Abstract
We screened radiation-sensitive yeast mutants for DNA damage checkpoint defects and identified Dot1, the conserved histone H3 Lys 79 methyltransferase. DOT1 deletion mutants (dot1Δ) are G1 and intra-S phase checkpoint defective after ionizing radiation but remain competent for G2/M arrest. Mutations that affect Dot1 function such as Rad6-Bre1/Paf1 pathway gene deletions or mutation of H2B Lys 123 or H3 Lys 79 share dot1Δ checkpoint defects. Whereas dot1Δ alone confers minimal DNA damage sensitivity, combining dot1Δ with histone methyltransferase mutations set1Δ and set2Δ markedly enhances lethality. Interestingly, set1Δ and set2Δ mutants remain G1 checkpoint competent, but set1Δ displays a mild S phase checkpoint defect. In human cells, H3 Lys 79 methylation by hDOT1L likely mediates recruitment of the signaling protein 53BP1 via its paired tudor domains to double-strand breaks (DSBs). Consistent with this paradigm, loss of Dot1 prevents activation of the yeast 53BP1 ortholog Rad9 or Chk2 homolog Rad53 and decreases binding of Rad9 to DSBs after DNA damage. Mutation of Rad9 to alter tudor domain binding to methylated Lys 79 phenocopies the dot1Δ checkpoint defect and blocks Rad53 phosphorylation. These results indicate a key role for chromatin and methylation of histone H3 Lys 79 in yeast DNA damage signaling.
Even a single DNA double-strand break (DSB) is potentially lethal in yeast cells and can activate a highly conserved signal transduction pathway to induce the DNA damage checkpoint response, which results in delayed cell cycle progression to allow time for DNA repair (25, 52). Similarly, checkpoint signaling in metazoan cells slows cell cycle progression, allowing damaged cells to undergo either DNA repair or apoptosis to eliminate potentially catastrophic mutations. Inborn or acquired defects in these pathways contribute to genomic instability and cancer (31, 58, 81).
Budding yeast display distinct G1, intra-S and G2/M DNA damage checkpoints, depending on the source and timing of damage (31, 37, 58). Many of the conserved yeast checkpoint regulators, such as members of the Mre11 nuclease complex (22), Rad24 clamp loader complex (12), Rad17/Mec3/Ddc1 clamp (38), ATR homolog Mec1 (65), 53BP1 ortholog Rad9 (66, 75), and Chk2 homolog Rad53 (76), participate in all of these checkpoint responses. Other factors such as Mrc1 and Chk1 have been ascribed roles in a specific cell cycle checkpoint (1, 53, 60).
Treating cells in G1 with ionizing radiation (IR) induces a dose-dependent delay before onset of DNA replication and bud emergence (16, 22). Like the well-studied yeast G2/M checkpoint, the G1 delay is dependent upon many of the classical checkpoint genes and associated with Rad53 phosphorylation and Rad9 activation. In turn, HO endonuclease-dependent DSBs formed in G1 recruit the Mre11 complex and ATM homolog Tel1 (45, 64). Based on studies in asynchronous cells, Mec1 recruitment to G1 DSBs would be dependent on its cofactor Ddc2 (59). Mec1 phosphorylation of Rad9 would promote binding and activation of the checkpoint kinase Rad53 (21). In contrast, G1 delay after UV irradiation depends upon the nucleotide excision repair protein Rad14 (46), perhaps via its role in recognition of UV-damaged sites and/or a direct molecular interaction with Ddc1 (19), a component of the yeast Rad17/Mec3/Ddc1 clamp (32). Rad14 is not required for G1 checkpoint arrest after IR (16).
The original description of DNA damage checkpoints has since yielded to a new understanding that involves chromatin proteins as active participants in signaling during the early response to genomic insults (2, 29, 54). Histone H2A Ser129 is rapidly phosphorylated after DNA damage (14, 64), assisting recruitment of chromatin modifying activities that may remodel chromatin at DSBs (13, 43, 72). Other posttranslational histone modifications and chromatin assembly and remodeling activities have also been implicated in DNA damage response (4, 9, 10, 13, 55, 71). Although histone methylation has traditionally been considered in the context of epigenetic transcriptional regulation (47, 49, 73), the histone methyltransferases Dot1 and Set1 have also been implicated in DNA damage response (11, 18), functioning in a single pathway of conserved histone modifications also important for checkpoint control (20). Methylation of H3 Lys 4 by Set1 (8, 33) and H3 Lys 79 by Dot1 (15, 35, 49, 73), each require H2B Lys 123 ubiquitination by Rad6/Bre1 (8, 51, 68). RAD6 encodes an E2 ubiquitin-conjugating enzyme targeted to histone H2B by the E3 ubiquitin ligase Bre1 (27, 28, 57, 79). H2B ubiquitination is promoted by a Paf1/RNA polymerase II protein complex that also directs Set1 to promoters of actively transcribed genes (23, 34, 48, 80).
Recent studies have linked histone methylation to recruitment of the checkpoint signaling protein 53BP1 and its paralogs to sites of DNA damage (26). The 53BP1 tandem tudor domains bind methylated histone H3 Lys 79 via residues conserved in both fission yeast Crb2 and budding yeast Rad9. Tudor domain mutations that impair methylated Lys 79 binding prevent accumulation of 53BP1 at DSB repair foci. In turn, the H4 methyltransferase Set9 has been implicated in fission yeast G2 checkpoint control via recruitment of Crb2 (61).
In the present study, we found that deletions of DOT1 and SET1 result in specific checkpoint defects in S. cerevisiae. Whereas Dot1 and Set1 methyltransferases are dispensable for the Rad9-dependent G2/M checkpoint, dot1Δ is profoundly defective for G1 and S phase delays after IR, while set1Δ is partially deficient in S phase checkpoint response after treatment with the DNA alkylator methyl methanesulfonate (MMS). Furthermore, dot1Δ mutants fail to induce phosphorylation of Rad9 or Rad53 after IR in G1 and are partially defective for recruitment of Rad9 to HO endonuclease cleavage sites. Mutating a conserved Rad9 tudor domain residue predicted to mediate binding to H3 methylated Lys 79 conferred defects in G1 checkpoint arrest, intra-S phase checkpoint delay, and activation of Rad53 after DNA damage in G1 but did not impair G2/M checkpoint response. Our results establish a surprising role for nucleosomes and their specific modifications at DNA damage sites as essential determinants of Rad9 function in both the G1 and intra-S phase DNA damage checkpoints.
MATERIALS AND METHODS
Yeast strains and plasmids.
Yeast strains used in the present study are listed in Table 1. Gene deletions and epitope tagging were performed by PCR-based gene modification (39). Yeast cells were grown in standard rich YPD (1% yeast extract, 2% peptone, 2% dextrose) and YPGal (1% yeast extract, 2% peptone, 2% galactose) media or selective synthetic complete media at 25°C. pRS315 (67) plasmids carrying DOT1 and dot1-Gly401Arg (73) were generous gifts of D. Gottschling. pRAD53F (pR316-RAD53-3xFLAG) and pD2-R53F (pRS316-DDC2-RAD53-3xFLAG) plasmids (36) were kindly provided by D. Stern.
TABLE 1.
Yeast strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| BY4730 | MATaleu2Δ0 met15Δ0 ura3Δ0 | 7 |
| W303-1A | MATaade2-1 can1-100 ura3-1 leu2-3,112 his3-11,15 trp1-1 | 69 |
| SKY2849 | W303-1A, dot1Δ::TRP1 | This study |
| SKY2850 | W303-1A, rad9Δ::kanMX6 | This study |
| SKY2851 | W303-1A, rad9Δ::URA3 | 16 |
| SKY2852 | W303-1A, rad9Δ::URA3 dot1Δ::TRP1 | This study |
| SKY2853 | W303-1A, rtf1Δ::his3MX6 | This study |
| SKY2854 | W303-1A, bre1Δ::kanMX6 | This study |
| SKY2855 | W303-1A, rad6Δ::kanMX6 | This study |
| SKY2856 | W303-1A, set1Δ::his3MX6 | This study |
| SKY2857 | W303-1A, set2Δ::kanMX6 | This study |
| SKY2858 | W303-1A, set1Δ::his3MX6 set2Δ::kanMX6 | This study |
| SKY2859 | W303-1A, dot1Δ::TRP1 set1Δ::his3MX6 | This study |
| SKY2860 | W303-1A, dot1Δ::TRP1 set2Δ::kanMX6 | This study |
| SKY2861 | W303-1A, dot1Δ::TRP1 set1Δ::his3MX6 set2Δ::kanMX6 | This study |
| SKY2862 | W303-1A, rad17Δ::LEU2 | 16 |
| SKY2863 | W303-1A, rad17Δ::LEU2 dot1Δ::TRP1 | This study |
| SKY2864 | W303-1A, RAD9::13Myc-kanMX6 | This study |
| SKY2865 | W303-1A, RAD9::13Myc-kanMX6 dot1Δ::TRP1 | This study |
| SKY2866 | W303-1A, RAD53::13Myc-kanMX6 | 16 |
| SKY2867 | W303-1A, RAD53::13Myc-kanMX6 dot1Δ::TRP1 | This study |
| SKY2868 | W303-1A, RAD53::13Myc-kanMX6 rad9Δ::kanMX6 | This study |
| Y131 | W303-1A, hta1-htb1Δ::LEU2 hta2-htb2Δ [pRS426-HTA1-HTB1::URA3] | 57 |
| Y132 | W303-1A, hta1-htb1Δ::LEU2 hta2-htb2Δ [pRS426-HTA1-htb1-Lys123Arg::URA3] | 57 |
| WZY42 | MATaade2-101 lys2-801 ura3-52 leu2-1 his3-200 trp1-63 hht1-hhf1::pWZ405-F2F9-LEU2 hht2-hhf2::pWZ403-F4F10-HIS3 [Ycp50-copyII HHT2-HHF2 (URA3 CEN ARS)] | 49 |
| WZY42 H3-Lys79Ala | MATaade2-101 lys2-801 ura3-52 leu2-1 his3-200 trp1-63 hht1-hhf1::pWZ405-F2F9-LEU2 hht2-hhf2::pWZ403-F4F10-HIS3 [Ycp50-copyII hht2-Lys 79 Ala-HHF2 (URA3 CEN ARS)] | 49 |
| QY364 | MATaho hmlΔ::ADE1 hmrΔ::ADE1 ade1-110 leu2,3-112 lys5 trp1::hisG ura3-52 ade3::GAL1,10:HO RAD9-HA::kanMX6 | This study |
| QY367 | MATaho hmlΔ::ADE1 hmrΔ::ADE1 ade1-110 leu2,3-112 lys5 trp1::hisG ura3-52 ade3::GAL1,10:HO RAD9-HA::kanMX6 dot1Δ::TRP1 | This study |
| QY363 | MATα ho hmlΔ::ADE1 hmrΔ::ADE1 ade1-110 leu2,3-112 lys5 trp1::hisG ura3-52 ade3::GAL1,10:HO RAD9-HA::kanMX6 | This study |
| QY368 | MATα ho hmlΔ::ADE1 hmrΔ::ADE1 ade1-110 leu2,3-112 lys5 trp1::hisG ura3-52 ade3::GAL1,10:HO RAD9-HA::kanMX6 dot1Δ::TRP1 | This study |
Site-directed mutagenesis.
To generate the rad9-Tyr798Gln mutation, an EcoRI-BamHI fragment encoding the 5′ region of RAD9 obtained by digesting pDL847, a pRS416-based vector carrying a full-length RAD9 clone (6), kindly provided by D. Lydall, was cloned into pUC18 and PCR mutagenized (30) to encode a Tyr798Gln substitution by using the primers YQMUT1 (5′-CGTGGAATTACAAATTTCAGCCGGGTATTTTATTGG-3′) and YQMUT2 (5′-CCAATAAAATACCCGGCTGAAATTTGTAATTCCACG-3′) (mutations underlined). The mutation was confirmed by sequencing and the fragment was recloned into pDL847 (6) to form pRS416-rad9-Tyr798Gln.
Cell cycle experiments.
Cell cycle synchronization, culture manipulation, and flow cytometry analysis of DNA content were as previously described (16, 30). To induce DNA damage, cells were irradiated at 30 Gy/min in a Gammacell 220 60Co source (Atomic Energy of Canada, Ltd.), treated with 0.03% MMS, exposed to 50 J/cm2 at 254 nm in a Stratalinker 1600 (Stratagene), or incubated in 25 μg of phleomycin (Sigma)/ml.
G1/S checkpoint function after IR damage was examined in yeast cells synchronized in 5 μM synthetic α-factor (WHWLQLKPGQPNleY) (56) at 106 cells per ml for 180 min, treated with 300 Gy or mock treated, and then released from arrest. To detect onset and kinetics of DNA replication, at 15-min intervals, 0.5-ml samples were collected, fixed with 70% ethanol for 60 min and treated with RNase (0.25 mg/ml) for 120 min at 50°C, stained with 2.5 μM SYTOX Green (Molecular Probes), and subjected to flow cytometry analysis of DNA content. The screen of radiation-sensitive yeast mutants for checkpoint defects will be described in detail elsewhere but, briefly, exponential-phase cultures of deletion strains from the Open Biosystems Yeast Knock Out collection of viable MATa haploid strains in the BY4730 background (7) were arrested, irradiated, released, fixed, and analyzed as described above. Mutants that displayed rapid onset and/or progression of DNA replication in flow cytometry were flagged as G1/S DNA damage checkpoint defective.
To determine the fraction of cells remaining arrested in G1, at 15-min intervals, 0.5-ml samples were collected, combined with 0.5 ml of trapping media (10 μM α-factor, 30 μg of nocodazole/ml), incubated for 90 min at 25°C, fixed, stained as described above, subjected to flow cytometry analysis of DNA content, and examined by phase microscopy to count cells displaying mating projections (G1 cells) or buds (post-G1 cells). To determine the effect of DNA damage on S phase progression, 106 cells/ml of α-factor-synchronized cells were released in the presence of 0.03% MMS or irradiated 15 min after release. Samples were collected at 20-min intervals, fixed, and analyzed by flow cytometry.
To analyze cell cycle delay at the G2/M transition, exponential-phase cultures at 106 cells/ml were arrested with 15 μg of nocodazole/ml for 180 min. Before release into fresh media, cells were irradiated with 300 Gy or left untreated. Aliquots were removed every 30 min, fixed, stained as for flow cytometry, and then examined by epifluorescence microscopy (Zeiss Axioskop, FITC filter set, 40×/0.75 NA objective) to score the percentage of binucleate large-budded cells. All cell cycle experiments were repeated a minimum of three times, and representative results are presented.
Western blot analysis and chromatin immunoprecipitation.
Protein extracts were prepared by glass bead disruption in phosphate-buffered saline (pH 7.4), 10% glycerol, 50 mM NaF, 100 mM β-glycerol phosphate, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Boehringer) and subjected to Western analysis as previously described (16). Rabbit polyclonal anti-Myc antibody (A14; Santa Cruz) was used to detect epitope-tagged Rad9 and Rad53.
Chromatin immunoprecipitation (ChIP) studies were performed on strains derived from JKM179 (64), kindly provided by J. Haber, a MATα strain carrying integrated GAL1,10::HO and lacking the HML and HMR loci. In QY363, the JKM179 chromosomal RAD9 coding sequence was carboxyl terminal tagged with 3HA::kanMX6 by one-step mutagenesis (39). In QY368, the DOT1 gene of QY363 was replaced with a dot1Δ::TRP1 cassette, obtained by PCR from the dot1Δ mutant SKY2849. Cell cycle specific studies were performed in MATa derivatives of these strains, QY364 and QY367, respectively, using 5 μM α-factor for G1 arrest and 15 μg of nocodazole/ml for mitotic arrest. Galactose induction of HO endonuclease, chromatin preparation, and immunoprecipitation performed on whole cells were previously described (13), using anti-HA 12CA5 (Roche); anti-H2A P-Ser129 (14), kindly provided by J. Downs; and anti-H3 diMeLys79 (15, 49), kindly provided by Y. Zhang. Sonication was performed by using a Bioruptor (Diagenode, Belgium) for 11 cycles of 30-s pulses with 60-s pauses at the highest setting (producing chromatin fragments ranging mostly from 200 to 500 bp). Totals of 1/2,000 of the input and 1/100 of the immunoprecipitated material were analyzed by LightCycler (Roche) real-time PCR with primers that were verified for efficiency or specificity of amplification over the range of template amounts used in these experiments. The occupancies of specific proteins or their modifications under different conditions were calculated as a ratio of immunoprecipitated to input material for loci surrounding the HO-cutting site and a control locus (large intergenic region on chromosome V). The predetermined efficiency factors per PCR cycle for each pair of primers were used in the calculations (ranging from 1.7 to 1.95). The efficiency of HO cleavage was measured at the different time points by real-time PCR with primers on each side of the cleavage site compared to the control locus.
RESULTS
To identify novel regulators of the DNA damage-induced G1/S checkpoint in budding yeast, we used flow cytometry-based screening to test a collection of deletion mutants for defects in G1 arrest and/or intra-S-phase delay after IR. We selected strains from a genome-wide gene deletion library in the S288c strain background (78) that were disrupted in genes previously identified in genome-wide screens for DNA damage sensitivity (3, 5, 17, 24, 77) and/or that had been otherwise reported in the literature as involved in DNA damage resistance. Our rationale was that failure to delay entry into S phase or rapid progression through DNA synthesis should lead to fixation of unrepaired damage and thereby lower DNA damage tolerance. Each strain was examined in an α-factor block-and-release experiment after 300 Gy irradiation in G1, a dose that yields ∼50% lethality in the control haploid wild-type strain, using flow cytometry to detect early entry into and/or rapid progression through S phase. Among the deletion strains that demonstrated marked deficiency in G1/S checkpoint delays after IR was dot1Δ, a mutant lacking the conserved histone H3 methyltransferase Dot1.
Histone H3 methyltransferase Dot1 is a bona fide DNA damage checkpoint protein.
To characterize a role for histone H3 methylation in the DNA damage response, we deleted the DOT1 gene in the W303 strain background (69) and analyzed the kinetics of cell cycle progression after IR. First, we determined the contribution of Dot1 to G1/S checkpoint delays. Wild-type, dot1Δ, and rad9Δ cells synchronized in G1 with α-factor were irradiated with 300 Gy before release into fresh media and incubated at 25°C. Wild-type cells showed very little increase in DNA content for the duration of the experiment, a finding consistent with arrest in G1 and/or early S phase. Irradiated dot1Δ mutants failed to perform this delay and instead progressed through the cell cycle with kinetics similar to the irradiated checkpoint-defective rad9Δ mutants and mock-irradiated wild-type cells (Fig. 1A). Treatment of G1-arrested wild-type, dot1Δ and rad9Δ cells with the radiomimetic drug phleomycin before release from α-factor yielded a similar pattern of G1/S checkpoint response defects (data not shown).
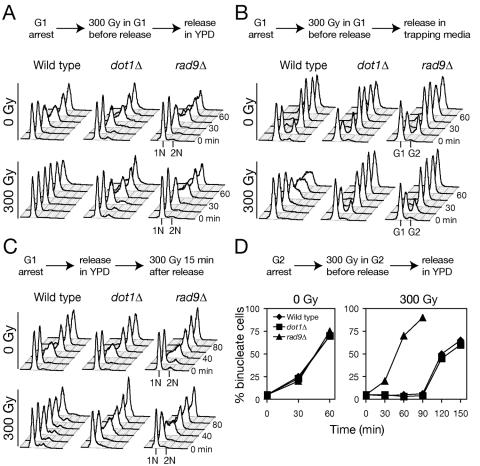
FIG. 1.
Histone H3 methyltransferase Dot1 is required for G1 phase and intra-S-phase checkpoint response to IR. (A) Arrest defect in dot1Δ after DNA damage in G1. α-Factor-arrested cells were irradiated with 300 Gy, released into fresh media, and analyzed at 15-min intervals for DNA content by flow cytometry. (B) Aliquots from the same experiment as in panel A were mixed with α-factor/nocodazole trapping media and incubated for an additional 90 min. The fraction of cells that remained in G1 was determined by flow cytometry. (C) Intra-S-phase checkpoint defect in dot1Δ. Cells were irradiated 15 min after release from G1 arrest, and DNA replication was monitored by flow cytometry. (D) The G2/M checkpoint is intact in dot1Δ. Wild-type (W303-1A), dot1Δ (SKY2849), and rad9Δ (SKY2851) cells were synchronized with nocodazole before irradiation and release into fresh media. Mitotic progression was determined by the percentage of large-budded cells with separated nuclei.
One potential pitfall of flow cytometry analysis is that early-S-phase cells may be mistaken for G1 cells. Therefore, we utilized an α-factor/nocodazole trap (16) to distinguish G1 arrest from early-S-phase arrest. Briefly, at intervals after the 300-Gy irradiation and release from α-factor, aliquots are transferred to media containing both α-factor and nocodazole and incubated for 90 min. Only bona fide G1 cells, which have not passed START, are rearrested by α-factor, whereas cells that have initiated S phase continue to synthesize DNA but arrest in G2/M due to the nocodazole. After the 90-min incubation, phase microscopy revealed two populations of cells, shmoos and budded cells, which correspond in flow cytometry to distinct 1N (G1) and ∼2N (S/G2/M) peaks. The α-factor/nocodazole trap assay revealed that irradiated wild-type cells remain α-factor sensitive for up to 45 min after release (Fig. 1B), a finding consistent with an intact G1 checkpoint. However, dot1Δ and rad9Δ lost α-factor sensitivity with the similar kinetics as nonirradiated cells, a finding consistent with a G1 checkpoint defect.
In turn, unlike the wild type, dot1Δ mutants irradiated in G1 failed to slow S phase progression (Fig. 1A), suggesting an intra-S-phase checkpoint defect. To directly assay S phase checkpoint function, wild-type, dot1Δ, and rad9Δ cells were irradiated with 300 Gy 15 min after release from G1 arrest, the time at which unperturbed cells pass Start and begin S phase (Fig. 1C). Although wild-type cells replicated slowly and did not appear to complete S phase during the experiment, both dot1Δ and rad9Δ completed S phase within 60 min whether irradiated or not. Thus, we concluded that Dot1 contributes to both G1 phase and intra-S-phase checkpoints in response to DNA DSBs. Finally, we examined the integrity of G2/M DNA damage checkpoint response in the dot1Δ mutant. Cells were synchronized at metaphase with nocodazole and irradiated with 300 Gy before release into fresh media. To monitor G2/M progression, we counted the number of binucleate large-budded cells. Wild-type and dot1Δ cells remained arrested at G2/M, whereas rad9Δ completed mitosis without delay (Fig. 1D).
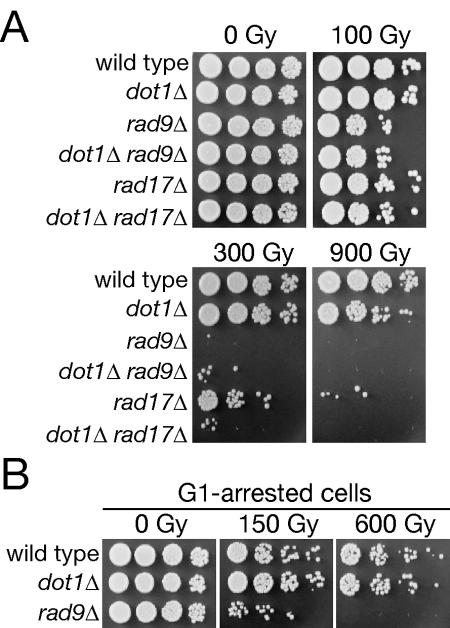
It has been proposed that DNA damage checkpoints slow cell cycle progression to provide time for proper DNA repair (40). Interestingly, dot1Δ and wild-type cells demonstrated similar DNA damage sensitivity, quite distinct from the marked radiation sensitivity of rad9Δ and rad17Δ mutations (Fig. 2A). Irradiation induces a similar G2/M checkpoint arrest in asynchronously growing cultures of wild-type and dot1Δ cells, potentially obscuring any effects on the more DNA damage-sensitive G1 and/or S phase cells. However, wild-type and dot1Δ cells irradiated in G1 and then released from α-factor arrest were again similarly sensitive (Fig. 2B). To test the model that redundant and/or downstream checkpoint regulator(s) might promote dot1Δ cell survival, we performed order-of-function analysis of Dot1 with respect to Rad9 and Rad17, checkpoint regulators with partially independent contributions to G2/M arrest (12). The rad9Δ and dot1Δ rad9Δ mutants displayed similar sensitivity to 100 and 300 Gy (Fig. 2A), suggesting epistasis, while a dot1Δ rad17Δ mutant displayed enhanced DNA damage sensitivity over rad17Δ alone, suggesting independent function (Fig. 2A).
FIG. 2.
DNA damage sensitivity and order-of-function between DOT1 and checkpoint genes RAD9 and RAD17. (A and B) Asynchronous (A) or G1-arrested (B) cultures were irradiated, serially diluted, and plated on rich media. Strains used were wild type (W303-1A), dot1Δ (SKY2849), rad9Δ (SKY2851), dot1Δ rad9Δ (SKY2852), rad17Δ (SKY2862), and dot1Δ rad17Δ (SKY2863).
Histone H3 methylation and H2B ubiquitination are required for G1/S checkpoints.
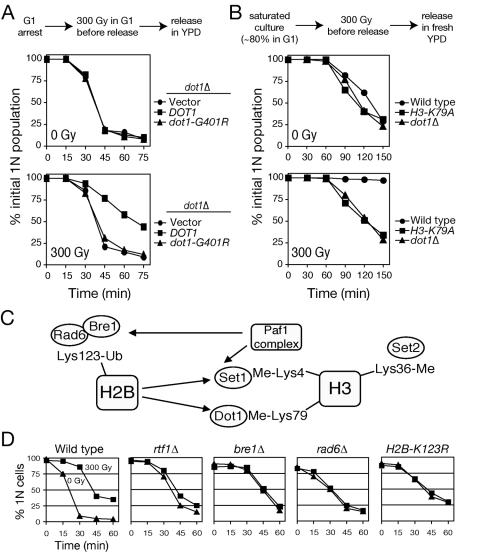
To test whether the methyltranferase activity of Dot1 is required for G1/S checkpoint, the dot1Δ mutant was transformed with a wild-type DOT1 gene or dot1-Gly401Arg, mutated within the binding site for the methyl donor S-adenosylmethionine (49, 73). In contrast to nearly complete suppression by plasmid-borne DOT1, dot1-Gly401Arg failed to restore G1/S checkpoint function to dot1Δ cells, indicating a requirement for Dot1 methyltransferase activity in yeast DNA damage checkpoint response (Fig. 3A). The only known Dot1 substrate is histone H3, which is methylated at Lys 79 in the globular domain (15, 49). Thus, we examined cell cycle progression after irradiation in a strain carrying histone H3 mutated to replace Lys 79 with Ala (49). Distinct from the dot1Δ mutation, histone H3 Lys 79 Ala confers α-factor resistance, reflecting its stronger effects on silencing (73). Thus, to obtain a G1-enriched sample, we used saturated cultures with a high content of unbudded cells (∼80%). After irradiation and release into fresh medium, many cells bearing wild-type histone H3 remained arrested in G1, whereas histone H3 Lys 79 Ala mutants entered the cell cycle and completed replication without delay (Fig. 3B). These data implicate methylation of histone H3 at Lys 79 as the critical mediator of Dot1 effects on cell cycle checkpoints.
FIG. 3.
Histone H3 methylation and histone H2B ubiquitination are involved in G1 DNA damage checkpoint. (A) DNA damage checkpoint dependence on Dot1 methyltransferase activity. The dot1Δ mutant (SKY2849) was transformed with an empty vector, wild-type DOT1, or catalytically defective dot1-Gly401Arg. Percentage of cells remaining in G1 was determined quantitatively from flow cytometry of DNA content using FlowJo 6.3.2 software. The 1N population on a two-dimensional scatter plot of side scatter versus DNA content was gated, and the counts were normalized to flow cytometry at t = 0 min. (B) Checkpoint role of H3-Lys 79. Wild type or Lys79Ala mutant H3 (WZY42) and dot1Δ mutant (SKY2849) were grown to saturation to increase G1 content, irradiated, released into fresh media, and analyzed by flow cytometry. Quantitation was performed as in panel A. (C) Scheme of Paf1/RNA polymerase II and H2B ubiquitination-dependent modifications of histone H3. (D) Dependence of G1 checkpoint on upstream regulators. Paf1-complex mutant rtf1Δ (SKY2853), H2B ubiquitination mutants bre1Δ (SKY2854) and rad6Δ (SKY2855), and histone H2B Lys123Ala mutant (Y132) were arrested in G1, irradiated, released into fresh media, and analyzed by α-factor/nocodazole trap assay to determine the percentage of cells that remained in G1. Triangles and squares represent mock-treated (0 Gy) and irradiated (300 Gy) samples, respectively.
That histone H2B monoubiquitination at Lys 123 is required for global histone H3 Lys 79 methylation by Dot1 (8, 51, 68) suggested a role for this modification in G1/S checkpoint function. H2B ubiquitination requires the E2 ubiquitin-conjugating enzyme Rad6 and the E3 ubiquitin ligase Bre1 (27, 57, 79) and the Paf1/RNA polymerase II protein complex, which consists of Paf1, Rtf1, Cdc73, Leo1 and Ctr9 (34, 48, 80) (Fig. 3C). Thus, histone H3 Lys 79 methylation is abolished in cells lacking Rad6, Bre1, Rtf1 or Paf1, or mutated at histone H2B Lys 123 (27, 57, 79). Indeed, checkpoint assays revealed G1/S checkpoint defects in all mutants examined that are deficient in H2B ubiquitination (Fig. 3D). These results confirmed a role for the conserved pathway of chromatin modifications linking histone H2B ubiquitination and histone H3 methylation in G1/S checkpoint response after IR.
Set1 contributes to cell cycle delays in S but not in G1 or G2 phase after DNA damage.
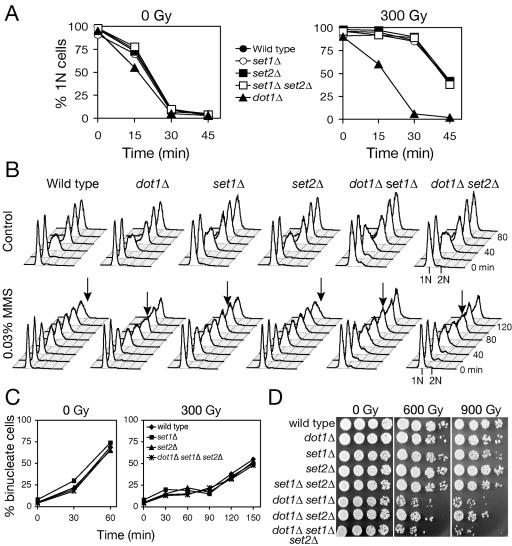
In addition to Dot1-dependent histone H3 methylation at Lys 79, ubiquitination of histone H2B is required for Set1-dependent methylation of histone H3 Lys4 but not Set2-dependent methylation of histone H3 Lys 36 (8, 51, 68) (Fig. 3C). Reflecting a role in regulated gene expression, Set1 is recruited to transcriptionally active genes by the Paf1/RNA polymerase II complex (23, 34, 50). We were interested in determining whether histone H3 Lys 4 and/or Lys 36 methylation also contribute to G1/S checkpoint response to IR. Single and double mutants lacking Set1 and/or Set2 remained arrested in G1 as long as wild-type cells after 300 Gy (Fig. 4A), confirming a unique role for Dot1 and methylation of histone H3 Lys 79 in G1 checkpoint function. To specifically probe intra-S phase checkpoint function, we examined replication kinetics in dot1Δ, set1Δ, and/or set2Δ treated with the DNA alkylating agent MMS, which generates DSBs during replication (Fig. 4B). Wild-type cells treated with MMS remained arrested in S phase for the duration of the experiment. The intra-S-phase checkpoint was partially compromised in the single dot1Δ and set1Δ mutants and not significantly more in the double dot1Δ set1Δ mutant. In turn, the set2Δ mutation alone did not confer any S phase checkpoint defect, whereas the dot1Δ set2Δ double mutant exhibited a defect similar to that of dot1Δ. Interestingly, neither dot1Δ, set1Δ, set2Δ, or any combination of these mutations affected G2/M checkpoint arrest (Fig. 4C). These data suggest that Set1 and Set2 may have overlapping roles in the S phase checkpoint response, which are independent of Dot1. Giannattasio et al. (20) recently implicated Dot1 and Set1 function in G1/S checkpoint responses to UV-mediated DNA damage. Using 254 nm UV light or the UV-mimetic 4-nitroquinoline N-oxide (4NQO) to induce base damage, leading to excision repair and a DNA damage signal, we confirmed their observations of G1 and S phase checkpoint defects in dot1Δ and set1Δ (data not shown). The distinct effects of mutations in histone methyltransferases on responses to single-strand damage versus DSBs may indicate divergent roles of histone methylations in damage recognition and/or checkpoint signaling.
FIG. 4.
Set1 contributes to intra-S but not to G1 or G2/M checkpoint after DNA damage. (A and B) The set1Δ mutant shows intact G1 checkpoint arrest (A) but a defective intra-S-phase checkpoint response (B). Cells arrested with α-factor were either irradiated with 300 Gy for G1 checkpoint analysis by α-factor/nocodazole trap assay (A) or released into 0.03% MMS and analyzed by flow cytometry (B). Arrows indicate the first time point at which a 2N population, consistent with completion of replication, is observed. (C) Analysis of G2/M checkpoint after IR. Cells synchronized in G2/M with nocodazole, treated with IR, and released into fresh media were analyzed for mitotic progression by microscopy. (D) Sensitivity to IR in combinations of dot1Δ, set1Δ, and set2Δ mutations. Asynchronous cultures were irradiated, serially diluted and plated on rich media. For panels A to D, the strains used were wild type (W303-1A), dot1Δ (SKY2849), set1Δ (SKY2856), set2Δ (SKY2857), set1Δ set2Δ (SKY2858), dot1Δ set1Δ (SKY2859), dot1Δ set2Δ (SKY2860) and dot1Δ set1Δ set2Δ (SKY2861).
The lack of DNA damage sensitivity in dot1Δ raised the question of whether Set1 and Set2 methylation might also contribute to DNA damage tolerance. Thus, we assayed survival after IR of the strains lacking dot1, set1, and/or set2 (Fig. 4D). Like dot1Δ mutants, set1Δ and set2Δ single mutants were similar to the wild type, but dot1Δ set1Δ, dot1Δ set2Δ, and dot1Δ set1Δ set2Δ mutants were markedly more sensitive to IR. Consistent with this, dot1Δ set1Δ and dot1Δ set2Δ double mutants were markedly more sensitive to MMS than dot1Δ, set1Δ, or set2Δ single mutants, while set1Δ set2Δ was as sensitive as set1Δ alone (data not shown). Taken together, these observations suggest that Dot1 functions in an independent pathway from Set1 and Set2, in which Set1 and Set2 may have overlapping roles.
Loss of Dot1 impairs Rad9 and Rad53 phosphorylation after IR in G1.
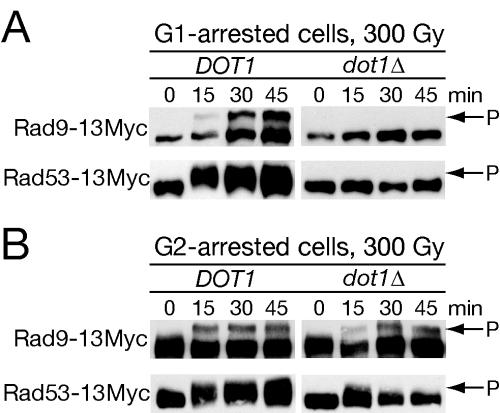
Given that dot1Δ rad9Δ cells are no more sensitive to IR than rad9Δ, Dot1 might function through Rad9. DNA damage induces the yeast ATR homolog Mec1 to phosphorylate Rad9 and thereby activate Rad53, leading to characteristic mobility shifts (21, 63). To test whether Rad9 phosphorylation required Dot1, we used Western analysis to monitor phosphorylation of Rad9-13Myc in wild-type and dot1Δ cells treated with IR during G1 arrest (Fig. 5A). In wild-type cells, the characteristic mobility shift of Rad9 phosphorylation was observed by 15 min after IR and persisted for the duration of the experiment. No Rad9 mobility shift was observed in dot1Δ. Rad9 phosphorylation is thought to lead to activation of the Rad53 kinase through trans-autophosphorylation (21, 63, 70), suggesting that Rad53 phosphorylation would also be defective in dot1Δ cells. Indeed, a mobility shift of Rad53-13Myc was observed in wild-type cells arrested in G1 with the same kinetics as Rad9 activation, whereas no shift was detected in the dot1Δ background (Fig. 5A).
FIG. 5.
G1-specific loss of Rad9 and Rad53 phosphorylation in dot1Δ cells. Wild-type (SKY2864 and SKY2866) and dot1Δ (SKY2865 and SKY2867) strains expressing Rad9-13Myc or Rad53-13Myc were synchronized in either G1 with α-factor (A) or G2/M with nocodazole (B) and irradiated, and arrest was maintained for the duration of the experiment. Aliquots were subjected to Western analysis to detect mobility shifts of Rad9 and Rad53.
Since dot1Δ mutants are not G2 checkpoint defective, we reasoned that dot1Δ deletion might not affect Rad9 and Rad53 phosphorylation in G2/M. Wild-type and dot1Δ cells carrying Rad9-13Myc or Rad53-13Myc were arrested in mitosis with nocodazole, treated with IR, and subjected to Western analysis (Fig. 5B). Rad9 appeared equally phosphorylated in response to DNA damage in both wild-type and dot1Δ cells. Surprisingly, Rad53 phosphorylation appeared qualitatively decreased in dot1Δ compared to the wild-type control. One reason that the lower level of Rad53 activation may not lead to G2/M checkpoint defects would be the contribution of Chk1 to mitotic arrest. Nonetheless, based on the intact checkpoint arrest and clear evidence of Rad9 and Rad53 activation, the G2/M checkpoint signaling pathway appears to be intact in dot1Δ cells.
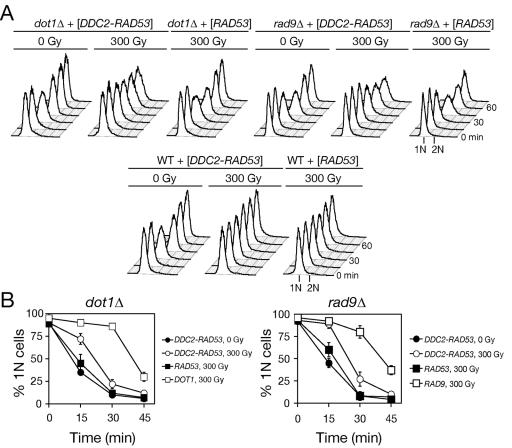
These results suggest that the dot1Δ defect may lead to a specific loss of Rad9 function in G1 and S phase. The Mec1 binding partner Ddc2 (ATRIP) mediates recruitment to DSBs and is considered to function upstream of Rad9 and Rad53 activation (59, 74). Consistent with this model, expression of a Ddc2-Rad53 fusion protein has been shown to bypass the requirement for Rad9 in Rad53 activation and phosphorylation after MMS treatment (36). Thus, the dot1Δ mutant was transformed with plasmids expressing DDC2-RAD53 or wild-type RAD53 as a control. Strikingly, expression of DDC2-RAD53 slowed S phase progression (Fig. 6A), placing the defect at the level of Rad9 function. Nonetheless, neither RAD53 nor DDC2-RAD53 could significantly restore G1 checkpoint arrest in dot1Δ when assayed in the α-factor/nocodazole trap assay. Similarly, DDC2-RAD53 failed to fully restore G1 checkpoint arrest in rad9Δ cells (Fig. 6B). These results may serve to genetically separate the G1 phase and intra-S-phase delays after IR.
FIG. 6.
Ddc2-Rad53 fusion protein restores the integrity of intra-S but not G1 checkpoint in cells lacking DOT1 or RAD9 after IR. Wild-type (W303-1A), dot1Δ (SKY2849), and rad9Δ (SKY2850) cells expressing DDC2-RAD53 or RAD53 were synchronized in G1, irradiated, released, and analyzed by flow cytometry (A) and α-factor/nocodazole trap assay (B).
Lack of histone H3 Lys 79 methylation confers a specific defect in Rad9 checkpoint function.
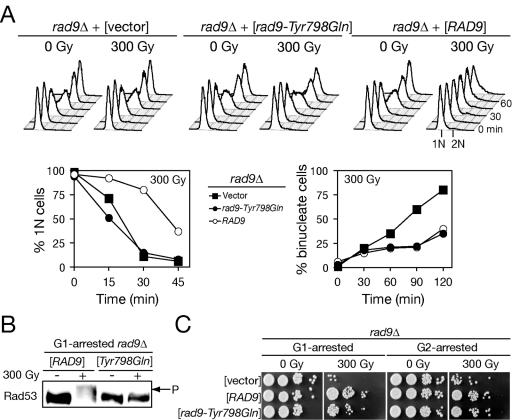
The apparent requirement for Dot1 function in the phosphorylation of Rad9 and Rad53 after DNA damage in G1 suggested that histone H3 Lys 79 may have a direct role in Rad9 recruitment and/or activation. The checkpoint regulator 53BP1 is considered the closest metazoan ortholog to yeast Rad9 (42). 53BP1 association with repair foci was recently shown to depend on binding to histone H3 methylated on Lys 79 by hDot1l (26). This binding was shown to occur via paired tudor domains as point mutations in contact residues specifically disrupt histone H3 binding and 53BP1 localization (26). A Rad9 construct including its tudor domains was also shown to interact with methylated histone H3 in vitro (26), suggesting a similar relationship may link Dot1 to Rad9 function in budding yeast. To recapitulate the 53BP1 results in yeast, we mutated a conserved residue in the tudor domain binding pocket, Tyr 798 to Gln. When expressed from a low-copy plasmid or via mutation of the genomic locus, rad9-Tyr798Gln could not restore G1 checkpoint function but fully complemented the G2/M checkpoint defect of rad9Δ (Fig. 7A). Consistent with results with dot1Δ, the mobility shift of Rad53-13Myc was absent after G1 irradiation in rad9-Tyr798Gln (Fig. 7B), and rad9-Tyr798Gln radiation sensitivity was comparable to that of the wild type when irradiated in either G1 or G2/M (Fig. 7C).
FIG. 7.
A rad9-Tyr798Gln tudor domain mutation phenocopies dot1Δ defects in G1 and intra-S checkpoint after IR. (A) Entry into S phase and DNA replication are not delayed in rad9Δ (SKY2850) cells expressing the rad9-Tyr798Gln allele in response to IR, whereas G2/M checkpoint remains intact. The integrity of G1, intra-S, and G2/M checkpoints in the rad9Δ transformants containing empty plasmid pRS416, pRS416-RAD9, or pRS416-rad9-Tyr798Gln was determined. (B) Failure of rad9-Tyr798Gln mutant to promote Rad53 phosphorylation after IR in G1. The rad9Δ strain expressing the Myc-tagged Rad53 was transformed with the wild-type RAD9 and the rad9-Tyr798Gln allele. Transformants were synchronized in G1 and treated with 300 Gy of IR to induce checkpoint response. Cells were harvested 30 min after IR for protein extracts and subjected to Western analysis with anti-Myc antibodies. (C) Effect of rad9-Tyr798Gln mutation on survival after IR. Aliquots of irradiated and mock-treated transformants were spotted onto YPD plates to determine the rate of survival after IR.
Dot1 function is necessary for normal Rad9 recruitment to DSBs.
The preceding data do not distinguish a requirement for Dot1 methylation of histone H3 Lys 79 either for Rad9 phosphorylation at a DSB or Rad9 localization to a DSB during G1. Previous work has demonstrated recruitment of Rad9 to chromatin adjacent to an HO break is Mec1 dependent (44). Suggesting that H3 Lys 79 methylation may be necessary, but cannot be sufficient, for Rad9 recruitment, up to 90% of nucleosomes may display constitutively methylated Lys 79 histone H3 during vegetative growth (73). Further, Western analysis showed no significant change in dimethylated Lys 79 after DNA damage in asynchronous wild-type cells and a similar lack of dimethylated Lys 79 in the dot1Δ mutant with or without damage (data not shown).
To directly evaluate the role of Dot1 in Rad9 recruitment, we performed ChIP in dot1Δ mutant and DOT1 control strains expressing Rad9-3HA. ChIP was performed to detect Rad9 association with irreparable DSBs formed at the mating type locus after induction of GAL1,10:HO (Fig. 8A). Briefly, raffinose-grown dot1Δ or control cells were transferred to galactose at time zero, and aliquots were collected at successive time points after HO endonuclease induction (20, 40, 60, and 120 min). In these experiments, HO cutting was 66 and 75% complete by 20 min in wild-type and dot1Δ cells, respectively, and >96% complete at subsequent time points (data not shown). Based on the lack of Rad9 phosphorylation after DNA damage, Lys 79 methylation might be critical for initial steps in DNA damage recognition leading to the recruitment of Mec1 and Tel1 checkpoint kinases. However, ChIP with anti-phospho-H2A (14) to detect histone H2A Ser 129 phosphorylation as a reporter for Mec1/Tel1 function at DSBs (14, 64) revealed similarly rapid and persistent increases in phosphorylation near the HO site in asynchronous dot1Δ or control cells (Fig. 8B, middle panel). In contrast, ChIP of these same samples with anti-methylated Lys 79 antibody revealed no significant change in histone H3 Lys 79 methylation at the HO site upon cleavage while, as expected, no signal was detected in dot1Δ mutant cells under any condition (Fig. 8B, lower panel). Similarly, histone H3 Lys 4 methylation also remained unchanged adjacent to an HO break (13).
FIG. 8.
Dot1-dependent association of Rad9 with the HO-induced DSB. (A) Association of Rad9 near the HO-induced DSB was determined for chromosomal sites 60bp (HO site), 0.5kb (HO + 0.5 kb), 1.5kb (HO + 1.5 kb), and/or 10kb (HO + 10 kb) from a HO-cutting site using the indicated pairs of primers and for a large intergenic region on chromosome V (control locus). (B) Recruitment of Rad9 to the HO-induced DSB is profoundly decreased in dot1Δ (QY368) cells despite the high level of histone H2A phosphorylation at Ser 129 near DSB. In addition, level of histone H3 methylation at Lys 79 is not altered near the HO-induced DSB in wild-type cells (QY363). ChIP assays were performed on asynchronous growing cells after formation of HO-induced DSB (180 min in YPGal medium) or without expression of HO endonuclease (YPD medium). (C) Kinetics of Rad9-HA recruitment to the HO-induced DSB in G1-arrested cells. The time course experiment was performed by incubating G1-synchronized wild-type (QY363) and dot1Δ (QY368) cells in YPGal medium for 0, 20, 40, 60, or 120 min before cross-link-ChIP and real-time PCR. The efficiency of HO cleavage at these time points was 0, 66, 96, 98, and 99% in wild-type cells, whereas 0, 75, 98, 99, and 99% in dot1Δ cells (data not shown). (D) Association of Rad9-HA with the HO-induced DSB is higher in G1- than in G2-arrested cells but is equally reduced in dot1Δ mutant cells. Wild-type (QY364) and dot1Δ (QY367) cultures were synchronized in either G1 or G2/M and incubated in galactose-containing medium for 180 min to induce DSB. (B to D) Data are presented as occupancy at specific loci based on the immunoprecipitation/input ratio obtained by real-time PCR (duplicate) after correction for efficiency of the specific pairs of primers over the range of PCR cycles used. All experiments were performed in JKM179 background with integrated GAL1,10:HO cassette and deleted HMR/HML loci (64).
Interestingly, assay of Rad9-HA association with the HO break in these asynchronous cells revealed an intermediate result, i.e., a significant decrease but not a complete loss of Rad9 recruitment to the DSB in dot1Δ versus controls (Fig. 8B, upper panel). One explanation would be that the Rad9 recruitment defect in dot1Δ is cell cycle dependent, like the checkpoint defect. When HO was induced in control cells arrested in G1 with α factor, enhanced localization of Rad9-HA to the HO site and adjacent chromatin were detected at the 20 min time point and continued to increase for the duration of the experiment (Fig. 8C). Significantly, in α factor-arrested dot1Δ cells, the initial phase of recruitment of Rad9 at 20 min was absent, but a subsequent increase in Rad9 localization was observed. Nonetheless, the absolute level of Rad9 recruitment in dot1Δ was significantly less than in wild-type cells and apparently localized closer to the break.
ChIP analysis has demonstrated persistent Rad9 association with an HO-induced DSB after prolonged arrest in G2 (44). Thus, we examined retention of Rad9 during G1 and G2 arrest (Fig. 8D). In wild-type cells, greater Rad9 retention was seen in G1 compared to G2. Dot1 was required for normal Rad9 retention in both cell populations. Based on these ChIP data, although the effect of Dot1 on Rad9 recruitment to a DSB is significant, the distinct pattern of Rad9 and Rad53 activation during G1 and S versus G2/M in the dot1Δ mutant suggests that histone H3 Lys 79 methylation may influence Rad9 activation as much as its recruitment.
DISCUSSION
We have demonstrated here that Dot1-dependent methylation of histone H3 Lys 79 is required for DNA damage checkpoint responses to IR in both G1 and S phase, but not G2 or M. We found that mutations in upstream elements of the pathway and the Dot1 target residue, histone H3 Lys 79, share DNA damage response phenotypes with mutants lacking Dot1. These defects are specific, insofar as neither the Set1 histone H3 Lys 4 methyltranferase nor Set2 histone H4 Lys 36 methyltransferase are similarly required for G1 checkpoint arrest in the face of DNA DSBs, a finding consistent with a specific role for histone H3 Lys 79 methylation in DNA damage sensing independent of replication forks. Although Dot1 and histone H3 Lys 79 methylation have been studied chiefly for their roles in silencing via Sir protein localization and function (47, 49, 73), Dot1 has previously been implicated by several groups in DNA damage tolerance and checkpoints (11, 18, 20).
The relative lack of DNA damage sensitivity of dot1Δ and rad9-Y798Q tudor domain mutant cells, although surprising, need not be overinterpreted. One model is that the lack of DNA damage sensitivity in the dot1Δ mutant may reflect pleiotropic effects on DNA repair. Perhaps dot1Δ cells relieve silencing and thereby upregulate DNA repair genes, increasing repair efficiency and thus allowing checkpoint defective cells to be insensitive to DNA damage. However, this seems improbable given that the rad9-Y798Q tudor domain mutant has identical checkpoint and DNA damage tolerance phenotypes to dot1Δ and yet almost certainly does not confer similar effects on chromatin and silencing. Furthermore, overexpression of DOT1 does not cause checkpoint defects but does cause silencing defects (73). Nonetheless, we cannot formally rule out that Dot1 has general effects on cell cycle progression mediated through its activation of Rad9 G1/S functions.
Thus, we interpret the lack of DNA damage sensitivity in the dot1Δ mutant as evidence of independent pathways mediating DNA damage response. Our data do show that Dot1-dependent G1/S checkpoint signaling can become important for DNA damage tolerance, since dot1Δ mutants enhance the DNA damage sensitivity of other checkpoint mutants such as rad17Δ and set1Δ. Most significantly, although dot1Δ cells are defective for checkpoint responses in G1/S, they are still competent for the G2 checkpoint. It is well known that yeast lacking homologous recombination (HR) are far more DNA damage sensitive than cells completely deficient for nonhomologous end joining (NHEJ). Thus, bypass of a G1 delay, perhaps leading to a partial deficit in NHEJ but incurring no loss of HR, might confer only negligible DNA damage sensitivity. Indeed, to date, no requirement for G1 checkpoint arrest in budding yeast DNA damage tolerance has been described. Although a role for intra-S-phase checkpoint response in DNA damage tolerance is well established, the defect in dot1Δ is not complete, suggesting that Dot1-independent pathways remain intact and thus can compensate for the dot1Δ defect. Determining in what contexts the Dot1 and Rad9-dependent signal that induces a G1 checkpoint arrest may contribute to DNA damage tolerance will likely contribute to a better understanding of how checkpoint delays enhance DNA damage repair and survival overall.
We found that in wild-type cells, Rad9 is rapidly recruited to chromatin adjacent to an HO break in G1 and that this domain may extend up to 10 kb from the site of damage. Our data suggest that global histone H3 Lys 79 methylation is unaffected by DNA damage, and Lys 79 methylation is neither induced nor decreased adjacent to DSBs. This would appear to rule out Dot1-dependent Lys 79 methylation as a signal for DNA damage. Although Lys 79 methylation may not be necessary for Rad9 recruitment to DSBs in G1 per se, both the initial phase of recruitment and the subsequent retention adjacent to the break were decreased in a dot1Δ mutant. In fact, our conservative approach of normalization of the ChIP data to the input signal, which significantly decreases because of end degradation at the HO break, may actually have led to a slight overestimation of the Rad9 recruitment. Nonetheless, another nucleosome modification such as histone H2A phosphorylation, a regulated histone acetylation, or remodeling may be the primary determinant of Rad9 chromatin binding and retention and/or cooperate with H3 Lys 79 methylation. Indeed, we observed a similarly large domain of enhanced H2A phosphorylation adjacent to G1 HO breaks. That Rad9 encodes paired BRCT domains that may recognize and mediate binding to phosphorylated H2A and/or to activated Rad9 suggests a simple model for Rad9 recruitment and assembly at DSBs after their recognition by Tel1 and/or Mec1. However, this fails to resolve the paradox that although Lys 79 methylation is not DNA damage regulated, it is necessary for G1/S activation of Rad9 and for normal Rad9 recruitment to an HO-induced DSB. Perhaps Rad9 must recognize a dual signal of H2A or another Tel1/Mec1-dependent phosphorylation via BRCT domains and Lys 79 methylation via tudor domains in order to be recruited, retained, and phosphorylated. Based on crystal structures, the Lys 79 side chain projects from a solvent-accessible loop of histone H3 and does not appear to make stable contacts with DNA or other histones in the octamer (41). Although the antigen is a specific mark of euchromatic regions and readily accessible in ChIP assays, higher-order chromatin structures might result in local Lys 79 inaccessibility via nucleosomal stacking (26). Thus, H2A phosphorylation or another chromatin modification may be required to expose methylated Lys 79 to allow interaction with the Rad9 tudor domains. It is striking that Tel1 and/or Mec1 can mediate phosphorylation of H2A in G1 independent of Lys 79 methylation but cannot similarly gain access to Rad9 as a substrate. In G2/M, where Dot1 is dispensable for Rad9 checkpoint function, DNA and chromatin modifications that facilitate HR may promote Rad9 activation. In either case, methylation at Lys 79 might be considered as permissive for DNA damage signaling or as an evolutionarily conserved licensing event allowing recruitment of checkpoint factors and establishment of the checkpoint.
We also studied the Ddc2-Rad53 fusion protein previously shown to bypass rad9Δ mutants for Rad53 activation, checkpoint function, and DNA damage tolerance (36). Ddc2 (ATRIP) normally recruits Mec1 (ATR) to RPA-single-stranded DNA complexes at sites of DNA damage (59). The Ddc2-Rad53 bypass has been used to ascribe Mec1 and Rad9 checkpoint functions to Rad53 recruitment and activation. Although Ddc2-Rad53 restored checkpoint function to dot1Δ and rad9Δ cells in S phase, the checkpoint function was not fully restored after DNA damage in G1. One provocative model is that neither Ddc2 nor Mec1 are rapidly recruited to DNA damage in G1, so that initial steps in the DNA damage response might depend on Tel1. Nonetheless, ddc2Δ cells have a mild G1/S checkpoint defect after IR (data not shown), a finding consistent with a role for Ddc2 in the G1 checkpoint pathway. Alternatively, insofar as our results appear to partly separate recruitment of Rad9 from its activation in G1, similar logic might suggest that recruitment of Rad53 may also be insufficient for activation in the absence of methylated histone H3 Lys 79.
Other histone H3 methylation events at Lys4 and Lys36 are necessary for DNA damage resistance and checkpoints. Two lines of evidence suggest that Set1 and Set2 modulate DNA damage checkpoint via a different mechanism than Dot1-dependent Rad9 recruitment to the site of DNA damage. First, that Set1 and Set2 are required for G1 checkpoint regulation after 4NQO (data not shown) but not IR induced damage suggests that methylation by Set1 or Set2 is not required for DSB response. Second, the enhanced sensitivity of set1Δ dot1Δ and set2Δ dot1Δ to IR and MMS suggest that Set1 and Set2 function in an independent pathway to Dot1. Given that dot1Δ rad17Δ double mutants are also more sensitive to DNA damage than rad17Δ single mutants, perhaps Set1, Set2, and Rad17 function in the same pathway regulating G1 and S phase checkpoints. Although it remains possible that the enhanced DNA damage sensitivity of set1Δ dot1Δ double mutants is exposing DNA repair or transcriptional defects, set1Δ actually suppresses mec3Δ via increased repair gene expression, suggesting that, if anything, repair genes are upregulated in SET mutants (62). Nonetheless, the checkpoint defects observed in set1Δ are likely to be Rad9 independent.
These results add unanticipated complexity to the role of chromatin modifications in checkpoint protein function in budding yeast. The histone code may be particularly important in allowing cells to respond to DNA damage rapidly and effectively, and in a manner that takes into account the absence or availability of repair templates through the cell cycle. Further analysis may establish adaptor proteins such as Rad9 as key translators of histone signals that then mediate proper responses in a cell cycle-dependent manner.
Acknowledgments
We thank C. D. Allis, J. Downs, D. E. Gottschling, J. Haber, D. Lydall, M. A. Osley, D. F. Stern, K. Struhl, and Y. Zhang for generously sharing strains, reagents, methods, and unpublished data.
This study was supported by grants from the NIH (R01 GM60443) and Ludwig Fund for Cancer Research to S.J.K. and the CIHR to J.C. A.J. is supported by the University of Chicago MSTP and an AHA Greater Midwest predoctoral fellowship. J.C. is a CIHR Investigator, and S.J.K. is a Leukemia and Lymphoma Society Scholar.
REFERENCES
- 1.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell. Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 2.Bassal, S., and A. El-Osta. 2005. DNA damage detection and repair, and the involvement of epigenetic states. Hum. Mutat. 25:101-109. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29:426-434. [DOI] [PubMed] [Google Scholar]
- 4.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 5.Birrell, G. W., G. Giaever, A. M. Chu, R. W. Davis, and J. M. Brown. 2001. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. USA 98:12608-12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blankley, R. T., and D. Lydall. 2004. A domain of Rad9 specifically required for activation of Chk1 in budding yeast. J. Cell. Sci. 117:601-608. [DOI] [PubMed] [Google Scholar]
- 7.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 8.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, W. L., F. B. Turner, T. Krishnamoorthy, B. Wolner, S. H. Ahn, M. Foley, J. A. Dorsey, C. L. Peterson, S. L. Berger, and C. D. Allis. 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in Saccharomyces cerevisiae. Curr. Biol. 15:656-660. [DOI] [PubMed] [Google Scholar]
- 10.Choy, J. S., and S. J. Kron. 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 22:8215-8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corda, Y., V. Schramke, M. P. Longhese, T. Smokvina, V. Paciotti, V. Brevet, E. Gilson, and V. Geli. 1999. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat. Genet. 21:204-208. [DOI] [PubMed] [Google Scholar]
- 12.de la Torre-Ruiz, M. A., C. M. Green, and N. F. Lowndes. 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17:2687-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger, N. Bouchard, S. J. Kron, S. P. Jackson, and J. Cote. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16:979-990. [DOI] [PubMed] [Google Scholar]
- 14.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001-1004. [DOI] [PubMed] [Google Scholar]
- 15.Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst, K. Struhl, and Y. Zhang. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12:1052-1058. [DOI] [PubMed] [Google Scholar]
- 16.FitzGerald, J. N., J. M. Benjamin, and S. J. Kron. 2002. Robust G1 checkpoint arrest in budding yeast: dependence on DNA damage signaling and repair. J. Cell. Sci. 115:1749-1757. [DOI] [PubMed] [Google Scholar]
- 17.Game, J. C., G. W. Birrell, J. A. Brown, T. Shibata, C. Baccari, A. M. Chu, M. S. Williamson, and J. M. Brown. 2003. Use of a genome-wide approach to identify new genes that control resistance of Saccharomyces cerevisiae to ionizing radiation. Radiat. Res. 160:14-24. [DOI] [PubMed] [Google Scholar]
- 18.Game, J. C., M. S. Williamson, and C. Baccari. 2005. X-ray survival characteristics and genetic analysis for nine Saccharomyces deletion mutants that show altered radiation sensitivity. Genetics 169:51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannattasio, M., F. Lazzaro, M. P. Longhese, P. Plevani, and M. Muzi-Falconi. 2004. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 23:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giannattasio, M., F. Lazzaro, P. Plevani, and M. Muzi-Falconi. 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280:9879-9886. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 22.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell. Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 23.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 24.Hanway, D., J. K. Chin, G. Xia, G. Oshiro, E. A. Winzeler, and F. E. Romesberg. 2002. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. USA 99:10605-10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartwell, L. H., and T. A. Weinert. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246:629-634. [DOI] [PubMed] [Google Scholar]
- 26.Huyen, Y., O. Zgheib, R. A. Ditullio, Jr., V. G. Gorgoulis, P. Zacharatos, T. J. Petty, E. A. Sheston, H. S. Mellert, E. S. Stavridi, and T. D. Halazonetis. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406-411. [DOI] [PubMed] [Google Scholar]
- 27.Hwang, W. W., S. Venkatasubrahmanyam, A. G. Ianculescu, A. Tong, C. Boone, and H. D. Madhani. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11:261-266. [DOI] [PubMed] [Google Scholar]
- 28.Kao, C. F., C. Hillyer, T. Tsukuda, K. Henry, S. Berger, and M. A. Osley. 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 18:184-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karagiannis, T. C., and A. El-Osta. 2004. Double-strand breaks: signaling pathways and repair mechanisms. Cell. Mol. Life Sci. 61:2137-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitazono, A. A., and S. J. Kron. 2002. An essential function of yeast cyclin-dependent kinase Cdc28 maintains chromosome stability. J. Biol. Chem. 277:48627-48634. [DOI] [PubMed] [Google Scholar]
- 31.Kolodner, R. D., C. D. Putnam, and K. Myung. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557, 2002. [DOI] [PubMed]
- 32.Kondo, T., K. Matsumoto, and K. Sugimoto. 1999. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol. Cell. Biol. 19:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogan, N. J., J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider, M. Johnston, and A. Shilatifard. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277:10753-10755. [DOI] [PubMed] [Google Scholar]
- 34.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 35.Lacoste, N., R. T. Utley, J. M. Hunter, G. G. Poirier, and J. Cote. 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277:30421-30424. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. J., J. K. Duong, and D. F. Stern. 2004. A Ddc2-Rad53 fusion protein can bypass the requirements for RAD9 and MRC1 in Rad53 activation. Mol. Biol. Cell 15:5443-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longhese, M. P., M. Clerici, and G. Lucchini. 2003. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat. Res. 532:41-58. [DOI] [PubMed] [Google Scholar]
- 38.Longhese, M. P., V. Paciotti, R. Fraschini, R. Zaccarini, P. Plevani, and G. Lucchini. 1997. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 16:5216-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 40.Lowndes, N. F., and J. R. Murguia. 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10:17-25. [DOI] [PubMed] [Google Scholar]
- 41.Min, J., Q. Feng, Z. Li, Y. Zhang, and R. M. Xu. 2003. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112:711-723. [DOI] [PubMed] [Google Scholar]
- 42.Mochan, T. A., M. Venere, R. A. DiTullio, Jr., and T. D. Halazonetis. 2004. 53BP1, an activator of ATM in response to DNA damage. DNA Repair 3:945-952. [DOI] [PubMed] [Google Scholar]
- 43.Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt, J. E. Haber, and X. Shen. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767-775. [DOI] [PubMed] [Google Scholar]
- 44.Naiki, T., T. Wakayama, D. Nakada, K. Matsumoto, and K. Sugimoto. 2004. Association of Rad9 with double-strand breaks through a Mec1-dependent mechanism. Mol. Cell. Biol. 24:3277-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakada, D., T. Shimomura, K. Matsumoto, and K. Sugimoto. 2003. The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 31:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neecke, H., G. Lucchini, and M. P. Longhese. 1999. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 18:4485-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625-33628. [DOI] [PubMed] [Google Scholar]
- 49.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 51.Ng, H. H., R. M. Xu, Y. Zhang, and K. Struhl. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277:34655-34657. [DOI] [PubMed] [Google Scholar]
- 52.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 53.Osborn, A. J., and S. J. Elledge. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17:1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peterson, C. L., and J. Cote. 2004. Cellular machineries for chromosomal DNA repair. Genes Dev. 18:602-616. [DOI] [PubMed] [Google Scholar]
- 55.Qin, S., and M. R. Parthun. 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22:8353-8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raths, S. K., F. Naider, and J. M. Becker. 1988. Peptide analogues compete with the binding of alpha-factor to its receptor in Saccharomyces cerevisiae. J. Biol. Chem. 263:17333-17341. [PubMed] [Google Scholar]
- 57.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 58.Rouse, J., and S. P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547-551. [DOI] [PubMed] [Google Scholar]
- 59.Rouse, J., and S. P. Jackson. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9:857-869. [DOI] [PubMed] [Google Scholar]
- 60.Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 61.Sanders, S. L., M. Portoso, J. Mata, J. Bahler, R. C. Allshire, and T. Kouzarides. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603-614. [DOI] [PubMed] [Google Scholar]
- 62.Schramke, V., H. Neecke, V. Brevet, Y. Corda, G. Lucchini, M. P. Longhese, E. Gilson, and V. Geli. 2001. The set1Δ mutation unveils a novel signaling pathway relayed by the Rad53-dependent hyperphosphorylation of replication protein A that leads to transcriptional activation of repair genes. Genes Dev. 15:1845-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 64.Shroff, R., A. Arbel-Eden, D. Pilch, G. Ira, W. M. Bonner, J. H. Petrini, J. E. Haber, and M. Lichten. 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14:1703-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siede, W., J. B. Allen, S. J. Elledge, and E. C. Friedberg. 1996. The Saccharomyces cerevisiae MEC1 gene, which encodes a homolog of the human ATM gene product, is required for G1 arrest following radiation treatment. J. Bacteriol. 178:5841-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siede, W., A. S. Friedberg, and E. C. Friedberg. 1993. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:7985-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 69.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 70.Toh, G. W., and N. F. Lowndes. 2003. Role of the Saccharomyces cerevisiae Rad9 protein in sensing and responding to DNA damage. Biochem. Soc. Trans. 31:242-246. [DOI] [PubMed] [Google Scholar]
- 71.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555-560. [DOI] [PubMed] [Google Scholar]
- 72.van Attikum, H., O. Fritsch, B. Hohn, and S. M. Gasser. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777-788. [DOI] [PubMed] [Google Scholar]
- 73.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 74.Wakayama, T., T. Kondo, S. Ando, K. Matsumoto, and K. Sugimoto. 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 76.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 77.Westmoreland, T. J., J. R. Marks, J. A. Olson, Jr., E. M. Thompson, M. A. Resnick, and C. B. Bennett. 2004. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryot. Cell 3:430-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 79.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 80.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739-34742. [DOI] [PubMed] [Google Scholar]
- 81.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]