Abstract
Transcription factor (TF) IIIB, the central transcription initiation factor of RNA polymerase III (pol III), is composed of three subunits, Bdp1, Brf1 and TATA-binding protein (TBP), all essential for normal function in vivo and in vitro. Brf1 is a modular protein: Its N-proximal half is related to TFIIB and binds similarly to the C-terminal stirrup of TBP; its C-proximal one-third provides most of the affinity for TBP by binding along the entire length of the convex surface and N-terminal lateral face of TBP. A structure-informed triple fusion protein, with TBP core placed between the N- and C-proximal domains of Brf1, has been constructed. The Brf1-TBP triple fusion protein effectively replaces both Brf1 and TBP in TFIIIC-dependent and -independent transcription in vitro, and forms extremely stable TFIIIB–DNA complexes that are indistinguishable from wild-type TFIIIB–DNA complexes by chemical nuclease footprinting. Unlike Brf1 and TBP, the triple fusion protein is able to recruit pol III for TATA box-directed transcription of linear and supercoiled DNA in the absence of Bdp1. The Brf1-TBP triple fusion protein also effectively replaces Brf1 function in vivo as the intact protein, creating a TBP paralogue in yeast that is privatized for pol III transcription.
Keywords: protein fusions, RNA polymerase III, promoter
The TATA-binding protein (TBP) is the common component of transcription by all three nuclear RNA polymerases. In the context of RNA polymerase (pol) III, which is the focus of the work that is presented here, TBP functions as one of three subunits of transcription factor (TF) IIIB; Brf1 and Bdp1 are the other two subunits. Budding yeast (Saccharomyces cerevisiae) Brf1 and TBP copurify as a complex that is also called B′; the less tightly associated Bdp1 separates during purification.
TFIIIB is the core transcription initiation factor of pol III, required for and sufficient for recruiting the enzyme to the promoter. Placement of TFIIIB at its canonical DNA site approximately two and one-half helical turns upstream of the transcriptional start site is secured by an assembly factor, the large six-subunit TFIIIC. The TBP subunit of TFIIIB also allows it to bind autonomously to the minority of S. cerevisiae pol III genes with strong TATA boxes. In these instances, TFIIIC is dispensable for transcription of DNA in vitro, although it remains essential for transcription of chromatin templates in vitro, and is also essential for pol III transcription in vivo (1).
All pol III-transcribed genes in yeast are transcribed by this unitary transcription apparatus composed of the polymerase, TFIIIA, B and C. Two kinds of elaboration have evolved in metazoans: In Drosophila, a TBP paralogue, TRF1, assumes the role of TBP in most and possibly all pol III transcription and associates tightly with Brf1; TRF1 probably also participates in some transcription by pol II (ref. 2; reviewed in ref. 3). The human pol III transcription system instead uses a Brf1 paralogue, Brf2, for transcription of a small group of genes with distinctive upstream promoter elements, including a TATA box. A TFIIIB-like complex composed of TBP, Brf2, and Bdp1 assembles at this TATA box subject to protein–protein interactions with other transcription initiation factors bound to a proximal promoter element and distal enhancer-like site (reviewed in ref. 4).
Brf1 is a hybrid protein: Its N-terminal half is related to TFIIB, whereas the C-terminal half has no counterpart in the pol II or pol I transcription apparatus. A conserved segment in the C-terminal one-third of Brf1 contributes most of its affinity for TBP. An additional, weaker site is located in the TFIIB-related N-terminal half (5–8). The C-proximal one-third of Brf1 also provides the primary binding site for Bdp1, but an additional Bdp1 site is located in the N-terminal half of Brf1. Thus, it is possible to assemble a TFIIIB–DNA complex in vitro with different domains of Brf1: a stable DNA complex is formed by the C-terminal one-third of Brf1, TBP, and Bdp1, but this complex is transcriptionally only very weakly active. Recruitment of Bdp1 and the N-terminal half of Brf1 to the TBP–DNA complex is co-dependent and yields an unstable TFIIIB–DNA complex that nevertheless can yield nearly wild-type levels of transcription (6).
Bdp1 is also modular. An extended N-terminal segment is dispensable for viability (9), but not without phenotype: Removing amino acids 1–240 disrupts the distinctive pattern of Ty1 retroposon integration upstream of pol III-transcribed genes, presumably because nucleosome positioning upstream of TFIIIB becomes less ordered (10). Splitting Bdp1 at amino acid 352 (within an internal dispensable segment) is also compatible with cell viability (9), with each separate Bdp1 segment able to assemble in vitro into a TFIIIB–DNA complex.
We are exploiting this modularity, in combination with information about the internal structure of TFIIIB, to explore ways of reconnecting the protein domains of TFIIIB. Here, we explore the in vitro properties and functional competence in vivo of Brf1-TBP fusions. One of these fusions places the highly conserved core of yeast TBP between the N- and C-terminal segments of Brf1 to create a protein that retains TBP and Brf1 function in vitro, retains Brf1 function in vivo, and creates a TBP paralogue in yeast that is dedicated to pol III transcription.
Materials and Methods
DNA, proteins, plasmids for overproduction of proteins in E. coli and strain construction in S. cerevisiae, and methods for Western and Northern blots are specified in detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Protein–DNA complexes for transcription, EMSA, and footprinting were formed at 20°C in 18–20 μl of a previously described reaction buffer (11) with specified concentrations of NaCl. For Fig. 1B, transcription complexes were formed for 60 min with 10 fmol of pol III in reaction buffer containing 60 mM NaCl followed by 30 min of multiple-round transcription as described in ref. 11. Samples were processed for denaturing gel electrophoresis and the phosphor image was quantified as described in ref. 12. For Figs. 1 C and D, protein–DNA complexes were formed for 60 min in reaction buffer containing 70 mM NaCl, followed by the addition of 2 μl of 2 mg/ml heparin where indicated. Native gel electrophoresis was performed as described in ref. 13, and two-dimensional MPE-Fe(II) footprinting followed ref. 11. For the experiment shown in Fig. 2, TFIIIB–TFIIIC–DNA complexes were formed for 40 min (A, C, and D) or 60 min (B) in reaction buffer containing 90 mM NaCl. For Figs. 2 A, C, and D, 2 μl of 5 fmol/μl pol III and 1 μl of 1 M NaCl were added to preformed TFIIIB–TFIIIC–TFIIIB–TFIIIC–DNA complexes, followed 20 min later by (i) 30 min of multiple-round transcription as described in ref. 11 or (ii) the addition of 2.5 μl of reaction buffer containing 2 mM each ATP and CTP and 250 μM [α-32P]UTP (20 cpm/fmol) for 5 min and subsequent addition of 2.5 μl of a mixture containing 2 mM GTP and 2 mg/ml heparin for 5 min to limit transcription to a single round.
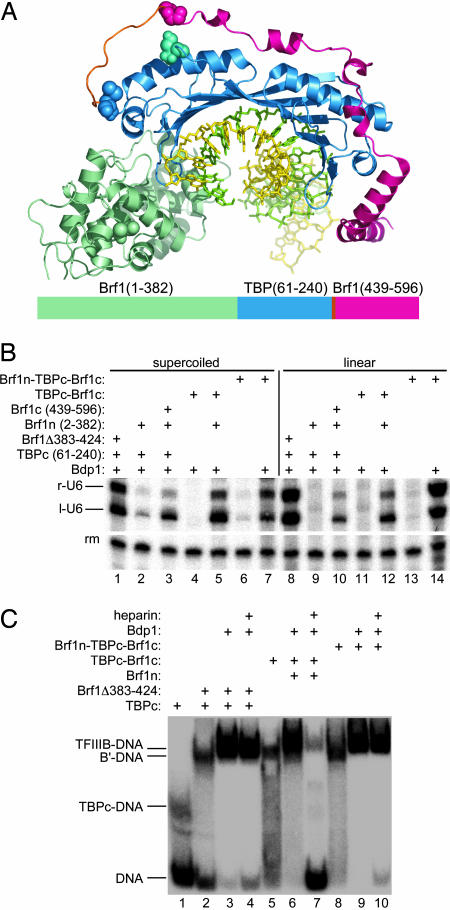
Fig. 1.
The Brf1n-TBPcore-Brf1c triple fusion can replace Brf1 and TBP for TFIIIC-independent transcription and for formation of heparin-resistant TFIIIB–DNA complexes. (A) A model of the Brf1 (1–382)-TBP core-Brf1 (439–596) triple fusion. DNA (yellow and green sticks), TBPc (blue ribbon), and the resolved Brf1 (439–506) segment (red ribbon) are from ref. 14. A possible path of a (GS)6 linker (orange) between TBP residue 240 and Brf1 residue 439 (space-filled) is shown. The Brf1 (76–273) segment modeled into the TFIIB–TBP–DNA crystal structure (8) is also shown (light green) with Brf1 residue 273 space-filled and the TBPc N-terminal residue 61 (space-filled in cyan) highlighted. A cartoon identifying the segments comprising the triple fusion is sketched out below the model [but the (GS)6 linker is inapparent on this scale]. (B) TFIIIC-independent transcription. Protein–DNA complexes were formed with 50 fmol of supercoiled plasmid DNA (lanes 1–7) or a 364-bp linear DNA fragment (lanes 8–14) and 200 fmol of the TFIIIB components designated above each lane. U6LboxB transcripts and a labeled DNA recovery marker (rm) are identified on the left. The weak transcripts with lower mobility than r-U6 and l-U6 RNA in lanes 9 and 11 do not depend on either Brf1 or Bdp1 (data not shown, but cf. 34). (C) EMSA. Protein–DNA complexes were formed with 200 fmol of the TFIIIB components designated above each lane and 8 fmol of a 57-bp TATA box-containing probe (specified in Materials and Methods) and were analyzed on a 4% native polyacrylamide gel.
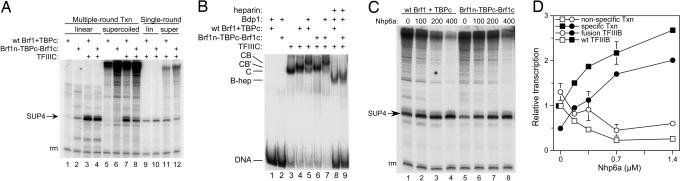
Fig. 2.
The Brf1n-TBPc-Brf1c triple fusion protein is competent for TFIIIC-dependent transcription and formation of a heparin-resistant TFIIIB–DNA complex. (A) TFIIIC-dependent transcription of the SUP4 tRNA gene variant TA-30 (50 fmol) as supercoiled plasmid (lanes 5–8, 11, 12) or 160-bp DNA fragment (lanes 1–4, 9, 10) with 50 fmol of TFIIIC, 100 fmol of Bdp1, and 25 fmol of wild-type Brf1 + TBPc or Brf1n-TBPc-Brf1c, as indicated above each lane. Multiple-round (lanes 1–8) and single-round (lanes 9–12) transcription was performed as described in Materials and Methods. The SUP4 transcript and recovery marker (rm) are identified on the left. (B) EMSA. Protein–DNA complexes were formed with 100 fmol each of TFIIIB component subunits and 50 fmol of TFIIIC on a TA-30 SUP4 DNA probe (1 fmol) as indicated above each lane. TFIIIC–TFIIIB–DNA (CB), TFIIIC–Brf1–TBPc–DNA (CB′), TFIIIC–DNA (C), and heparin-resistant TFIIIB–DNA (B-hep) complexes are identified on the left. (C) Nhp6a suppresses nonspecific transcription. Nhp6a (quantities, in nanograms, indicated above each lane) was mixed with TFIIIB assembled with wild-type Brf1 and TBPc (lanes 1–4) or Brf1n-TBPc-Brf1c (lanes 5–8), followed by the addition of TFIIIC and nonspecific competitor DNA for single-round transcription as specified for A.(D) Quantification of C, averaged with an additional experiment after normalization to the yield of SUP4 RNA with wild-type TFIIIB in the absence of Nhp6a. Average deviations exceeding symbol sizes are shown; the 800-ng Nhp6a points are from a single experiment. Open symbols, nonspecific transcription; filled symbols, specifically initiating transcription; squares, reference type TFIIIB; circles, TFIIIB assembled with Brf1n-TBPc-Brf1c.
Results
The Brf1 segment spanning amino acid residues 439–545 contains the major binding site for TBP as well as a major site of interaction with Bdp1 (6). The determination of structure of the Brf1 (439–596)–TBP core–DNA ternary complex (14) provides a ready framework for identifying Brf1 residues that are important for the assembly of Bdp1 into TFIIIB (unpublished work). Because the C terminus of TBP (M240; space-filled in Fig. 1 A) is located only ≈20 Å from Brf1 residue P439 (space-filled), we generated a fusion protein consisting of the highly conserved TBP core (TBPc) (amino acids 61–240) linked to Brf1c (amino acids 439–596) through a 12-aa alternating G–S connector (orange).
The C-terminal stirrup of TBP, which binds to TFIIB, is also the binding site of the N-terminal half of Brf1 (amino acids 1–282 or 1–365) (8). Brf1 accommodates ≈40 amino acid deletions between amino acid residues 365 and 425 without loss of activity (15). This region, which is predicted to be predominantly unstructured, is highly diverged among (and partly missing from) seven sensu stricto Saccharomyces and almost entirely missing in Brf1 from the yeast Kluyveromyces lactis. Because the N terminus of TBPc (S61; space-filled in cyan in Fig. 1 A) lies adjacent to the N terminus of the Brf1c segment (P439; space-filled), it occurred to us that fusing an N-terminal fragment of Brf1 (residues 1–382, Brf1n) directly to the N terminus of the TBPc-Brf1c fusion would generate a protein that retains the spatial relationship between the two halves of Brf1 and TBP while fusing the two subunits of the tightly associated TBP-Brf1 complex B′ (16) into a single polypeptide chain.
The ability of TBPc-Brf1c and Brf1n-TBPc-Brf1c to support TFIIIC-independent transcription by pol III is shown in Fig. 1B. Because TBP binds to its TATA box in either orientation (17, 18), the U6 small nuclear RNA gene-derived (SNR6) diagnostic template U6LboxB directs the synthesis of divergent transcripts (r-U6 and l-U6). Brf1Δ383–424 was chosen as the “reference type” protein because it most closely resembles the Brf1 segment in the Brf1n-TBPc-Brf1c fusion. Its ability to promote transcription on supercoiled and linear DNA templates is shown in lanes 1 and 8, respectively. Supercoiled DNA was poorly transcribed with Brf1n and TBPc (lane 2), and the linear template was not transcribed (lane 9); the addition of Brf1c allowed significant transcription of both templates (lanes 3 and 10). The TBPc-Brf1c fusion proved to be 2-fold more effective than its separate components in complementing Brf1n (lanes 5 and 12). Brf1n-TBPc-Brf1c effectively replaced both Brf1 and TBP for transcription of supercoiled and linear templates (lanes 7 and 14), and was comparably active for transcription of linear DNA (compare lanes 14 and 8) but only half as active for transcription of supercoiled DNA as Brf1Δ383–424 and TBPc (compare lanes 7 and 1). We ascribe this unusual difference (negatively supercoiled DNA is commonly seen to be the more active template) to a higher affinity of the Brf1n-TBPc-Brf1c fusion for imperfect TATA boxes (see below) and the reduction in the number of alternative binding sites in the significantly smaller PCR-derived linear DNA.
Surprisingly, the triple fusion protein was also capable of promoting a low level of transcription on both templates in the absence of Bdp1 (lanes 6 and 13). Bdp1-independent and specifically initiating transcription has previously been documented, but only on heteroduplex DNA templates with partially preopened promoters (15).
TFIIIC-independent DNA complex formation was also examined by EMSA (Fig. 1C) and methidiumpropyl-EDTA-Fe(II) footprinting. The triple fusion protein was comparable to Brf1Δ383–424 and TBPc in ability to form B′–DNA complexes (Fig. 1C, lanes 2 and 8), and also TFIIIB–DNA complexes (lanes 3 and 9) that were resistant to displacement by the polyanion heparin (lanes 4 and 10). The TBPc-Brf1c fusion formed a metastable complex with DNA with the same electrophoretic mobility as the corresponding B′–DNA complex (lane 5) but generated a TFIIIB–DNA complex with Brf1n and Bdp1 (lane 6) that was sensitive to heparin treatment (lane 7). The footprints of TFIIIB–DNA complexes assembled either with wild-type Brf1, TBPc, and Bdp1 or with Brf1n-TBPc-Brf1c and Bdp1 were indistinguishable, extending the TBP footprint of a TATA box-containing DNA probe both upstream and downstream by ≈10 bp (Fig. 5, which is published as supporting information on the PNAS web site).
The competence of Brf1n-TBPc-Brf1c for TFIIIC-dependent complex formation and transcription was also examined. The SUP4 tRNA gene derivative TA-30 (19), which embeds a partial TATA box (TATAAA) 30 bp upstream of the start site of transcription in a GC-rich sequence surround, retains TFIIIC-dependence of the assembly of wild-type TFIIIB at the promoter while forcing a unique positioning of TFIIIB that specifies initiation at bp + 1. Brf1n-TBPc-Brf1c and wild-type Brf1 + TBPc generated comparable levels of multiple-round, TFIIIC-dependent transcription of this template presented as a short DNA fragment (Fig. 2 A, compare lanes 3 and 4). However, the triple fusion generated higher levels of nonspecific transcription in the absence of TFIIIC (compare lanes 1 and 2), and the background of TFIIIC-independent SUP4 transcription also increased from 1.5% for wild-type Brf1 to 16% for the triple fusion. A similar result was obtained for transcription of supercoiled plasmid pTA-30 (lanes 5–8). The results of single-round transcription (which directly measures the relative number of active TFIIIB–DNA complexes) (lanes 9–12) reflected the outcome of multiple-round transcription (lanes 3, 4, 7, and 8). Because full transcriptional activity does not require stable TFIIIB complexes, TFIIIB–TFIIIC–DNA complex formation was examined separately (Fig. 2B). The Brf1n-TBPc-Brf1c triple fusion was indistinguishable from wild-type Brf1 and TBPc in ability to form B′–TFIIIC–DNA and TFIIIB–TFIIIC–DNA complexes as well as heparin-resistant, TFIIIC-dependent TFII-IB–DNA complexes (compare lanes 6, 7, and 9 with lanes 4, 5, and 8, respectively).
We surmised that the lower level of transcription achieved with the Brf1n-TBPc-Brf1c triple fusion on plasmid DNA templates relative to wild-type Brf1 and TBPc might result from an increased affinity of the triple fusion protein for imperfect TATA boxes (as indicated in Fig. 2 A, lanes 2 and 6), which would generate alternative TFIIIB–DNA complexes that compete for limiting pol III. The presence of naked DNA for direct TFIIIB binding is unique to in vitro studies that use fully recombinant TFIIIB with TFIIIC and pol III purified to near homogeneity, and distinct from the in vivo situation, where DNA is present as chromatin. The abundant HMG1-like yeast proteins Nhp6a and Nhp6b were used to examine the influence of naked DNA on specific transcription. Nhp6a and Nhp6b are closely related nonspecific DNA-binding proteins that are prevalent in partially purified yeast fractions. Yeast deleted for both genes are temperature-sensitive because of a defect in SNR6 (U6 small nuclear RNA gene) transcription by pol III, but other pol III-transcribed genes that have been examined are not affected. The SNR6 gene is unique in the grossly suboptimal separation (202 bp) of its box A and box B TFIIIC binding sites. Nhp6 severely bends DNA and it has been proposed that Nhp6 bound between box A and box B can bring these promoter elements into optimal range for simultaneous occupancy by TFIIIC (20–22). Nhp6a and Nhp6b also stimulate SNR6 transcription in vitro with partially purified proteins or with recombinant TFIIIB and highly purified pol III and TFIIIC. In contrast, no stimulation of transcription was observed on tRNA genes (with or without a TATA box) that contained optimally spaced box A and box B elements [in assays for which at least one transcription protein was only partially purified (20, 21)].
Fig. 2C compares the effect of Nhp6a on single-round transcription of the SUP4 variant TA-30 plasmid template with wild-type Brf1 and TBPc or the Brf1n-TBPc-Brf1c triple fusion, respectively. Nhp6a suppressed nonspecific transcription while favoring specific transcription of the SUP4 gene: Nonspecific transcription with wild-type Brf1 and TBPc was reduced ≈4-fold, and somewhat less (2- to 3-fold) with the triple fusion protein (Fig. 2D); specific transcription with the triple fusion protein increased 4-fold, somewhat more than with wild-type Brf1 and TBPc (≈2.5-fold). Nhp6a also increased the transcriptional reinitiation rate ≈2-fold in multiple rounds of transcription [performed in parallel with the experiments shown in Fig. 2D (data not shown)]. Nhp6b behaved similarly to Nhp6a (data not shown).
The ability of Brf1n-TBPc-Brf1c to form a heparin-resistant TFIIIB–DNA complex (Figs. 1C and 2B), and to support both TFIIIC-independent and TFIIIC-dependent transcription by pol III in vitro (Figs. 1B and 2 A), prompted us to examine whether the triple fusion protein could replace the essential function of Brf1 in vivo. The Brf1n-TBPc-Brf1c fusion was cloned into a pRS315-derived plasmid under the control of the galactose-inducible GAL1 promoter and transformed into a haploid brf1 deletion strain bearing a wild-type copy of BRF1 on a URA3-containing plasmid (see Materials and Methods). Plasmid shuffling by selection for resistance to 5-fluoroorotic acid yielded cells whose growth was strictly galactose-dependent, demonstrating that Brf1 function was provided by Brf1n-TBPc-Brf1c (Fig. 3; pGal-TF). A strain producing wild-type Brf1 under the control of the same galactose-inducible promoter (pGal-BRF) was constructed similarly. Substituting Brf1n-TBPc-Brf1c for Brf1 did not impose any detectable growth limitation (Fig. 3 and data not shown).
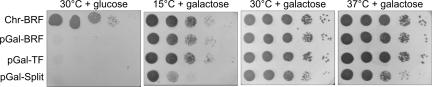
Fig. 3.
The Brf1n-TBPc-Brf1c triple fusion and the Brf1n-TBPc/Brf1c split fusion can replace Brf1 function in vivo. Ten-fold serial dilutions of the wild-type (chromosomal BRF1; Chr-BRF) strain DY9876 and strains dependent on plasmid-borne GAL1 promoter expression of Brf1 (pGal-BRF), Brf1n-TBPc-Brf1c (pGAL-TF), and the Brf1n-TBPc/Brf1c split (pGal-Split) were plated on yeast extract/peptone/dextrose or yeast extract/peptone plus galactose (serial dilutions of 107 cells per ml) and grown at 15°C, 30°C, and 37°C, as indicated.
Because Brf1 activity can be reconstituted in vitro by combining Brf1n and Brf1c and because Brf1n and TBPc-Brf1c can substitute for both Brf1 and TBP in transcription in vitro (Fig. 1B), we examined the possibility that Brf1 function in the pGal-TF strain might require proteolysis of Brf1n-TBPc-Brf1c (e.g., by cleavage of the flexible linker between TBPc and Brf1c). To this end, genes encoding the two halves of Brf1n-TBPc-Brf1c split either between Brf1n and TBPc-Brf1c or between Brf1n-TBPc and Brf1c were separately cloned under the control of the GAL1 promoter into pRS315 (LEU2)- and pRS313 (HIS3)-derived plasmids, both plasmids were transformed into the brf1-knockout strain, and LEU2+, HIS3+, and URA3+ transformants were isolated.
We similarly attempted to replace BRF1 with plasmids separately encoding Brf1n and Brf1c (without TBPc), and a plasmid encoding the Brf1n-TBPc fragment alone. Only coexpression of Brf1n-TBPc and Brf1c (pGal-Split) allowed the isolation of transformants whose growth was galactose-dependent: 16/16 (Brf1n-TBPc + Brf1c) isolates, compared with 0/88 (Brf1n + TBPc-Brf1c), 0/56 (Brf1n + Brf1c), and 0/29 Brf1n-TBPc isolates. These results support the idea that the major site of interaction of Brf1 on TBP is located in its C-terminal half (6) and that, although the N-terminal half of Brf1 provides an additional, lower affinity site for TBP (8), the assembly of the Brf1n fragment into a TFIIIB–DNA complex in vivo requires its fusion to TBP or its natural linkage to Brf1c. Brf1n-TBPc alone could not replace Brf1, consistent with the 100 C-terminal residues of Brf1 being essential for viability (23).
A comparison of growth at 15°C, 30°C, and 37°C for the wild-type (Chr-BRF; chromosomal BRF1), pGal-BRF, pGal-TF, and pGal-Split strains is shown in Fig. 3. As expected, only the wild-type strain grew on glucose-containing medium. All strains, except for pGal-Split, showed similar growth rates at 15°C, 21°C (data not shown), 30°C, and 37°C. The pGal-Split strain was partially low- and high-temperature-sensitive (growth defects at 15°C and 37°C) with slightly slower growth also detected at 30°C (data not shown), indicating that Brf1n-TBPc-Brf1c function in vivo does not depend on proteolytic cleavage between TBPc and Brf1c.
Western blotting confirmed the expression and integrity of the Brf1n-TBPc-Brf1c triple fusion in vivo (Fig. 6, which is published as supporting information on the PNAS web site). In fact, no proteolytic cleavage products of Brf1n-TBPc-Brf1c could be unambiguously identified. The fact that Brf1 readily becomes limiting under a variety of conditions (transition from logarithmic growth to stationary phase, mutations in genes encoding TFIIIC subunits that do not interact with Brf1, as well as box A and box B mutations) (24–28) argues strongly against the possibility that Brf1 function is provided so effectively in the pGal-TF strain by Brf1n-TBPc-Brf1c fragments that it displays no growth defect.
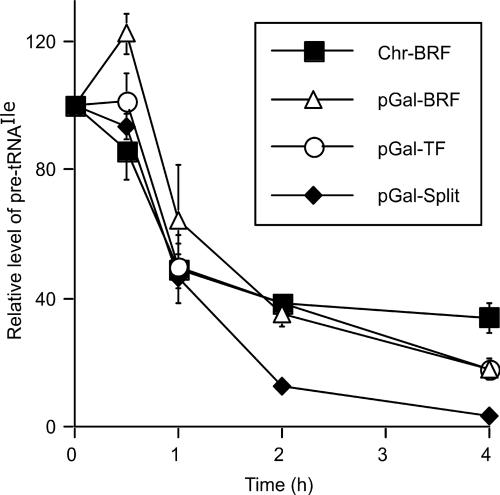
The different temperature sensitivities of pGal-TF and pGal-Split encouraged us to compare the responses of these two strains and the wild type to stress that is known to repress pol III transcription. Methanesulfonic acid methyl ester (MMS) represses pol III transcription in S. cerevisiae as a consequence of DNA damage (29), through a signaling pathway that involves Maf1, a repressor of pol III transcription that functions at the downstream end of multiple signaling pathways (30). Maf1 interacts with both Brf1 and pol III, preventing de novo TFIIIB complex formation and pol III recruitment to preformed TFIIIB–DNA complexes (31, 32). Because primary tRNA transcripts are rapidly processed, changes of abundance of a precursor tRNA provide an estimate of changed rate of synthesis. Fig. 4 shows the quantified results of Northern blots of RNA isolated from cells after 0–4 h of MMS treatment and probed with an oligonucleotide complementary to the intron of the isoleucine-tRNA genes tRNA_I(TAT) DR2 and LR1. The wild-type strain (Chr-BRF), pGal-BRF, and pGal-TF displayed similar time courses of repression of tRNA_I(TAT) synthesis upon MMS treatment, whereas the pGal-Split strain responded with more complete repression (≈8-fold after 2 h and nearly 30-fold after 4 h of treatment).
Fig. 4.
Repression of Ile tRNA I(TAT) transcription in response to DNA damage by methylmethane sulfonate (MMS). The steady-state levels of pre-tRNA I(TAT) were measured as a function of time of treatment with MMS by Northern blot analysis. Samples were collected before the addition of MMS and after 0.5, 1, 2, and 4 h of treatment in yeast strains producing Brf1 from the chromosomal (BRF1) gene (Chr-BRF, filled squares) or Brf1 (pGal-BRF1, open triangles), Brf1n-TBPc-Brf1c (pGal-TF, open circles), and the Brf1n-TBPc + Brf1c split (pGal-Split, filled diamonds) from the corresponding genes on centromeric plasmids under control of the GAL1 promoter. The hybridization signal for the pre-tRNA was normalized to the stable pol II transcript U4 and is plotted as fraction of the initial level of pre-tRNA (mean and average deviation of two independent experiments).
The absence of major proteolytic products of Brf1n-TBPc-Brf1c, the temperature sensitivity of the GAL-split strain, and its different response to MMS compared with the pGal-TF strain establish that the triple fusion protein, and not its degradation products, provides the essential function of Brf1 in vivo.
Discussion
We have exploited the modularity of Brf1 and available information about the structure of the TFIIIB–DNA complex to construct fusion proteins that rearrange the connectivity of subunits of the core transcription initiation factor TFIIIB. Conventional methods of gene construction are applied in this work, but it may be helpful to point out that an ingenious and general method for creating functional chimeras at the RNA level by nonspliceosomal trans-splicing has been devised (33).
TBPc-Brf1c was found to be functional for TFIIIC-independent TFIIIB–DNA complex formation (Fig. 1C) and transcription (Fig. 1B) when complemented with Brf1n. Although this TFIIIB–DNA complex was stable to native gel electrophoresis, it was stripped by heparin (Fig. 1C), in contrast to TFIIIB–DNA complexes formed with Brf1 split at amino acid residues 282/284, which are heparin-resistant (6). Because linking Brf1n to the N terminus of TBPc-Brf1c also generates heparin-resistant TFIIIB–DNA complexes, it is likely that Brf1n fails to establish contacts with TBPc and/or Brf1c that are maintained in intact Brf1, and that this defect may contribute to the inability of the Brf1n + TBPc-Brf1c split to replace Brf1 in vivo. In contrast, the Brf1n-TBPc + Brf1c split was partially competent in replacing Brf1 (Figs. 3 and 4). Unlike TFIIIB complexes formed with TBP and Brf1–282 (15, 34) or Brf1n (Fig. 1B), complexes formed with the Brf1n-TBPc fusion protein retained some competence for TFIIIC-independent transcription of linear DNA and TFIIIC-dependent transcription of supercoiled DNA (data not shown), probably as a result of being able to form a more stable DNA complex.
The propensity of the triple fusion protein for nonspecific transcription in vitro (Figs. 1B and 2 A) led us to anticipate that replacing Brf1 with Brf1n-TBPc-Brf1c would be deleterious in vivo, but no differences of growth parameters or response to stress caused by DNA damage (29, 30) were found (Figs. 3 and 4 and data not shown). This disparity between in vitro and in vivo phenotype prompted an examination of the possible role of nonspecific DNA-binding proteins in rectifying pol III transcription. Nhp6a substantially restored parity to transcription of a plasmid-borne tRNA gene with wild-type Brf1 and the triple fusion protein, respectively (Fig. 2 C and D). Nhp6a and TBP interact primarily with the DNA minor groove, insert amino acid side chains between stacked bases to sharply kink DNA, preferentially bind to more readily deformable DNA (35, 36), and are likely to compete for binding to partial TATA boxes. This role of Nhp6 in suppressing inappropriately initiating transcription in vitro may be nonspecific, but Nhp6a and/or Nhp6b also function as the previously described start site selection factor (37) that is present in partially purified preparations of Bdp1 (G.A.K. and D. F. Steiner, unpublished observations).
The Uses of TBP Fusions
Tethering the components of multiprotein complexes creates new possibilities for exploring the nature and detail of protein–protein interactions, and can also be used to create novel functional combinations. Here, we explore specific applications made possible by this work.
Pol III. The original impetus for tethering Brf1 (439–596) to the C terminus of TBPc stems from the “strung out” nature of the attachment of this segment of Brf1 to TBP (Fig. 1 A), which raises the concern that radical amino acid substitutions introduced into Brf1c to define its interaction surface with Bdp1 might adventitiously affect the Brf1c–TBP interaction. The ability to ameliorate unwanted effects of Brf1c mutations on TBP binding by structure-informed tethering of Brf1c to TBPc facilitates the identification of Brf1 mutants that directly affect Bdp1 assembly into the TFIIIB–DNA complex (G.A.K. and R.D., unpublished work). Brf1n-TBPc and Brf1n-TBPc-Brf1c fusions should allow the same approach to be applied to defining the amino acid residues important for Bdp1 interaction with the N-proximal half of Brf1. Clearly, linking Brf1n to TBPc will eliminate complications that arise from weak, co-dependent Brf1n-and-Bdp1 binding to the TBP–DNA complex (6).
The N-terminal half of Brf1 contains the major sites for interaction with the Tfc4 subunit of TFIIIC (38) and with pol III (39, 40). A library of Brf1n sequence substitutions in the Brf1n-TBP fusion protein also would be applicable to the dissection of these interactions. In this regard, the ability of the Brf1n-TBPc-Brf1c triple fusion to function for TFIIIC- and Bdp1-independent transcription on plasmid DNA templates (Fig. 1B) greatly simplifies the task of interpreting the effects of Brf1 sequence substitutions on pol III recruitment and transcription. The modular design of the Brf1n-TBPc-Brf1c plasmid construct makes it amenable to random mutagenesis of each segment separately for isolation of conditional mutants in yeast. Another possible benefit of the triple fusion lies in its potential for structure determination of TFIIIB–DNA complexes. Brf1n-TBPc-Brf1c is considerably more soluble than is wild-type Brf1 (concentrations of 200 μM soluble protein have been achieved; F. Saida and G.A.K., unpublished results) and reduces the complexity of the TFIIIB–DNA assembly to that of a ternary complex.
Pol II and Pol I. The Brfn-TBPc-Brfc fusion creates a TBP paralogue that is dedicated to pol III transcription. In wild-type S. cerevisiae, Brf1 is in slight excess over TBP and the concentrations of the two proteins are sufficiently high for complex formation in the absence of DNA (24). That Brf1 and factors involved in pol II transcription are in competition for binding to limiting TBP is indicated by the observation that some TBP mutants that are defective in pol III transcription elevate pol II transcription and vice versa (41). Surface-exposed amino acids of TBP that interact with Brf1 (5, 7, 8, 14, 41) at least partly overlap with TBP residues interacting with pol II factors TFIIB, Taf1, NC2, Mot1, and TFIIA (42–49) and with the (human) pol I TAF, TAFI48 (50). The Brf1c amino acid 439–596 segment covers much of the top convex surface and N-proximal lateral side of TBP (Fig. 1 A) (14), while Brf1n amino acids 1–382 shield at least the C-terminal stirrup of TBP (Fig. 1 A) (8). The linkage of Brf1n and Brf1c to TBPc (and the resulting large increase of effective local concentration) guarantees exclusion of competing ligands and effectively sequesters this large surface of TBP from access by components essential for pol II and pol I transcription.
Privatizing a TBP paralogue for pol III transcription frees the genetic analysis of TBP function in transcription by the other two nuclear RNA polymerases from a significant constraint. Thus, for example, rescreening preexisting libraries of yeast TBP mutants (e.g., refs. 41 and 51) in cells that replace Brf1 with the Brf1n-TBPc-Brf1c triple fusion may yield conditional phenotypes that can be unambiguously assigned to defects in pol II or pol I transcription [with the additional use of a plasmid that places 35S rRNA synthesis under the control of a pol II promoter (52)]. In addition, new conditional TBP mutants will probably be found among the 30% of TBP variants that were previously observed to be inviable (41).
Supplementary Material
Acknowledgments
We thank D. F. Steiner for skilful assistance; D. J. Stillman for valuable advice; G. P. Tocchini-Valentini for thought-provoking discussion; D. J. Stillman, Z. S. Juo, and R. C. Johnson for generously providing materials; and R. Haselkorn for helpful comments on the manuscript. This work was supported by the National Institute of General Medical Sciences.
Author contributions: G.A.K., E.S., and E.P.G. designed research; G.A.K., E.S., and R.D. performed research; G.A.K., E.S., and R.D. contributed new reagents/analytic tools; G.A.K., E.S., and E.P.G. analyzed data; and G.A.K., E.S., and E.P.G. wrote the paper.
Abbreviations: pol, RNA polymerase; TBP, TATA-binding protein; TBPc, TBP core; TF, transcription factor.
References
- 1.White, R. J. (2002) RNA Polymerase III Transcription (Landes Bioscience, Georgetown, TX).
- 2.Hansen, S. K., Takada, S., Jacobson, R. H., Lis, J. T. & Tjian, R. (1997) Cell 91, 71-83. [DOI] [PubMed] [Google Scholar]
- 3.Geiduschek, E. P. & Kassavetis, G. A. (2001) J. Mol. Biol. 310, 1-26. [DOI] [PubMed] [Google Scholar]
- 4.Schramm, L. & Hernandez, N. (2002) Genes Dev. 16, 2593-2620. [DOI] [PubMed] [Google Scholar]
- 5.Colbert, T., Lee, S., Schimmack, G. & Hahn, S. (1998) Mol. Cell. Biol. 18, 1682-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kassavetis, G. A., Kumar, A., Ramirez, E. & Geiduschek, E. P. (1998) Mol. Cell. Biol. 18, 5587-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen, Y., Kassavetis, G. A., Bryant, G. O. & Berk, A. J. (1998) Mol. Cell. Biol. 18, 1692-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schröder, O., Bryant, G. O., Geiduschek, E. P., Berk, A. J. & Kassavetis, G. A. (2003) EMBO J. 22, 5115-5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishiguro, A., Kassavetis, G. A. & Geiduschek, E. P. (2002) Mol. Cell. Biol. 22, 3264-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachman, N., Gelbart, M. E., Tsukiyama, T. & Boeke, J. D. (2005) Genes Dev. 19, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kassavetis, G. A., Letts, G. A. & Geiduschek, E. P. (2001) EMBO J. 20, 2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kassavetis, G. A., Riggs, D. L., Negri, R., Nguyen, L. H. & Geiduschek, E. P. (1989) Mol. Cell. Biol. 9, 2551-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun, B. R., Riggs, D. L., Kassavetis, G. A. & Geiduschek, E. P. (1989) Proc. Natl. Acad. Sci. USA 86, 2530-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juo, Z. S., Kassavetis, G. A., Wang, J., Geiduschek, E. P. & Sigler, P. B. (2003) Nature 422, 534-539. [DOI] [PubMed] [Google Scholar]
- 15.Kassavetis, G. A., Letts, G. A. & Geiduschek, E. P. (1999) EMBO J. 18, 5042-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kassavetis, G. A., Bartholomew, B., Blanco, J. A., Johnson, T. E. & Geiduschek, E. P. (1991) Proc. Natl. Acad. Sci. USA 88, 7308-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitehall, S. K., Kassavetis, G. A. & Geiduschek, E. P. (1995) Genes Dev. 9, 2974-2985. [DOI] [PubMed] [Google Scholar]
- 18.Cox, J. M., Hayward, M. M., Sanchez, J. F., Gegnas, L. D., van der Zee, S., Dennis, J. H., Sigler, P. B. & Schepartz, A. (1997) Proc. Natl. Acad. Sci. USA 94, 13475-13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joazeiro, C. A. P., Kassavetis, G. A. & Geiduschek, E. P. (1996) Genes Dev. 10, 725-739. [DOI] [PubMed] [Google Scholar]
- 20.Kruppa, M., Moir, R. D., Kolodrubetz, D. & Willis, I. M. (2001) Mol. Cell 7, 309-318. [DOI] [PubMed] [Google Scholar]
- 21.Lopez, S., Livingstone-Zatchej, M., Jourdain, S., Thoma, F., Sentenac, A. & Marsolier, M. C. (2001) Mol. Cell. Biol. 21, 3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin, M. P., Gerlach, V. L. & Brow, D. A. (2001) Mol. Cell. Biol. 21, 6429-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colbert, T. & Hahn, S. (1992) Genes Dev. 6, 1940-1949. [DOI] [PubMed] [Google Scholar]
- 24.Sethy-Coraci, I., Moir, R. D., López-de-León, A. & Willis, I. M. (1998) Nucleic Acids Res. 26, 2344-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sethy, I., Moir, R. D., Librizzi, M. & Willis, I. M. (1995) J. Biol. Chem. 270, 28463-28470. [DOI] [PubMed] [Google Scholar]
- 26.Rozenfeld, S. & Thuriaux, P. (2001) Mol. Genet. Genomics 265, 705-710. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre, O., Ruth, J. & Sentenac, A. (1994) J. Biol. Chem. 269, 23374-23381. [PubMed] [Google Scholar]
- 28.Jourdain, S., Acker, J., Ducrot, C., Sentenac, A. & Lefebvre, O. (2003) J. Biol. Chem. 278, 10450-10457. [DOI] [PubMed] [Google Scholar]
- 29.Ghavidel, A. & Schultz, M. C. (2001) Cell 106, 575-584. [DOI] [PubMed] [Google Scholar]
- 30.Upadhya, R., Lee, J. & Willis, I. M. (2002) Mol. Cell 10, 1489-1494. [DOI] [PubMed] [Google Scholar]
- 31.Desai, N., Lee, J., Upadhya, R., Chu, Y., Moir, R. D. & Willis, I. M. (2005) J. Biol. Chem. 280, 6455-6462. [DOI] [PubMed] [Google Scholar]
- 32.Pluta, K., Lefebvre, O., Martin, N. C., Smagowicz, W. J., Stanford, D. R., Ellis, S. R., Hopper, A. K., Sentenac, A. & Boguta, M. (2001) Mol. Cell. Biol. 21, 5031-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deidda, G., Rossi, N. & Tocchini-Valentini, G. P. (2003) Nat. Biotechnol. 21, 1499-1504. [DOI] [PubMed] [Google Scholar]
- 34.Kassavetis, G. A., Kumar, A., Letts, G. A. & Geiduschek, E. P. (1998) Proc. Natl. Acad. Sci. USA 95, 9196-9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allain, F. H., Yen, Y. M., Masse, J. E., Schultze, P., Dieckmann, T., Johnson, R. C. & Feigon, J. (1999) EMBO J. 18, 2563-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juo, Z. S., Chiu, T. K., Leiberman, P. M., Baikalov, I., Berk, A. J. & Dickerson, R. E. (1996) J. Mol. Biol. 261, 239-254. [DOI] [PubMed] [Google Scholar]
- 37.Andrau, J. C. & Werner, M. (2001) Eur. J. Biochem. 268, 5167-5175. [DOI] [PubMed] [Google Scholar]
- 38.Chaussivert, N., Conesa, C., Shaaban, S. & Sentenac, A. (1995) J. Biol. Chem. 270, 15353-15358. [DOI] [PubMed] [Google Scholar]
- 39.Ferri, M. L., Peyroche, G., Siaut, M., Lefèbvre, O., Carles, C., Conesa, C. & Sentenac, A. (2000) Mol. Cell. Biol. 20, 488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khoo, B., Brophy, B. & Jackson, S. P. (1994) Genes Dev. 8, 2879-2890. [DOI] [PubMed] [Google Scholar]
- 41.Cormack, B. P. & Struhl, K. (1993) Science 262, 244-248. [DOI] [PubMed] [Google Scholar]
- 42.Bryant, G. O., Martel, L. S., Burley, S. K. & Berk, A. J. (1996) Genes Dev. 10, 2491-2504. [DOI] [PubMed] [Google Scholar]
- 43.Tang, H., Sun, X., Reinberg, D. & Ebright, R. H. (1996) Proc. Natl. Acad. Sci. USA 93, 1119-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martel, L. S., Brown, H. J. & Berk, A. J. (2002) Mol. Cell. Biol. 22, 2788-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamada, K., Shu, F., Chen, H., Malik, S., Stelzer, G., Roeder, R. G., Meisterernst, M. & Burley, S. K. (2001) Cell 106, 71-81. [DOI] [PubMed] [Google Scholar]
- 46.Nikolov, D. B., Chen, H., Halay, E. D., Usheva, A. A., Hisatake, K., Lee, D. K., Roeder, R. G. & Burley, S. K. (1995) Nature 377, 119-128. [DOI] [PubMed] [Google Scholar]
- 47.Tsai, F. T. & Sigler, P. B. (2000) EMBO J. 19, 25-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, S., Hunziker, Y., Sargent, D. F. & Richmond, T. J. (1996) Nature 381, 127-151. [DOI] [PubMed] [Google Scholar]
- 49.Cang, Y., Auble, D. T. & Prelich, G. (1999) EMBO J. 18, 6662-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, S. & Hori, R. T. (2004) Gene 338, 177-186. [DOI] [PubMed] [Google Scholar]
- 51.Eriksson, P., Biswas, D., Yu, Y., Stewart, J. M. & Stillman, D. J. (2004) Mol. Cell. Biol. 24, 6419-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nogi, Y., Vu, L. & Nomura, M. (1991) Proc. Natl. Acad. Sci. USA 88, 7026-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.