Abstract
The Suppressor of the Hairy wing [Su(Hw)] binding region within the gypsy retrotransposon is the best known chromatin insulator in Drosophila melanogaster. According to previous data, two copies of the gypsy insulator inserted between an enhancer and a promoter neutralize each other's actions, which is indicative of an interaction between the protein complexes bound to the insulators. We have investigated the role of pairing between the gypsy insulators located on homologous chromosomes in trans interaction between yellow enhancers and a promoter. It has been shown that trans activation of the yellow promoter strongly depends on the site of the transposon insertion, which is evidence for a role of surrounding chromatin in homologous pairing. The presence of the gypsy insulators in both homologous chromosomes even at a distance of 9 kb downstream from the promoter dramatically improves the trans activation of yellow. Moreover, the gypsy insulators have proved to stabilize trans activation between distantly located enhancers and a promoter. These data suggest that gypsy insulator pairing is involved in communication between loci in the Drosophila genome.
The enhancer-mediated activation is the basic mechanism of gene regulation in eukaryotes. Enhancers can act over large distances to activate transcription independently of their orientation and position relative to the promoter without affecting adjacent genes (6, 11). Insulators represent a class of DNA sequences that restrain regulatory interactions within eukaryotic genomes (21, 35, 36, 53, 63, 65). These elements restrict the enhancer and silencer functions, contributing to the establishment of independent gene regulation within heterochromatic and euchromatic domains.
The best known insulator was identified in Drosophila melanogaster within the 5′ untranslated region of the gypsy retrotransposon (42). It consists of 12 binding sites for the Su(Hw) protein (44, 62). Recently, another protein, CP190, was shown to bind to the gypsy insulator (54). These DNA-binding proteins are important for the gypsy insulator function, as mutations in the su(Hw) and CP190 genes reverse the mutagenic effects of the gypsy retrotransposon (46, 54). Mutations in another gene, mod(mdg4), alter the phenotypes of the gypsy-induced mutations (17, 18, 22). Previous studies indicate that the Mod(mdg4)-67.2 protein isoform interacts with the Su(Hw) and CP190 proteins (14, 25, 54). The prevalent model suggests that boundary elements, or insulators, subdivide eukaryotic chromosomes into functionally and structurally autonomous domains (65). The insulators determine the limits of higher-order “looped” chromatin domains by interacting either with each other or with some nuclear structures (8, 36, 65). This interaction might be responsible, at least partially, for the establishment of independent chromatin domains. In this context, it is noteworthy that the gypsy insulators have been found to coalesce into a few “insulator bodies” located at the periphery of the nucleus (19), which are partially or completely disrupted in the nuclei of CP190 or su(Hw) or mod(mdg4) mutants (20, 54). The fact that duplication of the gypsy insulator neutralizes the enhancer-blocking activity is also indicative of the interaction between the protein complexes bound to the gypsy insulators (9, 51).
The properties of the gypsy insulator properties suggest its involvement in pairing between homologous chromosomes in germ and somatic cells. In dipterans, homologous chromosomes are intimately synapsed in somatic cells of different types (12, 13, 30, 31, 34, 57, 66) and massive pairing of sister chromatids and homologs is responsible for the precisely banded pattern of polytene chromosomes (40). In Drosophila, a number of loci have been found at which pairing significantly influences gene expression. Such an influence was first detected within the bithorax complex by E. B. Lewis, who coined the term “transvection” to describe it (39).
A useful system for studying the factors determining transvection is the yellow gene, which encodes the protein responsible for dark pigmentation of the cuticle in larvae and adult insects (52). The enhancers controlling yellow expression in the wings and body cuticle are located in the upstream gene region, whereas the enhancer controlling yellow expression in bristles resides in the intron (23, 43). The wing and body enhancers of one allele can trans activate the yellow promoter on the paired homologous chromosome (24, 47, 60). However, the cis preference of enhancers for their own promoter precludes their action in trans (49). According to recent studies, yellow transvection can occur at multiple genomic locations, and the Drosophila genome is generally permissive to the enhancer action in trans (10). It has also been shown that transvection between yellow alleles strongly depends on homologous pairing and does not take place between nonhomologous sites (10, 24).
In this study, we used the transvection at yellow as a model system to analyze the role of the interaction of gypsy insulators in pairing between homologous chromosomes. Taking into account that trans activation of the yellow promoter is more effective in the presence of gypsy insulators on both homologs, we supposed that the proteins bound to the gypsy insulator may be involved in this process. Moreover, the gypsy insulators have proved to stabilize trans activation between distantly located enhancers and a promoter.
MATERIALS AND METHODS
Plasmid construction.
The 8-kb fragment containing the yellow gene and the cDNA yellow clone were kindly provided by P. Geyer. The 3-kb SalI-BamHI fragment containing the yellow regulatory region (yr) was subcloned into BamHI- and XhoI-digested pGEM7 (yr plasmid). The 5-kb BamHI-BglII fragment containing the coding region (yc) was subcloned into CaSpeR2 (C2-yc) or CaSpeR3 (C3-yc). The 430-bp gypsy sequence containing the Su(Hw)-binding region was PCR amplified from the gypsy retrotransposon. To confirm its identity, the product after sequencing was subcloned into CaSpeR3 (C3-su) and pSK (pSK-su) and between the LOX and FRT sites (lox-su-lox and FRT-su-FRT).
(i) (E)(Y)W.
The yellow regulatory region includes the body enhancer and the wing enhancer located between bp −700 and −1868 and between bp −1868 and −2873 relative to the transcription start site of the yellow gene, respectively (23). The yellow enhancers were PCR amplified with primers 5′ TAT GCA ACT GAC GAT GGC TTA AG (between bp −2808 and −2786 relative to the yellow start site) and 5′ AAT TGG AAC TCG TGC TCG 3′ (between −711 and −728) and cloned between the FRT sites (y-en). The promoter and first exon of the yellow gene were PCR amplified with primers 5′ TAC AAG GAA ACA CCT GC 3′ (bp −702 and −686) and 5′ TCG TCT GTA CTA GAT TAA AAT 3′ (between bp +599 and +579) and cloned between the LOX sites (y-pr). The latter primer carried a point mutation in the final PCR product, which disrupted the site for SpeI endonuclease normally located in the yellow gene intron. The (y-pr) fragment was ligated into C2-yc cleaved with SpeI and XbaI [C2-yc-(y-pr)]. The (y-en) fragment was ligated into C2-yc-(y-pr) cleaved with XhoI and XbaI.
(ii) (E)(Y)SW.
The SphI-XbaI DNA fragment was cut from (E)(Y)W and ligated into C3-su cleaved with SphI and XbaI.
(iii) (E)(Y)WS.
The SphI-XbaI DNA fragment was cloned into EyeSYWS cleaved with SphI and XbaI. The EyeSYWS was earlier described by Muravyova et al. (51).
The fragment of the yellow wing and body enhancers SalI-Eco47III was cloned into FRT-su-FRT to obtain the FRT-E-su-FRT plasmid. The fragment FRT-E-su-FRT was ligated into yr which was digested with the SalI and Eco47III enzymes. Its correct orientation in the resulting yr-FRT-su-FRT plasmid was confirmed by PCR analysis.
(iv) (ES)Y(SW).
The single lox site was first ligated into C2-yc digested with NruI in the intron of the white gene. Then we cloned a lox-su fragment from the lox-su-lox plasmid between the yellow and white genes to produce C2-yc-lox-su-w-lox. Finally, we combined the C2-ys-lox-su-w-lox and yr-FRT-su-FRT fragments obtained with XbaI and BamHI endonucleases.
(v) (ES)(Y)W and (ES)(Y)SW.
The lox site was introduced into the SpeI site of the yellow gene intron within the C2-yc plasmid. We added another lox site or the lox site together with the gypsy insulator (su-lox) between the yellow and white genes to the BglII site to obtain plasmids C2-yc-lox-y-lox and C2-yc-lox-y-lox-su, respectively. To obtain the final constructs (ES)(Y)W and (ES)(Y)SW, the plasmids were cleaved with XbaI and BamHI and combined with the yr-FRT-su-FRT fragment.
Drosophila strains, transformation, and genetic crosses.
Flies were cultured on the standard yeast medium at 25°C. Five females were mated with two males in vials and brooded every second day. The temperature and crowding were carefully controlled, as both factors affect pigmentation. The mutant alleles and chromosomes used in this work and balancer chromosomes are described elsewhere (41).
The transposon constructs together with P25.7wc, the P element containing defectively inverted repeats that was used as a transposase source (32), were injected into y ac w1118 preblastoderm embryos. The resulting flies were crossed with the y ac w1118 flies, and the transgenic progeny were identified by their eye color. In all transgenic lines, the flies had dark-yellow to dark-orange eyes, which indicated that chromatin surrounding the transgenes was permissive to transcription. To check transposon integrity and copy number, the transformed lines were examined by Southern blot hybridization. Chromosome localization of various transgene insertions was determined by crossing the transformants with the y ac w1118 balancer stock containing dominant markers, In(2RL),CyO for chromosome 2, and In(3LR)TM3,Sb− for chromosome 3. The precise sites of transgene insertions were determined by inverse PCR (iPCR) (http://www.fruitfly.org/methods). Genomic DNA from two flies was digested by RsaI or FspBI. After heat inactivation of the endonucleases, DNA fragments were self-ligated at 4°C for 24 h in 0.4 ml reaction mixture. For iPCR, we used the primers from the P element, 5′ AAG ATT CGC AGT GGA AGG CTG CAC 3′ and 5′ TCC GCA CAC AAC CTT TCC TCT CAA C 3′. The successfully amplified products were cloned in a Bluescript plasmid (Stratagene, La Jolla, CA) and sequenced. We used fluorescence in situ hybridization (FISH) (37) to map the insertions that, according to iPCR data, were located in repetitive elements of the genome. The full-length yellow gene labeled by the Bionick labeling kit (Life Technologies) served as a probe.
The selected lines were crossed on a su(Hw)v/su(Hw)f and mod(mdg4)u1 mutant background (17) to determine the contribution of the gypsy insulator to the yellow phenotype.
The enhancerless or promoterless lines were obtained by crossing the flies bearing the transposons with the Flp (w1118; S2 CyO hsFLP ISA/Sco; +) or Cre (y1 wi; CyO P[w+,cre]/Sco; +) recombinase-expressing lines. A high level of F recombinase was produced by exposing late embryos and second or third instar larvae to heat shock at 37°C for 2 h. All excisions were confirmed by Southern blot hybridization and/or PCR analysis. The details of the crosses used for genetic analysis and for excision of functional elements are available upon request.
Analysis of yellow phenotypes.
Pigmentation of the wing blades and body cuticle in the abdominal stripes (below, referred to as wing and body pigmentation) was scored in 3- to 4-day-old females by using a five-grade pigmentation scale (48), with pigmentation scores of 1 and 5 corresponding to the null phenotype and the wild-type or almost wild-type state, respectively. Pigmentation scores were assigned by comparing the progeny of the flies obtained from parallel controls. Intermediate scores were determined relative to the pigmentation levels of y1#8/y1#8, y2/y2, y82f29/y1#8, and y2/y1#8 females, which corresponded to scores of 1, 1; 1, 1-2; 3, 3; and 4, 4 in the wing and body, respectively (10, 48). The pigmentation scores were independently determined by two investigators, who examined 30 to 50 female flies from each of the two independent crosses. Small variations in pigmentation depending on culture conditions did not exceed 0.5 points. The average phenotype was determined by averaging the pigmentation scores (http://www.igb.ac.ru/Kravchenko-Suppl.pdf).
RESULTS
The gypsy insulator facilitates trans activation of the yellow promoter.
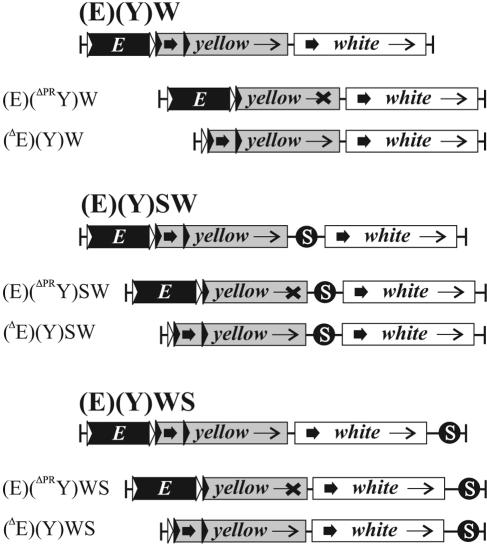
Previous studies showed that yellow transvection could occur at multiple genomic locations (10). The yellow region used in this work contained approximately 7.5 kb of 5′ flanking DNA and 2.1 kb of 3′ flanking DNA, which might include the sequences involved in homologous pairing. For this reason, we reexamined the trans action of the yellow enhancers without the flanking DNA. The yellow coding region was flanked by a 3-kb region that included only wing and body enhancers on the 5′ side and by a 200-bp region on the 3′ side. In the (E)(Y)W construct (Fig. 1), the wing and body enhancers were flanked by FRT sites, while the yellow promoter was flanked by LOX sites. The mini-white gene was used as a marker.
FIG. 1.
Structures of (E)(Y)WS, (E)(Y)SW, and (E)(Y)W transgenic constructs and their derivatives. The derivative pairs used in complementation tests for each construct are shown. Black boxes marked with an E are enhancers of the yellow gene. Coding regions of the yellow gene and the mini-white reporter gene are shown as gray and white boxes, respectively. Thick, short arrows in the boxes indicate promoters of yellow or mini-white. Thin arrows in the boxes indicate the direction of transcription. Black crossing indicates disruption of transcription resulting from excision of the promoter or part of the coding region. White and black triangles are FRT and LOX sites, respectively. Black circles marked with an S are the gypsy insulators.
Twelve transgenic lines with the (E)(Y)W transposon were established. Heterozygous (E)(Y)W/+ females (Table 1) displayed wing and body pigmentation ranging from almost wild type (score 4-5) to weak (score 2). The difference in yellow expression might be explained by the effect of surrounding chromatin.
TABLE 1.
Pigmentation scores in (E)(Y)W, (E)(Y)SW, and (E)(Y)WS transgenic lines and their transheterozygous enhancerless and promoterless derivatives
| Transgenic line (localization) | Cuticle pigmentation score (wing/body)a
|
|||
|---|---|---|---|---|
| (E)(Y)/+ | (ΔE)(Y)/(E)(ΔPRY)
|
|||
| +/+ | su(Hw)− | mod(mdg4)ul | ||
| (E)(Y)W | ||||
| 1 (3) | 5/4 | 3-4/3-4 | ND | ND |
| 2 (3) | 4-5/3-4 | 1/2-3 | ND | ND |
| 3c (3) | 4/4 | 3-4/3 | ND | ND |
| 4 (3) | 3-4/2-3 | 3-4/1-2 | ND | ND |
| 5 (3) | 3-4/2 | 2/2 | ND | ND |
| 6 (3) | 4/3-4 | 2-3/2 | ND | ND |
| 7 (3) | 4/3 | 3-4/4 | ND | ND |
| 8 (2) | 4/2-3 | 3/3 | 3/2-3 | 3/2 |
| 9 (3) | 3/2-3 | 1/2 | ND | ND |
| 10 (2) | 4-5/3-4 | 2/2 | 2/1-2 | 2/2 |
| 11 (3) | 4-5/4-5 | 1/2 | ND | ND |
| 12 (2) | 5/3-4 | 2-3/2 | 2-3/2 | 2/2 |
| APSb | 4.1/3.3 | 2.4/2.5 | ND | ND |
| (E)(Y)SW | ||||
| 13 28E9 | 5/5 | 3-4/3 | 2/2 | ND |
| 14 64B12 | 4-5/4 | 4/3-4 | ND | ND |
| 15 13A5 | 4-5/4 | 4-5/4 | 3-4/2-3 | ND |
| 16 (2) | 4/3-4 | 3-4/3-4 | 2/2 | ND |
| 17 52B2 | 4-5/4 | 3-4/3 | 2/2 | 2/2 |
| 18 64C9 | 3-4/3-2 | 3-4/3 | ND | ND |
| 19 (2) | 4-5/5 | 4-5/3-4 | 3/3 | 3-4/3 |
| 20 58Fd | 4/3-4 | 2-3/2-3 | 1-2/1-2 | ND |
| 21c (2) | 2-3/3 | 2-3/3 | ND | ND |
| 22 (3) | 5/4-5 | 5/4-5 | ND | ND |
| APS | 4.2/3.9 | 3.7/3.4 | 2.3/2.2 | ND |
| (E)(Y)WS | ||||
| 23 20E | 3-4/3-4 | 3/2 | ND | ND |
| 24 (3) | 4/3-4 | 4/3 | ND | ND |
| 25 23A3 | 4/3 | 3-4/3 | 1-2/1-2 | 3-4/2 |
| 26 39E3 | 5/5 | 5/4-5 | 4/3 | 5/4 |
| 27 29D4 | 5/5 | 5/3-4 | 3/2 | 3-4/2 |
| 28 75B5 | 4-5/4 | 4/3-4 | ND | ND |
| 29 100B3 | 5/4 | 4/3 | ND | ND |
| 30 68C13 | 4/3 | 3/2-3 | ND | ND |
| 31c 74A1 | 4/3 | 4/3 | ND | ND |
| 32 35DEd | 4/4 | 3-4/3-4 | 1/1 | 3-4/3-4 |
| 33 (3) | 4-5/4 | 3-4/3 | ND | ND |
| 34 28D2 | 4-5/4 | 4-5/3 | 3/2 | 4-5/3 |
| APS | 4.3/3.8 | 3.9/3.1 | 2.5/1.9 | 3.8/2.9 |
Pigmentation levels are shown for heterozygous (E)(Y)WS, (E)(Y)SW and (E)(Y)W females [(E)(Y)/+] and for transheterozygous females carrying the promoterless and enhancerless derivatives of the constructs on the +/+, su(Hw)−, or mod(mdg4)ul background [(ΔE)(Y)/(E)(ΔPRY)]. Bold data indicate complementation between y alleles. ND, not determined.
APS, average pigmentation scores assigned to each transgenic construct. Statistical data are available at http://www.igb.ac.ru/Kravchenko-Suppl.pdf.
Flies in these transgenic lines displayed variegated pigmentation of bristles.
Localization of these insertions was determined by FISH.
Enhancerless derivatives, (ΔE)(Y)W, were obtained by crossing (E)(Y)W flies with those carrying a Flp recombinase transgene and selecting the progeny with reduced cuticular pigmentation. All heterozygous (ΔE)(Y)W/+ females had yellow body cuticle and wing blades (a score of 1, 1) and wild-type bristle pigmentation, indicating that the yellow promoter was not influenced by the surrounding enhancers or repressive chromatin. Promoterless derivatives, (E)(ΔPRY)W, were produced by crossing (E)(Y)W flies with those carrying a Cre recombinase transgene and selecting progeny with a y1-like phenotype and pigmented eyes.
The flies carrying a (ΔE)(Y)W allele at an ectopic site were crossed with those carrying the corresponding (E)(ΔPRY)W allele at the same site; the level of the wing and body pigmentation in the female progeny was evaluated to determine whether interallelic complementation was possible (Table 1). At all 12 genomic sites, (ΔE)(Y)W/(E)(ΔPRY)W females had darker wing and body pigmentation than the heterozygous (ΔE)(Y)W/+ females. The pigmentation score of yellow phenotypes averaged 2.4/2.5 (wing/body). Interestingly, the strength of trans activation did not correlate with the level of the yellow activation in cis [Table 1, compare lines (E)(Y)W no. 3 and 8 with no. 11 and 12]. It seems possible that the trans activation level is influenced by homolog pairing efficiency depending on insertion site.
To check pairing between the gypsy insulators for the role in the improvement of activation in trans, the gypsy insulator was inserted between the yellow and mini-white genes in the (E)(Y)W construct (Fig. 1). Table 1 shows characteristics of 10 established (E)(Y)SW transgenic lines. The enhancerless and promoterless derivatives were obtained in the same ways as the (E)(Y)W transgenic lines. Pigmentation in the body cuticle and wings of all heterozygous (ΔE)(Y)W/+ females scored 1, 1 (with one exception: line no. 22 had the score 2-3, 2). The average score of y phenotypes for (ΔE)(Y)SW/(E)(ΔPRY)SW transheterozygous females was 3.7, 3.4 (wing, body). A comparison of the results obtained with (E)(Y)W and (E)(Y)SW transgenic lines showed that the pairing between the gypsy improved trans activation of the yellow promoter (Table 1).
The improvement of trans activation in the presence of the gypsy insulator could be explained by the blocking of cis interaction between the yellow enhancers and the white promoter. To check this assumption, we inserted the gypsy insulator into the (E)(Y)WS construct downstream from the white gene (Fig. 1). The enhancerless and promoterless derivatives were obtained for 12 transgenic lines. In a complementation test, transheterozygous (ΔE)(Y)WS/(E)(ΔPRY)WS females had the average pigmentation scores similar to that in the (ΔE)(Y)SW/(E)(ΔPRY)SW females carrying the insulator between the yellow and white genes: 3.9, 3.1 and 3.7, 3.4, respectively (Table 1). As the gypsy insulator did not separate the yellow and white genes in the (E)(Y)WS transgenic lines, trans activation improvement in its presence could not be explained by the block of interaction between the yellow enhancers and the white promoter in cis. It is noteworthy that the gypsy insulator located even at a distance of 9 kb from the yellow promoter is capable of improving trans activation.
To demonstrate the role of the gypsy insulator in trans activation, six pairs of the (E)(Y)SW and five pairs of the (E)(Y)WS derivatives located on the X and second chromosomes were tested on the su(Hw)− background. Inactivation of the su(Hw) gene had no influence on the pigmentation of the (ΔE)(Y)SW/+ and (ΔE)(Y)WS/+ females (data not shown). At the same time, the tested (ΔE)(Y)SW/(E)(ΔPRY)SW and (ΔE)(Y) WS/(E)(ΔPRY)WS females crossed on the su(Hw)− background displayed similar transvection as the (ΔE)(Y)W/(E)(ΔPRY)W females.
To test whether the su(Hw) mutation has a general effect, we crossed the flies of the three (E)(Y)W lines without the gypsy insulator on the su(Hw)− background. In a complementation test, no significant changes in trans activation were revealed in (ΔE)(Y)W/(E)(ΔPRY)W transheterozygous females (Table 1); therefore, the Su(Hw) protein is unlikely to have a general effect on trans activation.
The second component of the gypsy insulator, Mod(mdg4) protein, had a weaker effect on the transvection level. We observed only slight changes of pigmentation in transheterozygous (ΔE)(Y)SW/(E)(ΔPRY)SW and (ΔE)(Y)WS/(E)(ΔPRY)WS females on the mod(mdg4)u1 background (Table 1). Interestingly, all effects manifested in this case consisted in trans activation decrease.
The gypsy insulator promotes transvection between the yellow enhancer and promoter located in nonhomologous loci.
In complementation tests, the gypsy insulator proved to facilitate trans activation of the yellow promoter between homologous derivatives of the transgene, and the next question was whether it could stabilize transvection between nonhomologous derivatives. In the control experiment, we determined whether the transvection was supported by the enhancerless (ΔE)(Y)W allele and the promoterless (E)(ΔPRY)W allele from different ectopic sites. To this end, (ΔE)(Y)W males from twelve transgenic lines were mated to (E)(ΔPRY)W females from these lines. Phenotypic analysis of transheterozygous females obtained in 132 crosses revealed yellow pigmentation of the body cuticle and wing blades (data not shown). This result is in good agreement with those obtained by Chen et al. (10) and suggests that transvection between nonhomologous sites does not occur in the absence of the gypsy insulators.
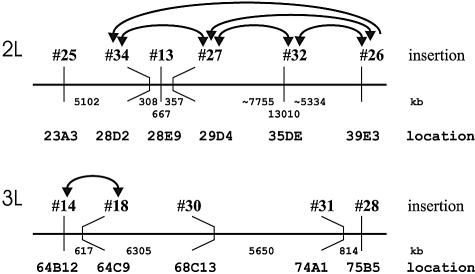
To analyze the effect of the gypsy insulator, we combined the nonhomologous enhancerless and promoterless derivatives of the 9 (E)(Y)SW and 12 (E)(Y)WS transgenic lines in an interallelic complementation test. Altogether, the phenotypes of 338 allele combinations were tested. Transheterozygous females obtained in 13 out of 338 crosses had visible activation of yellow expression, predominantly in the wings. In general, the transvection level was lower than that in complementation between the derivatives of the same insertion site. We observed one transvection event between nonhomologous insertions 14/18, both located on the third chromosome; one event between insertions 15/13 located on the X and second chromosomes, respectively; and five events between insertions located on the second chromosome (no. 27/34, no. 27/32, no. 27/26, no. 32/26, and no. 34/26) (Table 2). In most transvection events, trans activation occurred in both cases: the first allele provided the yellow promoter and the second provided the yellow enhancer, or vice versa. In the no. 34/26 event, however, trans activation occurred only between the no. 34 enhancerless and no. 26 promoterless derivatives. To clarify the circumstances of transvection, we determined the precise sites of insertions for most of the (E)(Y)SW and (E)(Y)WS transgenic lines (Table 1; Fig. 2). This analysis showed that insertions 34 and 26 were more than 20 Mb apart, which could account for limitation of the transvection to only one direction. Nevertheless, the relative distances between nonhomologous insertions could not be regarded as the determinants of tranvection efficiency. It was found that the complementation test between derivatives of no. 31 and no. 28 insertions located 814 kb away from each other did not give rise to yellow trans activation; conversely, comparable distances of 617 kb between insertions 14/18 and 667 kb between no. 27/34 did not prevent the tranvection of their enhancerless and promoterless derivatives. Likewise, a relatively short distance between insertions was not necessarily prerequisite for transvection. Thus, the no. 13 transgene insertion was between insertions 27 and 34 at distances of 308 and 359 kb from them, respectively. However, the no. 13 transgene derivatives could complement neither no. 27 nor no. 34 derivatives, whereas the latter two could support each other's transvections. It should also be noted that the no. 13 insertion did not represent a closed, inaccessible locus, as its derivatives show transvection with the no. 15 derivatives located on the X chromosome. In our opinion, the transvection efficiency depends mainly on the spatial arrangement of insertions within the nuclear architecture rather than on the linear chromosome distances between insertions sites. In a number of cases, transvection occurred when the distances between insertions were great: 7.16 Mb between no. 27/32, 5.33 Mb between no. 26/32, and 13.01 Mb between no. 27/26 (Fig. 2).
TABLE 2.
Complementation scores for the nonhomologous transheterozygotes of the (E)(Y)SW and (E)(Y)WS derivativesa
| Pairs of transgenic lines [(E)(ΔPRY)/(ΔE)(Y)] and distance between them, (kb) or locations | Cuticle pigmentation score (wing/body) [(ΔE)(Y)/(E)(ΔPRY)]
|
||
|---|---|---|---|
| +/+ | su(Hw)− | mod(mdg4)ul | |
| (E)(Y)SW | |||
| 14/18, 617 | 2-3/1 | ND | ND |
| 18/14 | 3/2 | ND | ND |
| 13/15, 2L and X | 3/1 | ND | ND |
| 15/13 | 3-4/1-2 | ND | ND |
| (E)(Y) WS | |||
| 27/34, 667 | 3/2 | 1-2/1 | ND |
| 34/27 | 3/2 | ND | ND |
| 26/32, ∼5,334 | 4/2 | 1/1 | 2-3/2 |
| 32/26 | 3-4/2 | ND | 3/2 |
| 27/32, ∼7,755 | 3-4/1-2 | 1/1 | 2/1 |
| 32/27 | 3-4/1-2 | ND | 2/1 |
| 27/26, 13,010 | 2-3/1 | 1-2/1 | 1-2/1 |
| 26/27 | 2-3/1 | 2/1 | 1-2/1 |
| 26/34, 13,677 | 3/1 | 1/1 | 1/1 |
Bold data indicate complementation between y alleles. ND, not determined.
FIG. 2.
trans activation between transposon derivatives of nonhomologous loci. Positions of the (E)(Y)WS and (E)(Y)SW insertions on the 2L and 3R chromosomes and distances between neighbors are shown. Arcs join the insertions whose derivatives can complement each other. Arrowheads at the ends of arcs indicate the direction of trans activation from enhancer to promoter.
To confirm the role of the gypsy insulator in trans activation between nonhomologous enhancerless and promoterless derivatives, we tested trans-activated allele combinations on the su(Hw)− or mod(mdg4)u1 background (Table 2). Both mutants had a strongly reduced level of trans activation. Surprisingly, the mod(mdg4)u1 mutation affected transvection between nonhomologous insertions much more strongly than transvection between homologous insertions (Tables 1 and 2); apparently, the long-range trans activation is more sensitive to the gypsy insulator's components. On the other hand, its effect on both homologous and nonhomologous trans activation was much less severe than that of the su(Hw)− mutation (Tables 1 and 2). This agrees with the extent of instability of insulator bodies on the su(Hw)− and mod(mdg4)u1 backgrounds: they are completely destroyed in the former case but only partially affected in the latter case (20).
Thus, the gypsy insulator supports transvection between nonhomologous loci, and the efficiency of trans activation mainly depends on the relative arrangement of the loci in the nuclear architecture rather than on the linear distances between them on the chromosomes.
Efficient pairing-dependent yellow activation requires the presence of the gypsy insulator in both homologous chromosomes.
The gypsy insulator contributes to various functions in the Drosophila nuclei, as follows from its interactions with GAGA factor or Mcp elements (45) and stimulation of the transcription of some genes (28, 64); moreover, there are multiple sites of the Su(Hw) protein localization on polytene chromosomes (20). Hence, the effect on trans activation caused by the su(Hw) mutation (see above) might be indirect, reflecting general changes in chromatin. To confirm that the Su(Hw) protein plays a role in transvection stabilization by participating in the direct interaction between the insulators located on different chromosomes, we created a construct yielding derivatives with or without the gypsy insulators.
In the (ES)Y(SW) transposon (Fig. 3), the yellow gene was flanked by the gypsy insulators inserted at −893 bp relative to the yellow transcription start and on the 3′ side of the yellow. The DNA fragment including the gypsy insulator at −893 bp and the yellow enhancers was flanked by FRT sites. The LOX sites flanked the DNA fragment containing the gypsy insulator located on the 3′ side of the yellow and the white promoter.
FIG. 3.
Complementation tests for (ES)Y(SW) construct and derivatives. Pairs of derivatives and pigmentation scores for seven different transgenic insertions are shown. Bold, italic numbers indicate the level of complementation between y alleles. For other designations, see Fig. 1.
We obtained seven (ES)Y(SW) transgenic lines with the gypsy insulator blocking the cis interaction between the yellow enhancers and the promoter, which accounted for the yellow pigmentation of the body cuticle and wing blades (Fig. 3). As the deletion of the yellow enhancers and the upstream gypsy insulator did not change the phenotype of flies, (ΔES)Y(SW) derivatives were selected by PCR analysis of individual flies obtained in the progeny of the cross with the flies carrying a Flp recombinase transgene (data not shown). The flies with the deleted 3′ gypsy insulator and the white promoter, (ES)Y(ΔSW) or with deletions of the two DNA fragments, (ΔES)Y(ΔSW) were selected by their white eyes and pigmented bristles. Pigmentation of the wings and body cuticle in heterozygous (ES)Y(SW)/+ flies and their derivatives was weak (approximately score 1) due to the blocking of yellow enhancers in the (ES)Y(SW) and (ES)Y(ΔSW) transgenes and their deletion in the (ΔES)Y(SW) and (ΔES)Y(ΔSW) transgenes.
In all seven transgenic lines, transheterozygous (ΔES)Y(SW)/(ES)Y(SW) and (ΔES)Y(SW)/(ES)Y(ΔSW) females displayed activation of the yellow gene (Fig. 3). As in (ES)Y(SW) and (ES)Y(ΔSW) alleles, the gypsy insulator blocked the yellow enhancer action on the yellow promoter in cis; we suggested that the yellow enhancers trans-activated the yellow promoter of the (ΔES)Y(SW) construct. As the gypsy insulators may interact in cis (51), it may appear that the level of transvection may be affected by cis interactions if one homolog carries two gypsy copies. In fact, this is not the case: transheterozygous (ΔES)Y(SW)/(ES)Y(SW) and (ΔES)Y(SW)/(ES)Y(ΔSW) females displayed the same level of yellow activation.
If only one of the paired y alleles contained one or two copies of the gypsy insulator, as in (ΔES)Y(ΔSW)/(ES)Y(SW) or (ΔES)Y(ΔSW)/(ES)Y(ΔSW) transheterozygotes, the wing and body pigmentations were lower than in the transheterozygotes with gypsy insulators in both y alleles (Fig. 3). This is evidence that gypsy insulators in both paired homologs are necessary for efficient transvection.
The gypsy insulator facilitates trans activation of the yellow promoter.
According to the results of studies on transvection between (ES)Y(SW) derivatives, the gypsy insulator inserted between the yellow enhancers and the promoter allowed the enhancers to activate the yellow promoter located on the homologous chromosome. This observation contradicts the previous data that the yellow enhancers isolated by a gypsy insertion in the y2 or y69 alleles failed in trans activation of the yellow promoter in the enhancerless y82f29 allele (47). The conflicting results might be explained by earlier finding that the pairing between homologous sequences sometimes promoted the gypsy insulator bypass in cis (48). Therefore, such kind of pairing between the (ES)Y(SW) derivatives may allow the yellow enhancers to bypass the gypsy insulator and act on the promoter in cis.
To verify this assumption, the (ES)(Y)SW and (ES)(Y)W transposons were constructed (Fig. 4). As in the previous (ES)Y(SW) transposon, the gypsy insulator at −893 bp and the yellow enhancers were flanked by the FRT sites. To inactivate the yellow function, the second exon of the yellow gene was flanked by LOX sites to promote its deletion from transgenic flies. To compare transvection in the presence or absence of the gypsy insulators in both homologs, the second gypsy insulator was inserted between the yellow and white genes in the (ES)(Y)SW construct [but not in the (ES)(Y)W construct].
FIG. 4.
Complementation tests for (ES)(Y)SW and (ES)(Y)W constructs and derivatives. For designations, see the legends to Fig. 1 and 3.
We obtained seven (ES)(Y)SW and five (ES)(Y)W transgenic lines. The body and wings were yellow due to the block of yellow enhancers by the gypsy insulator. Derivatives with deletions in the regulatory region were selected by PCR analysis as before. Derivatives with deletions in the coding region were selected by the yellow null phenotype. All genomic sites with the two series of transgenic lines had identical transvection levels in transheterozygotes carrying one y allele with the enhancer region deleted [(ΔES)(Y)SW or (ΔES)(Y)W] and the other allele with the functional yellow gene [(ES)(Y)SW or (ES)(Y)W], as well as in transheterozygotes carrying the same y allele with the enhancer region deleted together with the y allele containing a nonfunctional yellow gene [(ES)(YΔEX)SW or (ES)(YΔEX)W] (Fig. 4). This result provides evidence against the insulator bypass and suggests that the Su(Hw) insulator does not prevent the yellow enhancers from trans-activating the yellow promoter on the homologous chromosome. As a whole, the trans activation levels were higher in the allelic combinations derived from the (ES)(Y)SW transgenic lines than in those derived from the (ES)(Y)W lines (Fig. 4), thus confirming the role of pairing between the gypsy insulators located in homologous chromosomes in providing for efficient trans interaction between the yellow enhancers and the promoter.
Thus, we obtained objective evidence for trans activation in the cases of transposon insertions at many ectopic sites. In this context, previous data that the yellow enhancers isolated by a gypsy insertion into the y2 or y69 alleles failed in trans activation of the yellow promoter in the enhancerless y82f29 allele may be explained by some specific traits of the endogenous yellow locus. In particular, this locus contains the recently identified 1A-2 insulator, which is similar but not identical to the gypsy insulator (54). In addition, the y2 allele contains the whole gypsy retrotransposon, which makes the relative arrangement of regulatory elements in the y2/y82f29 allele combination different from that in our transgene derivatives.
DISCUSSION
We have demonstrated that the level of trans activation by the wing and body enhancers strongly depends on the site of insertion and does not correlate with the level of the yellow promoter activation in cis. Thus, the genomic regions do not provide identical yellow activation in trans that might be explained by their pairing strength between the homologs. Hence, there might be some specific elements facilitating the pairing between homologous chromosomes. The gypsy insulator inserted either 5 kb or 9 kb downstream from the yellow promoter improves its trans activation by the enhancers located on the homologous chromosome. The interaction between the gypsy insulators can improve the local pairing between homologous chromosomes. Recent cytological data (8) indicate that the gypsy insulators create chromatin loop domains by associating with the nuclear matrix. Two homologous chromosomes form only one loop (8), suggesting that the proteins present in the gypsy insulator and the nuclear matrix could maintain homologous chromosome pairing during the interphase.
It is noteworthy that the reported yellow sequences significant for efficient transvection between enhancerless and promoterless y alleles (10) include the 1A-2 insulator located on the 3′ side of the yellow gene (27, 55). The insulator containing two binding sites for the Su(Hw) protein was not present in our constructs. Thus, reliable and efficient trans activation observed previously (10) upon the transgene insertion at all seven genomic sites can be explained by the presence of the endogenous Su(Hw) insulator improving local homologous pairing. As the Su(Hw) protein binds to approximately 200 sites in the Drosophila genome (20), the Su(Hw) binding sites appear to play a role in the pairing between homologous chromosomes. Recent studies of the scs and scs′ insulators confirm that they interact with each other (7) and, therefore, may also be involved in this process.
Although the examples of transvection are many, the nature of chromosome pairing during the interphase is still obscure. Today, only the Zeste protein is known to be involved in some transvection effects. The zeste gene encodes the sequence- specific DNA-binding protein with the binding sites distributed throughout genome (3, 4, 58). Inactivation of Zeste disrupts allelic pairing, thus enhancing the heteroallelic mutant phenotype (33, 39). Zeste also supports transvection-like effects in the decapentaplegic (15, 16), white (1, 16), eyes absent (38), and Ubx genes (26).
Polycomb response elements (PREs) are another class of regulatory elements that may facilitate pairing between homologous chromosomes. PREs are short DNA segments initiating the assembly of silencing complexes composed of the Polycomb group (PcG) proteins (56). The silencing of a PRE-containing transposon construct is often dramatically enhanced in flies homozygous for the transposon insertion (61). The interaction between two copies of PREs on the homologous chromosomes is supposed to improve the stability and silencing power of the PcG complex. At the same time, the interaction between the PcG complexes may support homologous chromosome pairing. The combination of the binding sites for the proteins like Su(Hw), Zeste, and PcG may generate a unique code for making this process more efficient.
The Zeste, Su(Hw), and PcG proteins, along with having the ability to strengthen homologous effects, are involved in the formation of higher-order nuclear structures. Thus, Zeste can form high-order aggregates (5), which suggests that it can hold together certain DNA regions. Likewise, PcG proteins are organized into discrete nuclear bodies that may be the sites of the PRE-mediated silencing (59). The well-defined Fab-7 cellular memory module also leads to association of transgenes even when inserted into different chromosomes (2). The same was shown for the Mcp element, which contains an insulator and PRE (29, 50). Such long-distance interactions depend on the PcG proteins, at least partially (2, 50). Here, we have shown that the gypsy insulator provides for trans activation between selected genomic loci at distances exceeding 13 Mb. Together with previous data on the punctuated distribution of gypsy insulator proteins in the nucleus (20), the results of this study suggest their involvement in the arrangement of the chromatin fiber within the nucleus and in organization of communication between distant loci in the Drosophila genome.
Thus, the interaction between the gypsy insulators facilitates trans activation of the yellow promoter, which is evidence for the involvement of gypsy insulators in the regulation of homologous chromosome pairing and communication between distant loci. Further investigations are required to find out whether the transvection stabilization by gypsy insulator in homologous and distant locations relies on the same mechanism. The model system utilizing the effects of transvection between yellow transgenes provides a powerful tool for the analysis or identification of the proteins supporting interactions between loci.
Acknowledgments
We thank S. Demakov for his help in interpreting the results of FISH.
This work was supported by the Molecular and Cellular Biology Program of the Russian Academy of Sciences; the Russian Foundation for Basic Research (project no. 05-04-48911-a); the Volkswagen- Stiftung Foundation, Federal Republic of Germany; an International Research Scholar award from the Howard Hughes Medical Institute (to P.G.); a stipend from the Center for Medical Studies, University of Oslo (to E.S. and M.S.); and a fellowship from the Russian Science Support Foundation (to E.S.).
REFERENCES
- 1.Babu, P., and S. Bhat. 1980. Effect of zeste on white complementation. Basic Life Sci. 16:35-40. [DOI] [PubMed] [Google Scholar]
- 2.Bantignies, F., C. Grimaud, S. Lavrov, M. Gabut, and G. Cavalli. 2003. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 17:2406-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, M., and V. Pirrotta. 1987. The product of the Drosophila zeste gene binds to specific DNA sequences in white and Ubx. EMBO J. 6:1387-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, M., and V. Pirrotta. 1988. The Drosophila zeste protein binds cooperatively to sites in many gene regulatory regions: implications for transvection and gene regulation. EMBO J. 7:3907-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bickel, S., and V. Pirrotta. 1990. Self-association of the Drosophila zeste protein is responsible for transvection effects. EMBO J. 9:2959-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwood, E. M., and J. T. Kadonaga. 1998. Going the distance: a current view of enhancer action. Science 281:61-63. [DOI] [PubMed] [Google Scholar]
- 7.Blanton, J., M. Gaszner, and P. Schedl. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17:664-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd, K., and V. G. Corces. 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, H. N., and P. Shen. 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291:493-495. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J. L., K. L. Huisinga, M. M. Viering, S. A. Ou, C. T. Wu, and P. K. Geyer. 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc. Natl. Acad. Sci. USA 99:3723-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorsett, D. 1999. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9:505-514. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, I. W. 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36:521-556. [DOI] [PubMed] [Google Scholar]
- 13.Fung, J. C., W. F. Marshall, A. Dernburg, D. A. Agard, and J. W. Sedat. 1998. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J. Cell Biol. 141:5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gause, M., P. Morcillo, and D. Dorsett. 2001. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol. Cell. Biol. 21:4807-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelbart, W. M. 1982. Synapsis-dependent allelic complementation at the decapentaplegic gene complex in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 79:2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelbart, W. M., and C. T. Wu. 1982. Interactions of zeste mutations with loci exhibiting transvection effects in Drosophila melanogaster. Genetics 102: 179-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgiev, P., and M. Kozycina. 1996. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 142:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgiev, P. G., and T. I. Gerasimova. 1989. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet. 220:121-126. [DOI] [PubMed] [Google Scholar]
- 19.Gerasimova, T. I., K. Byrd, and V. G. Corces. 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6:1025-1035. [DOI] [PubMed] [Google Scholar]
- 20.Gerasimova, T. I., and V. G. Corces. 1998. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92:511-521. [DOI] [PubMed] [Google Scholar]
- 21.Gerasimova, T. I., and V. G. Corces. 2001. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 35:193-208. [DOI] [PubMed] [Google Scholar]
- 22.Gerasimova, T. I., D. A. Gdula, D. V. Gerasimov, O. Simonova, and V. G. Corces. 1995. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82:587-597. [DOI] [PubMed] [Google Scholar]
- 23.Geyer, P. K., and V. G. Corces. 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1:996-1004. [DOI] [PubMed] [Google Scholar]
- 24.Geyer, P. K., M. M. Green, and V. G. Corces. 1990. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J. 9:2247-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh, D., T. I. Gerasimova, and V. G. Corces. 2001. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 20:2518-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldsborough, A. S., and T. B. Kornberg. 1996. Reduction of transcription by homologue asynapsis in Drosophila imaginal discs. Nature 381:807-810. [DOI] [PubMed] [Google Scholar]
- 27.Golovnin, A., I. Birukova, O. Romanova, M. Silicheva, A. Parshikov, E. Savitskaya, V. Pirrotta, and P. Georgiev. 2003. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development 130:3249-3258. [DOI] [PubMed] [Google Scholar]
- 28.Golovnin, A., E. Melnick, A. Mazur, and P. Georgiev. 2005. Drosophila Su(Hw) insulator can stimulate transcription of a weakened yellow promoter over a distance. Genetics 170:1133-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruzdeva, N., O. Kyrchanova, A. Parshikov, A. Kullyev, and P. Georgiev. 2005. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol. Cell. Biol. 25:3682-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henikoff, S., and L. Comai. 1998. trans-sensing effects: the ups and downs of being together. Cell 93:329-332. [DOI] [PubMed] [Google Scholar]
- 31.Hiraoka, Y., A. F. Dernburg, S. J. Parmelee, M. C. Rykowski, D. A. Agard, and J. W. Sedat. 1993. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J. Cell Biol. 120:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karess, R. E., and G. M. Rubin. 1984. Analysis of P transposable element functions in Drosophila. Cell 38:135-146. [DOI] [PubMed] [Google Scholar]
- 33.Kaufman, T. C., S. E. Tasaka, and D. T. Suzuki. 1973. The interaction of two complex loci, zeste and bithorax in Drosophila melanogaster. Genetics 75: 299-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennison, J. A., and J. W. Southworth. 2002. Transvection in Drosophila. Adv. Genet. 46:399-420. [DOI] [PubMed] [Google Scholar]
- 35.Kuhn, E. J., and P. K. Geyer. 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15:259-265. [DOI] [PubMed] [Google Scholar]
- 36.Kuhn, E. J., M. M. Viering, K. M. Rhodes, and P. K. Geyer. 2003. A test of insulator interactions in Drosophila. EMBO J. 22:2463-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavrov, S., J. Dejardin, and G. Cavalli. 2004. Combined immunostaining and FISH analysis of polytene chromosomes. Methods Mol. Biol. 247:289-303. [DOI] [PubMed] [Google Scholar]
- 38.Leiserson, W. M., N. M. Bonini, and S. Benzer. 1994. Transvection at the eyes absent gene of Drosophila. Genetics 138:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis, E. B. 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88: 225-239. [Google Scholar]
- 40.Lifschytz, E., and D. Hareven. 1982. Heterochromatin markers: arrangement of obligatory heterochromatin, histone genes and multisite gene families in the interphase nucleus of D. melanogaster. Chromosoma 86:443-455. [DOI] [PubMed] [Google Scholar]
- 41.Lindsley, D. L., and G. G. Zimm. 1992. The genome of Drosophila melanogaster. Academic Press, New York, N.Y.
- 42.Marlor, R. L., S. M. Parkhurst, and V. G. Corces. 1986. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol. Cell. Biol. 6:1129-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin, M., Y. B. Meng, and W. Chia. 1989. Regulatory elements involved in the tissue-specific expression of the yellow gene of Drosophila. Mol. Gen. Genet. 218:118-126. [DOI] [PubMed] [Google Scholar]
- 44.Mazo, A. M., L. J. Mizrokhi, A. A. Karavanov, Y. A. Sedkov, A. A. Krichevskaja, and Y. V. Ilyin. 1989. Suppression in Drosophila: su(Hw) and su(f) gene products interact with a region of gypsy (mdg4) regulating its transcriptional activity. EMBO J. 8:903-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melnikova, L., F. Juge, N. Gruzdeva, A. Mazur, G. Cavalli, and P. Georgiev. 2004. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc. Natl. Acad. Sci. USA 101: 14806-14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modolell, J., W. Bender, and M. Meselson. 1983. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc. Natl. Acad. Sci. USA 80:1678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris, J. R., J. Chen, S. T. Filandrinos, R. C. Dunn, R. Fisk, P. K. Geyer, and C. Wu. 1999. An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics 151:633-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris, J. R., J. L. Chen, P. K. Geyer, and C. T. Wu. 1998. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc. Natl. Acad. Sci. USA 95:10740-10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris, J. R., P. K. Geyer, and C. T. Wu. 1999. Core promoter elements can regulate transcription on a separate chromosome in trans. Genes Dev. 13:253-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muller, M., K. Hagstrom, H. Gyurkovics, V. Pirrotta, and P. Schedl. 1999. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153:1333-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muravyova, E., A. Golovnin, E. Gracheva, A. Parshikov, T. Belenkaya, V. Pirrotta, and P. Georgiev. 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291:495-498. [DOI] [PubMed] [Google Scholar]
- 52.Nash, W. G., and R. J. Yarkin. 1974. Genetic regulation and pattern formation: a study of the yellow locus in Drosophila melanogaster. Genet. Res. 24:19-26. [DOI] [PubMed] [Google Scholar]
- 53.Oki, M., and R. T. Kamakaka. 2002. Blockers and barriers to transcription: competing activities? Curr. Opin. Cell Biol. 14:299-304. [DOI] [PubMed] [Google Scholar]
- 54.Pai, C. Y., E. P. Lei, D. Ghosh, and V. G. Corces. 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16:737-748. [DOI] [PubMed] [Google Scholar]
- 55.Parnell, T. J., M. M. Viering, A. Skjesol, C. Helou, E. J. Kuhn, and P. K. Geyer. 2003. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100: 13436-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pirrotta, V. 1998. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell 93:333-336. [DOI] [PubMed] [Google Scholar]
- 57.Pirrotta, V. 1999. Transvection and chromosomal trans-interaction effects. Biochim. Biophys. Acta 1424:M1-M8. [DOI] [PubMed] [Google Scholar]
- 58.Pirrotta, V., E. Manet, E. Hardon, S. E. Bickel, and M. Benson. 1987. Structure and sequence of the Drosophila zeste gene. EMBO J. 6:791-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saurin, A. J., C. Shiels, J. Williamson, D. P. Satijn, A. P. Otte, D. Sheer, and P. S. Freemont. 1998. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142:887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Savitsky, M., T. Kahn, E. Pomerantseva, and P. Georgiev. 2003. Transvection at the end of the truncated chromosome in Drosophila melanogaster. Genetics 163:1375-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sigrist, C. J., and V. Pirrotta. 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147:209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spana, C., D. A. Harrison, and V. G. Corces. 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2:1414-1423. [DOI] [PubMed] [Google Scholar]
- 63.Udvardy, A. 1999. Dividing the empire: boundary chromatin elements delimit the territory of enhancers. EMBO J. 18:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei, W., and M. D. Brennan. 2001. The gypsy insulator can act as a promoter-specific transcriptional stimulator. Mol. Cell. Biol. 21:7714-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 66.Wu, C. T., and J. R. Morris. 1999. Transvection and other homology effects. Curr. Opin. Genet. Dev. 9:237-246. [DOI] [PubMed] [Google Scholar]