Abstract
HEMA encodes glutamyl-tRNA reductase (GluTR), which catalyzes the first step specific for tetrapyrrole biosynthesis in plants, archaea, and most eubacteria. In higher plants, GluTR is feedback inhibited by heme and intermediates of chlorophyll biosynthesis. It plays a key role in controlling flux through the tetrapyrrole biosynthetic pathway. This enzyme, which in Chlamydomonas reinhardtii is encoded by a single gene (HEMA), exhibits homology to GluTRs of higher plants and cyanobacteria. HEMA mRNA accumulation was inducible not only by light but also by treatment of dark-adapted cells with Mg-protoporphyrin IX (MgProto) or hemin. The specificity of these tetrapyrroles as inducers was demonstrated by the absence of induction observed upon the feeding of protoporphyrin IX, the precursor of both heme and MgProto, or chlorophyllide. The HEMA mRNA accumulation following treatment of cells with light and hemin was accompanied by increased amounts of GluTR. However, the feeding of MgProto did not suggest a role for Mg-tetrapyrroles in posttranscriptional regulation. The induction by light but not that by the tetrapyrroles was prevented by inhibition of cytoplasmic protein synthesis. Since MgProto is synthesized exclusively in plastids and heme is synthesized in plastids and mitochondria, the data suggest a role of these compounds as organellar signals that control expression of the nuclear HEMA gene.
Tetrapyrroles, such as hemes and chlorophylls, are essential for the metabolism of all photosynthetic organisms. The first common precursor molecule of all tetrapyrroles is 5-aminolevulinic acid (ALA) which in plants, archaea, and most bacteria is synthesized from the C5 skeleton of glutamate (23). The C5 pathway consists of three enzymatic steps: the activation of glutamate to glutamyl-tRNA by glutamyl-tRNA synthetase, the reduction of glutamyl-tRNA to glutamate-1-semialdehyde by glutamyl-tRNA reductase (GluTR), and its transamination to ALA by glutamate-1-semialdehyde-2,1-aminotransferase (2). In plants, the early steps of tetrapyrrole biosynthesis up to protoporphyrinogen IX take place only in chloroplasts. A fraction of the protoporphyrinogen IX then, by unknown means, is transported into mitochondria (reviewed in reference 41). Protoporphyrinogen in chloroplasts and mitochondria is oxidized to protoporphyrin IX (Proto). The insertion of Fe2+ into Proto leads to the formation of heme; Mg2+ insertion into Proto gives rise to Mg-protoporphyrin IX (MgProto), the precursor of chlorophyll.
The formation of ALA is the rate-limiting step in tetrapyrrole biosynthesis (3). As a consequence, ALA synthesis is tightly regulated, and, because of its strategic position as the first committed step in tetrapyrrole biosynthesis, GluTR is viewed as the principal regulatory target (11). One end product of tetrapyrrole biosynthesis, heme, was assigned a prominent role in the regulation of ALA formation, since it was shown to inhibit ALA synthesis in vitro and in vivo (9, 17, 59). This inhibitory effect appears to be mediated through GluTR because exogenous heme can inhibit recombinant GluTR (42), acting on the N-terminal 30 amino acids of the enzyme (57).
A feedback control system mediated by the Mg-tetrapyrrole-synthesizing branch was uncovered by the analysis of Arabidopsis mutants defective in a gene named Flu that exhibited an upregulation of ALA synthesis and an overaccumulation of protochlorophyllide but not of heme (35). This nuclear-encoded regulator is tightly bound to plastid membranes and physically interacts with GluTR (35).
In higher plants, GluTR is encoded by a small family of genes that, in the cases studied, comprise two to three members (7, 22, 27, 28, 45, 50, 51). In most cases, these genes show strongly divergent patterns of expression. Thus, HEMA1 of Arabidopsis is highly expressed throughout seedling development and is light regulated at the transcriptional level through members of the red-far-red light (phytochrome) and blue light (cryptochrome) photoreceptor families (32, 33). In addition, HEMA1 is regulated by the endogenous clock (39) and requires a plastid signal (32). In contrast, HEMA2 of Arabidopsis is predominantly expressed in roots of seedlings and in flowers. It is not regulated by light or by plastid signaling pathways (27, 32, 53). This has led to the hypothesis that while both HEMA genes contribute to a basal rate of ALA synthesis to meet the normal cellular demands for tetrapyrroles, the highly regulated expression of HEMA1 appears to be tailored to meet an increased demand of the chloroplasts for chlorophyll (32).
In Chlamydomonas reinhardtii two chlorophyll precursors, MgProto and its methylester, have been shown to act as plastid factors in mediating the light induction of two nuclear-encoded HSP70 genes (24). Feeding of these two compounds to cell cultures in the dark activated transcription of the HSP70 genes. A mutant blocked in the H subunit of Mg-chelatase (brs-1) and thus unable to synthesize MgProto (10) showed no light induction of the HSP70 genes. However, feeding of MgProto to this mutant resulted in induction (24, 25).
When the expression of nuclear C. reinhardtii genes that were considered as candidates for regulation by tetrapyrroles was analyzed, only 4 genes out of 20 genes studied exhibited a response to MgProto feeding: in addition to the two HSP70 genes analyzed before, HSP70E, encoding a cytosolic chaperone (46), and HEMA were found to be induced by MgProto (50), supporting the concept that tetrapyrroles constitute one type of plastidic signal used for the control of nuclear genes (4, 5, 18, 48).
These results pointed to a role of tetrapyrroles in the regulation of the first gene specific for tetrapyrrole biosynthesis. Here we show that, in addition to MgProto, heme also appears to play a role as an organellar factor involved in the regulation of HEMA expression. While both heme and MgProto induce the accumulation of HEMA mRNA, enhanced levels of GluTR are seen only after heme treatment, suggesting posttranscriptional regulation of HEMA expression by Mg-tetrapyrroles.
MATERIALS AND METHODS
Strains and culture conditions.
C. reinhardtii wild-type strain CC-124 (mt−) (obtained from the Chlamydomonas Culture Collection at Duke University), used throughout, was grown photomixotrophically in Tris-acetate-phosphate medium (19) on a rotatory shaker either in the dark or under continuous irradiation with white light (50 μE m−2 · s−1) at 23°C.
DNA extraction and Southern blot analyses.
Total genomic DNA from Chlamydomonas was isolated as described previously (55). About 10 μg of DNA was digested with various restriction enzymes, separated by electrophoresis on 0.7% agarose gels, and transferred to Hybond-N membranes (Amersham, Freiburg, Germany). Hybridizations were performed for 20 h at 65°C in hybridization buffer I (50 mM Tris-HCl, pH 7.5, 1× SSC [0.15 M NaCl plus 0.015 M sodium citrate], 0.1% Na4P2O7, 1% sodium dodecyl sulfate [SDS], 10× Denhardt's solution, 10% dextran sulfate, 0.4% denatured herring sperm DNA) containing 60% formamide (vol/vol) or in hybridization buffer II (5× Denhardt's solution, 5× SSC, 0.5% SDS, 0.4% denatured herring sperm DNA) without formamide. After a first washing step with 1% SDS and 2× SSC at 65°C for 30 min, autoradiography was performed to estimate signal intensity and the remaining background. Subsequently, a second, more stringent wash step with 1% SDS and 0.1× SSC at 65°C was performed for 30 min, followed by autoradiography. A 560-bp fragment, corresponding to the coding region of the HEMA gene, was labeled with [α-32P]dCTP (15) and used as a probe.
RNA isolation and RNA blot analyses.
RNA isolation was performed as described previously (55). Ten micrograms of total RNA per lane was separated on formaldehyde-containing agarose gels and transferred to nylon membranes (Hybond-N+; Amersham), using 25 mM sodium phosphate buffer (pH 6.5). Prehybridizations and hybridizations were performed at 65°C in 0.1 M NaCl, 50 mM Tris-HCl (pH 7.5), 0.1% sodium pyrophosphate, 10× Denhardt's solution, 1% SDS, 10% dextran sulfate, 60% formamide, and 100 μg/ml of denatured herring sperm DNA. Radioactive DNA probes were prepared by the random priming technique (15), using [α-32P]dCTP (Amersham). The HEMA-specific probe was a 560-bp fragment corresponding to the coding region of HEMA. For a loading control, a 1-kb cDNA fragment of CBLP was employed (56). RNA blots were quantitated using a PhosphorImager (Bio-Rad Laboratories GmbH, Munich, Germany). Each experiment was repeated at least three times with RNA isolated from different cultures.
Protein extraction and immunoblot analyses.
For protein isolation, 10-ml cell suspensions were harvested by centrifugation and resuspended in 300 μl 0.1 M Na2CO3, 0.1 M dithiothreitol, 200 μl 5% SDS, and 30% sucrose. Cells were broken by ultrasonic treatment on ice. Protein concentrations were determined with amido black (43). After separation of whole-cell proteins by 9% SDS-polyacrylamide gel electrophoresis as described by Laemmli (29), they were transferred to polyvinylidene difluoride membranes (Hybond-P; Amersham). For transfer, a discontinuous buffer system was used. Two Whatman sheets were wetted with buffer T1 (20% methanol, 25 mM Tris-HCl, pH 10.4, 40 mM 6-aminocaproic acid) and placed onto the cathode. The gel was incubated briefly in buffer T2 (20% methanol, 25 mM Tris-HCl, pH 10.4) and placed on the Whatman sheets. The membrane was also incubated in buffer T2 and placed on the gel. Three Whatman sheets, wetted with buffer T3 (20% methanol, 300 mM Tris-HCl, pH 10.4), were placed on top of the membrane. Transfer was performed at 0.8 mA per cm2 for 1 h. Blots were developed by the enhanced chemiluminescence technique according to the manufacturer's protocol (Amersham), using anti-rabbit immunoglobulin G linked to horseradish peroxidase as the secondary antibody. Antisera for specific detection of C. reinhardtii glutamyl-tRNA reductase were kindly provided by S. Beale (Providence, Rhode Island). The level of the α subunit of plastidic ATP synthase was used as a loading control, employing an antibody kindly provided by O. Vallon (Paris, France). Signal intensities were quantified using the Quantity One quantification software (Bio-Rad).
Sources of porphyrins.
MgProto was prepared as described previously (24). For the synthesis of chlorophyllide (Chlide) and ZnChlide, we followed the protocol of Helfrich and Rüdiger (20). Its purity was checked by high-pressure liquid chromatography. Proto, ZnProto, and hemin were purchased from Sigma-Aldrich (Taufkirchen, Germany). The concentrations of MgProto and Proto were determined in ether by spectrophotometric measurement using extinction coefficients of 308 at 419 nm and 158 at 404 nm, respectively (14). The concentration of hemin was determined in a solvent consisting of 66.5% ethanol, 17% acetic acid, and 16.5% water (vol/vol/vol) using an extinction coefficient of 144 at 398 nm (52). Porphyrins were dissolved in dimethyl sulfoxide and used at the final concentrations indicated in Results. For feeding experiments, the compounds were added to cultures that had been preincubated in the dark for 20 h.
RESULTS
A conserved glutamyl-tRNA reductase in C. reinhardtii is encoded by a single-copy gene (HEMA).
Screening of the draft genome sequence for GluTR-encoding genes in C. reinhardtii revealed a single HEMA gene (gene model C_420027). This gene model is based on a HEMA cDNA sequence determined by R. D. Willows et al. (GenBank accession no. AF305613). The predicted gene exhibits a coding region of 1,449 bp, interrupted by a single intron of 116 bp. It is located on scaffold 42 of the second version of the Chlamydomonas genome sequence database (http://genome.jgi-psf.org/chlamy/).
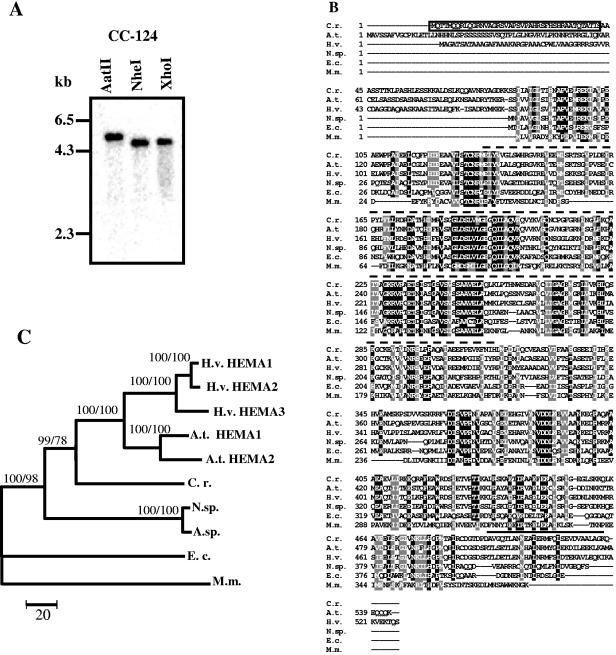
For all plants investigated, more than one HEMA gene has been observed. As a general rule, individual genes exhibit different expression patterns, suggesting specific roles for the isoenzymes (32, 50). In view of the still incomplete genome sequence, we employed Southern blot analyses, using as a probe a DNA fragment that corresponds to a region encoding the most highly conserved part of GluTR (Fig. 1B), to test whether Chlamydomonas may harbor more than one HEMA gene. Hybridization with this probe revealed a single fragment in genomic DNA cleaved with three different restriction enzymes (Fig. 1A). Also, at a reduced stringency no additional specific hybridization signals could be detected (data not shown), suggesting that C. reinhardtii indeed harbors only a single HEMA gene.
FIG. 1.
Test for the number of HEMA genes in the C. reinhardtii genome and relatedness of the C. reinhardtii HEMA product to those of other organisms. (A) Southern blot of C. reinhardtii genomic DNA. Genomic DNA (10 μg) from wild-type strain CC-124 was digested with either AatII, NheI, or XhoI. The sizes of marker fragments are shown on the left. (B) Multiple-sequence alignment of six HEMA proteins, generated by the ClustalW program. The deduced amino acid sequence of the C. reinhardtii HEMA (GenBank accession number AF305613) was compared with those of Arabidopsis thaliana (A.t. HEMA1; NP_176125), Hordeum vulgare (H.v. HEMA1; BAA25167), Nostoc sp. strain PCC7120 (N.sp; NP_485085), Escherichia coli K-12 (E.c; BAA36068), and Methanococcus maripaludis S2 (M.m; NP_987208). A black background indicates identical amino acids; residues highlighted with a gray background are similar. N/Q, D/E, R/K, S/T, F/Y, A/G, and V/I/L/M were considered similar. The transit peptide of C. reinhardtii HEMA, predicted by the ChloroP program (12), is boxed. The probe used for Northern blot and Southern blot analyses encodes the stretch of amino acids indicated by a dotted line above the sequences. A coiled-coil motif predicted for the C-terminal end is marked by a solid line above the sequences (6, 31). (C) The phylogenetic tree is based on the alignment of proteins described above and the following additional proteins: Arabidopsis thaliana HEMA2 (NP_172465), Hordeum vulgare HEMA2 (CAA60055) and HEMA3 (BAA25168), and Anabaena sp. (A.sp.; AAB58164). The phylogenetic tree was obtained by MEGA 2.1 distance-neighbor joining based on the number of amino acid substitutions. Bootstrap support is from consensus trees after 500 repetitions. HEMA sequences from E. coli and Methanococcus maripaludis were taken as outgroups. Distances are indicated by the scale bar. Numbers at nodes indicate bootstrap support values (first number from distance-neighbor joining, second from maximum parsimony).
This gene is predicted to encode a protein of about 57 kDa that at its N-terminal end harbors a chloroplast-targeting sequence of 42 amino acids (Fig. 1B). Multiple amino acid sequence alignments demonstrated high similarity of the C. reinhardtii GluTR to homologues from higher plants and cyanobacteria (Fig. 1B): there was 56% amino acid identity to the Arabidopsis thaliana HEMA1 product, 54% identity to the barley HEMA1 protein, and 46% identity to the Nostoc enzyme. A reduced degree of similarity to the bacterial GluTRs was detected: there was 30% identity to the Escherichia coli enzyme and 26% identity to Methanococcus GluTR. The N-terminal half of this protein is more highly conserved than its C terminus. The N-terminal part of GluTR, as deduced from the structure determined for the enzyme from Methanopyrus kandleri (37), harbors the catalytic domain. It is also necessary for inhibition of the enzyme by heme (57). At the very C terminus, a coiled-coil domain that may have a regulatory function is predicted by two different programs (6, 31) (Fig. 1B) (see Discussion).
The phylogenetic analysis of GluTRs from various organisms placed the C. reinhardtii GluTR within the green radiation between those from higher plants and cyanobacteria (Fig. 1C). Clearly, the two and three GluTRs observed for Arabidopsis and barley, respectively, are closely related to each other, suggesting that the additional HEMA genes in these species appeared after the divergence of monocotyledon and dicotyledon plants. The presence of a single HEMA gene in C. reinhardtii thus may reflect the ancestral situation in plants prior to this divergence. Clearly, though, the GluTRs of the higher plant species are more closely related to each other than to the C. reinhardtii protein.
We draw the conclusion that C. reinhardtii harbors a single HEMA gene that encodes a predicted plastid-localized GluTR with distinct homology to GluTRs of higher plants.
Induction of HEMA by light, MgProto, and hemin.
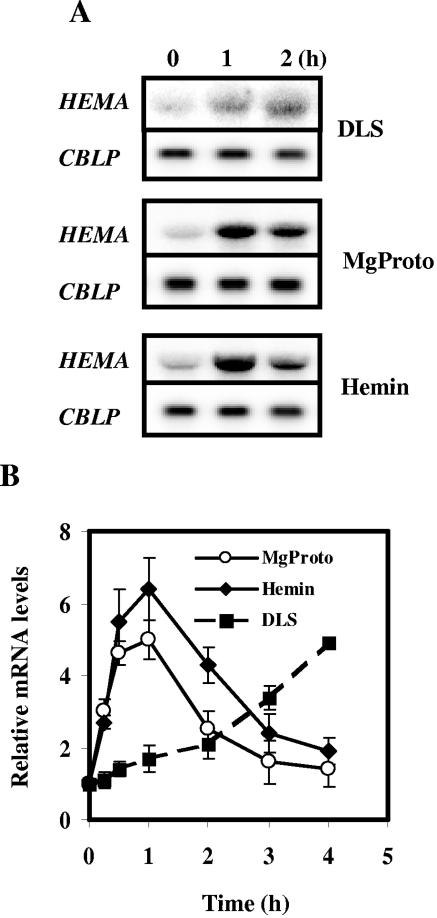
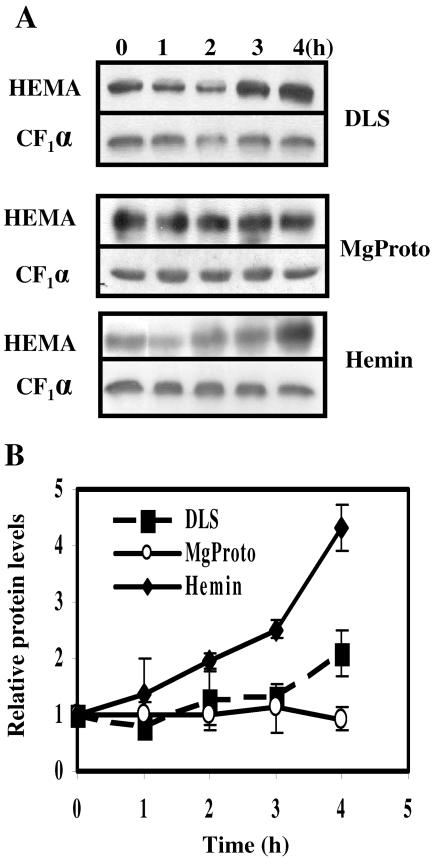
In organisms with two or more HEMA genes, one of the genes usually is expressed in a cell-type-specific manner while another is light inducible (33). The mRNA of the single HEMA gene of C. reinhardtii accumulated to a higher level when dark-adapted cell cultures were shifted into light (Fig. 2A). This accumulation proceeded at a low rate, since 2 h after light exposure, the mRNA level had only doubled (Fig. 2B). We have shown previously that Mg-porphyrins may regulate the expression of nuclear genes in C. reinhardtii (24, 54). Here we first analyzed the effect of a feeding of MgProto to cells kept in the dark. MgProto addition induced HEMA mRNA accumulation (Fig. 2A), and this increase was more rapid and more pronounced than that observed following light treatment (Fig. 2B). Thus, a peak in mRNA levels was observed 1 h after addition of the tetrapyrrole; light treatment for this time period elicited only a minor increase (Fig. 2B). A combined treatment of light and MgProto could not be tested due to the high toxicity of the tetrapyrrole in the light.
FIG. 2.
Induction of HEMA by light or by the exogenous addition of MgProto and hemin in the dark. (A) Prior to the treatment, cultures were incubated in the dark for 20 h. At time zero, cultures of strain CC-124 were either shifted from dark to light (fluence rate of white light, 50 μmol m−2 s−1) (DLS) or supplemented with MgProto (final concentration, 16 μM) or hemin (final concentration, 4 μM) in the dark. Samples were taken at the time points indicated. For RNA blot analyses, 10 μg of total RNA was hybridized with the HEMA probe described in the legend to Fig. 1. The constitutively expressed CBLP gene, encoding a Gβ-like protein (56), served as loading control. (B) Kinetics of HEMA transcript accumulation detected in total RNA isolated from cells at 0, 0.25, 0.5, 1, 2, 3, and 4 h after a dark-to-light shift (DLS) or from cells incubated in the dark after the addition of MgProto or hemin. Intensities of the HEMA hybridization signals from three independent experiments were quantified by phosphorimaging and corrected for unequal loading by the respective CBLP signal. Values are given in arbitrary units relative to the continuous dark level (zero time point), which was set to 1. Error bars indicate standard errors of the means.
The addition of hemin (which harbors an Fe3+ atom, conferring higher stability to the tetrapyrrole in solution than the Fe2+ containing heme) in the dark also activated HEMA expression (Fig. 2A). Similar to the case with MgProto treatment, HEMA mRNA accumulation was evident already 15 min after feeding (Fig. 2B). The kinetics of HEMA mRNA accumulation observed after the feeding of MgProto and hemin were similar. Both caused a transient five- to sixfold induction in which peaks were observed 1 hour after start of the treatment; about 3 h later, mRNA levels were close to the initial levels again.
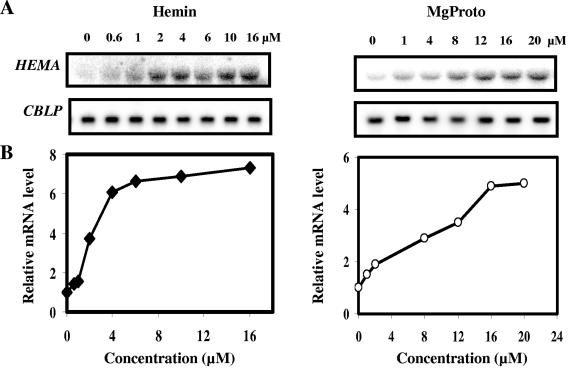
To further characterize the inducing activities of the two tetrapyrroles, different amounts of MgProto and hemin, ranging from 0.6 μM to 16 μM for hemin and from 1 μM to 20 μM for MgProto, were added to cells in the dark. The results revealed a correlation between the degree of HEMA mRNA accumulation and the concentration of the inducing tetrapyrrole (Fig. 3).
FIG. 3.
Dependence of HEMA mRNA accumulation on the concentration of MgProto or hemin. (A) Tetrapyrroles were added in the dark at the final concentrations indicated, and after 1 h of incubation, cells were harvested and RNA was isolated. RNA blot analyses were performed as described in the legend to Fig. 2. (B) Graphical presentation of the dependence of HEMA mRNA accumulation on the concentration of MgProto or hemin. HEMA hybridization signals were quantified by phosphorimaging and corrected for unequal loading by using CBLP as a control. The average from two independent experiments is presented in arbitrary units relative to the continuous dark level, which was set to 1.
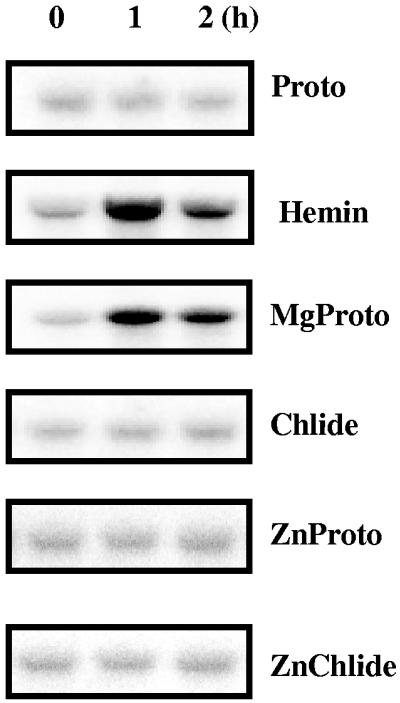
The concentrations of various tetrapyrroles in C. reinhardtii wild-type cells have previously been determined. They were shown to contain, per gram of dry weight, about 2 nmol of Proto (26), 0.5 nmol of MgProto (26), 0.5 nmol of the methylester of MgProto (26), and 27 nmol of protochlorophyllide (44). To test whether other tetrapyrroles besides heme and MgProto have the capacity for HEMA induction, Proto as well as Chlide was added to cells grown in the dark. These tetrapyrroles did not cause an upregulation of HEMA (Fig. 4). For Proto, an uptake by C. reinhardtii cells has previously been demonstrated (26). We assume but have no proof that Chlide, with physiochemical properties similar to those of MgProto, is also taken up by intact cells. The inducing ability conferred to Proto by insertion of the metal atom was investigated by using Proto containing a zinc atom. ZnProto did not elicit HEMA activation, nor did ZnChlide (Fig. 4).
FIG. 4.
Effects of different chlorophyll precursors and tetrapyrrole derivatives on HEMA mRNA abundance. After 20 h of incubation in the dark, cultures of CC-124 were supplemented with either Proto, MgProto, Chlide, ZnProto, or ZnChlide at a final concentration of 16 μM or with hemin at a concentration of 4 μM. Incubation was continued in the dark. Samples for RNA blot analysis were taken at the time points indicated and processed as described in the legend to Fig. 2.
We next assayed whether the observed accumulation of HEMA mRNA was followed by an increase in GluTR levels. Using antisera directed against C. reinhardtii GluTR, the protein levels were evaluated by immunoblot analysis and, using the same membrane, compared to the level in the α subunit of the plastidic ATPase CF1. With a delay of about 3 h, a small but significant increase in GluTR levels was seen when cultures were shifted from dark to light (Fig. 5). Compared to the time zero value, the average increase observed after 4 h was about twofold (Fig. 5B). Hemin feeding elicited a more pronounced increase in GluTR, which in three independent experiments after 4 h averaged about fourfold (Fig. 5).
FIG. 5.
Changes in levels of GluTR after the application of light or tetrapyrroles (MgProto and hemin). (A) Soluble protein was extracted from wild-type cells preincubated in the dark for at least 20 h and treated with either hemin (final concentration, 4 μM) or MgProto (final concentration, 16 μM) in the dark or transferred from dark to light (fluence rate, 50 μmol m−2 s−1) (DLS). In each lane 50 μg of protein was separated by SDS-polyacrylamide gel electrophoresis and, after transfer to a membrane, immunodecorated with antibodies directed against C. reinhardtii GluTR. Signals were detected and quantified as described in Materials and Methods. Differences in the intensities of time zero signals are caused by different exposure times of the films. (B) Changes in GluTR levels in soluble protein from cells before and after a dark-to-light shift (DLS) or the addition of MgProto or hemin. The signals from at least three independent experiments were quantified and corrected for unequal loading by the respective signal for the α subunit of ATP synthase (CF1α). Values are given in arbitrary units relative to the continuous dark level (zero time point), which was set to 1. Error bars indicate standard errors.
Surprisingly, no increase in GluTR levels was seen in four independent experiments when cells in the dark were fed with MgProto. Although HEMA mRNA levels increased after MgProto addition (Fig. 2), these elevated mRNA levels apparently did not manifest themselves in larger protein amounts. The increase seen in GluTR concentrations after hemin and light treatment, but not after MgProto feeding, suggests that both compounds have a similar effect at the transcriptional level but effect HEMA mRNA translation and/or GluTR stability in different ways.
Effect of inhibitors of protein synthesis on HEMA induction.
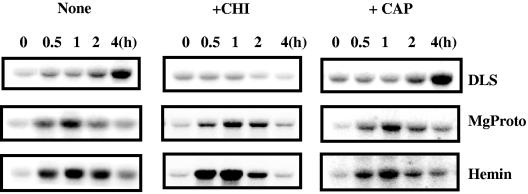
We next tested whether the induction by light or the tetrapyrroles requires de novo synthesis of proteins in cytoplasm or chloroplasts. To inhibit cytosolic protein synthesis, cycloheximide was added to cultures 30 min prior to treatment with light or the tetrapyrroles. As shown in Fig. 6, cycloheximide prevented the light-induced accumulation of HEMA mRNA. Upon prolonged incubation with the inhibitor, we observed a decrease in HEMA mRNA levels beyond those seen at the beginning of the treatment. The inhibitor, though, did not prevent an increased HEMA expression induced by MgProto or hemin. The kinetics of mRNA accumulation in the treated cells were similar to those in the untreated controls (Fig. 6). We conclude that cytoplasmic protein synthesis appears to be necessary for the induction of HEMA and its sustained expression after a shift from dark to light. The induction by MgProto or hemin, though, appears to be independent of de novo protein synthesis.
FIG. 6.
Effects of inhibitors of cytosolic and organellar protein synthesis on HEMA mRNA accumulation after light exposure (DLS) or upon feeding of MgProto (16 μM) or hemin (4 μM) in the dark. Cycloheximide (CHI) (0.035 mM) or chloramphenicol (CAP) (0.35 mM) was added 30 min prior to the shift to light or the beginning of tetrapyrrole treatment. Samples for RNA blots were taken at the time points indicated and processed as described in the legend to Fig. 2.
We also tested the effect of inhibitors of plastidic protein synthesis. Chloramphenicol, which prevents protein synthesis in plastids, was added 30 min prior to application of the inducers. Clearly, no effect on HEMA mRNA accumulation induced by light or MgProto was observed (Fig. 6). These results indicate that plastid protein synthesis is not required for HEMA induction. The same observation was made when lincomycin was used as an inhibitor (data not shown).
DISCUSSION
Chlamydomonas reinhardtii is shown here to possess a single HEMA gene, whose gene product exhibits about 50% identity to the corresponding enzymes of higher plants (Fig. 1). In a phylogenetic tree the C. reinhardtii protein is positioned between the GluTRs of higher plants and those of cyanobacteria.
For its very C-terminal end, a coiled-coil structure is predicted. This coiled-coil domain has been proposed to be a feature of higher-plant GluTRs, since it was not observed in bacterial enzymes (8). This domain of the Arabidopsis HEMA1-encoded enzyme has been shown to be essential for the interaction with the regulatory FLU protein, since its deletion abolished this interaction (16). In C. reinhardtii also, two FLU-like proteins (FLPs) that are the products of a single gene, produced by differential splicing, have recently been identified (13). The longer of the two FLPs has been demonstrated to interact with C. reinhardtii GluTR (13). The presence of a C-terminal coiled-coil motif in the C. reinhardtii enzyme, in analogy to the situation in higher plants (16), suggests that this domain may play a role in the interaction with the longer FLP.
The single C. reinhardtii HEMA gene is shown here to be induced by light and by two tetrapyrroles, heme and MgProto (Fig. 2). The feeding of tetrapyrroles took place in the dark, i.e., under conditions where photooxidative stress was absent (Fig. 2). These two tetrapyrroles, whose synthesis is assumed to take place in plastids (MgProto) and in plastids as well as mitochondria (heme), are proposed to act as organellar signals in controlling HEMA expression. This gene, whose product catalyzes the first and rate-limiting step of the pathway, thus occupies a special position, since none of the other tetrapyrrole biosynthesis genes tested were found to be regulated by MgProto (54) or heme (Z. Vasileuskaya and C. F. Beck, unpublished data).
In Arabidopsis seedlings the response of HEMA to changes in plastid signaling brought about by various chloroplast-damaging treatments has been studied in some detail. Treatment with the chlorosis-inducing herbicide norflurazon or the prolonged exposure of seedlings to far-red light irradiation severely inhibited the light-mediated accumulation of HEMA1 mRNA (28, 32, 34). Details on the signaling pathways involved in Arabidopsis were revealed by the analysis of genomes uncoupled (gun) mutants. These mutants retain partial Lhcb expression following norflurazon-mediated photooxidative damage (49). While GUN1 appears to represent one defined chloroplast-to-nucleus signaling pathway, GUN2, GUN3, GUN4, and GUN5 (encoding heme oxygenase, phytochromobilin synthase, a regulator of Mg-chelatase activity, and the H subunit of Mg-chelatase [CHLH], respectively [30, 36]), genes that are involved in tetrapyrrole synthesis, represent a second route for chloroplast communication. Both signaling pathways defined by these mutants of Arabidopsis appear to control HEMA1 expression, since gun1 alleviates the effects of treatment by norflurazon while in the gun5 mutant, the inhibition by extended far-red light treatment is alleviated (34). Participation of tetrapyrroles in the regulation of HEMA1 is also supported by the observation that this gene is fully rescued from the repressing effects of norflurazon and far-red light treatments in plants overexpressing NADPH:protochlorophyllide oxidoreductase (34). Previously, the repression of an Lhcb gene in norflurazon-treated plants was linked to the accumulation of MgProto (47). As an alternative to a direct involvement of MgProto in nuclear gene repression, it was proposed that the accumulation of phototoxic intermediates of the chlorophyll pathway generates signals that control the expression of nuclear genes (34).
A role for tetrapyrroles in HEMA regulation is also supported by studies with transgenic tobacco lines that, mediated by antisense RNAs specific for CHLH and CHLM, exhibited reduced levels of Mg-chelatase H subunit and Mg-protoporphyrin IX methyltransferase, respectively. In these lines (which have not been subjected to chloroplast-damaging treatments), lower pool levels of Mg-protoporphyrin IX monomethyl ester and heme correlated with a diminished ALA-synthesizing capacity, and this corresponded to reduced levels of HEMA transcripts (1, 40).
Analysis of HEMA induction by light, hemin, and MgProto in our experiments revealed an accumulation of mRNA under all three conditions but an accumulation of GluTR only after light and hemin treatment (compare Fig. 2 and 5). Evidently, the mRNA accumulated upon MgProto feeding did not result in larger amounts of enzyme, suggesting a regulatory step at the posttranscriptional level. In bacteria, modulation of GluTR by proteolytic degradation, possibly heme activated, has been reported (58). This points to an additional route for GluTR regulation in chloroplasts. Feeding of Proto results in the accumulation of Proto and Mg-tetrapyrroles, presumably within the plastids (26). The feeding of MgProto also results in elevated Mg-tetrapyrrole pool levels (J. Kropat, U. Oster, and C. F. Beck, unpublished data). It is tempting to speculate that these plastidal Mg-tetrapyrroles control the stability of GluTR, possibly via an interaction with the FLU/FLP proteins. Since hemin feeding is expected not to cause an elevated flux through the Mg-tetrapyrrole branch of the pathway, the specific effects of MgProto versus hemin on GluTR levels may thus be accounted for.
What is the role that heme and MgProto may play in the regulation of HEMA? Since a regulatory role of MgProto due to its divergent effects on HEMA expression at the RNA (Fig. 2) and protein (Fig. 5) levels is difficult to assess, we will focus on a possible role of heme as an organellar signal. We envision two scenarios. In the first scenario, heme may modulate the amount of HEMA mRNA in a light-independent manner with the goal of ensuring a sufficient capacity for tetrapyrrole biosynthesis. An increased expression of HEMA, activated by heme, may ensure the supply of chloroplasts with GluTR in the presence of elevated tetrapyrrole pool levels.
In the second scenario, heme may mediate the light induction of HEMA. However, levels of heme, in contrast to those of Proto and various Mg-tetrapyrroles, in light-grown cells are lower than those in dark-grown cells (10), which argues against a role of this compound in mediating light induction.
The inhibition of HEMA light induction by cycloheximide and the lack of such an inhibitory effect on tetrapyrrole-mediated HEMA induction (Fig. 6) point to the use of different signaling pathways by light and tetrapyrroles. However, as shown previously, cycloheximide also prevents the light-induced accumulation of tetrapyrroles and by this mechanism abrogates the light induction of HSP70A, while the induction by MgProto was unimpaired by the inhibitor (26). Thus, while the cycloheximide experiment does differentiate between the effects of light and tetrapyrroles, it does not provide us with the critical clues to decide whether the light signaling pathway employs tetrapyrroles or not.
Here, to our knowledge, we provide the first evidence for a role of heme in gene expression in plants. Heme in green algae, just as in higher plants, and assumed to be produced by both plastids and mitochondria, has previously been shown to play a role in regulating gene expression in yeasts and mammals. In these organisms it is synthesized in mitochondria and, by routes not yet elucidated, gains access to the cytosol or nucleus, where it modulates the DNA-binding activity of certain transcription factors (21, 38).
While the precise regulatory role of MgProto and heme in HEMA expression remains to be elucidated, the induction observed upon feeding of these compounds implies the existence of cytosolic or nuclear factors that specifically recognize these two tetrapyrroles. Tetrapyrroles that either lack Fe2+ or Mg2+ (Proto) or chlorophyllide had no inducing potential (Fig. 4). The inability of ZnProto to elicit induction was surprising. Mg2+ and Zn2+ both form pentcoordinate complexes with a square pyrimidal structure. Fe(II) complexes are able to form distorted octahedra with two extra ligands, one above and one below the porphyrin plane. Mg2+ but not Zn2+ may under some conditions (transition state) add two extra ligands just like Fe2+ (14). The recognition of tetrapyrroles by protein factors within the cytosol or nucleus may require the complex that has two extra ligands; in this case ZnProto is expected not to have inducing potential. Also, only a subfraction of MgProto is expected to have the conformation with two ligands. This may possibly contribute to the higher concentration of MgProto (16 μM) than of hemin (4 μM) that is required for induction of HEMA to about an equal extent (Fig. 3).
The induction of HEMA by MgProto and heme supports and extends the previously elaborated concept of a highly complex regulation of HEMA at both the gene and enzyme levels. A rise in Mg-tetrapyrrole and/or heme pool levels, which is known to cause an inhibition of GluTR activity within plastids by feedback mechanisms (16, 57), at the same time may induce an increase in HEMA mRNA levels. These seemingly conflicting effects may help the cells to cope with two requirements: to prevent the short-term accumulation of tetrapyrroles within the plastids and to enable the buildup of additional biosynthesis capacity on a longer time scale. The characterization of MgProto and heme as regulators of HEMA expression opens up new routes to investigate the roles of organelles in the synthesis of enzymes for tetrapyrrole biosynthesis and of intermediates in this biosynthesis chain in interorganellar signaling.
Acknowledgments
We thank Wolfgang Hess and Michael Schroda for stimulating input, and we thank both of them, as well as Bernhard Grimm, for a critical reading of the manuscript. We are grateful for the donation of antibodies by Samuel Beale (GluTR) and Olivier Vallon (ATPase CF1).
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to C.F.B.
REFERENCES
- 1.Alawady, A. E., and B. Grimm. 2005. Tobacco Mg-protoporphyrin IX methyltransferase is involved in inverse activation of Mg-porphyrin and protoheme synthesis. Plant J. 41:282-290. [DOI] [PubMed] [Google Scholar]
- 2.Beale, S. I. 1999. Enzymes of chlorophyll biosynthesis. Photosynth. Res. 60:43-73. [Google Scholar]
- 3.Beale, S. I., and J. D. Weinstein. 1991. Biochemistry and regulation of photosynthetic pigment formation in plants and algae, p. 155-235. In P. M. Jordan (ed.), Biosynthesis of tetrapyrroles. Elsevier, Amsterdam, The Netherlands.
- 4.Beck, C. F. 2001. Signalling pathways in chloroplast-to-nucleus communication. Protist 152:175-182. [DOI] [PubMed] [Google Scholar]
- 5.Beck, C. F., and B. Grimm. Involvement of tetrapyrroles in cellular regulation. In B. Grimm, R. J. Porra, W. Rüdiger, and H. Scheer (ed.), Biochemistry, biophysics and biological functions of chlorophylls, in press. Springer, Utrecht, The Netherlands.
- 6.Berger, B., D. B. Wilson, E. Wolf, T. Tonchev, M. Milla, and P. S. Kim. 1995. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bougri, O., and B. Grimm. 1996. Members of a low-copy number gene family encoding glutamyl-tRNA reductase are differentially expressed in barley. Plant J. 9:867-878. [DOI] [PubMed] [Google Scholar]
- 8.Brody, S. S., S. P. Gough, and C. G. Kannangara. 1999. Predicted structure and fold recognition for the glutamyl tRNA reductase family of proteins. Proteins 37:485-493. [DOI] [PubMed] [Google Scholar]
- 9.Castelfranco, P. A., and X. H. Zeng. 1991. Regulation of 5-aminolevulinic acid synthesis in developing chloroplasts 4. An endogenous inhibitor from the thylakoid membranes. Plant Physiol. 97:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chekounova, E., V. Voronetskaja, J. Papenbrock, B. Grimm, and C. F. Beck. 2001. Characterization of Chlamydomonas mutants defective in the H-subunit of Mg-chelatase. Mol. Gen. Genomics 266:363-373. [DOI] [PubMed] [Google Scholar]
- 11.Cornah, J. E., M. J. Terry, and A. G. Smith. 2003. Green or red: what stops the traffic in the tetrapyrrole pathway? Trends Plant Sci. 8:224-230. [DOI] [PubMed] [Google Scholar]
- 12.Emanuelsson, O., H. Nielsen, and G. von Heijne. 1999. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falciatore, A., L. Merendino, F. Barneche, M. Ceol, R. Meskauskiene, K. Apel, and J.-D. Rochaix. 2005. The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas. Genes Dev. 19:176-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falk, J. E. 1964. Porphyrins and metalloporphyrins: their general, physical and coordination chemistry, and laboratory methods. Elsevier Publishing Co., Amsterdam, The Netherlands.
- 15.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabelling DNA restriction endonuclease fragments to high activity. Anal. Biochem. 132:2-13. [DOI] [PubMed] [Google Scholar]
- 16.Goslings, D., R. Meskauskiene, C. Kim, K. P. Lee, M. Nater, and K. Apel. 2004. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 40:957-967. [DOI] [PubMed] [Google Scholar]
- 17.Gough, S. P., and C. G. Kannangara. 1977. Synthesis of δ-aminolevulinate by a chloroplast stroma preparation from greening barley leaves. Carlsberg Res. Commun. 42:459-464. [Google Scholar]
- 18.Gray, J. C. 2003. Chloroplast-to-nucleus signalling: a role for Mg-protoporphyrin. Trends Genet. 19:526-529. [DOI] [PubMed] [Google Scholar]
- 19.Harris, E. H. 1989. The Chlamydomonas source book: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 20.Helfrich, M., and W. Rüdiger. 1992. Various metallopheophorbides as substrates for chlorophyll synthesis. Z. Naturforsch. 47c:231-238. [Google Scholar]
- 21.Hon, T., A. Hach, D. Tamalis, Y. Zhu, and L. Zhang. 1999. The yeast heme-responsive transcriptional activator Hap1 is a preexisting dimer in the absence of heme. J. Biol. Chem. 274:22770-22774. [DOI] [PubMed] [Google Scholar]
- 22.Ilag, L. L., A. M. Kumar, and D. Söll. 1994. Light regulation of chlorophyll biosynthesis at the level of 5-aminolevulinate formation in Arabidopsis. Plant Cell 6:265-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahn, D., E. Verkamp, and D. Söll. 1992. Glutamyl-transfer RNA: a precursor of heme and chlorophyll biosynthesis. Trends Biochem. Sci. 17:215-218. [DOI] [PubMed] [Google Scholar]
- 24.Kropat, J., U. Oster, W. Rüdiger, and C. F. Beck. 1997. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc. Natl. Acad. Sci. USA 94:14168-14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kropat, J., G. Pöpperl, W. Rüdiger, and C. F. Beck. 1999. Identification of Mg-protoporphyrin IX as a chloroplast signal that mediates the expression of nuclear genes, p. 341-348. In E. Wagner, J. Normann, H. Greppin, J. H. P. Hackstein, R. G. Hermann, K. V. Kowallik, H. E. A. Schenk, and J. Seckbach (ed.), From symbiosis to eukaryotism. Endocytobiol VII. University of Geneva, Geneva, Switzerland.
- 26.Kropat, J., U. Oster, W. Rüdiger, and C. F. Beck. 2000. Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 24:523-531. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, A. M., G. Csankovszki, and D. Söll. 1996. A second and differentially expressed glutamyl-tRNA reductase gene from Arabidopsis thaliana. Plant Mol. Biol. 30:419-426. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, A. M., S. Chaturvedi, and D. Söll. 1999. Selective inhibition of HEMA gene expression by photooxidation in Arabidopsis thaliana. Phytochemistry 51:847-851. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Larkin, R. M., J. M. Alonso, J. R. Ecker, and J. Chory. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signalling. Science 299:902-906. [DOI] [PubMed] [Google Scholar]
- 31.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 32.McCormac, A. C., A. Fischer, A. M. Kumar, D. Söll, and M. J. Terry. 2001. Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana. Plant J. 25:549-561. [DOI] [PubMed] [Google Scholar]
- 33.McCormac, A. C., and M. J. Terry. 2002. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 32:549-559. [DOI] [PubMed] [Google Scholar]
- 34.McCormac, A.C., and M. J. Terry. 2004. The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signalling pathways during de-etiolation. Plant J. 40:672-685. [DOI] [PubMed] [Google Scholar]
- 35.Meskauskiene, R., M. Nater, D. Goslings, F. Kessler, R. op den Camp, and K. Apel. 2001. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98:12826-12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochizuki, N., J. A. Brusslan, R. Larkin, A. Nagatani, and J. Chory. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98:2053-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moser, J., W.-D. Schubert, V. Beier, I. Bringemeier, D. Jahn, and D. W. Heinz. 2001. V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J. 20:6583-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogawa, K., J. Sun, S. Taketani, O. Nakajima, C. Nishitani, S. Sassa, N. Hayashi, M. Yamamoto, S. Shibehara, H. Fujita, and K. Igarashi. 2001. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20:2835-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papenbrock, J., H.-P. Mock, E. Kruse, and B. Grimm. 1999. Expression studies in tetrapyrrole biosynthesis: inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta 208:264-273. [Google Scholar]
- 40.Papenbrock, J., H.-P. Mock, R. Tanaka, E. Kruse, and B. Grimm. 2000. Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 122:1161-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papenbrock, J., and B. Grimm. 2001. Regulatory network of tetrapyrrole biosynthesis—studies of intracellular signalling involved in metabolic and developmental control of plastids. Planta 213:667-681. [DOI] [PubMed] [Google Scholar]
- 42.Pontoppidan, B., and C. G. Kannangara. 1994. Purification and partial characterisation of barley glutamyl-tRNA(Glu) reductase, the enzyme that directs glutamate to chlorophyll biosynthesis. Eur. J. Biochem. 225:529-537. [DOI] [PubMed] [Google Scholar]
- 43.Popov, N., S. Schmitt, and H. Matthices. 1975. Eine störungsfreie Mikromethode zur Bestimmung des Proteingehalts in Gewebshomogenaten. Acta Biol. Germ. 31:1441-1446. [PubMed] [Google Scholar]
- 44.Pöpperl, G. 1996. Ph.D. thesis. University of Munich, Munich, Germany.
- 45.Sangwan, I., and M. R. O'Brian. 1999. Expression of a soybean gene encoding the tetrapyrrole-synthesis enzyme glutamyl-tRNA reductase in symbiotic root nodules. Plant Physiol. 119:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schroda, M. 2004. The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth. Res. 82:221-240. [DOI] [PubMed] [Google Scholar]
- 47.Strand, A., T. Asami, J. Alonso, J. R. Ecker, and J. Chory. 2003. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin IX. Nature 421:79-83. [DOI] [PubMed] [Google Scholar]
- 48.Surpin, M., R. M. Larkin, and J. Chory. 2002. Signal transduction between the chloroplast and the nucleus. Plant Cell 14(Suppl.):S327-S338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Susek, R. E., F. M. Ausubel, and J. Chory. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74:787-799. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka, R., K. Yoshida, T. Nakayashiki, T. Masuda, H. Tsuji, H. Inokuchi, and A. Tanaka. 1996. Differential expression of two hemA mRNAs encoding glutamyl-tRNA reductase proteins in greening cucumber seedlings. Plant Physiol. 110:1223-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka, R., K. Yoshida, T. Nakayashiki, H. Tsuji, H. Inokuchi, K. Okada, and A. Tanaka. 1997. The third member of the hemA gene family encoding glutamyl-tRNA reductase is primarily expressed in roots in Hordeum vulgare. Photosynth. Res. 53:161-171. [Google Scholar]
- 52.Thomas, J., and J. D. Weinstein. 1990. Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol. 94:1414-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ujwal, M. L., A. C. McCormac, A. Goulding, A. M. Kumar, D. Soll, and M. J. Terry. 2002. Divergent regulation of the HEMA gene family encoding glutamyl-tRNA reductase in Arabidopsis thaliana: expression of HEMA2 is regulated by sugars, but is independent of light and plastid signalling. Plant Mol. Biol. 50:83-91. [DOI] [PubMed] [Google Scholar]
- 54.Vasileuskaya, Z., U. Oster, and C. F. Beck. 2004. Involvement of tetrapyrroles in inter-organellar signaling in plants and algae. Photosynth. Res. 82:289-299. [DOI] [PubMed] [Google Scholar]
- 55.von Gromoff, E. D., U. Treier, and C. F. Beck. 1989. Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol. Cell. Biol. 9:3911-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Kampen, J., U. Nieländer, and M. Wettern. 1993. Stress-dependent transcription of a gene encoding a Gβ-like polypeptide from Chlamydomonas reinhardtii. J. Plant Physiol. 143:756-758. [Google Scholar]
- 57.Vothknecht, U. C., C. G. Kannangara, and D. von Wettstein. 1996. Expression of catalytically active barley glutamyl tRNAGlu reductase in Escherichia coli as a fusion protein with glutathione S-transferase. Proc. Natl. Acad. Sci. USA 93:9287-9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, L., M. Elliott, and T. Elliott. 1999. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J. Bacteriol. 181:1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinstein, J. D., and S. I. Beale. 1985. Enzymatic conversion of glutamate to delta-aminolevulinate in soluble extracts of the unicellular green alga, Chlorella vulgaris. Arch. Biochem. Biophys. 237:454-464. [DOI] [PubMed] [Google Scholar]