Abstract
Tocopherols are important antioxidants in lipophilic environments. They are synthesized by plants and some photosynthetic bacteria. Recent efforts to analyze and engineer tocopherol biosynthesis led to the identification of Synechocystis sp. strain PCC 6803 as a well-characterized model system. To facilitate the identification of the rate-limiting step(s) in the tocopherol biosynthetic pathway through the modulation of transgene expression, we established an inducible expression system in Synechocystis sp. strain PCC 6803. The nirA promoter from Synechococcus sp. strain PCC 7942, which is repressed by ammonium and induced by nitrite (S.-I. Maeda et al., J. Bacteriol. 180:4080-4088, 1998), was chosen to drive the expression of Arabidopsis thaliana p-hydroxyphenylpyruvate dioxygenase. The enzyme catalyzes the formation of homogentisic acid from p-hydroxyphenylpyruvate. Expression of this gene under inducing conditions resulted in up to a fivefold increase in total tocopherol levels with up to 20% of tocopherols being accumulated as tocotrienols. The culture supernatant of these cultures exhibited a brown coloration, a finding indicative of homogentisic acid excretion. Enzyme assays, functional complementation, reverse transcription-PCR, and Western blot analysis confirmed transgene expression under inducing conditions only. These data demonstrate that the nirA promoter can be used to control transgene expression in Synechocystis and that homogentisic acid is a limiting factor for tocopherol synthesis in Synechocystis sp. strain PCC 6803.
Vitamin E is a collective term that refers to the biological activity of a group of eight natural amphipathic compounds: α-, β-, γ-, and δ-tocopherol and α-, β-, γ-, and δ-tocotrienol (Fig. 1). Tocotrienols can be distinguished from their corresponding tocopherols by the presence of three isolated double bonds in their prenyl side chains. These compounds are synthesized by plants and certain photosynthetic bacteria and are well recognized as effective oxygen radical scavengers in lipophilic environments such as oils and the lipid bilayer of biological membranes (3, 4). In photosynthetic organisms, tocopherols are suggested to function as membrane-associated antioxidants and as constituents of the chloroplast membranes (17, 18, 27, 41). α-Tocopherol is an essential component in the mammalian diet and has the highest vitamin E activity among the isomers described above (3). Because of these health benefits and biological functions, there is considerable interest in engineering tocopherol biosynthesis to increase tocopherol levels and optimize their composition in agricultural crops.
FIG. 1.
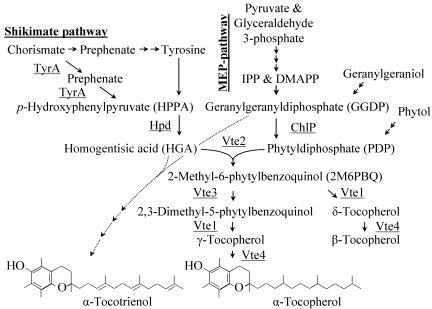
Schematic drawing of the tocopherol biosynthetic pathway. Abbreviations: DMAPP, dimithylallyldiphosphate; ChlP, geranylgeranyldiphosphate reductase; Hpd, p-hydroxyphenylpyruvate dioxygenase; IPP, isopentenyldiphosphate; MEP, methylerythritol phosphate; TyrA, bifunctional chorismate mutase and prephenate dehydrogenase; Vte1, tocopherol cyclase; Vte2, homogentisate phytyltransferase; Vte3, 2-methyl-6-phytylbenzoquinol methyltransferase; Vte4, γ-tocopherol methyltransferase.
The regulation and rate-limiting reactions in tocopherol biosynthesis are currently poorly understood. The first committed reaction of tocopherol biosynthesis is the prenylation of homogentisic acid (HGA), derived from the shikimate pathway, with phytyldiphosphate (PDP) derived from the 2C-methyl-d-erythritol 4-phosphate (MEP)-pathway, for the formation of 2-methyl-6-phytylbenzoquinol (Fig. 1). Tocotrienols are synthesized when geranylgeranyl diphosphate replaces PDP in the prenylation reaction of HGA, resulting in the formation of 2-methyl-6-geranylgeranylbenzoquinol. Therefore, the availability of HGA has an impact on tocopherol and tocotrienol biosynthesis. The cyanobacterium Synechocystis sp. strain PCC 6803 has been used as a model organism for elucidating the mechanisms of critical biological processes such as photosynthesis (14, 16), metabolic engineering of fatty acid saturation, and zeaxanthin biosynthesis (2, 21). A common strategy to define the rate-limiting steps of a biosynthetic pathway experimentally is to modulate expression of specific pathway enzymes and then monitor the corresponding changes of intermediates and end products. In this setting, it is advantageous to use inducible promoters for the controlled expression of target genes.
Cyanobacteria are prokaryotic organisms that perform oxygenic photosynthesis through the operation of a photosynthetic system very much like that present in the chloroplasts of higher plants. They preferentially use inorganic carbon, nitrogen, and mineral salts for growth. Nitrate and ammonium are excellent nitrogen sources for cyanobacteria (11). Nitrate is taken up into the cells and reduced to ammonium by sequential action of nitrate reductase and nitrite reductase. The genes encoding the nitrate transporter (nrtABCD) (29), nitrate reductase (narB) (32), and nitrite reductase (nirA) (22) form the nir operon (nirA-nrtABCD-narB) in Synechococcus sp. strain PCC 7942 (38). In all cyanobacteria tested to date, transcription of the nir operon is repressed by ammonium (20, 38). In contrast, the presence of nitrate in the culture medium enhances mRNA levels of the nir operon (12, 23, 24). This is further evidenced by discovering the requirement of the nitrate-promoted NtcB transcription factor for regulation of nir operon transcription in cyanobacteria (1, 12, 13, 19, 24, 38, 39). We hypothesized that the nirA promoter of Synechococcus can be used as a tight inducible system for controlling transgene expression in Synechocystis. Furthermore, by substitution of ammonium with nitrate for activation of the promoter this system would produce a minimal perturbation on the cellular processes in Synechocystis. Several studies have provided evidence that p-hydroxyphenylpyruvate dioxygenase (Hpd) limits tocopherol biosynthesis in plants (10, 15, 31, 42, 43). We reasoned, therefore, that transgenic expression of Arabidopsis thaliana hpd (hpdAt) under nirA promoter control would be an excellent model for developing a controlled Synechocystis expression system and to demonstrate tocopherol pathway engineering in Synechocystis.
MATERIALS AND METHODS
Construction of a nirAp-based expression plasmid.
Custom multiple cloning sites (MCS) were made by annealing the primers MCS2 (5′-AAGGCCTGACATATGTCGCGGCCGCTCTAGATTTAAATACTAGTCCCGGGCTAGCGA GCTCT-3′) and MCS2-rev.comp (5′-AGAGCTCGCTAGCCCGGGACTAGTATTTAAATCTAGAGCGGCCGCGACATATGTCAGGCCTT-3′). The annealed primers were cloned into the EcoRV site of pBluescript SK(+) (Stratagene, La Jolla, CA), yielding the plasmid pCER7.
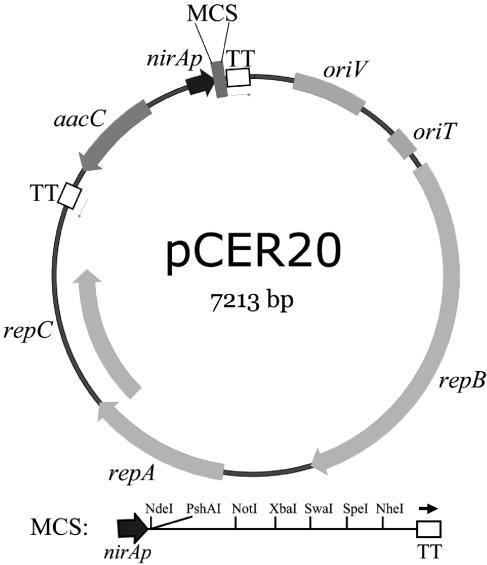
A 166-bp fragment upstream of the nirA operon was amplified by PCR from the genomic DNA of Synechococcus sp. strain PCC 7942 by using the primer pair nirA1 (5′-TAGGCCTTCCCTCTCAGCTCCAAAAAGT-3′) and nirA3 (5′-CTTGAGCCATATGTCCATCTGCCTAACA-3′). Underlined bases in the primer sequence indicate StuI and NdeI restriction sites that were added at the 5′ and 3′ termini of the nirA promoter element, respectively. The PCR product harboring nirAp was digested with StuI and NdeI, gel purified, and ligated into StuI- and NdeI-digested pCER7, resulting in the formation of pCER11. Promoter sequence integrity was confirmed by DNA sequence analysis. The amplified region possesses an NtcB-binding site (an inverted repeat with a LysR motif, TGCAN5TGCA), an NtcA-binding site (a palindromic structure with a conserved sequence signature, GTAN8TAC), and a TAN3T sequence fitting the −10 box of the Escherichia coli σ70 consensus promoter. This DNA segment has been showed to retain full responsiveness to nitrogen and transcription activity (24). To provide a selection marker for the transcription unit (nirAp plus MCS), the gentamicin resistance cassette (Gmr) from pUCGM (35) was cleaved as a SmaI-fragment, gel purified, and ligated into HindIII-digested and T4 DNA polymerase-blunt-ended pCER11. The resulting plasmids were screened and those that carried the Gmr cassette in the opposite orientation to the nirA promoter were selected and designated pCER12. Transcription terminators for the above expression cassettes were cloned from pHP45ω (30). They were PCR amplified from pHP45ω by using the primer pHP45-1 (5′-TAGGCCTGGATGACCTTTTGAATGACC-3′). The Omega cassette is flanked by two identical transcription terminators. The terminators are inverted relative to each other. The primer anneals to both terminators and extends outward during PCR. The final PCR product has the two terminators at the ends plus the rest of the plasmid (minus the spectinomycin cassette). The resulting PCR product was ligated to the blunt-ended XhoI- and EcoRI-digested Gmr::nirAp::MCS fragment of pCER12. This yielded the plasmid pCER18, which carries the final transcription module (Gmr::nirAp::MCS). The transcription module was excised with EcoRI, and the ends were filled in with T4 DNA polymerase. This DNA fragment was ligated to the 5.8-kb HincII-HincII fragment of plasmid RSF1010 (34), resulting in the new expression vector pCER20 (Fig. 2). RSF1010 and its derivatives replicate autonomously in cyanobacteria of the genera Synechococcus and Synechocystis (25, 28). All enzymes used were purchased from New England Biolabs (Beverly, MA).
FIG. 2.
Plasmid map of the inducible cyanobacterial expression vector pCER20. The expression of target gene(s) is under the transcriptional control of the nitrite reductase promoter, nirAp. The aacC gene, which confers resistance to gentamicin, serves as the selection marker. Bracketing nirAp and the selection marker are two transcriptional terminators (TT) from the plasmid, pHP45ω. The origin of replication (oriV) and replication proteins (repABC), as well as the origin of transfer (oriT) are derived from the broad-host-range plasmid RSF1010 (34).
Construction of a Synechocystis hpdAt expression vector.
For convenience of subsequent cloning steps, a DNA fragment containing multiple cloning sites (5′-GCGGCCGCGGGCCCTGATCATCTAGAGTCGACGGCCGGCCGAA TTCGCGGCCGCTCTAGA-3′) was inserted into NotI- and XbaI-digested pCER20. The resulting plasmid, designated pMON36546, was used as a vector control. To generate a nirAp::hpdAt expression vector, the full-length of hpdAt was PCR amplified from pMON36596 (43) (forward primer 5′-TGCTCTAGAACACAGGAGGACAGCCATGGGCCACCAAAACGCC-3′ and reverse primer 5′-ATAAGAATGCGGCCGCAAGCTTGTCGACTTCATCCCACTAACTGTTTG-3′). The Shine-Dalgarno sequence and ATG start codon are underlined in the forward primer. The resulting PCR fragment was XbaI and NotI digested and cloned into XbaI- and NotI-digested pMON36546, resulting in the formation of plasmid pMON36547.
Generation of Synechocystis sp. strain PCC 6803 Δslr0090.
The open reading frame slr0090 had been identified to encode the Synechocystis sp. strain PCC 6803 hpd gene (6). To create a hpd mutant, a knockout construct, pMON29153, was generated. This construct harbors the Synechocystis hpd interrupted by insertion of the nptII marker. Plasmid pMON29153 was constructed by digesting pMON29138 (43) with BstXI, filling in the sticky ends using Klenow fragment, and inserting the nptII expression cassette (blunted EcoRI fragment) from pUC4K (40). The resulting plasmid contained the nptII cassette inserted 647 bp downstream of the ATG start codon. This recombinant vector was transformed into wild-type (WT) Synechocystis sp. strain PCC 6803 by using the transformation procedure described by Williams (44). Homologous recombination led to the replacement of the WT slr0090 gene. Complete segregation of the mutant genome was obtained by restreaking single colonies several times on BG11 agar plates supplemented with kanamycin (25 μg/ml). PCR and Southern analysis of the Synechocystis sp. strain PCC 6803 Δslr0090 mutant were performed to confirm complete segregation of the mutant genome (data not shown).
Strains, growth conditions, and cell sample preparation.
Synechocystis sp. strain PCC 6803 (ATCC 27184) was obtained from the American Type Culture Collection. WT and recombinant cells of Synechocystis sp. strain PCC 6803 were cultivated photoautotrophically at 30°C under continuous illumination provided by fluorescent lamps (70 μE m−2 s−1). Liquid cultures were shaken at 225 rpm on a rotary shaker. The basal medium (BG110) was a nitrogen-free medium obtained by replacing NaNO3, Co(NO3)2, and ferric ammonium citrate in medium BG11 (37) with equimolar fractions of NaCl, CoCl2, and ferric citrate, respectively. Ammonium-containing medium (BG110NH4+) and nitrate-containing medium (BG110NO3−) were prepared by addition of 17.6 mM NH4Cl or 17.6 mM NaNO3, respectively, to the basal medium. Both media were buffered with 10 mM N-Tris-(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)-NaOH (pH 8.0). For growth on solid media, BG110NH4+ medium containing 1.5% (wt/vol) agar (Difco) was used. Plasmids replicating autonomously in Synechocystis were transformed into Synechocystis sp. strain PCC 6803 via triparental mating (9), and transformants were selected on medium supplemented with kanamycin at 25 μg ml−1 and/or gentamicin at 10 μg ml−1. Conjugated cells were spread on a 0.45-μm-pore-size cellulose nitrate membrane filter (Whatman) and placed on nonselective solid ammonium medium, incubated for 24 h as described above, and transferred to selective ammonium medium plates. Resistant colonies were used to inoculate 2 ml of liquid BG110NH4+ medium supplemented with gentamicin (and/or kanamycin) and incubated for 2 days. These cultures served as precultures for the final 150-ml liquid cultures. Cell density in the 150-ml cultures was monitored spectrophotometrically (SpectraMAX; Molecular Devices) at 730 nm (A730). Cells were harvested when the A730 of the cell culture reached 0.4 to 0.5 by 10 min of centrifugation at 25°C and 3,500 × g. The cell pellet was washed with 20 ml of BG110 and resuspended in 150 ml of fresh nitrate (BG110NO3−) or ammonium (BG110NH4+) medium. For promoter activation, cells were subsequently grown under the light and temperature conditions described above. Cell samples were harvested at various time intervals to measure tocopherol and tocotrienol content, gene expression, and enzyme activity.
Analysis of hpdAt transcript by reverse transcription-PCR (RT-PCR).
Nitrate (BG110NO3−) or ammonium (BG110NH4+) medium-grown Synechocystis harvested from three representative cultures was ground in liquid nitrogen. Total RNA was isolated as previously described (26), and any contaminating DNA was removed by treatment with RQ1-RNase-free DNase (Promega, Madison, WI). Three micrograms of total RNA for each sample was reverse transcribed to generate cDNA in two 50-μl reactions by using an Omniscript RT kit according to the manufacturer's recommendations (QIAGEN, Inc., Valencia, CA). An aliquot of cDNA corresponding to 200 ng of total RNA was subjected to 23 cycles of PCR with primers 5′-TTCCTTCGTCGCCTCCTATCG-3′ (forward) and 5′-ACTCCTTGATCTGATCATCGC-3′) (reverse), resulting in the amplification of a 600-bp fragment internal to the hpdAt gene. The reaction was done under the following thermocycle conditions: 5 min of incubation at 95°C, followed by 23 cycles of 1 min at 95°C, 1 min of annealing at 56°C, and a 2-min extension at 72°C. The amount of amplified DNA was estimated by DNA gel electrophoresis with ethidium bromide staining.
Tocopherol and tocotrienol analyses.
Tocopherols for WT and recombinant Synechocystis strains were measured by a normal-phase high-pressure liquid chromatography (HPLC) as described previously (33), but lyophilized cell pellets were used instead of fresh harvested cells. Tocotrienol content was analyzed by using the same procedure with tocotrienol standards purchased from Calbiochem (La Jolla, CA). Tocopherol content was normalized to the dry cell mass.
Protein analysis.
The presence of HpdAt protein in recombinant strains was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis with peptide-directed antibodies obtained by immunization of a rabbit with a synthetic peptide (CRTLREMRKRSSIGG). Peptide synthesis and antibody generation were performed by Sigma-Genosys, St. Louis, MO. Synechocystis sp. strain PCC 6803 cell extracts were prepared by six passages through a French press (Sim-Aminco Spectronic Instruments) at 20,000 lb/in2 with cells suspended in 50 mM Tris-HCl (pH 7.6) containing 5 mM dithiothreitol, 0.1% Triton X-100, 50 U of DNase I, and protease inhibitor cocktail tablets (Boehringer-Mannheim). The supernatant fraction from a 15-min centrifugation at 10,000 × g was used as a crude extract for protein analysis and enzyme assays. For protein gel analysis, 25 μg of total protein per lane was loaded on a 4 to 20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (Invitrogen), and polypeptides were separated according to the manufacturer's protocol. Immunodetection was performed with a 1:500 dilution of rabbit polyclonal peptide specific antibodies raised against a synthetic HpdAt peptide. An anti-rabbit immunoglobulin G-alkaline phosphatase conjugate was used as a secondary antibody (Sigma). The blots were developed by using nitroblue tetrazolium dye in conjunction with the alkaline phosphatase substrate BCIP (5-bromo-4-chloro-3-indolylphosphate; Sigma). Proteins were quantified with Bradford reagent (Bio-Rad) using bovine serum albumin as a standard.
Hpd assay.
Hpd assays were performed according to a modified procedure from Secor (36). Radiolabeled [U-14C]p-hydroxyphenylpyruvate ([U-14C]HPPA) was prepared enzymatically from [U-14C]tyrosine (449 mCi/mmol; Amersham Life Science) as described by Secor (36). The Hpd reaction mixture contained 50 mM potassium phosphate (pH.7.4), 0.1 mM unlabeled HPPA (freshly prepared and equilibrated for 2 h at room temperature), 0.5 μCi of [U-14C]HPPA, 5,000 U of catalase (Sigma), 100 μl of a freshly prepared 1:1 (vol/vol) mixture of 150 mM reduced glutathione (Sigma), 3 mM dichlorophenolindophenol (Sigma), and 250 μg of protein extract in a total volume of 500 μl. The reaction was incubated for 30 min at 30°C and terminated by the addition of 150 μl of 20% (wt/vol) perchloric acid. Precipitated protein was removed by 5 min of centrifugation at 14,000 rpm in an Eppendorf centrifuge. The analysis was performed on an HP1100 series HPLC system consisting of an HP G1329A automatic sampler, an HP G1311A quaternary pump, an HP G1315A diode array detector, and an HP G1321A fluorescence detector (Agilent Technologies, Englewood, CO) and a Packard Radiomatic 500TR flow scintillation analyzer (Hewlett-Packard, Meridien, CT). Hpd reaction products were separated by reversed-phase HPLC using a Vydac model 201HS54 C18 HPLC column (4.6 by 250 mm, 5 μm) coupled with an All-tech C18 guard column (P. J. Cobert Associates, Inc., St. Louis, MO) and a solvent system consisting of buffers A (0.1% H3PO4 in water) and B (0.1% H3PO4 in methanol). The solvent gradient raised buffer B from 0 to 15 min from 25 to 50% and held buffer B from 15 to 20 min constant at 50%. Compounds of interest were detected by diode array detector set at 210 and 254 nm and by the flow scintillation analyzer set at 156 KeV of ULD to detect 14C compounds. The sample injection volume was 20 μl, and the flow rate was set at 1.0 ml/min.
RESULTS AND DISCUSSION
Functional complementation of Synechocystis sp. strain PCC 6803 Δslr0090 with hpdAt driven by the nirA promoter.
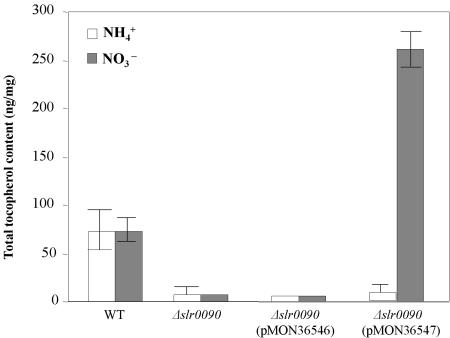
A targeted mutation in Synechocystis sp. strain PPC 6803 was created by homologous recombination (see Materials and Methods), and the resulting mutant, Synechocystis sp. strain PPC 6803 Δslr0090, was analyzed for changes in tocopherol content compared to WT cultures. Tocopherol levels in the mutant were below the limit of detection (<5 ng/mg dry cell mass) (Fig. 3) (6), indicating an essential role of Hpd in tocopherol biosynthesis. This result is consistent with the loss of Hpd activity in this mutant (data not shown). The mutant cell growth rates were comparable to that of WT cells, indicating that photosynthesis was not affected (6). Subsequently, the mutant was used to test the functionality of nirAp::hpdAt in complementation experiments.
FIG. 3.
Complementation of Synechocystis sp. strain PCC 6803 Δslr0090 with Arabidopsis hpd driven by the nirA promoter. Total tocopherol content was normalized to dry cell mass. Cells used for tocopherol analysis were harvested after an incubation period of 10 days. Tocopherol levels shown for strains Synechocystis Δslr0090 and Synechocystis Δslr0090(pMON36546) were below the limit of detection (<5 ng mg-1). Abbreviations: Δslr0090, Synechocystis hpd insertional inactivation mutant; pMON36546, empty vector control; pMON36547, nirAp::hpdAt.
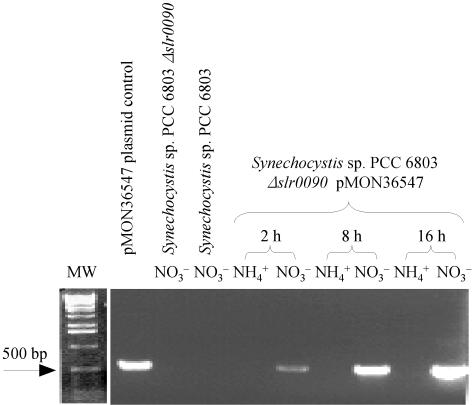
For complementation, Synechocystis sp. strain PCC 6803 Δslr0090 was transformed with pMON36547. This plasmid harbored a nirAp::hpdAt expression cassette. As shown in Fig. 3, complemented strains grown on BG110NO3− medium contained ∼3.5-fold more tocopherol than WT cells grown on the same medium. As expected, Synechocystis sp. strain PCC 6803 Δslr0090(pMON36547) failed to accumulate tocopherol when grown in BG110NH4+ medium (Fig. 3). To confirm hpdAt expression in BG110NO3−-grown cells, RT-PCR amplifications were performed. The hpdAt transcript was detected in Synechocystis sp. strain PCC 6803 Δslr0090(pMON36547) at 2 h after nitrate induction and continued to accumulate at 8 and 16 h postinduction (Fig. 4). In contrast, the transcript was not detected in Synechocystis sp. strain PCC 6803 Δslr0090-(pMON36547) cultivated in BG110NH4+ medium. These results support the induction of nirAp by nitrate in Synechocystis sp. strain PPC 6803 and the functionality of hpdAt complementing the Δslr0090 mutation.
FIG. 4.
RT-PCR expression analysis of hpdAt in Synechocystis sp. strain PCC 6803 Δslr0090(pMON36547). Three micrograms of total RNA prepared from BG110NO3−- or BG110NH4+-grown Synechocystis was subjected to RT-PCR (see Materials and Methods). The agarose gel shows a 1-kb DNA ladder (lane MW; Invitrogen Life Technologies), PCR of pMON36547 (nirAp::hpdAt) plasmid control, RT-PCR from Synechocystis sp. strain PCC 6803 Δslr0090 or wild-type Synechocystis sp. strain PCC 6803 incubated for 16 h in BG110NO3− medium, and that of pMON36S47-harboring Synechocystis sp. strain PCC 6803 Δslr0090 incubated for 2, 8, and 16 h in BG110NH4+ or BG110NO3− medium.
Nitrate-responsive expression of hpdAt under nirA promoter control in Synechocystis sp. strain PCC 6803.
To further demonstrate nitrate-dependent activation of the Synechococcus nirA promoter and assess its efficacy in induction of heterologous gene expression in Synechocystis, the expression vector pMON36547 and the empty vector control pMON36546 were conjugated into WT Synechocystis sp. strain PCC 6803. In order to analyze gene expression and tocopherol levels, transgenic and WT cells were grown in parallel and sampled at 2, 4, 10, and 12 days after inoculation. As shown in Fig. 5, utilization of different nitrogen sources in the culture media resulted in substantial differences in HpdAt polypeptide level and enzyme activity. Using anti-HpdAt peptide antibody, a single immunoreactive band was detected in an extract of Synechocystis sp. strain PCC 6803(pMON36547) expressing hpdAt when grown in BG110NO3− medium. Extracts of this recombinant strain grown in BG110NH4+ did not contain detectable immunoreactive proteins against anti-HpdAt peptide antibody (Fig. 5A). These results confirmed the induction of hpdAt in BG110NO3− and its repression by ammonium. The immunoreactive band was also not observed in extracts of WT cells growing in either BG110NH4+ or BG110NO3− medium. This result was consistent with the lack of cross-reactivity with the E. coli-expressed Synechocystis Hpd (data not shown).
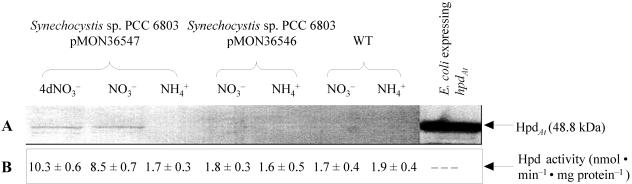
FIG. 5.
Induction of nirAp in BG110NO3− media. (A) Immunoblot analysis of HpdAt peptide in Synechocystis sp. strain PCC 6803 harboring pMON36547 (nirAp::hpdAt) or pMON36546 (empty vector control) incubated for 4 days in BG110NO3− medium (lane 1, 4dNO3−) or for 2 days in BG110NO3− or BG110NH4+ medium (remaining lanes). (B) Hpd enzyme activity detected in the corresponding crude cell extracts. Hpd enzyme activity in Synechocystis sp. strain PCC 6803(pMON36547) incubated in BG110NH4+ medium and in Synechocystis sp. strain PCC 6803(pMON36546) as well as in the WT control originates from endogenously expressed Synechocystis Hpd. Hpd activity in E. coli expressing hpdAt was typically 30 to 50 nmol min−1 mg of protein−1.
To evaluate whether the increase in polypeptide levels corresponds with increased enzyme activity, Hpd enzyme assays were carried out with crude extracts of the Synechocystis strains. As shown in Fig. 5B, the extract derived from nitrate-induced Synechocystis sp. strain PCC 6803(pMON36547) had ∼5-fold increased Hpd activity compared to extracts derived from the vector control [Synechocystis sp. strain PCC 6803(pMON36546)] or WT cell extracts. Such an increase in Hpd activity was not observed with cell extracts derived from Synechocystis sp. strain PCC 6803(pMON36547) incubated in ammonium-containing medium. In addition, the culture supernatant of hpdAt-expressing Synechocystis sp. strain PCC 6803(pMON3654) incubated in BG110NO3− medium turned a dark brown color after 10 days of incubation (Fig. 6). A similar phenomenon was observed in recombinant E. coli cultures expressing Arabidopsis or Synechocystis hpd (data not shown) and has been reported in other systems as well (7, 8). The brown color is thought to be the result of HGA accumulation and excretion into the culture medium (7). In aqueous medium HGA can be oxidized and polymerized to form a brown melanin-like pigment. Western blotting experiments, enzyme assay data, and color changes of the culture supernatant are consistent with transgenic hpdAt expression in Synechocystis sp. strain PCC 6803(pMON36547) incubated in BG110NO3−. Similarly, Western blot and enzyme assay data obtained with crude extracts of the same strain after incubation in BG110NH4+ did not provide any evidence for hpdAt expression, suggesting that the nirA promoter is inactive under such growth conditions.
FIG. 6.
Synechocystis culture supernatants. Culture supernatants shown above were collected by 25 min of centrifugation at 4,000 × g after 10 days of incubation at 30°C. The culture supernatant of Synechocystis sp. strain PCC 6803(pMON36547) incubated in BG110NO3− medium exhibits a characteristic brown color that is typical for bacteria excreting HGA (7).
Transgenic expression of hpdAt led to increased tocopherol levels.
To determine whether the tocopherol content and composition had been affected by transgenic expression of hpdAt, aliquots of the cell culture were taken at various time points for tocopherol analysis. Cultures of Synechocystis sp. strain PCC 6803 harboring pMON36547 that were incubated for 12 days in BG110NO3− had fivefold-increased tocopherol levels compared to the WT control, whereas cells from the same strain incubated in BG110NH4+ and cultures from the vector control had tocopherol levels comparable to the WT control (Fig. 7). Thus, tocopherol accumulation showed a positive correlation with the HpdAt polypeptide level and enzyme activity. These results confirm functionality of the nirA promoter for regulation of transgene expression via nitrate availability, demonstrate its utilization for engineering of the tocopherol metabolic pathway in Synechocystis, and support the hypothesis that Hpd catalyzes a rate-limited reaction in tocopherol biosynthesis (10, 15, 31, 42, 43).
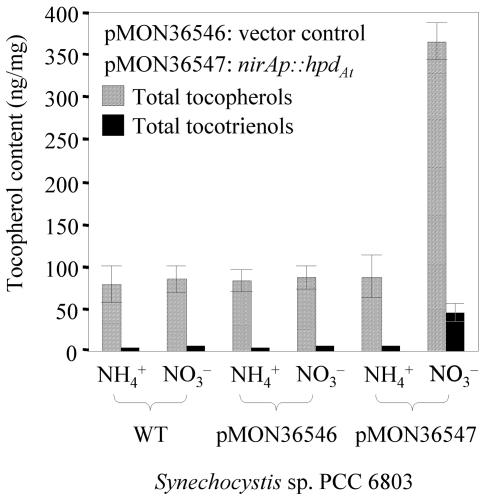
FIG. 7.
Tocopherol content and composition in Synechocystis sp. strain PCC 6803 cultures grown in BG110NH4+ and BG110NO3− media. Tocopherol and tocotrienol contents of Synechocystis cultures are normalized to dry cell mass. Cells used for this experiment were harvested after an incubation period of 12 days.
More interestingly, Synechocystis cells expressing hpdAt accumulated up to 20% of their total tocopherols as tocotrienols (Fig. 7). None of the WT or vector control Synechocystis samples contained detectable tocotrienol levels. As described above, the tocotrienol precursor, 2-methyl-6-geranylgeranylbenzoquinol, is formed via the condensation of HGA and geranylgeranyl diphosphate catalyzed by the homogentisate phytyltransferase (Vte2), whereas tocopherol synthesis utilizes HGA and PDP. The formation of tocotrienols by Synechocystis Vte2 is consistent with data provided by Collakova and DellaPenna (5) and suggests that 2-methyl-6-phytylbenzoquinol methyltransferase (Vte3), tocopherol cyclase (Vte1), and γ-methyltransferase (Vte4) in Synechocystis sp. strain PCC 6803 can utilize tocotrienol precursors as substrates as well. In addition, the formation of tocotrienols and accumulation of HGA provide evidence for a limitation of endogenous PDP, suggesting that experiments to engineer further tocopherol increases in Synechocystis should focus on this part of the pathway.
Conclusions.
A 166-bp DNA fragment from the 5′ untranslated region of the nirA operon was isolated from the genome of Synechococcus sp. strain PCC 7942. Sequence analysis revealed the presence of the consensus sequence (GTAN8TAC) for cyanobacterial NtcA-binding sites, an L1-like inverted repeat containing a LysR motif (TN11A), and the putative −10 element, showing a typical structure for a nitrogen-regulated promoter in cyanobacteria (12, 24, 38). By monitoring the expression level of hpdAt and its impact on tocopherol levels in Synechocystis sp. strain PCC 6803 or Synechocystis sp. strain PCC 6803 Δslr0090 harboring pMON36547, a nirAp::hpdAt expression construct, we were able to show that the nirA promoter is active in BG110NO3− medium and inactive in BG110NH4+ medium. More importantly, the results presented here are the first, to our knowledge, to demonstrate that the nitrate-inducible nirA promoter can be used to drive and regulate the transcription of transgenes in Synechocystis sp. strain PCC 6803 and that this regulation can lead to changes in metabolite flux for production of target chemicals, such as tocopherols. Therefore, the nirA promoter system provides a suitable tool for metabolic engineering in Synechocystis. Its ability to be inactivated in ammonium-containing medium makes it particularly suitable for the expression of potentially toxic genes or for engineering metabolic pathways that may produce toxic products or intermediates. In the present study, we obtained evidence that the upregulated hpdAt expression results in an increased synthesis of HGA, tocopherols, and tocotrienols in Synechocystis in vivo, indicating that Hpd plays an important role in tocopherol production and that additional enzymes such as geranylgeranyl diphosphate reductase may be required to overcome additional constraints for further enhancement of the tocopherol accumulation in this strain. The results obtained from this photosynthetic model system provide useful information for tocopherol metabolic engineering in other organisms.
Acknowledgments
We are grateful to Mylavarapu Venkatramesh, Dusty Post-Beittenmiller, and Kenneth J. Gruys (Monsanto Company) for helpful discussions during the experimental course of this study and the preparation of the manuscript. We thank Jeffrey M. Staub and Lisa M. Weaver (Monsanto Company) for critical reading of the manuscript. We also thank Yanping Zhu for skillful technical assistance with Synechocystis cell culture and Tatsuo Omata (Nagoya University) for Synechococcus sp. strain PCC 7942.
REFERENCES
- 1.Aichi, M. N., Takatani, and T. Omata. 2001. Role of NtcB in activation of nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 183:5840-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allakhverdiev, S. I., Y. Nishiyama, I. Suzuki, Y. Tasaka, and N. Murata. 1999. Genetic engineering of the unsaturation of fatty acids in membrane lipids alters the tolerance of Synechocystis to salt stress. Proc. Natl. Acad. Sci. USA 96:5862-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramley, P. M., I. Elmadfa, A. Kafatos, F. J. Kelly, Y. Manios, H. E. Roxborough, W. Schuch, P. J. A. Sheehy, and K. H. Wagner. 2000. Vitamin E. J. Sci. Food Agric. 80:913-938. [Google Scholar]
- 4.Burton, G. W., and M. G. Trader. 1990. Vitamin E: antioxidant activity, biokinetics, and bioavailability. Annu. Rev. Nutr. 10:357-382. [DOI] [PubMed] [Google Scholar]
- 5.Collakova, E., and D. DellaPenna. 2001. Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 127:1113-1124. [PMC free article] [PubMed] [Google Scholar]
- 6.Dähnhardt, D., J. Falk, J. Appel, T. A. W. van der Kooij, R., Schulz-Friedrich, and K. Krupinska. 2002. The hydroxyphenypyruvate dioxygenase from Synechocystis sp. PCC 6803 is not required for plastoquinone biosynthesis. FEBS Lett. 523:177-181. [DOI] [PubMed] [Google Scholar]
- 7.David, C., A. Daro, E. Szalai, T. Atarhouch, and M. Mergeay. 1996. Formation of polymeric pigments in the presence of bacteria and comparison with chemical oxidative coupling. II. Catabolism of tyrosine and hydroxyphenylacetic acid by Alcaligenes eutrophus CH34 and mutants. Eur. Polym. J. 32:669-679. [Google Scholar]
- 8.Denoya, C. D., D. D. Skinner, and M. R. Morgenstern. 1994. A Streptomyces avermitilis gene encoding a 4-hydroxyphenylpyruvic acid dioxygenase-like protein that directs the production of homogentisic acid and an ochronotic pigment in Escherichia coli. J. Bacteriol. 176:5312-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elhai, J., and C. P. Wolk. 1988. Conjugative transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed] [Google Scholar]
- 10.Falk, J., G. Andersen, B. Kernebeck, and K. Krupinska. 2003. Constitutive overexpression of barley 4-hydroxyphenylpyruvate dioxygenase in tobacco results in elevation of the vitamin E content in seeds but not in leaves. FEBS Lett. 540:35-40. [DOI] [PubMed] [Google Scholar]
- 11.Flores, E., and A. Herrero. 1994. Assimilatory nitrogen metabolism and its regulation, p. 487-517. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 12.Frias, J. E., E. Flores, and A. Herrero. 1997. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179:477-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frias, J. E., E. Flores, and A. Herrero. 2000. Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and BtcB (LysR family) transcription factors. Mol. Microbiol. 38:613-625. [DOI] [PubMed] [Google Scholar]
- 14.Gambetta, G. A., and J. C. Lagarias. 2001. Genetic engineering of phytochrome biosynthesis in bacteria. Proc. Natl. Acad. Sci. USA 98:10566-10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia, I., M. Rodgers, R. Pepin, T. F. Hsieh, and M. Matringe. 1999. Characterization and subcellular compartmentation of recombinant 4-hydroxyphenylpyruvate dioxygenase from Arabidopsis in transgenic tobacco. Plant Physiol. 119:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golden, S. 1994. Light-responsive gene expression and the biochemistry of the photosystem II reaction center, p. 693-714. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 17.Graßes, T., B. Grimm, O. Koroleva, and P. Jahns. 2001. Loss of α-tocopherol in tobacco plants with decreased geranylgeranyl reductase activity does not modify photosynthesis in optimal growth conditions but increases sensitivity to high-light stress. Planta 213:620-628. [DOI] [PubMed] [Google Scholar]
- 18.Havaux, M., C. Lütz, and B. Grimm. 2003. Chloroplast membrane photostability in chlP transgenic tobacco plants deficient in tocopherols. Plant Physiol. 132:300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko, T. S., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada. M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi, H., M. Aichi, I. Suzuki, and T. Omata. 1996. Positive regulation by nitrite of the nitrate assimilation operon in the cyanobacteria Synchococcus sp. strain PCC 7942 and Plectonema boryanum. J. Bacteriol. 178:5822-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagarde, D., L. Beuf, and W. Vermaas. 2000. Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 66:64-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luque, I., E. Flores, and A. Herrero. 1993. Nitrite reductase gene from Synechococcus sp. PCC 7942: homology between cyanobacterial and higher-plant nitrite reductase. Plant Mol. Biol. 21:1201-1205. [DOI] [PubMed] [Google Scholar]
- 23.Luque, I., E. Flores, and A. Herrero. 1994. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 13:2862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda, S.-I., Y. Kawaguchi, T. A. Ohe, and T. Omata. 1998. cis-Acting sequences required for NtcB-dependent, nitrite-responsive positive regulation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 180:4080-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marraccini, P., S. Bulteau, C. Cassier-Chauvat, P. Mermet-Bouvier, and F. Chauvat. 1993. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 23:905-909. [DOI] [PubMed] [Google Scholar]
- 26.Mohamed, A., and C. Jansson. 1989. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 13:693-700. [DOI] [PubMed] [Google Scholar]
- 27.Munné-Bosch, S., and L. Alegre. 2002. The function of tocopherols and tocotrienols in plants. Crit. Rev. Plant Sci. 21:31-57. [Google Scholar]
- 28.Ng, W.-O., R. Zentella, Y. Wang, J. S. A. Taylor, and H. B. Pakrasi. 2000. phrA, the major photoreactivating factor in the cyanobacterium Synechocystis sp. strain PCC 6803 codes for cyclobutane-pyrimidine-dimer-specific DNA photolyase. Arch. Microbiol. 173:412-417. [DOI] [PubMed] [Google Scholar]
- 29.Omata, T., X. Andriesse, and A. Hirano. 1993. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. strain PCC 7942. Mol. Gen. Genet. 236:193-202. [DOI] [PubMed] [Google Scholar]
- 30.Prentki, P., and H. M. Kirsch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 31.Rippert, P., C. Scimemi, M. Dubald, and M. Matringe. 2004. Engineering plant shikimate pathway for production of tocotrienol and improving herbicide resistance. Plant Physiol. 134:92-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubio, L. M., A. Herrero, and E. Flores. 1996. A cyanobacterial narB gene encodes a ferrodoxin-dependent nitrate reductase. Plant Mol. Biol. 30:845-850. [DOI] [PubMed] [Google Scholar]
- 33.Savidge, B., J. D. Weiss, Y.-H. H. Wong, M. W. Lassner, T. A. Mitsky, C. K. Shewmaker, D. Post-Beittenmiller, and H. E. Valentin. 2002. Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis PCC 6803 and Arabidopsis. Plant Physiol. 129:321-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, and M. Bagdasarian. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 35.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-833. [PubMed] [Google Scholar]
- 36.Secor, J. 1994. Inhibition of barnyardgrass 4-hydroxyphenylpyruvate dioxygenase by sulcotrione. Plant Physiol. 106:1429-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanier, R. Y., R. Kunisawa, M. Mandel, and G. Cohen-Bazire. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35:171-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, I., T. Sugiyama, and T. Omata. 1993. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 34:1311-1320. [Google Scholar]
- 39.Suzuki, I., H. Kikuchi, S. Nakanishi, Y. Fujita, T. Sugiyama, and T. Omata. 1995. A novel nitrite reductase gene from the cyanobacterium Plectonema boryanum. J. Bacteriol. 177:6137-6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trebst, A., B. Depka, and H. Holländer-Czytko. 2002. A specific role for tocopherol and of chemical singlet oxygen quenchers in the maintenance of photosystem II structure and function in Chlamydomonas reinhardtii. FEBS Lett. 516:156-160. [DOI] [PubMed] [Google Scholar]
- 42.Tsegaye, Y., D. K. Shintani, and D. DellaPenna. 2002. Overexpression of the enzyme p-hydroxyphenylpyruvate dioxygenase in Arabidopsis and its relation to tocopherol biosynthesis. Plant Physiol. Biochem. 40:913-920. [Google Scholar]
- 43.Valentin, H. E., and T. A. Mitzky. November 2002. TYRA genes and uses thereof. International patent application WO 2/089561.
- 44.Williams, J. G. K. 1988. Construction of specific mutations in Photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167:766-778. [Google Scholar]