Abstract
The large protein kinases, ataxia-telangiectasia mutated (ATM) and ATM-Rad3-related (ATR), coordinate the cellular response to DNA damage. In budding yeast, ATR homologue Mec1 plays a central role in DNA damage signaling. Mec1 interacts physically with Ddc2 and functions in the form of the Mec1–Ddc2 complex. To identify proteins interacting with the Mec1–Ddc2 complex, we performed a modified two-hybrid screen and isolated RFA1 and RFA2, genes that encode subunits of replication protein A (RPA). Using the two-hybrid system, we found that the extreme C-terminal region of Mec1 is critical for RPA binding. The C-terminal substitution mutation does not affect the Mec1–Ddc2 complex formation, but it does impair the interaction of Mec1 and Ddc2 with RPA as well as their association with DNA lesions. The C-terminal mutation also decreases Mec1 kinase activity. However, the Mec1 kinase-defect by itself does not perturb Mec1 association with sites of DNA damage. We also found that Mec1 and Ddc2 associate with sites of DNA damage in an interdependent manner. Our findings support the model in which Mec1 and Ddc2 localize to sites of DNA damage by interacting with RPA in the form of the Mec1–Ddc2 complex.
INTRODUCTION
The maintenance of genome stability is critical to cellular survival and proliferation in all organisms. Cells have evolved surveillance mechanisms that monitor genomic lesions and activate various DNA damage responses, including cell cycle arrest and transcriptional induction of DNA repair genes (Zhou and Elledge, 2000). This surveillance mechanism is called DNA damage checkpoint in eukaryotes. The checkpoint signals are initiated through two large protein kinases, ataxia-telangiectasia mutated (ATM) and ATM-Rad3-related (ATR) (Zhou and Elledge, 2000; Abraham, 2001). ATM and ATR are highly conserved among eukaryotes. ATR is closely related to Mec1 in the budding yeast Saccharomyces cerevisiae and Rad3 in the fission yeast Schizosaccharomyces pombe. ATM homologues are termed Tel1 in both budding and fission yeasts.
In the budding yeast S. cerevisiae, Mec1 plays a central role in DNA damage checkpoint control, whereas Tel1 plays a minor role (Morrow et al., 1995; Sanchez et al., 1996; Usui et al., 2001; Nakada et al., 2003b, 2004). Mec1 physically interacts with Ddc2 (also called Lcd1 and Pie1), a protein that exhibits homology to the ATR-interacting protein ATRIP and Rad3-interacting protein Rad26 (Edwards et al., 1999; Paciotti et al., 2000; Rouse and Jackson, 2000; Cortez et al., 2001; Wakayama et al., 2001). Mec1 and Ddc2 function in the form of the Mec1–Ddc2 complex, and both localize to sites of DNA damage (Kondo et al., 2001; Melo et al., 2001; Rouse and Jackson, 2002). Mec1 controls two downstream protein kinases Chk1 and Rad53, which are related to mammalian Chk1 and Chk2, respectively (Zhou and Elledge, 2000). Rad53 plays a central role in DNA damage checkpoints throughout the cell cycle (Longhese et al., 1998), whereas Chk1 acts in part at G2/M (Sanchez et al., 1999). Rad53 becomes phosphorylated and activated after DNA damage, and its activation leads to cell cycle arrest and induces the transcription of genes required for DNA damage repair (Elledge, 1996; Longhese et al., 1998).
Mec1 exhibits protein kinase activity because it contains similarity in the catalytic domain to phosphatidylinositol 3-kinase (PI3K) (Zhou and Elledge, 2000; Abraham, 2001). In contrast, the primary structure of Ddc2 does not provide an apparent clue to Ddc2 function. Replication protein A (RPA) is a conserved heterotrimer complex in eukaryotes, and its components are encoded by RFA1, RFA2, and RFA3 in budding yeast (Wold, 1997). RPA binds to single-stranded DNA (ssDNA), which is generated during a variety of DNA damage processes. Recent evidence provides a model in which Ddc2 recognizes DNA damage by interacting with RPA-coated ssDNA and enables the Mec1–Ddc2 complex to associate with sites of DNA damage (Zou and Elledge, 2003). Consistent with this model, generation of long ssDNA tracts facilitates Mec1 association with double-strand breaks (DSBs) (Nakada et al., 2004). However, it is not precisely determined whether Ddc2 by itself recognizes DNA damage. One report demonstrated that Ddc2 associates with sites of DNA damage independently of Mec1 (Rouse and Jackson, 2002), whereas another study showed that Ddc2 fails to accumulate at DNA lesions in the absence of Mec1 (Melo et al., 2001). Besides sensing DNA damage, Ddc2 seems to contribute to substrate recognition of Mec1 kinase. Recent in vitro studies showed that Mec1 phosphorylates a peptide substrate poorly in the absence of Ddc2 (Takata et al., 2004). Thus, Mec1 and Ddc2 might collaborate in recognition of target proteins. However, no protein has been described that interacts specifically with the Mec1–Ddc2 complex.
In this study, we describe the isolation of RFA1and RFA2 in a two-hybrid screen searching for proteins that interact with Mec1 in a Ddc2-dependent manner. We show that the extreme C terminus of Mec1 is important for its interaction with RPA and localization to sites of DNA damage. We also show that Mec1 and Ddc2 associate interdependently with sites of DNA damage. Our results support the idea that Mec1 and Ddc2 cooperate in DNA damage recognition.
MATERIALS AND METHODS
Plasmids
To construct YEpT-ADH1-DDC2, the coding region of DDC2 gene was amplified by PCR and cloned into a TRP1-marked YEp plasmid containing the ADH1 promoter (a gift from A. Yamagishi, Nagoya University). To construct the plasmid pBD-MEC1-85, the C-terminal region of MEC1 was amplified by PCR replacing Met-Tyr-Ile at position 2360–2362 with Ala. The resulting PCR fragment was treated with NheI and SalI and then cloned into NheI-SalI-treated pBD-MEC1(2-2368) (Wakayama et al., 2001). The plasmids pBD-MEC1(2-1860) and pBD-MEC1(2-2140), derived from pBD-MEC1(2-2368) were obtained from T. Wakayama (Nagoya University). The FLAG-tagged RFA1 construct was generated as follows. The C-terminal coding region of RFA1 was amplified by PCR and digested with SacI and BamHI. The resulting fragment was cloned into pRS304-FLAG (Green et al., 2000), generating YIpT-RFA1-FLAG. To construct the hemagglutinin (HA)-tagged DDC2 gene, the 5′ noncoding and amino-terminal regions of DDC2 were amplified by PCR and treated with SacI-NdeI or EcoRI-SalI, respectively. These two fragments were sequentially inserted into SacI-NdeI and EcoRI-SalI sites of pRS304-HA (a gift from T. Kondo, Nagoya University), creating YIpT-DDC2-HA. The plasmid YCp-DDC2-HA was constructed by replacing a SacI-HindIII fragment of YCp-PIE1 (Wakayama et al., 2001) with the corresponding fragment from YIpT-DDC2-HA. The YCpA-GAL-HO, YCpT-RAD53-HA, pBD-MEC1(2-500), and pBD-MEC1(2-938) plasmids were described previously (Wakayama et al., 2001; Nakada et al., 2003a). The tagged constructs expressed appropriate-sized proteins from their own promoters and complemented their null mutations with regard to sensitivity to DNA-damaging agents.
Strains
The RFA1-FLAG::URA3 strains were obtained by transforming YIpT-RFA1-FLAG after treatment with EcoRI and subsequently replacing the TRP1 marker with URA3 (Cross, 1997). The DDC2-HA::TRP1 strains were obtained by transforming YIpT-DDC2-HA after treatment with PstI. The mec1-85::ura3 and mec1-87::ura3 strains were constructed as follows. Diploids carrying sml1Δ mutation (Wakayama et al., 2001) were transformed with PCR fragments amplified using pWJ1077 as a template (Reid et al., 2002), introducing mec1-85::URA3 or mec1-87::URA3 into the cells. After isolation of Ura+ haploid cells, Ura– cells were selected out on a 5-fluoroorotic acid plate (Boeke et al., 1987). Other strain constructs were described previously (Kondo et al., 2001; Wakayama et al., 2001; Nakada et al., 2003a; Naiki et al., 2004). All the strains used in this study are isogenic to KSC1178 (Wakayama et al., 2001) and are listed in Table 1.
Table 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| KSC1302 | MATaDDC2-myc::TRP1 mec1Δ::LEU2 |
| KSC1304 | MATaDDC2-myc::TRP1 |
| KSC1560 | MATa-inc |
| KSC1561 | MATa-inc mec1Δ::LEU2 |
| KSC1635 | MATa-inc MEC1-HA::TRP1 |
| KSC1636 | MATa-inc MEC1-HA::TRP1 ddc2Δ::LEU2 |
| KSC1641 | MATa-inc mec1-85::ura3 |
| KSC1642 | MATa-inc MEC1-HA::TRP1 DDC2-myc::TRP1 |
| KSC1643 | MATa-inc mec1-85-HA::TRP1 DDC2-myc::TRP1 |
| KSC1644 | MATa-inc DDC2-myc::TRP1 |
| KSC1645 | MATa-inc mec1-KN-HA::TRP1::URA3 |
| KSC1646 | MATa-inc mec1-85-HA::TRP1 |
| KSC1647 | MATa-inc DDC2-HA::TRP1 mec1-85::ura3 |
| KSC1649 | MATa-inc mec1-87-HA::TRP1 |
| KSC1651 | MATα DDC2-HA::TRP1 |
| KSC1652 | MATα DDC2-HA::TRP1 RFA1-FLAG::URA3 |
| KSC1653 | MATα DDC2-HA::TRP1 RFA1-FLAG::URA3 mec1Δ::LEU2 |
| KSC1654 | MATα DDC2-HA::TRP1 RFA1-FLAG::URA3 mec1-85::ura3 |
| KSC1655 | MATα RFA1-FLAG::URA3 |
| KSC1656 | MATα MEC1-HA::TRP1 RFA1-FLAG::URA3 |
| KSC1657 | MATα mec1-85-HA::TRP1 RFA1-FLAG::URA3 |
| KSC1658 | MATα MEC1-HA::TRP1 |
| KSC1659 | MATα mec1-85-HA::TRP1 |
| KSC1717 | MATa-inc DDC2-HA::TRP1 |
| KSC1727 | MATa-inc DDC2-HA::TRP1 mec1Δ::LEU2 |
All the strains are isogenic to KSC1178 (MATa sml1Δ::LEU2 ade1 his2 leu2 trp1 ura3) (Wakayama et al., 2001). The listed strains contain ADH4cs::HIS2 except KSC1302 and KSC1304.
Two-Hybrid Screening
Yeast two-hybrid screening of an S. cerevisiae genomic library was carried out using PJ69-4A cells carrying both pBD-MEC1(2-2368) and YEpT-ADH1-DDC2 as described previously (Wakayama et al., 2001). After transformation with the library, cells growing on medium containing 10 mM 3-aminotriazole (AT) were selected and further characterized as positive clones. The library plasmids were recovered from the positive cells and retransformed into PJ69-4A cells carrying both pBD-MEC1(2-2368) and YEpT-ADH1-DDC2 or pBD-MEC1(2-2368) alone. Ten library plasmids were found to support proliferation only when cells carried both of the pBD-MEC1(2-2368) and YEpT-ADH1-DDC2 plasmid. Restriction and sequence analyses followed by a DNA database search revealed that two of them contained RFA1 and eight contained RFA2. RFA1 and RFA2 encode proteins with 621 and 273 amino acids, respectively. Both of the RFA1 plasmids contained fragments coding 97–621 amino acid residues of Rfa1 and were designated as pAD-RFA1. All the RFA2 plasmids were named pAD-RFA2 because they contained insertions corresponding to 17–273 amino acid residues of Rfa2.
Immunoblotting and Immunoprecipitation
Immunoblotting analysis was carried out using anti-FLAG, anti-HA, or anti-myc antibodies as described previously (Nakada et al., 2004). Immunoprecipitation was performed as follows (Naiki et al., 2001; Nakada et al., 2003a). Cells were suspended in 200 μl of lysis buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 15 mM p-NO2-phenylphosphate, 40 mM β-glycerophosphate) supplemented with Protease Inhibitor Cocktail (Roche Diagnostics, Indianapolis, IN) and physically disrupted with glass beads. Extracts were incubated with protein A-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) bound with anti-HA antibodies or with M2-Agarose (Sigma-Aldrich, St. Louis, MO).
UV Radiation and Drug Sensitivities
Cell viability after exposure to methyl methane sulfonate (MMS) and UV light was determined as described previously (Wakayama et al., 2001). To monitor Rad53 phosphorylation, cells were arrested at G2/M with nocodazole and then irradiated with a 254-nm UV lamp at 75 J/m2 (Nakada et al., 2004). After UV irradiation, cells were maintained in an arrested state in medium containing nocodazole.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation assay was carried out as described previously (Nakada et al., 2004). PCR reaction was performed in a nonsaturating condition in which the rate of PCR amplification is proportional to the concentration of substrate and cycling.
Other Methods
In vitro Mec1 kinase assay was performed with immunoprecipitated Mec1 proteins using PHAS-1 as a substrate (Nakada et al., 2003b). Nuclear and cytoplasmic fractionation was described previously (Wakayama et al., 2001). DNA degradation at DSB ends was monitored as described previously (Nakada et al., 2003a; Naiki et al., 2004).
RESULTS
Two-Hybrid Screening of Proteins Interacting with Mec1 in a Ddc2-dependent Manner
Mec1 and Ddc2 interact physically and act in the form of the complex. To identify protein(s) that interact with the Mec1–Ddc2 complex, we conducted a modified two-hybrid screen using the BD-MEC1(2-2368) construct as a bait under DDC2 overexpression. BD-MEC1(2-2368) consists of a kinase-negative version of MEC1, containing the whole coding sequence except the initiation codon (Wakayama et al., 2001). We isolated positive clones carrying RFA1 or RFA2 and found that their interaction with Mec1 depends on DDC2 overexpression in the two-hybrid system (Figure 1A; our unpublished data). Both RFA1 and RFA2 encode components of RPA in budding yeast. Recent evidence indicates that Ddc2 itself binds to RPA-coated ssDNA (Zou and Elledge, 2003). It is therefore possible that the observed Mec1–RPA interaction could result from two independent Mec1–Ddc2 and Ddc2–RPA interactions. To exclude this possibility, we mapped the Mec1 region that contributes to interaction with RPA using the two-hybrid system. We previously showed that the N terminus of Mec1 interacts with Ddc2 in a two-hybrid assay; the BD-MEC1(2-500), BD-MEC1(2-938), and BD-MEC1(2-2368) constructs established its interaction with Ddc2 (Wakayama et al., 2001). In addition, the BD-MEC1(2-1860) and BD-MEC1(2-2140) constructs supported the Mec1–Ddc2 interaction in the two-hybrid system (our unpublished data). However, only the MEC1(2-2368) construct established the Mec1–Rfa1 interaction in the modified two-hybrid system (Figure 1B). Similar results were obtained in the Mec1–Rfa2 interaction (our unpublished data). These results raise the possibility that RPA interacts physically with the Mec1–Ddc2 complex and that the interaction between RPA and the Mec1–Ddc2 complex requires the amino acid residues (2140–2368) at the extreme C terminus of Mec1.
Figure 1.
Interaction between Mec1 and Rfa1 in a modified two-hybrid assay. (A) Requirement of DDC2 overexpression for the Mec1–Rfa1 interaction. Strain PJ69-4A carrying pBD-MEC1(2-2368) was transformed with pAD-RFA1 and YEpT-ADH1-DDC2 or their control vector. Transformants were streaked on selectable minimum medium containing 10 mM AT. (B) Identification of the Mec1 region required for its Ddc2-dependent interaction with Rfa1. Strain PJ69-4A carrying both pAD-RFA1 and YEpT-ADH1-DDC2 was transformed with pBD-MEC1(2-2368), pBD-MEC1(2-500), pBD-MEC1(2-938), pBD-MEC1(2-1860), pBD-MEC1(2-2140), or pBD-MEC1-85. The kinase domain (filled in dots) and the extreme C-terminal region (black bar) are indicated. Stars denote substitution mutations in mec1-85 (see Figure 2A). Transformants were streaked on a selectable plate containing 10 mM AT. Interaction with Rfa1 was assessed by the growth of transformants.
Role of the Extreme Mec1 C Terminus in Cellular Response to DNA Damage
ATM and ATR family proteins possess a kinase domain homologous to PI3K in the C-terminal half (Abraham, 2001). Apart from the kinase domain, they share significant similarity at the extreme C terminus (Figure 2A). This extreme C-terminal region, named the FATC domain (Bosotti et al., 2000), extends to other PI3K-related protein kinases, including TOR and DNA-PK proteins. To uncover the significance of this region, we constructed two substitution mutations in which the amino acid residues 2360–2362 and 2367–2368 were all replaced with alanine. These mutations were termed mec1-85 and mec1-87, respectively (Figure 2A). Similar to mec1Δ mutation, mec1-85 and mec1-87 mutations caused viability loss, which can be rescued by sml1Δ mutations (Zhao et al., 1998; our unpublished data). Because the mec1-85 and mec1-87 mutations behave like a null mutation, we examined the expression level of their gene products (Figure 2B). The mec1-87 strains have fourfold lower Mec1 protein levels compared with wild-type cells, whereas the mec1-85 mutation did not significantly affect the protein expression. We hereafter characterized the mec1-85 mutation.
Figure 2.
Function of the Mec1 C-terminal region. (A) Substitution mutation of Mec1 at the conserved C-terminal region. Mec1 possesses a kinase domain (filled in dots) within the C-terminal portion. The aligned sequences are derived from the extreme C-terminal regions of ATR family proteins (Mec1, ATR, fission yeast Rad3, and fruit fly Mei-41), ATM family proteins (Tel1 and ATM), TOR proteins (budding yeast Tor1 and mammalian TOR), and DNA-PK. Identical amino acid residues are boxed in black. Related amino acid residues are highlighted in gray. The mec1-85 and mec1-87 mutations change the amino acid residues into alanine at positions 2360–2362 and 2367–2368 (indicated by asterisks), respectively. (B) Expression level of Mec1 in mec1-85 and mec1-87 mutants. Cells expressing Mec1-HA (KSC1635), Mec1-85-HA (KSC1646), or Mec1-87-HA (KSC1649) were subjected to immunoblotting analysis with anti-HA or anti-tubulin antibodies. All the strains contained an sml1Δ mutation, which suppresses the lethality associated with mec1 mutations. (C) Effect of mec1-85 mutation on cell viability after UV irradiation. Viability was determined after UV irradiation at the indicated dosages. Strains used were wild-type (KSC1560), mec1Δ (KSC1561), and mec1-85 (KSC1641) cells. (D) Effect of mec1-85 mutation on Rad53 phosphorylation after UV irradiation. The same strains as in C were transformed with YCpT-RAD53-HA. Transformants were arrested at G2/M with nocodazole and then exposed to UV light. Cells were collected 30 min after UV irradiation and analyzed by immunoblotting with anti-HA antibodies.
We first examined the DNA damage sensitivity of mec1-85 mutants. The mec1-85 mutation conferred essentially the same sensitivity to UV light and MMS as a mec1Δ mutation did (Figure 2C and Supplemental Figure S1). Mec1 controls phosphorylation and activation of Rad53 after DNA damage, and its phosphorylation status is well correlated with activation of DNA damage checkpoint (Elledge, 1996; Longhese et al., 1998). We then monitored UV-induced Rad53 phosphorylation in mec1-85 mutant cells (Figure 2D). Rad53 phosphorylation can be detected as a slowly migrating form on immunoblots (Wakayama et al., 2001). Cells expressing HA-tagged Rad53 were grown to log phase and then arrested at G2/M with nocodazole. After arrest, cells were irradiated with UV light and subjected to immunoblotting analysis with anti-HA antibodies. Rad53 phosphorylation occurred in wild-type cells, whereas no phosphorylation was detected in mec1Δ or mec1-85 mutants. The DNA damage sensitivity and Rad53 phosphorylation defect of mec1-85 mutants were fully suppressed by the introduction of the wild-type MEC1 gene (our unpublished data), indicating that mec1-85 is a recessive mutation. These results indicate that the mec1-85 mutation eliminates most Mec1 functions.
Role of the Extreme Mec1 C Terminus in Its Interaction with RPA
We next addressed whether the mec1-85 mutation affects the interaction with RPA. The mec1-85 construct failed to support the Mec1–Rfa1 interaction in the modified two-hybrid system (Figure 1B). Again, the mec1-85 mutation did not affect the Mec1–Ddc2 interaction in the two-hybrid assay (Wakayama et al., 2001; our unpublished data). To confirm the above-mentioned findings obtained in the two-hybrid assays, we monitored the interaction between the Mec1–Ddc2 complex and RPA by coimmunoprecipitation experiments. We first examined whether Mec1-85 mutant and Ddc2 proteins interact physically (Figure 3A). Extracts prepared from cells expressing Ddc2-myc and Mec1-HA or Mec1-85-HA were subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitates were then examined by immunoblotting analysis with anti-HA or anti-myc antibodies. Ddc2-myc was similarly detected in the immunoprecipitates of Mec1-HA and Mec1-85-HA, indicating that the mec1-85 mutation does not affect the Mec1–Ddc2 complex formation. We next examined the effect of mec1-85 mutation on the Mec1–RPA interaction by coimmunoprecipitation experiments (Figure 3B). Extracts were prepared from cells expressing HA-tagged Mec1 or Mec1-85 and FLAG-tagged Rfa1 proteins and were subjected to immunoprecipitation with anti-FLAG antibodies. The immunoprecipitates were then analyzed by immunoblotting with anti-HA or anti-FLAG antibodies. Mec1 and Rfa1 were found to interact physically, consistent with the previous study (Kim and Brill, 2003). However, the Mec1–Rfa1 interaction was not detected in mec1-85 mutants. We further examined whether Ddc2 and Rfa1 interact physically in mec1-85 mutants (Figure 3C). Extracts were prepared from wild-type, mec1Δ, or mec1-85 cells expressing Ddc2-HA and Rfa1-FLAG and were subjected to immunoprecipitation and subsequent immunoblotting analysis as described above. Although the Ddc2–Rfa1 interaction was observed in wild-type cells, its interaction was undetectable in mec1-85 mutants. Moreover, the Ddc2–Rfa1 interaction was undetectable in mec1Δ mutants (Figure 3C). The Mec1–Rfa1 interaction has been shown to become defective in ddc2Δ mutants (Kim and Brill, 2003). These observations support the model in which the Mec1–Ddc2 complex interacts with RPA more efficiently than does Mec1 or Ddc2 alone, and they indicate that its interaction with RPA requires the extreme C terminus of Mec1.
Figure 3.
Effect of mec1-85 mutation on interaction among Mec1, Ddc2, and Rfa1. (A) Effect of mec1-85 mutation on the Mec1–Ddc2 complex formation. Extracts prepared from cells expressing tagged or untagged Mec1 and Ddc2 proteins were immunoprecipitated with anti-HA antibodies. Immunoprecipitate (IP) and extract were subjected to immunoblotting analysis with anti-HA or anti-myc antibodies. Strains used here were MEC1-HA (KSC1635), MEC1-HA DDC2-myc (KSC1642), mec1-85-HA DDC2-myc (KSC1643), and DDC2-myc (KSC1644). (B) Effect of mec1-85 mutation on the interaction between Mec1 and Rfa1. Extracts were prepared from cells expressing tagged or untagged Mec1 and Rfa1, and immunoprecipitated with anti-FLAG antibodies. Immunoprecipitate (IP) and extract were then subjected to immunoblotting analysis with anti-FLAG or anti-HA antibodies. Strains used were RFA1-FLAG (KSC1655), MEC1-HA RFA1-FLAG (KSC1656), mec1-85-HA RFA1-FLAG (KSC1657), MEC1-HA (KSC1658), and mec1-85-HA (KSC1659). (C) Effect of mec1-85 mutation on the interaction between Ddc2 and Rfa1. Extracts were prepared from cells expressing tagged or untagged Ddc2 and Rfa1 and immunoprecipitated with anti-FLAG antibodies. IP and extract were then subjected to immunoblotting analysis with anti-FLAG or anti-HA antibodies. Strains used were RFA1-FLAG (KSC1655), DDC2-HA RFA1-FLAG (KSC1652), mec1Δ DDC2-HA RFA1-FLAG (KSC1653), mec1-85 DDC2-HA RFA1-FLAG (KSC1654), and DDC2-HA (KSC1651).
Kinase Activity Associated with Mec1-85 Mutant Protein
Because the mec1-85 mutation is located near the PI3K-related kinase domain, we considered that its mutation might affect Mec1 kinase activity. To address this possibility, we performed in vitro kinase assay with Mec1 wild-type and Mec1-85 mutant proteins (Figure 4). As a negative control, we included the kinase-negative Mec1-KN mutant protein, which possesses two substitutions in the conserved kinase domain (Wakayama et al., 2001). We have shown that kinase activities associated with Mec1-KN are undetectable and that the mec1-KN mutation confers the same phenotypes as mec1Δ mutation does (Wakayama et al., 2001; Nakada et al., 2003b). Extracts were prepared from cells expressing Mec1-HA, Mec1-85-HA, or Mec1-KN-HA and were immunoprecipitated with anti-HA antibodies. The immunoprecipitates were then subjected to in vitro kinase assay using phosphorylated heat- and acid-stable protein (PHAS)-1 as a substrate. PHAS-1 was efficiently phosphorylated by immunoprecipitates containing Mec1-HA (Nakada et al., 2003b). However, phosphorylation by Mec1-85-HA was significantly decreased, very similar to that observed with Mec1-KN-HA. Thus, the extreme C terminus of Mec1 is essential for its kinase activity.
Figure 4.

Effect of mec1-85 mutation on Mec1 kinase activity. Cells expressing Mec1-HA (KSC1635), Mec1-85-HA (KSC1646) or Mec1-KN-HA (KSC1645) were harvested for preparation of extracts. Extracts were subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitated Mec1 proteins were subjected to in vitro kinase assay using PHAS-1 as a substrate. The amount of 32P incorporation into PHAS-1 was detected by autoradiography (top). The amount of Mec1 protein used for the kinase assay was examined by immunoblotting (bottom).
Effect of mec1-85 Mutation on Mec1 and Ddc2 Association with Sites of DNA Damage
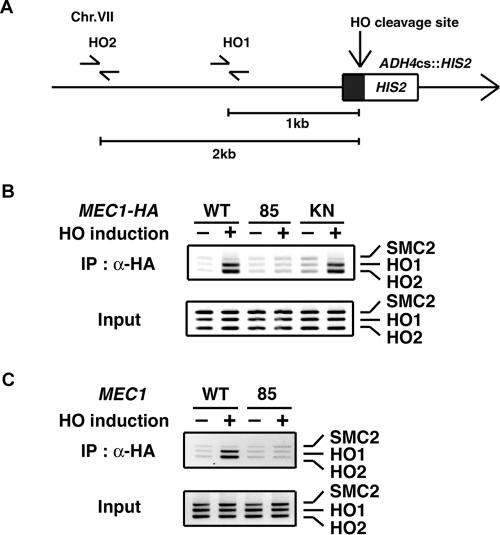
Because the mec1-85 mutation impairs interaction of the Mec1–Ddc2 complex with RPA, we expected that Mec1-85 mutant proteins fail to localize to sites of DNA damage. To test this hypothesis, we used an experimental system in which cells contained a single HO cleavage site at the ADH4 locus (Figure 5A), and HO is expressed from the GAL-HO plasmid after incubation with galactose (Nakada et al., 2003a). The HO-induced DSB at the ADH4 locus is not efficiently repaired by homologous recombination and thereby activates the DNA damage checkpoint pathway at G2/M (Toczyski et al., 1997). We have shown that Mec1 associates with the HO-induced DSB at G2/M (Nakada et al., 2004). Because Mec1-85 mutant proteins are defective in kinase activity, we also monitored the association of Mec1-KN mutant proteins as a control (Figure 5B). Cells expressing HA-tagged Mec1 were transformed with the GAL-HO plasmid. Transformants were grown initially in sucrose to repress HO expression and then transferred to medium containing nocodazole to arrest at G2/M. After arrest, one-half of the culture was maintained in sucrose, and the other half was incubated for 3 h with galactose to induce HO expression. Extracts prepared after formaldehyde cross-linking were sonicated and subjected to immunoprecipitation with anti-HA antibodies. Coprecipitated DNA was extracted and amplified by PCR using a primer set corresponding to regions near the HO-restriction site or primers for the SMC2 locus containing no cleavage site. Mec1 associated with the HO-induced DSB; PCR amplified sequences near the HO-restriction site after incubation with galactose, but not the control site in the SMC2 locus. However, the association of Mec1-85 mutant proteins was not detected. This Mec1-85 association defect was not due to impaired kinase activity, because Mec1-KN mutant proteins associated with the DSB as efficiently as wild-type Mec1 proteins did. Similar to mec1Δ mutants, mec1-85 mutants exhibited no defect in generating ssDNA at DSB ends (Naiki et al., 2004; Supplemental Figure S2), excluding the possibility that the impaired Mec1-85 association might result from decreased ssDNA generation at the DSB ends. We also examined association of Ddc2 with the HO-induced DSB in mec1-85 mutants (Figure 5C). Cells expressing Ddc2-HA were transformed with the GAL-HO plasmid and analyzed as described above. The Ddc2 association was observed in wild-type cells, but not in mec1-85 mutants. Thus, the extreme C terminus of Mec1 is required for localization of both Mec1 and Ddc2 to sites of DNA damage.
Figure 5.
Effect of mec1-85 mutation on association of Mec1 and Ddc2 with the HO-induced DSB. (A) Schematic of the HO cleavage site at the ADH4 locus (ADH4cs). An HO cleavage site, marked with HIS2, was introduced at the ADH4 locus on chromosome VII. The primer pairs were designed to amplify regions 1 and 2 kb apart from the HO cleavage site. An arrow represents telomere. (B) Effect of mec1-85 mutation on Mec1 association with HO-induced DSBs. Cells expressing Mec1-HA (KSC1635), Mec1-85-HA (KSC1646), or Mec1-KN-HA (KSC1645) were transformed with YCpA-GAL-HO plasmid. Transformed cells were initially grown in sucrose and arrested at G2/M with nocodazole. The culture was then incubated with galactose to induce HO expression for 3 h, whereas part of the culture was maintained in sucrose to repress HO expression. Cells were collected and subjected to chromatin immunoprecipitation. PCR was carried out with the primers for the HO cleavage site at the ADH4 locus and for the control SMC2 locus (see A). PCR products from the respective input extracts are shown below. (C) Effect of mec1-85 mutation on Ddc2 association with HO-induced DSBs. Wild-type (KSC1717) and mec1-85 (KSC1647) cells expressing Ddc2-HA were transformed with YCpA-GAL-HO plasmid and were analyzed as in B.
Interdependent Association of Mec1 and Ddc2 with DNA Lesions
As discussed above, the interaction between the Mec1–Ddc2 complex and RPA is functionally coupled to its localization to sites of DNA damage. Moreover, RPA interacts with the Mec1–Ddc2 complex more efficiently than Ddc2 or Mec1 alone. If this were the case, Mec1 and Ddc2 proteins should cooperate in association with sites of DNA damage. We then examined the association of Mec1 or Ddc2 with the HO-induced DSB in ddc2Δ or mec1Δ mutants, respectively (Figure 6, A and B). Mec1 association with the HO-induced DSB was significantly decreased in ddc2Δ mutants (Figure 6A), and the Ddc2 association was undetectable in mec1Δ mutants (Figure 6B). These association defects are not because of delocalization of Mec1 and Ddc2 proteins from nucleus. We have shown that ddc2Δ mutation does not affect the cellular distribution of Mec1 (Wakayama et al., 2001). Moreover, mec1Δ mutation did not alter the nuclear localization of Ddc2 (Figure 6C). These results are consistent with the idea that Mec1 and Ddc2 localize to DNA lesions in the form of the Mec1–Ddc2 complex. However, our results do not agree with the model in which Ddc2 has abilities to recognize DNA damage and recruits the Mec1–Ddc2 complex to the damage sites (Rouse and Jackson, 2002; Zou and Elledge, 2003). We hypothesized that Ddc2 by itself might have relatively weak abilities to associate with sites of DNA damage, and a high dosage of Ddc2 could restore its association with DSBs to mec1Δ mutants. We thus compared Ddc2 association with HO-induced DSBs in DDC2-HA mec1Δ cells carrying YCp-DDC2-HA or the control vector (Figure 6D). The YCp-DDC2-HA plasmid expresses DDC2 from its own promoter (see Materials and Methods). Immunoblotting analysis revealed that cells carrying the DDC2-HA plasmid expressed Ddc2-HA proteins threefold more abundantly than those carrying the control vector (Figure 6D). Consistent with the hypothesis, Ddc2 association with DSBs became detectable in mec1Δ mutant cells carrying the DDC2-HA plasmid, although this Ddc2 association was much less efficient than that detected in the wild-type cells (Figure 6D). Altogether, these results support the model in which Mec1 and Ddc2 form a complex and cooperate in localization to sites of DNA damage.
Figure 6.
Interdependent association of Mec1 and Ddc2 with the HO-induced DSB. (A) Effect of ddc2Δ mutation on Mec1 association with HO-induced DSBs. Wild-type (KSC1635) and ddc2Δ (KSC1636) cells expressing Mec1-HA were analyzed as in Figure 5B. All the strains contained an sml1Δ mutation, which suppresses the lethality associated with ddc2Δ mutation (Wakayama et al., 2001). (B) Effect of mec1Δ mutation on Ddc2 association with HO-induced DSBs. Wild-type (KSC1717) and mec1Δ (KSC1727) cells expressing Ddc2-HA were analyzed as in A. (C) Intracellular localization of Ddc2 in wild-type and mec1Δ cells. Wild-type (KSC1304) and mec1Δ (KSC1302) cells expressing Ddc2-myc were harvested and spheroplasted. Spheroplasts were homogenized to prepare whole cell extract (W) and then separated into the cytoplasmic (C) and nuclear (N) fractions. Aliquots were analyzed on immunoblots with anti-HA, anti-glucose-6-P dehydrogenase (G6PDH), and anti-nuclear pore complex (NPC) antibodies. (D) Top, dosage effect on Ddc2 association with HO-induced DSBs. Wild-type (KSC1717) and mec1Δ (KSC1727) cells expressing Ddc2-HA were transformed with YCp-DDC2-HA or the control vector and were analyzed as in A. Bottom, to monitor the expression level of Ddc2-HA, the extract was also analyzed by immunoblotting analysis.
DISCUSSION
Recognition of DNA damage by the checkpoint machinery is the key to initiation of checkpoint signaling. Mec1 and Ddc2 form a complex and localize to sites of DNA damage. However, the molecular mechanism of how the Mec1–Ddc2 complex recognizes DNA damage is not completely understood. We searched for proteins that might interact with the Mec1–Ddc2 complex in a modified two-hybrid screen, and we identified RPA subunits Rfa1 and Rfa2 as interacting proteins. Subsequent deletion and mutagenesis analyses revealed that the C terminus of Mec1 is essential for both interaction with RPA and localization to DSBs, but not for complex formation with Ddc2. The Mec1 C-terminal region is also required for its kinase activity. However, kinase-negative Mec1 mutant proteins associate efficiently with DSBs, suggesting that Mec1 kinase activity is dispensable for its localization to sites of DNA damage. Moreover, Mec1 and Ddc2 interdependently associate with DSBs. Our present results support the model in which Mec1 and Ddc2 cooperate in interacting with RPA and thereby localize to sites of DNA damage in the form of the complex.
The two-hybrid experiment revealed that the extreme C-terminal region of Mec1 is critical for its interaction with RPA in a Ddc2-dependent manner. The extreme C terminus called the FATC domain (Bosotti et al., 2000) is highly conserved among PI3K-related protein kinase, which includes ATR proteins, ATM proteins, TOR proteins, and DNA-PK (Figure 2A). Previous studies showed that the FATC domain is indispensable for TOR kinase activity (Peterson et al., 2000). Similarly, a substitution mutation in the Mec1 domain, mec1-85, decreases its kinase activity. The mec1-85 mutation impairs interaction with RPA and localization to DSBs, but it does not affect the complex formation with Ddc2. Mec1 kinase activity seems to be dispensable for its localization to sites of DNA damage; Mec1-KN proteins associate efficiently with DSBs, although we cannot exclude the possibility that weak residual kinase activity in Mec1-KN might contribute to its association with DSBs. Thus, the FATC domain of Mec1 possesses two separate roles; one in phosphorylating substrates and another in associating with sites of DNA damage. The mec1-85 mutation behaves like mec1Δ mutation, probably because of its impaired kinase activity. It remains to be determined whether strains defective in Mec1 interaction with RPA or localization to sites of DNA damage exhibit the same phenotypes as mec1Δ mutants.
The three-dimensional structure analysis of DNA-PKcs indicates that the FATC domain protrudes from the overall structure (Rivera-Calzada et al., 2005). In addition, the FATC domain shows the greatest conformational difference between various analysis methods of any part of the structure, suggesting that the FATC domain is highly flexible (Rivera-Calzada et al., 2005). Relative motion of the protruded FATC domain could directly influence the kinase domain nearby. Thus, the FATC domain is suggested to act as a sensor that couples conformation changes to directly activate the catalytic center (Bosotti et al., 2000; Rivera-Calzada et al., 2005). Similarly, the Mec1 FATC domain might mediate conformation changes to increase its kinase activity or promote substrate recognition. At the moment, it is not clear how the Mec1 FATC domain regulates interaction with RPA or localization to DNA lesions. One possibility could be that the Mec1 FATC domain directly interacts with RPA. Alternatively, the FATC domain might promote the Ddc2–RPA interaction. It seems less likely that the M-Y-I/L sequence in the FATC domain contributes specifically to association with DNA lesions, because the sequence is relatively conserved in other PI3K-related proteins, including ATM and TOR proteins (Figure 2A). In contrast to the C-terminal regions, the N-terminal regions of ATR family proteins are not homologous to those of other PI3K-related proteins. We have shown that the N terminus of Mec1 is involved in complex formation with Ddc2 (Wakayama et al., 2001). However, it remains possible that the N terminus contributes to RPA binding as well.
We showed that Mec1 and Ddc2 localize to DNA lesions interdependently. Consistently, one study demonstrated that Ddc2 associates with sites of DNA damage through a MEC1-dependent mechanism, although this study did not determine whether Mec1 localizes to the damage site in ddc2Δ mutants (Melo et al., 2001). However, another report showed that Ddc2 (Lcd1) associates with sites of DNA damage independently of Mec1 (Rouse and Jackson, 2002). One explanation could be that the Mec1-independent Ddc2 association might result from moderate overexpression of Ddc2 in cells. In this study, the assay was done with cells transformed with a YCp plasmid containing a tagged DDC2 (LCD1) gene (Rouse and Jackson, 2002). The YCp plasmids are present at very low copy numbers, ranging from one to two per cell, but possibly a little more (Clarke and Carbon, 1980; Futcher and Carbon, 1986). We showed that when Ddc2 was threefold overproduced, we became able to detect, albeit weakly, the Ddc2 association with DSBs in mec1Δ mutants. Thus, Ddc2 by itself seems to possess a weak ability to localize to sites of DNA damage. Consistent with this view, cytological studies have reported that weak Ddc2 focus formation occurs independently of Mec1 after DNA damage (Melo et al., 2001; Lisby et al., 2004), and biochemical experiments have demonstrated that Ddc2 alone can bind to RPA-coated ssDNA (Zou and Elledge, 2003). This damage recognition ability is more or less conserved in the Ddc2 homologues, Rad26 in fission yeast, and ATRIP in mammals; Rad26 and ATRIP by themselves were shown to localize to foci after DNA damage (Wolkow and Enoch, 2003; Itakura et al., 2004). Our studies support a model in which Mec1 and Ddc2 localize to sites of DNA damage by interacting with RPA as a complex. At the moment, however, we cannot rule out the possibility that Ddc2 first associates with the damage sites and that its association becomes stabilized by Mec1 function. Our assay might not be sensitive enough to detect low levels of Ddc2 at DNA lesions in cells lacking Mec1. Although the Mec1 association with DSBs was not detected in ddc2Δ mutants, Mec1 might associate transiently or weakly with DNA lesions in the absence of Ddc2. Several groups have investigated biochemical properties of human ATR and ATRIP on binding to DNA and RPA-covered DNA. ATR was found to bind DNA and RPA-coated DNA without the aid of ATRIP (Unsal-Kacmaz and Sancar, 2004). Another report showed that ATRIP alone possesses an ability to bind to RPA-coated DNA (Zou and Elledge, 2003). A separate study showed that the ATR–ATRIP complex binds to DNA with higher affinity in the presence of RPA, but with lower affinity in the absence of RPA (Bomgarden et al., 2004). Some discrepancies might result from difference between the reaction conditions in these biochemical studies. Alternatively, these discrepancies might suggest that some components are missing in these in vitro reactions. Although RPA-coated ssDNA seems to be a key structure, it is possible that proteins other than RPA might be required to recruit the ATR or Mec1 complex to sites of DNA damage.
In addition to DNA damage response, Mec1 contributes to telomere maintenance (Ritchie et al., 1999; Chan et al., 2001). Recent evidence indicated that Mec1 localizes to telomere ends (Takata et al., 2004). In contrast with association with DNA lesions, association of Mec1 with telomere ends depends on its own kinase activity (Takata et al., 2004). Mec1 phosphorylates RPA in vivo and in vitro (Brush et al., 1996; Brush and Kelly, 2000; Kim and Brill, 2003; Mallory et al., 2003). Several biochemical studies have indicated that RPA phosphorylation alters its interaction with DNA (Binz et al., 2003; Oakley et al., 2003). Mec1-dependent phosphorylation might stabilize the RPA binding to telomere sequences, thereby promoting the Mec1 association with telomeres. Alternatively, Mec1 might associate with telomere ends through a distinct, RPA-independent mechanism.
In summary, our results provide evidence indicating that Mec1 and Ddc2 recognize sites of DNA damage in the form of the Mec1–Ddc2 complex. However, it is not precisely determined how the Mec1–Ddc2 complex activates signaling at sites of DNA damage. Future experiments will be aiming at elucidating the biochemical properties of the Mec1–Ddc2 complex when interacting with damaged DNA.
Supplementary Material
Acknowledgments
We especially thank Tatsushi Wakayama for initial endeavor in screening and plasmid construction. We also thank John Kang for critical reading; Steve Brill, Tae Kondo, Noel Lowndes, Rodney Rothstein, Atsushi Yamagishi for materials; Steve Brill and Takahiro Naiki for technical suggestions; and Carol Newlon and laboratory members for helpful discussion. This work was supported by National Institutes of Health Grant R01 GM-073876 and New Jersey Commission on Cancer Research research grant.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–05–0405) on September 7, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Abraham, R. T. (2001). Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15, 2177–2196. [DOI] [PubMed] [Google Scholar]
- Binz, S. K., Lao, Y., Lowry, D. F., and Wold, M. S. (2003). The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J. Biol. Chem. 278, 35584–35591. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987). 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164–175. [DOI] [PubMed] [Google Scholar]
- Bomgarden, R. D., Yean, D., Yee, M. C., and Cimprich, K. A. (2004). A novel protein activity mediates DNA binding of an ATR-ATRIP complex. J. Biol. Chem. 279, 13346–13353. [DOI] [PubMed] [Google Scholar]
- Bosotti, R., Isacchi, A., and Sonnhammer, E. L. (2000). FAT: a novel domain in PIK-related kinases. Trends Biochem. Sci. 25, 225–227. [DOI] [PubMed] [Google Scholar]
- Brush, G. S., and Kelly, T. J. (2000). Phosphorylation of the replication protein A large subunit in the Saccharomyces cerevisiae checkpoint response. Nucleic Acids Res. 28, 3725–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush, G. S., Morrow, D. M., Hieter, P., and Kelly, T. J. (1996). The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc. Natl. Acad. Sci. USA 93, 15075–15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S. W., Chang, J., Prescott, J., and Blackburn, E. H. (2001). Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11, 1240–1250. [DOI] [PubMed] [Google Scholar]
- Clarke, L., and Carbon, J. (1980). Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287, 504–509. [DOI] [PubMed] [Google Scholar]
- Cortez, D., Guntuku, S., Qin, J., and Elledge, S. J. (2001). ATR and ATRIP: partners in checkpoint signaling. Science 294, 1713–1716. [DOI] [PubMed] [Google Scholar]
- Cross, F. R. (1997). Marker swap plasmids: convenient tools for budding yeast molecular genetics. Yeast 13, 647–653. [DOI] [PubMed] [Google Scholar]
- Edwards, R. J., Bentley, N. J., and Carr, A. M. (1999). A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1, 393–398. [DOI] [PubMed] [Google Scholar]
- Elledge, S. J. (1996). Cell cycle checkpoints: preventing an identity crisis. Science 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- Futcher, B., and Carbon, J. (1986). Toxic effects of excess cloned centromeres. Mol. Cell. Biol. 6, 2213–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, C. M., Erdjument-Bromage, H., Tempst, P., and Lowndes, N. F. (2000). A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 10, 39–42. [DOI] [PubMed] [Google Scholar]
- Itakura, E., Takai, K. K., Umeda, K., Kimura, M., Ohsumi, M., Tamai, K., and Matsuura, A. (2004). Amino-terminal domain of ATRIP contributes to intranuclear relocation of the ATR-ATRIP complex following DNA damage. FEBS Lett. 577, 289–293. [DOI] [PubMed] [Google Scholar]
- Kim, H.-S., and Brill, S. J. (2003). MEC1-dependent phosphorylation of yeast RPA in vitro. DNA Repair 2, 1321–1335. [DOI] [PubMed] [Google Scholar]
- Kondo, T., Wakayama, T., Naiki, T., Matsumoto, K., and Sugimoto, K. (2001). Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 5543, 867–870. [DOI] [PubMed] [Google Scholar]
- Lisby, M., Barlow, J. H., Burgess, R. C., and Rothstein, R. (2004). Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713. [DOI] [PubMed] [Google Scholar]
- Longhese, M. P., Foiani, M., Muzi-Falconi, M., Lucchini, G., and Plevani, P. (1998). DNA damage checkpoint in budding yeast. EMBO J. 17, 5525–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, J. C., Bashkirov, V. I., Trujillo, K. M., Solinger, J. A., Dominska, M., Sung, P., Heyer, W. D., and Petes, T. D. (2003). Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Repair 2, 1041–1064. [DOI] [PubMed] [Google Scholar]
- Melo, J. A., Cohen, J., and Toczyski, D. P. (2001). Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 21, 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, D. M., Tagle, D. A., Shiloh, Y., Collins, F. S., and Hieter, P. (1995). TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82, 831–840. [DOI] [PubMed] [Google Scholar]
- Naiki, T., Kondo, T., Nakada, D., Matsumoto, K., and Sugimoto, K. (2001). Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol. 21, 5838–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki, T., Wakayama, T., Nakada, D., Matsumoto, K., and Sugimoto, K. (2004). Association of Rad9 with double-strand breaks through a Mec1-dependent mechanism. Mol. Cell. Biol. 24, 3277–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, D., Hirano, Y., and Sugimoto, K. (2004). Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 24, 10016–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, D., Matsumoto, K., and Sugimoto, K. (2003a). ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17, 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, D., Shimomura, T., Matsumoto, K., and Sugimoto, K. (2003b). The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 31, 1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, G. G., Patrick, S. M., Yao, J., Carty, M. P., Turchi, J. J., and Dixon, K. (2003). RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry 42, 3255–3264. [DOI] [PubMed] [Google Scholar]
- Paciotti, V., Clerici, M., Lucchini, G., and Longhese, M. P. (2000). The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14, 2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Peterson, R. T., Beal, P. A., Comb, M. J., and Schreiber, S. L. (2000). FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J. Biol. Chem. 275, 7416–7423. [DOI] [PubMed] [Google Scholar]
- Reid, R. J., Lisby, M., and Rothstein, R. (2002). Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 350, 258–277. [DOI] [PubMed] [Google Scholar]
- Ritchie, K. B., Mollory, J. C., and Petes, T. D. (1999). Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Calzada, A., Maman, J. P., Spagnolo, L., Pearl, L. H., and Llorca, O. (2005). Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Structure 13, 243–255. [DOI] [PubMed] [Google Scholar]
- Rouse, J., and Jackson, S. P. (2000). LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 19, 5793–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, J., and Jackson, S. P. (2002). Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9, 857–869. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Bachant, J., Wang, H., Hu, F., Liu, D., Tetzlaff, M., and Elledge, S. J. (1999). Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286, 1166–1171. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Desany, B. A., Jones, W. J., Liu, Q., Wang, B., and Elledge, S. J. (1996). Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271, 357–360. [DOI] [PubMed] [Google Scholar]
- Takata, H., Kanoh, Y., Gunge, N., Shirahige, K., and Matsuura, A. (2004). Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell 14, 515–522. [DOI] [PubMed] [Google Scholar]
- Toczyski, D. P., Galgoczy, D. J., and Hartwell, L. H. (1997). CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90, 1097–1106. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz, K., and Sancar, A. (2004). Quaternary structure of ATR and effects of ATRIP and replication protein A on its DNA binding and kinase activities. Mol. Cell. Biol. 24, 1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui, T., Ogawa, H., and Petrini, J. H. (2001). A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Wakayama, T., Kondo, T., Ando, S., Matsumoto, K., and Sugimoto, K. (2001). Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold, M. S. (1997). Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92. [DOI] [PubMed] [Google Scholar]
- Wolkow, T. D., and Enoch, T. (2003). Fission yeast Rad26 responds to DNA damage independently of Rad3. BMC Genet. 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Muller, E.G.D., and Rothstein, R. (1998). A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pool. Mol. Cell 2, 329–340. [DOI] [PubMed] [Google Scholar]
- Zhou, B.-B.S., and Elledge, S. J. (2000). The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- Zou, L., and Elledge, S. J. (2003). Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.