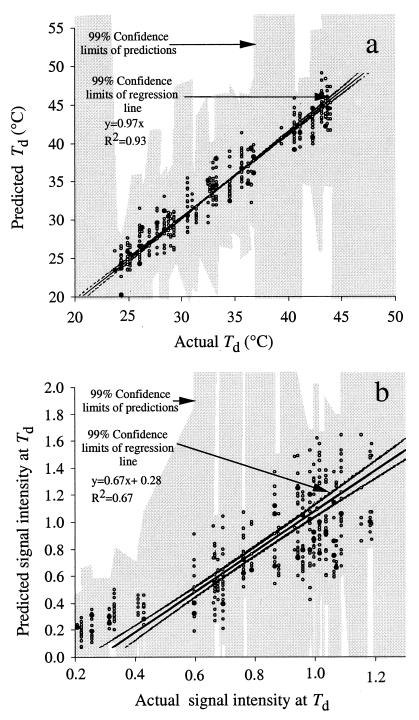
Comparison of the predicted versus the actual Td of three trained NNs yielded similar results with high regression coefficients (R2 = 0.93). Figure 6a shows the relationship between predicted and actual Tds for one of the three NNs. The trained NNs were able to predict the Td given the position of the mismatch, the type of mismatch, and the formamide concentration in the washing buffer within approximately 2°C from the actual Td (mean errors ± standard deviations: NN 1, 1.4 ± 1.2°C; NN 2, 1.4 ± 1.1°C; NN 3, 1.4 ± 1.2°C; n = 386).
FIG. 6.
Relationship between actual and predicted Td (a) and signal intensity (b) as determined with an NN. Each datum represents a single sample. Open circles, data used to train the NN (n = 347); closed circles, data used to test the NN (n = 39). Confidence limits of the predictions (shaded) were calculated with a sliding window of 10 sampling points. The confidence limits of regression line and the predictions (shaded) were based on training and test data (n = 386).
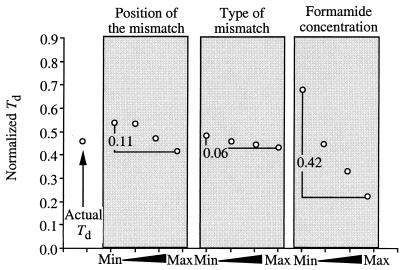
The analysis of the sensitivity of individual inputs to the Td was repeated for three separately trained NNs to determine the contribution of position of the mismatch, type of the mismatch and formamide concentration to the Td. Figure 7 shows the results of the sensitivity analysis of one sample from one trained NN. In these experiments, the concentration of formamide had a much greater effect on Td than the position of the mismatch or the type of mismatch. Table 4 provides a summary of the sensitivity analysis for three separately trained NNs. Sensitivity analysis was performed on several trained NNs because NNs with low correlations between actual and predicted outputs may yield inconsistent results. However, all three sensitivity analyses yielded similar and consistent results. More than 75% of the variation in the data set was explained by the formamide concentration, consistent with the expectation that formamide played a key role in predicting the Td. Approximately 19% of the variation of the data was explained by the position of the mismatches, and less than 6% of the variation was explained by the type of mismatch.
FIG. 7.
Sensitivity analysis of one sample (position = 5, type = 1, and formamide = 10 and corresponding normalized Td) showing the change in Td as a function of increasing position, from its minimum value of 0 to its maximum value of 5, type, from its minimum value of 0 to its maximum value of 1, and formamide, from its minimum value of 0 to its maximum value of 30. Formamide concentration had the largest change in normalized Td, followed by position of the mismatch and then type of mismatch. The sum of the changes in Td for all inputs is 0.59 (0.11 + 0.06 + 0.42). The corresponding relative sensitivity of position is the change in Td/sum of the changes in Td for all inputs, which in this case is 0.11/0.59, or 0.19. The corresponding relative sensitivities of type of mismatch and formamide concentration are 0.10 and 0.71, respectively.
TABLE 4.
Contribution of sources to changes in normalized Td based on sensitivity analyses of three independently trained NNs
| Source | Relative sensitivitya by NN
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| Position of the mismatch for 5′ end | 0.20 ± 0.02 | 0.19 ± 0.02 | 0.19 ± 0.02 |
| Type of mismatch | 0.06 ± 0.03 | 0.05 ± 0.01 | 0.06 ± 0.03 |
| Formamide concentration | 0.74 ± 0.03 | 0.76 ± 0.04 | 0.75 ± 0.03 |
Each sensitivity value is based on the contribution of the variable to changing the normalized Td. Values are means ± standard deviations (n = 386).
Comparison of the predicted and actual signal intensities of three trained NNs yielded similar results with moderate regression coefficients (R2 = 0.67). Figure 6b shows the relationship between predicted and actual Tds for one of the three NNs. The trained NNs were able to predict the signal intensity at low signal intensities (as depicted by the 99% confidence limits of the predictions). However, at high signal intensities the predictability decreased significantly, presumably due to increased noise in the signal intensity. All data points having actual signal intensities greater than 0.9 U in Fig. 6b represented samples treated with 0 to 10% formamide. More predictable signal intensities were obtained in samples containing 20 to 30% formamide. No other input factors were correlated with the variability of the predictions.
It is also important to note that the relationship between predicted and actual signal intensities does not appear to be linear (Fig. 6b). This finding suggests that the NNs had difficulties in finding patterns in the data presumably due to intrinsic noise in the signal intensity, particularly in samples having low concentrations of formamide in the washing buffer. Noise in the signal intensity clearly limited the ability of the NN to make reproducible predictions.
The analysis of the sensitivity of individual inputs to the signal intensity at the Td was repeated for three separately trained NNs to determine the contribution of the position of the mismatch, the type of the mismatch and concentration of formamide to the signal intensity. Table 5 is a summary of the sensitivity analysis conducted with three separately trained NNs. All three sensitivity analyses yielded similar and consistent results. It is important to note that we did not anticipate identical results from the separately trained NN, since random weights used during the first training step are different for each NN and the weights are individually adjusted during training. Rather, we anticipated that different NNs would yield similar and consistent results if they were able to adequately recognize patterns in the data. More than 69% of the variation in the data set was explained by the formamide concentration, indicating that the formamide in the washing buffer played a key role in determining the signal intensity. Approximately 20% of the variation in the data was explained by the position of the mismatch, and less than 7% of the variation was explained by the type of mismatch.
TABLE 5.
Contribution of sources to changes in signal intensity at Td based on sensitivity analyses of three independently trained NNs
| Source | Relative sensitivitya by NN
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| Position of the mismatch for 5′ end | 0.23 ± 0.04 | 0.23 ± 0.06 | 0.19 ± 0.06 |
| Type of mismatch | 0.08 ± 0.04 | 0.07 ± 0.05 | 0.07 ± 0.04 |
| Formamide concentration | 0.69 ± 0.05 | 0.71 ± 0.09 | 0.74 ± 0.07 |
Each sensitivity value is based on the contribution of the variable to changing the signal intensity at Td. Values are means ± standard deviations (n = 386).
DISCUSSION
The use of DNA probes in any format for bacterial or gene identification relies on good discrimination between probe-target duplexes with perfect matches and duplexes with mismatched base pairing. Since complete discrimination between perfectly matched duplexes and duplexes with mismatches is often difficult to achieve, the rules governing oligonucleotide duplex stability must be determined in order to establish the relative contribution of nonspecific hybridizations to signal intensity and to facilitate the development of probes having good discriminating capabilities. We chose Td as the discriminating measure in this study because Td has been extensively used in studies employing conventional membrane hybridization techniques (21, 29, 30, 37), and because calculating the Td using the nonequilibrium dissociation approach (i.e., melting profiles) and oligonucleotide microarrays has been successfully demonstrated (16). Moreover, determining the Td by using microarrays provides rapid and reproducible data, which facilitates rigorous statistical analyses.
NN and sensitivity analyses.
The application of NNs to the analysis of complex data in microbiology is relatively new (2), and to our knowledge, no study has used back-propagation of NNs to analyze microarray data. NNs have been used to identify the restriction enzyme profiles of E. coli O157:H7 (7), the pyrolysis mass spectra of Mycobacterium tuberculosis complex species (9), the promoter sites of E. coli (13), protein-DNA binding sites (25), fatty acid profiles of microbial communities (24), nifH-specific binding patterns from denaturing gradient gel electrophoresis analysis (26), and stable low-molecular-weight rRNA profiles (23). However, there is a paucity of studies which have clearly demonstrated the utility of NNs over conventional statistical approaches, and even fewer studies have demonstrated the utility of sensitivity analysis.
NNs are able to recognize nonlinear patterns in complex data which cannot be discerned by using conventional statistical approaches (18). Two independent studies have recently shown that NNs outperformed and provided better predictive power than conventional statistical approaches (22, 24). Based on these studies, we reasoned that better recognition of patterns should improve our ability to predict outputs (such as Td), provided that the trained NN has been appropriately optimized (as defined in reference 6). In this study, trained NNs accurately predicted the Td and signal intensity at Td given the position and type of mismatch and concentration of formamide. However, despite the considerable correspondence in the results of linear (e.g., ANOVA) and nonlinear (e.g., NN) approaches, we were unable to clearly resolve any differences. This finding is presumably due to the limited range of input factors (i.e., position of mismatch, mismatch type, and formamide concentration) used to predict the outputs (i.e., Td or signal intensity at Td) and/or because all NNs were optimized to produce generalized predictions and their performance is ultimately limited by the intrinsic noise of the data (6). Nonetheless, on all occasions the NNs yielded results which were consistent with those obtained by ANOVA.
Studies now in progress are considering more complicated interactions, such as the contribution of stacking energies, secondary structure of DNA duplexes, GC content, and length of the probes to Td prediction. These studies will likely yield more definitive differences between linear and nonlinear approaches and demonstrate that NNs provide more detailed examination of the data than conventional linear statistical approaches.
Sensitivity analysis of trained NNs yielded information on which individual inputs (in this study, the position of the mismatch, the type of mismatch, and the formamide concentration) significantly contributed to the target output (i.e., Td and signal intensity at Td). To our knowledge, this is the second study that has demonstrated the utility of sensitivity analysis. A previous study by Noble et al. (24) compared the results of sensitivity analysis to those obtained by principal-component analysis (a linear analysis method). They showed that sensitivity analysis was reproducible and attributed differences in the results to the nonlinearity of the data. In this study, the results of sensitivity analyses were consistent and reproducible, demonstrating that sensitivity analysis was statistically valid and robust.
Effects of signal intensity on Td.
An unexpected finding of this study was the relative importance of signal intensity for determining the Td. As shown in Fig. 6b, the NNs had considerable difficulties predicting signal intensities at low formamide concentrations, presumably owing to increased noise in the signal intensity, as clearly depicted in Fig. 4b. High variability in signal intensities occurred at 0 and 10% formamide. Yet the NNs were able to successfully predict the Td regardless of noise in the signal intensities (Fig. 6a). Although Td predictions were not affected in this study, it is possible that intrinsic noise in signal intensity could affect Td predictions when low concentrations of formamide are used in the washing buffer. Future studies will need to consider signal intensity as an important factor contributing to the variability in Td predictions. It is also important to emphasize that variability in the signal intensity can be minimized by using high concentrations of formamide (Fig. 4b). Hence, adding formamide to the washing buffer may improve Td prediction.
Rules governing probe-target duplexes having single-base-pair mismatches.
Currently, this technology has limited use in routine environmental microbiology because we know little about the extent of nonspecific hybridizations of probe-target duplexes and the diversity of 16S rRNA and other gene targets in nature. Clarifying the rules governing probe-target duplexes which have perfect matches and those with a single-base-pair mismatch near or at the 5′ terminus is a necessary and fundamental task for the development of DNA microarray technology.
One rule discovered in this study is that position of the mismatch is more important than the type of mismatch for duplexes with a near-terminus or terminus single-base-pair mismatch. This finding is important for probe design for two reasons: (i) probes having a single-base-pair mismatch in the first or second position from the 5′ terminus may provide an effective means of identifying closely related species or identifying individual species which have high variability in a specific region of the 16S rRNA, and (ii) probes with a single-base-pair mismatch at positions 3 to 5 have greater discriminating abilities than those with a mismatch at or near the 5′ terminus. We anticipate that these are general rules. However, further studies using additional targets and probes are needed to refine and expand these findings.
Another rule discovered in this study is that formamide in the washing buffer decreased noise in the signal intensity. Minimizing noise in the signal intensity is very important for the development of microarray technology because too much noise increases the uncertainty of hybridization events and clouds our ability to accurately discriminate between probe-target duplexes with perfect matches and duplexes with mismatches. This is particularly relevant for analysis of melting profiles because of the problems associated with pixel saturation. Pixel saturation may affect Td determination in samples containing low concentrations of formamide because of significant differences between initial and maximum signal intensities (Fig. 1). This problem may be minimized by using appropriate exposure times. However, if this format is to be used for environmental samples, one can expect that the dynamic range will be much larger than our experimental results. Consequently, pixel saturation may be a substantial problem that needs to be addressed in future studies.
Effect of formamide in the washing buffer.
It is well recognized that differentiation between probe-target duplexes with perfect matches and those with mismatches is affected by the washing conditions (e.g., buffers, salt concentration, and temperature) (29, 41). We examined the effects of formamide in the washing buffer because formamide, being a mild denaturant for nucleic acids, disrupts hydrogen bonding and increases the specificity of hybridizations (38). Thus, we originally hypothesized that formamide should have a greater effect on the type of single-base-pair mismatch than the position of the mismatch in internal mismatches. However, our hypothesis was not supported by the data on near-terminus or terminal mismatches, since we found that the position of the mismatch was more important than the compositional effects of the mismatch, at least for mismatches occurring at and near the terminus of the probe. Nonetheless, formamide decreased the Td of G or C mismatches at a slightly higher rate than A or T type mismatches.
Formamide decreased the Td at a rate of 0.6°C per 1% formamide (Fig. 4a) in our microarray. This rate is similar but not identical to that reported by McConaughy et al. (19), who showed that Td decreases at a rate of 0.7°C per 1% formamide in solution. Salt concentration and subtle differences in duplex stability such as length, GC content, secondary structure, and composition and number of mismatches of probe-target duplexes presumably account for the observed differences.
In summary, oligonucleotide microarrays coupled with NN and sensitivity analysis were used to assess the effects of single-base-pair mismatches located at and near the 5′ termini of probes on discrimination of probe-target duplexes. NNs were able to accurately predict the Td given the position and type of mismatch and concentration of formamide. The relevant contributions of formamide concentration, position of the mismatch, and type of mismatch for the NN predictions were identified by sensitivity analysis.
Acknowledgments
We thank G. Yershov, A. Kukhtin, and A. Gemmell for their efforts in manufacturing the oligonucleotide microarrays and S. Surzhikov for synthesis of the oligonucleotide probes. We thank E. Tribou for his computer expertise. We also thank the College Inn Environmental Microbiology Journal Club, who critically reviewed the manuscript.
This work was supported by grants from DARPA and NASA to D.A.S. and NSF DEB-0088879 to P.A.N.
REFERENCES
- 1.Aleksander, I., and H. Morton. 1991. An introduction to neural computing, p.1–20. Chapman & Hall, Ltd., London, United Kingdom.
- 2.Almeida, J. S., and P. A. Noble. 2000. Neural computing in microbiology. J. Microbiol. Methods 43:1–2. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 4.Basheer, I. A., and M. Hajmeer. 2000. Artificial neural networks: fundamentals, computing, design, and application. J. Microbiol. Methods 43:3–31. [DOI] [PubMed] [Google Scholar]
- 5.Bavykin, S. G., J. P. Akowski, V. M. Zakhariev, V. E. Barsky, A. N. Perov, and A. D. Mirzabekov. 2001. Portable system for microbial sample preparation and oligonucleotide microarray analysis. Appl. Environ. Microbiol. 67:922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, C. M. 1995. Neural networks for pattern recognition. Oxford Press, Oxford, United Kingdom.
- 7.Carson, C. A., J. M. Keller, K. K. McAdoo, D. Wang, B. Higgins, C. W. Bailey, J. G. Thorne, B. J. Payne, M. Skala, and A. W. Hahn. 1995. Escherichia coli O157:H7 restriction pattern recognition by artificial neural networks. J. Clin. Microbiol. 33:2894–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, Y. J., D. Glasner, F. R. Blattner, and E. W. Triplett. 2001. Genomic interspecies microarray hybridization: rapid discovery of three thousand genes in the maize endophyte Klebsiella pneumoniae 342 by microarray hybridization with Escherichia coli K-12 open reading frames. Appl. Environ. Microbiol. 67:1911–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman, R., R. Goodacre, P. R. Sisson, J. G. Magee, A. C. Ward, and N. F. Lightfoot. 1994. Rapid identification of species within the Mycobacterium tuberculosis complex by artificial neural network analysis of pyrolysis mass spectra. J. Med. Microbiol. 40:170–173. [DOI] [PubMed] [Google Scholar]
- 10.Guschin, D., G. Yershov, A. Zaslavsky, A. Gemmell, V. Shick, D. Proudnikov, P. Arenkov, and A. Mirzabekov. 1997. Manual manufacturing of oligonucleotide, DNA, and protein microchips. Anal. Biochem. 250:203–211. [DOI] [PubMed] [Google Scholar]
- 11.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagan, M. T., H. B. Demuth, and M. Beale. 1996. Neural network design. PWS Publishing Co., Boston, Mass.
- 13.Horton, P. B., and M. Kanehisa. 1992. An assessment of neural networks and statistical approaches for prediction of E. coli promoter sites. Nucleic Acids Res. 20:4331–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huebner, J., and D. A. Goldmann. 1999. Coagulase-negative staphylococci: role as pathogens. Annu. Rev. Med. 50:223–236. [DOI] [PubMed] [Google Scholar]
- 15.Koops, H.-P., B. Böttcher, U. C. Möller, A. Pommerening-Röser, and G. Stehr. 1991. Classification of 8 new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J. Gen. Microbiol. 137:1689–1699. [Google Scholar]
- 16.Liu, W.-T., A. D. Mirzabekov, and D. A. Stahl. 2001. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol. 3:619–629. [DOI] [PubMed] [Google Scholar]
- 17.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masters, T. 1993. Practical neural network recipes in C++. Academic Press, New York, N.Y.
- 19.McConaughy, B. L., C. D. Laird, and B. J. McCarthy. 1969. Nucleic acid reassociation in formamide. Biochemistry 8:3289–3295. [DOI] [PubMed] [Google Scholar]
- 20.Miller, R. G. 1966. Simultaneous statistical inference. McGraw-Hill, Inc., New York, N.Y.
- 21.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moschetti, G., G. Blaiotta, F. Villani, S. Coppola, and E. Parente. 2001. Comparison of statistical methods for identification of Streptococcus thermophilus, Enterococcus faecalis, and Enterococcus faecium from randomly amplified polymorphic DNA patterns. Appl. Environ. Microbiol. 67:2156–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble, P. A., K. D. Bidle, and M. Fletcher. 1997. Natural microbial community compositions compared by a back-propagating neural network and cluster analysis of 5S rRNA. Appl. Environ. Microbiol. 63:1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noble, P. A., J. S. Almeida, and C. R. Lovell. 2000. Application of neural computing methods for interpreting phospholipid fatty acid profiles of natural microbial communities. Appl. Environ. Microbiol. 66:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Neill, M. C. 1998. A general procedure for locating and analyzing protein-binding sequence motifs in nucleic acids. Proc. Natl. Acad. Sci. USA 95:10710–10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piceno, Y. M., P. A. Noble, and C. R. Lovell. 1999. Spatial and temporal assessment of diazotroph assemblage composition in vegetated salt marsh sediments using denaturing gradient gel electrophoresis analysis. Microb. Ecol. 38:157–167. [DOI] [PubMed] [Google Scholar]
- 27.Proudnikov, D., E. Timofeev, and A. Mirzabekov. 1998. Immobilization of DNA in polyacrylamide gel for the manufacture of DNA and DNA-oligonucleotide microchips. Anal. Biochem. 259:34–41. [DOI] [PubMed] [Google Scholar]
- 28.Rao, V. B., and H. V. Rao. 1993. C++ neural networks and fuzzy logic. MIS Press, New York, N.Y.
- 29.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittmann, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors using oligonucleotide probe hybridizations. Appl. Environ. Microbiol. 60:1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richmond, C. S., J. D. Glasner, R. Mau, H. F. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rumelhart, D. E., G. E. Hinton, and R. J. Williams. 1986. Learning internal representation by error back propagation, p.318–362. In D. E. Rumelhart and J. L. McCleland (ed.), Parallel distributed processing. MIT Press, Cambridge, Mass.
- 33.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467–470. [DOI] [PubMed] [Google Scholar]
- 34.Schütz, E., and N. von Ahsen. 1999. Spreadsheet software for thermodynamic melting point prediction of oligonucleotide hybridization with and without mismatches. BioTechniques 27:1218–1222, 1224. [DOI] [PubMed] [Google Scholar]
- 35.Service, R. F. 1998. Microchip arrays put DNA on the spot. Science 282:396–399. [DOI] [PubMed] [Google Scholar]
- 36.Sokal, R. R., and F. J. Rohlf. 1981. Biometry, 2nd ed. W. H. Freeman and Co., New York, N.Y.
- 37.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p.205–248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 39.Szostak, J. W., J. I. Stile, B.-K. Tye, P. Chiu, F. Sherman, and R. Wu. 1979. Hybridization with synthetic oligonucleotide. Methods Enzymol. 68:419–428. [DOI] [PubMed] [Google Scholar]
- 40.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tijssen, P. 1993. Hybridization with nucleic acid probes. Part 1. Theory and nucleic acid preparation. In P. C. van der Vliet (ed.), Laboratory techniques in biochemistry and molecular biology. Elsevier, Amsterdam, The Netherlands.
- 42.Timofeev, E., S. V. Kochetkova, A. D. Mirzabekov, and V. L. Florentiev. 1996. Regioselective immobilization of short oligonucleotides to acrylic copolymer gels. Nucleic Acids Res. 24:3142–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, M., G. Rath, H.-P. Koops, and K.-H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251–264. [Google Scholar]
- 44.Yershov, G., V. Barsky, A. Belgovskiy, E. Kirillov, E. Kreindlin, I. Ivanov, S. Parinov, D. Guschin, A. Drobishev, S. Dubiley, and A. Mirzabekov. 1996. DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl. Acad. Sci. USA 93:4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zart, D., and E. Bock. 1998. High rate of aerobic nitrification and denitrification by Nitrosomonas eutropha grown in a fermentor with complete biomass retention in the presence of gaseous NO2 or NO. Arch. Microbiol. 169:282–286. [DOI] [PubMed] [Google Scholar]