Abstract
Small interfering RNA (siRNA) has become a powerful tool for selectively silencing gene expression in cultured mammalian cells. Because different siRNAs of the same gene have variable silencing capacities, RNA interference with synthetic siRNA is inefficient and cost intensive, especially for functional genomic studies. Here we report the use of Escherichia coli RNase III to cleave double-stranded RNA (dsRNA) into endoribonuclease-prepared siRNA (esiRNA) that can target multiple sites within an mRNA. esiRNA recapitulates the potent and specific inhibition by long dsRNA in Drosophila S2 cells. In contrast to long dsRNA, esiRNA mediates effective RNA interference without apparent nonspecific effect in cultured mammalian cells. We found that sequence-specific interference by esiRNA and the nonspecific IFN response activated by long dsRNA are independent pathways in mammalian cells. esiRNA works by eliciting the destruction of its cognate mRNA. Because of its simplicity and potency, this approach is useful for analysis of mammalian gene functions.
Double-stranded RNA interference (RNAi) has become a powerful genetic tool for selectively silencing gene expression in many eukaryotes (1, 2).
In the RNAi reaction, the cellular RNase III enzyme Dicer cleaves the double-stranded RNA (dsRNA) silencing trigger into 21- to 25-nt RNA called siRNA (small interfering RNA) (3, 4). siRNA pairs with its cognate mRNA, leading to degradation of target mRNA and amplification of gene-specific silencing signals (1–5). Although RNAi has also been observed in mouse oocytes, embryos, embryonic stem cells, and embryonal carcinoma cell lines, dsRNA triggers nonspecific inhibition of gene expression in most mammalian cell lines (6–8). In mammalian cells, dsRNAs longer than 30 bp can activate the dsRNA-dependent kinase PKR and 2′-5′-oligoadenylate synthetase, normally induced by IFN (9). By virtue of its small size, synthetic siRNA avoids activation of the IFN response. The activated PKR inhibits general translation by phosphorylation of the translation factor eukaryotic initiation factor 2α, whereas 2′-5′-oligoadenylate synthetase causes nonspecific mRNA degradation via activation of RNase L (9).
In contrast to the nonspecific effect of long dsRNA, siRNA can mediate selective gene silencing in the mammalian system (10, 11). Hairpin RNA with a short loop and 19–27 bp in the stem also selectively silences expression of genes that are homologous to the sequence in the double-stranded stem (12, 13). Mammalian cells can convert short hairpin RNA into siRNA to mediate selective gene silencing (12, 13). Although many mammalian cells can also convert long dsRNA into siRNA, long dsRNA is incapable of triggering RNAi in these cells (7). The inability of long dsRNA to elicit RNAi in vertebrates has been generally attributed to nonspecific activation of the IFN response (7, 8). However, the relationship between the IFN signaling pathway and RNA interference has not been addressed definitively.
Although siRNA provides a promising tool for assessing the consequences of suppressing gene expression in cultured mammalian cells, RNAi with synthetic siRNA is limited because siRNAs to different sequences within a gene have dramatically varied inhibitory ability (14, 15). Therefore, each mRNA must be screened for an efficient siRNA, a laborious and costly process. However, processing of long dsRNAs should generate a great variety of siRNAs capable of interacting with multiple sites on target mRNAs, increasing the chance that at least one siRNA will pair with its target sequence. Thus, the power of siRNA as a genetic tool in the mammalian system could be greatly enhanced by using siRNA processed from dsRNA.
Although Dicer is involved in the dsRNA cleavage in vivo, using Dicer to prepare siRNA in vitro may be problematic because dsRNA cleavage by Dicer is very inefficient, particularly for short dsRNAs (3, 16). In contrast, Escherichia coli RNase III (EC3.1.24) can digest dsRNA very efficiently into short pieces with the same end structures as siRNA, 5′ phosphate/3′ hydroxyl termini and 2- to 3-nt 3′ overhangs (17). These end structures of siRNA are reported to be important for RNAi activity (18). In addition, large amounts of soluble recombinant E. coli RNase III protein can be obtained (17). These attributes make E. coli RNase III a promising enzyme for preparing siRNAs in vitro.
Exhaustive cleavage of dsRNA by E. coli RNase III leads to duplex products averaging 12–15 bp in length (17). These short dsRNA are unable to trigger an RNAi response in mammalian cells (ref. 6 and D.Y., unpublished observation). To obtain siRNA of appropriate length we performed limited RNase III digestion of dsRNA, efficiently generating 20- to 25-bp siRNA. These siRNAs recapitulated the potent and sequence-specific gene silencing by long dsRNA in Drosphila S2 cells. More importantly, they also mediated effective RNAi without nonspecific effects in mammalian cells. siRNA produced by the method successfully inhibited various endogenous genes in different mammalian cell lines. Because it is relatively quick and simple, this method may prove to be useful in using siRNA for analysis of gene functions in cultured mammalian cells.
Materials and Methods
Protein Expression and Purification.
The E. coli RNase III coding sequence was amplified with PCR from the bacterial strain DH5α genomic DNA with the upstream primer cgc gga tcc aac ccc atc gta att aat cgg ctt ca and downstream primer gac gtc cga cga tgg caa t and cloned into BamHI and SmaI of pGEX-2T (Amersham Pharmacia). To produce glutathione S-transferase (GST)-RNase III protein, the bacterial strain BL21(DE3) carrying the expression vector was grown at 37°C to an OD600 of 0.5 and induced with 1 mM isopropyl β-d-thiogalactoside for 2 h. GST-RNase III was purified with glutathione-agarose beads according to the manufacturer's instruction (Amersham Pharmacia), dialyzed against 20 mM Tris⋅HCl, 0.5 mM EDTA, 5 mM MgCl2, 1 mM DTT, 140 mM NaCl, 2.7 mM KCl, 30% glycerol, pH 7.9 overnight, and kept at −20°C. Approximately 1 mg of RNase III was purified from 1 liter of culture. There was no significant loss of enzyme activity after 6 months of storage.
Single-Strand RNA Synthesis and Preparation of dsRNA.
The siRNAs CS1 and CS2 were synthesized chemically (Dharmacon, Lafayette, CO). siRNAs, T1 to T6, and short RNA hairpins, H1 to H6, were synthesized by using the MEGAshortscript kit (Ambion, Austin, TX) from oligo DNA templates carrying a phage T7 promoter at one end. All siRNAs were 21 nt long and had 2-nt 3′ overhangs. All short hairpin RNAs had a 24-bp stem with a UCU loop and an extra GGGA at the 5′ end for efficient transcription in vitro. Other RNA strands were individually synthesized by using the MEGAscript kit (Ambion) from PCR-derived linear templates carrying a phage T7 promoter at both ends. dsRNA was formed by annealing as described (16).
The ends of the dsR-457 RNAs corresponded to 1200–1656 in pRL-CMV (Promega). The firefly luciferase (F-luc) RNAs corresponded to the following positions in pEGFPLuc (CLONTECH): dsF-592, 1455–2046; CS1, 1520–1538; CS2, 1900–1918; T1, 603–621; T2, 624–642; T3, 2448–2466; T4, 2994–3012; T5, 3013–3031; T6, 3049–3067; H1, 989-1012; H2, 1520–1543; H3, 2529–2552; H4, 2601–2624; H5, 2906–2929; H6, 2947–2970. Templates for clathrin light chain a (LCa) and Cdk1 dsRNA represented the full coding sequences of their genes. Templates for the 681-bp c-myc dsRNA were amplified with forward primer gactcaacgttagcttcaccaaca and reverse primer ggactccgtcgaggagagcaga. All of these primers had appended 18-base phage promoters.
Production and Purification of Short RNA Duplexes.
To prepare endoribonuclease-prepared siRNAs (esiRNAs) for F-luc and Renilla luciferase (R-luc), 100 μg of dsRNAs was digested by 1 μg of recombinant RNase III in a 200 μl reaction buffer (same as dialysis buffer except 5% glycerol) for 15 min at 37°C. Reactions were terminated by adding EDTA to 20 mM and the products were separated on 12% polyacrylamide gel, 1 × 89 mM Tris·HCl (pH 8.4)/89 mM boric acid/2 mM EDTA. A 10-bp DNA marker was used to estimate the migration of RNA duplexes. Short RNAs of appropriate sizes were eluted from gel slices by soaking in 1 M (NH4)2AC at 37°C overnight and recovered by ethanol precipitation. The precipitate was dissolved in 10 mM Tris·HCl (pH 7.4) and 0.5 mM EDTA at 1.0 μg/μl. For other genes, 100 μg of dsRNAs was digested with 0.2 μg of RNase III for 1 h at 21°C, and reactions were loaded onto QIAquick spin columns after being supplemented with 5 vol of PN buffer (QIAquick nucleotide removal kit, Qiagen, Chatsworth, CA). Flow-through usually contained RNA from 15 to 25 bp, which was precipitated by ethanol and dissolved in TE buffer. The concentration of esiRNA was determined by UV260.
Cell Culture, Nucleic Acid Transfections, and Luciferase Assays.
C33A human cervical carcinoma cell line, HeLa human cervical epithelioid carcinoma cell line, and 293 transformed human embryonic kidney cells were obtained from the American Type Culture Collection and grown in DMEM. The hTERT-RPE1 cell line (CLONTECH) is a human retinal pigment epithelial (RPE) cell line that stably expresses the human telomerase reverse transcriptase subunit (hTERT). hTERT-RPE1 cells were permanently transfected with a F-luc expression construct to express F-luc from chromatin templates and grown in DEME/F12 medium. Drosophila S2 cells were grown in Schneider medium. All media were from Life Technologies, Grand Island, NY, and supplemented with 10% FBS (vol/vol). Mammalian cells were cultured at 37°C; S2 cells were at room temperature.
To silence luciferase expression from plasmid templates, plasmid DNA and esiRNAs were cotranfected by using Superfect (Qiagen). Cells were transfected at 50–70% confluence in 24-well plates for 2.5 h and then changed to fresh medium. Unless otherwise described, transfections used 1 μg of DNA per well in 0.5 ml transfection medium (0.5 μg pEGFPLuc/0.1 μg pRL-SV40/0.4 μg pUC19/47 nM esiRNA). Data were expressed as mean ratio of relative luciferase activities (F-luc/R-luc or R-luc/F-luc) and normalized to that in cells transfected with DNA only. Error bars equal ± one SD. To silence F-luc expression from chromatin templates in RPE cells, 0.2 μg esiRNA was transfected into each well of a 6-well plate by using Lipofectamine 2000 for 3 h and then changed to the fresh medium. Luciferase activity was measured as described (16). Protein concentrations were measured by Bradford assays (Bio-Rad). To silence endogenous gene expression, cells were cotransfected with 1 μg esiRNA by using Lipofectamine 2000.
In Vitro RNAi Assay.
Preparation of cell extracts and in vitro RNAi were carried out as described (19). The HeLa cells (2 × 107) were washed in PBS and resuspended in a hypotonic buffer (10 mM Hepes, pH 7.0/2 mM MgCl2/6 mM mercaptoethanol) and lysed. Cell lysates were centrifuged at 20,000 × g for 30 min. We stored supernatants at −80°C. As described (16), mRNAs were produced by using a MAXIscript kit (Ambion) and uniformly labeled during the transcription reaction with 32P-labeled UTP. For in vitro RNAi, 10 μl of extracts was incubated for indicated times at 37°C with 100 nM esiRNA in a 20 μl of reaction containing 20 mM Hepes (pH 7.0), 1 mM MgOAC, 100 mM K2 (OAC), 5 mM DTT, 500 μM NTP, and 2 units of RNasin (Promega).
Western Blotting.
For analysis of clathrin light chain and heavy chain, cells extracts were prepared as described (20, 21). For c-myc, Cdk1, and Cdk2, transfected cells grown in 6-well plates were harvested in PBS buffer containing 1% Nonidet P-40 and mixed with an equal volume of SDS sample buffer. Equal amounts of total protein were separated on 12.5% polyacrylamide gels and transferred to nitrocellulose. Standard immunostaining was carried out by using ECL (Amersham Pharmacia). Signals were quantified by using Molecular Dynamics equipment. Rabbit serum, which recognizes both LCa and clathrin light chain b (LCb), and monoclonal TD.1 against clathrin heavy chain have been described (20, 21). Anti-c-Myc antibody, N262, and antibodies for cyclin A and Cdk2 were from Santa Cruz Biotechnology, and antiactin antibody was from Sigma.
Results and Discussion
Processing Long dsRNAs into Short RNA Duplexes with E. coli RNase III.
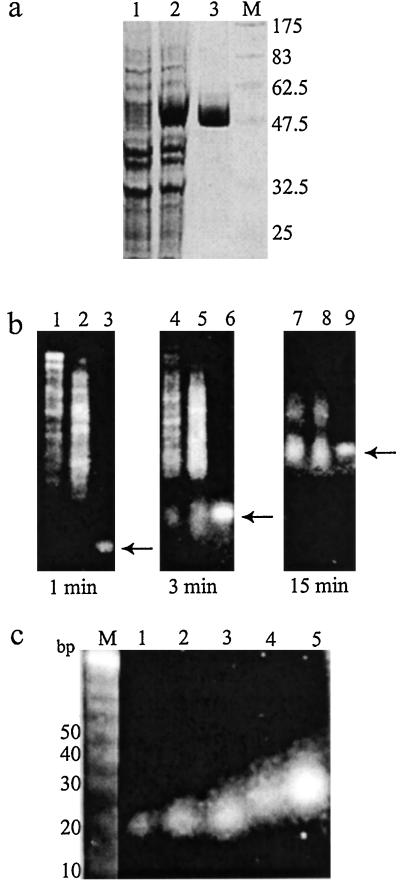
To prepare a heterogeneous siRNA population that could potentially target multiple sites per mRNA molecule for inhibition, we used E. coli RNase III to digest long dsRNAs representing the mRNA targets. We overexpressed E. coli RNase III as a GST fusion protein in E. coli and purified it to homogeneity (Fig. 1a). To make a 592-bp dsRNA (dsF-592) corresponding to the F-luc and a 457-bp dsRNA (dsR-457) of R-luc, we performed in vitro transcription from PCR products that had appended phage promoters at each end (Fig. 2e). We found that GST-RNase III fusions were highly active in cleaving these dsRNAs at 37°C. Even at low RNase III concentrations, cleavage products were visible within 1 min incubation, and molecules approximately 15–30 bp in length were accumulated during longer incubation and became the main products (Fig. 1b). Similar digestion patterns were also observed with dsRNAs representing more than 20 other genes (data not shown). After optimization, we found that limited RNase III digestion of dsRNA at room temperature for 1 h yielded ample amounts of esiRNA for inhibition of most genes. Efficiently processing dsRNA with high sequence complexity into short species is consistent with the lack of sequence specificity in substrate recognition and cleavage by E. coli RNase III (22). We separated RNase III digestion products on polyacrylamid gels and purified the RNAs corresponding to approximately 21–23, 24–26, and 27–30 bp (Fig. 1c and data not shown). For simplicity, we named siRNA prepared by RNase III digestion esiRNA.
Figure 1.
Preparation of siRNA from dsRNA by hydrolysis with E. coli RNase III. (a) Overexpression and purification of GST-RNase III fusion. Lanes 1 and 2, E. coli extracts before and after isopropyl β-d-thiogalactoside induction; lane 3, purified GST-RNase III fusion; lane M, protein molecular weight marker. (b) Time course of RNase III digestion of dsRNA. DsR-457 or dsF-592 RNA was incubated with RNase III for the indicated time, and then separated by electrophoresis in a 4% agarose gel. Lanes 1, 4, and 7 are dsR-457; lanes 2, 5, and 8 are dsF-592; lanes 3, 6, and 9 are 21-bp siRNA marked by an arrow. (c) Agarose gel analysis of purified short RNA species processed from dsF-592. Lane M, 10-bp DNA marker; lanes 1 and 2 are chemically synthesized siRNAs; lane 3, 21–23 bp; lane 4, 24–26 bp; lane 5, 27–30 bp.
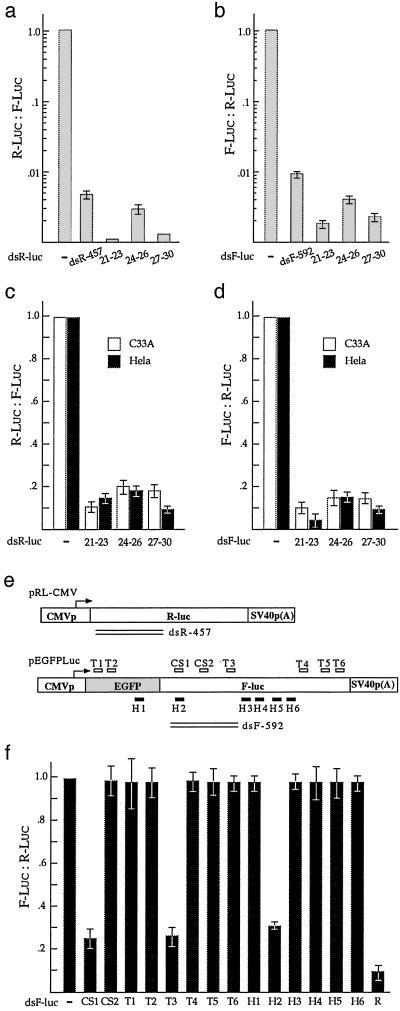
Figure 2.
siRNA-mediated gene silencing in insect and mammalian cells. The effect of esiRNA on R-luc (a and c) or F-luc (b and d) expression was determined in Drosophila S2 cells (a and b) and mammalian cell lines, HeLa and C33A (c and d). Plasmid DNAs with or without various sized dsRNAs were cotransfected into cells. (e) Reporter plasmids, siRNAs, dsRNAs, and short hairpin RNAs. The diagram illustrates DNA constructs used to produce in vivo mRNA and in vitro dsRNA for F-luc and R-luc genes. The positions of dsRNAs, chemically synthesized siRNAs (CS1 and CS2), in vitro-transcribed siRNAs (T1 to T6), and short hairpin RNAs (H1 to H6) are indicated. (f) The variable effect of synthetic siRNAs or short hairpin RNAs on different sequences within a gene. Plasmid DNAs with or without the various indicated short RNA species were cotransfected into HeLa cells. R is the F-luc esiRNA shorter than 30 bp. transfected with pGL-3-control (Promega) and pRL-CMV with or without 21- to 23-bp esiRNA corresponding to either the cytomegalovirus (CMV) promoter or R-luc. Luciferase activity was measured 24–96 h after cotransfection. (e and f) Relationship between RNAi and the IFN signaling pathway. DsF-592 and F-luc esiRNA (21–23 bp) were transfected individually or together into HeLa cells to silence F-luc expression from plasmid templates. Luciferase activity was measured 24–48 h after cotransfection. Data are expressed as either F-luc units (e) or relative F-luc activity (f), normalized to that of DNA-only transfections.
esiRNA Mediates Effective RNAi Against Reporter Gene Expression in Cultured Insect and Mammalian Cells.
To test whether esiRNA has RNAi activity, we cotransfected short RNA duplexes along with expression constructs for F-luc and R-luc into Drosophila S2 cells. Inhibition was estimated by measuring the relative F-luc (F-luc/R-luc) or relative R-luc (R-luc/F-luc) activity as described (16). We observed that the 21- to 23-bp esiRNA, processed from dsR-457, caused about 900-fold inhibition of R-luc activity (Fig. 2a). The similarly sized esiRNA of F-luc exerted 500-fold of inhibition of F-luc activity (Fig. 2b). esiRNAs of 24–26 bp and 27–30 bp showed similarly potent inhibition of their cognate luciferase activity (Fig. 2 a and b). The unprocessed dsRNAs, dsR-457 and dsF-592, were 2- to 10-fold less potent than esiRNA (Fig. 2 a and b). Recapitulation of RNAi activity of long dsRNA with its esiRNA is consistent with the presumption that siRNA is the bona fide mediator of the RNAi reaction.
We further tested whether esiRNA is a specific gene silencer in mammalian cells. We observed that 21- to 23-bp esiRNA of R-luc caused 85–90% inhibition of relative R-luc expression in both HeLa and C33A cells (Fig. 2c). Similarly sized esiRNAs of F-luc inhibited 90–95% of relative F-luc expression (Fig. 2d). This reciprocal effect indicates that the short RNA duplexes acted in a sequence-specific manner without affecting expression from the noncomplementary templates. Similarly, 24- to 26-bp and 27- to 30-bp esiRNAs exerted sequence-specific inhibition, consistent with a published report that the synthetic enhanced green fluorescent protein siRNAs ranging from 21 to 27 nt all have RNAi activity (Fig. 2 c and d and ref. 11). Similar results were obtained with column-purified esiRNAs (Fig. 2f). We compared inhibitory ability of siRNA and esiRNA. We found that different siRNAs of F-luc made by either chemical synthesis or in vitro transcription had dramatically varied silencing ability and were less potent inhibitors than esiRNA (Fig. 2 d and f). This was also true for short RNA hairpins. Of the 14 siRNAs and short hairpin RNAs for F-luc, only three showed reasonable RNAi activity (Fig. 2f). The synthetic siRNAs or hairpin RNAs recognized only one sequence element, whereas esiRNAs covered comprehensive sequence elements of the target mRNA. Thus, esiRNA likely increases the chance that at least one siRNA can pair with and lead to destruction of its target.
Similar strong and specific inhibition by esiRNA was observed in IMR-90, CHO, hTERT-RPE1, MEF, and 293 cells, suggesting the approach is generally applicable for mammalian cell lines (data not shown). As expected, long dsRNA, either dsF-592 or dsR-457, caused sequence-independent inhibition of both F-luc and R-luc expression, suggesting activation of the IFN signaling pathway (Fig. 3 e and f and data not shown). In contrast, esiRNA did not cause significant nonspecific inhibition of control luciferase expression in these mammalian cell lines.
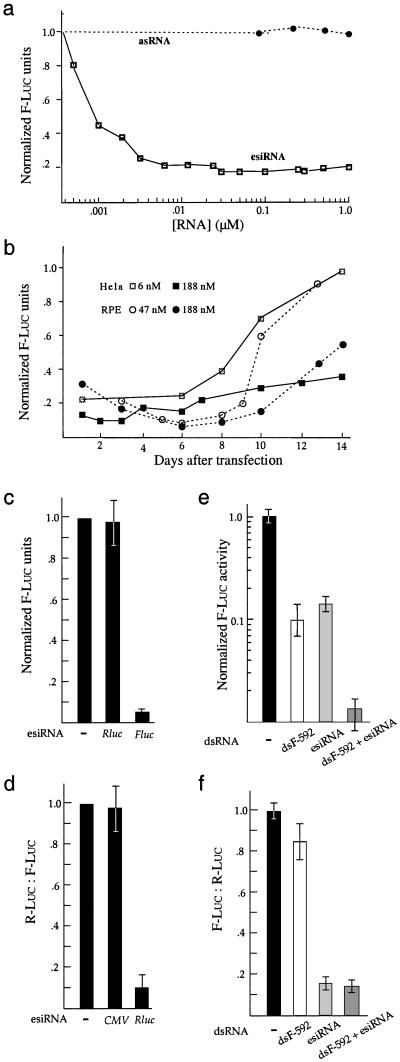
Figure 3.
Characteristics of RNAi. (a) Dose–response for antisense RNA (asRNA) and esiRNA against F-luc. Indicated amounts of F-luc esiRNA (21–23 bp) or antisense strand of CS1 were transfected into RPE cells to target F-luc expression from chromatin templates, and luciferase activities were measured at 24 h after transfection. Data represent the average of 3–6 experiments and are expressed as relative luciferase activity after normalization to the luciferase units/μg of protein observed in control cells transfected with R-luc esiRNA. (b) Duration of esiRNA-mediated inhibition. F-luc esiRNA (21–23 bp) was transfected into RPE cells, and luciferase activities were measured at the indicated time period. When HeLa cells were used, F-luc target was expressed from cotransfected plasmid DNA. (c) esiRNA silences gene expression from mRNA templates. HeLa cells were transfected with 0.5 μg F-luc mRNA with or without 0.1 μg of esiRNA (21–23 bp) for either F-luc or R-luc for 3 h and then collected for luciferase assays. Data are expressed as arbitrary luciferase units and normalized to that produced in cells transfected with mRNA only. (d) esiRNA corresponding to the promoter region has no RNAi activity. HeLa cells were
Dose Dependence of RNAi Mediated by esiRNA in Mammalian Cells.
Cotransfection assays do not allow accurate calculation of the minimal effective esiRNA concentration. To study dose dependence of esiRNA-induced inhibition, we used F-luc esiRNA to inhibit gene expression in the hTERT-RPE1 cells that were stably transfected with an F-luc expression construct. We observed a saturated inhibition when the transfection buffer contained at least 3 nM of esiRNA (Fig. 3a). Inhibition decreased at subsaturating esiRNA doses and was undetectable when the esiRNA dose was below 0.5 nM (Fig. 3a). The antisense strand of the chemically synthesized siRNA CS1 was incapable of inhibiting F-luc activity even at 10- to 20-fold higher concentrations than the effective concentration used for its double-stranded form, suggesting siRNAs are more potent inhibitors than antisense RNAs (Figs. 2f and 3a).
Rapid Onset and Prolonged Duration of Inhibition.
To further characterize the RNAi reaction, we determined the duration of F-luc esiRNA-mediated inhibition of chromatin templates in the above hTERT-RPE1 cells. We observed 50–65% inhibition of F-luc activity at 8 h after transfection (data not shown), suggesting that silencing was established rapidly. Silencing reached the maximum 5–6 days after transfection and then declined (Fig. 3b). When plasmid DNA was used to express targets for inhibition in HeLa cells, we observed that the inhibition could last 2 weeks if high esiRNA doses were used. Low doses of esiRNA caused less potent inhibition that was sustained for several days (Fig. 3b). Thus, the longevity of esiRNA-mediated inhibition at least partially depended on esiRNA dose with either episomal or chromatin templates. The rapidity and longevity of esiRNA-mediated inhibition suggest that esiRNAs could be used to inactivate genes whose protein products are relatively stable.
esiRNA Elicits mRNA Decay.
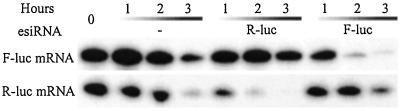
A key feature of RNAi in Caenorhabditis elegans and Drosophila is that it exerts its effect by eliciting the destruction of targeted mRNA (1). To test whether siRNAs also act posttranscriptionally in the mammalian system, we asked whether esiRNA could directly inhibit expression from its cognate mRNA. We cotransfected esiRNA and F-luc mRNA into HeLa cells. Three hours after transfection, control cells that had received F-luc mRNA produced 20,000–50,000 units of luciferase activity. However, cotransfected esiRNAs for F-luc but not R-luc caused 15-fold inhibition of F-luc activity, suggesting that siRNA-mediated gene silencing is directed at mRNA in mammalian cells (Fig. 3c). Consistent with this result, we found that the esiRNA corresponding to a promoter region had no RNAi activity (Fig. 3d). To test whether esiRNA elicits destruction of the target mRNA, we used an extract of HeLa cells to perform an in vitro RNAi assay identical to that used to characterize RNAi responses in Drosophila embryo extracts (19). We found that esiRNAs for either F-luc or R-luc accelerated the degradation of their cognate mRNA 4- to 8-fold (Fig. 4). This finding is consistent with previous reports on the effect of siRNA (10, 14).
Figure 4.
esiRNA induces degradation of its cognate mRNA. 32P-labeled F-luc or R-luc mRNA was incubated in extracts of HeLa cells with or without the cognate esiRNAs for the indicated time periods and then analyzed in a 4% sequence gel.
Nonspecific Gene Silencing by Long dsRNA and Sequence-Specific Interference by siRNA Are Independent Pathways.
To test the effect of the IFN signaling pathway on RNAi, we first asked whether we could detect RNAi activity of siRNA in the presence of a nonspecific IFN response activated by long dsRNA in HeLa cells. We found that dsF-592 caused 10- to 20-fold inhibition of both F-luc and R-luc expression, suggesting activation of the nonspecific IFN response (Fig. 3 e and f). F-luc esiRNA selectively inhibited F-luc expression as observed before. When esiRNA and dsF-592 were cotransfected, we observed additive inhibition of F-luc expression (Fig. 3e). However, esiRNA inhibited relative F-luc activity equally with or without cotransfected dsF-592, suggesting that activation of nonspecific IFN response has no effect on the potency of specific RNAi inhibition (Fig. 3f).
We then tested whether inactivation of the IFN response suffices to activate specific gene silencing of long dsRNA. We found that mouse embryonic fibroblast cells lacking either PKR or both PKR and RNase L were defective in both nonspecific and specific gene silencing triggered by dsF-592, although siRNA-mediated RNAi could be detected (data not shown). We concluded that sequence-specific RNAi mediated by siRNA is independent of dsRNA-triggered nonspecific gene silencing.
esiRNA Selectively Silences the Expression of Endogenous Genes.
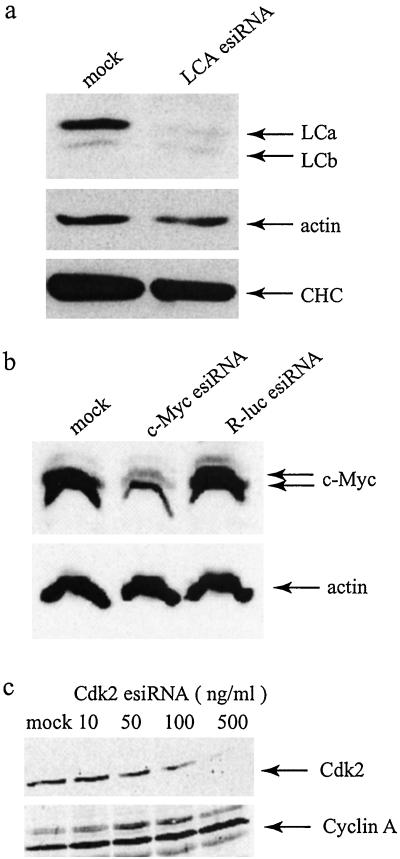
To test whether the expression of endogenous genes could be silenced we targeted a wide variety of genes, including those with either abundant long-lived or rare short-lived transcripts/proteins. We first attempted to silence the expression of the LCa in HeLa cells. We examined the level of LCa protein 5 days after esiRNA transfection to allow turnover of the protein, whose half-life is 24 h (20). Western blot analysis revealed that the LCa chain was reduced up to 90% (Fig. 5a). LCb was unaffected although it has 66% homology in DNA sequence with LCa and was recognized by the same antibody. A cross-silencing between LCa and LCb was not expected because these two genes do not share any uninterrupted DNA sequence longer than 21 nt in length (data not shown). LCa turns over independently of the other subunits of clathrin. Accordingly, inhibition of LCa expression did not affect the expression of clathrin heavy chain (Fig. 5a). We found that esiRNA-treated cells grew normally despite dramatically reduced LCa protein. This finding is presumably caused by functional redundancy between LCa and LCb. For example, complete loss of LCa in PC12 cells has been reported to have little effect on clathrin-mediated endocytosis and secretion (20).
Figure 5.
Silencing of endogenous mammalian genes with esiRNA. (a) Specific inhibition of expression of LCa. Western blotting was used to analyze HeLa cells that had been mock-transfected or transfected with LCa esiRNA. Three antibodies were used: rabbit serum that recognizes both LCa and LCb, mAb against actin, and monoclonal TD.1 against the clathrin heavy chain (CHC). (b) Selective reduction of c-Myc expression; 293 cells were transfected with buffer, c-myc esiRNA, or R-luc esiRNA. Western blotting was performed with antibodies against c-myc and actin 72 h after transfection. (c) Dose-dependent inhibition of Cdk2 expression by esiRNA; 293 cells were transfected with indicated doses of Cdk2 esiRNA. Western blotting was performed with antibodies against Cdk2 and cyclin A 5 days after transfection.
We next asked whether esiRNA could be used to inhibit genes whose mRNA and protein products are low in abundance and relatively unstable. As an example, we transfected c-myc esiRNA into 293 cells. We observed that the level of c-myc protein was reduced by about 70% (Fig. 5b). Inhibition was sequence-specific because actin expression was not affected and unrelated R-luc esiRNA failed to reduce c-myc expression. In addition, we observed specific reduction of cyclin-dependent kinases, Cdk1 and Cdk2, in response to treatment with their corresponding esiRNA (Fig. 5c and data not shown). The RNase III/esiRNA protocol was also used to specifically reduce expression of chaperon p23 (K. Yamamoto, personal communication). To date, we have successfully used esiRNA to inhibit the expression of seven diverse genes in cell culture. These examples established esiRNA as a valuable tool for selective depletion of gene products in mammalian cells.
Our results demonstrate that the short RNAs produced by hydrolysis with E. coli RNase III can specifically silence gene expression in cultured mammalian cells, in a manner similar to that observed with synthetic siRNA. There are several advantages to the esiRNA protocol presented here. Because esiRNA can potentially target multiple sites within an mRNA, the requirement of screening efficient siRNA for individual genes is eliminated. Targeting of multiple sites would also be important in efforts to reduce viral replication because it can restrict the emergence of siRNA-resistant strains produced by base pair mismatches. The protocol is technically simple and quick, and it is effective on a wide range of proteins in different mammalian cell lines. Large-scale functional genomic studies with dsRNA have already been successful in C. elegans, but comparable analysis is still lacking in vertebrates (23, 24). Because of its attributes, RNAi with esiRNA could be easily adapted for analysis of gene functions at the genome scale in cultured mammalian cells. A reverse genetics approach with esiRNA could be particularly useful for identifying genes essential for viability because it is difficult to make stable mutants for those genes. The technique should also be useful in the study of cancer. Cancer cells are genetically different from their normal cell counterparts, often having undergone at least a half-dozen mutations (25). esiRNA could be used to search for genes whose down-regulation specifically kills tumor cells, an approach that would simultaneously identify and validate appropriate new drug targets.
Acknowledgments
We thank Larry Chasin and James W. Erickson for reviewing the manuscript, John Bell and Raul Andino for PKR−/− and RNase L−/− mouse embryonic fibroblasts, Tim Tian for helpful discussion, and Sue Kim and Kevin Hill for critical reading of the manuscript. This work was supported by National Institutes of Health Grant CA44338 (to J.M.B.), National Institutes of Health Grant GM38093 (to F.M.B.), and the G. W. Hooper Research Foundation. D.Y. is supported by a Postdoctoral Fellowship from the Susan G. Komen Breast Cancer Fund. F.B. was a Fellow of the Leukemia and Lymphoma Society. A.G. is supported by a Postdoctoral Research Fellowship for Physicians from the Howard Hughes Medical Institute. C.-Y.C. is supported by a Postdoctoral Fellowship from the Arthritis Foundation.
Abbreviations
- RNAi
RNA interference
- dsRNA
double-stranded RNA
- siRNA
small interfering RNA
- esiRNA
endoribonuclease-prepared siRNA
- F-luc
firefly luciferase
- R-luc
Renilla luciferase
- GST
glutathione S-transferase
- RPE
retinal pigment epithelial
- LCa/LCb
clathrin light chain a/b
References
- 1.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 2.Bosher J M, Labouesse M. Nat Cell Biol. 2000;2:E31–E36. doi: 10.1038/35000102. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature (London) 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir S M, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishikura K. Cell. 2001;16:415–418. doi: 10.1016/s0092-8674(01)00581-5. [DOI] [PubMed] [Google Scholar]
- 6.Paddison P J, Caudy A A, Hannon G J. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang S, Tutton S, Pierce E, Yoon K. Mol Cell Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wianny F, Zernicka-Goetz M. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 9.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 10.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 11.Caplen N J, Parrish S, Imani F, Fire A, Morgan R A. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paddison P J, Caudy A A, Bernstein E, Hannon G J, Conklin D S. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brummelkamp T R, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 14.Holen T, Amarzguioui M, Wiiger M T, Babaie E, Prydz H. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harborth J, Elbashir S M, Bechert K, Tuschl T, Weber K. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 16.Yang D, Lu H, Erickson J W. Curr Biol. 2000;10:1191–1200. doi: 10.1016/s0960-9822(00)00732-6. [DOI] [PubMed] [Google Scholar]
- 17.Amarasinghe A K, Calin-Jageman I, Harmouch A, Sun W, Nicholson A W. Methods Enzymol. 2001;342:143–158. doi: 10.1016/s0076-6879(01)42542-0. [DOI] [PubMed] [Google Scholar]
- 18.Elbashir S M, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 20.Action S L, Wong D H, Parham P, Brodsky F M, Jackson A P. Mol Biol Cell. 1993;4:647–660. doi: 10.1091/mbc.4.6.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nathke I S, Heuser J, Lupas A, Stock J, Turck C W, Brodsky F M. Cell. 1992;68:899–910. doi: 10.1016/0092-8674(92)90033-9. [DOI] [PubMed] [Google Scholar]
- 22.Zhang K, Nicholson A W. Proc Natl Acad Sci USA. 1997;94:13437–13441. doi: 10.1073/pnas.94.25.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonczy P, Echeverri G, Oegema K, Coulson A, Jones S J, Copley R R, Duperon J, Oegema J, Brehm M, Cassin E, et al. Nature (London) 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 24.Fraser A G, Kamath R S, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Nature (London) 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]