Abstract
Downstream elements are a newly appreciated class of core promoter elements of RNA polymerase II-transcribed genes. The downstream core element (DCE) was discovered in the human β-globin promoter, and its sequence composition is distinct from that of the downstream promoter element (DPE). We show here that the DCE is a bona fide core promoter element present in a large number of promoters and with high incidence in promoters containing a TATA motif. Database analysis indicates that the DCE is found in diverse promoters, supporting its functional relevance in a variety of promoter contexts. The DCE consists of three subelements, and DCE function is recapitulated in a TFIID-dependent manner. Subelement 3 can function independently of the other two and shows a TFIID requirement as well. UV photo-cross-linking results demonstrate that TAF1/TAFII250 interacts with the DCE subelement DNA in a sequence-dependent manner. These data show that downstream elements consist of at least two types, those of the DPE class and those of the DCE class; they function via different DNA sequences and interact with different transcription activation factors. Finally, these data argue that TFIID is, in fact, a core promoter recognition complex.
Promoters transcribed by RNA polymerase II are complex and composed of different classes of elements, one of which consists of the core promoter elements represented by the TATA box, the TFIIB response elment (BRE), the initiator, the downstream promoter element (DPE), and other newly recognized elements (reviewed in reference 73). One view of core promoters is that they are fairly ubiquitous and that they play only a trivial role in transcriptional regulation. According to this view, the uniqueness and specificity inherent in a promoter derive from the various DNA binding activators and coactivators that recognize the regulatory elements usually located upstream of the TATA box and/or initiator. The small number of core promoter elements and their apparent lack of diversity compared to that of the staggering numbers of activators have supported such a view. Also contributing to this view is the fact that core promoters have been rather poorly studied in their natural context, and thus, regulatory phenomena may have been missed.
Yet there are a few examples where the core promoter apparently plays a very specific role. Early examples include studies of the myoglobin and simian virus 40 TATA boxes (86) and the hsp70 TATA box (72) that indicated a requirement for TATA boxes of a particular sequence. The TdT promoter was also shown to become inactive upon the inclusion of a TATA box (20). Lastly, some activators display preferences for particular core promoter architectures (7, 18, 56).
This perception of core promoters lacking diversity is being revised with the findings of other core promoter elements such as the BRE (41) and elements located downstream of the transcriptional start site such as the DPE (5, 6, 40, 89), the downstream core element (DCE) (43), and the motif 10 element (MTE) (48). The most studied of these downstream elements is the DPE. The DPE was discovered during the analysis of TFIID interactions with several Drosophila promoters (5, 6). The DPE is centered at approximately +30 relative to the transcriptional start site. Thus far, it has been shown to function only in TATA-less promoter contexts and is at least partially redundant in the context of a TATA box (6). Additional indications of core promoter specificity arose from studies showing that specific enhancers require a specific core promoter structure containing either a TATA box or DPE (7, 56).
A second class of downstream elements which are architecturally distinct from the DPE exists. Initially discovered in the human β-globin promoter, the DCE consists of three subelements. Point mutations in each subelement are found in human β-thalassemia patients (this paper and references 2, 8, 24, 31, 57, and 90). As with the DPE, experiments suggested that TFIID recognized the DCE in vitro (43). Here we confirm and extend these observations by showing that the adenovirus major late promoter (Ad MLP), Ad E3, Ad IX, and the herpes simplex virus (HSV) UL38 promoters contain functional DCE sequences, thus demonstrating that the DCE is not simply restricted to the promoters of the β-globin locus. Computer analysis of promoter databases indicates that DCE sequences are found in a variety of promoters, further suggesting that the DCE represents a novel, widely distributed core promoter element. We demonstrate that DCE function can be recapitulated in a highly purified transcription system and requires the general transcriptional machinery together with the transcription activation factor (TAF) subunits of the TFIID complex. We also show that TAF1 is the major TAF species that can be cross-linked to wild-type but not mutant DCE subelements. Our results show that the DCE represents a distinct class of downstream elements and underscores the intricacies in transcriptional regulation at the level of core promoter elements.
MATERIALS AND METHODS
In vitro transcriptions.
Nuclear extracts were prepared as described previously (15). Primer extensions and crude nuclear extract in vitro transcriptions were as described previously (43, 44). Affinity-purified TFIID was purified as described previously (34, 43). Reconstituted in vitro transcription assays contained recombinant TFIIA (rTFIIA), rTFIIB, rTFIIE, and rTFIIF (49). TFIIH is a partially purified phenyl-Superose fraction (49). RNA polymerase II was purified to homogeneity as described previously (49). The TATA box-binding component of TFIID (TBP) and TFIID concentrations used in the in vitro transcriptions were determined empirically by titration and correspond to saturating amounts of each. Transcription reaction mixtures contained 25 ng of DNA, 10 mM Tris-HCl, pH 7.9, 10 mM HEPES-KOH, pH 8.0, 4 mM MgCl2, 10 mM (NH4)2SO4, 100 μg/ml bovine serum albumin, 10 mM dithiothreitol, and 250 μM recombinant nucleoside triphosphates. Ad E3 in vitro transcription mixtures also contained baculovirus-expressed highly purified Sp1 and a partially purified mediator fraction in order to increase transcription signals (45, 65, 66).
Template construction.
MLP scanning mutants, a mutant TATA box, and +7/9 β-globin mutants were constructed as described previously (43). The mutations are as follows: +1 to +3 (+1/3) ACT to GGG, +4/6 CTC to GGG, +7/9 TTC to GGG, +10/12 CGC to AAA, +13/15 ATC to GGG, +16/18 GCT to AAA, +19/21 GTC to AAA, +22/24 TGC to AAA, +25/27 GAG to CCC, +28/30 GGC to TTT, and +31/33 CAG to TTT; the β-globin mutant TATA box was changed from CATAA to CGCGC, and β-globin +7/9 GCT was changed to AAA. All other β-globin templates are from reference 43. MLP DNA PCR upstream and downstream primers contained XbaI and XhoI restriction sites, which were used to insert the MLP DNA into the XbaI-XhoI-digested pSP72 vector (Promega). Ad E3 and IX promoter regions were amplified from the adenovirus type 2 genome by PCR using primers containing 5′ EcoRI and 3′ XhoI adapters (5′-CGGAATTCCGGGAAGTGAAATCTGAATAAT-3′ and 5′-CCGCTCGAGCGGCTT CGGTAATAACACCTCCG-3′), digested with EcoRI and XhoI, and subcloned into the pSP72 vector (Promega) linearized with the same enzymes. To generate the mutant E3 promoter, wild-type E3 promoter in the pSP72 vector was amplified using the following two sets of primers (SP6 primer and 5′-ACCAAGAGAGGAAAACACCGACTCGTC-3′, and T7 and 5′-GACGAGTCGGTGTTTTCCTCTCTTGGT-3′). Those PCR products were used as templates for another PCR with SP6 and T7 primers, digested with EcoRI and XhoI, and subcloned into the pSP72 vector (Promega) linearized with the same enzymes. The IX mutant promoter was amplified using the following sets of primers (SP6 and 5′-AAACGAGTTGGCAAACATGGCGGCGGC-3′ and T7 and 5′-GCCGCCGCCATGTTTGCCAACTCGTTT-3′).
Statistical analysis.
A total of 1,871 nonredundant human promoter sequences 600 bp long (−499 to +100 bp around the TSS) from Eukaryotic Promoter Database (EPD) release 75 (4, 68) (http://www.epd.isb-sib.ch/), and 8,793 promoters sequences 1,200 bp long (−1,000 to +200 bp) from the Database of Transcriptional Start Sites (DBTSS) (59, 74, 75) (http://dbtss.hgc.jp/index.html) were used for statistical analyses. The software package Promoter Classifier (available at our website, http://bmi.osu.edu/∼ilya/promoter_classifier/) (21, 22) was used for statistical analysis. To divide the promoter databases into TATA+/TATA− and Inr+/Inr− subsets, the respective position weight matrices were applied (4). Since there are no matrices for DPE, we matched five out of five letters for the DPE consensuses (73).
To define an interval (window) for a functional position of the TATA box, Inr, and DPE core elements, we considered the distribution of the element's occurrence frequencies along the promoters. For both databases, we found the unambiguous maximums for the occurrence frequencies of the centers of the TATA and Inr elements at positions −28 and +1, respectively, which is consistent with the known functional positions of these elements. The occurrence frequency of the TATA box is essentially larger in the window (from −33 to −23 bp) than in the surrounding area. We consider this window the functional window for the TATA box. For the Inr element, the functional window is −5 to +6 bp since the +1 position in the EPD is defined with an accuracy of ±5 bp. Since DPE works in cooperation with an Inr if positioned 27 bp downstream from it (5), we applied the window (28 − 5) -(28 +5) bp for the DPE. A more detailed description of the statistical analysis procedure can be found elsewhere (21).
To calculate the statistical significance of the occurrence frequency of a subelement in the functional window, we calculated a parameter (statistical significance [dS] measured in units of standard deviation [SD] as 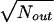 ) and dS as (Nin − Nout)/
) and dS as (Nin − Nout)/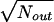 , where Nin is the number of promoters containing an element or combination inside its functional window and Nout is a statistically expected number obtained for the promoter sequence segments outside the functional window.
, where Nin is the number of promoters containing an element or combination inside its functional window and Nout is a statistically expected number obtained for the promoter sequence segments outside the functional window.
Site-specific protein-DNA photo-cross-linking.
Purification of FLAG-tagged TFIID complexes was essentially as described previously (11). To generate single-stranded mutant DNAs for photo-cross-linking, Ad MLP in pSP72 was digested with XhoI, filled in at the recessed end, and digested with XbaI. M13mp19 RF DNA was digested with EcoRI, filled in at the recessed end, and digested with XbaI. DNAs were ligated and transformed into XL1-Blue cells. Single-stranded template DNAs were prepared as described in Sambrook et al. (66a) Derivatized promoter DNA fragments containing cross-linking agent at positions +9, +19, and +31 of the template strand and radiophosphorylated, derivatized oligodeoxyribonucleotides (10 pmol) were prepared essentially as described previously (39). Photo-cross-linking reactions were performed as described previously (39). Assignment of cross-linked product to TAF1/TAFII250 was confirmed by immunoprecipitation with monoclonal antibody to TAF1/TAFII250 (Santa Cruz) as previously described (39). TFIID/TFIIA binding reactions were as described previously (43). The TFIID used was identical to that used for the in vitro transcription assays.
RESULTS
The adenovirus major late promoter contains a DCE.
Our initial work demonstrated the existence of a unique downstream element in the human β-globin promoter (43); however, we did not know how prevalent the DCE was in other promoters. In order to search for additional downstream elements, we chose to study the Ad MLP. Like the β-globin promoter, the Ad MLP has been well studied, and a considerable body of evidence indicated that the Ad MLP contained a downstream element. Deletion of sequences downstream of the start site from +7 to +33 resulted in a severe reduction in Ad MLP transcription during viral infection (46). Similar results were obtained from in vitro transcription experiments using HeLa nuclear extracts (1). Second, TFIID is well known to protect the Ad MLP from DNase I digestion, from the TATA box at −30, downstream of the transcriptional start site at +1, to approximately +40 (52, 67, 80, 92). Despite this preponderance of data, no firm conclusions on the presence of a downstream element in the Ad MLP were made. Yet the Ad MLP seemed a likely candidate promoter containing a downstream element.
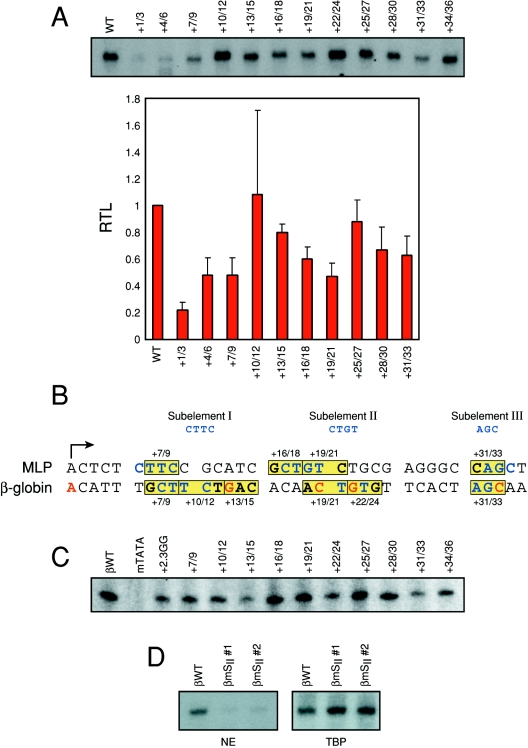
In order to accurately define the Ad MLP downstream element, we conducted a scanning mutagenesis from +1 to +36 (Fig. 1A). The standard errors for transcription assays using nuclear extracts are typically in the 15 to 20% range (43, 44). Thus, it is not possible to distinguish any mutants giving rise to ∼80 to 85% of wild-type levels of transcription. Applying this statistical cutoff to Fig. 1A excludes mutations at positions +10/12, +13/15, and +25/27. However, reductions from mutations at positions +1/3, +4/6, +7/9, +16/18, +19/21, +28/30, and +31/33 all indicate the presence of regulatory elements. As expected, mutations within the Inr element (+1/3 and +4/6) decreased Ad MLP transcription (35, 58). However, the mutagenesis revealed that three other regions, in addition to the Inr element, are important for optimal transcription: +7 to +9, +16 to +21, and +28 to +33 (Fig. 1A). Although quantifications of the +22/24 mutant are not shown, transcription of this mutant was near wild-type levels in two experiments.
FIG. 1.
The adenovirus major late promoter contains a DCE type of downstream element. A. Scanning mutagenesis of the adenovirus MLP from +1 to +36. Triplet mutations from +1 to +36 were inserted into the Ad MLP templates and assayed by in vitro transcriptions using crude HeLa nuclear extracts. Transcription products were detected by primer extension. Relative transcription levels (RTL) represent the mutant transcription levels relative to the wild-type transcription level (WT). Mean values and standard deviations are calculated from the relative transcription levels (mutant versus wild type) from three to four experiments. Quantifications of the +22/24 and +34/36 mutants were approximately 80 to 90% of the wild-type MLP (n = 2) (data not shown). B. Sequence alignment of the wild-type human β-globin and adenovirus major late promoters. Boxed sequences, arrows, and numbering indicate positions of deleterious mutations in each promoter (panels A and C and reference 43). Bases in red indicate the positions of known β-thalassemia point mutations in the human β-globin promoter (2, 8, 24, 31, 57, 90), except for +13 (13). Blue-colored bases indicate the sequences representing DCE subelements. C. The β-globin DCE extends to positions +7/9 and does not show any erythroid cell specificity. Triplet mutations of the human β-globin promoter (43) were assayed by in vitro transcription using crude HeLa nuclear extracts. βWT, wild-type β-globin; 2.3GG, deleterious mutation in the human β-globin Inr element (44); mTATA, defective TATA box created by the replacement of the wild-type β-globin TATA box sequence with four TATA box β-thalassemia point mutations. D. Mutation of β-globin subelement II (βmSII) CTGT to AAAA. Two different preparations of a β-globin template containing mutations in all four base pairs of subelement II were assayed in an in vitro transcription assay using HeLa nuclear extracts (NE). The same templates were assayed using TBP in the reconstituted in vitro transcription assay as a negative control (see the text and Fig. 2).
Comparison of the β-globin (43) (Fig. 1B) and Ad MLP mutants with decreased transcription indicated significant similarities between the promoter sequences affected (Fig. 1B). The sequences common to both sets of mutations in the Ad MLP and β-globin promoters are CTTC, CTGT, and AGC (Fig. 1B). Furthermore, the sequence alignment predicted that, due to the overlap between the two promoters, positions +7 to +9 (CT of CTTC) of the human β-globin promoter would compromise transcription when mutated, and we found that this was indeed the case (Fig. 1C) (in our initial work, mutagenesis analysis had started at +10 [43]). Since β-globin gene expression is restricted to erythroid cells and we had previously analyzed DCE function in erythroid nuclear extracts, we examined whether the β-globin DCE functioned in nonerythroid extracts. Transcription reactions performed in vitro show that there is no apparent erythroid cell specificity to the β-globin DCE, since the mutant defects are recapitulated with HeLa nuclear extracts (Fig. 1C).
We have observed some quantitative differences when comparing the mouse erythroid and HeLa nuclear extract data of the β-globin scanning mutants. This was especially noticeable with mutations at +19/21 and +22/24. Quantifications of the HeLa data were 0.79 ± 0.22 and 0.65 ± 0.18 (means ± standard deviations) for mutants with mutations at +19/21 and +22/24, respectively, while our previous work gave 0.60 ± 0.009 and 0.43 ± 0.04, respectively. We were concerned that the subelement II (SII) sequence may either behave differently in the two extracts or that we needed to revisit our conclusion that CTGT represents a functional cis element. This is especially a concern when looking at the magnitude of the +19/21 mutant in HeLa extracts (0.79 ± 0.22), which approaches wild-type levels (the +19/21 mutant contains a mutation in the CT positions of the CTGT SII sequence). Therefore, we next mutated all four positions of SII (Fig. 1D) (CTGT to AAAA). We examined two independent mutant templates in transcription assays performed in vitro using HeLa nuclear extracts. The quadruple mutant is considerably decreased to a degree greater than either scanning mutant was by itself. Reconstituted in vitro transcriptions with TBP showed no difference, indicating that this was not a general nonspecific defect in the promoter DNA (see below). This result is consistent with our interpretation that the CTGT sequence represents a functional cis element. Moreover, the differences observed between the extract and TBP-reconstituted transcription system further stress the idea that the CTGT element is functionally important for optimal transcription of the (two) promoters analyzed.
From the sequence comparison and the effects of the mutations on transcription, we conclude that the β-globin and Ad major late promoters each contain a DCE. We refer to each of the three regions of the DCE as subelements and infer minimal specific sequences for each, based on the alignment of the two promoters: SI is CTTC, SII is CTGT, and SIII is AGC (Fig. 1B).
TFIID is necessary and sufficient for DCE function.
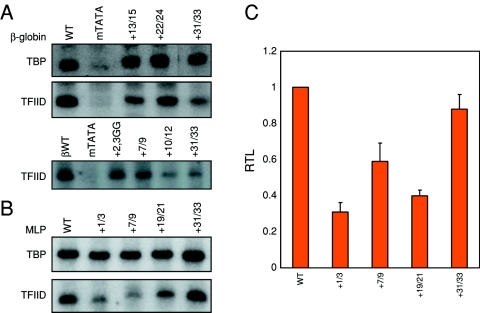
The next step in the analysis of the DCE was to ask whether we could recapitulate DCE function using a highly purified transcription system in vitro. This system contains recombinant TFIIA, TFIIB, TFIIE, TFIIF, highly purified TFIIH, RNA polymerase II, and either recombinant TBP or affinity-purified TFIID. Previous results indicated that TFIID physically contacts DCE subelements in the β-globin promoter (43). If TFIID is responsible for DCE function, then we would expect TFIID but not TBP to reconstitute the observed differences between the wild-type and mutant templates. TBP-dependent transcriptions of β-globin subelements II and III (+22/24 and +31/33, respectively) and the +13/15 mutant were indistinguishable from that of the wild-type template (Fig. 2A, top panel, and 1D). However, a TATA box mutation was severely compromised, as expected. Replacing TBP with TFIID showed reductions with all three subelement mutants as well as the TATA box mutant (middle panel of Fig. 2A). We have also reconstituted SI function with TFIID (bottom panel in Fig. 2A) (+7/9 and +10/12). These two SI mutants (+7/9 and +10/12) showed decreased transcription relative to that of the wild-type template, when TFIID was used. Two positive controls, an Inr mutant (2,3GG) and the SIII mutant (+31/33), also showed a decrease in transcription.
FIG. 2.
TFIID functionally recapitulates β-globin and MLP DCE activity in vitro. A. TFIID is necessary for β-globin DCE function. Using the highly purified in vitro transcription system, the three major β-globin DCE subelement mutants (Fig. 1B and C and reference 43) show decreased transcription using affinity-purified TFIID (two bottom panels) but not recombinant TBP (top panel). The mutant TATA box (mTATA) serves as a positive control. The second TFIID panel illustrates decreases in transcription using an Inr mutant (2,3GG), two mutations in SI (+7/9 and +10/12), and the SIII mutation (+31/33). WT and βWT, wild-type β-globin. B. MLP Inr mutant (+1/3) and DCE subelement mutants (Fig. 1A and B) show decreased transcription in vitro using the highly purified transcription system and affinity-purified TFIID but not recombinant TBP. C. Quantifications of the MLP TFIID-dependent transcriptions. Mean relative transcription levels (RTL) and standard deviations are calculated for three to four experiments.
We observed similar effects with reconstitutions of the Ad MLP. TBP-dependent transcription assays showed no differences in transcription between wild-type Ad MLP and Ad MLP with a mutation in the Inr element (+1/3), which is known to be TFIID dependent (9, 36, 37, 81, 83). We also did not observe any differences between the wild-type Ad MLP and mutations in any of the three DCE subelements (+7/9, +19/21, and +31/33) (top panel, Fig. 2B). However, we did observe defects in transcription from three of the four templates when TBP was replaced with TFIID (bottom panel, Fig. 2B), and these closely resembled the transcription levels observed using nuclear extracts (Fig. 1A). The only exception is the Ad MLP +31/33 mutant, which shows only a 20% reduction, and we do not understand yet why this is so. However, we offer additional data below showing further reconstitutions of subelement III with TFIID (Fig. 3).
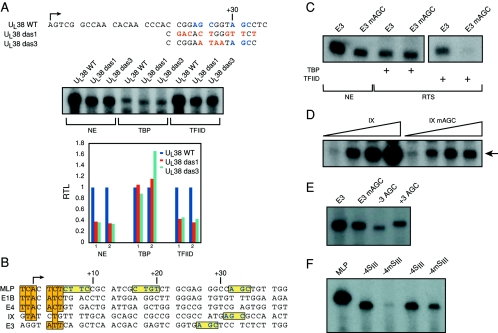
FIG. 3.
DCE subelement III can function independently of subelements I and II. A. HSV UL38 wild-type (WT) and mutant promoter sequences. Predicted SIII sequences are in blue, and mutated bases are in red (26). In vitro transcriptions of the UL38 WT promoter and DAS mutants 1 and 3 (das1 and das3) were performed using HeLa nuclear extracts (NE) or a highly purified transcription system containing either TBP or affinity-purified TFIID. Below the in vitro transcription are quantifications of two sets of the in vitro transcription experiments. B. Adenovirus promoter sequences containing DCE sequences. Shown are several promoters from adenovirus 5 (GenBank accession numbers BK000408 and X02996). The promoters are indicated on the left. The arrow indicates the +1 transcriptional start site. The vertical boxes indicate positions that fit the Inr consensus sequence (YYANA/TYY [73]). The three boxes and blue lettering indicate, in the case of the MLP, experimentally derived positions of the three DCE subelements (Fig. 1). In the case of the remaining promoters, the boxes and blue lettering indicate predicted DCE subelements. C. In vitro transcription analysis of the wild-type adenovirus E3 promoter and a mutation of the predicted SIII (E3 mAGC; AGC is mutated to TTT). Shown are transcriptions using HeLa nuclear extracts (NE) or using the highly purified reconstituted transcription system (RTS) containing either recombinant TBP or affinity-purified TFIID. D. Titration of the adenovirus IX promoter containing either the wild-type promoter sequences (IX) or a mutation of the predicted SIII AGC sequence (IX mAGC; AGC is mutated to TTT). Shown are transcriptions using HeLa nuclear extracts. E. DCE subelement III function has severe spatial constraints. Wild-type and mutant E3 in vitro transcriptions using HeLa nuclear extracts are compared to the deletion or insertion of 3 base pairs between the subelement III AGC and the start site of transcription (−3AGC and +3AGC, respectively; the insertion of AAA was at +24 for +3AGC, and deletion of the TCG sequence was at +21 to +23 for −3AGC). F. Analysis of Ad MLP spacing mutants. The Ad MLP SIII was moved 4 bp closer to SII to establish a 10-bp interval between SII and SIII as in the β-globin promoter (−4SIII; the GGCC from +28 to +31 was deleted). In this context, SIII was also mutated to TTT (−4mSIII). Shown are results of duplicate in vitro transcriptions (using HeLa NE) of these two templates, compared to the wild-type Ad MLP (MLP).
From these results, we conclude that TFIID is necessary for the functional recapitulation of DCE activity in vitro and we speculate that one or more TAF components of TFIID interact with the DCE subelements in a sequence-specific manner (see below). This study also demonstrates that DCE function can be recapitulated in a minimal reconstituted RNA polymerase II transcription system and is the first such demonstration of downstream element function using a highly reconstituted transcription system.
Several viral promoters contain DCE subelement III.
One question arising from this analysis is whether any individual subelement can function independently of the others. Previous work showed that a region downstream of the start site of the HSV UL38 and US11 promoters, termed the downstream activation sequence (DAS), was necessary for the proper expression of these promoters, both during viral infection and in vitro (26, 27). Our sequence analysis suggested that the DAS contains the AGC of SIII of the DCE (Fig. 3A). We analyzed two UL38 promoter DAS mutants, das1 and das3, using nuclear extracts in vitro and confirmed the results of Guzowski et al. (26), who showed that these mutants were defective for UL38 transcription (Fig. 3A, NE lanes). In the das3 mutant, two out of three positions of the AGC (at +26 to 28) were mutated along with two adjacent guanines (AGCGG to AATAA [Fig. 3A]). These results suggest that the HSV UL38 DAS element is a DCE subelement III and that this subelement can function independently of SI and SII of the DCE. If the DAS is actually DCE subelement III, then we would expect to be able to reconstitute its activity with TFIID but not TBP, as we observed previously. TBP-dependent transcriptions using the highly purified reconstituted in vitro system showed no differences between the wild-type UL38 promoter and das1 and das3 (Fig. 3A, TBP lanes). However, when we substituted TFIID for TBP, we observed that the das1 and das3 mutants were now defective transcriptionally (Fig. 3A, TFIID lanes), and closely resembled the defects observed with nuclear extracts (compare the lanes with NE with those of TFIID in Fig. 3). Thus, by sequence identity, position, and requirement for TFIID, we conclude that the DAS element is DCE SIII.
Since the Ad MLP contains a DCE, we asked whether any other adenovirus promoters also contained DCEs. The TATA box-containing E1b and E4 promoters do not have DCE sequences (Fig. 3B), and this correlates with the absence of TFIID DNase I protections downstream of the TATA box on these promoters (12, 32, 92). Although we did not find a promoter that contained all three subelements, the TATA box-containing IX and E3 promoters included DCE subelement III sequences in the expected neighborhood of +30 (Fig. 3B). We also did not find in these promoters any sequences resembling a DPE.
We amplified and cloned the Ad E3 and Ad IX promoter DNA fragments from adenovirus genomic DNA and then mutated only the predicted subelement III AGC sequence in both promoters. Transcriptions using nuclear extracts in vitro were used to compare transcription levels from wild-type and mutant promoters (Fig. 3C and D). As predicted, mutation of the AGC in both the E3 (0.35 ± 0.12) and IX (0.019 ± 0.01) promoters resulted in significant reductions in RNA levels in vitro. These data are significant in several respects. They suggest that our initial derivation of the sequence of subelement III is correct in that it accurately predicted the presence of subelement III in two other promoters. Second, it is clear from the UL38, E3, and IX promoters that subelement III can function as a separate distinct element.
If these are DCE subelements, then we would expect SIII function to be recapitulated using the highly purified reconstituted transcription system in vitro. As with the UL38 promoter, we found that the function of Ad E3 subelement III is recapitulated not with TBP, but only with the TFIID complex (Fig. 3C). These data strongly suggest that one or more TAF components of TFIID are capable of a sequence-specific interaction with SIII (see below).
DCE subelement III spacing requirements.
We had previously established that the insertion of 5 nucleotides between the subelements in the β-globin promoter had very deleterious effects on transcription in vitro (43). In a similar vein, we moved SIII in the Ad E3 promoter 3 base pairs closer or further from its original position. In this case, the effects were equally deleterious, severely decreasing transcription (Fig. 3E). We suggest then that the possible critical spacing parameter established by TFIID is not the relative distance between SII and SIII, which is one interpretation of the spacing data (43). Instead, the critical factor may be the distance of SIII from the rest of the core promoter, either the TATA box or Inr or some combination of the two. This is especially apparent in the Ad E3 promoter, where there are no other subelements. Similar spacing constraints have been observed for both DPE and MTE downstream elements and the Inr (5, 48).
To further address the spacing properties of the DCE subelement III, we reexamined the Ad MLP. We have previously noted that in the β-globin promoter, SII and SIII are separated by 10 bp but that in the Ad MLP, the distance is approximately 14 base pairs. Is it possible by moving the Ad MLP SIII AGC closer to SII to increase its contribution to the magnitude of transcription? Note that this also serves to test our interpretation of the E3 spacing mutants. Is it important to maintain the spacing between SII and SIII or, instead, the spacing between SIII and either the TATA box and/or Inr?
We constructed two Ad MLP templates; in one, 4 nucleotides were removed between SII and SIII (−4SIII). The second is the corresponding mutation in SIII (AGC to TTT) in this mutant (−4mSIII). We compared these and the wild-type constructs to assess the function of SIII in this new spacing context (Fig. 3F). We found that −4SIII shows a decrease in transcription relative to that of the wild-type Ad MLP. The experiment also shows that −4SIII still contains a functional SIII, since mutation of it (−4mSIII) further abrogates transcription. Since moving SIII closer to SII did not increase transcription and instead showed less transcription than the wild-type configuration, we conclude that it is the spacing between SIII and the remaining core promoter elements (TATA box and Inr) that is critical for SIII function. We suggest that this optimal spacing resides in the neighborhood of +32. While an SIII positioned away from its optimal position is capable of functioning, transcription levels are compromised.
TAF1/TAFII250 photo-cross-links to the three DCE subelements in a sequence-specific manner.
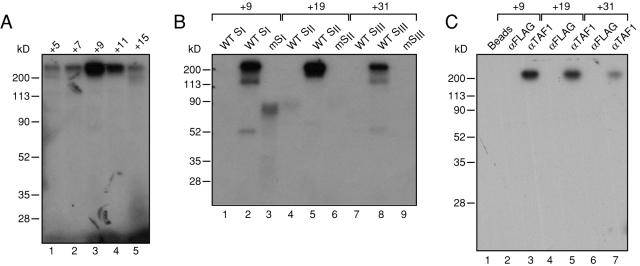
The functional data (Fig. 2) and previous work (43) suggested a physical, sequence-specific interaction of TFIID with the DCE. To identify the TAF(s) responsible for this interaction, we utilized a photo-cross-linking protocol developed previously (39, 41, 42). Briefly, a photo-cross-linking adduct, which protrudes from the sugar-phosphate backbone, was incorporated into the Ad MLP DNA probe at the site of interest, and the DNA was labeled with 32P immediately adjacent to the adduct position. The resulting Ad MLP DNA probe was incubated with affinity-purified TFIID (in the presence of TFIIA to facilitate TFIID binding to promoter sequences [36]) and exposed to UV irradiation to cross-link the DNA to a neighboring protein(s). Following nuclease digestion, the “cross-linked” labeled proteins were analyzed by denaturing polyacrylamide gel electrophoresis (PAGE) and autoradiography. Figure 4A shows the results of an initial scan in the neighborhood of subelement I, where we detected an ∼200-kDa species that cross-linked to positions +9 and +11. We next extended this analysis to the two remaining subelements. Again, a predominant species of ∼200 kDa was cross-linked in a UV-dependent manner to wild-type Ad MLP promoter sequences containing the photo-cross-linking adduct at position +9, +19, or +31, which represent the positions of DCE subelements I, II, and III, respectively (Fig. 4B). In each case, no protein was detected when we used Ad MLP templates containing mutations in each of the three subelements (mSI, mSII, and mSIII), which were shown to be transcriptionally defective (Fig. 1). We also note the presence of several minor (cross-linked) proteins of ∼120 kDa and 50 kDa; the nature of these proteins remains to be elucidated, but they possibly represent minor interactions of TAFs with the three subelements.
FIG. 4.
TAF1/TAFII250 photo-cross-linking is observed with wild-type but not mutant MLP DCE subelements in the adenovirus MLP. A. Photo-cross-linking adducts were placed at the indicated positions relative to the transcriptional start site on the template strand of the wild-type adenovirus MLP and incubated with affinity-purified TFIID and rTFIIA (see Materials and Methods). PAGE and autoradiography were performed on the resulting cross-linked products. B. Photo-cross-linking adducts were placed at positions +9, +19, and +31 of the template strand of the adenovirus MLP wild-type or mutant DCE subelements (WT SI, mSI, etc.) The mutant subelement sequences are identical to those used in Fig. 1A. PAGE and autoradiography were performed on the resulting cross-linked products, with (lanes 2, 3, 5, 6, 8, 9) or without (lanes 1, 4, and 7) irradiation with UV light. Binding reactions were as described for panel A. C. TFIIA/TFIID photo-cross-linked products (at positions +9, +19, and +31 of the template strand) were dissociated and immunoprecipitated with either protein A-agarose beads alone (lane 1), an anti-FLAG antibody (αFLAG; Sigma) plus protein A-agarose beads (lanes 2, 4, and 6), or an anti-TAF1/TAFII250 antibody plus protein A-agarose beads (lanes 3, 5, and 7). The immunoprecipitates were then analyzed by PAGE and autoradiography.
The presence of the large 200-kDa protein detected by the photo-cross-linking immediately suggested itself as TAF1/TAFII250, as it is the only TAF in this size range. To determine whether this was in fact TAF1, we performed photo-cross-linking using a wild-type Ad MLP DNA probe containing the photo-cross-linking adduct at +9 (SI), +19 (SII), and +31 (SIII). After cross-linking, these complexes were DNase treated, denatured (39), and immunoprecipitated using an antibody directed against TAF1. The immunoprecipitates were analyzed by PAGE followed by autoradiography (Fig. 4C). As predicted, the radiolabeled cross-linked species observed in the initial experiments was immunoprecipitated by the TAF1 antibody but not by protein A-agarose beads alone or the unrelated anti-FLAG antibody. These results demonstrate that TAF1 cross-linking is abrogated by mutations in the three DCE subelements.
Statistical analysis of human promoter databases.
To begin to establish the prevalence of the DCE in the mammalian genome, we analyzed two human promoter databases and searched for DCE subelements (see Materials and Methods). We utilized the Eukaryotic Promoter Database (EPD) and the Database of Transcriptional Start Sites (DBTSS). Importantly, the EPD specifically restricts entries to promoters whose start sites have been determined experimentally (4, 68). The DBTSS start sites were determined from full-length cDNAs (59, 74, 75), and a similar strategy was used to construct the Drosophila database used in the initial identification of the MTE (48, 55). Such a strategy is valid, not only because of the identification of the MTE, unknown at the time of that analysis, but also because searches of that database revealed TATA- and DPE-containing promoters (55). As indicated below, our analysis of the EPD and DBTSS yielded similar results, thus indicating that the DBTSS has as accurate a representation of promoter sequences as the EPD, whose transcription start sites were determined with traditional methods (4, 59). The advantage of our approach was demonstrated with a statistical analysis of core promoter elements and their combinations using databases of human promoters (21).
To examine the distribution of subelements, we calculated their actual number, their percentage, and the statistical significance of promoter sequences containing a corresponding motif (SI, SII, or SIII) at the expected positions for different subsets of promoters (Table 1). The promoter database was divided into the following sets (see Materials and Methods): TATA+/TATA− (promoters with/without a TATA box at any position in the window from −33 to −23 bp from +1 [sections 1 and 2 in Table 1]); Inr+/Inr− (promoters with/without the Inr element at any position in the window from −5 to +6 bp [sections 3 and 4]); DPE+/DPE− (promoters with/without DPE at any position in the window from +23 to +28 bp [sections 5 and 6]); TATA+ Inr− (promoters with TATA but without Inr elements [section 7]); Inr+ TATA− (promoters with Inr but without TATA elements [section 8]); and TATA+ Inr+/TATA− Inr− (promoters with/without the TATA and Inr elements with an optimal spacing between them [sections 9 and 10]). A promoter is considered as having an optimal combination of the TATA box and Inr element if the distance between their centers is 25 to 30 bp (17, 58). For comparison, we also calculated the averaged percentage of each motif in a randomly generated DNA sequence (section 11). We considered two scenarios of SI and SII motifs: exact matches to the experimentally derived sequences (i.e., CTTC for SI and CTGT for SII) (Fig. 1) and motifs with any 3 out of 4 bases matching the experimentally derived sequences.
TABLE 1.
Absolute numbers, percentages, and statistical significance of promoter sequences containing one of the DCE subelementsa
| Row | DCE subelement | 1. TATA+
|
2. TATA−
|
3. Inr+
|
4. Inr−
|
5. DPE+
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | dS | n | % | dS | n | % | dS | n | % | dS | n | % | dS | ||
| 1 | CTTC (4 of 4) | 39, 40 | 9.6, 4.4 | 7.50, 2.1 | 39, 244 | 2.7, 3.1 | −1.2, −0.1 | 50, 145 | 5.5, 3.4 | 4.1, 1.0 | 28, 139 | 2.9, 3.1 | −0.5, −0.2 | 21, 76 | 4.4, 3.5 | 1.6, 1.0 |
| 2 | CTTC (3 of 4) | 159, 315 | 39.0, 34.4 | 4.4, 3.5 | 394, 2,148 | 26.9, 27.2 | −0.9, −1.7 | 299, 1,297 | 32.6, 30.4 | 2.6, 2.3 | 254, 1,166 | 26.6, 25.7 | −0.9, −3.0 | 128, 619 | 27.1, 28.4 | −0.4, 0.3 |
| 3 | CTGT (4 of 4) | 5, 30 | 1.2, 3.3 | −1.6, 1.1 | 46, 211 | 3.1, 2.7 | 2.00, 1.2 | 28, 128 | 3.1, 3.0 | 1.3, 1.8 | 23, 113 | 2.4, 2.5 | 0.10, 0.4 | 9, 54 | 1.9, 2.5 | −0.9, 0.0 |
| 4 | CTGT (3 of 4) | 116, 272 | 28.5, 29.7 | 0.3, 0.3 | 420, 2,340 | 28.6, 29.7 | 0.9, 3.0 | 266, 1,304 | 29.0, 30.6 | 0.8, 2.7 | 270, 1,308 | 28.3, 28.8 | 0.5, 1.5 | 130, 633 | 27.5, 29.1 | −0.2, 0.9 |
| 5 | AGC (+30-+34) | 53, 117 | 13.0, 12.7 | 2.5, 2.9 | 203, 1,011 | 13.8, 12.8 | 5.1, 8.5 | 129, 577 | 14.0, 13.5 | 4.3, 7.5 | 127, 551 | 13.3, 12.1 | 3.7, 5.2 | 44, 188 | 9.3, 8.7 | 0.01, −1.5 |
| 6 | AGC (+24-+27) | 64, 125 | 15.7, 13.6 | 6.1, 6.23 | 175, 897 | 11.9, 11.3 | 5.6, 10.9 | 130, 556 | 14.1, 13.0 | 7.0, 11.6 | 109, 466 | 11.4, 10.2 | 4.1, 5.9 | 27, 139 | 5.7, 6.4 | −1.4, −2.3 |
n, number of times the subelement occurs; %, percentage of sequences with the subelement. The first value in every cell (except in the last section) depicts the percentage of promoters in the EPD database, and the second value depicts the percentage in the DBTSS database. The definitions of the positions of each subelement are as follows: SI (rows 1 and 2), positions from +6 to +11 bp for sections 1 to 8 and 10 and positions from +5 to +10 bp from the position of the center of Inr for section 9; SII (rows 3 and 4), positions from +16 to +21 bp for sections 1 to 8 and 10 and positions from +15 to +20 bp from the position of the center of Inr for section 9; SIII (row 5), positions from +30 to +34 bp for sections 1 to 8 and 10 and positions from +29 to +33 bp from the position of the center of Inr for section 9; SIII (row 6), positions from +24 to +27 bp for sections 1 to 8 and 10 and positions from +23 to +26 bp from the position of the center of Inr for section 9. The last section shows the average percentage of the respective motif in the randomly generated sequence with the same percentage of each of 4 nucleotides as in the averaged promoter sequences from EPD database, namely, with A averaging 20.6%, C averaging 29.4%, G averaging 29.4%, and T averaging 20.6%.
From Table 1 (rows 1 and 2) one can see that the percentage of promoters having the SI motif at any position from +6 to +11 (here and hereinafter the position indicates the 5′ edge of the motif) is considerably larger in the TATA+, Inr+, and especially the TATA+ Inr+ subsets of promoters than in the respective TATA−, Inr−, and TATA− Inr− subsets. This overrepresentation of SI is also confirmed by the high statistical significance of their occurrence frequencies (see sections 1, 3, and 9).
Although the occurrence frequency of SII downstream of +1 is larger than in regions upstream of +1 when the entire EPD or DBTSS is analyzed, there is no indication of a local occurrence frequency maximum at the expected positions from +16 to +21 relative to +1 (data not shown). However, the presence of SII has weak preference for the TATA−, Inr+, DPE−, and Inr+ TATA− subsets of promoters (Table 1, rows 3 and 4 in sections 2, 3, 5, and 8).
The occurrence frequency of subelement SIII is above a random distribution in the proximal downstream area, especially at positions from +24 to +34 bp when the entire EPD or DBTSS is examined. The statistical significance of the SIII presence in this interval of positions is extremely high (8.4 SD for EPD and 40.3 SD for DBTSS), suggesting the functional significance of this element in many promoters. The percentages of promoters having subelement SIII at any of those positions are 26.5% and 25.3% for the EPD and DBTSS, respectively. As we showed above, SIII functions as part of the DCE at positions from +30 to +34 and can stand alone at positions from +24 to +27. Therefore, we calculated the percentage of promoter sequences containing SIII in these two intervals for different subsets of promoters (Table 1, rows 5 and 6). From Table 1 (row 5, AGC [+30 to +34]), we see that the presence of SIII correlates strongly with the presence of an Inr element (compare sections 3 and 4). Similar to the correlation between SI and the optimal spacing of the TATA box and Inr, the promoters having the TATA+ Inr+ optimal combination are more likely to have SIII (compare sections 9 and 10). For SIII at positions +24 to +27, the dependence on the presence of the TATA box and Inr element is more definite (compare section 1 to section 2 for the TATA box and section 3 to section 4 for the Inr). The percentage of SIII in TATA+ Inr+ (row 6, AGC [+24 to +27]) promoters is essentially higher than in the TATA− Inr− subset of promoters (sections 9 and 10). The presence of the DPE is not compatible with the presence of SIII in both considered intervals of positions (compare sections 5 and 6 in rows 5 and 6). The conclusion of this analysis is that SIII sequences alone are found in a statistically significant number of promoters at the position predicted from our experimental data.
The positional distribution of all three subelements is graphically presented in Fig. S1 in the supplemental material. We also examined how many promoters have various combinations of subelements (see Table S2 in the supplemental material) and the degeneracy of the DCE subelements in the two promoter databases (see Table S3 in the supplemental material).
DISCUSSION
The data that we present here are significant in several respects. First, they provide conclusive evidence that the DCE represents a novel core promoter element that is found in several promoters transcribed by RNA polymerase II. Even more importantly, we provide the first examples of the functional recapitulation of transcription of several promoters containing a downstream element using a highly purified in vitro transcription system. These studies demonstrate that DCE function is solely dependent on the TAFs within the TFIID complex. We also show that one DCE subelement can function independently of the others: subelement III is found by itself in the HSV UL38, adenovirus E3, and IX promoters. SIII alone resides in a significant number of promoters, and the derivation of AGC as an accurate sequence representation is confirmed by mutagenesis of the Ad E3 and Ad IX promoters. The enormous overrepresentation of SIII in the downstream area in promoters from both databases (Table 1 and the supplemental material) indicates the biological importance of this element in a number of promoters. In comparison, 54/205 promoters (26%) in Drosophila contained a DPE (in an Inr-only context) (40). Most importantly, we have found that human TAF1 can be cross-linked to wild-type but not mutant DCE subelement sequences. The combination of the TFIID-dependent reconstitution experiments and the UV cross-linking identification exceeds the instructiveness of DNase I protection experiments using TFIID. The reconstituted in vitro transcriptions offer convincing functional evidence of the TFIID/TAF requirements. The photo-cross-linking experiments not only show TFIID-dependent interactions that are sequence specific and correlate with mutations in the DCE subelements but also achieve a superior resolution relative to DNase I protection assays.
A second interpretation of the TAF1 photo-cross-linking data are that alterations in DCE sequences might disrupt TAF cross-linking indirectly. These sequence changes may alter DNA bending (which is also sequence specific) such that the TFIID (TAF1)-DNA complex is affected, which in turn perturbs transcription. It is also possible that the TAF1 cross-links are simply proximity cross-linking and do not reflect a sequence-specific interaction of TAF1 and DCE DNA sequences. Instead, as mentioned above, the DNA structure might be altered or there may be another TAF responsible for DNA binding. The inability of this hypothetical TAF(s) to bind a mutated DCE sequence might then result in additional structural changes in TFIID such that the TAF1 cross-linking is lost.
A comparison of the DCE and DPE reveals several important differences. The DPE consists of one element centered around +30 (and possible additional sequence information at +24), whose consensus sequence is A/G G A/T C/T GT (40). The DCE is strikingly different in sequence, architecture, and factor requirements. The DCE consists of three subelements, and each is distinct from the DPE sequence: SI is CTTC, SII is CTGT, and SIII is AGC. SI resides approximately from +6 to +11, SII from +16 to +21, and SIII from +30 to +34. SIII may display more positional variability since it is found from +26 to +28 in the HSV UL38 and Ad E3 promoters. Note that the presence of SI and SIII does not correlate with the presence of DPE (compare rows 2 to 6 in sections 5 and 6 in Table 1), suggesting that the functions of these two subelements and the DPE are mutually exclusive. We also amend the conclusions in reference 43 to now suggest that the β-globin DCE is more extensive than previously recognized. SI seems to comprise positions +8 to +11, and it is not clear whether to consider the +13/15 mutation a defect in SI or whether it represents an additional subelement.
Photo-cross-linking data indicate that TAF1 interacts with the DCE in a sequence-dependent manner. These data can be contrasted with the observations of Burke and Kadonaga (5), who concluded that dTAFII40/60 (TAF9/TAF6) are necessary for sequence-specific recognition of the DPE. This contrast between the DCE and DPE shows that downstream elements will function by contacting distinct TAFs, and we suggest that other TAFs will recognize additional, as-yet-undiscovered core promoter elements (for example, the recently described MTE [48]).
Despite the expanded definition of the subelements, the distances between the subelements are maintained at 10-base-pair intervals (43). This is most readily seen in the spacing between the β-globin subelements II and III and the spacing of SI and SII of the Ad MLP DCE. The exception is SIII of the Ad MLP, which is ∼14 bp from SII. However, SIII function in the β-globin, UL38, and E3 promoters is accurately recapitulated with TFIID. Previous work indicated that the spacing between the subelements may be functionally relevant, as 5-base-pair insertions between each of the three subelements was deleterious to transcription (43). Our analysis here though indicates that another interpretation is more likely. We found that movement of SIII in the Ad E3 and Ad MLP promoters was deleterious for promoter function (Fig. 3). Since Ad E3 (or Ad IX) does not appear to have SI and SII, we argue that it is much more likely to be the distance from either the TATA box or the Inr element to SIII that is important. In any case, it is apparent that TFIID and the resident TAFs impose architectural constraints on the position of the DCE subelements.
The derivation of sequences of the DCE subelements explains additional data as well. There are two β-thalassemia mutations at +10 (−T) (2) and +20 (C to G) (24) and these can now be explained as mutations in SI and SII. The sequence derivations also help to explain the intermediate effects seen in the scanning mutagenesis of both the β-globin and Ad major late promoters as partial defects in a subelement. Indeed, similar partial defects can be seen in the analysis of the MTE (48), and we suggest that these are likely characteristics of downstream elements in general. The presence of a DCE in the Ad MLP explains the observations of several groups that TFIID has extended interactions from the TATA box at −30, downstream to +40 but that the TFIID interaction with the adenovirus E4 promoter, which does not have any DCE subelements (Fig. 3B), is centered only around the TATA box (12, 52, 67, 92).
Finally, the identification of a TAF1 interaction with the DCE has several significant implications. Our data support the interpretation that TFIID serves as a core promoter recognition complex (9, 25, 28, 36, 37, 81, 82). Additionally, it offers a mechanistic explanation as to why certain core promoter regions are TAF1 responsive (29, 47, 53, 64, 70, 71, 76, 78, 79, 84, 85), and we suggest that some of these promoters will contain DCE subelements. Interestingly, HSV late gene expression, including the UL38 gene, is markedly reduced in a TAF1 temperature-sensitive cell line at the nonpermissive temperature (14). We suggest that this down-regulation is due to the presence of DCE subelements in these late gene promoters. Others have noted that several such late genes do contain DAS elements, within which we find DCE SIII (Fig. 3), and these were proposed to be the basis for the regulation of late gene expression (38, 63, 88).
The photo-cross-linking of TAF1 to wild-type DCE DNA but not mutant DCE DNA suggests that TAF1 contains distinct DNA-binding domains in addition to its kinase, acetyltransferase, and ubiquitinase activities (16, 51, 60), as well as having bromodomains which bind acetylated proteins (33, 50). Several reports have predicted that TAF1 contains an HMG box with extensive similarities to HMG1 (30, 64, 69, 87). Some members of this family of HMG proteins bind nonspecifically to DNA and instead recognize DNA structural features such as bent sequences (HMG1/2), while others in this family do recognize specific DNA sequences (LEF-1, SRY) (3).
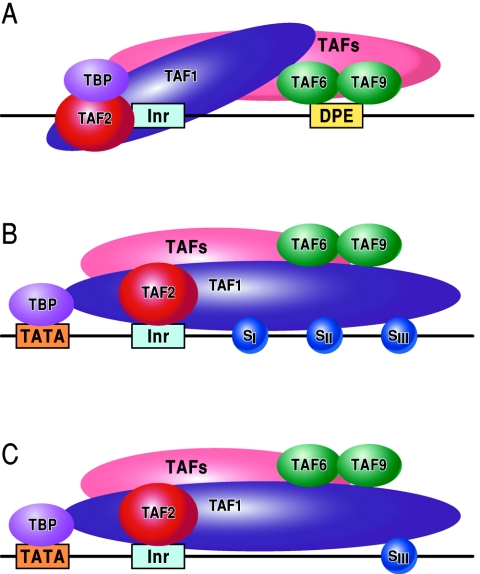
In Fig. 5 we propose a model of downstream element function that explains the distinctions between the DCE and DPE. The DCE is readily explained simply via its physical and functional interactions with TFIID via TAF1. TAF1 is capable of sequence-specific DNA binding and is the third example of TAF/sequence-specific core promoter DNA recognition (TAF1/TAF2 Inr recognition and TAF6/9 DPE recognition) (5, 9, 36, 37, 81, 83), although other TAF/promoter DNA contacts have been described previously (54), especially downstream contacts involving TAF1 and TAF2 (19, 23, 61, 62, 77, 91). In contrast, the DPE is recognized by TAF6/TAF9 (5). In order to accommodate core promoters containing a DCE or DPE, we suggest that TFIID assumes different conformations on the DCE- and DPE-dependent promoters (compare Fig. 5A to B and C). These different TFIID conformations lead to the formation of unique PICs (PICDCE and PICDPE), which, due to the architecture of the PIC, may require additional factors to attain a productive transcription initiation complex (as is the case with PICDPE [45]). Alternatively, others have suggested that different TFIID complexes with differing TAF contents exist (10).
FIG. 5.
Models depicting the interaction of TFIID with the DPE and DCE. In the three models, TAF1 and TAF2 are jointly responsible for Inr element recognition (for a review, see reference 73). A. DPE sequence recognition is established by TAF6/TAF9 components of TFIID. This interaction results in a unique TFIID conformation that in turn results in the formation of a DPE-specific PIC (PICDPE). B. TFIID interacts with the DCE subelements via TAF1. Again, this results in a TFIID conformation that is different than a TFIID/DPE interaction. This also leads to the formation of a DCE-specific PIC (PICDCE). C. A similar interaction of TFIID occurs with promoters containing only SIII of the DCE. In all three cases, these unique PICs consist of their own unique set of factors and cofactors that, in the end, manifest themselves as different regulatory phenomena.
What then are the functions of downstream core promoter elements and these postulated unique PICs? We suggest that downstream core promoter elements contribute a novel specificity to the promoter. But why is it necessary to make such differing factor requirements? Why do different core promoter architectures exist? These differences may explain the observations that TATA- and DPE-dependent promoters are specific for particular enhancers (7, 56). These differences in core promoter factor requirements may also explain the context dependency of the TATA box and the preferences of activators for specific core promoter architectures (18, 20, 72, 86). Lastly, the presence of a DCE or DPE might be indicative of an architecture designed for specific regulatory networks, such as the regulation of housekeeping promoters versus tissue-specific promoters (or other highly regulated promoters) or the regulation of subsets of viral promoters. In any case, the study of core promoter elements remains a fruitful avenue of investigation and continues to expand and contribute to our understanding of promoter-specific transcriptional regulation.
Supplementary Material
Table 1a.
| 6. DPE−
|
7. TATA+ Inr−
|
8. Inr+ TATA−
|
9. TATA+ Inr+
|
10. TATA− Inr−
|
11. Random DNA sequence | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | dS | n | % | dS | n | % | dS | n | % | dS | n | % | dS | |
| 57, 208 | 4.1, 3.1 | 1.9, 0.04 | 10, 13 | 6.5, 3.4 | 2.4, 0.4 | 21, 118 | 3.2, 3.2 | 0.1, 0.1 | 22, 19 | 12.5, 5.1 | 7.2, 2.1 | 56, 265 | 3.3, 3.1 | 0.3, 0.1 | 2.2 |
| 425, 1,844 | 30.3, 27.8 | 1.6, −0.7 | 59, 120 | 38., 31.3 | 2.4, 1.3 | 199, 1,102 | 29.9, 29.5 | 0.8, 1.2 | 68, 140 | 38.8, 37.2 | 2.8, 3.2 | 485, 2,323 | 28.6, 27.6 | 0.4, −1.2 | 29.8 |
| 42, 187 | 3.0, 2.8 | 1.7, 1.8 | 3, 10 | 1.9, 2.6 | −0.4, 0.01 | 26, 108 | 3.9, 2.9 | 2.6, 1.4 | 1, 16 | 0.6, 4.3 | −1.6, 1.9 | 50, 225 | 2.9, 2.67 | 1.6, 1.1 | 2.2 |
| 406, 1,979 | 29.0, 29.8 | 1.2, 2.9 | 43, 109 | 27.7, 28.4 | −0.1, −0.1 | 193, 1,141 | 29.0, 30.6 | 0.7, 2.7 | 53, 120 | 30.2, 31.9 | 0.6, 0.9 | 483, 2,492 | 28.4, 29.6 | 0.7, 2.8 | 29.8 |
| 212, 940 | 15.1, 14.2 | 6.5, 11.1 | 14, 35 | 9.0, 9.1 | −0.1, −0.3 | 90, 495 | 13.5, 13.2 | 3.1, 6.5 | 29, 58 | 16.5, 15.4 | 3.1, 3.3 | 227, 1,070 | 13.3, 12.7 | 5.0, 8.5 | 8.9 |
| 212, 883 | 15.1, 13.3 | 9.8, 15.4 | 22, 50 | 14.1, 13.0 | 3.1, 3.8 | 88, 481 | 13.2, 12.9 | 4.9, 10.6 | 35, 57 | 20.0, 15.1 | 6.0, 4.8 | 204, 965 | 12.0, 11.4 | 6.3, 11.6 | 7.1 |
Acknowledgments
We thank Lynne Vales for critical reading of the manuscript, Edward Wagner for supplying the HSV UL38 promoter constructs, Cheng-Ming Chiang and Robert Roeder for providing the FLAG-tagged TBP HeLa cell line, Laszlo Tora for the 3G3 TBP monoclonal antibody, and Thomas E. Shenk for providing adenovirus genomic DNA plasmids. B.A.L. thanks Stuart Orkin for his continued support and interest.
B.A.L. was supported by NIH/NIDDK grant K01 DK60001, and D.R. was supported by NIH grant GM 64844. D.R. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abmayr, S., J. Workman, and R. Roeder. 1988. The pseudorabies immediate early protein stimulates in vitro transcription by facilitating TFIID: promoter interactions. Genes Dev. 2:542-553. [DOI] [PubMed] [Google Scholar]
- 2.Athanassiadou, A., A. Papachatzopoulou, N. Zoumbos, G. M. Maniatis, and R. Gibbs. 1994. A novel beta-thalassaemia mutation in the 5′ untranslated region of the beta-globin gene. Br. J. Haematol. 88:307-310. [DOI] [PubMed] [Google Scholar]
- 3.Bewley, C. A., A. M. Gronenborn, and G. M. Clore. 1998. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu. Rev. Biophys. Biomol. Struct. 27:105-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucher, P. 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212:563-578. [DOI] [PubMed] [Google Scholar]
- 5.Burke, T. W., and J. T. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII 60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke, T. W., and J. T. Kadonaga. 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10:711-724. [DOI] [PubMed] [Google Scholar]
- 7.Butler, J. E., and J. T. Kadonaga. 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15:2515-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, S. P., B. Eng, W. H. Francombe, N. F. Olivieri, A. G. Kendall, J. S. Waye, and D. H. Chui. 1992. Two novel beta-thalassemia mutations in the 5′ and 3′ noncoding regions of the beta-globin gene. Blood 79:1342-1346. [PubMed] [Google Scholar]
- 9.Chalkley, G. E., and C. P. Verrijzer. 1999. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 18:4835-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Z., and J. L. Manley. 2003. Core promoter elements and TAFs contribute to the diversity of transcriptional activation in vertebrates. Mol. Cell. Biol. 23:7350-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang, C., and R. G. Roeder. 1995. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267:531-536. [DOI] [PubMed] [Google Scholar]
- 12.Chiang, C. M., H. Ge, Z. Wang, A. Hoffmann, and R. G. Roeder. 1993. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 12:2749-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowie, A., and R. M. Myers. 1988. DNA sequences involved in transcriptional regulation of the mouse beta-globin promoter in murine erythroleukemia cells. Mol. Cell. Biol. 8:3122-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar, S., and J. P. Weir. 2000. Herpes simplex virus 1 late gene expression is preferentially inhibited during infection of the TAF250 mutant ts13 cell line. Virology 270:190-200. [DOI] [PubMed] [Google Scholar]
- 15.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dikstein, R., S. Ruppert, and R. Tjian. 1996. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell 84:781-790. [DOI] [PubMed] [Google Scholar]
- 17.Emami, K. H., A. Jain, and S. T. Smale. 1997. Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev. 11:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emami, K. H., W. W. Navarre, and S. T. Smale. 1995. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol. Cell. Biol. 15:5906-5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emanuel, P. A., and D. S. Gilmour. 1993. Transcription factor TFIID recognizes DNA sequences downstream of the TATA element in the Hsp70 heat shock gene. Proc. Natl. Acad. Sci. USA 90:8449-8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garraway, I., K. Semple, and S. T. Smale. 1996. Transcription of the lymphocyte-specific terminal deoxynucleotidyltransferase gene requires a specific core promoter structure. Proc. Natl. Acad. Sci. USA 93:4336-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gershenzon, N. I., and I. P. Ioshikhes. 2005. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics 21:1295-1300. [DOI] [PubMed] [Google Scholar]
- 22.Gershenzon, N. I., G. D. Stormo, and I. P. Ioshikhes. 2005. Computational technique for improvement of the position-weight matrices for the DNA/protein binding sites. Nucleic Acids Res. 33:2290-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilmour, D. S., T. J. Dietz, and S. C. R. Elgin. 1990. UV cross-linking identifies four polypeptides that require the TATA box to bind to the Drosophila hsp70 promoter. Mol. Cell. Biol. 10:4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Redondo, J. M., T. A. Stoming, A. Kutlar, F. Kutlar, K. D. Lanclos, E. F. Howard, Y. J. Fei, M. Aksoy, C. Altay, A. Gurgey, et al. 1989. A C→T substitution at nt 101 in a conserved DNA sequence of the promotor region of the beta-globin gene is associated with “silent” beta-thalassemia. Blood 73:1705-1711. [PubMed] [Google Scholar]
- 25.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 26.Guzowski, J. F., J. Singh, and E. K. Wagner. 1994. Transcriptional activation of the herpes simplex virus type 1 UL38 promoter conferred by the cis-acting downstream activation sequence is mediated by a cellular transcription factor. J. Virol. 68:7774-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzowski, J. F., and E. K. Wagner. 1993. Mutational analysis of the herpes simplex virus type 1 strict late UL38 promoter/leader reveals two regions critical in transcriptional regulation. J. Virol. 67:5098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hampsey, M., and D. Reinberg. 1997. Transcription: why are TAFs essential? Curr. Biol. 7:R44-R46. [DOI] [PubMed] [Google Scholar]
- 29.Hilton, T. L., and E. H. Wang. 2003. Transcription factor IID recruitment and Sp1 activation. Dual function of TAF1 in cyclin D1 transcription. J. Biol. Chem. 278:12992-13002. [DOI] [PubMed] [Google Scholar]
- 30.Hisatake, K., S. Hasegawa, R. Takada, Y. Nakatani, M. Horikoshi, and R. G. Roeder. 1993. The p250 subunit of native TATA box-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature 362:179-181. [DOI] [PubMed] [Google Scholar]
- 31.Ho, P. J., J. Rochette, C. A. Fisher, B. Wonke, M. K. Jarvis, A. Yardumian, and S. L. Thein. 1996. Moderate reduction of B-globin gene transcript by a novel mutation in the 5′ untranslated region: a study of its interaction with other genotypes in two families. Blood 87:1170-1178. [PubMed] [Google Scholar]
- 32.Horikoshi, M., T. Hai, Y. S. Lin, M. R. Green, and R. G. Roeder. 1988. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell 54:1033-1042. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 34.Jacq, X., C. Brou, Y. Lutz, I. Davidson, P. Chambon, and L. Tora. 1994. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell 79:107-117. [DOI] [PubMed] [Google Scholar]
- 35.Javahery, R., A. Khachi, K. Lo, B. Zenzie-Gregory, and S. T. Smale. 1994. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol. 14:116-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufmann, J., and S. T. Smale. 1994. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 8:821-829. [DOI] [PubMed] [Google Scholar]
- 37.Kaufmann, J., C. P. Verrijzer, J. Shao, and S. T. Smale. 1996. CIF, an essential cofactor for TFIID-dependent initiator function. Genes Dev. 10:873-886. [DOI] [PubMed] [Google Scholar]
- 38.Kibler, P. K., J. Duncan, B. D. Keith, T. Hupel, and J. R. Smiley. 1991. Regulation of herpes simplex virus true late gene expression: sequences downstream from the US11 TATA box inhibit expression from an unreplicated template. J. Virol. 65:6749-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, T. K., R. H. Ebright, and D. Reinberg. 2000. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288:1418-1422. [DOI] [PubMed] [Google Scholar]
- 40.Kutach, A. K., and J. T. Kadonaga. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20:4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagrange, T., A. N. Kapanidis, H. Tang, D. Reinberg, and R. H. Ebright. 1998. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagrange, T., T. K. Kim, G. Orphanides, Y. W. Ebright, R. H. Ebright, and D. Reinberg. 1996. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photo-cross-linking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc. Natl. Acad. Sci. USA 93:10620-10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis, B. A., T. K. Kim, and S. H. Orkin. 2000. A downstream element in the human beta-globin promoter: evidence of extended sequence-specific transcription factor IID contacts. Proc. Natl. Acad. Sci. USA 97:7172-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis, B. A., and S. H. Orkin. 1995. A functional initiator element in the human beta-globin promoter. J. Biol. Chem. 270:28139-28144. [DOI] [PubMed] [Google Scholar]
- 45.Lewis, B. A., R. J. Sims III, W. S. Lane, and D. Reinberg. 2005. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol. Cell 18:471-481. [DOI] [PubMed] [Google Scholar]
- 46.Lewis, E. D., and J. L. Manley. 1985. Control of adenovirus late promoter expression in two human cell lines. Mol. Cell. Biol. 5:2433-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, X. Y., S. R. Bhaumik, X. Zhu, L. Li, W. C. Shen, B. L. Dixit, and M. R. Green. 2002. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 12:1240-1244. [DOI] [PubMed] [Google Scholar]
- 48.Lim, C. Y., B. Santoso, T. Boulay, E. Dong, U. Ohler, and J. T. Kadonaga. 2004. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 18:1606-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maldonado, E., R. Drapkin, and D. Reinberg. 1996. Purification of human RNA polymerase II and general transcription factors. Methods Enzymol. 274:72-100. [DOI] [PubMed] [Google Scholar]
- 50.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 51.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima, N., M. Horikoshi, and R. G. Roeder. 1988. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol. Cell. Biol. 8:4028-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Brien, T., and R. Tjian. 1998. Functional analysis of the human TAFII250 N-terminal kinase domain. Mol. Cell 1:905-911. [DOI] [PubMed] [Google Scholar]
- 54.Oelgeschlager, T., C.-M. Chiang, and R. G. Roeder. 1996. Topology and reorganization of a human TFIID-promoter complex. Nature 382:735-738. [DOI] [PubMed] [Google Scholar]
- 55.Ohler, U., G. C. Liao, H. Niemann, and G. M. Rubin. 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3:RESEARCH0087. [DOI] [PMC free article] [PubMed]
- 56.Ohtsuki, S., M. Levine, and H. N. Cai. 1998. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12:547-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oner, R., S. Agarwal, A. J. Dimovski, G. D. Efremov, G. H. Petkov, C. Altay, A. Gurgey, and T. H. J. Huisman. 1991. The G-A mutation at position +22 3′ to the cap site of the β-globin as a possible cause for a β-thalassemia mutation. Hemoglobin 15:67. [DOI] [PubMed] [Google Scholar]
- 58.O'Shea-Greenfield, A., and S. T. Smale. 1992. Roles of TATA and initiator elements in determining the start site location and direction of RNA polymerase II transcription. J. Biol. Chem. 267:6450. [PubMed] [Google Scholar]
- 59.Ota, T., Y. Suzuki, T. Nishikawa, T. Otsuki, T. Sugiyama, R. Irie, A. Wakamatsu, K. Hayashi, H. Sato, K. Nagai, K. Kimura, H. Makita, M. Sekine, M. Obayashi, T. Nishi, T. Shibahara, T. Tanaka, S. Ishii, J. Yamamoto, K. Saito, Y. Kawai, Y. Isono, Y. Nakamura, K. Nagahari, K. Murakami, T. Yasuda, T. Iwayanagi, M. Wagatsuma, A. Shiratori, H. Sudo, T. Hosoiri, Y. Kaku, H. Kodaira, H. Kondo, M. Sugawara, M. Takahashi, K. Kanda, T. Yokoi, T. Furuya, E. Kikkawa, Y. Omura, K. Abe, K. Kamihara, N. Katsuta, K. Sato, M. Tanikawa, M. Yamazaki, K. Ninomiya, T. Ishibashi, H. Yamashita, K. Murakawa, K. Fujimori, H. Tanai, M. Kimata, M. Watanabe, S. Hiraoka, Y. Chiba, S. Ishida, Y. Ono, S. Takiguchi, S. Watanabe, M. Yosida, T. Hotuta, J. Kusano, K. Kanehori, A. Takahashi-Fujii, H. Hara, T. O. Tanase, Y. Nomura, S. Togiya, F. Komai, R. Hara, K. Takeuchi, M. Arita, N. Imose, K. Musashino, H. Yuuki, A. Oshima, N. Sasaki, S. Aotsuka, Y. Yoshikawa, H. Matsunawa, T. Ichihara, N. Shiohata, S. Sano, S. Moriya, H. Momiyama, N. Satoh, S. Takami, Y. Terashima, O. Suzuki, S. Nakagawa, A. Senoh, H. Mizoguchi, Y. Goto, F. Shimizu, H. Wakebe, H. Hishigaki, T. Watanabe, A. Sugiyama, et al. 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36:40-45. [DOI] [PubMed] [Google Scholar]
- 60.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 61.Purnell, B. A., P. A. Emanuel, and D. S. Gilmour. 1994. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 8:830-842. [DOI] [PubMed] [Google Scholar]
- 62.Purnell, B. A., and D. S. Gilmour. 1993. Contribution of sequences downstream of the TATA element to a protein-DNA complex containing the TATA-binding protein. Mol. Cell. Biol. 13:2593-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romanelli, M. G., P. Mavromara-Nazos, D. Spector, and B. Roizman. 1992. Mutational analysis of the ICP4 binding sites in the 5′ transcribed noncoding domains of the herpes simplex virus 1 UL 49.5γ2 gene. J. Virol. 66:4855-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruppert, S., E. H. Wang, and R. Tjian. 1993. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature 362:175-179. [DOI] [PubMed] [Google Scholar]
- 65.Ryu, S., and R. Tjian. 1999. Purification of transcription cofactor complex CRSP. Proc. Natl. Acad. Sci. USA 96:7137-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 66a.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 67.Sawadogo, M., and R. G. Roeder. 1985. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell 43:165-175. [DOI] [PubMed] [Google Scholar]
- 68.Schmid, C. D., V. Praz, M. Delorenzi, R. Perier, and P. Bucher. 2004. The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res. 32(Database issue):D82-D85. [DOI] [PMC free article] [PubMed]
- 69.Sekiguchi, T., Y. Nohiro, Y. Nakamura, N. Hisamoto, and T. Nishimoto. 1991. The human CCG1 gene, essential for progression of the G1 phase, encodes a 210-kilodalton nuclear DNA-binding protein. Mol. Cell. Biol. 11:3317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen, W. C., S. R. Bhaumik, H. C. Causton, I. Simon, X. Zhu, E. G. Jennings, T. H. Wang, R. A. Young, and M. R. Green. 2003. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 22:3395-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen, W.-C., and M. R. Green. 1997. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90:615-624. [DOI] [PubMed] [Google Scholar]
- 72.Simon, M. C., T. M. Fisch, B. J. Benecke, J. R. Nevins, and N. Heintz. 1988. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell 52:723-729. [DOI] [PubMed] [Google Scholar]
- 73.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki, Y., and S. Sugano. 2003. Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method. Methods Mol. Biol. 221:73-91. [DOI] [PubMed] [Google Scholar]
- 75.Suzuki, Y., R. Yamashita, S. Sugano, and K. Nakai. 2004. DBTSS, database of transcriptional start sites: progress report 2004. Nucleic Acids Res. 32(Database issue):D78-D81. [DOI] [PMC free article] [PubMed]
- 76.Suzuki-Yagawa, Y., M. Guermah, and R. G. Roeder. 1997. The ts13 mutation in the TAF(II)250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol. Cell. Biol. 17:3284-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sypes, M. A., and D. S. Gilmour. 1994. Protein/DNA crosslinking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res. 22:807-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsukihashi, Y., M. Kawaichi, and T. Kokubo. 2001. Requirement for yeast TAF145 function in transcriptional activation of the RPS5 promoter that depends on both core promoter structure and upstream activating sequences. J. Biol. Chem. 276:25715-25726. [DOI] [PubMed] [Google Scholar]
- 79.Tsukihashi, Y., T. Miyake, M. Kawaichi, and T. Kokubo. 2000. Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol. Cell. Biol. 20:2385-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Dyke, M. W., R. G. Roeder, and M. Sawadogo. 1988. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science 241:1335-1338. [DOI] [PubMed] [Google Scholar]
- 81.Verrijzer, C. P., J. L. Chen, K. Yokomori, and R. Tjian. 1995. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 81:1115-1125. [DOI] [PubMed] [Google Scholar]
- 82.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338-342. [PubMed] [Google Scholar]
- 83.Verrijzer, C. P., K. Yokomori, J. L. Chen, and R. Tjian. 1994. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science 264:933-941. [DOI] [PubMed] [Google Scholar]
- 84.Wang, E. H., and R. Tjian. 1994. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science 263:811-814. [DOI] [PubMed] [Google Scholar]
- 85.Wang, E. H., S. Zou, and R. Tjian. 1997. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 11:2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wefald, F. C., B. H. Devlin, and R. S. Williams. 1990. Functional heterogeneity of mammalian TATA-box sequences revealed by interaction with a cell-specific enhancer. Nature 344:260-262. [DOI] [PubMed] [Google Scholar]
- 87.Weinzierl, R. O., B. D. Dynlacht, and R. Tjian. 1993. Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature 362:511-517. [DOI] [PubMed] [Google Scholar]
- 88.Weir, J. P., and P. R. Narayanan. 1990. Expression of the herpes simplex virus type 1 glycoprotein C gene requires sequences in the 5′ noncoding region of the gene. J. Virol. 64:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Willy, P. J., R. Kobayashi, and J. T. Kadonaga. 2000. A basal transcription factor that activates or represses transcription. Science 290:982-984. [DOI] [PubMed] [Google Scholar]
- 90.Wong, C., C. E. Dowling, R. K. Saiki, R. G. Higuchi, H. A. Erlich, and H. H. Kazazian, Jr. 1987. Characterization of beta-thalassaemia mutations using direct genomic sequencing of amplified single copy DNA. Nature 330:384-386. [DOI] [PubMed] [Google Scholar]
- 91.Wu, C.-H., L. Madabusi, H. Nishioka, P. Emanuel, M. Sypes, I. Arkhipova, and D. S. Gilmour. 2001. Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol. Cell. Biol. 21:1593-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou, Q., P. M. Lieberman, T. G. Boyer, and A. J. Berk. 1992. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 6:1964-1974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.