Abstract
Ligand-activated receptor tyrosine kinases undergo endocytosis and are transported via endosomes to lysosomes for degradation. This “receptor down-regulation” process is crucial to terminate the cell proliferation signals produced by activated receptors. During the process, ubiquitination of the receptors serves as a sorting signal for their trafficking from endosomes to lysosomes. Here, we describe the role of a deubiquitinating enzyme UBPY/USP8 in the down-regulation of epidermal growth factor (EGF) receptor (EGFR). Overexpression of UBPY reduced the ubiquitination level of EGFR and delayed its degradation in EGF-stimulated cells. Immunopurified UBPY deubiquitinated EGFR in vitro. In EGF-stimulated cells, UBPY underwent ubiquitination and bound to EGFR. Overexpression of Hrs or a dominant-negative mutant of SKD1, proteins that play roles in the endosomal sorting of ubiquitinated receptors, caused the accumulation of endogenous UBPY on exaggerated endosomes. A catalytically inactive UBPY mutant clearly localized on endosomes, where it overlapped with EGFR when cells were stimulated with EGF. Finally, depletion of endogenous UBPY by RNA interference resulted in elevated ubiquitination and accelerated degradation of EGF-activated EGFR. We conclude that UBPY negatively regulates the rate of EGFR down-regulation by deubiquitinating EGFR on endosomes.
INTRODUCTION
Growth factor-induced activation of receptor tyrosine kinases (RTKs) on the cell surface triggers the proliferation of cells by inducing tyrosine autophosphorylation of RTKs and recruiting Src homology 2 domain-containing signaling molecules to RTKs. Simultaneously, activated RTKs are rapidly internalized into cells by endocytosis and eventually transported to lysosomes, where they are degraded by acid hydrolases. This process, known as receptor down-regulation, plays a crucial role in terminating the cell proliferation signals and in preventing the overgrowth of the cell.
Ligand-activated RTKs are incorporated into clathrin-coated vesicles from the plasma membrane and are transported to early endosomes. They are subsequently incorporated into lumenal vesicles of the endosomes that are formed by inward budding of the endosomal limiting membrane. Endosomes containing such vesicles are called multivesicular bodies (MVBs), and they mature to or fuse with late endosomes. Eventually, the late endosomes fuse with lysosomes and release the RTK-containing vesicles into the lumen of the lysosomes (Katzmann et al., 2002). In this RTK trafficking, conjugation of ubiquitin (Ub) to RTKs is implicated in two steps. By analogy to yeast cells in which monoubiquitination of cell surface receptors such as the α-mating factor receptor Ste2 is required for their ligand-induced internalization from the plasma membrane, ubiquitination of RTKs has been suggested to serve as an endocytosis signal that directs their internalization from the plasma membrane (Hicke, 2001). The role of ubiquitination in this step, however, is controversial because several studies show that ubiquitination is dispensable for endocytosis (Longva et al., 2002; Duan et al., 2003). The second, and better-understood, role of RTK ubiquitination is as a sorting signal at early endosomes that directs RTKs for trafficking to lysosomes. Namely, the ubiquitination is a signal for RTKs to be incorporated into the lumenal vesicles of MVBs (Katzmann et al., 2002). Endocytosed receptors that are not ubiquitinated, such as those for low-density lipoprotein and transferrin, escape the incorporation into the MVB vesicles and are returned to the cell surface via recycling endosomes (Gruenberg, 2001).
Endosomal sorting of ubiquitinated RTKs is initiated by a complex of two Ub-binding proteins, hepatocyte growth factor-regulated substrate (Hrs) and signal-transducing adaptor molecule (STAM), which localizes on the cytoplasmic face of the early endosomal membrane (Komada et al., 1997; Bache et al., 2003b; Mizuno et al., 2003, 2004). Depletion of Hrs or STAM using RNA interference (RNAi) or gene disruption inhibits RTK down-regulation in mammalian cells (Bache et al., 2003b; Hammond et al., 2003; Kanazawa et al., 2003; Lu et al., 2003) and in Drosophila (Lloyd et al., 2002). The budding yeast Saccharomyces cerevisiae also has orthologues of Hrs and STAM, which are Vps27 (vacuolar protein sorting 27) and Hse1 (Hbp/STAM/EAST 1), respectively. Yeast mutants lacking Vps27 or Hse1 exhibit defects in the trafficking of monoubiquitinated membrane proteins, such as endocytosed a-mating factor receptor Ste3 and newly synthesized vacuolar carboxypeptidase S precursor, from endosomes to the lumen of vacuoles, a counterpart of mammalian lysosomes (Bilodeau et al., 2002; Shih et al., 2002). Thus, the Hrs–STAM complex is proposed to be the sorting receptor that recognizes the Ub moieties of activated RTKs and introduces them into MVB vesicles on early endosomes in mammalian cells (Komada and Kitamura, 2005).
c-Cbl is a RING finger-type E3 Ub ligase responsible for the ubiquitination of activated RTKs (Thien and Langdon, 2001). It is recruited to specific phosphotyrosine residues of activated RTKs via its tyrosine kinase-binding domain, leading to monoubiquitination of RTKs at multiple lysine residues (Haglund et al., 2003; Mosesson et al., 2003). v-Cbl is an oncogenic counterpart of c-Cbl that consists only of the tyrosine kinase-binding domain (Thien and Langdon, 2001). In v-Cbl-overexpressing cells, it dominant negatively inhibits the ubiquitination of RTKs mediated by endogenous c-Cbl, resulting in impaired RTK down-regulation and cellular transformation (Levkowitz et al., 1998). A mutant of c-Met, an RTK for hepatocyte growth factor, that is deficient in c-Cbl binding also escapes from ubiquitination-dependent down-regulation and transforms cells (Peschard et al., 2001). These observations indicate a crucial role for c-Cbl-mediated RTK ubiquitination in the termination of growth factor signaling. However, if activated RTKs are down-regulated too rapidly, it is expected that cells cannot fully respond to growth factor stimulation due to insufficient activation of downstream signaling molecules. Therefore, the rate of RTK down-regulation must be appropriately regulated.
Ub-specific protease Y (UBPY) also designated as Ub-specific protease 8 (USP8), is a deubiqutinating enzyme that belongs to the Ub-specific protease (UBP) family of cysteine proteases (Naviglio et al., 1998; Baker et al., 1999). Previous in vitro experiments have shown that UBPY binds to the Src homology 3 (SH3) domain of STAM, a component of the Hrs-STAM endosomal sorting complex for ubiquitinated RTKs (Kato et al., 2000). Here, we provide evidence that UBPY deubiquitinates ligand-activated epidermal growth factor (EGF) receptor (EGFR) on endosomes and negatively regulates its down-regulation.
MATERIALS AND METHODS
cDNA Expression Vectors and Transfection
The expression vectors for FLAG- and hemagglutinin (HA)-tagged mouse wild-type UBPY, UBPYC748A, UBPYΔSBM, and human AMSH were con-structed as described previously (Kato et al., 2000). The construction of expression vectors for human EGFR (Morino et al., 2004), HA-tagged mouse Hrs (Komada et al., 1997), and green fluorescent protein (GFP)-tagged mouse SKD1E235Q (Yoshimori et al., 2000) was also described previously. The FLAG-tagged monoubiquitin expression vector and the c-Cbl expression vector were provided by Dr. T. Suzuki (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan) and Dr. K. Yokote (Chiba University, Chiba, Japan), respectively. The expression vectors were transfected into cells for 2 d using the FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN).
Immunoprecipitation and Immunoblotting
Cell lysates were prepared by solubilizing cells with lysis buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.5% NP-40, 1 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A) for 30 min on ice and collecting the supernatants after centrifugation at 12,000 × g for 15 min at 4°C. To analyze proteins in EGF-stimulated cells, cells were grown in the presence of 0.5% fetal bovine serum for 24 h and subsequently incubated with EGF (100 ng/ml; PeproTech, Rocky Hill, NJ) in the presence of 10 μg/ml cycloheximide at 37°C. The lysates were used directly for immunoblotting, or immunoprecipitated with anti-EGFR (0.1 μg; MBL, Nagoya, Japan), anti-FLAG (1 μg; Sigma-Aldrich, St. Louis, MO), anti-STAM1 (5 μl; Mizuno et al., 2004), anti-UBPY (5 μl; Kato et al., 2000), or anti-HA (1 μg; Roche Diagnostics) antibody. Immunoblot analysis was performed by standard procedures. Primary antibodies used were anti-FLAG (4 μg/ml; Sigma-Aldrich), anti-EGFR (0.5 μg/ml; MBL), anti-Ub (5 μg/ml; Covance, Princeton, NJ), anti-UBPY (1:500; Kato et al., 2000), anti-Hrs (1:200; Komada and Kitamura, 1995), anti-STAM1 (1:200; Mizuno et al., 2004), anti-HA (0.4 μg/ml; Roche Diagnostics), and anti-α-tubulin (1 μg/ml; Sigma-Aldrich) antibodies. Secondary antibodies were horseradish peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG antibodies (GE Healthcare, Piscataway, NJ). Blots were detected using the enhanced chemiluminescence reagent (GE Healthcare). To quantify the intensity of bands in immunoblot membranes, the NIH Image analysis program ImageJ was used.
In Vitro Deubiquitination Assay
COS-7 cells were transfected with FLAG-tagged UBPY, UBPYC748A, or AMSH and lysed with lysis buffer without EDTA. The lysates were immunoprecipitated with anti-FLAG antibody conjugated to agarose beads (anti-FLAG M2 affinity gel; Sigma-Aldrich). Precipitated recombinant proteins were eluted by incubation with 100 μl of phosphate-buffered saline (PBS), pH 7.0, containing 150 μg/ml FLAG peptide (Sigma-Aldrich). The purity and concentration of recombinant proteins in the eluates were assessed by Coomassie brilliant blue (CBB) staining after SDS-PAGE using purified bovine serum albumin (BSA) as a standard. Ubiquitinated EGFR was immunoprecipitated with anti-EGFR antibody from COS-7 cells transfected with EGFR, c-Cbl, and FLAG-Ub and stimulated with EGF for 15 min. K48- and K63-linked Ub chains (Ub2-7) were purchased from Affiniti Research Products (Exeter, United Kingdom) and Boston Biochem (Cambridge, MA), respectively. The FLAG peptide-eluted recombinant proteins (∼0.3 μM) were incubated with the Ub chains (0.5 μg) or ubiquitinated EGFR (immunoprecipitated from cells in a 6-cm dish) in PBS, pH 7.0, containing 5 mM MgCl2 and 2 mM dithiothreitol at 37°C for 18 h (Ub chains) or 1 h (EGFR). Reaction products were separated by SDS-PAGE and detected by immunoblotting with anti-Ub or anti-FLAG antibody.
Immunofluorescence Staining
Cells were fixed with 4% paraformaldehyde in PBS or with methanol for 10 min on ice, permeabilized with 0.2% Triton X-100 in PBS, and stained with rabbit polyclonal anti-UBPY (1:500; Kato et al., 2000), rabbit polyclonal anti-FLAG (0.4 μg/ml; Sigma-Aldrich), mouse monoclonal anti-HA (4 μg/ml; Roche Diagnostics), rabbit polyclonal anti-HA (1 μg/ml; Sigma-Aldrich), rabbit polyclonal anti-Hrs (1:1000; Komada and Kitamura, 1995), rabbit polyclonal anti-LAMP1 (1:1000; Carlsson et al., 1988; provided by Dr. M. Fukuda, The Burnham Institute, La Jolla, CA), mouse monoclonal anti-EGFR (10 μg/ml; MBL), and mouse monoclonal anti-EEA1 (1 μg/ml; BD Transduction Laboratories, Lexington, KY) antibodies using standard procedures. Secondary antibodies were Alexa488- and Alexa594-conjugated anti-rabbit IgG and anti-mouse IgG antibodies (Molecular Probes, Eugene, OR). Fluorescence images were captured with a confocal microscope (Axiovert 200M; Carl Zeiss, Oberkochen, Germany) using the LSM5 PASCAL system (Carl Zeiss).
RNAi
Using the siRNA expression vector pSilencer 1.0-U6 (Ambion, Austin, TX), a vector that allows the production of a small interfering RNA (siRNA) for human UBPY mRNA was constructed. It targets the nucleotide residues 3607–3627 (5′-AATCTTCAGCAGCTTATATCC-3′) from the translation initiation codon. The Hrs siRNA expression vector was also constructed using pSilencer 1.0-U6 as described previously (Mizuno et al., 2004). These vectors or an empty mock vector were transfected into HeLa cells twice at 36-h intervals. To express other constructs in these cells, the expression vectors were cotransfected with the siRNA vectors in the second round of transfection.
RESULTS
UBPY Overexpression Reduces the Level of Ligand-induced EGFR Ubiquitination
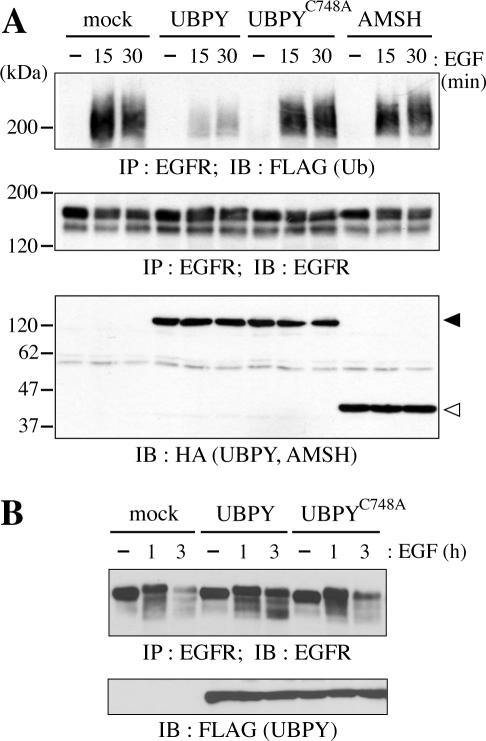
We first examined whether overexpression of UBPY affects the ubiquitination level of EGFR in EGF-stimulated cells. COS-7 cells were transfected with HA-tagged UBPY or its mutant, together with EGFR, c-Cbl, and FLAG-tagged Ub to enhance the receptor ubiquitination, and stimulated with EGF for 15 or 30 min. Ubiquitination of EGFR was detected by immunoprecipitation of EGFR from these cells followed by immunoblotting with anti-FLAG antibody (Figure 1A, top). In mock-transfected cells, EGFR ubiquitination was induced by 15 min of EGF stimulation. In cells overexpressing wild-type UBPY, the level of EGFR ubiquitination was significantly reduced at both 15 and 30 min after EGF treatment. No such effect was observed on overexpression of UBPYC748A, a catalytically inactive mutant in which cysteine 748 in the Cys box is replaced by alanine (Naviglio et al., 1998), indicating that the Ub isopeptidase activity of UBPY is required for the effect.
Figure 1.
Effect of UBPY overexpression on EGFR down-regulation. (A) COS-7 cells were transfected with mock, HA-UBPY, HA-UBPYC748A, or HA-AMSH expression vector together with EGFR, c-Cbl, and FLAG-Ub, and stimulated with EGF for 15 or 30 min. EGFR was immunoprecipitated (IP) from the cells and immunoblotted (IB) with anti-FLAG (top) or anti-EGFR (middle) antibody. The expression levels of the UBPY constructs and AMSH were assessed by immunoblotting of the total cell lysates with anti-HA antibody (bottom). Closed and open arrowheads indicate HA-UBPY and HA-AMSH, respectively. (B) COS-7 cells were transfected with mock, FLAG-UBPY, or FLAG-UBPYC748A expression vector together with EGFR and c-Cbl and stimulated with EGF for 1 or 3 h. EGFR was immunoprecipitated from these cells and immunoblotted with anti-EGFR antibody (top). The expression levels of UBPY and UBPYC748A were assessed by immunoblotting of the total cell lysates with anti-FLAG antibody (bottom).
In addition to UBPY, the SH3 domain of STAM binds to AMSH (associated molecule with the SH3 domain of STAM), a protein that contains the JAB1/MPN/Mov34 metalloenzyme (JAMM) motif (Tanaka et al., 1999). Recently, AMSH was shown to possess deubiquitinating activity toward EGFR in vitro (McCullough et al., 2004). We therefore examined whether AMSH affects the ubiquitination level of EGFR. In cells overexpressing HA-tagged AMSH, however, EGFR was ubiquitinated to a similar level to that in mock-transfected cells after EGF stimulation (Figure 1A, top). EGFR was not drastically degraded by 30 min of EGF treatment (Figure 1A, middle). Different UBPY constructs and AMSH were expressed at similar levels (Figure 1A, bottom).
UBPY Overexpression Delays the Rate of Ligand-induced EGFR Degradation
To examine whether the reduced ubiquitination of ligand-activated EGFR in UBPY-overexpressing cells affects the down-regulation of the receptor, COS-7 cells were transfected with FLAG-tagged UBPY or UBPYC748A together with EGFR and c-Cbl and then stimulated with EGF for 1 or 3 h. EGFR was immunoprecipitated with anti-EGFR antibody and detected by immunoblotting with the same antibody (Figure 1B, top). After 3 h of EGF stimulation, EGFR was mostly degraded in mock- as well as in UBPYC748A-transfected cells. In contrast, a significant amount of EGFR was still detectable in cells transfected with wild-type UBPY, indicating that the reduced EGFR ubiquitination in these cells resulted in delayed down-regulation of the receptor.
UBPY Deubiquitinates EGFR Directly
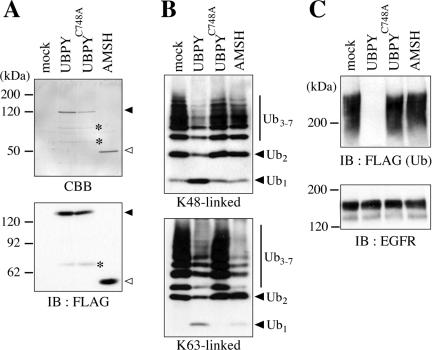
Next, we examined using an in vitro assay whether the reduced ubiquitination of EGFR in UBPY-overexpressing cells is due to a direct action of UBPY on EGFR. FLAG-tagged UBPY, UBPYC748A, and AMSH were expressed in COS-7 cells, immunoprecipitated with anti-FLAG antibody, and eluted with the FLAG competing peptide. The purity of the recombinant proteins in eluted fractions was assessed by CBB staining after SDS-PAGE. In addition to a major band that corresponds to intact UBPY (Figure 2A, top, closed arrowhead), two smaller minor bands were detected in the UBPY and UBPYC748A fractions (Figure 2A, top, asterisks). They most likely represent degradation products of UBPY because they were not detected in the mock and AMSH fractions and one of them was detected by anti-FLAG immunoblotting (Figure 2A, bottom, asterisks). In the AMSH fraction, no protein but intact AMSH was detected by CBB staining as well as immunoblotting with anti-FLAG antibody (Figure 2A, open arrowheads). No band was detected in the fraction prepared from mock-transfected cells (Figure 2A). Using purified BSA as a standard in CBB staining, the yield of the recombinant proteins was roughly estimated to be 2.5 μg for UBPY and AMSH and 1 μg for UBPYC748A from transfected cells in a 6-cm dish (our unpublished data).
Figure 2.
Deubiquitination of EGFR by UBPY in vitro. (A) FLAG-UBPY, FLAG-UBPYC748A, and FLAG-AMSH were expressed in COS-7 cells, immunoprecipitated with anti-FLAG antibody, and eluted with an excess amount of the FLAG peptide. Proteins in the eluates were separated by SDS-PAGE and detected by CBB staining (top) or immunoblotting with anti-FLAG antibody (bottom). Closed and open arrowheads indicate intact FLAG-UBPY and FLAG-AMSH, respectively. Faint bands designated by asterisks are probably degradation products of UBPY. (B) Immunopurified UBPY, UBPYC748A, and AMSH were incubated with K48-linked (top) or K63-linked (bottom) Ub chains, and the reaction products were detected by immunoblotting with anti-Ub antibody. Positions of the Ub monomer (Ub1), dimer (Ub2), and trimer-heptamer (Ub3–7) are indicated. (C) Immunopurified UBPY, UBPYC748A, and AMSH were incubated with ubiquitinated EGFR that was immunoprecipitated from EGF-stimulated COS-7 cells transfected with FLAG-Ub. The reaction products were detected by immunoblotting with anti-FLAG (top) or anti-EGFR (bottom) antibody.
In poly-Ub chains, Ub is mainly conjugated to lysine 48 (K48) or lysine 63 (K63) of another Ub molecule by an isopeptide bond. It has been shown that bacterially expressed glutathione S-transferase (GST)-UBPY fusion protein cleaves the isopeptide bonds in K48-linked Ub chains (McCullough et al., 2004). When K48- and K63-linked Ub chains (0.5 μg) were incubated with our immunopurified UBPY fraction (∼0.3 μM UBPY), the amounts of Ub chains (Ub2 and Ub3-7) were reduced and the amount of Ub monomer (Ub1) was increased, compared with their levels when Ub chains were incubated with the mock fraction, indicating that the UBPY fraction retains the Ub isopeptidase activity (Figure 2B). The UBPYC748A fraction cleaved neither K48- nor K63-linked Ub chains, excluding the possibility that the enzymatic activity in the wild-type UBPY fraction is due to other UBPY-associated or contaminating proteases (Figure 2B). As reported for bacterially expressed GST-AMSH fusion protein (McCullough et al., 2004), the AMSH fraction exhibited isopeptidase activity toward K63-linked but not K48-linked Ub chains (Figure 2B). It is unclear why more K63-linked Ub dimers (Ub2) than monomers (Ub1) were accumulated after incubation with the AMSH fraction (Figure 2B, bottom).
We then examined the deubiquitinating activity of these fractions toward EGFR. COS-7 cells were transfected with EGFR, c-Cbl, and FLAG-Ub and stimulated with EGF for 15 min. Ubiquitinated EGFR was immunoprecipitated from these cells and incubated with the UBPY and AMSH fractions (∼0.3 μM recombinant protein) for 1 h. Whereas the wild-type UBPY fraction completely deubiquitinated EGFR, the UBPYC748A and AMSH fractions exhibited undetectable deubiquitinating activity on EGFR in this assay (Figure 2C, top). The amount of EGFR was unchanged by the incubation (Figure 2C, bottom).
UBPY Binds to Ligand-activated EGFR in an Hrs- and STAM-dependent Manner
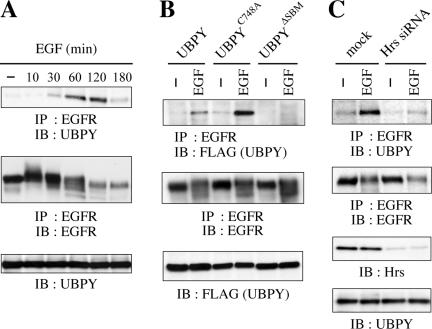
The deubiquitination of EGFR by UBPY suggested that UBPY might bind to ubiquitinated EGFR stably. We examined this possibility by coimmunoprecipitation experiments. Endogenous EGFR was immunoprecipitated from HeLa cells stimulated with EGF for up to 3 h. Immunoblotting of the precipitates with anti-UBPY antibody showed that EGF induced the binding of endogenous UBPY to EGFR (Figure 3A, top). The binding was detectable after 30 min of stimulation and reached a maximal level in ∼1 to 2 h.
Figure 3.
UBPY binding to EGFR. (A) HeLa cells were stimulated with EGF for indicated periods. EGFR was immunoprecipitated from the cells and immunoblotted with anti-UBPY (top) or anti-EGFR (middle) antibody. The expression level of UBPY was assessed by immunoblotting of the total cell lysates with anti-UBPY antibody (bottom). (B) HeLa cells were transfected with FLAG-UBPY, FLAG-UBPYC748A, or FLAG-UBPYΔSBM and stimulated with or without EGF for 1 h. EGFR was immunoprecipitated from the cells and immunoblotted with anti-FLAG (top) or anti-EGFR (middle) antibody. The expression levels of the UBPY constructs were assessed by immunoblotting of the total cell lysates with anti-FLAG antibody (bottom). (C) HeLa cells were transfected with mock or Hrs siRNA expression vector and stimulated with or without EGF for 1 h. EGFR was immunoprecipitated from the cells and immunoblotted with anti-UBPY (top) or anti-EGFR (second panel from the top) antibody. The expression levels of Hrs and UBPY were assessed by immunoblotting of the total cell lysates with anti-Hrs (third panel from the top) and anti-UBPY (bottom) antibodies.
We next examined the binding of the mutants, UBPYC748A and UBPYΔSBM, to EGFR. UBPYΔSBM harbors mutations in the two STAM-binding motifs (SBMs) composed of nine amino acids PX(V/I)(D/N)RXXKP and lacks the ability to interact with the Hrs–STAM complex (Kato et al., 2000). HeLa cells were transfected with FLAG-tagged UBPY constructs and stimulated with EGF for 1 h. EGFR was immunoprecipitated from these cells, and bound UBPY proteins were detected by anti-FLAG immunoblotting. UBPYC748A bound to EGFR more efficiently than the wild-type protein after EGF treatment (Figure 3B, top). UBPYΔSBM, in contrast, did not bind to EGFR (Figure 3B, top).
To test whether the binding of UBPY to EGFR requires the Hrs–STAM complex, we examined the effect of RNAi-mediated depletion of Hrs on the binding. We have previously shown that the Hrs depletion results in the mislocalization of STAM proteins to the cytoplasm and in the reduction of their levels within cells, leading to the depletion of the Hrs–STAM complex on the endosomal membrane (Mizuno et al., 2004). In Hrs siRNA-transfected cells, EGF-induced binding of UBPY to EGFR was drastically reduced (Figure 3C, top). The levels of EGFR (Figure 3C, second panel from the top) and UBPY (Figure 3C, bottom) were unchanged by Hrs depletion. These results, together with the inability of UBPYΔSBM to bind to EGFR (Figure 3B), suggest that the interaction with the Hrs–STAM complex is required for UBPY to bind to EGFR within cells.
UBPY Is Not Stably Associated with the Hrs–STAM Complex
The Hrs–STAM complex binds to ubiquitinated EGFR (Morino et al., 2004; Sigismund et al., 2005), raising the possibility that UBPY and EGFR are not directly associated but are linked by the Hrs–STAM complex. To test this possibility, lysates of HeLa cells stimulated with EGF for up to 60 min were immunoprecipitated with antibody against STAM1, one of the two STAM family proteins with redundant function (Komada and Kitamura, 2005). However, coprecipitation of endogenous UBPY with endogenous STAM1 was undetectable irrespective of EGF stimulation (Figure S1A, top). Hrs, in contrast, was continuously associated with STAM1 (Figure S1A, middle). In a converse experiment, anti-UBPY antibody did not coprecipitate STAM1 from unstimulated or EGF-stimulated cells (Figure S1B). The failure to coprecipitate UBPY and STAM1 is consistent with the low affinity of the UBPY SBMs to the STAM SH3 domain (Kd = 27 μM; Kaneko et al., 2003). These results therefore exclude the possibilities that UBPY is stably associated with the Hrs–STAM complex and that the UBPY-EGFR interaction is mediated by the presence of the Hrs–STAM complex between them.
UBPY Undergoes EGF-induced Ubiquitination
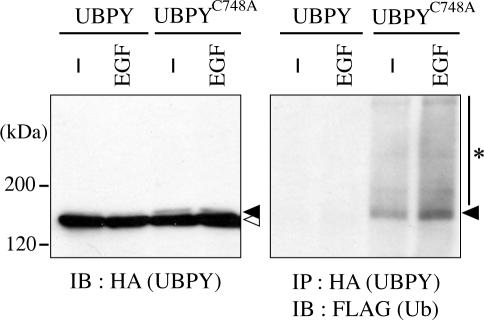
When HA-tagged UBPYC748A, but not wild-type UBPY, was expressed in HeLa cells together with FLAG-Ub, a faint band that migrates slightly more slowly than the major band was detected by anti-HA immunoblotting (Figure 4, left, closed and open arrowheads). We examined whether this shift-up is due to the ubiquitination of UBPYC748A. Immunoprecipitation of HA-UBPYC748A followed by immunoblotting with anti-FLAG antibody showed that UBPYC748A was indeed ubiquitinated to some extent in unstimulated cells (Figure 4, right). Moreover, stimulation of the cells with EGF for 15 min elevated the ubiquitination level of the protein (Figure 4, right). Ubiquitinated UBPYC748A was mainly detected as a single band (Figure 4, right, arrowhead) with the same mobility as the faint band detected by anti-HA antibody, suggesting that UBPY is mostly monoubiquitinated. Smeared bands detected above monoubiquitinated UBPYC748A by anti-FLAG antibody (Figure 4, right, asterisk) could represent polyubiquitinated UBPY or UBPY-associated ubiquitinated proteins. In contrast, ubiquitination of wild-type UBPY was undetectable in unstimulated or EGF-stimulated cells (Figure 4, right), suggesting a possibility that UBPY deubiquitinates itself, thereby regulating its own function.
Figure 4.
Ubiquitination of UBPY. HeLa cells were transfected with HA-UBPY or HA-UBPYC748A together with FLAG-Ub and stimulated with or without EGF for 15 min. Lysates of the cells were immunoblotted with anti-HA antibody (left), or immunoprecipitated with anti-HA antibody and immunoblotted with anti-FLAG antibody (right). Closed and open arrowheads indicate monoubiquitinated and unmodified UBPY proteins, respectively. The asterisk in the right panel indicates polyubiquitinated UBPYC748A or UBPY-associated ubiquitinated proteins.
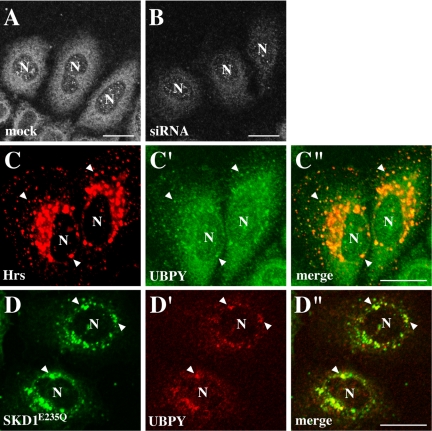
UBPY Functions at Endosomes
To elucidate the subcellular localization of UBPY, we performed immunofluorescence staining of HeLa cells with anti-UBPY antibody. As a negative control, we used cells in which UBPY was depleted by RNAi. In cells transfected with an siRNA expression vector that targets human UBPY, UBPY expression was significantly reduced (Figure 7A, third panel from the top). Staining of mock-transfected cells with anti-UBPY antibody exhibited a cytoplasmic localization pattern that was not characteristic of any single organelle (Figure 5A). This staining was mostly lost in siRNA-transfected cells (Figure 5B), indicating that it represents the UBPY localization but not background staining. However, when HA-tagged Hrs was exogenously expressed, UBPY colocalized with Hrs on enlarged endosomes generated by Hrs overexpression (Figure 5, C–C″, arrowheads). SKD1 is a mammalian orthologue of yeast Vps4, an AAA-type ATPase that also participates in the endosomal sorting of ubiquitinated membrane proteins (Katzmann et al., 2002). Overexpression of SKD1E235Q, a dominant-negative SKD1 mutant lacking ATPase activity, causes the accumulation of endosomal proteins on morphologically aberrant endosomes (Bishop and Woodman, 2000; Yoshimori et al., 2000). In SKD1E235Q-overexpressing cells, UBPY also localized to the SKD1E235Q-positive aberrant endosomes (Figure 5, d–d″, arrowheads).
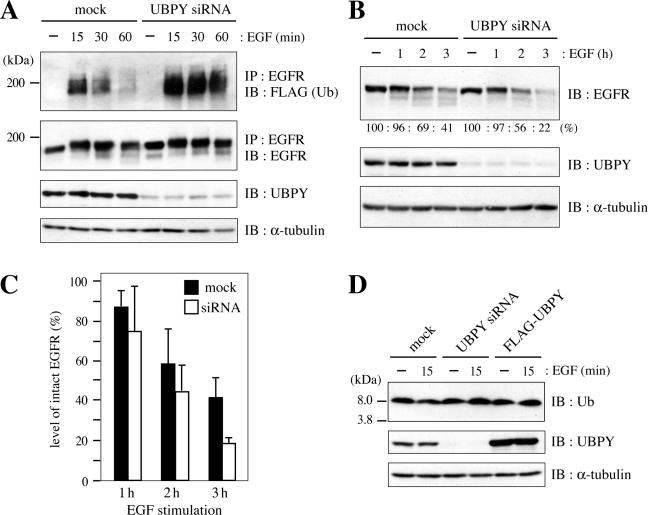
Figure 7.
Effect of UBPY depletion on EGFR down-regulation. (A) HeLa cells were transfected with mock or UBPY siRNA expression vector together with FLAG-Ub and stimulated with EGF for indicated periods. EGFR was immunoprecipitated from the cells and immunoblotted with anti-FLAG (top) or anti-EGFR (second panel from the top) antibody. The expression level of UBPY was assessed by immunoblotting of the total cell lysates with anti-UBPY antibody (third panel from the top). As a loading control, the total lysates were also immunoblotted with anti-α-tubulin antibody (bottom). (B) HeLa cells were transfected with mock or UBPY siRNA expression vector and stimulated with EGF for indicated periods. Lysates of the cells were immunoblotted with anti-EGFR (top), anti-UBPY (middle), and anti-α-tubulin (bottom) antibodies. The intensity of bands for intact EGFR in EGF-stimulated cells, relative to that in unstimulated cells, are indicated as percentages below the top panel. (C) The experiment shown in B was repeated three times, and the mean ± SD of the percentages of intact EGFR remaining in EGF-stimulated cells are shown. (D) HeLa cells were transfected with mock, UBPY siRNA, or FLAG-UBPY expression vector, and stimulated with or without EGF for 15 min. Lysates of the cells were immunoblotted with anti-Ub (top), anti-UBPY (middle), and anti-α-tubulin antibodies.
Figure 5.
Subcellular localization of endogenous UBPY. (A and B) HeLa cells were transfected with mock (A) or UBPY siRNA (B) expression vector and stained with anti-UBPY antibody. (C–C″) HeLa cells were transfected with HA-tagged Hrs and double-stained with anti-HA (C) and anti-UBPY (C′) antibodies. (D–D″) HeLa cells were transfected with GFP-tagged SKD1E235Q and stained with anti-UBPY (D′) antibody. D shows the localization of SKD1E235Q detected by the GFP fluorescence. C″ and D″ are merged images. Arrowheads indicate colocalization of UBPY with transfected Hrs (C–C″) and SKD1E235Q (d–d″). N indicates the nucleus. Bar, 20 μm.
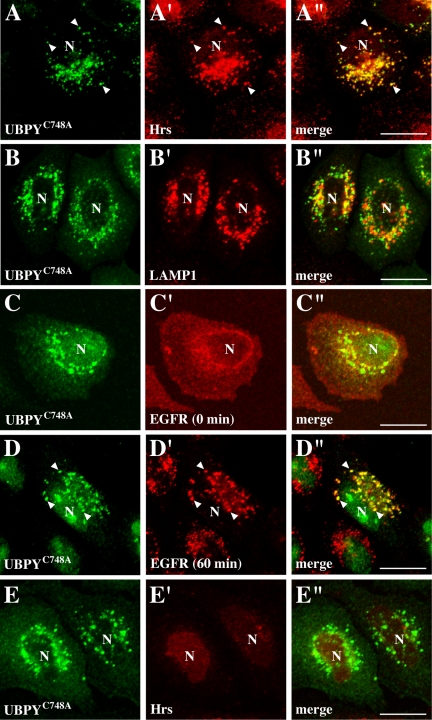
Exogenously expressed UBPY localized diffusely in the cytoplasm but was similarly accumulated on aberrant endosomes when Hrs or SKD1E235Q was overexpressed (Figure S2, A–A″ and C–C″). In contrast, HA-UBPYC748A localized on punctate structures even in the absence of overexpressed Hrs or SKD1E235Q (Figure 6, A and B) and overlapped mostly with endogenous Hrs (Figure 6, A′ and A″) and partially with a late endosome marker LAMP1 (Figure 6, B′ and B″). EGFR localized on the cell surface in these cells when they were unstimulated (Figure 6C′). After 60 min of EGF stimulation, however, clear colocalization of endocytosed EGFR with UBPYC748A was observed on the punctate structures (Figure 6, D–D). These results suggest that UBPY partially localizes and deubiquitinates EGFR on endosomes.
Figure 6.
Subcellular localization of UBPYC748A. (A–A″, B–B″) HeLa cells were transfected with HA-UBPYC748A and stained with anti-HA antibody (A and B), together with anti-Hrs (A′) or anti-LAMP1 (B′) antibody. (C–C″, D–D″) HeLa cells were transfected with HA-UBPYC748A, stimulated with (D–D″) or without (C–C″) EGF for 60 min, and double-stained with anti-HA (C and D) and anti-EGFR (C′ and D′) antibodies. (E–E″) HeLa cells were cotransfected with HA-UBPYC748A and Hrs siRNA expression vectors and double-stained with anti-HA (E) and anti-Hrs (E′) antibodies. A″, B″, C″, D″, and E″ are merged images. Arrowheads indicate colocalization of UBPYC748A with Hrs (A–A″) and EGFR (D–D″). N, nucleus. Bar, 20 μm.
Endosomal Localization of UBPY Is Not Mediated by the Hrs–STAM Complex
In cells overexpressing Hrs or SKD1E235Q, UBPYΔSBM localized on aberrant endosomes (Figure S2, B–B″ and D–D″). This localization pattern was essentially the same as that observed for wild-type UBPY (Figure S2, A–A″ and C–C″), suggesting that the endosomal localization of UBPY is independent of its interaction with the Hrs–STAM complex. To further test this possibility, we examined the effect of Hrs depletion, which results in the depletion of the endosomal Hrs–STAM complex, on the localization of UBPYC748A. In Hrs siRNA-transfected cells, UBPYC748A exhibited a punctate localization (Figure 6, E–E″), which was similar to that in untransfected cells (Figure 6, A–A″). The UBPYC748A-positive punctate structures in Hrs-depleted cells were mostly positive for an early endosome marker EEA1 (Figure S3, A–A″) and partially positive for LAMP1 (Figure S3, B–B″), strongly suggesting that the endosomal localization of UBPY is regulated by an Hrs- and STAM-independent mechanism.
UBPY Depletion Elevates the Level of EGFR Ubiquitination and Accelerates Its Down-Regulation
We last examined the effects of RNAi-mediated UBPY depletion on EGFR down-regulation as well as the cytoplasmic free Ub level. HeLa cells were transfected with the mock or UBPY siRNA expression vector together with FLAG-Ub, and stimulated with EGF for up to 60 min. Immunoblot analysis of the total cell lysates with anti-UBPY antibody showed that transfection of the siRNA resulted in a significant reduction in the UBPY level (Figure 7A, third panel from the top). EGFR was immunoprecipitated from these cells and immunoblotted with anti-FLAG antibody. The ubiquitination level of ligand-activated EGFR was higher in siRNA-transfected cells than in mock-transfected cells in all periods, especially at 30 and 60 min, after EGF stimulation, suggesting that UBPY deubiquitinates ligand-activated EGFR in normal cells (Figure 7A, top). The level of EGFR was similar between these cells until 60 min of stimulation (Figure 7A, second panel from the top). To examine the rate of EGFR down-regulation in UBPY-depleted cells, cells were stimulated with EGF for longer periods. After 2 and 3 h of stimulation, less EGFR molecules remained in siRNA-transfected cells than in mock-transfected cells (Figure 7B, top). Quantification of the intensity of bands corresponding to intact EGFR indicated that 69% (mock) versus 56% (siRNA) of EGFR remained after 2 h stimulation, and 41% (mock) versus 22% (siRNA) remained after 3 h stimulation. This experiment was repeated three times, and the mean ± SD of the relative EGFR levels remaining in EGF-stimulated cells are shown in Figure 7C. At 3 h after stimulation, the level of EGFR in UBPY-depleted cells was consistently reduced to ∼50% of that in mock-transfected cells, suggesting that the EGFR down-regulation is accelerated in the absence of UBPY.
Doa4, a yeast deubiquitinating enzyme homologous to mammalian UBPY, is implicated in maintaining the level of free Ub in the cytoplasm by recycling Ub molecules from proteasome- and vacuole-targeted ubiquitinated proteins (Swaminathan et al., 1999; Amerik et al., 2000). We therefore examined the effect of depleting or overexpressing UBPY on the cytoplasmic free Ub level in unstimulated cells as well as in cells stimulated with EGF for 15 min. However, immunoblot analysis of the lysates of cells transfected with the mock, UBPY siRNA, and FLAG-UBPY expression vectors with anti-Ub antibody showed that the level of Ub monomer was similar among these cells irrespective of EGF stimulation (Figure 7D).
DISCUSSION
The human genome encodes >50 deubiquitinating enzymes, suggesting that each enzyme possesses unique substrate specificity (Wing, 2003; Amerik and Hochstrasser, 2004; Soboleva and Baker, 2004). For many of them, however, substrate proteins as well as biological significance remain unclear. In this study, we demonstrated that UBPY/USP8 deubiquitinates ligand-activated EGFR and regulates the rate of its down-regulation.
UBPY Is a Deubiquitinating Enzyme for Activated EGFR
The ubiquitination level of EGF-activated EGFR was reduced in UBPY-overexpressing cells (Figure 1) and elevated in UBPY-depleted cells (Figure 7). FLAG-tagged UBPY, which was immunopurified from COS-7 cells, efficiently deubiquitinated EGFR in vitro (Figure 2). In addition, EGF induced the binding of UBPY to EGFR within cells (Figure 3). Together, we conclude that UBPY is the deubiquitinating enzyme for EGFR in EGF-stimulated cells. EGFR undergoes monoubiquitination at multiple lysine residues upon EGF treatment (Haglund et al., 2003; Mosesson et al., 2003). Therefore, our results suggest that UBPY acts on monoubiquitinated substrates, in addition to poly-Ub chains as reported previously (Naviglio et al., 1998; McCullough et al., 2004). UBPYC748A bound to activated EGFR more efficiently than the wild-type protein (Figure 3). This might suggest that UBPY dissociates from substrate proteins after cleaving their Ub moieties, and the dissociation is prevented when the enzyme is catalytically inactive. Such a “substrate trap” effect is also demonstrated for catalytically inactive mutants of protein tyrosine phosphatase 1B that dephosphorylates tyrosine-phosphorylated EGFR (Flint et al., 1997; Liu and Chernoff, 1997). The binding of UBPY to EGFR was induced after 30 min of EGF stimulation (Figure 3), and the EGFR ubiquitination was largely affected also after 30 min of ligand stimulation in UBPY-depleted cells (Figure 7). Inconsistent with these, EGFR was maximally deubiquitinated by 15 min of EGF stimulation in UBPY-overexpressing cells (Figure 1). This is most likely due to an ectopic action of overexpressed UBPY, because it localized diffusely in the cytoplasm (Figure S2). Namely, UBPY is leaked to the cytoplasm when overexpressed and then deubiquitinates activated EGFR before the receptor is transported to endosomes from the cell surface.
UBPY also binds to several other proteins: CDC25Mm, a GDP/GTP exchange factor for Ras (Gnesutta et al., 2001); GRAIL, an E3 Ub ligase involved in anergy induction in CD4+ T-cells (Soares et al., 2004); and Nrdp1, another E3 ligase that regulates the steady-state level of ErbB3 and ErbB4 RTKs (Wu et al., 2004). UBPY is implicated in deubiquitinating and regulating the stability of these proteins (Gnesutta et al., 2001; Soares et al., 2004; Wu et al., 2004). Therefore, through its deubiquitinating activity, UBPY probably participates in several different cellular processes by regulating the degradation or stability of distinct substrate proteins.
UBPYC748A, but not wild-type UBPY, was mono- and possibly polyubiquitinated within cells at detectable levels (Figure 4). Moreover, the level of its ubiquitination was elevated when cells were stimulated with EGF (Figure 4). These observations suggest that the function of UBPY is regulated by its ubiquitination and that the ubiquitination level is in turn regulated by the self-deubiquitinating activity of UBPY. Because UBPY acts on ligand-activated EGFR, its activity is possibly regulated by EGF. We detected no tyrosine phosphorylation of UBPY in EGF-stimulated cells (our unpublished data). Therefore, EGF-induced UBPY ubiquitination might regulate its function by changing the protein stability, enzymatic activity, or subcellular localization.
Subcellular Site of UBPY-mediated EGFR Deubiquitination
Immunostaining of cells with anti-UBPY antibody exhibited a cytoplasmic localization that was not typical of any particular organelle (Figure 5). This might indicate that UBPY localizes to several different subcellular sites because it is implicated in the deubiquitination of several unrelated proteins (Gnesutta et al., 2001; Soares et al., 2004; Wu et al., 2004). When endosomes were exaggerated by impairing membrane traffic via the organelle, however, UBPY was accumulated on the aberrant endosomes. First, overexpression of Hrs causes the enlargement of early endosomes (Komada et al., 1997). Cell surface receptors such as EGFR and transferrin receptor, as well as other ubiquitinated proteins, are accumulated on these endosomes (Komada et al., 1997; Bishop et al., 2002; Raiborg et al., 2002; Morino et al., 2004). UBPY colocalized with overexpressed Hrs on the aberrant endosomes (Figure 5). Second, yeast Vps4 and its mammalian orthologue SKD1 are AAA-type ATPases dysfunction of which results in the accumulation of endocytosed receptors as well as proteins of the endosomal Ub-sorting machinery on aberrant endosomes (Bishop and Woodman, 2000; Yoshimori et al., 2000; Katzmann et al., 2002). UBPY was also accumulated on aberrant endosomes in cells overexpressing SKD1E235Q, a dominant-negative SKD1 mutant lacking ATPase activity (Figure 5). Moreover, UBPYC748A mostly localized on Hrs-positive but not LAMP1-positive endosomes, where it overlapped with EGFR when cells were stimulated with EGF (Figure 6). Together, UBPY is suggested to deubiquitinate EGFR on early endosomes. Wild-type UBPY, when exogenously expressed, did not exhibit an endosomal localization like UBPYC748A (Figure S2). Therefore, the localization of UBPYC748A might be exaggerated due to its substrate trap effect that immobilizes unidentified substrate proteins as well as the enzyme itself on the site of its action. In cells overexpressing Hrs or SKD1E235Q, UBPYΔSBM that cannot interact with the Hrs–STAM complex was also accumulated on aberrant endosomes similarly to wild-type UBPY (Figure S2), suggesting that in these cells, UBPY was concentrated on endosomes due to impaired morphology and function of the organelle without direct interaction with the Hrs–STAM complex. Moreover, UBPYC748A normally localized on early endosomes in cells in which the endosomal Hrs–STAM complex was depleted by RNAi for Hrs (Figures 6 and S3). Therefore, it is likely that the endosomal localization of UBPY is not mediated by its interaction with the Hrs–STAM complex but is regulated by a distinct unknown mechanism.
EGF-induced binding of UBPY to EGFR reached a maximal level after 1 h of stimulation (Figure 3), suggesting that UBPY binds to endocytosed EGFR on the endosomal membrane. In contrast, UBPYΔSBM did not bind to EGFR (Figure 3). In addition, binding of UBPY to EGFR was reduced in Hrs-depleted cells (Figure 3). However, the binding of UBPY to EGFR is not linked via the Hrs–STAM complex because UBPY is not stably associated with the Hrs-STAM complex (Figure S1). We therefore speculate that ubiquitinated EGFR is first bound by the Hrs–STAM complex on endosomes and subsequently transferred to UBPY and that a transient interaction between STAM and UBPY is required for UBPY to receive EGFR from the Hrs–STAM complex.
Role of UBPY in EGFR Down-Regulation
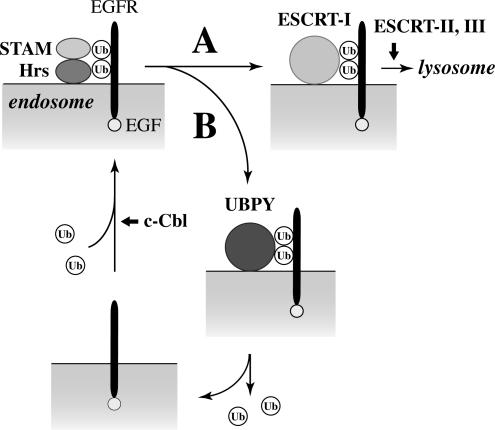
The EGF-induced degradation of EGFR was delayed in cells overexpressing UBPY (Figure 1) and accelerated in cells in which UBPY was depleted (Figure 7). These results suggest that UBPY negatively regulates the rate of EGFR down-regulation by deubiquitinating EGFR and delaying its degradation in EGF-stimulated cells. Based on the results presented in this study, we propose the following mode of action for UBPY (Figure 8). On transport from the cell surface to endosomes, ubiquitinated EGFR is recognized by the Hrs–STAM complex. EGFR is then transferred to endosomal sorting complex required for transport (ESCRT)-I, a protein complex that also binds to ubiquitinated cargo proteins and sorts them into MVB lumenal vesicles in collaboration with ESCRT-II and ESCRT-III (Katzmann et al., 2001, 2002; Bache et al., 2003a; Lu et al., 2003; Figure 8, route A). However, a certain proportion of EGFR is transferred to UBPY from the Hrs–STAM complex instead, via transient interaction between STAM and UBPY (Figure 8, route B). This leads to the deubiquitination of EGFR and prevents it from being recognized by ESCRT-I and trafficking to lysosomes. It is unclear whether deubiquitinated EGFR is recycled back to the cell surface or stays on the endosomal membrane. In either case, deubiquitinated EGFR, which remains activated, is again ubiquitinated by c-Cbl, recognized by the Hrs–STAM complex, and eventually transported to lysosomes. Namely, EGFR proteins that take route A in Figure 8 undergo rapid degradation, whereas those that take route B undergo delayed degradation. When the binding of UBPY to EGFR reached the maximal level (∼1 to 2 h after EGF stimulation), a substantial amount of EGFR had already undergone degradation (Figure 3). This is consistent with the model that the EGFR molecules that are not bound by UBPY undergo rapid degradation. The role of UBPY might be to help EGF-stimulated cells generate sufficient intracellular downstream signals to trigger cell proliferation by preventing a too-rapid EGFR down-regulation. Although the ubiquitination of RTKs is also implicated in their ligand-induced internalization from the cell surface, the internalization of EGFR was unaffected in UBPY-depleted cells as assessed by immnunostaining of EGFR in these cells (our unpublished data). This observation also supports our model that UBPY functions at endosomes.
Figure 8.
Model for the mode of UBPY action. At endosomes, ubiquitinated EGFR is first recognized by the Hrs–STAM complex. EGFR is then transferred to ESCRT-I for further sorting and trafficking to lysosomes (route A). However, a certain proportion of EGFR is transferred to UBPY instead, deubiquitated, and recycled back to the cell surface or stays activated on endosomes (route B). Deubiquitinated EGFR is again ubiquitinated by c-Cbl, recognized by the Hrs-STAM complex, and eventually takes route A for degradation. EGFR proteins that take route A undergo rapid down-regulation, whereas those that take route B undergo delayed down-regulation.
Whether UBPY deubiquitinates and regulates the down-regulation of other RTKs remains to be addressed in future studies. For example, EGFR belongs to the ErbB family of RTKs. Because the proteins in this family are structurally related, UBPY might also regulate the ubiquitination-dependent down-regulation or the steady-state levels of other ErbB family proteins. Stimulation of cells with EGF induces the heterodimerization of EGFR with other family members (Alroy and Yarden, 1997). It has been shown that the heterodimerization of EGFR with ErbB2 and ErbB3 inhibits the down-regulation of EGFR (Lenferink et al., 1998; Wang et al., 1999; Worthylake et al., 1999). Therefore, it is possible that UBPY also indirectly regulates the down-regulation of EGFR by controlling the cellular levels of its heterodimerization partners, ErbB2 and/or ErbB3.
Among deubiquitinating enzymes in S. cerevisiae, Doa4 shares the highest sequence homology with UBPY in the Cys- and His-boxes of the catalytic domain as well as in the rhodanese domain, a functionally uncharacterized domain found only in three of 16 yeast UBP family proteins. Doa4 deubiquitinates ubiquitinated cargo proteins on the endosomal membrane before they are incorporated into the lumenal vesicles of MVBs (Dupre and Haguenauer-Tsapis, 2001; Katzmann et al., 2001; Losko et al., 2001). In doa4 mutant yeast cells, the cytoplasmic free Ub pool is depleted, suggesting that Doa4 retrieves Ub molecules to the cytoplasm before they are cotransported with cargo proteins to and degraded in vacuoles (Swaminathan et al., 1999; Amerik et al., 2000). In cells in which UBPY was depleted or overexpressed, however, the level of free Ub was unchanged compared with that in normal cells (Figure 7). In addition, the SBMs in UBPY are not conserved in Doa4, suggesting that Doa4 does not interact with the SH3 domain of Hse1, a yeast orthologue of STAM proteins. Therefore, UBPY is likely to have a distinct function from Doa4.
Through the SH3 domain, STAM proteins also bind to AMSH, a protein containing the Zn2+-binding JAMM motif (Tanaka et al., 1999). The JAMM motif is also present in Rpn11/POH1, a component of the 19S regulatory complex of the proteasome. Rpn11/POH1 exhibits a JAMM motif-associated metalloprotease activity toward the Ub isopeptide bond (Verma et al., 2002; Yao and Cohen, 2002). Recently, McCullough et al. (2004) demonstrated that GST-fused AMSH purified from Escherichia coli also exhibits a JAMM motif-associated Ub isopeptidase activity toward K63-linked Ub chains as well as ubiquitinated EGFR in vitro. We showed in this study that FLAG-tagged AMSH immunopurified from mammalian cells similarly cleaves K63-linked Ub chains to Ub dimers and monomers (Figure 2). However, it did not exhibit detectable EGFR-deubiquitinating activity in our experimental conditions (Figure 2). Whereas ubiquitinated EGFR was incubated with UBPY or AMSH for only 1 h in our assay, McCullough et al. (2004) incubated EGFR with GST-AMSH for 18 h. It is therefore possible that our incubation period was too short to detect the deubiquitinating activity of AMSH. Regardless, our results, together with the observation that overexpression of AMSH did not reduce the ubiquitination level of EGFR in EGF-stimulated cells (Figure 1), suggest that UBPY might be more responsible for the deubiquitination of activated EGFR than AMSH. When AMSH is depleted by RNAi, however, ligand-induced EGFR degradation is accelerated in HeLa cells (McCullough et al., 2004). Because the ubiquitination level of activated EGFR in AMSH-depleted cells has not been examined, it is unclear whether AMSH regulates the EGFR degradation by directly deubiquitinating the protein within cells. Further study is therefore necessary to understand whether the two deubiquitinating enzymes UBPY and AMSH regulate the EGFR down-regulation in a redundant manner or by distinct mechanisms.
Supplementary Material
Acknowledgments
We thank Dr. Kato for providing various UBPY and AMSH expression vectors. We also thank Drs. Suzuki, Yokote, and Fukuda for providing the FLAG-Ub expression vector, the c-Cbl expression vector, and the anti-LAMP1 antibody, respectively. E. M. is a research fellow of the Japan Society for the Promotion of Science. This work was supported by Grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan to E. M. (No. 16004686), to N. K. (No. 15370053), and to M. K. (No. 16044213), and a research grant from the Naito Foundation to M. K.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–06–0560) on August 24, 2005.
Abbreviations used: CBB, Coomassie brilliant blue; EGFR, epidermal growth factor receptor; ESCRT, endosomal sorting complex required for transport; MVB, multivesicular body; RNAi, RNA interference; RTK, receptor tyrosine kinase; SH3, Src homology 3; SBM, STAM-binding motif; siRNA, small interfering RNA; Ub, ubiquitin; UBP, ubiquitin-specific protease.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alroy, I., and Yarden, Y. (1997). The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand-receptor interactions. FEBS Lett. 410, 83–86. [DOI] [PubMed] [Google Scholar]
- Amerik, A. Y., and Hochstrasser, M. (2004). Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695, 189–207. [DOI] [PubMed] [Google Scholar]
- Amerik, A. Y., Nowak, J., Swaminathan, S., and Hochstrasser, M. (2000). The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11, 3365–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache, K. G., Brech, A., Mehlum, A., and Stenmark, H. (2003a). Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 162, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache, K. G., Raiborg, C., Mehlum, A., and Stenmark, H. (2003b). STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278, 12513–12521. [DOI] [PubMed] [Google Scholar]
- Baker, R. T., Wang, X.-W., Woollatt, E., White, J. A., and Sutherland, G. R. (1999). Identification, functional characterization, and chromosomal localization of USP15, a novel human ubiquitin-specific protease related to the UNP oncoprotein, and a systematic nomenclature for human ubiquitin-specific proteases. Genomics 59, 264–274. [DOI] [PubMed] [Google Scholar]
- Bilodeau, P. S., Urbanowski, J. L., Winistorfer, S. C., and Piper, R. C. (2002). The Vps27p-Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4, 534–539. [DOI] [PubMed] [Google Scholar]
- Bishop, N., Horman, A., and Woodman, P. (2002). Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 157, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, N., and Woodman, P. (2000). ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell 11, 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson, S. R., Roth, J., Piller, F., and Fukuda, M. (1988). Isolation and characterization of human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2. J. Biol. Chem. 263, 18911–18919. [PubMed] [Google Scholar]
- Duan, L., et al. (2003). Cbl-mediated ubiquitylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J. Biol. Chem. 278, 28950–28960. [DOI] [PubMed] [Google Scholar]
- Dupre, S., and Haguenauer-Tsapis, R. (2001). Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of Doa4p ubiquitin isopeptidase. Mol. Cell. Biol. 21, 4482–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, A. J., Tiganis, T., Barford, D., and Tonks, N. K. (1997). Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 94, 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnesutta, N., Ceriani, M., Innocenti, M., Mauri, I., Zippel, R., Sturani, E., Borgonovo, B., Berruti, G., and Martegani, E. (2001). Cloning and characterization of mouse UBPy, a deubiquitinating enzyme that interacts with the Ras guanine nucleotide exchange factor CDC25Mm/Ras-GRF1. J. Biol. Chem. 276, 39448–39454. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J. (2001). The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell. Biol. 2, 721–730. [DOI] [PubMed] [Google Scholar]
- Haglund, K., Sigismund, S., Polo, S., Szymkiewicz, I., Di Fiore, P. P., and Dikic, I. (2003). Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5, 461–466. [DOI] [PubMed] [Google Scholar]
- Hammond, D. E., Carter, S., McCullough, J., Urbe, S., Vande Woude, G., and Clague, M. J. (2003). Endosomal dynamics of Met determine signaling output. Mol. Biol. Cell 14, 1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke, L. (2001). A new ticket for entry into budding vesicles-ubiquitin. Cell 106, 527–530. [DOI] [PubMed] [Google Scholar]
- Kanazawa, C., Morita, E., Yamada, M., Ishii, N., Miura, S., Asao, H., Yoshimori, T., and Sugamura, K. (2003). Effects of deficiencies of STAMs and Hrs, mammalian class E Vps proteins, on receptor downregulation. Biochem. Biophys. Res. Commun. 309, 848–856. [DOI] [PubMed] [Google Scholar]
- Kaneko, T., Kumasaka, T., Ganbe, T., Sato, T., Miyazawa, K., Kitamura, N., and Tanaka, N. (2003). Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain. J. Biol. Chem. 278, 48162–48168. [DOI] [PubMed] [Google Scholar]
- Kato, M., Miyazawa, K., and Kitamura, N. (2000). A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J. Biol. Chem. 275, 37481–37487. [DOI] [PubMed] [Google Scholar]
- Katzmann, D. J., Babst, M., and Emr, S. D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Katzmann, D. J., Odorizzi, G., and Emr, S. D. (2002). Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 3, 893–905. [DOI] [PubMed] [Google Scholar]
- Komada, M., and Kitamura, N. (1995). Growth factor-induced tyrosine phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol. Cell. Biol. 15, 6213–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada, M., and Kitamura, N. (2005). The Hrs/STAM complex in the down-regulation of receptor tyrosine kinases. J. Biochem. 137, 1–8. [DOI] [PubMed] [Google Scholar]
- Komada, M., Masaki, R., Yamamoto, A., and Kitamura, N. (1997). Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J. Biol. Chem. 272, 20538–20544. [DOI] [PubMed] [Google Scholar]
- Lenferink, A.E.G., Pinkas-Kramarski, R., van de Poll, M.L.M., van Vugt, M.J.H., Klapper, L. N., Tzahar, E., Waterman, H., Sela, M., van Zoelen, E.J.J., and Yarden, Y. (1998). Differential endocytic routing of homo- and heterodimeric ErbB tyrosine kinases confers signaling superiority to receptor heterodimers. EMBO J. 17, 3385–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz, G., Waterman, H., Zamir, E., Kam, Z., Oved, S., Langdon, W. Y., Beguinot, L., Geiger, B., and Yarden, Y. (1998). c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12, 3663–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., and Chernoff, J. (1997). Protein tyrosine phosphatase 1B interacts with and is tyrosine phosphorylated by the epidermal growth factor receptor. Biochem. J. 327, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, T. E., Atkinson, R., Wu, M. N., Zhou, Y., Pennetta, G., and Bellen, H. J. (2002). Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108, 261–269. [DOI] [PubMed] [Google Scholar]
- Longva, K. E., Blystad, F. D., Stang, E., Larsen, A. M., Johannessen, L. E., and Madshus, I. H. (2002). Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losko, S., Kopp, F., Kranz, A., and Kolling, R. (2001). Uptake of the ATP-binding cassette (ABC) transporter Ste6 into the yeast vacuole is blocked in the doa4 mutant. Mol. Biol. Cell 12, 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q., Hope, L. W., Brasch, M., Reinhard, C., and Cohen, S. N. (2003). TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA 100, 7626–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough, J., Clague, M. J., and Urbe, S. (2004). AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166, 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, E., Kawahata, K., Kato, M., Kitamura, N., and Komada, M. (2003). STAM proteins bind ubiquitinated proteins on the early endosome via the VHS domain and ubiquitin-interacting motif. Mol. Biol. Cell 14, 3675–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, E., Kawahata, K., Okamoto, A., Kitamura, N., and Komada, M. (2004). Association with Hrs is required for the early endosomal localization, stability, and function of STAM. J. Biochem. 135, 385–396. [DOI] [PubMed] [Google Scholar]
- Morino, C., Kato, M., Yamamoto, A., Mizuno, E., Hayakawa, A., Komada, M., and Kitamura, N. (2004). A role for Hrs in endosomal sorting of ligand-stimulated and unstimulated epidermal growth factor receptor. Exp. Cell Res. 297, 380–391. [DOI] [PubMed] [Google Scholar]
- Mosesson, Y., Shtiegman, K., Katz, M., Zwang, Y., Vereb, G., Szollosi, J., and Yarden, Y. (2003). Endocytosis of receptor tyrosine kinases is driven by monoubiquitination, not polyubiquitination. J. Biol. Chem. 278, 21323–21326. [DOI] [PubMed] [Google Scholar]
- Naviglio, S., Matteucci, C., Matoskova, B., Nagase, T., Nomura, N., Di Fiore, P. P., and Draetta, G. F. (1998). UBPY: a growth-regulated human ubiquitin isopeptidase. EMBO J 17, 3241–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschard, P., Fournier, T. M., Lamorte, L., Naujokas, M. A., Band, H., Langdon, W. Y., and Park, M. (2001). Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol. Cell 8, 995–1004. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., Bache, K. G., Gillooly, D. J., Helene Madshus, I., Stang, E., and Stenmark, H. (2002). Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4, 394–398. [DOI] [PubMed] [Google Scholar]
- Shih, S. C., Katzmann, D. J., Schnell, J. D., Sutanto, M., Emr, S. D., and Hicke, L. (2002). Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4, 389–393. [DOI] [PubMed] [Google Scholar]
- Sigismund, S., Woelk, T., Puri, C., Maspero, E., Tacchetti, C., Transidico, P., Di Fiore, P. P., and Polo, S. (2005). Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA 102, 2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, L., Seroogy, C., Skrenta, H., Anandasabapathy, N., Lovelace, P., Chung, C. D., Engleman, E., and Fathman, C. G. (2004). Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat. Immunol. 5, 45–54. [DOI] [PubMed] [Google Scholar]
- Soboleva, T. A., and Baker, R. T. (2004). Deubiquitinating enzymes: their functions and substrate specificity. Curr. Protein Pept. Sci. 5, 191–200. [DOI] [PubMed] [Google Scholar]
- Swaminathan, S., Amerik, A. Y., and Hochstrasser, M. (1999). The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10, 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, N., Kaneko, K., Asao, H., Kasai, H., Endo, Y., Fujita, T., Takeshita, T., and Sugamura, K. (1999). Possible involvement of a novel STAM-associated molecule “AMSH” in intracellular signal transduction mediated by cytokines. J. Biol. Chem. 274, 19129–19135. [DOI] [PubMed] [Google Scholar]
- Thien, C.B.F., and Langdon, W. Y. (2001). Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell. Biol. 2, 294–305. [DOI] [PubMed] [Google Scholar]
- Verma, R., Aravind, L., Oania, R., McDonald, W. H., Yates Spaceiiiqq, J. R., Koonin, E. V., and Deshaies, R. J. (2002). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Zhang, L., Yeung, T. K., and Chen, X. (1999). Endocytosis deficiency of epidermal growth factor (EGF) receptor-ErbB2 heterodimers in response to EGF stimulation. Mol. Biol. Cell 10, 1621–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing, S. S. (2003). Deubiquitinating enzymes - the importance of driving in reverse along the ubiquitin-proteasome pathway. Int. J. Biochem. Cell Biol. 35, 590–605. [DOI] [PubMed] [Google Scholar]
- Worthylake, R., Opresko, L. K., and Wiley, H. S. (1999). ErbB-2 amplification inhibits down-regulation and induces constitutive activation of both ErbB-2 and epidermal growth factor receptors. J. Biol. Chem. 274, 8865–8874. [DOI] [PubMed] [Google Scholar]
- Wu, X., Yen, L., Irwin, L., Sweeney, C., and Carraway Spaceiiiqq, K. L. (2004). Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 24, 7748–7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, T., and Cohen, R. E. (2002). A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407. [DOI] [PubMed] [Google Scholar]
- Yoshimori, T., Yamagata, F., Yamamoto, A., Mizushima, N., Kabeya, Y., Nara, A., Miwako, I., Ohashi, M., Ohsumi, M., and Ohsumi, Y. (2000). The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell 11, 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.