Abstract
To study the tempo and pattern of mitochondrial gene loss in plants, DNAs from 280 genera of flowering plants were surveyed for the presence or absence of 40 mitochondrial protein genes by Southern blot hybridization. All 14 ribosomal protein genes and both sdh genes have been lost from the mitochondrial genome many times (6 to 42) during angiosperm evolution, whereas only two losses were detected among the other 24 genes. The gene losses have a very patchy phylogenetic distribution, with periods of stasis followed by bursts of loss in certain lineages. Most of the oldest groups of angiosperms are still mired in a prolonged stasis in mitochondrial gene content, containing nearly the same set of genes as their algal ancestors more than a billion years ago. In sharp contrast, other plants have rapidly lost many or all of their 16 mitochondrial ribosomal protein and sdh genes, thereby converging on a reduced gene content more like that of an animal or fungus than a typical plant. In these and many lineages with more modest numbers of losses, the rate of ribosomal protein and sdh gene loss exceeds, sometimes greatly, the rate of mitochondrial synonymous substitutions. Most of these mitochondrial gene losses are probably the consequence of gene transfer to the nucleus; thus, rates of functional gene transfer also may vary dramatically in angiosperms.
Mitochondrial genomes exhibit a 20-fold range in protein gene content, from only three in the virtually extinct mtDNA of Plasmodium and other apicomplexans to 61 in Reclinonomas (1, 2). However, even Reclinomonas mtDNA encodes but a small fraction of the proteins encoded by the bacterial progenitor of the mitochondrion (3). This finding implies that the great majority of the original set of mitochondrial genes was either transferred to the nucleus or lost entirely from the cell early in eukaryotic evolution, before the emergence of essentially all extant lineages of eukaryotes. Mitochondrial gene loss and functional gene transfer to the nucleus all but ceased in the common ancestor of animals, more than 600 million years ago, as the many sequenced animal mtDNAs all contain the same 13 protein genes (occasionally 12 genes; ref. 4). Although functional gene transfer has ceased in animals, pseudogenes of mitochondrial origin are common in animal nuclear genomes (reviewed in ref. 5). The protein gene content of fungal mtDNAs is nearly as reduced (with two additional genes) and as invariant as in animals (1, 2, 6). Only one case of fungal gene transfer is known (7), and gene loss is largely restricted to the loss in certain yeasts of all NADH dehydrogenase genes (1, 6) through loss of the protein complex, rather than gene transfer.
Plant mtDNAs contain many more protein genes than do animals or fungi, 30–39 in the case of the three sequenced plant mtDNAs (8–10). Within angiosperms, mitochondrial gene loss and gene transfer are relatively frequent, ongoing phenomena. The completely sequenced mtDNAs of Arabidopsis and sugar beet are each missing, entirely or in part, 9–10 genes that are present in other angiosperm mtDNAs (9, 10). In Arabidopsis, most of these genes have been relocated to the mostly sequenced nuclear genome (ref. 11, see Discussion). Southern hybridizations reveal additional gene losses in several other flowering plants (12–19), many of which result from gene transfer to the nucleus.
To comprehensively examine the tempo and pattern of gene loss in angiosperm mtDNAs, we recently undertook a Southern blot hybridization survey of DNAs from 280 genera of flowering plants, representing 191 families (20). This survey was made possible by the very low rate of nucleotide substitutions in almost all angiosperm mtDNAs (20–22). So far, we have discovered remarkably frequent loss of all four mitochondrial genes surveyed, the ribosomal protein genes rps10 (23) and rpl2 (24) and the succinate dehydrogenase genes sdh3 and sdh4 (25). Here, we greatly expand this survey to include the other 36 protein-coding genes that have been found in the mtDNA of at least one angiosperm. We find that all 14 ribosomal protein genes have been lost frequently, but that sdh3 and sdh4 are anomalous among respiratory genes, with only two losses detected among the other 19 respiratory genes. Our results also reveal a surprisingly punctuated tempo of mitochondrial gene loss (and probably gene transfer to the nucleus) in angiosperms. Certain lineages have rapidly lost most or all of their 16 ribosomal protein and sdh genes, at rates that greatly exceed the mitochondrial synonymous substitution rate, whereas other lineages have maintained a constant set of mitochondrial genes for hundreds of millions of years.
Materials and Methods
Total DNAs were extracted as in ref. 26. Several sets of pseudoreplicate filter blots were made, each set containing one digest (with either BamHI or HindIII) of each of the DNAs. DNAs from cotton, tomato, and soybean were surveyed separately from the other 277 plants. Details of blotting and hybridization procedures are in ref. 25.
Methods used for isolating and sequencing cox1 genes from Lachnocaulon, Goodenia, and Phlox are given as supporting information on the PNAS web site, www.pnas.org, as are accession numbers of nuclear rps1 and rps19 sequences extracted from the National Center for Biotechnology Information expressed sequence tag (EST) databases.
KL, defined as the number of gene losses per locus in a pairwise comparison of taxa, was calculated as follows: For a given pair of taxa, the total number of separate losses (i.e., those absences not resulting from a shared loss in their common ancestor) of ribosomal protein and sdh genes in both species was divided by the number of these genes present in the last common ancestor of the species pair. The pairwise proportion of synonymous substitutions (KS) in the mitochondrial cox1 gene was calculated by using mega version 2.0 (www.megasoftware.net), with a Jukes–Cantor correction for multiple hits by the Nei–Gojobori distance method.
Results
Ribosomal Protein and sdh Genes Are Lost Frequently, Other Respiratory-Related Genes Rarely.
Southern blots containing total DNA from 280 angiosperms were hybridized with probes for the 40 protein genes known to be present in at least one angiosperm mtDNA (Table 1; for details of the probes, see Table 3, which is published as supporting information on the PNAS web site). As with most respiratory gene probes, the cox2 probe shown in Fig. 1 hybridized well to all DNAs shown, i.e., the lane-to-lane variations in hybridization intensity were reproducible across these probes (also see refs. 23–25). We therefore interpret these variations as reflecting differences in the amount of mtDNA present in each lane and conclude that each mtDNA probably contains an intact copy of cox2. In contrast, the six probes shown for ribosomal protein genes failed to hybridize to several to most lanes (Fig. 1). Mitochondrial gene loss was inferred if there was no detectable or severely diminished hybridization on an overexposed autoradiograph relative to two controls: good hybridization to the DNA in question on the same filter by using other probes and good hybridization to other DNAs with the probe in question. This inference assumes that mitochondrial genomes are in high copy number and conserved in sequence (see ref. 20 for two exceptionally divergent genomes) relative to nuclear genes of single or low copy number. This approach was validated by comparing to sequenced mtDNAs—blots and sequences show the same seven complete gene losses for Arabidopsis (9) and nine losses for sugar beet (10)—and by examining hybridization intensities to known mitochondrial pseudogene fragments (see below). Also, follow-up sequencing of the nuclear rps10 gene from 16 lineages of inferred mitochondrial gene loss did not reveal any mitochondrial copies of rps10 (23).
Table 1.
Inferred numbers of mitochondrial gene losses among 280 angiosperms
| Gene | # of losses | Gene | # of losses |
|---|---|---|---|
| rpl2 | 41 | atp1 | 0 |
| rpl5 | 19 | atp6 | 0 |
| rpl16 | 15 | atp8 | 1 |
| rps1 | 33 | atp9 | 0 |
| rps2 | 8* | ccb2 | 0 |
| rps3 | 7 | ccb3 | 0 |
| rps4 | 7 | ccb6c | 0 |
| rps7 | 42 | ccb6n | 0 |
| rps10 | 26 | cob | 0 |
| rps11 | 14* | cox1 | 0 |
| rps12 | 6 | cox2 | 1 |
| rps13 | 30 | cox3 | 0 |
| rps14 | 27 | nad1 | 0 |
| rps19 | 39 | nad2 | 0 |
| nad3 | 0 | ||
| sdh3 | 40 | nad4 | 0 |
| sdh4 | 19 | nad4L | 0 |
| nad5 | 0 | ||
| matR | 0 | nad6 | 0 |
| mtt2 | 0 | nad7 | 0 |
| orf25 | 0 | nad9 | 0 |
Includes one deep loss (see text).
Figure 1.
Southern blot hybridizations of 91 angiosperm DNAs (of 280 examined in total) with probes for seven mitochondrial genes.
The inferred gene absences were plotted on a molecular phylogeny of the 280 plants to infer the number and phylogenetic timing of gene losses (Fig. 2; Table 1). A single gene loss was inferred in the common ancestor of each clade whose members all are missing the gene in question. The 40 mitochondrial genes fall into two groups with respect to frequency of loss (Table 1). Twenty-four genes, 21 of which function in respiration either directly or indirectly (right half of Table 1), show no losses other than the well-studied loss of cox2 in legumes (12, 13, 27, 28) and the loss of atp8 in Allium. These include all 14 respiratory genes that are usually, if not invariantly, present in mtDNA across eukaryotes (1, 2). In stark contrast, all 14 ribosomal protein genes and both sdh genes have been lost several to many times (6–42 losses per gene, mean = 23.3, median = 22.5).
Figure 2.
Inferred losses of mitochondrial ribosomal protein and sdh genes among 280 angiosperms (species names are listed at www.bio.indiana.edu/∼palmerlab.html). Bullets indicate absence of hybridization to the probe in question. Numbers on the branches of the tree indicate inferred numbers of losses per internode. The three Gs indicate hybridizations scored as gains after ancient losses (see Results for explanation). Vertical names with brackets indicate plant orders. (A) Magnoliids (Bottom, Ceratophyllum and below), monocots (Middle, Acorus through Hordeum), and basal eudicots (Top, Dicentra and above). (B) Rosids (Middle and Top, Mytilaria and above) and basal eudicots (Bottom, Tetracentron and below). (C) Asterids (Middle and Top, Cornus and above) and caryophyllids (Bottom, Bougainvillea and below). The tree of the 280 examined angiosperms was constructed by consulting various recent molecular systematics studies. The following were the primary sources of interfamilial relationships: asterids (48) except for Lamiales (49), rosids and caryophyllids (50), basal eudicots (51), monocots (52), and magnoliids (53). Intra-familial relationships were based on various sources listed at www.bio.indiana.edu/∼palmerlab.html. rps10 data are from ref. 23. rpl2 data are for the 5′ portion of the gene (24). sdh3 and sdh4 data are from ref. 25.
Phylogenetic Depth and Pattern of Mitochondrial Gene Losses.
The inferred gene losses display a range of phylogenetic depth among angiosperms (Fig. 2). Most losses are quite restricted in phylogenetic distribution and are thus relatively recent (Fig. 2). Fully 74% of the 373 losses shown in Fig. 2 occur on terminal branches (i.e., are restricted to a single species surveyed). Likewise, most of the losses are restricted to a single family or even a single genus within a multiply sampled family. Only a small number of losses are moderately deep and broad, encompassing several related families, e.g., rpl2 throughout the Apiales and Asterales (Fig. 2C Top), rps10 in a large portion of the Caryophyllales (Fig. 2C Bottom), and sdh3 throughout the Asparagales and also the Poales (Fig. 2A Middle and Top).
Some of the general patterns of gene losses are well illustrated within the Poales (Fig. 2A, second group from top). The most striking example of gene losses is found in Lachnocaulon, inferred to have lost all 16 ribosomal protein and sdh genes. Maize (Zea) has lost eight of these 16 genes, compared with 3–6 losses among the other three grasses examined. In contrast, Typha has lost only one gene. There is a deep loss of sdh3, encompassing the entire order, whereas most losses (20 of 26) are restricted to a single species.
There appear to be two very deep, ancient losses, both with the same curious twist. Probes for rps2 and rps11 did not hybridize to the DNAs of virtually all core eudicots (a group comprising 179 of the 280 angiosperms in our survey), suggesting ancient loss of both genes (Fig. 2 B and C). Mitochondrial-derived copies of both rps2 and rps11 have been discovered in the nucleus of Arabidopsis, soybean, and tomato (ref. 29; Table 4, which is published as supporting information on the PNAS web site), suggesting that gene loss followed functional transfer to the nucleus. The curious twist: Despite the lack of hybridization to virtually all core eudicot DNAs, the rps2 probe did hybridize strongly to DNA from Actinidia, a highly derived eudicot (Fig. 2C), as did the rps11 probe to DNAs from Betula (Fig. 2B) and Lonicera (Fig. 2C). We have investigated Actinidia and found that its mtDNA does have an rps2 sequence, but that this sequence may have been regained, by lateral transfer from a distantly related, noneudicot mtDNA (unpublished data). We tentatively interpret all three very isolated cases of rps2 and rps11 hybridization as subsequent regains (marked as G in Fig. 2) by these plants after an early loss; this inference makes for a conservative interpretation of the numbers of losses of these two genes.
All but one high-loss genes were lost with similar frequency (within a factor of 2) in rosids and asterids, the two major groups of eudicots (Fig. 2, Table 5, which is published as supporting information on the PNAS web site). Rps13 is the glaring exception: 24 losses of rps13 were inferred among the 84 rosids examined vs. a single loss among the 77 asterids. We have shown that the many rps13 losses in rosids probably all trace back to a single event—a duplication of the nuclear gene for chloroplast RPS13 and substitution of its gene product for RPS13 in the mitochondrion—in the common ancestor of most rosids (54).
Variable Tempo of Gene Losses: High Rates of Loss in Some Lineages, Stasis in Others.
The gene absences are not distributed evenly across angiosperms. Seventy-five percent of the plants are missing only 0–4 of the 40 protein genes from their mtDNAs, and many of these are missing only the two genes (rps2 and rps11) that were lost very deeply in eudicot evolution. The magnoliids, which include all of the earliest angiosperms, show very little loss; 27 of the 33 magnoliids have not lost any genes, and the other six have lost but one or two genes. In contrast, 25 of 280 plants (9%) are missing eight or more genes, and 10 plants (4%), representing seven very distinct lineages, have lost 12 or more genes (Fig. 2).
The gene losses appear to be nonrandomly clustered on the angiosperm evolutionary tree (Fig. 2; Table 6, which is published as supporting information on the PNAS web site). Among the asterids, a total of 106 gene losses were inferred to account for the observed distribution of gene absences. Of these losses, fully 42 (40%) map to but six of the 136 branches on the asterid portion of the tree (Fig. 2C). These are the terminal branches leading to Phlox (11 losses), Goodenia (eight losses), Callitriche (seven losses), Clerodendrum (six losses), Daucus (five losses), and Cyrilla (five losses). In contrast, 91 (67%) of the asterid branches show no losses and 25 (18%) have one loss. The monocots reveal an even starker contrast. Of 85 gene losses in monocots, fully 34 (40%) map to but three of the 91 monocot branches (Fig. 2A). These are the terminal branches leading to Lachnocaulon (13 losses) and Allium (11 losses), and the two-steps-subterminal branch leading to Anacharis/Vallisneria/Echinodorus (10 losses). Conversely, 55 (60%) of the monocot branches show no losses and 20 (22%) have only one loss.
An illuminating perspective on the frequency of gene loss comes from comparing relative rates of mitochondrial gene loss and synonymous substitution. By analogy to the commonly used parameter KS (the number of substitutions per synonymous site in a pairwise comparison), we calculated KL (the number of losses per gene in a pairwise comparison) for the 16 ribosomal protein and sdh genes for various pairs of taxa and compared this result to KS calculated for the same taxa for the mitochondrial gene cox1. Cox1 is representative of plant mitochondrial genes in silent substitution rate (21) and was chosen because it is widely sequenced among angiosperms. The well-known (21, 22) rarity of synonymous substitutions in almost all angiosperm mtDNAs is illustrated in Fig. 3 for the cox1 gene relative to the frequent loss of these 16 mitochondrial genes.
Figure 3.
Comparison of patterns of mitochondrial gene absences and synonymous site differences among selected taxa. Shown are 85 4-fold degenerate, third-position silent sites in cox1 codons specifying amino acids that are invariant among the 26 angiosperms compared. Dots indicate nucleotides identical to the top sequence (Magnolia). The distribution of gene absences and the tree topology are extracted from Fig. 2. Eup. indicates Euphorbiaceae, Ros. indicates Rosales, and b.e. indicates basal eudicots.
Pairs of taxa for KL/KS comparisons were selected from various groups of angiosperms and include taxa with many losses, few or no losses, and a moderate number of losses (Fig. 3). For all but three of the 42 pairs of taxa selected, KL is higher than KS (Table 2); each of these three pairs had a total of only one or no derived losses. In comparing two high-loss plants, especially within an order or family, the KL/KS ratio was often very high (≈10–36). Thus, in all but a few of the comparisons, the per-gene rate of ribosomal protein and sdh gene loss exceeds, often greatly, the per-site rate of silent nucleotide substitution. In other words, in these lineages, if a particular silent site in a ribosomal protein or sdh gene is considered, it is less likely that the site will undergo a nucleotide substitution than that the gene itself will be lost (and transferred to the nucleus, see Discussion). Even if these comparisons are extended from the 16 high-loss genes to all 40 mitochondrial genes surveyed, the KL/KS ratios is reduced by only a factor of 2.5 and KL still exceeds KS in most cases shown in Table 2.
Table 2.
Relative rates of gene losses (KL) compared to silent substitutions (KS)
| KL | KS | KL/KS | |
|---|---|---|---|
| Monocots | |||
| Zea–Lachnocaulon | 1.39 | 0.14 | 9.6 |
| Zea–Maranta | 0.50 | 0.11 | 4.5 |
| Lachnocaulon–Maranta | 0.88 | 0.10 | 9.0 |
| Musa–Maranta | 0.06 | 0.13 | 0.5 |
| Philodendron–Maranta | 0.31 | 0.06 | 5.2 |
| Lachnocaulon–Philodendron | 1.31 | 0.10 | 13.4 |
| Acorus–Philodendron | 0.56 | 0.31 | 1.8 |
| Zea–Philodendron | 0.81 | 0.09 | 8.7 |
| Asterids | |||
| Phlox–Goodenia | 1.57 | 0.16 | 9.9 |
| Phlox–Hebe | 0.79 | 0.12 | 6.7 |
| Goodenia–Ilex | 0.86 | 0.16 | 5.5 |
| Goodenia–Hydrocotyle | 0.98 | 0.16 | 6.3 |
| Ilex–Hydrocotyle | 0.21 | 0.04 | 5.1 |
| Clerodendrum–Phlox | 1.36 | 0.13 | 10.7 |
| Goodenia–Lamium | 1.36 | 0.19 | 7.0 |
| Phlox–Ilex | 0.71 | 0.08 | 8.5 |
| Lamium–Ilex | 0.50 | 0.08 | 6.0 |
| Hydrocotyle–Lamium | 0.71 | 0.07 | 10.3 |
| Ilex–Sanchezia | 0.0 | 0.08 | 0.0 |
| Rosales | |||
| Elaeagnus–Rhamnus | 0.71 | 0.03 | 28.6 |
| Rhamnus–Hovenia | 0.21 | 0.04 | 5.6 |
| Hovenia–Elaeagnus | 0.64 | 0.06 | 11.5 |
| Lamiales | |||
| Lamium–Scutellaria | 0.36 | 0.03 | 14.6 |
| Scutellaria–Clerodendrum | 0.55 | 0.03 | 21.8 |
| Clerodendrum–Lamium | 0.91 | 0.03 | 36.4 |
| Hebe–Callitriche | 0.54 | 0.03 | 15.8 |
| Sanchezia–Clerodendrum | 0.64 | 0.04 | 16.9 |
| Hebe–Sanchezia | 0.07 | 0.05 | 1.5 |
| Callitriche–Lamium | 1.07 | 0.04 | 24.9 |
| Euphorbiaceae | |||
| Hevea–Croton | 0.43 | 0.03 | 14.3 |
| Croton–Euphorbia | 0.36 | 0.04 | 8.3 |
| Euphorbia–Hevea | 0.36 | 0.01 | 27.5 |
| Angiosperms | |||
| Buxus–Grevillea | 0.38 | 0.06 | 6.7 |
| Musa–Grevillea | 0.06 | 0.20 | 0.3 |
| Beta–Rhamnus | 0.64 | 0.13 | 5.1 |
| Lachnocaulon–Gardenia | 1.88 | 0.26 | 7.2 |
| Elaeagnus–Clerodendrum | 1.21 | 0.02 | 9.6 |
| Goodenia–Elaeagnus | 1.43 | 0.16 | 9.0 |
| Grevillea–Ilex | 0.13 | 0.08 | 1.6 |
| Scutellaria–Hevea | 0.29 | 0.09 | 3.1 |
| Zea–Grevillea | 0.50 | 0.12 | 4.0 |
| Beta–Phlox | 1.21 | 0.21 | 5.8 |
Strengths and Limitations of the Southern Hybridization Survey: The Pseudogene Problem.
By using Southern blots, we were able to survey mtDNAs from a few hundred diverse angiosperms for the presence or absence of 40 protein genes. Although genome sequencing obviously would give more definitive insights into which genes are either intact, present as pseudogenes, or entirely absent from these mtDNAs, sequencing is entirely unfeasible with so many and such diverse plants. At several hundred kb per genome (9, 10, 30), 280 angiosperm mtDNAs are roughly equivalent to the entire length of the Arabidopsis nuclear genome, and purifying enough mtDNA for sequencing would be difficult, if not impossible, for many of these plants.
The major weakness of the Southern approach involves mitochondrial pseudogenes. These occasionally occur in plants and are difficult to score properly by a hybridization approach. Pseudogenes that are nearly intact, such as rps14 and sdh4 in Arabidopsis (9, 31), will be counted as “intact” genes by our scoring and could be properly diagnosed only by sequencing. Conversely, but much less problematic for our purposes, pseudogenes that are present as only small fragments of a gene will be scored as absent in Fig. 2 (no or significantly reduced hybridization). In some cases, trace pseudogenes were “successfully” detected as such by noting substantially reduced hybridization to DNA from a species known to contain only a fragment of a gene in the mitochondrion [e.g., rps12 in Oenothera (32) and rps19 in Arabidopsis (9, 33)].
Depending on the phylogenetic context, undetected pseudogenes can lead to either overestimates or underestimates of the number of functional gene losses. Gene losses will be overestimated when separate losses as inferred from blots trace back to a single deeper loss of mitochondrial gene function, with one or more taxa within the group in question having a nearly intact pseudogene scored as present. Rps1 in tomato mtDNA is a good candidate for such a pseudogene (see next section); if so, then the three separate losses of rps1 scored for Capsicum, Nicotiana, and Petunia (Fig. 2C) would collapse, at minimum, to a single loss in the common ancestor of these four Solanaceae species. An extreme and probably exceptional example of retained pseudogenes and potential overestimates of gene losses involves the 3′ end of rpl2, as described (24). Conversely, gene losses will be underestimated when nearly intact pseudogenes occur within a group, all of whose members were scored positively for the gene in question. Examples here are rps14 and sdh4 in Arabidopsis (9, 31). Undetected pseudogenes probably lead to inflated KL/KS estimates within certain families or orders (e.g., Euphorbiaceae, Lamiales, and Rosales), but to deflated estimates on a broader scale, across monocots, asterids, and all angiosperms.
The problem of undetected pseudogenes means that all of our estimates of gene loss are tinged with a level of uncertainty. We do not, however, regard this as a serious problem that compromises any of the major conclusions of this study, both because of the offsetting nature of the misestimates of gene loss they cause and because pseudogenes do not appear to be that common relative to functional genes (9, 10, 23). Limited sequencing might be performed in the future to diagnose pseudogenes in clades for which the loss patterns raise the possibility of pseudogenes (e.g., see ref. 24).
rps19 and rps1 Have Been Transferred to the Nucleus in Diverse Angiosperms.
The large number of losses of all 14 ribosomal protein genes raises the possibility that each gene may have been transferred to the nucleus many times, as recently shown for one ribosomal protein gene (rps10; ref. 23) and both sdh genes (25). Here we evaluate fresh data on transferred sequences for two other ribosomal protein genes.
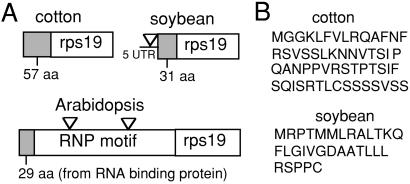
The ribosomal protein gene rps19 is inferred to have been lost 39 times among the surveyed angiosperms (Table 1; Fig. 2). Rps19 was isolated previously from the nucleus of Arabidopsis and encodes an N-terminal extension of S19 that is homologous to small, glycine-rich RNA-binding proteins (33). We discovered transferred, nuclear genes (see additional Results, which are published as supporting information on the PNAS web site) for rps19 in maize, cotton, and two legumes by searches of National Center for Biotechnology Information EST databases. Each of these lineages represents a phylogenetically separate loss of rps19 from mtDNA. Soybean and cotton rps19 encode predicted targeting presequences of different lengths that show no evidence of homology to each other (Fig. 4), to the Arabidopsis rps19 presequence, or to any other sequences in the National Center for Biotechnology Information databases. Most likely, therefore, as explained in ref. 25, these distinct targeting sequences from three rosids were acquired from different source sequences during separate activations and, probably, separate transfers. Although the predicted presequence of maize rps19 shows no evidence of homology to those of the three rosids (Fig. 4), in itself this is fairly meaningless because orthologous mitochondrial presequences of monocot and eudicot genes have often diverged to the point of nonalignability. However, taken together with the likely separate transfers among the three rosid lineages, and the highly disjunct loss of mitochondrial rps19 in maize relative to the rosid losses (Fig. 2), the maize nuclear rps19 is probably also the result of a transfer separate from those in the rosids. Overall, then, we conclude that each of the four rps19 genes depicted in Fig. 4 is probably the result of a separate transfer event, although, as discussed in ref. 25 for similar data for transferred sdh genes, other scenarios involving only one or two, more ancient transfers cannot be ruled out.
Figure 4.
Nuclear rps19 gene structures and predicted presequences. (A) Gene structure diagrams. Triangles indicate positions of introns. The Arabidopsis and soybean sequences are genomic; that of cotton is a cDNA. UTR, untranslated region. (B) Presequences of rps19 from cotton and soybean are predicted by mitoprot (34).
Rps1 is inferred to have been lost 33 times among the surveyed angiosperms. Searches of the National Center for Biotechnology Information EST databases revealed transferred, nuclear (see additional Results on the PNAS web site) genes for rps1 in tomato, cotton, and three legumes. Tomato RPS1 and Medicago RPS1 have putative mitochondrial targeting presequences of 38 aa and 13 aa, respectively, as predicted by mitoprot (34). These presequences are not alignable, suggesting they may have arisen by means of separate acquisitions and that the genes may be the products of independent transfer events. Because the cotton rps1 EST appears to be incomplete, lacking the 5′ end, including a potential targeting element, the independence of its transfer is more difficult to assess.
Interestingly, cotton, soybean, and Medicago all retain a strongly hybridizing rps19 sequence in their mtDNAs, and tomato and Medicago mtDNAs both have a strongly hybridizing rps1 sequence (Fig. 2). Whether these sequences are intact, much less expressed, in any of these mitochondria requires further study. Soybean and various related legumes are already known to contain intact copies of the cox2 gene in both the mitochondrial and nuclear genomes, with both genes expressed in a subset of these taxa (28).
Discussion
Do the Many Gene Losses Reflect Many Gene Transfers to the Nucleus?
Any one of the approximately 375 mitochondrial gene losses inferred in this study could be explained by either transfer of the gene to the nucleus, functional substitution by a related protein, or loss of the protein and its function from the plant. Evidence is building, that many, probably most, of the gene losses result from gene transfer to the nucleus. Most impressively, in the case of maize, all eight genes that were inferred to be lost from its mtDNA by our blot surveys have been discovered as recently transferred genes in the nucleus (refs. 23–25 and 35; this study; Table 4). The mostly sequenced nuclear genome of Arabidopsis (11) contains transferred copies of eight angiosperm mitochondrial genes (refs. 11, 24, 25, 29, 33, 36, and 37; Table 4), two of which also still reside in its mtDNA as pseudogenes, but lacks transferred copies of two missing mitochondrial genes (see next paragraph). For each of four other, disparately related plants (tomato, cotton, soybean, and rice) for which extensive EST sequences are available, several transferred genes also have been identified (Table 4). Finally, in the case of rps10, the most extensively characterized of the high-loss genes, transferred copies of the gene have been recovered from all 16 of its 26 identified loss lineages that were examined for a nuclear copy of rps10 (23), and multiple transferred genes have been documented for a few other high-loss genes (see below).
For only three genes do we have evidence to suspect that some losses may not reflect transfer, but either substitution or loss of the protein. One gene is rps7, which despite 41 mitochondrial losses has not yet been isolated from the nucleus of any angiosperm, including two mitochondrial loss lineages (soybean and tomato) for which extensive EST data are available. Also, concerted efforts to isolate rps7 from the nucleus of two loss lineages (soybean and Podophyllum) have failed (ref. 38; L. Bonen, personal communication). A second gene with many losses, rps1, has not been identified in the mostly sequenced nuclear genome of Arabidopsis (11). Although rps1 has been found transferred to the nucleus in tomato, legumes, and cotton (see Results), it may have suffered a different fate in Arabidopsis. Finally, as described in Results, the unusual concentration of losses of rps13 in rosids (including Arabidopsis) probably reflects a single gene substitution event in the common ancestor of rosids. This is the only known case of gene substitution in the evolution of these 16 high-loss genes and contrasts with the many case of gene transfer documented for these genes.
Although gene transfer seems clearly to be the predominant explanation for mitochondrial gene loss in angiosperms, the question still remains as to how many transfer events are responsible for the hundreds of losses detected in this study. At one extreme, a single relatively ancient transfer, say in the common ancestor of angiosperms, could have given rise to all of the many subsequent, hence dependent, mitochondrial gene losses inferred for a high-loss gene. At the other extreme, the losses could all be independent, resulting from separate events of gene transfer. For rps10, we have shown that many, if not all, of its 26 losses are the result of separate, independent transfers to the nucleus, each occurring relatively recently during angiosperm evolution (23). There is also evidence for four separate transfers of sdh3 (25) and rps19 (ref. 33; this study), three separate transfers of sdh4 (25), rpl2 (24), rps14 (refs. 35, 37, and 39; this study, see Table 4), and rps1 (this study), and two separate transfers of rps11 (40, 41). Given the many additional, phylogenetically scattered and recent losses of these genes and other ribosomal protein genes, it is likely that most of the 16 high-loss genes have been transferred to the nucleus repeatedly during the course of angiosperm evolution. This being the case, then the patterns and rates of mitochondrial gene loss depicted and summarized in Figs. 2 and 3 and Tables 1 and 2 can reasonably be used as a rough approximation for patterns and rates of functional gene transfer to the nucleus.
Stasis in Gene Content During Much of Plant Evolution and Bursts of Recent Losses and Transfers.
The 33 basal angiosperms (bottom taxa in Fig. 2A), which represent multiple ancient lineages, exhibit extreme stasis in mitochondrial gene content. Only six losses (Fig. 2A) have occurred among these many old lineages, and most of these plants have kept all of their mitochondrial genes, i.e., they have preserved the set of ≈40 protein genes that characterized the common ancestor of all angiosperms. This angiosperm stasis is just part of a prolonged stasis extending all of the way through land plant and green algal evolution—specifically in the lineage leading to land plants, but by no means in all green algae, some of which have sustained massive loss of mitochondrial genes (1, 2, 42)—to their common ancestry with red algae at least 1.2 billion years ago (43). One can infer that this common ancestor contained 46 protein genes in its mitochondrial genome (42, 44). Only four of these genes were lost in the 750+ million years leading to the origin of land plants, and only two more were lost in the first ca. 300 million years of land plant evolution leading to angiosperms. Nonvascular plants, insofar as represented by the fully sequenced mitochondrial genome of Marchantia, are also static. Marchantia has functionally lost and transferred to the nucleus only one of the protein genes inferred to be present in the common ancestor of land plants, and this apparently only recently, leaving a pseudogene copy in the mitochondrion (8, 45).
The contrast is striking between this prolonged stasis throughout one branch of land plant/green algal evolution, a stasis that continues in many lineages of angiosperms, and the sudden and rapid loss of many or even all of the 14 ribosomal protein genes and two sdh genes in a number of recent lineages. Some plants, such as Lachnocaulon, Allium, and Erodium, have suddenly become rather animal-like or fungal-like with respect to mitochondrial gene content. These plants have lost most or all of their ribosomal protein and sdh genes, genes that were transferred wholesale to the nucleus in the common ancestor of animals and fungi, but have retained all of the respiratory genes that also remain in animal mtDNAs [plus 10 other genes, four of which (the ccb genes) are missing from animal and fungal mtDNAs because of loss of the entire CCB complex and its functional replacement by a different pathway for cytochrome c biogenesis (46)].
Rates of gene loss and, most likely, gene transfer (see preceding section) are (or have been) extraordinarily high in these and certain other lineages. For the 16 high-loss genes, rates of loss often exceed, sometimes by an order of magnitude or more, rates of synonymous substitutions in protein genes (Table 2; Fig. 3). This comparison is admittedly skewed in the sense that almost all angiosperm mtDNAs have very low substitution rates (21, 22); still, these rates of gene loss and gene transfer are at least comparable to synonymous substitution rates in chloroplast and plant nuclear genomes (21, 22). Furthermore, it is entirely possible that greatly increased sampling within the high-loss lineages (often represented here by but a single taxa) will reveal that episodes of gene loss and transfer have occurred even more furiously than can presently be recognized.
The remarkably punctuated pattern of evolution of mitochondrial gene content in angiosperms probably is driven largely by major episodic surges in the rate of functional gene transfer. The underlying mechanism(s) responsible for highly elevated rates of gene transfer will be fascinating to elucidate. Functional gene transfer is a complex, multistep process thought to involve reverse transcription of a mitochondrial mRNA (at least for most mitochondrial genes in angiosperms; e.g., refs. 12, 23, 27, 32, and 36), mRNA or cDNA movement to the nucleus, chromosomal integration, gain of a nuclear promoter and other regulatory elements, and, usually, gain of a mitochondrial targeting presequence. Changes in the rate of any of these processes could, in principle, drive an increased rate of gene transfer.
A high rate of gene transfer could be driven by an elevated rate of reverse transcription (either inside or outside the mitochondrion), an increased supply of mitochondrial targeting peptides and regulatory elements (through either nuclear genomewide duplication, i.e., polyploidy, or specific, high-copy number amplification of one or a few nuclear genes that encode mitochondrial proteins), an increased propensity for mitochondria to fuse with the nucleus or to lyse and spill their contents into the cytoplasm, or various other factors. The discovery of many different lineages of plants that have or are experiencing high rates of gene transfer both increases the likelihood of success in unraveling underlying factors in at least certain high-rate lineages and also offers the opportunity for comparison of underlying mechanism between different lineages. Periods of pronounced stasis in mitochondrial gene content could, in theory, reflect both the absence of forces, such as a high rate of reverse transcription, that actively promote gene transfer and the elaboration of genetic incompatibilities (e.g., the evolution of nonstandard genetic codes in the mitochondrion, or of highly divergent codon usage patterns between the two genomes) that prevent functional gene transfer. Although divergence in genetic code is almost certainly a major contributor to the long-term lack of gene transfer in animals and fungi (see Introduction), there is no evidence so far that it has played a role in the long-term plant stasis that has been recently shattered in many derived lineages of angiosperms.
Gene-Specific Patterns of Mitochondrial Gene Loss.
We have documented a stark contrast in angiosperms between many losses of 16 ribosomal protein and sdh genes vs. virtually no losses of 21 other respiratory-related genes and three other genes. A commonly held explanation—the so-called “hydrophobicity hypothesis”—for why many respiratory genes are so refractory to gene transfer is that their hydrophobic, integral-membrane protein products are very difficult to successfully import and correctly assemble into mitochondria (see ref. 20 and references therein). Conversely, according to this view, relatively small, soluble proteins such as ribosomal proteins should be relatively easy to import. Some respiratory proteins may require special targeting sequences for successful import and sorting (55), whereas ribosomal proteins seem to make use of just about any potential targeting sequence (Fig. 4, e.g., refs. 23 and 40). Although hydrophobicity and, more generally, importability are probably important constraints on the transfer of some, perhaps many respiratory proteins (see ref. 56 for important recent data on cox2), other factors are undoubtedly also involved (57, 58), and the relative contributions of these factors must vary between genes and over time (see below). The succinate dehydrogenase genes sdh3 and sdh4 stand out as exceptions to the general pattern of rare transfer of respiratory genes in angiosperms (25) and have also been lost and presumably transferred many times across eukaryotic evolution (1, 2). Among ribosomal protein genes, all of which have been lost relatively frequently in angiosperms, there nonetheless seems to be a gradient/hierachy with respect to likelihood of transfer, both within angiosperms (Table 1) and on the broad scale of eukaryotic evolution (44). As with respiratory genes, this gradient is likely to reflect a gene- and lineage-specific interplay of multiple factors.
The angiosperm pattern of frequently vs. infrequently lost mitochondrial genes largely holds up across the broad sweep of eukaryotes (1, 2), but with several exceptions. All of the 16 high-loss genes in angiosperms have also been repeatedly lost in other mitochondrial lineages. Similarly, many of the respiratory genes that are invariantly present in angiosperm mtDNA are also rarely if ever lost from other mtDNAs. Most notably, cox1 and cob are present in every examined mtDNA, and nad1, nad4, and nad5 are universally mtDNA-encoded in all organisms that retain complex I in their electron transfer chain. The exceptional genes, invariantly present in angiosperm mtDNAs but lost from nonplant mtDNAs about as frequently as ribosomal protein genes (1, 2), are the four ccb genes and several respiratory genes (most prominently atp1 and nad7, and to a lesser extent nad4L and nad9). Some, perhaps most, of the ccb gene losses across eukaryotes reflect loss of the entire suite of ccb genes from these organisms and the employment of an alternate pathway for cytochrome c biogenesis (46). For atp1 (e.g., ref. 47) and the nad genes, however, the many mitochondrial losses across eukaryotes probably reflect gene transfer to the nucleus. Future studies could attempt to identify the factors that so constrain the functional transfer of these respiratory genes in plants relative to other eukaryotes.
Supplementary Material
Acknowledgments
We thank Brandon Gaut, Michael Gray, Patrick Keeling, and Ken Wolfe for critical reading of the manuscript, Claude dePamphilis for sharing the Elaeagnus cox1 sequence, and Rich Cronn for providing cotton DNA. This study was supported by National Institutes of Health Grants R01 GM-35087 (to J.D.P.) and F32 GM-17923 (to Y.-L.Q.) and United States Department of Agriculture Fellowship 95–38420-2214 and Indiana University Floyd and Ogg fellowships (to K.L.A.).
Abbreviation
- EST
expressed sequence tag
Footnotes
References
- 1.Gray M W, Lang B F, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn T G, et al. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lang B F, Gray M W, Burger G. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 3.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. Nature (London) 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 4.Boore J L. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bensasson D, Zhang D, Hartl D L, Hewitt G M. Trends Ecol Evol. 2001;16:314–321. doi: 10.1016/s0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- 6.Paquin B, Laforest M-J, Forget L, Roewer I, Wang Z, Longcore J, Lang B F. Curr Genet. 1997;31:380–395. doi: 10.1007/s002940050220. [DOI] [PubMed] [Google Scholar]
- 7.van den Boogaart P, Samallo J, Agsteribbe E. Nature (London) 1982;298:187–189. doi: 10.1038/298187a0. [DOI] [PubMed] [Google Scholar]
- 8.Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T, et al. J Mol Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- 9.Unseld M, Marienfeld J R, Brandt P, Brennicke A. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 10.Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, Mikami T. Nucleic Acids Res. 2000;28:2571–2576. doi: 10.1093/nar/28.13.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 12.Nugent J M, Palmer J D. Cell. 1991;66:473–481. doi: 10.1016/0092-8674(81)90011-8. [DOI] [PubMed] [Google Scholar]
- 13.Nugent J M, Palmer J D. In: Plant Mitochondria. Brennicke A, Kuck U, editors. New York: VCH; 1993. pp. 163–170. [Google Scholar]
- 14.Wahleithner J A, Wolstenholme D R. Nucleic Acids Res. 1988;16:6897–6913. doi: 10.1093/nar/16.14.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanlungo S, Quinones V, Moenne A, Holuigue L, Jordana X. Plant Mol Biol. 1994;25:743–749. doi: 10.1007/BF00029612. [DOI] [PubMed] [Google Scholar]
- 16.Knoop V, Ehrhardt T, Lattig K, Brennicke A. Curr Genet. 1995;27:559–564. doi: 10.1007/BF00314448. [DOI] [PubMed] [Google Scholar]
- 17.Zhuo D G, Bonen L. Mol Gen Genet. 1993;236:395–401. doi: 10.1007/BF00277139. [DOI] [PubMed] [Google Scholar]
- 18.Kubo N, Ozawa K, Hino T, Kadowaki K-I. Plant Mol Biol. 1996;31:853–862. doi: 10.1007/BF00019472. [DOI] [PubMed] [Google Scholar]
- 19.Vaitilingom M, Stupar M, Grienenberger J-M, Gualberto J M. Mol Gen Genet. 1998;258:530–537. doi: 10.1007/s004380050764. [DOI] [PubMed] [Google Scholar]
- 20.Palmer J D, Adams K L, Cho Y, Parkinson C L, Qiu Y-L, Song K. Proc Natl Acad Sci USA. 2000;97:6960–6966. doi: 10.1073/pnas.97.13.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfe K H, Li W-H, Sharp P M. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laroche J, Li P, Maggia L, Bousquet J. Proc Natl Acad Sci USA. 1997;94:5722–5727. doi: 10.1073/pnas.94.11.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams K L, Daley D O, Qiu Y-L, Whelan J, Palmer J D. Nature (London) 2000;408:354–357. doi: 10.1038/35042567. [DOI] [PubMed] [Google Scholar]
- 24.Adams K L, Ong H C, Palmer J D. Mol Biol Evol. 2001;18:2289–2297. doi: 10.1093/oxfordjournals.molbev.a003775. [DOI] [PubMed] [Google Scholar]
- 25.Adams K L, Rosenblueth M, Qiu Y-L, Palmer J D. Genetics. 2001;158:1289–1300. doi: 10.1093/genetics/158.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu Y-L, Cho Y, Cox J C, Palmer J D. Nature (London) 1998;394:671–674. doi: 10.1038/29286. [DOI] [PubMed] [Google Scholar]
- 27.Covello P S, Gray M W. EMBO J. 1992;22:3815–3820. doi: 10.1002/j.1460-2075.1992.tb05473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams K L, Song K, Roessler P, Nugent J, Doyle J L, Doyle J J, Palmer J D. Proc Natl Acad Sci USA. 1999;96:13863–13868. doi: 10.1073/pnas.96.24.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrotta G, Grienenberger J M, Gualberto J M. In: Plant Mitochondria: From Gene to Function. Moller I M, Gardestrom P, Glimelius K, Glaser E, editors. Leiden: Backhuys; 1998. pp. 37–41. [Google Scholar]
- 30.Levings C S, III, Brown G G. Cell. 1989;56:171–179. doi: 10.1016/0092-8674(89)90890-8. [DOI] [PubMed] [Google Scholar]
- 31.Geigé P, Knoop V, Brennicke A. Curr Genet. 1998;34:313–317. doi: 10.1007/s002940050401. [DOI] [PubMed] [Google Scholar]
- 32.Grohmann L, Brennicke A, Schuster W. Nucleic Acids Res. 1992;20:5641–5646. doi: 10.1093/nar/20.21.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez H, Fester T, Kloska S, Schroder W, Schuster W. EMBO J. 1996;15:2138–2149. [PMC free article] [PubMed] [Google Scholar]
- 34.Claros M G, Vincens P. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 35.Figueroa P, Gomez I, Holuigue L, Araya A, Jordana X. Plant J. 1999;18:601–609. doi: 10.1046/j.1365-313x.1999.00485.x. [DOI] [PubMed] [Google Scholar]
- 36.Wischmann C, Schuster W. FEBS Lett. 1995;375:152–156. doi: 10.1016/0014-5793(95)01100-s. [DOI] [PubMed] [Google Scholar]
- 37.Figueroa P, Gomez I, Carmona R, Holuigue L, Araya A, Jordana X. Mol Gen Genet. 1999;262:139–144. doi: 10.1007/s004380051068. [DOI] [PubMed] [Google Scholar]
- 38.Deiderick H. Ph.D. dissertation. Bloomington: Indiana University; 1999. [Google Scholar]
- 39.Kubo N, Harada K, Hirai A, Kadowaki K-I. Proc Natl Acad Sci USA. 1999;96:9207–9211. doi: 10.1073/pnas.96.16.9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadowaki K, Kubo N, Ozawa K, Hirai A. EMBO J. 1996;15:6652–6661. [PMC free article] [PubMed] [Google Scholar]
- 41.Kubo N, Harada K, Kadowaki K-I. In: Plant Mitochondria: From Gene to Function. Moller I M, Gardestrom P, Glimelius K, Glaser E, editors. Leiden: Backhuys; 1998. pp. 25–27. [Google Scholar]
- 42.Turmel M, Lemieux C, Burger G, Lang B F, Otis C, Plante I, Gray M W. Plant Cell. 1999;11:1717–1730. doi: 10.1105/tpc.11.9.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butterfield N J. Paleobiology. 2000;26:386–404. [Google Scholar]
- 44.Gray M W. Curr Opin Genet Dev. 1999;9:678–687. doi: 10.1016/s0959-437x(99)00030-1. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi Y, Knoop V, Fukuzawa H, Brennicke A, Ohyama K. Mol Gen Genet. 1997;256:589–592. doi: 10.1007/pl00008616. [DOI] [PubMed] [Google Scholar]
- 46.Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- 47.Bowman E J, Knock T E. Gene. 1992;114:157–163. doi: 10.1016/0378-1119(92)90569-b. [DOI] [PubMed] [Google Scholar]
- 48.Albach D A, Soltis P S, Soltis D E, Olmstead R G. Ann Mo Bot Gard. 2001;88:163–212. [Google Scholar]
- 49.Olmstead R G, dePamphilis C W, Wolfe A D, Young N D, Elisons W J, Reeves P A. Am J Bot. 2001;88:348–361. [PubMed] [Google Scholar]
- 50.Soltis D E, Soltis P S, Chase M W, Mort M E, Albach D C, Zanis M, Savolainen V, Hahn W H, Hoot S B, Fay M F, et al. Bot J Linnean Soc. 2000;133:381–461. [Google Scholar]
- 51.Hoot S B, Magallon S, Crane P R. Ann Mo Bot Gard. 1999;86:1–32. [Google Scholar]
- 52.Chase M W, Soltis D E, Soltis P S, Rudall P J, Fay M F, Hahn W H, Sullivan S, Joseph J, Molvray M, Kores P J, et al. In: Monocots: Systematics and Evolution. Wilson K L, Morrison D A, editors. Collingwood, Australia: Commonwealth Scientific & Industrial Research Organization; 2000. pp. 3–16. [Google Scholar]
- 53.Qiu Y L, Lee J, Bernasconi-Quadroni F, Soltis D E, Soltis P S, Zanis M, Zimmer E A, Chen Z, Savolainen V, Chase M W. Nature (London) 1999;402:404–407. doi: 10.1038/46536. [DOI] [PubMed] [Google Scholar]
- 54.Adams K L, Daley D O, Whelan J, Palmer J D. Plant Cell. 2002;14:931–943. doi: 10.1105/tpc.010483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daley D O, Adams K L, Clifton R, Qualmann S, Millar A H, Palmer J D, Pratje E, Whelan J. Plant J. 2002;30:11–21. doi: 10.1046/j.1365-313x.2002.01263.x. [DOI] [PubMed] [Google Scholar]
- 56. Daley, D. O., Clifton, R. & Whelan, J. (2002) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 57.Race H L, Herrmann R G, Martin W. Trends Genet. 1999;15:364–370. doi: 10.1016/s0168-9525(99)01766-7. [DOI] [PubMed] [Google Scholar]
- 58.Martin W, Schnarrenberger C. Curr Genet. 1997;32:1–18. doi: 10.1007/s002940050241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.