Abstract
Genome sequences are available for many bacterial strains, but there has been little progress in using these data to understand the molecular basis of pathogen emergence and differences in strain virulence. Serotype M3 strains of group A Streptococcus (GAS) are a common cause of severe invasive infections with unusually high rates of morbidity and mortality. To gain insight into the molecular basis of this high-virulence phenotype, we sequenced the genome of strain MGAS315, an organism isolated from a patient with streptococcal toxic shock syndrome. The genome is composed of 1,900,521 bp, and it shares ≈1.7 Mb of related genetic material with genomes of serotype M1 and M18 strains. Phage-like elements account for the great majority of variation in gene content relative to the sequenced M1 and M18 strains. Recombination produces chimeric phages and strains with previously uncharacterized arrays of virulence factor genes. Strain MGAS315 has phage genes that encode proteins likely to contribute to pathogenesis, such as streptococcal pyrogenic exotoxin A (SpeA) and SpeK, streptococcal superantigen (SSA), and a previously uncharacterized phospholipase A2 (designated Sla). Infected humans had anti-SpeK, -SSA, and -Sla antibodies, indicating that these GAS proteins are made in vivo. SpeK and SSA were pyrogenic and toxic for rabbits. Serotype M3 strains with the phage-encoded speK and sla genes increased dramatically in frequency late in the 20th century, commensurate with the rise in invasive disease caused by M3 organisms. Taken together, the results show that phage-mediated recombination has played a critical role in the emergence of a new, unusually virulent clone of serotype M3 GAS.
Keywords: superantigen‖streptococcal pyrogenic exotoxin‖comparative genomics
A common theme in medical microbiology is that most species of bacterial pathogens are characterized by substantial genetic variation, far in excess of that present in eukaryotic organisms (1, 2). Strains of pathogenic bacteria also are well known to differ in biomedically relevant phenotypes such as antimicrobial agent resistance and virulence (2). Contemporary investigative methods such as chromosomal sequencing and DNA microarray analysis permit strains of pathogens to be studied rapidly at the whole-genome level (3–5). However, despite the accumulation of vast amounts of bacterial genome sequence information, there has been little progress in using these data to understand the molecular basis of pathogen emergence and link strain-specific gene content with differences in biomedically relevant phenotypes such as virulence.
Group A Streptococcus (GAS) is a human bacterial pathogen that causes pharyngitis, cellulitis, sepsis, necrotizing fasciitis, and poststreptococcal acute rheumatic fever (6, 7). GAS strains commonly are classified on the basis of serologic differences in M protein, an anti-phagocytic cell-surface molecule. Although more than 130 M types have been identified, epidemiologic studies have repeatedly identified nonrandom associations of M serotypes and infection types (6–10). For example, serotype M1 strains are common causes of pharyngitis and invasive infections, and serotype M18 strains have been associated with acute rheumatic fever outbreaks in the United States (5, 7).
Several observations stimulated us to study serotype M3 GAS. These strains are associated with unusually severe infections and a high mortality rate, but the molecular basis of this phenotype is unknown (7, 8, 10). Comparison of the genome sequences of serotype M1 and M18 organisms recently identified 290 ORFs uniquely present in either strain, and provided many new avenues for pathogenesis research (5). Proteome analysis of a serotype M3 strain identified many previously uncharacterized extracellular proteins (11), suggesting that M3 organisms express distinct arrays of virulence factors. The sequenced M1 strain lacks the phage-encoded gene (speA) for streptococcal pyrogenic exotoxin A, a potent superantigen made by many GAS strains causing invasive infections, and implicated in virulence (8, 12). Serotype M3 strains also have the gene (ssa) for streptococcal superantigen A (SSA), a bacterial superantigen (13–15). This gene is thought to be horizontally transmitted in natural populations, but the mechanism is not known (14). The identification of superantigens encoded by the genome of serotype M1 and M18 strains suggests that uncharacterized virulence factors exist in other GAS genomes (5, 12). Taken together, these observations led us to hypothesize that genome-scale analysis of serotype M3 strains would provide new information bearing on the unusually virulent phenotype of these organisms. To test this hypothesis, we sequenced the genome of strain MGAS315, a contemporary strain representative of M3 organisms isolated from patients with invasive disease in the United States, Canada, Europe, and Japan.
Materials and Methods
Bacterial Strain Used for Sequencing.
Strain MGAS315 has been characterized extensively (8, 11, 16–19). The organism was recovered in the late 1980s from a patient in the United States with streptococcal toxic shock syndrome (8).
Genome Sequencing, Closure, and Annotation.
The genome of strain MGAS315 was sequenced by the random-sheered library method (5). Regions of low sequence quality in the assembled genome were identified by consed (http://www.genome.washington.edu) and directed sequencing was performed to increase the minimum consensus base quality to Q40. Proteins predicted to be secreted were identified with SIGNALP V2.0 and other bioinformatic methods (20–23).
Cloning of sla, ssa, and speK Genes.
The genes encoding mature Sla (amino acid residues 28–191), SpeK (27–259), and SSA (45–191) were cloned from strain MGAS315 with paired primers by standard techniques. Recombinant Sla (rSla) and SpeK (rSpeK) made by the clones had 11 amino acid residues MHHHHHHLETM fused to the amino terminus of the mature proteins.
Purification of rSla, rSpeK, and rSSA.
rSla and rSpeK were purified from Escherichia coli BL21 (DE3) containing recombinant plasmids.
Assay for Superantigen Activity.
The mitogenicity of rSpeK was determined with [3H]thymidine incorporation assays using rabbit splenocytes and peripheral blood mononuclear cells (PBMC) (24).
Analysis of T Cell Repertoire.
Stimulation of T cell expansion and Vβ receptor repertoire use by rSpeK was conducted by flow cytometry (25).
Pyrogenicity and Lethality Models of Toxic Shock Syndrome.
American Dutch belted rabbits were used to assess the pyrogenicity and toxicity of rSpeK and rSSA (25, 26).
Phospholipase A2 (PLA2) Assay.
PLA2 activity was assayed with a commercially available kit (Cayman Chemical, Ann Arbor, MI) that measures the hydrolysis of phospholipids at the sn-2 position, yielding a free fatty acid and a lysophospholipid (27). Bee venom PLA2 was used as positive control.
Detection and Chromosomal Mapping of speK and sla.
PCR was used to screen GAS strains for the presence of genes encoding SpeK and Sla in serotype M3 strains from diverse localities and times (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org).
Detection of Anti-SSA, -SpeK, and -Sla Antibodies in Sera Obtained from Humans with GAS Infections.
Western immunoblotting was used to test for the presence of specific antibodies to rSSA, rSpeK, and rSla in paired acute and convalescent sera from patients with GAS pharyngitis and invasive episodes (31).
A complete description of this section can be found in Supporting Materials and Methods, which is published on the PNAS web site.
Results
Overview of the Genome Sequence of Strain MGAS315 and Comparison with the Genomes of GAS Strain SF370 (Serotype M1) and MGAS8232 (Serotype M18).
The genome of strain MGAS315 is a circular chromosome of 1,900,521 bp (see Fig. 7, which is published as supporting information on the PNAS web site) with a G+C content of 38.6%, a value essentially identical to the genomes of GAS strains SF370 (38.5%) and MGAS8232 (38.6%). There are 1,865 predicted coding sequences (CDS), oriented predominately in the direction of DNA replication (see Table 4, which is published as supporting information on the PNAS web site). Coding sequence accounts for 1,657 kb (87.2%) of the genome. The average G+C content of the CDS (38.5%) is similar to the overall genome, but differs from the 39.1% reported for SF370 CDS. The inferred proteins vary in size from 26 to 1,465 amino acid residues, with an average of 258 amino acid residues.
The genome of strain MGAS315, SF370, and MGAS8232 have a collinear structure, and ≈1.7 Mbp (90%) of each genome is conserved in content and context (see Table 5, which is published as supporting information on the PNAS web site). This “core” genome encodes many well studied proven and putative virulence factors such as streptolysin O, streptococcal cysteine protease, M protein, streptolysin S, and hyaluronic acid capsule (6). The genome of strain MGAS315 is slightly larger than the genome of M1 strain SF370 and M18 strain MGAS8232 (see Table 4). Six rRNA operons account for 33 kb or 1.7% of the genome of strain MGAS315, and these are highly conserved in sequence and chromosomal location among the three sequenced GAS genomes (see Fig. 7).
Mobile Genetic Elements and Genome Plasticity.
Six regions of the genome of strain MGAS315 are composed of phages or phage-like elements, each 34.4–41.8 kb in length, a size range common for phages present in low G+C Gram-positive bacteria (see Figs. 1 and 7). (For ease of description and discussion, each of these six elements will be referred to as phages, with the understanding that none of them has been documented to be a bona fide phage.) Each phage has a hyaluronidase gene (32). The distribution of the phages in the chromosome is apparently nonrandom, with five of six located in the half of the chromosome distal to the origin of replication. All of the phages are integrated in the chromosome such that the majority of CDS are transcribed in the direction of DNA replication. Phage DNA constitutes 235 kb (12.4%) of the genome of strain MGAS315, a larger proportion than the phages in the genome of strain SF370 (130 kb, 7.1%) or strain MGAS8232 (204 kb, 10.8%), which have four and five phages, respectively (refs. 5 and 12; see Table 6, which is published as supporting information on the PNAS web site).
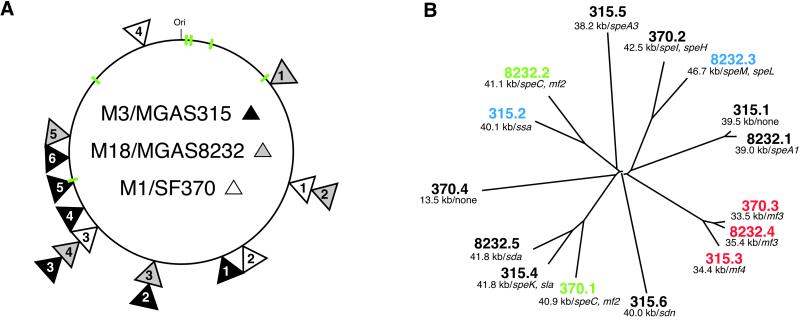
Figure 1.
GAS phages. (A) Schematic of GAS core genome and phage integration sites. Shown is the core genome (≈1.7 Mbp) derived by deleting phage sequence from the genome sequence of strains SF370 (serotype M1), MGAS8232 (serotype M18), and MGAS315 (serotype M3). Phage integration sites are indicated with triangles that are shaded to match the source GAS strain. Stacked triangles indicate that the phages are integrated at the same chromosomal site. The numbers in each triangle are designations that refer to the clockwise order of the phages. The six rRNA operons that are conserved in all three genomes are shown in green. (B) Relationships among GAS phages. Phage sequences present in the three GAS genomes were aligned with clustalw (http://inn-prot.weizmann.ac.il/software) and an unrooted tree was generated with the drawtree application in phylip (http://evolution.genetics.washington.edu/phylip.html). Phage size (kb) and proven or putative virulence factors encoded by each phage are indicated. Phages that are integrated at the same chromosomal location are color coded in B. mf2, mitogenic factor 2; mf3, mitogenic factor 3; mf4, mitogenic factor 4; sda, streptodornase alpha; sdn, streptodornase.
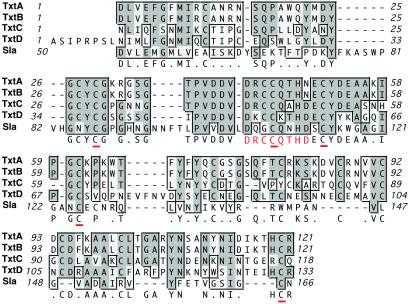
Phages are the major source of variation in gene content between the three strains, and they differ in number, composition, and chromosomal integration position (see Table 6). Estimates of the overall relationships among the 15 phages present in these three strains are shown in Fig. 1. Twelve of the 15 phages cluster into one of five lineages containing two to three phages per genetic group. No genetic group contains more than one phage from the same strain, a result that may be due to immunity to phage superinfection. Phage Φ315.1 is >90% homologous to ΦspeA in the genome of MGAS8232; however, Φ315.1 lacks a speA gene or a gene encoding homologues of other putative or proven GAS virulence factors (see Table 4). Phage Φ315.2 is a T12-like phage that encodes the ssa gene and is inserted at the predicted T12att site, similar to phage ΦspeL/M recently identified in the genome of strain MGAS8232 (5). Phage Φ315.3 is closely related to phages in strains SF370 and MGAS8232 that encode mitogenic factor 3 (MF3), and has a gene encoding a mitogenic factor homologue (designated MF4) (see Figs. 8 and 9, which are published as supporting information on the PNAS web site). This phage is integrated at the same chromosomal location in all three GAS strains, suggesting that it is an ancestral condition in these organisms. Phage Φ315.4 is related to Φsda in strain MGAS8232 and the speC-containing phage in strain SF370. However, this phage encodes a homologue of streptococcal pyrogenic exotoxins (designated SpeK) and a protein with a region of conserved amino acids found in some PLA2 enzymes, including toxins made by venomous snakes (see Figs. 2 and 9A). This protein, designated Sla (streptococcal phospholipase), has a typical Gram-positive secretion signal sequence (ref. 20; see below). Sla is most closely related to the C subunit of textilotoxin, a phospholipase neurotoxin made by the Australian common brown snake, Pseudonaja textiles (Fig. 2; ref. 33). Sla has amino acid residues that are well conserved with the PLA2 active site residues in the four subunits of textilotoxin. Moreover, like the textilotoxin subunits, Sla has many conserved cysteine residues, and a high content of aromatic amino acids (Fig. 2).
Figure 2.
Amino acid alignment of Sla (streptococcal phospholipase A2 homologue) and textilotoxin subunits. The carboxy terminal 116 amino acid residues of Sla were aligned with the four subunits of textilotoxin with clustalw, using default parameters. Amino acid residues that match the consensus sequence are shaded in dark gray, and similar amino acid residues are shaded in light gray. The textilotoxin phospholipase A2 active site consensus sequence is shown in red, and conserved Cys residues are underlined.
The genome of strain MGAS315 has two other phages, but these lack homologues in strains SF370 and MGAS82332. Phage Φ315.5 encodes the variant of SpeA (SpeA3) found in contemporary isolates of serotype M3 strains recovered from patients with invasive disease and pharyngitis in many geographic areas (16). However, this phage is highly divergent from the speA-containing phage in strain MGAS8232 (5). Phage Φ315.6 encodes a streptodornase homologue, and has a higher G+C content than other GAS phages. Together, 93% (106/114) of CDS uniquely present in strain MGAS315 are encoded by phages, and Φ315.5 and Φ315.6 contain 71% (75/106) of all ORFs found in the genome of neither strain SF370 nor strain MGAS3823. The majority of the unique ORFs do not have a known homologue or encode a homologue of a protein with unknown function. Many of the proteins encoded by these ORFs presumably participate in phage biology.
To summarize, the genome of strain MGAS315 contains six phages, including five that encode homologues of proven or putative virulence factors. Two of the phages (Φ315.5 and Φ315.6) are present in the genome of strain MGAS315, but not strain SF370 or MGAS8232, and account for 71% of the unique CDS in the genome of strain MGAS315.
Insertion sequences (transposons) also are an abundant source of variation in gene content. Twenty-two CDS present in the genome of strain MGAS315 have homology to insertion sequence (IS) elements, including three copies of IS1548 (34), four copies of IS861 (35), and two copies each of IS904 and IS905 (36). In contrast to strains SF370 and MGAS8232, strain MGAS315 does not have IS1239 or IS1562 (37, 38). The occurrence of three copies of IS1548 in the chromosome of strain MGAS315 and the lack of IS1239 are consistent with data reported previously for serotype M3 strains (18, 38).
Characterization of Three Unique Extracellular Secreted Proteins.
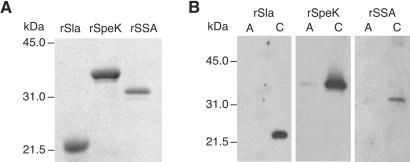
Given the importance of many phage-encoded extracellular proteins in GAS–host interactions (6), we elected to clone the genes encoding SSA, SpeK, and Sla, purify the proteins, and test their ability to elicit responses likely to contribute to pathogenesis. The gene segment encoding the inferred mature form of each protein was cloned and the recombinant proteins were purified to apparent homogeneity (Fig. 3).
Figure 3.
SDS/PAGE and Western immunoblot analysis of rSla, rSpeK, and rSSA. (A) SDS/PAGE gel of purified recombinant proteins. The purified recombinant proteins were analyzed with a 15% polyacrylamide gel. (B) Representative Western immunoblot using sera obtained humans infected with serotype M3 strains of GAS. A, acute-phase sera; C, convalescent-phase sera.
Mitogenicity and Vβ Profiling of rSpeK.
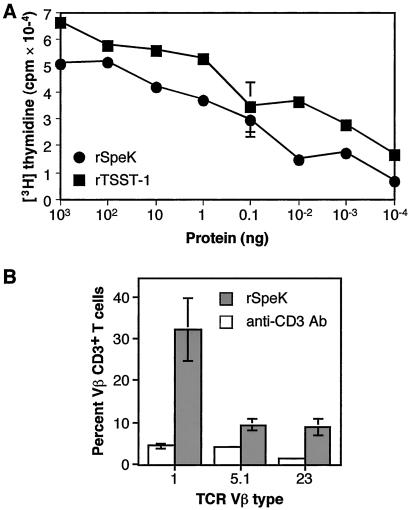
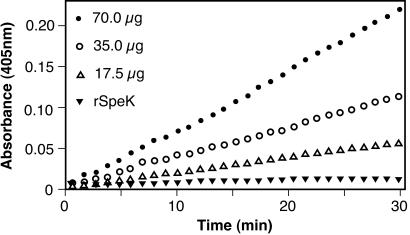
The ability to induce clonal proliferation of T cells is a characteristic of superantigens (15, 21). SSA was shown previously to have potent mitogenic activity for human T cells (15), therefore we focused our analysis on rSpeK. This protein caused proliferation of rabbit splenocytes and human PBMC at concentrations as low as 1 × 10−4 ng/ml (Fig. 4). The results observed for rSpeK were similar to those obtained for recombinant toxic shock syndrome toxin-1, a documented superantigen.
Figure 4.
Mitogenic activity of rSpeK. (A) Human peripheral blood mononuclear cells were incubated with purified rSpeK at 37°C for 3 days. [3H]thymidine was added to the medium, cells were harvested after 24 h, and cpm were determined by scintillation counting. Experiments done in quadruplicate, and error bars indicate the standard error of the mean. TSST-1, a known superantigen, was used as a positive control. (B) T cell TCR Vβ stimulation by rSpeK. PBMC from five human donors were studied. The percentage of T cells expressing the listed Vβ TCRs are shown. Only the Vβs with statistically significant stimulation (as determined by the Student's t test; P < 0.01) are shown.
To further investigate the superantigenicity of rSpeK, the Vβ profile of human PBMC treated with rSpeK was determined (Fig. 4). rSpeK significantly stimulated T cells with Vβ1, Vβ5.1, and Vβ23 regions (P < 0.01). rSpeK induced a very substantial expansion of T cells with Vβ1 (32%).
Pyrogenicity and Lethality of rSSA and rSpeK.
SPEs are known to induce fever and enhance the susceptibility of rabbits to endotoxin (25, 26, 39). To determine whether rSpeK and rSSA have these characteristics, sublethal concentrations of the proteins were injected intravenously into three rabbits, and their temperatures were recorded at 4 h (Table 1). A temperature increase of 0.5°C or greater is considered to be significant in this model (39). Average temperature increases of 1.3, 1.4, and 0.9°C were observed in rabbits given rSpeK, rSSA, and rTSST-1, respectively. Moreover, rSpeK and rSSA each enhanced the susceptibility of rabbits to endotoxic shock (Table 1). The lethality of rSpeK and rSSA also was assessed with the miniosmotic pump model of STSS, which causes death in rabbits in the absence of exogenous endotoxin (25, 26). Two of the three rabbits infused with 500 μg rSpeK died, but negative control rabbits did not die after 15 days (Table 1). The three rabbits that were given 500 μg rSpeC (positive control) also died.
Table 1.
Pyrogenicity and lethality of rSpeK and rSSA
| Toxin | Pyrogenicity* (Avg. change, °C) | Models of lethality (No. dead per no. total animals)
|
|
|---|---|---|---|
| Endotoxin† enhancement | Miniosmotic‡ pump | ||
| rSpeK | +1.3 | 3/3 | 2/3 |
| rSSA | +1.4 | 3/3 | 3/3 |
| rTSST-1 | +0.9 | 3/3 | 3/3 |
| PBS | −0.1 | 0/3 | ND§ |
Animals were injected intravenously with 5 μg/kg of recombinant protein at 0 h, and temperatures were recorded at 4 h. Three rabbits were used for all assays with the exception of rTSST-1, in which six were used.
Mortality was recorded for a 48-h period after 10 μg/kg endotoxin injection.
Mortality was recorded for 15 days after subcutaneous implantation of pump.
Not determined.
Phospholipase A2 Activity of Purified rSla.
To test the hypothesis that rSla had phospholipase A2 activity, we investigated the ability of the protein to hydrolyze phospholipids at the sn-2 position by using a standard colorimetric assay (27). rSla was catalytically active in this assay in a dose-dependent fashion (Fig. 5).
Figure 5.
PLA2 activity of rSla. PLA2 activity was assayed with a commercially available kit that measures the hydrolysis of phospholipids at the sn-2 position. The assay uses the 1,2-dithio analog of diheptanoyl phosphatidylcholine as a chromogenic substrate. Bee venom PLA2 was used as positive control (data not shown) and rSpeK was used as negative control.
Human Antibody Response to rSpeK, rSSA, and rSla.
Western immunoblotting was used to determine whether SpeK, SSA, and Sla were produced in vivo during human GAS infections. Convalescent sera (but not acute sera) obtained from patients with pharyngitis and invasive infections was reactive to these three proteins (Fig. 3), indicating that SpeK, SSA, and Sla are made in humans infected with GAS.
Temporal and Geographic Distribution, and Chromosomal Mapping of the speK and sla Genes.
To assess the prevalence of speK and sla among serotype M3 GAS strains from diverse localities and decades, we screened 121 isolates by PCR with primers specific for each gene. All 68 isolates recovered from the 1920s to 1984 lacked these two genes, including organisms that were geographically widely distributed (United States, Canada, and Europe). In striking contrast, regardless of locality, 50 of 53 serotype M3 strains isolated from patients in recent years (late 1987 to present) had these two genes (see Table 2). All serotype M3 strains studied that contained speK and speL had these genes located contiguously with phage Φ315.4-specific DNA. Moreover, PCR analysis indicated that these genes were integrated at the same chromosomal position in all serotype M3 strains studied (see Table 2).
Discussion
Common Themes of Bacterial Interstrain Variation.
Complete genome sequences are now available for three GAS strains, including serotype M1, M3, and M18 organisms, that together are responsible for extensive human morbidity and mortality (5, 7–10, 40, 41). Multiple genome sequences are available in public databases for several other important human bacterial pathogens, including Chlamydia trachomatis, Staphylococcus aureus, Mycobacterium tuberculosis, Helicobacter pylori, Neisseria meningitidis, and E. coli (3). There is considerable variance in the magnitude of genetic diversity among these organisms. For example, strains of S. aureus, H. pylori, and E. coli can differ in gene content by greater than 25% (3). In contrast, strains of C. trachomatis and M. tuberculosis are far more conserved, differing mainly by relatively few large chromosomal areas and single nucleotide polymorphisms. Our results demonstrate that among bacterial species with multiple strains sequenced, GAS is unique in the magnitude to which phages account for genome diversification. Inasmuch as virtually all of the GAS phages have genes encoding proven and putative virulence factors, it is likely that they contribute to the differences in phenotypic characteristics observed among GAS strains.
Chimeric Phages and Virulence Factor Distribution.
One of the unexpected discoveries documented by the genome sequences of strains SF370, MGAS8232, and MGAS315 is that chimeric phages are common among GAS isolates, an observation consistent with the idea of modular phage evolution (44). Many of the epidemiologic features of GAS infections currently lack molecular explanation. For example, rapid changes in disease frequency and severity in local geographic areas are common. In the case of infections caused by serotype M1 organisms, evidence has been presented that rapid selection of variants of the streptococcal inhibitor of complement protein plays a key role in perpetuating the length of these epidemics and increasing the number of individuals affected (45, 46). Given the relative frequency of occurrence of chimeric phages, it is reasonable to speculate that generation of new subclones marked by M protein serotypes also may contribute. Under this hypothesis, creation of new subclones by recombination events involving phage-borne toxin genes provides a ready mechanism to generate a virulent or otherwise unusually fit new subclone. Inasmuch as genome sequencing has identified many new phage-encoded toxins in three GAS strains in less than 2 years, there are >130 distinct M protein serotypes, and relatively few strains from areas outside of North America and Europe have been examined extensively, it is likely that additional novel toxins, or alleles of toxins remain to be discovered in natural populations of GAS.
Streptococcal PLA2.
We demonstrated that the PLA2 homologue encoded by phage Φ315.4 was expressed in the course of human–GAS interactions, as assessed by Western immunoblot analysis. Moreover, purified rSla had PLA2 enzymatic activity. Sla has several structural similarities to snake venom toxins, including textilotoxin, a very potent molecule made by the Australian common brown snake. Although the physiologic and pathogenesis roles of Sla are unknown, PLA2 enzymes can have profound biologic functions. Most notably, by releasing arachidonic acid, eukaryotic PLA2 play a central role in the proinflammatory cascade. PLA2 present in snake venoms also are well known to have neurotoxic, myotoxic, proinflammatory, and anticoagulant activity. Inasmuch as GAS pharyngeal infections commonly are characterized by inflammation, and patients with invasive episodes may have disseminated intravascular coagulation, platelet consumption, and extensive myonecrosis, it is reasonable to speculate that Sla participates in one or more of these detrimental processes. Clearly, additional investigation is needed to dissect the contribution of Sla in GAS pathogenesis.
Insights Into the Unusually High Virulence of Contemporary Serotype M3 Strains in Human Infections.
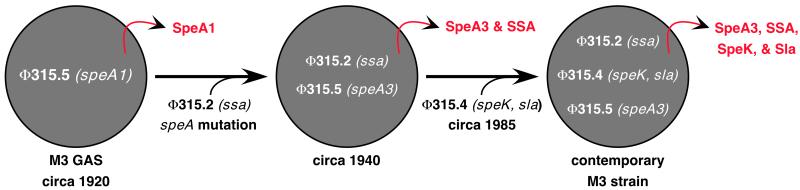
In addition to being an abundant cause of pharyngitis and invasive disease episodes, in some patient populations serotype M3 strains are significantly more likely to cause necrotizing fasciitis and death than GAS strains expressing other M protein serotypes (10). The molecular basis for the more severe disease caused by serotype M3 strains is not known. However, the MGAS315 genome sequence, when integrated with published observations (13, 14, 16, 47), provides critical insight into this issue (Fig. 6). Contemporary isolates of serotype M3 express the SpeA3 variant of streptococcal pyrogenic exotoxin A (16). This variant is approximately 50% more mitogenic in vitro than the SpeA1 variant made by serotype M3 strains recovered before the resurgence of severe invasive disease episodes beginning in the 1980s (47). Contemporary serotype M3 isolates have the phage encoding SpeK and Sla, whereas this phage and these genes were not present in M3 strains recovered before 1987. Similarly, contemporary isolates contain the ssa gene and express SSA, but many old M3 isolates lack the ssa gene (13). Taken together, the data indicate that the unusually high virulence observed among contemporary serotype M3 strains is closely linked to very recent emergence and widespread dissemination of a serotype M3 subclone that expresses a unique combination of phage-encoded virulence factors. Creation of distinct arrays of potent virulence factors by phage-mediated recombination events may very well contribute to the geographically localized bursts of disease activity caused by strains marked with particular M protein serotypes.
Figure 6.
Hypothesis to explain the emergence of a new, unusually virulent subclone of serotype M3 GAS. Under this hypothesis, all isolates shown are multilocus enzyme electrophoretic type 2 (8). Relatively early in the 20th century, an ancestral strain acquired phage Φ315.2, a T12-like phage that encodes SSA (13). Subsequently, a single nucleotide mutation resulting in a single amino acid replacement (Val106Ile) produced the SpeA3 allele (16) encoded by phage Φ315.5. This ssa-positive, speA3-positive strain gained phage Φ315.4 that encodes Sla and SpeK, and disseminated widely in the mid/late 1980s. The hypothesis is based on data presented in this paper and published previously (8, 13, 14, 16, 28).
Genome-Scale Pathogenesis Studies.
Most species of pathogenic bacteria, including GAS, are well known to have a large amount of variation in gene content and allelic diversity (1, 3, 48). This genetic variation is responsible for the occurrence of strain differences in biomedically relevant phenotypes such as virulence. Traditionally, it has been very difficult or impossible to gain very rapid insight into the molecular basis of important phenotypic differences between strains, or newly emerged clones. Bacterial genome sequencing is now relatively inexpensive, and subsequent downstream investigations also can be conducted rapidly and efficiently with microarray techniques and new protein analysis technologies. Hence, in recent years we have entered a new era in clinical investigation of human bacterial pathogenesis. In the new era, large numbers of clones of bacterial species nonrandomly associated with specific infection types (2) will be sequenced and clinical syndromes studied in a genome-specific manner. This strategy already has led to a fundamental change in how many studies of microbial pathogenesis are conducted, and will undoubtedly lead to enhanced understanding of clone emergence, and accelerated development of improved diagnostics, therapies, control measures, and vaccines.
Supplementary Material
Acknowledgments
We are indebted to V. Kapur, Q. Zhang, T. M. Smith, and colleagues, for helpful discussions and assistance with the genome sequencing project; and M. Lewis, A. Dunlap, and R. Cole for technical assistance.
Abbreviations
- GAS
group A Streptococcus
- SSA
streptococcal superantigen A
- Spe
streptococcal pyrogenic exotoxin
- rSpe
recombinant Spe
- CDS
coding sequences
- Sla
streptococcal phospholipase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE014074).
References
- 1.Blaser M J, Musser J M. J Clin Invest. 2001;107:391–392. doi: 10.1172/JCI11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musser J M. Emerging Infect Dis. 1996;2:1–17. doi: 10.3201/eid0201.960101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzgerald J R, Musser J M. Trends Microbiol. 2001;9:547–553. doi: 10.1016/s0966-842x(01)02228-4. [DOI] [PubMed] [Google Scholar]
- 4.Dziejman M, Balon E, Boyd D, Fraser C M, Heidelberg J F, Mekalanos J J. Proc Natl Acad Sci USA. 2002;99:966–971. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smoot J C, Barbian K D, Van Gompel J J, Smoot L M, Chaussee M S, Sylva G L, Sturdevant D E, Ricklefs S M, Porcella S F, Parkins L D, et al. Proc Natl Acad Sci USA. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham M W. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musser J M, Krause R M. In: Emerging Infections. Krause R M, editor. New York: Academic; 1998. pp. 185–218. [Google Scholar]
- 8.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beall B, Facklam R, Hoenes T, Schwartz B. J Clin Microbiol. 1997;35:1231–1235. doi: 10.1128/jcm.35.5.1231-1235.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharkawy A, Low D E, Saginur R, Gregson D, Schwartz B, Jessamine P, Green K, McGeer A and the Ontario Group. Clin Infect Dis. 2002;34:454–460. doi: 10.1086/338466. [DOI] [PubMed] [Google Scholar]
- 11.Lei B, Mackie S, Lukomski S, Musser J M. Infect Immun. 2000;68:6807–6818. doi: 10.1128/iai.68.12.6807-6818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferretti J J, McShan W M, Ajdic D, Savic D J, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov A N, Kenton S, et al. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reda K B, Kapur V, Mollick J A, Lamphear J G, Musser J M, Rich R R. Infect Immun. 1994;62:1867–1874. doi: 10.1128/iai.62.5.1867-1874.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reda K B, Kapur V, Goela D, Lamphear J G, Musser J M, Rich R R. Infect Immun. 1996;64:1161–1165. doi: 10.1128/iai.64.4.1161-1165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens K R, Van M, Lamphear J G, Orkiszewski R S, Ballard K D, Cook R G, Rich R R. J Immunol. 1996;157:2479–2487. [PubMed] [Google Scholar]
- 16.Musser J M, Kapur V, Kanjilal S, Shah U, Musher D M, Barg N L, Johnston K H, Schlievert P M, Henrichsen J, Gerlach D, et al. J Infect Dis. 1993;167:337–346. doi: 10.1093/infdis/167.2.337. [DOI] [PubMed] [Google Scholar]
- 17.Kapur V, Maffei J T, Greer R S, Li L-L, Adams G J, Musser J M. Microb Pathog. 1994;16:443–450. doi: 10.1006/mpat.1994.1044. [DOI] [PubMed] [Google Scholar]
- 18.Hoe N, Nakashima K, Grigsby D, Pan X, Dou S J, Naidich S, Garcia M, Kahn E, Bergmire-Sweat D, Musser J M. Emerging Infect Dis. 1999;5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukomski S, Nakashima K, Abdi I, Cipriano V J, Ireland R M, Reid S D, Adams G G, Musser J M. Infect Immun. 2000;68:6542–6553. doi: 10.1128/iai.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen H, Engelbrecht S, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Rice P, Longden I, Bleasby A. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 22.Janulczyk R, Rasmussen M. Infect Immun. 2001;69:4019–4026. doi: 10.1128/IAI.69.6.4019-4026.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutcliffe I C, Russell R R B. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick J K, Tripp T J, Olmstead S B, Matsuka Y V, Gahr P J, Ohlendorf D H, Schlievert P M. J Immunol. 2000;165:2306–2312. doi: 10.4049/jimmunol.165.4.2306. [DOI] [PubMed] [Google Scholar]
- 25.Dinges M M, Schlievert P M. Infect Immun. 2001;69:7169–7172. doi: 10.1128/IAI.69.11.7169-7172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee P K, Deringer J R, Kreiswirth B N, Novick R P, Schlievert P M. Infect Immun. 1991;59:879–884. doi: 10.1128/iai.59.3.879-884.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds L J, Hughes L L, Dennis E A. Anal Biochem. 1992;204:190–197. doi: 10.1016/0003-2697(92)90160-9. [DOI] [PubMed] [Google Scholar]
- 28.Musser J M, Nelson K, Selander R K, Gerlach D, Huang J C, Kapur V, Kanjilal S. J Infect Dis. 1993;167:759–762. doi: 10.1093/infdis/167.3.759. [DOI] [PubMed] [Google Scholar]
- 29.Bynoe E T. Can J Public Health. 1943;34:272–281. [Google Scholar]
- 30.Kohler W, Gerlach D, Knoll H. Zentralbl Bakteriol Mikrobiol Hyg A. 1987;266:104–115. doi: 10.1016/s0176-6724(87)80024-x. [DOI] [PubMed] [Google Scholar]
- 31.Lei B, DeLeo F R, Hoe N P, Graham M R, Mackie S M, Cole R L, Liu M, Hill H R, Low D E, Federle M J, et al. Nat Med. 2001;7:1298–1305. doi: 10.1038/nm1201-1298. [DOI] [PubMed] [Google Scholar]
- 32.Hynes W L, Hancock L, Ferretti J J. Infect Immun. 1995;63:3015–3020. doi: 10.1128/iai.63.8.3015-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson J A, Tyler M I, Retson K V, Howden M E H. Biochim Biophys Acta. 1993;1161:223–229. doi: 10.1016/0167-4838(93)90217-f. [DOI] [PubMed] [Google Scholar]
- 34.Granlund M, Oberg L, Sellin M, Norgren M. J Infect Dis. 1998;177:967–976. doi: 10.1086/515233. [DOI] [PubMed] [Google Scholar]
- 35.Rubens C E, Heggen L M, Kuypers J M. J Bacteriol. 1989;171:5531–5535. doi: 10.1128/jb.171.10.5531-5535.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dodd H M, Horn N, Gasson M J. J Bacteriol. 1994;176:3393–3396. doi: 10.1128/jb.176.11.3393-3396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berge A, Rasmussen M, Bjorck L. Infect Immun. 1998;66:3449–3453. doi: 10.1128/iai.66.7.3449-3453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapur V, Reda K B, Li L-L, Ho L-J, Rich R R, Musser J M. Gene. 1994;150:135–140. doi: 10.1016/0378-1119(94)90872-9. [DOI] [PubMed] [Google Scholar]
- 39.Schlievert P M, Watson D W. Infect Immun. 1978;21:753–763. doi: 10.1128/iai.21.3.753-763.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muotiala A, Seppala H, Huovinen P, Vuopio-Varkila J. J Infect Dis. 1997;175:392–399. doi: 10.1093/infdis/175.2.392. [DOI] [PubMed] [Google Scholar]
- 41.Smoot J C, Korgenski E K, Daly J A, Veasy L G, Musser J M. J Clin Microbiol. 2002;40:1805–1810. doi: 10.1128/JCM.40.5.1805-1810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musser J M, Kapur V, Szeto J, Pan X, Swanson D S, Martin D R. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser J M. J Clin Invest. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Botstein D. Ann NY Acad Sci. 1980;354:484–490. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- 45.Hoe N P, Nakashima K, Lukomski S, Grigsby D, Liu M, Kordari P, Dou S-J, Pan X, Vuopio-Varkila J, Salmelinna S, et al. Nat Med. 1999;5:924–929. doi: 10.1038/11369. [DOI] [PubMed] [Google Scholar]
- 46. Hoe, N. P., Ireland, R. M., DeLeo, F. R., Gowen, B. B., Dorward, D. W., Voyich, J. M., Liu, M., Burns, E. H., Jr., Culnan, D. M., Bretscher, A. & Musser, J. M. (2002) Proc. Natl. Acad. Sci. USA., in press. [DOI] [PMC free article] [PubMed]
- 47.Kline J B, Collins C M. Infect Immun. 1996;64:2122–2129. doi: 10.1128/iai.64.6.2122-2129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid S D, Hoe N P, Smoot L, Musser J M. J Clin Invest. 2001;107:393–399. doi: 10.1172/JCI11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.