Abstract
α1-Adrenergic receptors mediate several biological effects of catecholamines, including the regulation of myocyte growth and contractility and transcriptional regulation of the atrial natriuretic factor (ANF) gene whose promoter contains an α1-adrenergic response element. The nuclear pathways and effectors that link receptor activation to genetic changes remain poorly understood. Here, we describe the isolation by the yeast one-hybrid system of a cardiac cDNA encoding a novel nuclear zinc finger protein, Zfp260, belonging to the Krüppel family of transcriptional regulators. Zfp260 is highly expressed in the embryonic heart but is downregulated during postnatal development. Functional studies indicate that Zfp260 is a transcriptional activator of ANF and a cofactor for GATA-4, a key cardiac regulator. Knockdown of Zfp260 in cardiac cells decreases endogenous ANF gene expression and abrogates its response to α1-adrenergic stimulation. Interestingly, Zfp260 transcripts are induced by α1-adrenergic agonists and are elevated in genetic models of hypertension and cardiac hypertrophy. The data identify Zfp260 as a novel transcriptional regulator in normal and pathological heart development and a nuclear effector of α1-adrenergic signaling.
The endogenous catecholamines epinephrine and norepinephrine are key regulators of numerous physiologic functions, including learning, memory, and cardiovascular and endocrine homeostasis. Their dysregulation has been implicated in human conditions such as depression and addiction and in cardiovascular and metabolic diseases. Their effects are mediated by three classes of adrenergic receptors (ARs), β, α1, and α2, each comprised of three distinct gene products. They all belong to the superfamily of seven transmembrane G-protein-coupled receptors. α1-ARs are critical for a variety of catecholamine actions such as the control of blood pressure, smooth muscle contraction, myocardial function, and glycogenolysis. The importance of α1-ARs in physiology and pathophysiology is evidenced by the wide clinical use of α1-AR agonists and antagonists for the treatment of cardiovascular disease, flu and allergy symptoms, and benign prostate hyperplasia (40, 43). Paradoxically, the molecular mechanisms underlying α1-AR action remain undefined.
Historically, the role of α1-ARs in different biologic systems was largely inferred from pharmacologic studies, but the development of transgenic mice with targeted deletion or overexpression of specific α1-AR subtypes has further confirmed the essential role of specific α1-ARs in regulation of physiologic processes (reviewed in references 40 and 44). For example, α1-null mice rapidly develop hyperinsulinemia, insulin resistance, and obesity in response to high-fat feeding, confirming the important role of α1-AR in the regulation of glucose homeostasis (7). The use of genetically altered mice also confirmed the essential role of α1-ARs in mediating the effects of some psychostimulants and opiates and, more generally, their involvement in the regulation of various aspects of behavior (3, 4, 24, 51).
The phenotypes of mice with genetically altered α1-AR levels further clarified the important role of different α1-AR subtypes in cardiovascular homeostasis. Mice lacking α1b- or α1d-ARs have decreased pressure and contractile responses (8, 45), whereas mice lacking the α1a-receptor subtype are hypotensive (42). At the level of the heart, α1-ARs are involved in mediating both contractile and growth promoting effects of catecholamines and have been linked to the pathogenesis of cardiac hypertrophy. Consistent with this, overexpression of α1b-AR under its own promoter or cardiac-specific expression of a constitutively active α1b-AR mutant produce cardiac hypertrophy (30, 55). Recently, the development of mice lacking both α1a- and α1b-ARs revealed an essential role for α1-AR signaling in physiologic cardiac hypertrophy (36). This finding is consistent with previous reports demonstrating the essential role of catecholamines in embryonic cardiac development (47).
The profound effects of α1-ARs on cell growth and differentiation involve changes in gene expression. Unfortunately, knowledge of transcriptional regulation by α1-ARs remains limited. Transcriptome analysis in whole brains of mice overexpressing the α1b-AR, which suffer from apoptotic neurodegeneration (55), has revealed alterations in genes associated with calcium homeostasis, apoptosis and neuronal signaling (53). Whether any of these genes represent direct α1-AR targets is not known. In other α1-AR target organs such as kidney, liver, and skeletal or smooth muscle, the repertoire of α1-AR downstream genes remains largely unknown.
In contrast, several transcriptional targets of α1-ARs have been identified in cardiac cells, where α1-AR stimulation induces transcriptional changes in an ensemble of cardiac genes, many of which are associated with cardiac hypertrophy. This includes upregulation of immediate-early genes and reinduction of a set of fetal genes such as α-skeletal actin, β-myosin heavy chain, and ventricular atrial natriuretic factor (ANF) which is the hallmark of genetic changes associated with cardiac hypertrophy (reviewed in reference 10). The intracellular signaling cascades and the nuclear factors involved in the growth response of cardiomyocytes to α1-AR stimulation are starting to be elucidated but are not fully understood. α1-AR can activate numerous signaling cascades (52), and α1-induced hypertrophy can be transduced through multiple signaling pathways (reviewed in reference 32). For example, agonist stimulation of α1-AR induces the phospholipase C/protein kinase C (PKC) pathway, the mitogen-activated protein kinase (MAPK) pathway, and phosphatidylinositol 3-kinase and calcium/calmodulin signaling, all of which have been implicated in α1-AR-dependent myocyte hypertrophy. Treatment of cardiomyocytes with α1-AR agonists has also identified a few transcription factors whose expression or activity is targeted by α1-AR. These include c-Jun, c-Fos, and EGR1 (reviewed in reference 1), as well as the transcriptional corepressor CARP (28), which are all activated at the transcriptional level. The α1-agonist phenylephrine (PE) also causes phosphorylation of CREB (29), RTEF-1 (49), GATA-4 (10, 27, 33), and the coactivators CBP and p300 (20). Although some of these transcription factors have been found to bind to and activate α1-inducible promoters (9, 23, 28, 33, 48), their involvement in mediating nuclear α1-AR action remains unclear with the exception of GATA-4, which was shown to be essential for PE response of cardiomyocytes (2, 10).
A critical step in establishing the pathway by which extracellular signals regulate gene transcription in the nucleus is the identification of DNA regulatory elements and nuclear proteins that are required for the transcriptional responses. We previously identified a novel α1-AR regulatory element in the 5′-flanking sequences of the ANF gene; this sequence termed PERE (PE response element) is necessary for maximal transcriptional activation of ANF in response to α1 agonists. The location and sequence of the PERE element are perfectly conserved between ANF genes of different species, suggesting an important role for this element in the regulation of ANF promoter activity. Moreover, PERE elements are also present on the promoter of other α1-inducible cardiac genes. Preliminary characterization of the DNA-binding proteins which interact with the PERE sequence suggested that the PERE protein complexes (PEXs) correspond to as-yet-uncharacterized Sp1-related, zinc-dependent DNA-binding proteins (1).
We now report the isolation, using the yeast one-hybrid interaction system, of a cardiomyocyte-derived cDNA encoding a novel transcription factor consisting of multiple zinc fingers (ZFs) of the Krüppel family, that we termed PEX1. In silico analysis revealed that PEX1 is the rat homolog of the human Zfp260 gene whose protein product and function have not been characterized. PEX1 mRNAs are expressed in a tissue-restricted manner, are highly enriched in the heart, and are developmentally regulated. PEX1 levels are upregulated in response to α1 agonists and are elevated in genetic models of hypertension and cardiac hypertrophy. The PEX1 protein localizes to cardiac cell nuclei, where it acts as a sequence-specific transcriptional activator of the ANF gene. Moreover, PEX1 physically and functionally interacts with GATA-4, a key cardiac regulator. Knockdown of PEX1 using an antisense strategy in cardiomyocytes abrogates the endogenous ANF gene response to α1-AR stimulation. Thus, PEX1 appears to be a novel regulator of cardiac transcription and an effector of α1-adrenergic signaling.
MATERIALS AND METHODS
Plasmids.
The rat ANF reporter plasmids and GATA-4 expression vectors were detailed previously (9, 19). Mutations of the PERE sequence in the ANF promoter constructs were performed by the Altered Sites In Vitro Mutagenesis System (Promega). pcDNA3-PEX1 and pCGN-PEX1 and MBP-PEX1 constructs were generated by subcloning a KpnI/BamHI PCR fragment containing the entire PEX1 coding sequence into the KpnI/BamHI sites of pcDNA3 (Invitrogen) or pCGN. All constructs were confirmed by sequencing.
Cardiomyocyte cultures.
Unless specified otherwise, experiments were performed with primary cultures of cardiac myocytes prepared from 4-day-old Sprague-Dawley rats as previously described (9). Cardiomyocytes were plated at a density of 26,316 cells/cm2 in Primeria six-well plates or petri dishes (Falcon) and cultured for 16 to 20 h in Dulbecco modified Eagle medium containing 10% fetal bovine serum. On the morning of day 2, the medium was replaced by serum-free hormone-free medium. Transfections and luciferase activity determination were carried out by using calcium phosphate precipitation as previously described (9). When specified, cardiomyocytes were stimulated with 0.1 mM PE or vehicle (serum-free hormone-free medium) for the required period.
Reporter constructs for library screen.
Oligonucleotides containing the PERE binding site and EcoRI linkers were annealed, ligated in three tandem repeats, and subcloned into the yeast reporter plasmids pLacZi and pHISi-1 (Clontech). The reporter constructs were sequentially integrated into the same yeast strain YM4271 at different loci, URA3 and HIS3, respectively, yielding YM4271::PERE::lacZ::His3. This dual reporter yeast strain was used as host for the library screen.
Screening of the cDNA library.
A 1-day-old rat ventricular cardiomyocyte cDNA library fused to the GAL4 activation domain was constructed by using the HybriZap two-hybrid cDNA synthesis kit (Stratagene) according to the recommendations of the supplier. The yeast reporter strain YM4271::PERE::lacZ::His3 was transformed with the cDNA library by the lithium acetate-polyethylene glycol method. Approximately 17 × 104 transformants were plated per 150-mm dish containing His− Leu− minimal selective medium supplemented with 6 mM 3-aminotriazole. The positive clones were then subjected to the filter replica method with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) to test their β-galactosidase activities. Positive plasmids were recovered from the yeast, transformed into Escherichia coli, and sequenced.
Generation of anti-PEX1 polyclonal antibodies.
Polyclonal anti-PEX1 antibodies were generated by inoculating rabbits with a purified, bacterially expressed MBP-PEX1 fusion protein encoding amino acids 1 to 115 of the PEX1 protein. The immunoglobulin fraction was purified by using CNBr-activated Sepharose A. The antibody specificity was tested by Western blotting. It did not recognize the related protein OZF.
Indirect immunofluorescence.
HeLa cells were plated on glass coverslips at a density of 30,000 cells/cm2 in 12-well plates (Falcon) in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and then transfected with a the hemagglutinin-PEX1 expression plasmid (pCGN-PEX1) or with pCGN. Cardiomyocytes were plated on glass coverslips at a density of 105 cells/cm2 in 12-well plates. Cells were fixed in 4% paraformaldehyde for 10 min and assayed for PEX1 expression by using the anti-HA antibody (1:500) or the anti-PEX1 antibody (1:500), followed by biotinylated anti-rabbit antibody (1:250) and fluorescein isothiocyanate-avidin (1:500). To differentiate cardiac myocytes from fibroblasts, cells were costained with mouse antidesmin antibody and revealed by rhodamine-conjugated anti-mouse antibody.
Western blotting.
For overexpression studies, HeLa cells were plated at a density of 1 million cells/100-mm-diameter plates (Falcon), and 20 μg of pCGN or pCGN-PEX1 was transfected as described above. At 36 h postransfection, cells were harvested, and nuclear extracts were prepared as described previously (19). Untransfected cardiomyocytes were also used for preparation of nuclear extracts. A total of 20 μg of nuclear extracts was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a Hybond polyvinylidene difluoride membrane, and immunoblotted by using the Renaissance chemiluminescence system (NEN Life Sciences) as described by the manufacturer. Rabbit polyclonal anti-rat PEX1 antibody was used at a dilution of 1/500 and was revealed with an anti-rabbit horseradish peroxidase-conjugated antibody (Sigma) at a dilution of 1/100,000.
Electrophoretic mobility shift assay (EMSA).
Binding reactions were carried out at room temperature for 30 min with 3 μg of cardiomyocyte nuclear extracts in a 20-μl reaction mixture containing 60 mM KCl, 10 mM Tris-HCl (pH 7.9), 5 mM MgCl2, 1 mM ZnCl2, 1 mM EDTA, 1 mM dithiothreitol, 4% Ficoll, 1 mg of poly(dI-dC), 25,000 cpm of radiolabeled double-stranded probe and, when appropriate, 2 μl of rabbit immunoglobulin G or anti-rat PEX1 antibody was added on ice 30 min before incubation with the probe (1). Reactions were then loaded on a 4% polyacrylamide gel and run at 200 V at room temperature in 0.25× Tris-borate-EDTA.
Pull-down assays.
Recombinant MBP and in vitro-translated proteins were produced as previously described (34). Pull-down assays with MBP-PEX1 were carried out essentially as described previously with MBP-LacZ and MBP-SRF as negative and positive controls, respectively, for GATA-4 interaction (34), except that the binding buffer contained 1 mM ZnCl2.
Rats and treatments.
Male Wistar-Kyoto rats (WKY) and spontaneously hypertensive rats (SHR) were obtained from Taconic farms. Rats were maintained in standard rat diet and water ad libitum and kept in a 12-h light-dark cycle. WKY and SHR of 15 weeks of age were randomly divided into three groups (n = 4 to 6 per group): untreated normotensive (WKY), untreated hypertensive, and hypertensive treated with hydralazine (25 mg/kg/day; Sigma) for 3 weeks in the rat chow. One day before sacrifice, systolic blood pressure (SBP) was measured by the tail-cuff method in conscious warmed animals. The hearts were removed and weighed. Atria and ventricles were carefully dissected, frozen in liquid nitrogen, and stored at −80°C until RNA extraction. Tissue samples from age- and weight-matched animals (n = 6) were pooled in two batches for RNA extraction. All animal procedures were approved by the IRCM Animal Care Committee and conducted according to the recommendations of the Canadian Council on Animal Care.
RNA analysis.
Total RNA was isolated from cardiomyocytes or from rat tissues with TRIzol (Invitrogen). Northern blotting and semiquantitative reverse transcription-PCR were carried out as previously described (19). Rat cDNA probes for PEX1, ANF, 18S, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were used for Northern blot. QPCR was carried out on cDNA generated with the Omniscript RT Kit (QIAGEN, Inc.) with the Quantitect SYBR Green PCR kit (QIAGEN) in a MX4000 real-time PCR machine (Stratagene). The oligonucleotides were designed to have a melting temperature of 60°C and were used with an annealing temperature of 58°C. The oligonucleotides used for QPCR were 5′-CCGATAGATCTGCCCTCTTG-3′ (forward) and 5′-TCCAGGAGGGTATTCACCAC-3′ (reverse) for ANF and 5′-TCTGGGCAAGGAGAGATTTG-3′ (forward) and 5′-CCGCCAAACTTCTTGGATTC-3′ (reverse) for 40S ribosomal protein S16.
Immunohistochemistry.
Mouse embryos of 9.5, 10.5 and 14.5 day postcoitum (dpc), and 17.5 dpc mouse fetal hearts, stomach and intestine, as well as 5 days postnatal hearts and lungs, and 150 days old adult wild-type and AT1R transgenic heart with cardiac hypertrophy (38) were dissected, paraformaldehyde-fixed and paraffin-embedded. Immunohistochemistry was performed as previously described (2). The anti-PEX1 antibody was used at 1:200 dilution.
Adenovirus preparation and infections.
Two recombinant replication-deficient type 5 adenoviruses expressing antisense regions directed specifically toward PEX1 (AS-PEX1 and HA-AS-PEX1) were generated by using the AdEasy XL Adenoviral Vector System (Stratagene) developed by the laboratory of Bert Vogelstein. Briefly, the HA-AS-PEX1 adenovirus was generated by first subcloning a 442-bp KpnI/BglII fragment containing proximal part of 5′-untranslated region (UTR) and the two first ZFs of rat PEX1 gene into KpnI/BglII in Ad5 shuttle vector pAdTrack-CMV (generously provided by Bert Vogelstein), and the adenovirus was generated by recombination with the pAdEasy-1 as described previously (21). The other adenovirus, AS-Pex1, was generated by first subcloning a 366-bp DNA fragment containing of 5′UTR sequence into BglII/HindIII of pShuttle-CMV (Stratagene), and then the shuttle vector was linearized with PmeI and transformed into BJ5183-AD-1-competent cells. Transformants were selected for kanamycin resistance, and recombinants were subsequently identified by restriction digestion. Once a recombinant was identified, it was produced in bulk by using the recombination-deficient XL10-Gold strain. Purified recombinant adenovirus plasmid DNA was digested with PacI to expose its inverted terminal repeats and was then used to transfect AD-293 cells where deleted viral assembly genes are complemented in vivo. The viruses were produced, and titers were determined as previously described (9) or by using the BD Adeno-X virus purification and titer kits (Clontech). Cardiomyocytes were infected by incubation overnight with 10 PFU of HA-AS-PEX1 or 10 to 50 infectious units of AS-PEX1 per cell in the culture medium. The following day, the medium was replaced with fresh medium.
Statistics.
The data are reported as means ± the standard errors of the mean (SEM). A Student unpaired t test was used to compare two groups. Multiple group comparisons were made by using the one-way analysis of variance test, by the Student-Newman-Keuls test. In all cases, differences were considered to be statistically significant when the P value was <0.05.
RESULTS
The PERE element contributes to both basal and PE-induced ANF promoter activity.
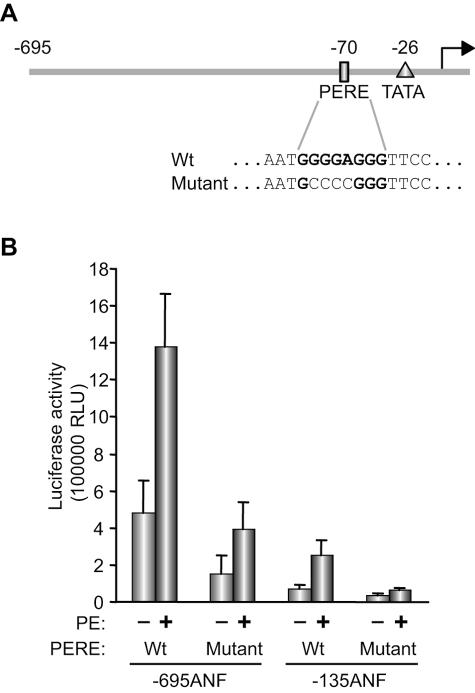
ANF is the major secretory product of the heart and its promoter has served as a paradigm for the elucidation of the regulatory networks controlling cardiac transcription (46). The ANF promoter contains several regulatory elements required for cell specificity and hormone response. We previously showed that an evolutionary conserved sequence, termed PERE, within the proximal promoter was essential for α1-agonist (PE) stimulation of ANF promoter activity (1). The effect of a mutation in the PERE sequence that abolishes in vitro interaction with cardiac nuclear proteins was evaluated in primary cardiomyocyte cultures. The introduced mutation (Fig. 1A) was generated in the context of both the full-length (−695 bp) and the proximal (−135 bp) ANF promoters. Transfection experiments in ventricular cardiomyocytes showed that, in both contexts, basal promoter activity was reduced by ca. 40 to 50% compared to that of the corresponding wild-type constructs (Fig. 1B). Moreover, the response of the mutant promoters to PE stimulation was reduced by ca. 50%, confirming the importance of the PERE element for basal, as well as PE-inducible, ANF transcription.
FIG. 1.
The PERE element affects both basal and PE-induced promoter activity. (A) Schematic representation of the −695 ANF promoter with the sequence of the wild-type (Wt) and mutated PERE. (B) Cardiomyocytes were transfected with wild-type and mutated PERE −695 ANF promoter luciferase reporter constructs and stimulated for 48 h with 0.1 mM PE. The data shown represent the mean ± the SEM of at least six independent determinations.
Isolation of a novel cardiac cDNA clone encoding a PERE interacting protein.
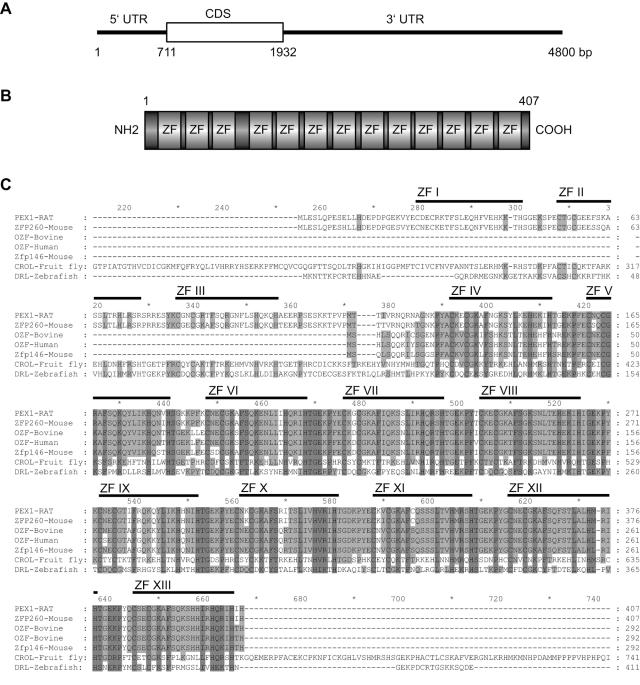
The yeast one-hybrid strategy was used to screen a 1-day-old rat cardiomyocyte cDNA library. Three tandem copies of the PERE element were ligated together and subcloned upstream of the minimal promoter of the pHISi-1 and pLacZi reporter plasmids and integrated into the yeast genome of YM4271. For a more stringent library screening, we constructed a dual reporter strain by sequentially integrating the HIS3 and lacZ reporters into the same yeast genome at different loci. Approximately 2.5 × 106 clones were screened in one transformation. Based on large colony size and rapid growth, a total of 130 histidine positive clones were selected. Eighteen of these clones were positive in the β-galactosidase assay and were all sequenced. One cDNA was found to encode a putative transcription factor with multiple ZF motifs and was further characterized. In silico sequence searching in the databases revealed that this cDNA was the rat ortholog of mouse Zfp260, a gene whose function has not been elucidated (accession number U56862) (6). This cDNA termed PEX1 for PERE complex 1 rescued growth of yeast on His− selective media but was less potent to drive rapid growth of transformant yeast for the mutant construct.
The 4.8-kb PEX1 cDNA contains a 1221-bp open reading frame predicted to encode a 407-amino-acid protein composed of 13 ZFs of the C2H2-type and H/C links (Fig. 2A and B) which would belong to the Krüppel subfamily of ZF proteins (5). In silico sequence analysis did not show any conventional transactivation domain in the coding region. However, the protein possesses several putative phosphorylation sites for PKC, PKA, and casein kinase II.
FIG. 2.
Characteristics of PEX1 mRNA and protein. (A and B) Schematic representation of PEX1 mRNA (A) and protein (B). (C) Alignment of protein sequence of rat PEX1 with homologous proteins. The sequence of PEX1 ZFs are depicted by bold lines and the ZFs are identified by ZF-I to ZF-XIII. CDS, coding sequence. Identical and conserved residues are highlighted by dark and light gray shading, respectively.
Comparison of the amino acid sequences of the rat and murine PEX1 showed a high degree of homology (95%). Searching in databases revealed a PEX1-related protein in mice and humans: OZF, also named Zfp146 (6, 25), whose function is not yet determined. Mouse PEX1 is larger than mouse OZF, with three additional N-terminal ZFs (ZFs I to III). PEX1 and OZF share high homology in the region containing ZFs IV to XIII (Fig. 2C). The Drosophila rerio protein draculin (accession number NP571052.1) and the D. melanogaster protein CROL (accession number AAF53121.1) are also highly homologous to PEX1 in ZFs IV to XIII region (Fig. 2C). Draculin is expressed during early patterning of the zebra fish embryo (22), and crooked legs is required for leg morphogenesis and ecdysone-regulated gene expression during Drosophila metamorphosis (13, 14).
PEX1 is an early α1-adrenergic target.
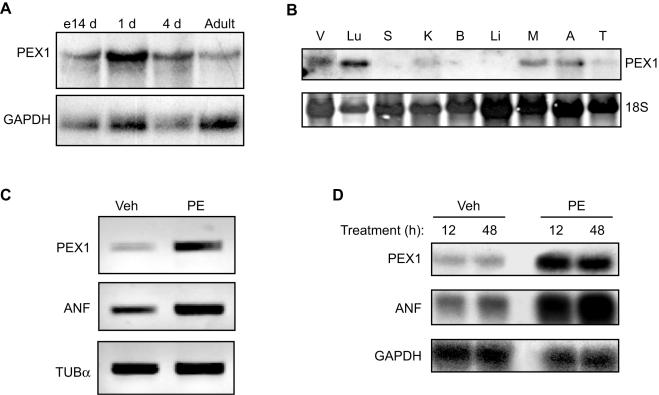
We analyzed the expression of the rat PEX1 gene by Northern analysis. A single transcript of ca. 4.8 kb was detected in total RNA from embryonic day 14 (E14) heart and from adult heart, as well as from cultured cardiomyocytes isolated from 1- or 4-day-old rat hearts (Fig. 3A and B). PEX1 transcripts were also detected in other tissues, notably in lung, skeletal muscle, and adrenal glands (Fig. 3B). Interestingly, most of these tissues are well known α1-AR targets that express α1-ARs (18). In addition to spatial regulation, PEX1 expression was regulated by α1-adrenergic agonists. PE stimulation of primary cardiomyocyte cultures significantly increased PEX1 mRNA levels as early as 6 h after PE treatment; this induction, which was accompanied by an increase of ANF mRNA (Fig. 3C and D), reached fourfold and was sustained for 48 h (the maximal time examined).
FIG. 3.
Expression and regulation of PEX1 mRNA. (A) PEX1 and GAPDH mRNA levels were analyzed by Northern blot in total RNA isolated from the heart at E14 and from adult heart, as well as from primary cardiomyocyte cultures prepared from neonatal (1- and 4-day-old) rats. (B) Tissue distribution of PEX1 mRNA on total RNA from adult rat tissues. V, ventricles; Lu, lung; S, stomach; K, kidney; B, brain; Li, liver; M, skeletal muscle; A, adrenal glands; T, testis. (C) PEX1, ANF, and tubulin-α mRNA levels were determined by reverse transcription-PCR with total RNA extracted from cardiomyocytes isolated from 4-day-old rats treated or not treated with PE for 6 h. (D) PEX1, ANF, and GAPDH mRNA levels were analyzed by Northern blotting with total RNA extracted from cardiomyocytes isolated from 4-day-old rats treated with PE for longer times. Veh, vehicle.
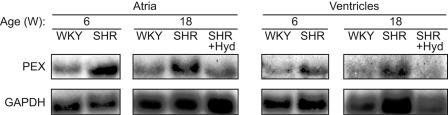
In light of these results and since α1-AR-mediated sympathetic hyperactivity is well documented in SHR (41), we analyzed cardiac PEX1 mRNA expression in SHR at 6 and 18 weeks of age. As shown in Table 1, the SBP is increased at both ages in SHR compared to the controls; additionally, older SHR animals develop cardiac hypertrophy. PEX1 mRNA levels were increased twofold in both cardiac compartments of 6- and 18-week-old SHR (Fig. 4), suggesting that increased PEX1 expression correlates with high blood pressure. This was further confirmed by administration of hydralazine, an arterial vasodilator that reduces blood pressure without affecting cardiac hypertrophy. As expected, 3 weeks of treatment with hydralazine at 25 mg/kg/day led to a significant decrease of SBP but did not affect cardiac hypertrophy (Table 1, HW/BW). Hydralazine treatment also blunted the increase in PEX1 mRNA levels in SHR (Fig. 4). Thus, both in vitro and in vivo PEX1 expression is upregulated by activation of α1-adrenergic receptors.
TABLE 1.
Hydralazine reduces the increase in SBP without altering the development of cardiac hypertrophy in SHRa
| Group(s) | Age (wk) | n | Mean value ± SEM
|
|||
|---|---|---|---|---|---|---|
| BW (g) | HW (g) | HW/BW (mg/g) | SBP (mmHg) | |||
| WKY | 6 | 6 | 176.0 ± 4.1 | 1.51 ± 0.07 | 8.59 ± 0.37 | 105.0 ± 4.3 |
| SHR | 6 | 6 | 137.7 ± 4.0† | 1.16 ± 0.02† | 8.42 ± 0.24 | 133.3 ± 2.5† |
| WKY | 18 | 6 | 510.3 ± 9.3 | 2.16 ± 0.05 | 4.24 ± 0.11 | 114.2 ± 27 |
| SHR | 18 | 4 | 363.5 ± 8.7† | 1.72 ± 0.03† | 4.76 ± 0.16* | 191.2 ± 8.2† |
| SHR + Hyd | 18 | 6 | 386.7 ± 17.8† | 1.87 ± 0.05† | 4.85 ± 0.12* | 167.2 ± 3.9†‡ |
Body weight (BW), heart weight (HW), and SBP were determined in WKY, SHR, and SHR treated with hydralazine (Hyd) as described in Materials and Methods. * and †P < 0.05 and P < 0.01 versus WKY; ‡, P <0.05 versus SHR.
FIG. 4.
The increase in PEX1 mRNA level in the atria and ventricles of SHR correlates with high blood pressure. The levels of PEX1 and GAPDH mRNA were analyzed by Northern blot in total RNA isolated from the atria and ventricles of 6- and 18-week-old control (WKY) and SHR treated or not with the antihypertensive agent hydralazine (Hyd) for 3 weeks. W, weeks. Each lane corresponds to RNA extracted from a pool of three animals.
Spatial and temporal regulation of PEX1 in embryonic and postnatal development.
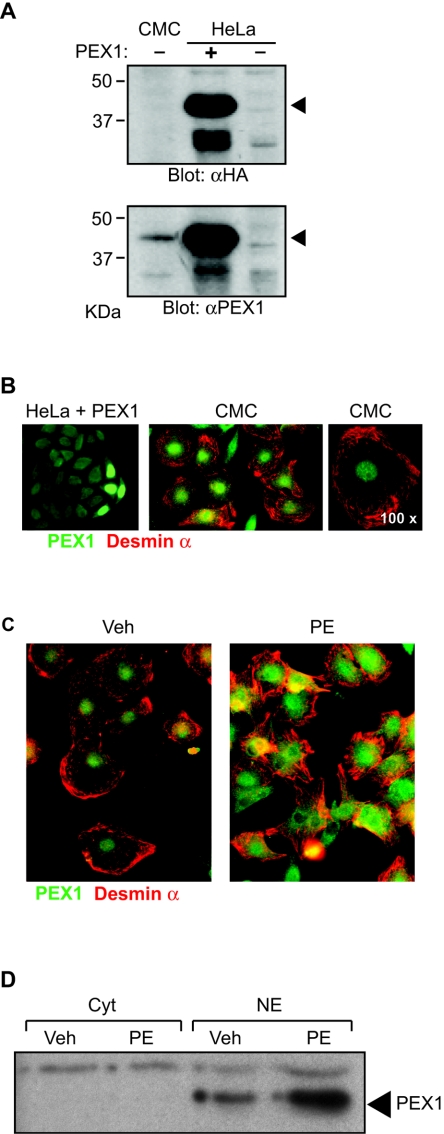
To determine the ontogeny, cell type specificity, and subcellular localization of PEX1 protein, we generated an anti-PEX1 antibody against residues 1 to 115 of PEX1, thus avoiding cross-reactivity with the related OZF protein. In Western blots, anti-PEX1 antibody detected ectopically expressed HA-PEX1 in HeLa cells, which was also detected by the anti-HA antibody (Fig. 5A). The in vitro-translated PEX1 but not OZF, as well as endogenous PEX1, in cardiomyocyte nuclear extracts was also detected by the anti-PEX antibody (Fig. 5A and data not shown). Immunocytofluorescence revealed the presence of both transfected and endogenous PEX1 exclusively in the nuclei (Fig. 5B). Consistent with the observed changes at the transcript level (Fig. 3), protein analysis also revealed that PEX1 level was increased in cardiomyocytes stimulated with PE (Fig. 5C and D).
FIG. 5.
Expression and regulation of PEX1 protein in cardiomyocytes. (A) Western blots were generated with nuclear extracts isolated from cardiomyocytes, HeLa cells transfected with HA-tagged PEX1 expression vector, or an empty vector. The membranes were incubated with anti-HA (αHA) or anti-PEX1 (αPEX1) antibodies. (B) PEX1 was detected by indirect immunofluorescence with the anti-PEX1 antibody in HeLa cells transfected with an HA-tagged PEX1 expression vector (left panel) and in postnatal cardiomyocytes. The right panel shows labeling in cardiomyocytes at a 2.5-fold-higher magnification. (C) PEX1 protein level was determined by immunofluorescence in cardiomyocytes treated with vehicle (Veh) or 0.1 mM PE for 48 h. (B) and (C) The cardiomyocytes were costained with an anti-desmin α antibody. (D) The level of PEX1 protein was determined by Western blotting with the anti-PEX1 antibody in nuclear (NE) and cytoplasmic (Cyt) extracts from cardiomyocytes stimulated (PE) or not stimulated (Veh) with 0.1 mM PE for 48 h. CMC, cardiomyocytes.
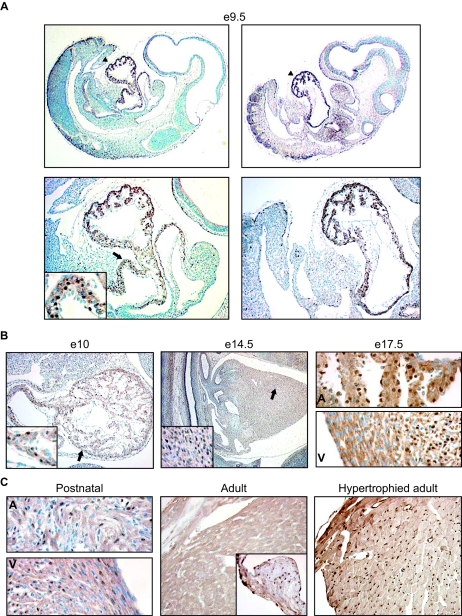
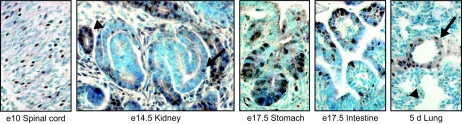
Next, we used immunohistochemistry to study the developmental expression of PEX1 in mouse hearts at different embryonic stages and in postnatal and adult hearts. PEX1 is detected in cardiomyocyte nuclei as early as E9.5, and the heart is the predominant site of PEX1 expression at this stage (Fig. 6A). PEX1 expression is maintained throughout embryonic development in the atria and in the ventricular walls and trabeculae (Fig. 6B). Labeled cells are also present in the outflow tract, the truncus arteriosus, the developing atrioventricular valve, and the cushion mesenchyme (data not shown). PEX1 expression appeared to decrease after E14 and by E17.5; it was spatially redistributed, with highest levels in subendocardial myocytes and the septum and no expression in epicardial and apical myocytes (Fig. 6B, right panel and data not shown). PEX1 was also strongly expressed in the atrioventricular valve (data not shown). During postnatal development, PEX1 expression decreased in both atria and ventricles (Fig. 6C). In the adult mouse heart, PEX1 expression was found in the aortic valve in scattered cells and in the atria, ventricles, and septum (Fig. 6C and data not shown). Interestingly, PEX1 immunoreactivity was markedly upregulated in hypertrophied adult ventricles of transgenic mice overexpressing the angiotensin II receptor (Fig. 6C, right panel). Thus, PEX1 expression appears to be highly regulated during embryonic and postnatal cardiac development. The pattern of PEX1 protein expression paralleled the findings obtained at the mRNA levels (Fig. 3 and data not shown). Outside the heart, PEX1 immunoreactivity was found in embryonic and postnatal vascular smooth muscle cells and in epithelial cells of the lung, gut, and kidney at sites of epithelial morphogenesis and in the spinal cord (Fig. 7 and data not shown).
FIG. 6.
Developmental pattern of PEX1 expression in the mouse heart. The expression of PEX1 was determined by immunohistochemistry with the anti-PEX1 antibody on sections from embryos of E9.5, E10, E14, and E17.5 (A and B); from postnatal and adult mouse hearts (C); and from a mouse model of angiotensin II-induced cardiac hypertrophy (38). (A) The two upper panels show expression of PEX1 in the heart in whole embryos. The portion indicated with arrowheads is magnified four times and shown below. (A) and (B) The areas indicated with arrows are magnified four times and shown in the inserts. (C) The insert in the middle panel shows a portion of the aortic valve. A, atria, V, ventricles.
FIG. 7.
Extracardiac expression of PEX1 in embryonic development. Immunohistochemical staining of tissue sections with the anti-PEX1 antibody revealed PEX1 presence in embryonic gut (stomach and intestine), spinal cord, and postnatal lung. The arrow and arrowhead in the lung panel indicate PEX1-positive smooth muscle and epithelial cells, respectively. The arrow and arrowhead in the kidney panel indicate PEX1 positive tubular and S-shaped body cells, respectively.
PEX1 is a transcriptional regulator and a GATA-4 cofactor.
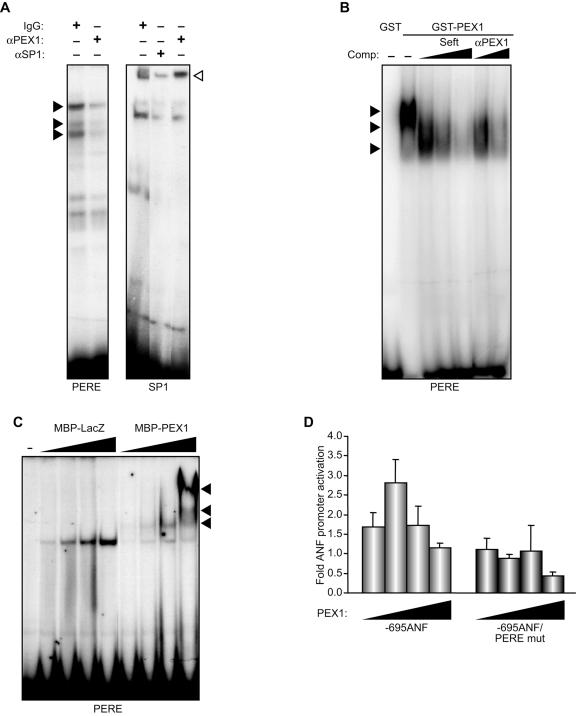
The isolation of PEX1 using a one-hybrid strategy reflected the ability of PEX1 to bind to the PERE element. To confirm that endogenous PEX1 is part of the DNA-binding complex detected over PERE, we performed an EMSA and tested the effect of anti-PEX1 antibody on the binding of cardiac nuclear protein extracts on the PERE probe. As previously described (1), incubation of the PERE probe with cardiomyocyte nuclear extracts lead to the formation of three specific complexes (Fig. 8A, closed arrowheads). Addition of the anti-PEX1 antibody abrogated complex formation, suggesting that PEX1 is part of these complexes and that its presence is required for formation of a DNA-binding complex (Fig. 8A). The specificity of the PEX antibody was demonstrated by its inability to displace SP1 binding over its probe (Fig. 8A, open arrowhead). We also directly confirmed the ability of PEX1 to bind to the PERE element using gel shifts with bacterially expressed GST-PEX1 protein. Recombinant PEX1 strongly bound to the PERE probe, and this binding was competed for by excess cold probe and was blocked by the anti-PEX1 antibody (Fig. 8B). Interestingly, displacement experiments suggested that PEX1 binding may be multimeric as faster migrating complexes appeared at lower doses of cold competitor. Consistent with this, recombinant MBP-PEX1 also produced increasingly larger multimeric complexes in a dose-dependent manner (Fig. 8C).
FIG. 8.
PEX1 is a transcriptional regulator of the ANF promoter. (A) EMSA was performed with nuclear extracts prepared from cardiomyocytes on PERE and SP1 probes. Binding disruption with antibodies to PEX1 (αPEX1), SP1 (αSP1), or immunoglobulin G were used to confirm the identity of the PERE (closed arrowheads) and SP1 (open arrowhead) DNA-binding complex in cardiomyocytes. (B and C) Binding of bacterially produced PEX1 on the PERE probe. In panel B, GST-PEX1 was used; competition with cold PERE (self) was done at a 100- to 500-fold excess, and antibody blocking was performed with increasing amount of the anti-PEX1 antibody (αPEX1). In panel C, increasing amounts of MBP-PEX1 or MBP-LacZ were used with the PERE probe; the close arrowheads indicate the position of the DNA-binding complexes specifically obtained with the recombinant PEX1 protein. (D) Cardiomyocytes were cotransfected with wild-type and mutant ANF luciferase reporters and increasing amounts of PEX1 expression vector. The data shown represent the mean ± the SEM of at least six independent determinations.
Next, we analyzed the ability of PEX1 to modulate ANF promoter activity. Reporter constructs driven by the intact or PERE-mutated ANF promoter were cotransfected with increasing doses of PEX1 expression vector into different cell types. PEX1 activated the ANF promoter in a dose-dependent manner. This effect was dependent on the presence of the PERE element as its mutation abrogated promoter activation (Fig. 8D). These results indicate that PEX1 is a transcriptional activator of ANF.
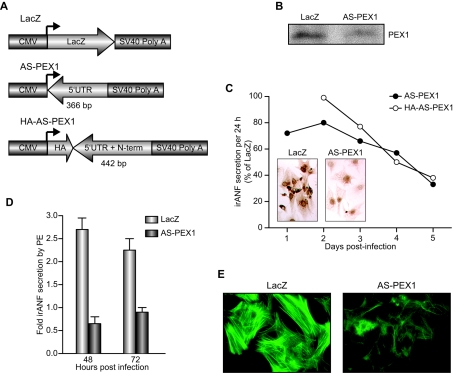
To further confirm the role of PEX1 in basal and/or α1-AR-induced transcription, we generated two different adenovirus vectors expressing two antisense PEX1 transcripts (Fig. 9A) and used them to infect primary cardiomyocyte cultures. As a control, cardiomyocytes were infected with an adenovirus expressing LacZ. The effect of the antisense-PEX1 vector on endogenous PEX1 levels was monitored by Western blotting and immunohistochemistry. A 2.5- to 3-fold reduction in PEX1 protein was achieved 4 days after infection with 10 PFU/cell (Fig. 9B). Under these conditions, ANF levels were consistently reduced by 40 to 50% (Fig. 9C, open circles). The effect of PEX1 depletion on ANF expression was assessed by using quantitative PCR and immunohistochemistry and by measuring secreted immunoreactive ANF (irANF) in the culture media. Since ANF is constitutively secreted from postnatal ventricular myocytes, measurements of irANF in the medium faithfully reflect ANF gene transcription and allow longitudinal assays. As shown in Fig. 9C, the two distinct PEX1 antisense vectors produced a significant, time-dependent decrease in secreted irANF (50 to 60%). The effect was dose dependent, and maximal inhibition was observed after 5 days of adenoviral infection (Fig. 9C and data not shown), a time course highly similar to the one reported previously for GATA-4 effect by using the same approach (9). As expected, cardiomyocytes infected with AS-PEX1 adenovirus had decreased endogenous ANF content and mRNA levels compared to LacZ-infected cardiomyocytes (Fig. 9C, inset, and data not shown). Next, we assessed the effect of PEX1 knockdown on ANF upregulation in response to PE. As shown in Fig. 9D, the presence of the AS-PEX1 adenovirus completely blocked the response to PE stimulation. Interestingly, at a lower virus titer, AS-PEX1 blocked PE-induced ANF expression without altering basal levels (data not shown). Thus, the genetic response to α1-adrenergic agonist appeared exquisitely sensitive to intact PEX1 expression.
FIG. 9.
PEX1 regulates the endogenous ANF gene in cardiomyocytes. (A) Schematic representation of adenovirus constructs expressing LacZ, a PEX1 antisense containing 366 bp of the 5′UTR (AS-PEX1), and another PEX1 antisense containing 442 bp of the 5′UTR and the N-terminal (N-term) coding sequence (HA-AS-PEX1). All adenovirus constructs are driven by the cytomegalovirus (CMV) promoter and have a simian virus 40 (SV40) poly(A) sequence to stabilize the RNA. (B) PEX1 protein levels in cardiomyocytes infected with the AS-PEX1 or the control LacZ adenoviruses as detected by Western blotting. The anti-PEX1 antibody was used with nuclear extracts prepared 4 days postinfection. (C) ANF expression was determined in ventricular cardiomyocytes by using immunohistochemistry, and secreted ANF levels were determined by radioimmunoassay and represent accumulations over 24 h. (D) The level of secreted ANF in the media was determined by radioimmunoassay at 48 and 72 h postinfection with LacZ or AS-PEX1 adenoviruses and chronic treatment with vehicle or 0.1 mM PE. (E) Actin filament organization was examined by using phalloidin-fluorescein isothiocyanate staining in cardiomyocytes infected with LacZ or AS-PEX1 adenoviruses and treated for 48 h with 0.1 mM PE. Note how the cells infected with AS-PEX1 fail to reorganize the myofibrils in response to PE.
Another feature of α1-AR stimulation of cardiomyocytes is cytoskeletal reorganization (10). PEX1 knockdown also interfered with PE-induced myofibrillar reorganization (Fig. 9E). These effects on ANF gene expression and on the genetic and cytoskeletal response to PE were highly reminiscent of the ones observed in cardiomyocytes in which GATA-4 levels were downregulated by using a similar approach (10, 11). We checked whether inhibition of GATA-4 might account for the AS-PEX1 phenotype. Using quantitative PCR analysis, we were unable to detect any decrease in GATA-4 expression in cardiomyocytes infected with the AS-PEX1 adenovirus; conversely, PEX1 transcripts were not decreased in cardiomyocytes infected with antisense GATA-4 adenovirus (data not shown). Thus, the similar phenotype elicited by downregulation of either GATA-4 or PEX1 was not due to a hierarchical relationship between the two proteins.
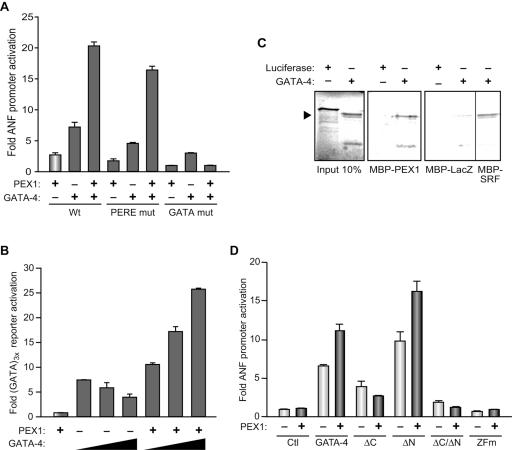
We then tested the possibility that the similar effects elicited by inhibiting PEX1 or GATA-4 reflect cooperative interaction between the two transcription factors. Using the proximal ANF promoter, we found that PEX1 and GATA-4 functionally cooperate to activate transcription (Fig. 10A). GATA-4/PEX1 synergy depends on the presence of the GATA binding site but does not require the PERE element, although maximal synergy is achieved when both elements are present. Thus, PEX1/GATA-4 synergy requires GATA-4 binding to DNA, whereas PEX1 may be recruited to the promoter through interaction with GATA-4. Indeed, GATA sites on the (GATA)3x-Luc reporter are sufficient to mediate GATA-4/PEX1 synergy (Fig. 10B). Consistent with this, GATA-4 and PEX1 were found to physically interact in a pull-down assay (Fig. 10C), probably through direct physical interaction (Fig. 10B). Structure-function analysis indicates that the C-terminal domain of GATA-4, as well as an intact DNA-binding domain, is required for functional cooperation with PEX1. Thus, PEX1 appears to be a novel GATA-4 collaborator, and an epistatic relationship between the two factors is suggested. Together, the data are consistent with an important role for PEX1 in basal and α1-adrenergic-induced cardiac transcription. Moreover, a dual mode for PEX1 action is revealed, one involving direct binding to DNA via PERE elements and the other involving recruitment to promoter bound GATA-4 via protein-protein interactions.
FIG. 10.
PEX1 is a GATA-4 cofactor. (A) Mapping of the DNA elements required for GATA-4/PEX1 synergy. HeLa cells were cotransfected with wild-type (Wt), mutated PERE (PERE mut), and mutated GATA (GATA mut) −695-bp ANF promoter luciferase constructs and PEX1, GATA-4, or both expression vectors. (B) GATA elements are sufficient to mediate GATA-4/PEX1 synergy. HeLa cells were cotransfected with a minimal BNP promoter driven by multimerized GATA binding sites (GATA3x) and increasing amount of GATA-4 in presence or absence of PEX1 expression vector. (C) PEX1 directly interacts with GATA-4 in vitro. Luciferase and GATA-4 were translated and 35S labeled; LacZ, SRF, and PEX1 were produced in bacteria as MBP fusion, and the in vitro pull-down assays were performed as described in Materials and Methods. MBP-SRF and MBP-LacZ were used as positive and negative controls for GATA-4 interaction. (D) Mapping of GATA-4 domains required for PEX1 synergy. HeLa cells were cotransfected with the −695 ANF promoter luciferase reporter and different GATA-4 expression vectors: wild-type GATA-4 (1 to 440), C-terminal-deleted GATA-4 (ΔC, 1 to 332), N-terminal-deleted GATA-4 (ΔN, 210 to 440), and N- and C-terminal-deleted GATA-4 (ΔC/ΔN, 210 to 332) or with a point mutation in the second zinc finger (ZFm), with or without PEX1. All GATA-4 constructs were described previously (9). In panels A, B, and D, the data show the means ± the SEM of at least four independent determinations.
DISCUSSION
The α1 subfamily of adrenergic receptors mediates several of the biological effects of endogenous catecholamines on the visceral, endocrine, nervous, and cardiovascular systems. They also transduce the actions of some psychostimulants and are therefore linked to behavioral processes such as addiction. Despite their evident relevance to physiology and pathophysiology, the mechanisms by which α-AR profoundly alter cell fate and behavior remain undefined. Although it is well accepted that α1-ARs regulate gene transcription, the nuclear signaling pathways and transcription factors that mediate α1-AR actions remain poorly understood. We now report the isolation of a novel transcription factor enriched in the heart, PEX1/Zfp260, that mediates at least some of the effects of α1-ARs. Our data show that PEX1, a member of the Krüppel family of ZF proteins, acts as a transcriptional regulator of the ANF gene and functionally cooperates with GATA-4, a key cardiac regulator. The results also suggest that PEX1/GATA-4 interaction is critical for transducing the nuclear and cytoskeletal effects of α1-adrenergic agonists. In addition to identifying a novel regulator of cardiac gene expression, the study reported will help elucidate the signaling cascade linking membrane activation of α1-ARs to nuclear changes in the heart and in other α1-AR target organs.
PEX1 shows high similarity to another Krüppel protein, OZF/Zfp146, which is also expressed in the heart and other tissues (25). The two are contiguously present on mouse chromosome 7 and human 19q13 within a region with frequent rearrangements and amplifications in tumors (12). The function of OZF is not known, but a role in cellular proliferation could be inferred from the findings that it is overexpressed or amplified in human pancreatic cancer (16), as well as in pancreatic carcinoma cell lines (12). The regulation of PEX1 and its role in mediating α1-AR effects on cytoskeletal organization raise the possibility for a role in cell growth. First, PEX1 levels in the heart peak between E9.5 and E13.5 which is the period of most rapid growth of this organ (37). Postnatally, as cardiomyocyte proliferative ability ceases, PEX1 levels are dramatically downregulated but are upregulated in two models of cardiac hypertrophy. It is noteworthy that this expression pattern is similar to that of ANF, a marker for the hypertrophy genetic program. In cardiomyocyte cultures, PEX1 is required for α1-induced ANF transcription, cytoskeletal reorganization, and myocyte hypertrophy. Together, the results suggest that, at least in the heart, PEX1 may play an important role in transducing the growth effects of catecholamines. In addition to their well-established functions, catecholamines play important roles in development by acting as morphogens and growth-promoting agents in embryogenesis (39). Indeed, several studies have provided evidence for expression of catecholamine-synthesizing enzymes (15), catecholamines (47, 54), and α1-ARs (50) in the heart and other structures during embryonic development. Moreover, targeted disruption of the tyrosine hydroxylase or dopamine β-hydroxylase genes lead to embryonic lethality, apparently of cardiac failure (47, 54). In both cases, cardiomyocyte cell size was decreased, and they were disorganized, resulting in atrophied hearts as early as E10.5 (47). Interestingly, the peak of PEX1 expression in the embryonic heart matches the transient burst of phenylethanolamine N-methyltransferase, the final enzyme in catecholamine synthesis which takes place between E9.5 and E13.5 (15). Together with its demonstrated function as a transcriptional regulator and an effector of α1-adrenergic signaling in cardiomyocytes, these data raise the intriguing possibility that PEX1 may have a role in the control of cardiomyocyte proliferation and/or the response of cardiac cells to catecholamines during development. In this respect, it is noteworthy that, besides the heart, PEX1 is expressed in catecholamine-synthesizing tissues and in α1-AR target cells, notably vascular smooth muscle, where α1-AR have been shown to mediate proliferative growth (17).
Finally, knockdown of PEX1 in postnatal myocytes decreased endogenous ANF gene expression and interfered with the genetic and cytoskeletal response to α1-adrenergic stimulation. This phenotype was highly reminiscent of the one obtained with loss of GATA-4 function in cardiomyocytes by using a similar antisense approach (9) or by overexpressing dominant-negative GATA-4 isoform (26) and prompted us to further investigate the functional relationship between PEX1 and GATA-4. We found that the two proteins functionally and physically interact to synergistically activate transcription. Synergy required GATA-4 binding to its DNA element, and GATA elements were sufficient to support synergy. Thus, PEX1 may function as an α1-inducible GATA-4 cofactor, which could explain the similar phenotype obtained by ablating one or the other factor. Alternatively, GATA-4 and PEX1 may act as nuclear effectors of different converging signaling cascades activated by α1-ARs. Such possibility is supported by the results of reporter gene analysis in PC12 cells showing that, although the activity of different regulatory elements (AP1, SRE, and NFAT) is induced by PE, they apparently mediate distinct α1-ARs downstream signals as evidenced by differential sensitivities to specific kinase inhibitors (31). GATA-4 has been shown to be a nuclear target and effector of MAPK signaling (10, 27). Whether PEX1 acts as a nuclear effector of MAPK or other signaling cascades activated by α1-AR will need to be investigated, but the presence of multiple, conserved PKC sites on PEX1 is noteworthy given the documented involvement of PKC in α1-AR signaling.
In conclusion, PEX1 and GATA-4 are presently two of only a few transcription factors known to be required for nuclear and cytoskeletal response to α1 agonists. Further analysis of PEX1 regulation and mode of action in α1-target organs will provide molecular insight into α1-adrenergic receptor function. Finally, given the coexpression of PEX1 with other members of the GATA family (35), it is tempting to speculate on the role of GATA-PEX1 interactions in development and transcriptional regulation by α1-adrenergic receptors.
Acknowledgments
We are grateful to Gérard Goubin for the gift of OZF antibody and for sharing data, Gaétan Thibault for help with radioimmunoassay, André Turgeon for help with blood pressure measurements, Lynda Robitaille and Chantal Lefebvre for excellent technical assistance, Lise Laroche for secretarial help, Annie Vallée for the tissue sections, Jacques Lavigne for DNA sequencing, and Bert Vogelstein for the plasmid and bacteria required for generation of the first recombinant adenovirus. We thank Stéphanie Monnier for bioinformatics and members of the Nemer laboratory for helpful discussions.
S.D. received a fellowship from the Fondation de la Recherche Médicale (France), and M.M. of a Bourse d'Excellence from the Education Ministry of Québec. This study was supported by a grant from the Canadian Institutes of Health Research (MT13056). M.N. holds a Canada Research Chair in Molecular Biology.
REFERENCES
- 1.Ardati, A., and M. Nemer. 1993. A nuclear pathway for a1-adrenergic receptor signaling in cardiac cells. EMBO J. 12:5131-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aries, A., P. Paradis, C. Lefebvre, R. J. Schwartz, and M. Nemer. 2004. Essential role of GATA-4 in cell survival and drug-induced cardiotoxicity. Proc. Natl. Acad. Sci. USA 101:6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auclair, A., S. Cotecchia, J. Glowinski, and J. P. Tassin. 2002. d-Amphetamine fails to increase extracellular dopamine levels in mice lacking α1b-adrenergic receptors: relationship between functional and nonfunctional dopamine release. J. Neurosci. 22:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battaglia, G., F. Fornai, C. L. Busceti, G. Lembo, F. Nicoletti, and A. De Blasi. 2003. Alpha-1B adrenergic receptor knockout mice are protected against methamphetamine toxicity. J. Neurochem. 86:413-421. [DOI] [PubMed] [Google Scholar]
- 5.Bellefroid, E. J., P. J. Lecocq, A. Benhida, D. A. Poncelet, A. Belayew, and J. A. Martial. 1989. The human genome contains hundreds of genes coding for finger proteins of the Kruppel type. DNA 8:377-387. [DOI] [PubMed] [Google Scholar]
- 6.Blottiere, L., F. Apiou, D. Ferbus, C. Guenzi, B. Dutrillaux, M. T. Prosperi, and G. Goubin. 1999. Cloning, characterization, and chromosome assignment of Zfp146 the mouse ortholog of human ZNF146, a gene amplified and overexpressed in pancreatic cancer, and Zfp260 a closely related gene. Cytogenet. Cell Genet. 85:297-300. [DOI] [PubMed] [Google Scholar]
- 7.Burcelin, R., M. Uldry, M. Foretz, C. Perrin, A. Dacosta, M. Nenniger-Tosato, J. Seydoux, S. Cotecchia, and B. Thorens. 2004. Impaired glucose homeostasis in mice lacking the α1b-adrenergic receptor subtype. J. Biol. Chem. 279:1108-1115. [DOI] [PubMed] [Google Scholar]
- 8.Cavalli, A., A. L. Lattion, E. Hummler, M. Nenniger, T. Pedrazzini, J. F. Aubert, M. C. Michel, M. Yang, G. Lembo, C. Vecchione, M. Mostardini, A. Schmidt, F. Beermann, and S. Cotecchia. 1997. Decreased blood pressure response in mice deficient of the α1b-adrenergic receptor. Proc. Natl. Acad. Sci. USA 94:11589-11594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charron, F., P. Paradis, O. Bronchain, G. Nemer, and M. Nemer. 1999. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol. Cell. Biol. 19:4355-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charron, F., G. Tsimiklis, M. Arcand, L. Robitaille, Q. Liang, J. D. Molkentin, S. Meloche, and M. Nemer. 2001. Tissue-specific GATA factors are transcriptional effectors of the small GTPase RhoA. Genes Dev. 15:2702-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charron, J., H. Richard-Foy, D. S. Berard, G. L. Hager, and J. Drouin. 1989. Independent glucocorticoid induction and repression of two contiguous responsive genes. Mol. Cell. Biol. 9:3127-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis, L. J., Y. Li, M. Gerbault-Seureau, R. Kuick, A. M. Dutrillaux, G. Goubin, J. Fawcett, S. Cram, B. Dutrillaux, S. Hanash, and M. Muleris. 1998. Amplification of DNA sequences from chromosome 19q13.1 in human pancreatic cell lines. Genomics 53:42-55. [DOI] [PubMed] [Google Scholar]
- 13.D'Avino, P. P., and C. S. Thummel. 1998. Crooked legs encodes a family of zinc finger proteins required for leg morphogenesis and ecdysone-regulated gene expression during Drosophila metamorphosis. Development 125:1733-1745. [DOI] [PubMed] [Google Scholar]
- 14.D'Avino, P. P., and C. S. Thummel. 2000. The ecdysone regulatory pathway controls wing morphogenesis and integrin expression during Drosophila metamorphosis. Dev. Biol. 220:211-224. [DOI] [PubMed] [Google Scholar]
- 15.Ebert, S. N., J. M. Baden, L. H. Mathers, B. J. Siddall, and D. L. Wong. 1996. Expression of phenylethanolamine n-methyltransferase in the embryonic rat heart. J. Mol. Cell. Cardiol. 28:1653-1658. [DOI] [PubMed] [Google Scholar]
- 16.Ferbus, D., A. Flechon, M. Muleris, Y. Li, S. Hanash, B. Terris, P. Hammel, L. Pibouin, B. Dutrillaux, and G. Goubin. 1999. Amplification and overexpression of OZF, a gene encoding a zinc finger protein, in human pancreatic carcinomas. Int. J. Cancer 80:369-372. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Cabrera, P. J., T. Shi, J. Yun, D. F. McCune, B. R. Rorabaugh, and D. M. Perez. 2004. Differential regulation of the cell cycle by α1-adrenergic receptor subtypes. Endocrinology 145:5157-5167. [DOI] [PubMed] [Google Scholar]
- 18.Graham, R. M., D. M. Perez, J. Hwa, and M. T. Piascik. 1996. α1-Adrenergic receptor subtypes: molecular structure, function, and signaling. Circ. Res. 78:737-749. [DOI] [PubMed] [Google Scholar]
- 19.Grépin, C., L. Dagnino, L. Robitaille, L. Haberstroh, T. Antakly, and M. Nemer. 1994. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol. Cell. Biol. 14:3115-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gusterson, R., B. Brar, D. Faulkes, A. Giordano, J. Chrivia, and D. Latchman. 2002. The transcriptional coactivators CBP and p300 are activated via phenylephrine through the p42/p44 MAPK cascade. J. Biol. Chem. 277:2517-2524. [DOI] [PubMed] [Google Scholar]
- 21.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbomel, P. 1999. Spinning nuclei in the brain of the zebra fish embryo. Curr. Biol. 9:R627-R628. [DOI] [PubMed] [Google Scholar]
- 23.Jin, Y., F. Sheikh, K. A. Detillieux, and P. A. Cattini. 2000. Role for early growth response-1 protein in α1-adrenergic stimulation of fibroblast growth factor-2 promoter activity in cardiac myocytes. Mol. Pharmacol. 57:984-990. [PubMed] [Google Scholar]
- 24.Kyprianou, N., and C. M. Benning. 2000. Suppression of human prostate cancer cell growth by α1-adrenoceptor antagonists doxazosin and terazosin via induction of apoptosis. Cancer Res. 60:4550-4555. [PubMed] [Google Scholar]
- 25.Le Chalony, C., M. T. Prosperi, R. Haluza, F. Apiou, B. Dutrillaux, and G. Goubin. 1994. The OZF gene encodes a protein consisting essentially of zinc finger motifs. J. Mol. Biol. 236:399-404. [DOI] [PubMed] [Google Scholar]
- 26.Liang, Q., L. J. De Windt, S. A. Witt, T. R. Kimball, B. E. Markham, and J. D. Molkentin. 2001. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 276:30245-30253. [DOI] [PubMed] [Google Scholar]
- 27.Liang, Q., R. J. Wiese, O. F. Bueno, Y. S. Dai, B. E. Markham, and J. D. Molkentin. 2001. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol. Cell. Biol. 21:7460-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda, T., J. Sepulveda, H. H. Chen, and A. F. Stewart. 2002. α1-Adrenergic activation of the cardiac ankyrin repeat protein gene in cardiac myocytes. Gene 297:1-9. [DOI] [PubMed] [Google Scholar]
- 29.Markou, T., M. Hadzopoulou-Cladaras, and A. Lazou. 2004. Phenylephrine induces activation of CREB in adult rat cardiac myocytes through MSK1 and PKA signaling pathways. J. Mol. Cell Cardiol. 37:1001-1011. [DOI] [PubMed] [Google Scholar]
- 30.Milano, C. A., P. C. Dolber, H. A. Rockman, R. A. Bond, M. E. Venable, L. F. Allen, and R. J. Lefkowitz. 1994. Myocardial expression of a constitutively active α1B-adrenergic receptor in transgenic mice induces cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 91:10109-10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minneman, K. P., D. Lee, H. Zhong, A. Berts, K. L. Abbott, and T. J. Murphy. 2000. Transcriptional responses to growth factor and G protein-coupled receptors in PC12 cells: comparison of α1-adrenergic receptor subtypes. J. Neurochem. 74:2392-2400. [DOI] [PubMed] [Google Scholar]
- 32.Molkentin, J. D., and G. W. Dorn, I. I. 2001. Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol. 63:391-426. [DOI] [PubMed] [Google Scholar]
- 33.Morimoto, T., K. Hasegawa, S. Kaburagi, T. Kakita, H. Wada, T. Yanazume, and S. Sasayama. 2000. Phosphorylation of GATA-4 is involved in α1-adrenergic agonist-responsive transcription of the endothelin-1 gene in cardiac myocytes. J. Biol. Chem. 275:13721-13726. [DOI] [PubMed] [Google Scholar]
- 34.Morin, S., P. Paradis, A. Aries, and M. Nemer. 2001. Serum response factor-GATA ternary complex required for nuclear signaling by a G-protein-coupled receptor. Mol. Cell. Biol. 21:1036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemer, G., and M. Nemer. 2003. Transcriptional activation of BMP-4 and regulation of mammalian organogenesis by GATA-4 and -6. Dev. Biol. 254:131-148. [DOI] [PubMed] [Google Scholar]
- 36.O'Connell, T. D., S. Ishizaka, A. Nakamura, P. M. Swigart, M. C. Rodrigo, G. L. Simpson, S. Cotecchia, D. G. Rokosh, W. Grossman, E. Foster, and P. C. Simpson. 2003. The alpha(1A/C)- and alpha(1B)-adrenergic receptors are required for physiological cardiac hypertrophy in the double-knockout mouse. J. Clin. Investig. 111:1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmiter, R. D. 1996. New adrenergic cells stimulate heart research. Nat. Med. 2:1194-1195. [DOI] [PubMed] [Google Scholar]
- 38.Paradis, P., N. Dali-Youcef, F. W. Paradis, G. Thibault, and M. Nemer. 2000. Overexpression of angiotensin II type 1 receptor in cardiomyocytes induces cardiac hypertrophy and remodeling. Proc. Natl. Acad. Sci. USA 97:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pendleton, R. G., A. Rasheed, R. Roychowdhury, and R. Hillman. 1998. A new role for catecholamines: ontogenesis. Trends Pharmacol. Sci. 19:248-251. [DOI] [PubMed] [Google Scholar]
- 40.Philipp, M., and L. Hein. 2004. Adrenergic receptor knockout mice: distinct functions of nine receptor subtypes. Pharmacol. Ther. 101:65-74. [DOI] [PubMed] [Google Scholar]
- 41.Reja, V., A. K. Goodchild, and P. M. Pilowsky. 2002. Catecholamine-related gene expression correlates with blood pressures in SHR. Hypertension 40:342-347. [DOI] [PubMed] [Google Scholar]
- 42.Rokosh, D. G., and P. C. Simpson. 2002. Knockout of the alpha 1A/C-adrenergic receptor subtype: the alpha 1A/C is expressed in resistance arteries and is required to maintain arterial blood pressure. Proc. Natl. Acad. Sci. USA 99:9474-9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart, J. M., J. Munoz, and A. Weldon. 2002. Clinical and physiological effects of an acute α1 adrenergic agonist and a β1 adrenergic antagonist in chronic orthostatic intolerance. Circulation 106:2946-2954. [DOI] [PubMed] [Google Scholar]
- 44.Tanoue, A., T. A. Koshimizu, K. Shibata, Y. Nasa, S. Takeo, and G. Tsujimoto. 2003. Insights into alpha1 adrenoceptor function in health and disease from transgenic animal studies. Trends Endocrinol. Metab. 14:107-113. [DOI] [PubMed] [Google Scholar]
- 45.Tanoue, A., Y. Nasa, T. Koshimizu, H. Shinoura, S. Oshikawa, T. Kawai, S. Sunada, S. Takeo, and G. Tsujimoto. 2002. The α1D-adrenergic receptor directly regulates arterial blood pressure via vasoconstriction. J. Clin. Investig. 109:765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Temsah, R., and M. Nemer. 2005. GATA factors and transcriptional regulation of cardiac natriuretic peptide genes. Regul. Pept. 128:177-185. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, S. A., A. M. Matsumoto, and R. D. Palmiter. 1995. Noradrenaline is essential for mouse fetal development. Nature 374:643-646. [DOI] [PubMed] [Google Scholar]
- 48.Tsoporis, J. N., A. Marks, L. J. Van Eldik, D. O'Hanlon, and T. G. Parker. 2003. Regulation of the S100B gene by alpha 1-adrenergic stimulation in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 284:H193-H203. [DOI] [PubMed] [Google Scholar]
- 49.Ueyama, T., C. Zhu, Y. M. Valenzuela, J. G. Suzow, and A. F. Stewart. 2000. Identification of the functional domain in the transcription factor RTEF-1 that mediates alpha 1-adrenergic signaling in hypertrophied cardiac myocytes. J. Biol. Chem. 275:17476-17480. [DOI] [PubMed] [Google Scholar]
- 50.Wang, R. X., and L. E. Limbird. 1997. Distribution of mRNA encoding three α2-adrenergic receptor subtypes in the developing mouse embryo suggests a role for the alpha 2A subtype in apoptosis. Mol. Pharmacol. 52:1071-1080. [DOI] [PubMed] [Google Scholar]
- 51.Weinshenker, D., N. S. Miller, K. Blizinsky, M. L. Laughlin, and R. D. Palmiter. 2002. Mice with chronic norepinephrine deficiency resemble amphetamine-sensitized animals. Proc. Natl. Acad. Sci. USA 99:13873-13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiang, Y., and B. K. Kobilka. 2003. Myocyte adrenoceptor signaling pathways. Science 300:1530-1532. [DOI] [PubMed] [Google Scholar]
- 53.Yun, J., R. J. Gaivin, D. F. McCune, A. Boongird, R. S. Papay, Z. Ying, P. J. Gonzalez-Cabrera, I. Najm, and D. M. Perez. 2003. Gene expression profile of neurodegeneration induced by α1B-adrenergic receptor overactivity: NMDA/GABAA dysregulation and apoptosis. Brain 126:2667-2681. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, Q. Y., C. J. Quaife, and R. D. Palmiter. 1995. Targeted disruption of the tyrosine hydroxylase gene reveals that catecholamines are required for mouse fetal development. Nature 374:640-643. [DOI] [PubMed] [Google Scholar]
- 55.Zuscik, M. J., S. Sands, S. A. Ross, D. J. Waugh, R. J. Gaivin, D. Morilak, and D. M. Perez. 2000. Overexpression of the α1B-adrenergic receptor causes apoptotic neurodegeneration: multiple system atrophy. Nat. Med. 6:1388-1394. [DOI] [PubMed] [Google Scholar]