Abstract
Mitochondria are essential organelles of eukaryotic cells. Inheritance and maintenance of mitochondrial structure depend on cytoskeleton-mediated organelle transport and continuous membrane fusion and fission events. However, in Saccharomyces cerevisiae most of the known components involved in these processes are encoded by genes that are not essential for viability. Here we asked which essential genes are required for mitochondrial distribution and morphology. To address this question, we performed a systematic screen of a yeast strain collection harboring essential genes under control of a regulatable promoter. This library contains 768 yeast mutants and covers approximately two thirds of all essential yeast genes. A total of 119 essential genes were found to be required for maintenance of mitochondrial morphology. Among these, genes were highly enriched that encode proteins involved in ergosterol biosynthesis, mitochondrial protein import, actin-dependent transport processes, vesicular trafficking, and ubiquitin/26S proteasome-dependent protein degradation. We conclude that these cellular pathways play an important role in mitochondrial morphogenesis and inheritance.
INTRODUCTION
Mitochondria supply the cell with ATP generated by oxidative phosphorylation (Saraste, 1999), they are involved in the biosynthesis of many cellular metabolites (Scheffler, 2000), they play a key role in the assembly of iron/sulfur clusters (Lill and Mühlenhoff, 2005), and they are central regulators of programmed cell death (Desagher and Martinou, 2000). Budding yeast Saccharomyces cerevisiae is an excellent model organism to study mitochondrial biogenesis and function. Because S. cerevisiae is a facultative anaerobic yeast capable of satisfying its energy requirements with ATP generated by fermentation, only few mitochondrial proteins are essential for viability in this organism. Among these are several factors required for biogenesis of iron/sulfur clusters that serve as cofactors for a number of mitochondrial and extramitochondrial enzymes (Lill and Mühlenhoff, 2005). Other essential mitochondrial proteins are subunits of the molecular machines that mediate import, folding, and assembly of nuclear-encoded mitochondrial proteins (Neupert and Brunner, 2002; Pfanner et al., 2004; Rehling et al., 2004). Recently, it was reported that a soluble ATP-binding cassette protein, Rli1, carries iron/sulfur clusters. Rli1 is a factor required for nuclear export of newly assembled subunits of cytosolic ribosomes and thus is essential for cell viability. As the biogenesis of iron/sulfur clusters requires the mitochondrial assembly machinery, the dependence of Rli1 on iron/sulfur clusters is sufficient to explain the essential nature of mitochondria in yeast (Kispal et al., 2005; Yarunin et al., 2005).
Because mitochondria cannot be generated de novo, they must be inherited during cell division (Warren and Wickner, 1996). Inheritance of the organelles is mediated by cytoskeleton-mediated transport, and maintenance of mitochondrial structure and function depends on continuous fusion and fission events (Bereiter-Hahn and Vöth, 1994; Yaffe, 1999). In recent years, an increasing number of proteins essential for mitochondrial inheritance and structure have been identified in yeast. These include factors of the mitochondrial membrane fusion and division machineries as well as proteins involved in structural maintenance of the organelle and cytoskeleton-dependent motility (Hermann and Shaw, 1998; Jensen et al., 2000; Boldogh et al., 2001b; Dimmer et al., 2002; Shaw and Nunnari, 2002; Scott et al., 2003). Surprisingly, all components specifically required for mitochondrial dynamics are encoded by nonessential genes. However, some synthetic lethal relationships have been revealed. Deletion of the genes encoding the mitochondrial inheritance components Mdm10, Mdm12, and Mmm1 is synthetically lethal with deletion of the genes encoding prohibitin family members (Berger and Yaffe, 1998); and deletion of the MDM10, MDM12, MMM1, and MMM2 genes is synthetically lethal with deletion of the MDM31 and MDM32 genes (Dimmer et al., 2005). Furthermore, overexpression of Mdm33, an inner membrane protein required for mitochondrial morphogenesis, is lethal (Messerschmitt et al., 2003). These genetic findings underscore the fact that inheritance of mitochondria is an essential process.
To identify essential genes required for mitochondrial distribution and morphology in yeast, we screened a library containing mutants harboring essential genes that have been placed under control of a regulatable promoter (Mnaimneh et al., 2004). Assignment of mutants with aberrant mitochondrial morphology to functional classes identified the essential cellular pathways that are important for mitochondrial morphogenesis.
MATERIALS AND METHODS
Yeast Genetic Methods
S. cerevisiae was cultivated and manipulated according to standard procedures (Sherman, 1991). Yeast Tet regulated ORF clone collection and update, the strain collections used for expression of essential genes from a regulatable promoter (Gari et al., 1997; Mnaimneh et al., 2004), were obtained from BioCat (Heidelberg, Germany). Repression of the TetO7 promoter was achieved by the addition of 10 μg/ml doxycycline to the medium and incubation over night or for up to 3 d.
Screening for Essential Genes Involved in Mitochondrial Morphogenesis
After thawing 96-well plates containing frozen stocks of the yeast strains, cells were transferred to yeast extract/peptone/glucose (YPD) plates without or with 10 μg/ml doxycycline using a sterile pinning tool and incubated at 30°C over night. Media lacking doxycycline allowed expression of essential genes from the TetO7 promoter, and these cultures served as negative controls. Media containing 10 μg/ml doxycycline (YPD/Dox) efficiently repress the TetO7 promoter (Gari et al., 1997; Mnaimneh et al., 2004). About one third of the strains failed to grow on the YPD/Dox plate. Strains that did grow were subjected to two additional rounds of replica plating on fresh YPD/Dox plates using a pinning tool. About half of the strains were still able to grow on the third YPD/Dox plate. For these strains, growth could be observed even after seven rounds of replica plating.
For screening for mitochondrial morphology mutants, strains were inoculated in 0.5 ml YPD or YPD/Dox and grown over night at 30°C. YPD cultures were inoculated from YPD plates. YPD/Dox cultures were inoculated from YPD plates for strains that failed to grow on the first YPD/Dox plate, or from the first YPD/Dox plate for strains that failed to grow on the second YPD/Dox plate, or from the second YPD/Dox plate for strains that failed to grow on the third YPD/Dox plate, or from the third YPD/Dox plate for all remaining strains. The next morning, 3 ml fresh medium was added, and incubation was continued for at least 3 h. Mitochondria were stained by the addition of 0.1 μM rhodamine B hexyl ester (Molecular Probes, Eugene, OR) and inspected by fluorescence microscopy. Strains (n = 183) that showed aberrant mitochondrial morphology on YPD/Dox media, or that did not stain well, were transformed with pYX142-mtGFP expressing mitochondria-targeted GFP (Westermann and Neupert, 2000). These transformants were subjected to another round of screening to identify clones that reproducibly do not exhibit wild-type-like mitochondria under promoter shut off conditions. All rounds of screening were performed without prior reference to strain identity. To exclude the possibility that mitochondrial phenotypes were indirect consequences of cell death in the presence of doxycycline, cultures were stained with FUN 1 (LIVE/DEAD yeast viability kit, Molecular Probes) to score for viability of cells (Millard et al., 1997). With very few exceptions (as indicated in Supplementary Table 1) 70–100% of the cells grown in YPD/Dox media were alive and metabolically active under screening conditions.
Staining of Cellular Structures and Microscopy
Mitochondria were stained in living cells with 0.1 μM rhodamine B hexyl ester (Molecular Probes) or mitochondria-targeted GFP expressed from pYX142-mtGFP (Westermann and Neupert, 2000; Dimmer et al., 2002). The actin cytoskeleton was stained with rhodamine-phalloidin (Molecular Probes) as described (Amberg, 1998). Differential interference contrast (DIC) and epifluorescence microscopy was performed using an Axioplan 2 microscope equipped with a Plan-Neofluar 100×/1.30 Ph3 oil objective (Carl Zeiss Jena GmbH, Oberkochen, Germany). Images were recorded with an Evolution VF Mono Cooled monochrome camera (Intas, Göttingen, Germany) and processed with Image ProPlus 5.0 and ScopePro 4.5 software (Media Cybernetics, Silver Spring, MD).
RESULTS AND DISCUSSION
Identification of Essential Cellular Functions Required for Mitochondrial Morphogenesis
To identify essential genes required for establishment and maintenance of normal mitochondrial distribution and morphology, we systematically screened a promoter shut off yeast strain collection that covers approximately two thirds of all essential yeast genes (Mnaimneh et al., 2004). The 768 strains contained in this collection were incubated under permissive and restrictive conditions, stained with mitochondria-specific probes and screened for mutants that showed aberrant mitochondrial morphology upon depletion of essential gene products. A total number of 119 mutants (corresponding to 15% of the strains present in the collection) were identified that reproducibly showed severe defects in the organization of their mitochondria (see Supplementary Table 1 for a complete list).
Based on information obtained from the Saccharomyces Genome Database (Christie et al., 2004) and the Comprehensive Yeast Genome Database (Güldener et al., 2005) and manual annotation, the strains present in the collection were grouped into functional categories (see Supplementary Table 1). The following five functional categories were identified that contained a high proportion of mutants with aberrant mitochondria: (1) ergosterol biosynthesis, with 100% of the screened mutants being affected; (2) mitochondrial protein import, 70% affected; (3) actin cytoskeleton-associated proteins, 67% affected; (4) vesicular transport and secretion, 61% affected; and (5) ubiquitin/26S proteasome-dependent protein degradation, 32% affected. In contrast, only 7% of the mutants lacking essential proteins of other functions showed aberrant mitochondria. A similar number, 8%, was obtained for mutants lacking proteins of unknown function. These results are summarized in Figure 1, Table 1, and Supplementary Table 1. We conclude that mitochondrial morphogenesis and inheritance rely on the presence of a complete complement of ergosterol biosynthetic enzymes and intact machineries of mitochondrial protein import, actin cytoskeleton-dependent motility, vesicular transport, and proteasome-dependent protein degradation.
Figure 1.
Essential cellular processes required for maintenance of mitochondrial morphology. Promoter shutoff strains (n = 768) lacking essential gene products were screened for mutants with aberrant mitochondrial morphology. Of these strains, 15% showed strong defects in mitochondrial morphology and distribution (average, indicated by dotted line). Black columns indicate functional classes that contained a high percentage of mutants with disorganized mitochondria. In case of protein degradation factors, genes encoding proteasome subunits were counted separately (white column). Light gray columns represent genes of unknown function, and genes of known functions other than that included in the black columns. A complete list of screened yeast strains can be found in Supplementary Table 1.
Table 1.
Essential cellular pathways, protein complexes, and proteins required for mitochondrial morphogenesis in yeast
| Function | Complex | Protein |
|---|---|---|
| Ergosterol biosynthesis | Erg1, Erg7, Erg8, Erg10, Erg12, Erg13, Erg25, Erg26, Erg27, Mvd1, Ncp1 | |
| Mitochondrial protein import and assembly | TOM complex | Mim1, Tom22 |
| SAM complex | Sam35, Sam50 | |
| Tim23 complex | Mge1, Pam18, Zim17 | |
| Vesicular trafficking/protein secretion | Bfr2, Dsl1, Ret2, Sec2, Sec3, Sec4, Sec5, Sec8, Sec10, Sec13, Sec15, Sec14, Sec17, Sec18, Sec20, Sec21, Sec26, Sec27, Sec31, Sec53, Sec61, Sec63, Sec65, Sed5, Sly1, Srp14, Srp21, Srp68, Srp72, Srp101, Srp102, Trs20, Trs120, Use1, Yip1 | |
| Actin cytoskeleton-dependent transport | ARP2/3 complex | Arc35, Arc40, Arp2 |
| CCT complex | Cct4, Cct6 | |
| Myosins | Mlc1, Myo2 | |
| Other | Cof1, Iqg1, Pfy1 | |
| Ubiquitin/26S proteasome-dependent protein degradation | Proteasome | Pre1, Pre3, Pre5, Pre6, Rpn8, Rpt2, Rpt4 |
| SCF ubiquitin ligase | Cdc34, Cdc53 | |
| Other | Uba1, Ufd1 |
Only standard names of proteins are indicated. A complete list including systematic gene names can be found in Supplementary Table 1.
Role of Ergosterol Biosynthesis in Maintenance of Mitochondrial Morphology
Mutants lacking enzymes required for the biosynthesis of ergosterol showed clumped and swollen mitochondria (Table 1 and Supplementary Table 1). This is exemplified for erg7 and erg8 (Figure 2), whereas the other mutants looked very similar. When the strains were grown in the absence of doxycycline, i.e., under permissive conditions, they exhibited the characteristic mitochondrial tubular network of wild-type cells (Figure 2). Addition of doxycycline to the growth medium did not affect mitochondrial morphology of wild-type cells, indicating that the observed defects were consequences of depletion of essential gene products (Figure 2).
Figure 2.
Yeast cells lacking ergosterol biosynthetic enzymes harbor aberrant mitochondria. Strains expressing mitochondria-targeted GFP were grown in the absence (- Dox) or presence (+ Dox) of doxycycline in YPD medium to logarithmic growth phase and analyzed by fluorescence microscopy. Left, differential interference contrast (DIC) image; middle, mitochondrial morphology of a representative cell; right, merged image. WT, wild type. Bar, 5 μm.
The fact that all strains with down-regulated ergosterol biosynthetic enzymes showed severe mitochondrial morphology defects points to an essential role of this membrane lipid for maintenance of the structure of membrane-bounded organelles. Interestingly, ergosterol has been reported to be required for the priming step of homotypic vacuole fusion in yeast (Kato and Wickner, 2001). Because vacuolar membranes have relatively low ergosterol content, the role of this lipid in membrane fusion presumably is not the modulation of the physical properties of the membrane. Rather, it has been suggested to specifically activate and/or rearrange membrane proteins required for the vacuolar fusion reaction (Kato and Wickner, 2001; Fratti et al., 2004). The ergosterol content of mitochondrial membranes (6 μg ergosterol per mg of organellar protein in the outer membrane and 25 μg/mg in the inner membrane) is even lower than that of vacuolar membranes (49 μg/mg; Zinser et al., 1993). Thus, it is conceivable that also in the case of mitochondria, ergosterol may have a modulatory function on the machinery of membrane fusion and/or fission.
Link between the Mitochondrial Protein Import/Assembly Machinery and Mitochondrial Morphology
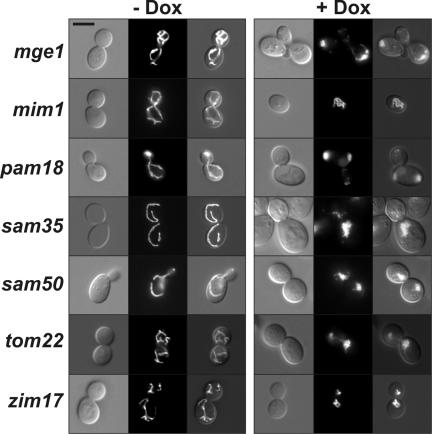
Many mutants lacking proteins involved in mitochondrial protein translocation, sorting, and assembly showed severe defects in mitochondrial morphology. These include essential subunits of the TOM complex (translocase of the outer membrane), the SAM complex (sorting and assembly machinery of the outer membrane), and the TIM23 complex (translocase of the inner membrane).1 The following proteins were found to be required for normal mitochondrial morphology: Tom22, a subunit of the TOM complex; Mim1, a protein required for TOM complex assembly; Sam35 and Sam50, two subunits of the SAM complex; and Pam18, Zim17, and Mge1, three components or cofactors of the TIM23 translocase and its associated import motor (Figure 3 and Table 1). Strains that showed normal mitochondria in the presence of doxycycline include mas1, lacking a subunit of the mitochondrial processing peptidase; tim50, lacking a subunit of the TIM23 translocase; and tim22, lacking a subunit of the TIM22 translocase. It is presently unclear whether residual amounts of these proteins are sufficient to maintain normal mitochondrial shape or whether these proteins are not required for mitochondrial morphogenesis.
Figure 3.
Yeast cells lacking components of the mitochondrial protein import and sorting machinery harbor aberrant mitochondria. Yeast strains were analyzed as in Figure 2. Bar, 5 μm.
Because 70% of the strains with down-regulated components of the mitochondrial protein sorting machinery showed aberrant mitochondria, we conclude that intact TOM, SAM, and TIM23 complexes are required for mitochondrial morphogenesis. This is in accordance with our earlier observation that a nonessential TOM component, Tom7, is required for wild-type-like mitochondrial distribution and morphology (Dimmer et al., 2002). In case of Sam50, similar mitochondrial defects have been reported in a recent study using temperature-sensitive sam50 mutants. It has been suggested that Sam50 together with Mas37 and the mitochondrial morphology protein Mdm10 is required for assembly of Tom40 and its association with Tom22 to generate a functional TOM complex (Meisinger et al., 2004). Most of the known proteins required for maintenance of mitochondrial structure are located in the mitochondrial outer and inner membrane. Because these proteins have to be imported via the TOM complex and then sorted to the mitochondrial subcompartments, it is conceivable that defects of the mitochondrial protein sorting machinery lead to aberrant mitochondrial morphology as a secondary consequence.
Dependence of Mitochondrial Morphology on the Actin Cytoskeleton
Mitochondrial transport in S. cerevisiae is dependent on actin filaments (Hermann and Shaw, 1998; Jensen et al., 2000; Boldogh et al., 2001b). Therefore, we expected that depletion of actin cytoskeleton-related factors would result in mitochondrial morphology defects. Mutants of 15 essential yeast genes required for integrity or function of the actin cytoskeleton are contained in the strain collection. We examined the organization of mitochondria and the actin cytoskeleton in all of these mutants by fluorescence microscopy. Based on their phenotypes, the mutants were grouped into four classes: class A mutants have aberrant mitochondria and disorganized actin filaments; class B mutants have aberrant mitochondria and normal actin filaments; class C mutants have normal mitochondria and disorganized actin filaments; and class D mutants have normal mitochondria and normal actin filaments (Figure 4). Class A comprises subunits of the ARP2/3 complex (Arc35, Arc40, Arp2), subunits of the CCT chaperone, which is required for assembly of actin and tubulin (Cct4, Cct6), a myosin-related motor protein (Myo2), and other factors required for organization of the actin cytoskeleton (Cof1/cofilin, Iqg1, Pfy1/profilin). Class B contains Mlc1, a myosin light chain associated with Myo2. Class C comprises a myosin-related motor protein (Myo1), and a component of cortical actin patches (Las17). Class D comprises three proteins involved in actin cytoskeleton assembly (Mss4, Pan1, Sda1). Class C and D components apparently do not play an important role in mitochondrial morphogenesis.
Figure 4.
(facing page). Yeast cells lacking components linked to the actin cytoskeleton harbor aberrant mitochondria. Strains expressing mitochondria-targeted GFP were grown in the absence (- Dox) or presence (+ Dox) of doxycycline in YPD medium to logarithmic growth phase, fixed, stained with rhodamine phalloidin, and analyzed by fluorescence microscopy. First image, differential interference contrast (DIC) image; second image, mitochondrial morphology (mito); third image, organization of filamentous actin (a reversed fluorescence image is shown to better visualize faint actin cables); and fourth image, merged image of reversed actin fluorescence and mitochondrial fluorescence. On the right hand side, the mutant class is indicated: A, aberrant mitochondria and disorganized actin cytoskeleton; B, aberrant mitochondria and normal actin cytoskeleton; C, normal mitochondria and disorganized actin cytoskeleton; D, normal mitochondria and normal actin cytoskeleton. WT, wild type. Bar, 5 μm.
The large number of actin cytoskeleton mutants with aberrant mitochondria found in classes A and B underscores the fundamental importance of actin-dependent transport for mitochondrial distribution and morphology in yeast. Although the mitochondrial phenotype seen in cells lacking Cof1, Iqg1, Pfy1, and CCT subunits is most likely an indirect consequence of disturbed microfilament organization, a direct role in mitochondrial binding to actin has been suggested for the ARP2/3 complex (Boldogh et al., 2001a; Fehrenbacher et al., 2004) and Myo2 (Itoh et al., 2002; Boldogh et al., 2004; Itoh et al., 2004). It is still a matter of debate whether ARP2/3 complex-driven motility or myosin-related motor proteins are of major importance for mitochondrial movement (Boldogh et al., 2001a, 2004; Itoh et al., 2004). Here we found that the mlc1 mutant harbors intact actin cables, but highly aberrant mitochondria. Intriguingly, mitochondrial defects were also apparent in mlc1 and myo2 mutants in the absence of doxycycline (Figure 4), conditions that likely lead to nonphysiological expression levels of these proteins. The fact that mitochondrial defects are manifest in cells that display a relatively normal actin cytoskeleton is in favor of a direct role of Myo2 and its associated light chain, Mlc1, in linking the organelle to the cytoskeleton.
Connection of Vesicular Transport and Mitochondrial Morphology
Components of the vesicular trafficking system constitute the largest group of essential yeast proteins that were identified as mitochondrial biogenesis components. Thirty-five proteins mediating protein translocation into the ER and vesicle budding and fusion were found to be required for maintenance of mitochondrial morphology (Table 1 and Supplementary Table 1). On depletion of these proteins, mitochondria appeared fragmented and/or aggregated. This phenotype was very similar in all mutants. Examples shown in Figure 5 are Sec17 and Sec18, homologues of mammalian alpha-SNAP and NSF, which are required for priming of the vesicular fusion machinery, Sec63, a subunit of the SEC61 translocon, and Srp101, the alpha subunit of the signal recognition particle receptor.
Figure 5.
Yeast cells lacking components of the protein secretion and vesicular trafficking machinery harbor aberrant mitochondria. Yeast strains were analyzed as in Figure 2. Bar, 5 μm.
Previous observations of mitochondria in mutants of the secretory pathway have yielded ambiguous results. For example, temperature-sensitive sec18 mutants did not show defects in mitochondrial fusion (Nunnari et al., 1997), whereas temperature-sensitive srp101 mutants contained fragmented mitochondria (Prinz et al., 2000), similar to the aberrant organelles observed here (Figure 5). It is thought that defects in mitochondrial structure do not affect the ER, because mutants such as fzo1-1, Δmdm30, Δmdm31, Δmdm32, Δmdm33, mmm1-1, Δmmm2, and Δugo1 maintain a normal ER structure (Prinz et al., 2000; Sesaki and Jensen, 2001; Fritz et al., 2003; Messerschmitt et al., 2003; Youngman et al., 2004; Dimmer et al., 2005). In contrast, the majority of vesicular transport mutants analyzed here showed strong defects in mitochondrial morphology. We conclude that the ER and secretory pathway play an important and general role in maintenance of a normal mitochondrial reticulum. We consider it likely that this role involves the supply of mitochondria with lipids.
Role of the Ubiquitin/26S Proteasome System for Mitochondrial Biogenesis
A large fraction, 32%, of the mutants lacking proteins of the ubiquitin/26S proteasome system showed strong defects in mitochondrial morphology. When only proteasome subunits are considered, this number increases to 64% (Figure 1), whereas strains lacking cell cycle-specific protein degradation factors, such as components of the anaphase-promoting complex, showed no mitochondrial defects (see Supplementary Table 1). In addition to seven proteasome subunits (Table 1) the following components were found to be required for normal mitochondrial structure: two subunits of the SCF ubiquitin ligase complex (Cdc34 and Cdc53/cullin), a ubiquitin-activating enzyme (Uba1), and a protein involved in recognition of ubiquitinated proteins (Ufd1). Mitochondria generally appeared highly fragmented or aggregated in these mutants, as shown for cdc34, cdc53, pre1, and rpn8 (Figure 6). These results are in accordance with earlier observations that ubiquitin (Fisk and Yaffe, 1999), the 26S proteasome (Rinaldi et al., 1998) and SCF-dependent protein degradation (Fritz et al., 2003) are involved in maintenance of mitochondrial morphology.
Figure 6.
Yeast cells lacking components of the ubiquitin/26S proteasome system harbor aberrant mitochondria. Yeast strains were analyzed as in Figure 2. Bar, 5 μm.
Proteins of Unknown Function
Four mutants lacking proteins of unknown function showed severe mitochondrial defects, ymr134, ydr339c, ynl149c (and its overlapping ORF ynl150w), and yor060c (Figure 7). These ORFs might encode novel essential proteins required for mitochondrial morphogenesis. However, the percentage of mutants showing aberrant mitochondria is very similar for ORFs of unknown function (8%) and for the group of all remaining mutants (7%; Supplementary Table 1). We consider it likely that the latter mutants show aberrant mitochondria due to pleiotropic effects of their mutations or general defects in cell physiology. It remains to be determined whether the novel components of unknown function might contribute to mitochondrial inheritance directly or indirectly.
Figure 7.
Yeast cells lacking components of unknown function harbor aberrant mitochondria. Yeast strains were analyzed as in Figure 2. Bar, 5 μm.
CONCLUSIONS
With the screening of 4794 deletion mutants lacking nonessential genes (Dimmer et al., 2002) and the screening of 768 promoter shutoff strains reported here we have now systematically analyzed 5562 of the ca. 6000 yeast genes for their role in mitochondrial distribution and morphology. Most of the known components mediating mitochondrial fusion, fission, and maintenance of mitochondrial structure are encoded by nonessential genes (Hermann and Shaw, 1998; Jensen et al., 2000; Boldogh et al., 2001b; Dimmer et al., 2002; Shaw and Nunnari, 2002; Scott et al., 2003). The present work allows us to define the essential cellular pathways that contribute to the complex process of mitochondrial inheritance. In particular, our results have revealed an important role of ergosterol biosynthesis, mitochondrial protein import, and vesicular trafficking. Apparently, these processes are much more important for mitochondrial morphogenesis than previously anticipated. Moreover, our results underscore the fundamental importance of the actin cytoskeleton and the proteasome for mitochondrial biogenesis.
Obviously, many of the identified components affect mitochondrial morphology in a rather indirect way, as, for example, subunits of the mitochondrial protein translocation machinery or the vesicular transport system. Moreover, depletion of essential gene products for several days may lead to pleiotropic defects, including aberrant mitochondrial morphology. It should be noted that such pleiotropic defects might play a role in all classes of proteins reported here. On the other hand, some of the components identified (e.g., Dpm1, Mot1, Ncp1, Prp31, Sec4, and Sec63) were recently found in the mitochondrial proteome (Sickmann et al., 2003) although they were previously localized to other cellular compartments. This opens the exciting possibility that these proteins have a dual localization and might play a more direct role in mitochondrial biogenesis. Clearly, much more work is required to reveal the molecular interactions that link mitochondrial morphogenesis with cellular processes such as synthesis and exchange of membrane lipids, mitochondrial protein import, cytoskeleton-dependent organelle transport, and protein degradation.
Supplementary Material
Acknowledgments
We thank Jan Fiedler for his contributions to some experiments and the Deutsche Forschungsgemeinschaft for financial support through grants We 2174/2–4 and We 2174/3–1.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0678) on August 31, 2005.
Abbreviations used: Dox, doxycycline; YPD, yeast extract/peptone/glucose.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
Footnotes
Many components of mitochondrial protein sorting machineries have alternative names. Here, we use the standard gene names according to the Saccharomyces Genome Database.
References
- Amberg, D. C. (1998). Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol. Biol. Cell 9, 3259-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn, J., and Vöth, M. (1994). Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 27, 198-219. [DOI] [PubMed] [Google Scholar]
- Berger, K. H., and Yaffe, M. P. (1998). Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 4043-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I. R., Ramcharan, S. L., Yang, H. C., and Pon, L. A. (2004). A type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol. Biol. Cell 15, 3994-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I. R., Yang, H.-C., Nowakowski, W. D., Karmon, S. L., Hays, L. G., Yates Spaceiiiqq, J. R., and Pon, L. A. (2001a). Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc. Natl. Acad. Sci. USA 98, 3162-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I. R., Yang, H.-C., and Pon, L. A. (2001b). Mitochondrial inheritance in budding yeast. Traffic 2, 368-374. [DOI] [PubMed] [Google Scholar]
- Christie, K. R. et al. (2004). Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 32, D311-D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desagher, S., and Martinou, J. C. (2000). Mitochondria as the central control point of apoptosis. Trends Cell Biol. 10, 369-377. [DOI] [PubMed] [Google Scholar]
- Dimmer, K. S., Fritz, S., Fuchs, F., Messerschmitt, M., Weinbach, N., Neupert, W., and Westermann, B. (2002). Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer, K. S., Jakobs, S., Vogel, F., Altmann, K., and Westermann, B. (2005). Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J. Cell Biol. 168, 103-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher, K. L., Yang, H. C., Gay, A. C., Huckaba, T. M., and Pon, L. A. (2004). Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr. Biol. 14, 1996-2004. [DOI] [PubMed] [Google Scholar]
- Fisk, H. A., and Yaffe, M. P. (1999). A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145, 1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti, R. A., Jun, Y., Merz, A. J., Margolis, N., and Wickner, W. (2004). Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J. Cell Biol. 167, 1087-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz, S., Weinbach, N., and Westermann, B. (2003). Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol. Biol. Cell 14, 2303-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari, E., Piedrafita, L., Aldea, M., and Herrero, E. (1997). A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13, 837-848. [DOI] [PubMed] [Google Scholar]
- Güldener, U. et al. (2005). CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 33, D364-D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G. J., and Shaw, J. M. (1998). Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 14, 265-303. [DOI] [PubMed] [Google Scholar]
- Itoh, T., Toh, E. A., and Matsui, Y. (2004). Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J. 23, 2520-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, T., Watabe, A., Toh-e, A., and Matsui, Y. (2002). Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 7744-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R. E., Aiken Hobbs, A. E., Cerveny, K. L., and Sesaki, H. (2000). Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc. Res. Tech. 51, 573-583. [DOI] [PubMed] [Google Scholar]
- Kato, M., and Wickner, W. (2001). Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. EMBO J. 20, 4035-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal, G. et al. (2005). Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 24, 589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill, R., and Mühlenhoff, U. (2005). Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30, 133-141. [DOI] [PubMed] [Google Scholar]
- Meisinger, C. et al. (2004). The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell 7, 61-71. [DOI] [PubMed] [Google Scholar]
- Messerschmitt, M., Jakobs, S., Vogel, F., Fritz, S., Dimmer, K. S., Neupert, W., and Westermann, B. (2003). The inner membrane protein Mdm33 controls mitochondrial morphology in yeast. J. Cell Biol. 160, 553-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard, P. J., Roth, B. L., Thi, H. P., Yue, S. T., and Haugland, R. P. (1997). Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 63, 2897-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh, S. et al. (2004). Exploration of essential gene functions via titratable promoter alleles. Cell 118, 31-44. [DOI] [PubMed] [Google Scholar]
- Neupert, W., and Brunner, M. (2002). The protein import motor of mitochondria. Nat. Rev. Mol. Cell. Biol. 3, 555-565. [DOI] [PubMed] [Google Scholar]
- Nunnari, J., Marshall, W.F., Straight, A., Murray, A., Sedat, J. W., and Walter, P. (1997). Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8, 1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner, N., Wiedemann, N., Meisinger, C., and Lithgow, T. (2004). Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 11, 1044-1048. [DOI] [PubMed] [Google Scholar]
- Prinz, W. A., Grzyb, L., Veenhuis, M., Kahana, J. A., Silver, P. A., and Rapoport, T. A. (2000). Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150, 461-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling, P., Brandner, K., and Pfanner, N. (2004). Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell. Biol. 5, 519-530. [DOI] [PubMed] [Google Scholar]
- Rinaldi, T., Ricci, C., Porro, D., Bolotin-Fukuhara, M., and Frontali, L. (1998). A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol. Biol. Cell 9, 2917-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, M. (1999). Oxidative phosphorylation at the fin de siècle. Science 283, 1488-1493. [DOI] [PubMed] [Google Scholar]
- Scheffler, I. E. (2000). A century of mitochondrial research: achievements and perspectives. Mitochondrion 1, 3-31. [DOI] [PubMed] [Google Scholar]
- Scott, S. V., Cassidy-Stone, A., Meeusen, S. L., and Nunnari, J. (2003). Staying in aerobic shape: how the structural integrity of mitochondria and mitochondrial DNA is maintained. Curr. Opin. Cell Biol. 15, 482-488. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R. E. (2001). UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 152, 1123-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, J. M., and Nunnari, J. (2002). Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12, 178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- Sickmann, A. et al. (2003). The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 100, 13207-13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, G., and Wickner, W. (1996). Organelle inheritance. Cell 84, 395-400. [DOI] [PubMed] [Google Scholar]
- Westermann, B., and Neupert, W. (2000). Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421-1427. [DOI] [PubMed] [Google Scholar]
- Yaffe, M. P. (1999). The machinery of mitochondrial inheritance and behavior. Science 283, 1493-1497. [DOI] [PubMed] [Google Scholar]
- Yarunin, A., Panse, V. G., Petfalski, E., Dez, C., Tollervey, D., and Hurt, E. C. (2005). Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 24, 580-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman, M. J., Aiken Hobbs, A. E., Burgess, S. M., Srinivasan, M., and Jensen, R. E. (2004). Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 164, 677-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser, E., Paltauf, F., and Daum, G. (1993). Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J. Bacteriol. 175, 2853-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.