Abstract
One of many protein–protein interactions modulated upon DNA damage is that of the single-stranded DNA-binding protein, replication protein A (RPA), with the p53 tumor suppressor. Here we report the crystal structure of RPA residues 1–120 (RPA70N) bound to the N-terminal transactivation domain of p53 (residues 37–57; p53N) and, by using NMR spectroscopy, characterize two mechanisms by which the RPA/p53 interaction can be modulated. RPA70N forms an oligonucleotide/oligosaccharide-binding fold, similar to that previously observed for the ssDNA-binding domains of RPA. In contrast, the N-terminal p53 transactivation domain is largely disordered in solution, but residues 37–57 fold into two amphipathic helices, H1 and H2, upon binding with RPA70N. The H2 helix of p53 structurally mimics the binding of ssDNA to the oligonucleotide/oligosaccharide-binding fold. NMR experiments confirmed that both ssDNA and an acidic peptide mimicking a phosphorylated form of RPA32N can independently compete the acidic p53N out of the binding site. Taken together, our data suggest a mechanism for DNA damage signaling that can explain a threshold response to DNA damage.
Keywords: DNA binding, protein–protein interaction, structural analysis, ssDNA mimicry
Upon DNA damage, the p53 tumor suppressor is activated and orchestrates a cellular response by transcriptional regulation of genes involved in cell cycle arrest and apoptosis (1, 2). p53 protein is central to an extensive network of DNA damage sensing protein–protein and protein–nucleic acid interactions. As yet, however, details of how this network is regulated are unclear. One component of the network is replication protein A (RPA), the major single-stranded (ss) DNA-binding protein of the eukaryotic nucleus (3–5). The interaction of p53 with RPA mediates suppression of homologous recombination (6) and modulates Werner syndrome helicase activity (7). It is also linked with DNA repair and disruption of p53 and RPA complexes after DNA damage is thought to coordinate DNA repair with the p53-dependent checkpoint control (8).
Because the ability of p53 to bind specific DNA target sequences via its DNA-binding core (9) (Fig. 1,) is blocked when the protein is complexed with RPA it follows that UV-mediated disruption of the complexes is predicted to favor p53 transactivation functions (10). p53–RPA complex formation is affected by the presence of various lengths of ssDNAs, because RPA, when bound to these ssDNAs, is unable to interact with p53 (10). UV radiation of cells also reduces p53–RPA complexes by a second mechanism, because hyperphosphorylated RPA does not associate with p53 (8). Thus p53–RPA interaction is subject (i) to the presence of ssDNA molecules and also (ii) to the phosphorylation status of the RPA protein.
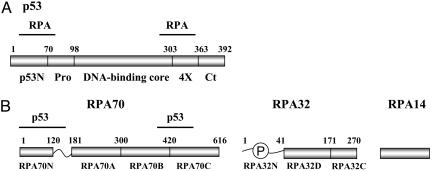
Fig. 1.
Binding regions for p53 and RPA70. Schematic representation of p53 (A) and RPA (B) with indicated borders of the protein domains. The major structural domains of p53 indicated are as follows: p53N, N-terminal domain; Pro, proline-rich domain; DNA binding core, central core domain; 4×, tetramerisation domain; Ct, C-terminal domain. The major structural domains of RPA indicated are as follows: RPA70N, N-terminal domain; RPA70A, RPA70B, RPA70C, RPA32D, DNA-binding domain A, B, C, and D, respectively. P, unstructured phosphorylated N terminus of subunit RPA32 (RPA32N). Horizontal lines above the schematics indicate previously identified sites of interaction.
RPA is a heterotrimer (RPA70, RPA32, and RPA14; Fig. 1B) involved in many aspects of DNA metabolism such as replication, recombination, and repair (11, 12). The largest subunit, RPA70, is a tandem repeat of four oligonucleotide/oligosaccharide-binding (OB) folds (13) comprising RPA70N, important for protein–protein interactions, and three ssDNA binding domains, RPA70A, RPA70B, and RPA70C (Fig. 1B; also see ref. 12). Weak interaction of RPA70N with ssDNA has recently been demonstrated in ref. 14. The RPA32 subunit is substrate for at least three PI3-kinases, ATM, ATR, and DNA-PK, which hyperphosphorylate the N-terminal domain of RPA32 (RPA32N) (15–20). Hyperphosphorylated, but not unphosphorylated, RPA32N is capable of binding the RPA70N subunit (21).
Two distinct fragments of p53 and two distinct fragments of RPA synergize to give the strong binding (Fig. 1 A and B). The N-terminal 70 residues of p53 (transactivation domain) are involved, with some contribution from multiple subdomains at the C terminus (4, 5, 22). Mutational analysis indicated that aromatic amino acids Trp-53 and Phe-54 of p53, flanked by negatively charged residues, are important for binding (8, 22). The interaction requires the N-terminal third of RPA70 (RPA70N) with some contribution from the C-terminal third (23). More recently, the direct interaction between residues 39–59 of the p53 transactivation domain and a fragment of RPA (residues 1–168) was demonstrated by using NMR relaxation experiments (24). These data suggested that RPA may induce local folding in the p53 peptide upon binding.
Here, we demonstrate that residues 37–57 of the p53 transactivation domain bind residues 1–120 of RPA70 in an interaction that is coupled with a coil-to-helix transition in the p53 peptide. The 1.6-Å crystal structure of a chimeric protein containing the N-terminal 120 residues of RPA70, followed by p53 residues 33–60, showed that an acidic, amphipatic helix comprising residues 47–57 of p53 structurally mimics ssDNA binding to an OB fold. NMR experiments confirmed that both ssDNA and an acidic peptide mimicking a phosphorylated form of RPA32N can independently compete p53N out of the binding site.
Materials and Methods
Constructs, Proteins, and Peptides. Human p53 constructs GST-p53 (1–73) and GST-p53 (20–73) (8) were kindly provided by R. Li (University of Virginia, Charlottesville). Human RPA fragments coding for amino acids 1–120, 1–129, and 1–168 of RPA70 and a chimera comprising RPA70 amino acids 1–121 followed by p53 amino acids 33–60 were cloned into pET15b vector (Novagen) expressed in Escherichia coli and purified according to the manufacturer's instructions. 15N- and 13C/15N-labeled p53 fragments (amino acid 1–73 and 38–58) were cloned into a GST-2TK vector (Amersham Pharmacia) and expressed in CodonPlus E. coli (Stratagene). GST-fusion protein was grown on defined M9 media supplemented with 15N-ammonium chloride and d-glucose. For the double-labeled samples, d-glucose was replaced by 13C6-d-glucose. Proteins were purified as described, and GST fusion was removed. Synthetic RPA32-Asp (amino acid 1–34; Ser/Thr 8, 13, 21, 23, 29, and 33 replaced with Asp), RPA32wt (amino acid 1–34) peptides, and ssDNA (11 nt, CGCCGGATCAG) were purchased from Dalton Chemical and ACGT Corps (Toronto), respectively.
In Vitro Transcription and Translation and GST Pull-Down Assay. Plasmid DNA constructs coding for RPA domains were linearized by appropriate restriction enzyme digest downstream of the RPA domain coding sequence. In vitro transcription reaction mixture that contained 30 units of T7 RNA polymerase, 40 mM Tris·HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM UTP, 25 μM GTP, 10 mM DTT, 0.5 mM RNA cap, (m7G5′ppp5′G), and 1 μg of linearized plasmid DNA was incubated at 37°C for 30 min. GTP was then added to a final concentration of 0.5 mM and incubation at 37°C was continued for a further 30 min. The RNAs were isolated by phenol/chloroform extraction and ethanol precipitation. The RNA pellets were washed with 70% ethanol, resuspended in RNase-free water, and stored at -80°C. In vitro translation reactions were carried out by using rabbit reticulocyte lysate supplemented with a 1 mM amino acid mixture lacking methionine (Promega). A typical 50-μl translation reaction consisted of 38 μl of rabbit reticulocyte lysate (including amino acids), 3 μl of water, and 4 μl [35S]methionine (Amersham Pharmacia, specific activity of 15 μCi/μl; 1 Ci = 37 GBq) to a final concentration of 0.75 mM. The mRNA was heated for 7 min at 67°C, and the translation mix was prewarmed at 30°C for 3 min. The mRNA was then added to the translation mix, and the reaction was incubated at 30°C for 1 h. The efficiency of translation reactions was assessed by scintillation counts of the radioactivity incorporated into the synthesized protein. E. coli BL21 strain was transformed with recombinant pGEX-2TK plasmids, and GST or GST-fused proteins were purified on GST-agarose beads (Novagen) according to manufacturer's conditions. Beads coupled with fusion proteins were aliquoted into 500-ng fractions and used for subsequent binding experiments for 2 h at 4°C in buffer containing 25 mM Tris·HCl (pH 8.0), 200 mM NaCl, 0.5% Nonidet P-40, 5 mM DTT, and EDTA-free protease inhibitor mixture (Roche Applied Science). After extensive wash and removal of the unbound material, proteins associated with beads were recovered by boiling in Laemmli buffer, separated by electrophoresis on a 15% SDS/PAGE, and analyzed by autoradiography.
NMR Data Collection. All spectra were performed on Varian Inova-600 and 500 MHz spectrometers equipped with a pulsed field gradient triple-resonance 1H/13C/15N probe. All experiments were conducted at 25°C and at pH 7.5. All NMR samples were prepared at pH 7.5 by using 25 mM Tris/250 mM NaCl/0.5 mM Tris (2-carboxyethyl)phosphine hydrochloride/2 mM benzamidine hydrochloride/1 mM PMSF/0.5 mM EDTA. The final NMR samples contained 90% H2O/10% 2DO with a protein concentration ranged between 0.5 and 0.8 mM. Two-dimensional, gradient-enhanced heteronuclear sequential quantum correlation (HSQC) spectra were acquired on uniformly 15N or 13C/15N-labeled p53 (38–58). Linear prediction in the 13C and 15N dimensions was used to improve the digital resolution. The spectra were processed with nmrpipe software package (25) and analyzed with xeasy (26). spscan (www.molebio.uni-jera.de/~rwg/spscan) was used to convert nmrpipe-formatted spectra into xeasy format. The backbone nuclear resonances (1HN, 15N, 13Cα, 13C′) and 13Cβ nuclear resonances were assigned for p53 in complex with RPA (1–120) by using data from a combination of the 3D HNCACB, 3D CBCA-(CO)NH, 3D HNCO, CCCTOCSY, and CCNNHTOCSY experiments. Greater than 85% of the backbone atoms were assigned. (all p53 residues, except two prolines and 3 amino acid residues, Gly, Ser, and His remained from the tag). The backbone nuclear resonances (1HN and 15N) of p53 obtained upon addition of RPA32-Asp and ssDNA to the p53/RPA70 complex, were tentatively assigned by assuming a minimal change in chemical shift. Using the minimal change criterion, it was possible to make 1H and 15N resonance assignments for Leu45, Ser46, Asp48, Ile50, Trp53, and Thr55.
NMR Titration Experiments. For competition titration experiments, labeled p53 peptide (0.5–0.8 mM) was titrated with an excess of unlabeled RPA70N until no further changes in chemical shifts were detected in the HSQC spectrum. Then, four aliquots of ssDNA (11 nt; CGCCGGATCAG) were added to the p53/RPA complex until the final concentration of ssDNA was 6 mM. Similarly, aliquots of RPA32N-Asp and RPA32wt were added to final oligonucleotide concentrations of 2.2 mM and 2 mM, respectively. 1H-15N HSQC spectra were acquired and analyzed after each addition of ssDNA and/or RPA32. The pH of the NMR sample before and after one of the titrations was directly measured and did not change when ssDNA added.
Crystallization and X-Ray Structure Solution. Crystals of the RPA70 (1–120), RPA (1–129), and RPA70 (1–120)-p53 (33–60) were grown at 20°C by vapor diffusion as hanging drops prepared by mixing 1 μl of protein (concentration of ≈10 mg/ml)/1 μl of the crystallization buffer; for RPA70 (1–120) and (1–129), 1.8 M ammonium sulfate/100 mM Tris, pH 8.5; for RPA/p53, 0.82 M trisodium citrate/100 mM Hepes, pH 7.9. Derivatives (HgCl and 4-chloromercuribenzoic acid) for RPA70 (1–120) were prepared by soaking the crystals within the crystallization buffer containing 5- to 10-mM compounds for 30 min. Data from flash-cooled crystals at 100°K were collected on Rigaku-H3R/Raxis-IV (Rigaku/MSC), integrated, and scaled by using denzo/scalepack (27). For the RPA (1–120) structure, Hg sites in HgCl derivative were manually located, and the experimental map was generated by using phases (28). All other structures were solved by molecular replacement by using RPA (1–120) structure as a search model for molrep (29). All structures were initially traced by arp/warp (30) and then manually rebuilt in o (31). Final refinement was performed by using refmac (32). Additional crystallographic statistics are given in Table 1, which is published as supporting information on the PNAS web site. The drawings are generated with ribbons (33) and pymol (DeLano Scientific, South San Francisco, CA).
Results
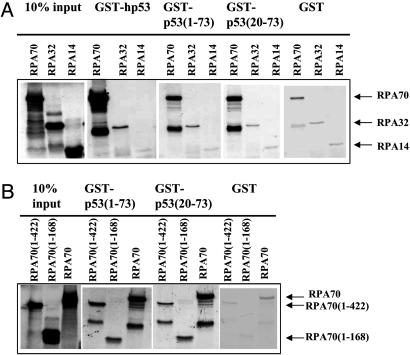
RPA70N Binds the p53 Transactivation Domain. Previous reports demonstrated synergy between different domains of p53 and RPA involved in the binding of these two proteins. The N-terminal part of p53, particularly amino acids 20 to 73, has been shown to be the major determinant of binding, with additional affinity provided by C-terminal residues (3, 8, 22). However, relative contribution of these determinants was not clearly characterized. We sought to characterize the smallest regions of each protein involved in the interaction. The GST-p53 fusion proteins comprising full-length p53 (GST-p53), its first 73 residues [GST-p53 (1–73)] and residues 20–73 [GST-p53 (20–73)] were mixed with the RPA proteins that were produced by in vitro translation and labeled with [S35]Met. After washing off the unbound fraction, p53-bound RPA proteins were quantified by scintillation counting and/or visualized by SDS/PAGE followed by autoradiography.
In agreement with previous reports, full-length p53 was most efficient in binding RPA70 (Fig. 2A). There was also some inefficient, most likely nonspecific, binding to RPA32 and no detectable binding to the RPA14. The interaction between p53 and RPA70 involves the N terminus of p53, residues 20–73.
Fig. 2.
Mapping of bindings regions for p53 and RPA70. (A)[35S]-labeled RPA subunits (RPA70, RPA32, and RPA14) were mixed with corresponding GST-fused p53 proteins [GST-p53, GST-p53 (1–73), GST-p53 (20–73), and GST]. Pull-down fractions were analyzed by 15% SDS/PAGE, followed by the autoradiography. Approximately 25–30% of the input RPA70 was bound to the GST-p53 beads. (B) [35S]-labeled RPA70-derived proteins [RPA70, RPA70 (1–422), and RPA70 (1–168)] were mixed with corresponding GST-fused derivatives of the p53 N-terminal part [GST-p53 (1–73), GST-p53 (20–73), and GST] and analyzed by 15% SDS/PAGE of the pull-down fractions, followed by the autoradiography.
Next, GST-fused p53 fragments were mixed with the radiolabeled RPA70 and its derivatives, RPA70 (1–168) and RPA70 (1–422). The GST-pull-down assay showed that amino acids 1–168 of RPA70 mediate the binding with the N-terminal part of p53 (Fig. 2B). The presence of the RPA70A and RPA70B did not increase the binding efficiency. Consistent with this observation, pull-down with the RPA70 (181–422) did not result in any detectable binding (data not shown). Together, these data indicate that the RPA70N domain is important, and the RPA70C domain is dispensable, for binding with the N-terminal transactivation domain of p53.
Residues 1–120 of RPA70 Comprise a Globular OB-Fold Domain. The NMR solution structure of RPA70 (1–169) revealed a folded domain with the OB-fold topology (residues 1–105) and a flexible tail, residues 106–169, that was poorly defined by the experimental NMR restraints (34). We attempted to crystallize RPA70 (1–168) but were unsuccessful, possibly because of the long, flexible C-terminal tail. Based on the NMR data, we generated two protein constructs that comprise residues 1–105 and 1–116. Surprisingly, these proteins had low solubility and could not be used for structural analysis. In contrast, longer constructs comprising residues 1–120 and 1–129 both had good solubility and yielded well diffracting crystals. The structure of RPA70 (1–120) was solved by the Multiple Isomorphous Replacement with Anomalous Scattering method by using two derivatives, HgCl and 4-chloromercuribenzoic acid, and refined against 1.6-Å data to R = 21.8% and Rfree = 26.1%. The structure of RPA70 (1–129) was solved by molecular replacement with RPA70 (1–120) as a search model and refined at 1.9 Å (data not shown).
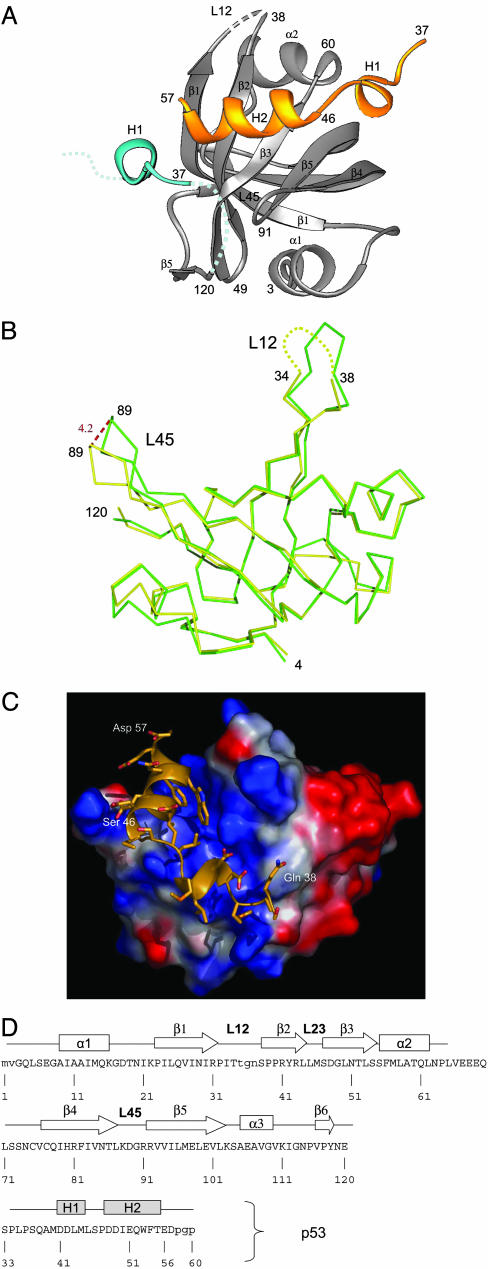
The N-terminal 120 amino acids in the two crystal structures were very similar (RMS deviation of 0.4 Å with 113 Cα atoms), and N-terminal 105 amino acids were similar to the NMR structure (34) (RMS deviation of 2.0 Å with 72 Cα atoms). Residues 106–120 form a loop and small β-strand that were not detected in the NMR structure. Residues 121–129 in the RPA70 (1–129) structure were disordered, leading us to conclude that the globular domain of RPA70N comprises amino acids 1–120. This domain has an OB fold similar to the other OB folds in RPA and features a prominent basic “oligosaccharide/nucleotide” binding cleft between loops L12 and L45 (the loops between β-strands 1 and 2 and between β-strands 4 and 5) (34), a likely site for intermolecular interactions in RPA70N (data not shown).
Structure of the RPA70N/p53N Complex. Attempts at cocrystallizing the p53 (38–57) peptide with RPA70N were unsuccessful. As an alternative strategy, we constructed a number of chimeric proteins with different p53 fragments fused to the C terminus of RPA70 (1–120). The chimera comprising residues 1–120 of RPA70 followed by residues 33–60 of p53 produced crystals that diffracted to 1.6 Å. The structure was solved by molecular replacement with the RPA70 (1–120) structure as a search model and refined to R = 20.4% and Rfree = 23.9%. Relevant crystallographic statistics are given in Table 1 23.9%. Continuous experimental density covers the whole protein except residues 35–37 of RPA70N (the tip of the L12 loop) and the two N-terminal and three C-terminal residues.
The p53 peptide binds in the basic cleft of RPA70N (Fig. 3), and binding induces significant conformational changes in the L12 and L45 loops (Fig. 3B). For example, Asp-89 in the L45 loop shows a 4.2-Å displacement between the bound and unbound structures, whereas the residues not in the L12/L45 loops have only an rms deviation of 0.7 Å with 109 Cα atoms included. Characteristically, flexibility of L12 and L45 loops mediates ssDNA binding in the case of RPA and BRCA2 OB folds (12, 35). For comparison, the ssDNA binding-mediated displacement of the L45 loop in RPA70B was 6.3 Å (12).
Fig. 3.
Structure of RPA70N/p53N complex. (A) Ribbons presentation of the structure. The RPA70N is in gray, and the fusion p53 peptide is in blue. P53 from the symmetry-related molecule is in gold. H1 and H2 are helices within p53. Only the H1 helix of the blue p53 peptide is shown for clarity. (B) Conformational change in RPA70N induced by p53 binding. A superposition of free and bound RPA70N was generated as discussed in the text. Shown is the Cα trace of the free (green) and bound (yellow) domain. Important amino acids are labeled. Maximal shift of the L45 loop are indicated with dashed lines, and the size of the shift is indicated in angstroms (large numbers). The disordered L12 loop is represented by a dotted line. (C) pymol surface potential rendering of RPA70N. Positive and negative charge potential surfaces are blue and red, respectively. (D) Secondary structure elements of RPA70N and p53. RPA70N elements are referred to as in agreement with the nomenclature of the OB fold. The β-strands are indicated by arrows and the α-helices by boxes. The residues disordered in the structure are shown with a dashed line.
The p53 fragment is folded into two helices, H1 (residues 41–44) and H2 (residues 47–55) (Figs. 3 A and D and 4 A and B). The linker area (residues 45–46) makes a sharp turn such that the p53 helices appear to wrap around RPA (Fig. 4B). Interestingly, in the crystals, each RPA70N molecule interacts with two separate p53 peptides. Two separate H1 helices, one from the internal fusion partner of RPA70N (blue in Figs. 3A and 4A) and the other (gold) from a neighboring molecule (Figs. 3A and 4B), interact with two separate sites of RPA. Each H1 helix of p53, in turn, interacts with both its covalently fused RPA and a symmetry-related RPA within the crystal lattice. In solution, the most likely mode of interaction is that observed for the gold peptide in Fig. 3A.
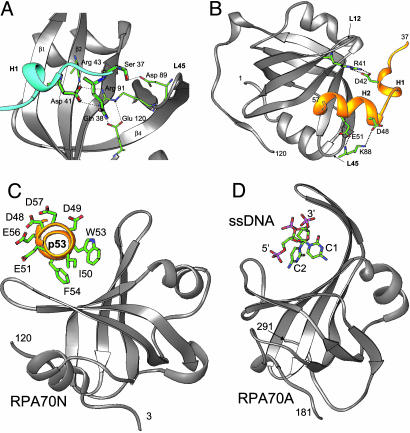
Fig. 4.
Structural mimicry in the p53N/RPA70N complex. Structural details of the p53 interaction with the fusion partner RPA70N (A) and symmetry-related RPA70N (B). (C) Aromatic side chains of p53 helix H2 mediate the interaction with the binding cleft of RPA70N and acidic side chains are exposed to solvent (Helix H1 is omitted for clarity). (D) Structure of RPA70A (ribbon diagram) bound to ssDNA (stick model). Bases C1 and C2 mediate interaction in the binding cleft and acidic phosphates exposed to solvent. RPA70N and p53 are colored as in Fig. 3A; the interacting residues are represented as thick sticks for p53, and thin sticks for RPA70N colored as per atom type: blue for nitrogen, red for oxygen, and green for carbon.
Helix H2 has the most extensive buried surface area and, therefore, appears to be the major determinant of the interaction. It comes from a neighboring symmetry related chimera in the crystal lattice (gold in Fig. 3A and 4B) and binds in the deep basic cleft corresponding to the nucleic acid-binding pocket of the OB fold (Fig. 3C). The orientation of this helix is stabilized by electrostatic interactions of negatively charged side chains of p53 (Asp-48 and Glu-51) with main chain and side chain atoms of the L45 loop of RPA70N (Lys-88), as well as hydrophobic interactions between the buried surface of the p53 helix (Ile-50, Trp-53, and Phe-54) and the hydrophobic floor of the OB-fold binding cleft (Fig. 4B).
Helix 1 has a smaller interaction surface and smaller changes in NMR resonance frequencies upon binding to RPA70N, suggesting that H1 plays a secondary role in the interaction. This notion is consistent with recent data from Vise et al. (24) in which an Arg to Glu mutation at residue 41 in RPA70, which forms a hydrogen bond with ASP42 of p53, significantly reduced, but did not eliminate the p53–RPA70N interaction.
We were able to directly observe changes in resonance frequency for both p53 and RPA70N upon binding. The changes for p53 residues 38–57 in complex with RPA70 (1–120) indicate the formation of two α-helices upon binding to RPA70N (Fig. 6, which is published as supporting information on the PNAS web site).
The RPA/p53 interface has similarities with those of the MDM2 oncoprotein bound to the p53 transactivation domain (residues 17–29) (36). In both structures, the interface is mediated by two aromatic and one hydrophobic residues of p53 (Phe-19, Trp-23, and Leu-26 in MDM2/p53). For both interactions, p53 forms a helix that is folded upon binding. However, despite these gross similarities, RPA does not bind the MDM2 binding site on p53 (residues 17–29) (24, 36) nor do the RPA and MDM2 binding sites overlap.
Strikingly, the mode of p53 binding to RPA is similar to that of ssDNA binding to the DNA-binding domains of RPA (Fig. 3D and 4C) (37). For example, in the RPA70A–ssDNA complex, the negative phosphate groups of DNA are exposed to solvent and the aromatic bases are involved in stacking or hydrophobic interactions with the base of the binding cleft. In the RPA70N–p53 complex, at least five negatively charged side chains are exposed to solvent (Asp-48, Asp-49, Glu-51, Glu-56, and Asp-57), whereas the aromatic residues, Trp-53 and Phe-54 are reminiscent of bases C1 and C2 in the RPA70A/ssDNA complex. Conservation of the binding mechanism, flexible L12 and L45 loops, further extends the similarity.
The RPA-binding region of p53 contains two functionally important phosphorylation sites, Ser-37 and Ser-46 (Fig. 4A and 3B, respectively). Phosphorylation of p53 at Ser-37 is involved in the response to DNA damage (38, 39). Ser-46 is phosphorylated in response to UV damage and is associated with p53-mediated apoptosis (39, 40). Phosphorylation of these residues could potentially modulate the p53–RPA interaction. Alternatively, because of the specific conformation of the p53 peptide when bound to RPA, the ability of Ser-37 and/or Ser-46 to interact productively with a kinase active site could be affected.
ssDNA Competes with p53N for Binding to RPA70N. RPA70N has previously been shown to have weak binding affinity for ssDNA (14). NMR chemical shift mapping suggested that ssDNA interacts with a region that largely overlaps with the p53 binding site of RPA70N (24). Furthermore, it is known that ssDNA disrupts the RPA70–p53 interaction (10). One interpretation of these results is that p53 and ssDNA are in direct competition for the binding site in RPA70N.
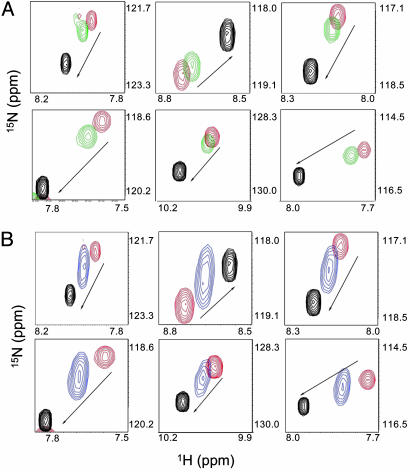
To test this hypothesis, we performed 15N-HSQC NMR experiments in which the 15N-p53/RPA complex was titrated with ssDNA. (Fig. 5A). When increasing amounts of ssDNA were added, the NMR signals of p53 reverted from their chemical shift values in the p53/RPA complex back to the values of unbound p53. This result shows direct competition between ssDNA and p53 and confirms the hypothesis that ssDNA and p53 are in direct competition for a binding site on RPA70N. However, it should be noted that a large molar excess of ssDNA is needed to fully displace all of the p53, indicating that the affinity of p53 for RPA70N is greater than that of ssDNA. It would be reasonable to speculate that, in the full RPA trimer, local concentrations of ssDNA around RPA70N will be elevated upon RPA binding to DNA, and additional events such as phosphorylation of p53 may also alter the affinity.
Fig. 5.
Displacement of p53N from RPA70N by addition of ssDNA and RPA32-Asp. The black peaks represent NMR resonances for unbound 15N-p53N and red peaks for 15N-p53N bound to RPA70N. Changes in resonance frequency are shown for specific p53 residues in the presence of ssDNA (green) (A) and RPA32-Asp (blue) (B). Peaks are labeled according to their residue-specific assignments. With the addition of either ssDNA or a RPA32-Asp peptide, the peaks migrate toward the position of unbound p53 as indicated by the arrows.
A Peptide Mimetic of Hyperphosphorylated RPA32N Can Displace p53N Out of the Binding Site. Several experimental facts taken together suggested a possibility for yet another competition. First, in response to DNA damage, the N terminus of RPA32 (RPA32N) is extensively phosphorylated (15, 16, 41, 42). Second, a negatively charged hyperphosphorylation mimetic peptide derived from the RPA32N (but not its unphosphorylated form) has been reported to bind to RPA70N (43). Third, p53N fragment 32-LSPLPSQA-39 and RPA32N fragment 28-GSPAPSQA-35 have surprising similarity. To test whether such a peptide can compete with p53 for binding to RPA70N, we used a peptide corresponding to residues 1–34 of RPA32 in which Ser/Thr residues that have been reported to be phosphorylated in vivo (Ser-8, Ser-13, Thr-21, Ser-23, Ser-29, and Ser-33) (44) were replaced with Asp (RPA32N-Asp). Interestingly, in addition to nine serines/threonines, at least six of which are phosphorylated in vivo, this peptide also contains six aromatic residues and could potentially form one or more amphipathic helices.
Both a “wild-type peptide” and the hyperphosphorylation mimetic, RPA32N-Asp, were titrated into a 15N-p53(38–58)/RPA70N complex, and 15N-HSQC NMR was used again to monitor changes in the amide resonances of 15N-p53. Although no substantial changes were observed with the wild-type peptide (data not shown), titration of RPA32N-Asp caused chemical shift perturbations in the p53 peptide, progressively changing from the RPA-bound values to those of the unbound p53 peptide (Fig. 5B). This data indicates that a negatively charged RPA32N-Asp peptide (mimicking that after DNA damage) can effectively compete with p53 for binding to RPA70N. As with ssDNA, a molar excess of RPA32-Asp peptide was needed to displace p53, but not as much as for the ssDNA titration experiment.
From the titration experiments, we estimate that the binding affinities of the three species for RPA70N are in the range of 10–150 μM, with ssDNA having ≈10-fold lower affinity than p53N and RPA32-Asp having an intermediate affinity. However, it should be noted that many factors could influence these relative affinities, such as the effects of increased effective local concentrations of interacting components due to oligomerization (p53) and complex formation with other regions of trimeric RPA (RPA32/ssDNA).
Discussion
In this work, we detail a molecular interface between p53 and RPA, two proteins essential for a normal response to DNA damage in mammalian cells. We reveal that crucial to the regulation of this protein–protein interaction is the OB-fold domain of RPA70N and residues 37–57 of p53, a region associated with induction of apoptosis in response to UV damage (40, 45). Our results also indicate a mechanism by which ssDNA, a primary product of DNA damage, can provide positive feedback to dissociate p53/RPA complexes, thus releasing each protein to function independently.
A simplified mechanistic model has been proposed in ref. 46. According to this model, p53 (and many other proteins) interact with RPA via a “two-point interaction” in which two distinct areas of the p53 (p53N and p53C) contact two distinct areas of RPA (RPA70N and RPA70C). After DNA binding, the N and C termini of RPA70 move apart, allosterically switching the two-point interaction. Recently, a more comprehensive model has been suggested (24). ssDNA (a product of DNA damage) has been hypothesized to position RPA70N into an orientation favorable for binding ssDNA. The linkage effect increases the affinity of RPA70N to ssDNA by increasing the local concentration of ssDNA. The increased affinity will result in the dissociation of p53 from RPA (24). Our new data are in agreement with this comprehensive model and provides it with structural basis. The interaction between RPA70C and the C-terminal domain of p53 remains more speculative.
It is generally assumed that the overall cellular response to DNA damage is in part determined by the degree of damage sustained. DNA damage induces hyperphosphorylation of RPA32 (15–19). Our data suggest a possible role for RPA32N in the regulation of RPA70N binding to either p53 or ssDNA. We show that a peptide mimetic of phosphorylated RPA32N is able to dissociate a p53 peptide from its OB binding site on RPA70N. Thus, it is possible that ssDNA and/or phosphorylated RPA32, both consequences of DNA damage, can structurally compete with p53 (and one another) for binding to the OB fold of RPA70N. The OB fold of RPA70N thus represents a structural entity endowing dual affinity (i) for acidic, amphipathic peptide helices and (ii) for ssDNA. Binding is mutually exclusive and may be hierarchical.
The binding mode and regulatory mechanisms described in this work may be of a general nature. For example, the N-terminal acidic peptide of Rad51 and ssDNA compete for binding to the ssDNA-binding domain, RPA70A, which is a canonical OB fold (47). The interaction between RPA70B and hyperphosphorylated RPA32N has been proposed to play a role in modulating the cellular pathways by altering the RPA70B-mediated RPA–DNA and RPA–protein interactions (48). The acidic transactivation domain of BRCA2 binds RPA. Although the precise domain of RPA has not been mapped, the cancer predisposing mutation of Tyr-42 to Cys (aromatic residue) significantly compromises this interaction (49). Many other acidic transactivators may share the same regulatory mechanisms, and many other OB folds and those of RPA may also be subject to similar competition between proteins and ssDNA.
Supplementary Material
Acknowledgments
This work was supported by research grants from the National Institute of General Medical Science (to A.B.), the National Cancer Institute of Canada (NCIC) with Funds from the Canadian Cancer Society (to C.H.A.), and the Yorkshire Cancer Research (to J.M.). A.A. was the recipient of a fellowship from the NCIC, and L.K. is the recipient of a fellowship from the Leukemia and Lymphoma Society of Canada.
Author contributions: J.M., C.H.A., and A.B. designed research; E.B., L.K., A.A., G.-S.Y., Y.L., A.P.-L., J.L., and A.L.O. performed research; C.H.A. and A.B. analyzed data; and J.M., C.H.A., and A.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HSQC, heteronuclear sequential quantum correlation; OB, oligonucleotide/oligosaccharide-binding; RPA, replication protein A.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org/pdb (PDB ID codes 2B29 and 2B3G).
References
- 1.Ko, L. J. & Prives, C. (1996) Genes Dev. 10, 1054-1072. [DOI] [PubMed] [Google Scholar]
- 2.Levine, A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 3.Dutta, A., Ruppert, J. M., Aster, J. C. & Winchester, E. (1993) Nature 365, 79-82. [DOI] [PubMed] [Google Scholar]
- 4.He, Z., Brinton, B. T., Greenblatt, J., Hassell, J. A. & Ingles, C. J. (1993) Cell 73, 1223-1232. [DOI] [PubMed] [Google Scholar]
- 5.Li, R. & Botchan, M. R. (1993) Cell 73, 1207-1221. [DOI] [PubMed] [Google Scholar]
- 6.Romanova, L. Y., Willers, H., Blagosklonny, M. V. & Powell, S. N. (2004) Oncogene 23, 9025-9033. [DOI] [PubMed] [Google Scholar]
- 7.Sommers, J. A., Sharma, S., Doherty, K. M., Karmakar, P., Yang, Q., Kenny, M. K., Harris, C. C. & Brosh, R. M., Jr. (2005) Cancer Res. 65, 1223-1233. [DOI] [PubMed] [Google Scholar]
- 8.Abramova, N. A., Russell, J., Botchan, M. & Li, R. (1997) Proc. Natl. Acad. Sci. USA 94, 7186-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265, 346-355. [DOI] [PubMed] [Google Scholar]
- 10.Miller, S. D., Moses, K., Jayaraman, L. & Prives, C. (1997) Mol. Cell. Biol. 17, 2194-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wold, M. S. (1997) Annu. Rev. Biochem. 66, 61-92. [DOI] [PubMed] [Google Scholar]
- 12.Bochkarev, A., Bochkareva, E. &. (2004) Curr. Opin. Struct. Biol. 14, 36-42. [DOI] [PubMed] [Google Scholar]
- 13.Murzin, A. G. (1993) EMBO J. 12, 861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daughdrill, G. W., Ackerman, J., Isern, N. G., Botuyan, M. V., Arrowsmith, C., Wold, M. S. & Lowry, D. F. (2001) Nucleic Acids Res. 29, 3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, V. F. & Weaver, D. T. (1993) Mol. Cell. Biol. 13, 7222-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brush, G. S., Anderson, C. W. & Kelly, T. J. (1994) Proc. Natl. Acad. Sci. USA 91, 12520-12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Block, W. D., Yu, Y. & Lees-Miller, S. P. (2004) Nucleic Acids Res. 32, 997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brush, G. S., Morrow, D. M., Hieter, P. & Kelly, T. J. (1996) Proc. Natl. Acad. Sci. USA 93, 15075-15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta, A. & Stillman, B. (1992) EMBO J. 11, 2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, H., Guan, J., Wang, H., Perrault, A. R., Wang, Y. & Iliakis, G. (2001) Cancer Res. 61, 8554-8563. [PubMed] [Google Scholar]
- 21.Binz, S. K., Sheehan, A. M. & Wold, M. S. (2004) DNA Repair (Amst.) 3, 1015-1024. [DOI] [PubMed] [Google Scholar]
- 22.Leiter, L. M., Chen, J., Marathe, T., Tanaka, M. & Dutta, A. (1996) Oncogene 12, 2661-2668. [PubMed] [Google Scholar]
- 23.Lin, Y. L., Chen, C., Keshav, K. F., Winchester, E. & Dutta, A. (1996) J. Biol. Chem. 271, 17190-17198. [DOI] [PubMed] [Google Scholar]
- 24.Vise, P. D., Baral, B., Latos, A. J. & Daughdrill, G. W. (2005) Nucleic Acids Res. 33, 2061-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277-293. [DOI] [PubMed] [Google Scholar]
- 26.Bartles, C., Xia, T., Billeter, M., Guntert, P. & Wuthrich, K. (1995) J. Biomol. NMR 5, 1-10. [DOI] [PubMed] [Google Scholar]
- 27.Otwinoski, Z. & Minor, W. (1997) in Macromolecular Crystallography, Part A., eds. Carter, C. W., Jr., & Sweet, R. M. (Academic, Orlando, FL), pp. 307-326.
- 28.Furey, W. & Swaminathan, S. (1997) in Methods in Enzymology: Macromolecular Crystallography, Part B., eds. Carter, C. W. J. & Sweet, R. M. (Academic, Orlando, FL), pp. 590-620.
- 29.Vagin, A. & Teplyakov, A. (1997) J. Appl. Crystallogr. 30, 1022-1025. [Google Scholar]
- 30.Perrakis, A., Morris, R. & Lamzin, V. S. (1999) Nat. Struct. Biol. 6, 458-463. [DOI] [PubMed] [Google Scholar]
- 31.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov, G., Vagin, A. & Dodson, E. (1996) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 33.Carson, M. (1997) in Macromolecular Crystallography, Part B., eds. Carter, C. W., Jr., & Sweet, R. M. (Academic, Orlando, FL), pp. 493-505.
- 34.Jacobs, D. M., Lipton, A. S., Isern, N. G., Daughdrill, G. W., Lowry, D. F., Gomes, X. & Wold, M. S. (1999) J. Biomol. NMR 14, 321-331. [DOI] [PubMed] [Google Scholar]
- 35.Yang, H., Jeffrey, P. D., Miller, J., Kinnucan, E., Sun, Y., Thoma, N. H., Zheng, N., Chen, P. L., Lee, W. H. & Pavletich, N. P. (2002) Science 297, 1837-1848. [DOI] [PubMed] [Google Scholar]
- 36.Kussie, P. H., Gorina, S., Marechal, V., Elenbaas, B., Moreau, J., Levine, A. J. & Pavletich, N. P. (1996) Science 274, 948-953. [DOI] [PubMed] [Google Scholar]
- 37.Bochkarev, A., Pfuetzner, R. A., Edwards, A. M. & Frappier, L. (1997) Nature 385, 176-181. [DOI] [PubMed] [Google Scholar]
- 38.Dohoney, K. M., Guillerm, C., Whiteford, C., Elbi, C., Lambert, P. F., Hager, G. L. & Brady, J. N. (2004) Oncogene 23, 49-57. [DOI] [PubMed] [Google Scholar]
- 39.Bulavin, D. V., Saito, S., Hollander, M. C., Sakaguchi, K., Anderson, C. W., Appella, E. & Fornace, A. J., Jr. (1999) EMBO J. 18, 6845-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oda, K., Arakawa, H., Tanaka, T., Matsuda, K., Tanikawa, C., Mori, T., Nishimori, H., Tamai, K., Tokino, T., Nakamura, Y., et al. (2000) Cell 102, 849-862. [DOI] [PubMed] [Google Scholar]
- 41.Cheng, X., Cheong, N., Wang, Y. & Iliakis, G. (1996) Radiother. Oncol. 39, 43-52. [DOI] [PubMed] [Google Scholar]
- 42.Brush, G. S. & Kelly, T. J. (2000) Nucleic Acids Res. 28, 3725-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binz, S. K., Lao, Y., Lowry, D. F. & Wold, M. S. (2003) J. Biol. Chem. 278, 35584-35591. [DOI] [PubMed] [Google Scholar]
- 44.Zernik-Kobak, M., Vasunia, K., Connelly, M., Anderson, C. W. & Dixon, K. (1997) J. Biol. Chem. 272, 23896-23904. [DOI] [PubMed] [Google Scholar]
- 45.Latonen, L., Taya, Y. & Laiho, M. (2001) Oncogene 20, 6784-6793. [DOI] [PubMed] [Google Scholar]
- 46.Bochkareva, E., Belegu, V., Korolev, S. & Bochkarev, A. (2001) EMBO J. 20, 612-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stauffer, M. E. & Chazin, W. J. (2004) J. Biol. Chem. 279, 25638-25645. [DOI] [PubMed] [Google Scholar]
- 48.Liu, Y., Kvaratskhelia, M., Hess, S., Qu, Y. & Zou, Y. (2005) J. Biol. Chem. 280, 32775-32783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong, J. M., Ionescu, D. & Ingles, C. J. (2003) Oncogene 22, 28-33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.