Abstract
Brucella abortus is a facultative intracellular gram-negative bacterial pathogen that infects humans and animals by entry mainly through the digestive tract. B. abortus causes abortion in pregnant cattle and undulant fever in humans. The immunogenic B. abortus ribosomal protein L7/L12 is a promising candidate antigen for the development of oral live vaccines against brucellosis, using food-grade lactic acid bacteria (LAB) as a carrier. The L7/L12 gene was expressed in Lactococcus lactis, the model LAB, under the nisin-inducible promoter. Using different signals, L7/L12 was produced in cytoplasmic, cell-wall-anchored, and secreted forms. Cytoplasmic production of L7/L12 gave a low yield, estimated at 0.5 mg/liter. Interestingly, a secretable form of this normally cytoplasmic protein via fusion with a signal peptide resulted in increased yield of L7/L12 to 3 mg/liter; secretion efficiency (SE) was 35%. A fusion between the mature moiety of the staphylococcal nuclease (Nuc) and L7/L12 further increased yield to 8 mg/liter. Fusion with a synthetic propeptide (LEISSTCDA) previously described as an enhancer for heterologous protein secretion in L. lactis (Y. Le Loir, A. Gruss, S. D. Ehrlich, and P. Langella, J. Bacteriol. 180:1895-1903, 1998) raised the yield to 8 mg/liter and SE to 50%. A surface-anchored L7/L12 form in L. lactis was obtained by fusing the cell wall anchor of Streptococcus pyogenes M6 protein to the C-terminal end of L7/L12. The fusions described allow the production and targeting of L7/L12 in three different locations in L. lactis. This is the first example of a B. abortus antigen produced in a food-grade bacterium and opens new perspectives for alternative vaccine strategies against brucellosis.
Brucellosis remains a worldwide zoonosis that causes abortion and infertility in cattle. It also causes undulant fever, endocarditis, arthritis and osteomyelitis in humans (for a review, see reference 3). Infection in humans occurs through direct contact with infected animals and from ingestion of contaminated dairy products. Brucella abortus, a facultative intracellular gram-negative bacterium, is the causative agent (49). Vaccination against brucellosis in cattle and in wild ungulates relies on live attenuated strains such as B. abortus strain 19 (6, 7) or strain RB51 (6). Although efficient in promoting a protective immune response, strain 19 is pathogenic for humans and provokes abortion when administered to pregnant cattle (6). Although strain RB51 is considered the vaccine of choice against brucellosis for cattle in the United States, it is derived from a virulent B. abortus 2308 strain and is not, like strain 19, recommended for pregnant animals (6, 18, 43). Furthermore, there are no human vaccines currently available, and the use of a food-grade oral vaccine expressing a common antigen from B. abortus and Brucella melitensis would be an alternative approach to immunize humans and animals against brucellosis.
Alternative vaccination strategies using B. abortus antigens with characterized immunogenic properties have recently been investigated (1, 29, 36, 44). Live vaccine strategies are being optimized using isolated antigens produced in either recombinant attenuated B. abortus strains (44, 45) or in a live Escherichia coli strain (16, 31). An effective vaccine against intracellular pathogens requires a cell-mediated immune response. An antigen of B. abortus that elicits a cell-mediated immune response and confers protective immunity in mice is the ribosomal protein L7/L12 (30).
The use of live, food-grade, noninvasive, nonpathogenic lactic acid bacteria (LAB) as antigen delivery vehicles is a promising vaccine strategy. This strategy could overcome potential problems due to the use of live B. abortus strains as antigen delivery vehicles (such as cross-reaction with diagnostic test, residual virulence, and reversion risks), and it provides a means for large scale and low-cost vaccine administration. To date, several bacterial and viral antigens have already been produced in Lactococcus lactis, the model LAB (15, 17, 21, 34, 48); in some cases, immunogenicity was demonstrated (12, 27, 35). These encouraging results suggest the feasibility of the LAB-based vaccine approach. Note that a protective immune response depends not only on the antigen and the delivery vehicle, but also on the location of the antigen (25). In some cases, antigen export may be of interest since it allows a direct contact between the antigen and the immune system.
In this work, we constructed L. lactis strains that produce recombinant B. abortus L7/L12 antigen. High L7/L12 levels were obtained using an inducible promoter (9). Three different expression vectors were used to target the protein to cytoplasm, surface, or culture medium. In the case of exported L7/L12, both secretion efficiency and yield were improved by its fusion to the staphylococcal nuclease (Nuc), a well-secreted reporter enzyme (38), or to a synthetic propeptide (23).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and DNA manipulations.
L. lactis NZ9000 (19) was grown in M17 medium (Difco [41]) supplemented with 1% glucose or in brain heart infusion (Difco) at 30°C without agitation. E. coli TG1 (14) was grown in aerated Luria-Bertani (37) medium at 37°C. Unless otherwise indicated, plasmid constructions were first established in E. coli and then transferred to L. lactis. Electrotransformation was performed as previously described (21). Plasmids were selected by addition of antibiotics as follows (concentrations in micrograms per milliliter): for L. lactis, erythromycin (5) or chloramphenicol (10); and for E. coli, ampicillin (100) or chloramphenicol (10). When both erythromycin and ampicillin were used for E. coli, concentrations were 75 μg/ml for each antibiotic. DNA manipulations were performed as described (37).
Cloning of the L7/L12 gene under control of nisin-inducible promoter PnisA.
The different L7/L12 expression cassettes are described in Fig. 1 and plasmids used in this work are listed in Table 1. Constructions were checked by DNA sequencing. Dideoxynucleotide chain termination DNA sequencing reactions were carried out on double-stranded DNA with the Taq Dye Primer Cycle Sequencing kit (Applied Biosystems, San Jose, Calif.) using a Perkin-Elmer PCR apparatus. Sequencing reactions were primed with fluorescent oligonucleotides and analyzed on a model 370A DNA sequencer (Applied Biosystems).
FIG. 1.
Expression cassettes for L7/L12 production and export using the nisin-inducible promoter (PnisA) and the lactococcal signal peptide SPUsp. Schematic structures of the fusion proteins carried by the indicated plasmids. For details of plasmid construction, see Materials and Methods and Table 1. PnisA, nisin-inducible promoter; RBS, ribosomal binding site of the usp45 gene; SPUsp, signal peptide of Usp45; Nuc, staphylococcal nuclease; Leiss, sequence encoding the LEISSTCDA synthetic propeptide; black bar, 11 first amino acids of Usp45; L7/L12, DNA fragment encoding the mature L7/L12 protein; CWA, sequence encoding the cell wall M6 protein; T1T2, termination signal of L. lactis tryptophan gene (no to scale).
TABLE 1.
Plasmids used in this study
| Plasmid | Characteristics | Reference or source |
|---|---|---|
| pMal:L7/L12 | Apr; pMal-c2 carrying the L7/L12 gene | 27 |
| pCYT:Nuc | pGK:Cmr; mature Nuc expressed under the control of the nisin-inducible promoter PnisA | 12 |
| pSEC:Nuc | pGK:Cmr; expression vector containing a fusion between the signal peptide of usp45 gene (SPUsp) and mature Nuc expressed under the control of PnisA | 12 |
| pSEC:LEISS:Nuc | pGK:Cmr; mature Nuc fused at 5" end with sequence of propeptide LEISSTCDA under the control of PnisA | P. Langellaa |
| pVE3753 | pBS SKII+:Apr; carrying nuc gene | S. Nouaillea |
| pVE3655 | pGK:Cmr; expression vector containing the PnisA | P. Langellaa |
| pVE5547 | pIL252:Eryr; mature Nuc fused at 3" end with sequence of cell wall protein M6 under the control of PnisA | S. Usaia |
| pCYT:L7/L12 | pGK:Cmr; pCYT:Nuc in which mature Nuc was replaced by DNA fragment encoding L7/L12 | This work |
| pSEC:L7/L12 | pGK:Cmr; pSEC:Nuc in which mature Nuc was replaced by DNA fragment encoding L7/L12 | This work |
| p5547:L7/L12 | pIL252:Eryr; pVE5547 in which mature Nuc was replaced by DNA fragment encoding L7/L12 | This work |
| pSEC:Nuc:L7/L12 | pGK:Cmr; mature L7/L12 fused at 5" end with mature Nuc | This work |
| pSEC:LEISS:L7/L12 | pGK:Cmr; pSEC:LEISS:Nuc in which mature Nuc was replaced by DNA fragment encoding L7/L12 | This work |
Unité de Recherches Laitières et Génétique Appliquée, Institut National de la Recherche Agronomique, Jouy en Josas, France.
Plasmids pCYT:Nuc, pSEC:Nuc, pSEC:LEISS:Nuc, and pVE5547 (Table 1) were used to clone the L7/L12 gene under the transcriptional control of lactococcal nisin-inducible promoter PnisA (9). pSEC:Nuc and pCYT:Nuc are derivatives of pNZ8010 in which the gus gene is under the transcriptional control of PnisA. To construct pSEC:Nuc, the gus gene was replaced by a BamHI-XhoI-cut DNA fragment encoding the ribosomal binding site (RBSUsp) and signal peptide (SPUsp) of the usp45 gene and the mature part of Nuc. To construct pCYT:Nuc, the DNA fragment encoding SPUsp was deleted by reverse PCR from plasmid pSEC:Nuc. An NsiI site was introduced at the 3" end of RBSUsp to allow replacement of the nuc gene by a DNA fragment encoding a heterologous protein. pSEC:LEISS:Nuc was constructed by inserting a DNA fragment having NsiI ends encoding LEISSTCDA synthetic propeptide between SPUsp and Nuc into NsiI-linearized pSEC:Nuc. pCYT:L7L12, pSEC:L7L12 and pSEC:LEISS:L7L12 were constructed by replacing the nuc gene with the L7/L12 gene in pCYT:Nuc, pSEC:Nuc and pSEC:LEISS:Nuc, respectively (Fig. 1) (see below for the restriction sites used).
The B. abortus L7/L12 ribosomal gene was amplified by PCR from E. coli expression vector pMal-L7/L12 (28), which encodes an in-frame fusion between L7/L12 and maltose binding protein (MBP). Primers containing one artificial restriction site at each end were constructed according to the L7/L12 nucleotide sequence (GenBank accession number L19101) (Table 2, oligonucleotides 1 and 2). To secrete L7/L12 protein, we used sense primer (5"-GGATGCATCAGCTGATCTCGCAAAG-3") (the NsiI site is underlined) in which CA (in boldface type) was added just after the NsiI restriction site; the amplified L7/L12 fragment can be cloned in the same reading frame as SPUsp.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide no. | Direction | Sequence (restriction enzyme)a |
|---|---|---|
| 1 | Sense | 5"-GGATGCATGCTGATCTCGCAAAG-3" (NsiI) |
| 2 | Antisense | 5"-GGGATATCTTACTTGAGTTCAAC-3" (EcoRV) |
| 3 | Sense | 5"-GGGTCGACGCTGATCTCGCAAAG-3" (SalI) |
| 4 | Antisense | 5"-GGGATATCCTTGAGTTCAACCTTGGC-3" (EcoRV) |
Restriction enzyme sites are underlined.
To produce L7/L12 protein in the cytoplasm or to secrete it in the extracellular medium, the amplified fragment containing L7/L12 was digested with NsiI and EcoRV restriction endonucleases and ligated to NsiI-EcoRV-digested pCYT:Nuc and pSEC:Nuc, respectively. In the first construction (pCYT:L7/L12; Fig. 1A), L7/L12 is produced in the cytoplasm. In the latter construction (pSEC:L7/L12; Fig. 1B), L7/L12 protein may be secreted via SPUsp, resulting in the pSEC:L7/L12 plasmid (Fig. 1B).
To obtain fusion protein Nuc:L7/L12, the NsiI-digested fragment containing nuc was purified from plasmid pVE3753 and ligated to NsiI-digested pSEC:L7/L12. The resulting plasmid pSEC:Nuc:L7/L12 contains the nuc:L7/L12 gene fusion (Fig. 1C). For fusion protein LEISS:L7/L12, the pSEC:L7/L12 plasmid was digested with NsiI and ClaI enzymes, and the resulting NsiI-ClaI fragment containing L7/L12 was then cloned into NsiI-ClaI-digested pSEC:LEISS:Nuc. The resulting plasmid was named pSEC:LEISS:L7/L12 (Fig. 1D).
To obtain a cell-wall-anchored form of L7/L12, L7/L12 was cloned into pVE5547 (Stefania Usai, personal communication) using primers 3 and 4 (Table 2). This vector contains the DNA fragment encoding the Streptococcus pyogenes M6 protein cell wall anchor region, CWAM6. CWAM6 corresponds to the 138 C-terminal amino acids of the mature M6 protein. The PCR fragment containing L7/L12 was digested with SalI and EcoRV enzymes and cloned directly into the SalI-EcoRV-digested pVE5547, resulting in p5547:L7/L12 encoding an in-frame fusion of L7/L12 with CWAM6 (Fig. 1E).
Purification of L7/L12 protein.
L7/L12 expression was induced by adding 0.6 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to a culture of an E. coli strain containing pMal-L7/L12 (28). Protein extracts were prepared after induction and diluted 1:5 with phosphate-buffered saline (PBS) (pH 8.4). The suspension was then loaded onto an amylose resin column (New England Biolabs) and washed 10 times with PBS, and the fusion protein was eluted with PBS plus 10 mM maltose. The purified fusion protein MBP-L7/L12 was cleaved with the Xa factor that recognizes a specific amino acid sequence between MBP and L7/L12. Although the expected products were observed, Xa digestion appeared to be partial.
Nisin induction.
PnisA induction for L7/L12 expression in the different lactococcal constructs was performed as follows: overnight cultures of L. lactis NZ9000 derivative strains were used to inoculate fresh medium at a 1:50 dilution. After 3 h of incubation (corresponding to an optical density at 600 nm of around 0.4), nisin A (Sigma) was added to cultures at a final concentration of 1 ng/ml. Growth was continued for 1 more hour before performing protein extractions.
Protein extraction and Western blot experiments.
For cell fractionation, 2 ml of L. lactis cultures was centrifuged for 5 min at 6,000 × g at 4°C. Protein extracts were then prepared from exponentially growing cultures according to Le Loir et al. (23).
Western blotting was performed as described (37). After blotting, nonspecific protein binding sites were blocked with a solution containing 1% bovine serum albumin in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.5% Tween 20. The nylon membranes were incubated 2 h with a 1/2,000 dilution of L7/L12 antibodies (Eurogentec). After washing, membranes were incubated for 45 min with protein G-horseradish peroxidase conjugate (Bio-Rad) and signals were detected using an enhanced chemiluminescence (ECL) kit (Dupont-NEN). After ECL detection, different nonsaturated film exposures were scanned by a Scanjet II (Hewlett-Packard) using Deskscan II and ImageQuant programs, and average values were determined. For quantification, signals were compared to those of known amounts of purified L7/L12.
In the case of Nuc:L7/L12 fusions, membranes were dehybridized by washing twice at 65°C, for 30 min, in 200 mM glycine-0.5% Tween 20-0.12 N HCl. Hybridization was then performed with Nuc antibodies kindly provided by J. R. Miller (Lilly Corporate Center, Indianapolis, Ind.).
RESULTS
Cytoplasmic production of L7/L12 in L. lactis.
A cytoplasmic form of L7/L12 was expressed in L. lactis NZ9000 using pCYT:L7/L12 (Fig. 1A; Table 1). L7/L12 production was analyzed by Western blot using L7/L12 antibodies (Fig. 2). Proteins from whole cells (noninduced and induced cultures) and supernatants were fractionated. Strain NZ9000(pVE3655) was used as a negative control. No L7/L12 signal was detected in the absence of nisin in strain NZ9000(pCYT:L7/L12) extracts. In induced cultures, a faint band with an apparent molecular mass of ∼28 kDa was present in the cell fraction only (Fig. 2). This is about twice the size expected for cytoplasmic L7/L12, and might correspond to either aberrant protein migration or the persistence of a sodium dodecyl sulfate (SDS)-resistant dimer of the cytoplasmic L7/L12 protein. This result shows that cytoplasmic production of L7/L12 with a yield estimated at 0.5 mg/liter can be achieved in L. lactis.
FIG. 2.
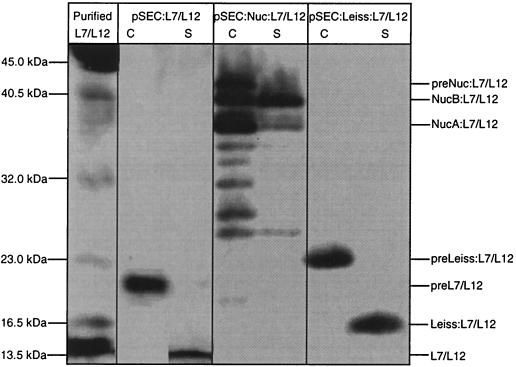
Intra- and extracellular production of L7/L12 protein in L. lactis. L7/L12 production was estimated by Western blot analysis on exponential-phase cultures of lactococcal strains containing pVE3655 (negative control), pCYT:L7/L12 (encoding the cytoplasmic form) and pSEC:L7/L12 (encoding the secreted form). Protein extractions were prepared on cellular (C) and supernatant (S) fractions of these cultures induced with nisin for 1 h. Purified L7/L12 protein was loaded as standard. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunodetection was performed with anti-L7/12 antibodies. Arrows indicate migration positions of precursor form (preL7/L12) and mature form (L7/12).
SPUsp can drive the secretion of the ribosomal protein L7/L12 in L. lactis.
L7/L12 is normally a cytoplasmic protein. Extracellular expression of L7/L12 was obtained by its fusion to the Usp45 signal peptide, SPUsp (Usp45 is the major L. lactis- secreted protein) (42). The construct encoding this fusion, pSEC:L7/L12 (Fig. 1B), was introduced into strain NZ9000, and L7/L12 secretion was evaluated by Western blotting (Fig. 2). Two bands were revealed: (i) one major band in the cell fraction corresponds to the preL7/L12 precursor form (preL7/L12) (migration position around 21 kDa for an expected size around 17 kDa); and (ii) a weaker band in the supernatant fraction migrates at the expected size of L7/L12 mature protein (around 14 kDa). This result shows that L. lactis can secrete the cytoplasmic protein L7/L12. The total protein yield of the secreted form of L7/L12 (3 mg/liter) was about sixfold greater than that for the cytoplasmic form with a secretion efficiency (SE; percentage of secreted L7/L12 detected in the supernatant) of 35%. The above results show the feasibility of secreting a naturally cytoplasmic protein in L. lactis.
L7/L12 SE is low regardless of induction levels.
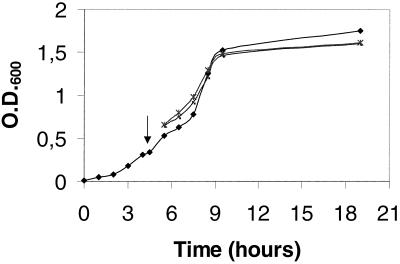
The precursor accumulation observed above with the secreted L7/L12 form could result from a secretion bottleneck in L. lactis due to saturation of the secretion machinery by high-level nisin induction (1 ng/ml) (in this case, processing might be more efficient at lower induction levels) or from incompatibility between intrinsic features of the preL7/L12 precursor and the L. lactis secretion machinery (in this case, precursor processing might be hampered regardless of the induction level). To address these hypotheses, growth and L7/L12 production of secreting strain NZ9000(pSEC:L7/L12) were monitored after a 1-h induction with two different concentrations of nisin (0.1 and 1 ng/ml; Fig 3). Growth of the control strain NZ9000(pVE3655) was not affected by nisin addition (data not shown). Also, growth of strain NZ9000(pSEC:L7/L12) cultures with or without nisin was similar, suggesting that L7/L12 precursor production did not slow down growth. Western blots showed that precursor accumulation occurs even at low induction levels (data not shown). This result suggests that the poor SE of preL7/L12 is probably due to intrinsic features of the precursor leading to poor recognition and/or processing by the lactococcal secretion machinery, favoring the second hypothesis.
FIG. 3.
Influence of L7/L12 production level on the growth of L. lactis. Growth curves of L7/L12-secreting strains NZ9000(pSEC:L7/L12) after a 1-h induction with different concentrations of nisin (0.1 and 1 ng/ml). The arrow indicates the time of nisin induction (optical density at 600 nm [O.D.600] = 0.345). Symbols: ⧫, without nisin; ▪, with nisin at 1 ng/ml; ∗, with nisin at 0.1 ng/ml.
Production yield of secreted L7/L12 is increased by a fusion with Nuc.
To improve the SE of L7/L12, the nuc gene encoding the Staphylococcus aureus nuclease (Nuc) was inserted between the DNA fragment encoding the fusion between SPUsp and L7/L12. Nuc is a highly stable, naturally secreted protein that has been successfully used as an export reporter in protein fusions (2, 20, 32-33). Nuc is synthesized as an intracellular precursor that is cleaved in two mature forms, NucB and NucA (23). The plasmid pSEC:Nuc:L7/L12 (Fig. 1C), which encodes the fusion protein Nuc:L7/L12, was transferred into L. lactis NZ9000. The resulting strain was streaked on a solid medium containing nisin and tested for Nuc activity (22). All Nuc:L7/L12-secreting L. lactis colonies displayed a clear Nuc+ phenotype (data not shown). Noninduced cells showed weak Nuc activity probably due to background PnisA promoter activity in colonies after overnight incubation (data not shown). Yields of Nuc:L7/L12 were examined by Western blots using either Nuc (data not shown) or L7/L12 antibodies (Fig. 4). In NZ9000- induced cultures, the total amount of fusion protein is estimated to be 2.5-fold higher for pSEC:Nuc:L7/L12 than for pSEC:L7/L12 (8 versus 3 mg/liter, respectively). In the cell fraction, three major bands were detected for Nuc:L7/L12: (i) the highest band corresponds to the preNuc:L7/L12 precursor form; (ii) the second and third bands correspond to mature NucB:L7/L12 and NucA:L7/L12 forms. Several bands of lower molecular weight present in the cell fraction are likely to correspond to degradation products.
FIG. 4.
Improvement of the secretion of L7/L12 protein in L. lactis. Recombinant L7/L12 production was analyzed by Western blot analysis on induced cultures of L. lactis NZ9000 strains containing pSEC:L7/12 (encoding the secreted form), pSEC:Nuc:L7/12 (encoding the fusion protein Nuc:L7/L12), or pSEC:LEISS:L7/L12 (encoding the fusion protein Leiss:L7/L12). Protein extractions were prepared on cellular (C) and supernatant (S) fractions of these cultures induced with nisin for 1 h. Purified L7/L12 protein was loaded as standard. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunodetection was performed with anti-L7/12 antibodies. Note that migration of all bands is slower than expected for preNuc:L7/L12 precursor and NucB:L7/L12 and NucA:L7/L12 mature forms (migrating as 46, 44, and 40 kDa for expected sizes around 36, 33, and 30 kDa).
The supernatant fraction contained a major band corresponding to mature NucB:L7/L12. Only a few weak bands corresponding to degradation products were detected in the supernatant, indicating that the secreted fusion is not subject to extracellular proteolytic degradation. Similar patterns were obtained using anti-Nuc antibodies (data not shown). The SEs of Nuc:L7/L12 and L7/L12 were similar (38 and 35%, respectively). These results show that Nuc mature moiety acts as a protein carrier that mainly enhances the production yield of L7/L12.
Synthetic propeptide enhances both SE and production yield.
We previously showed that the synthetic propeptide, LEISSTCDA (hereafter called LEISS), enhanced secretion of heterologous proteins in L. lactis (20, 23-24). The effect of LEISS on the SE of L7/L12 protein was examined. LEISS was fused between SPUsp and L7/L12 protein as expressed from pSEC:LEISS:L7/L12 plasmid (Fig. 1D). A comparison of L7/L12 and LEISS:L7/L12 secretion was made by Western blotting (Fig. 4). The SE of LEISS:L7/L12 was significantly improved compared to that observed for L7/L12 (50% for LEISS:L7/L12 compared to 35% for L7/L12 in repeated experiments). Compared to L7/L12 protein alone, production yield increased from 3 to 8 mg/liter. A major effect of LEISS was the increase in the amount of mature LEISS:L7/L12 protein present in the medium compared to L7/L12 protein alone (4 versus 1 mg/liter). Thus, L7/L12 production in L. lactis can be optimized by fusion of either a heterologous secreted protein or a synthetic propeptide at the N terminus of the mature protein.
Cell wall anchoring of L7/L12 in L. lactis.
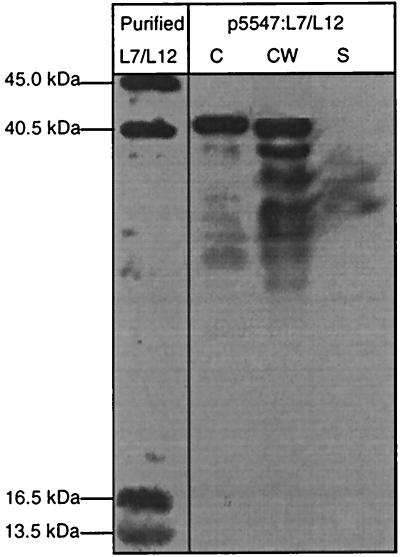
A cell-wall-anchored form of L7/L12 was obtained by creating a fusion between L7/L12 and the CWAM6 region of S. pyogenes M6 protein (expressed from plasmid p5547:L7/L12; Fig 1E). Production of L7/L12:CWAM6 in L. lactis was analyzed by Western blots in protoplast, cell wall and supernatant fractions (Fig. 5). About 40% of L7/L12:CWAM6 was localized in the cell wall, while 60% remained unprocessed in the protoplast as a band corresponding to preL7/L12:CWAM6. Cell-wall-anchored L7/L12 appeared as several closely spaced bands, a characteristic of cell-wall-anchored proteins; this is attributed to cell surface proteolysis (10). Bands corresponding to smaller proteins may also reflect degradation of anchored L7/L12 by cytoplasmic or surface housekeeping proteases. Two very weak bands were detected in the supernatant, and may correspond to L7/L12:CWAM6 released from the cell wall (Fig. 5). The cell-wall-anchored L7/L12 protein obtained here may be a particularly useful candidate for oral vaccine against B. abortus.
FIG. 5.
Inducible cell wall anchoring of L7/L12 protein in Lactococcus lactis. L7/L12 production was estimated by Western blot analysis on exponential-phase cultures of lactococcal strains containing p5547:L7/12 (encoding the fusion protein L7/L12:CWAM6). Protein extractions were performed in cell lysates (C), supernatants (S), and cell wall (CW) fractions of these cultures induced with nisin for 1 h. Purified L7/L12 protein was loaded as standard. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunodetection was performed with anti-L7/12 antibodies. Note that preL7/L12:CWAM6 migrates at 38 kDa for an expected size of 30 kDa.
DISCUSSION
Efficient targeting and production of the B. abortus protective ribosomal antigen L7/L12 (29) were attained for the first time in the food-grade LAB L. lactis. The constructed strains will allow us to determine whether recombinant L. lactis can be used as L7/L12 delivery vehicles to elicit both mucosal and systemic immune response against brucellosis and which antigen location will induce the most efficient response.
L. lactis has been previously reported to successfully target tetanus toxin fragment C (TTFC) model antigen to cytoplasm, cell wall, and extracellular medium (47, 48). TTFC was shown to elicit an immune and protective response, suggesting that L. lactis could be used as a successful delivery vector (26, 35, 48). Furthermore, secretion of interleukin-10 by L. lactis was recently shown to have biological activity in mice, although secretion levels are reportedly low (40). These encouraging results suggest that protein export from L. lactis may indeed be a feasible mode of antigen delivery.
Since L. lactis is not a colonizing commensal bacterium, the chosen approach was to preload the organism in vitro with high levels of heterologous antigen prior to immunization (4, 48). In this work, the L7/L12 gene was expressed under the control of PnisA, the nisin-inducible lactococcal promoter, to preload the bacteria with antigen before administration. This expression system is now frequently used to achieve high-level heterologous protein production in L. lactis (2, 5, 12).
Five L. lactis strains were constructed to produce L7/L12 in three different forms: cytoplasmic, anchored and secreted. The cytoplasmic production can protect the antigen from degradation in the upper digestive tract. During intestinal transit, L. lactis will then be lysed (11), and the accumulated antigen will thus be released. However, in strain NZ9000(pCYT:L7/L12), L7/L12 was detected in the cellular fraction at only a very low level (0.5 mg/liter). Our hypothesis was that ClpP, the major housekeeping intracellular protease of L. lactis (13), degraded the cytoplasmic form of L7/L12. pCYT:L7/L12 was then introduced in a clpP mutant of NZ9000 but no significant increase was observed, suggesting that ClpP was not involved in this putative proteolysis (data not shown).
To target a protein at a precise cell location requires specific signals that do not guarantee successful targeting, particularly for cytoplasmic proteins reportedly difficult to secrete (39). When we fused L7/L12 with SPUsp, aiming to direct the secretion of the cytoplasmic protein L7/L12, the production in strain NZ9000(pSEC:L7/L12) was enhanced sixfold compared to intracellular production. After fusion with SPUsp, the same increase was recently observed in the rate of production in L. lactis of the nonstructural protein NSP4 of bovine rotavirus (12), the E7 oncoprotein of human papillomavirus (2) and the bovine β-lactoglobulin protein (5). To explain this enhancement, our hypothesis is that the recognition of the preL7/L12 by the L. lactis secretion machinery could allow it to escape intracellular proteolysis even if no degradation band was observed. However, the SE was low (35%), possibly due to inefficient translocation of a cytoplasmic protein, or to inefficient precursor processing; inefficient export of a cytoplasmic protein was previously observed in E. coli (8).
To enhance production and secretion of L7/L12 in L. lactis, we fused L7/L12 with the staphylococcal nuclease (Nuc). We showed that the fusion with Nuc resulted in a 2.5-fold enhancement of the total amount of fusion protein with a similar SE compared to the secreted L7/L12. Nuc, reportedly strongly immunogenic, could also reveal some carrier activity towards the immunogenicity of L7/L12 and the capacity of L7/L12 to elicit an immune response will be compared when fused or not to Nuc protein.
The synthetic propeptide LEISS described as a secretion enhancer (23, 24) was also used to improve the L7/L12 secretion. LEISS alters the N terminus of the mature fusion protein by introducing two negative charges at positions +2 and +8 (23). Analysis of the peptidic sequence of the N-terminal part of L7/L12 revealed a net global charge of −2 in the first 10 amino acid residues. The observed SE of LEISS:L7/L12 was 50%, compared to 35% for L7/L12. However, the major effect of LEISS was the 2.7-fold increase in the amount of mature LEISS-L7/L12 protein secreted in the extracellular medium in comparison to L7/L12 protein alone.
The fusion of L7/L12 with the cell wall anchor (CWA) region of the Streptococcus pyogenes M6 protein led to an efficient anchoring of L7/L12. This construction will be interesting for in vivo tests since the cell wall anchoring of L7/L12 may be advantageous because the bacterial cell may provide an adjuvant activity that would enhance the immunological response of hosts (46). Another advantage is that the anchored L7/L12 may be less exposed to degrading or denaturing agents such as proteinases or acid-rich environments such as the stomach of man and animals.
Our results provide a means of improving yields and stability of a protein for in vitro and possibly in vivo presentation. They indicate that L. lactis is able to produce recombinant L7/L12 protein that possesses antigenic properties. These strains will be very useful to explore new strategies of vaccination against brucellosis. Such a system would represent a good alternative to the commonly studied live bacterial vectors that are mostly derived from pathogenic invasive microorganisms such as Salmonella spp., Vibrio cholerae, Mycobacterium spp., and B. abortus.
At present, immunization assays are under investigation with these constructions; this will allow definition of the construction that will give the highest immune response and protection against challenge with B. abortus.
Acknowledgments
We are grateful to Stefania Usai, who kindly gave us the plasmid pVE5547. We also thank Sébastien Nouaille, Jacqueline Commissaire, Jean-Jacques Gratadoux, and Anderson Miyoshi for the scientific discussion and their friendly support.
This research was supported by grants from Ministère de l'Education Nationale, de la Recherche de la Technologie (MENRT, France), and from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasilia, Brazil).
REFERENCES
- 1.Baloglu, S., T. E. Toth, G. G. Schurig, N. Sriranganathan, and S. M. Boyle. 2000. Humoral immune response of BALB/C mice to a vaccinia virus recombinant expressing Brucella abortus GroEL does not correlate with protection against a B. abortus challenge. Vet. Microbiol. 76:193-199. [DOI] [PubMed] [Google Scholar]
- 2.Bermúdez-Humarán, L. G., P. Langella, A. Miyoshi, A. Gruss, R. Tamez Guerra, R. Montes de Oca-Luna, and Y. Le Loir. 2002. Production of human papillomavirus type 16 E7 protein in Lactococcus lactis. Appl. Environ. Microbiol. 68:917-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boschiroli, M.-L., V. Foulongne, and D. O'Callaghan. 2001. Brucellosis: a worldwide zoonosis. Curr. Opin. Microbiol. 4:58-64. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain, L., J. M. Wells, R. Robinson, K. Schofield, and R. Le Page. 1997. Mucosal immunization with recombinant Lactococcus lactis, p. 83-106. In G. Pozzi and J. M. Wells (ed.), Gram-positive bacteria as vaccine vehicles for mucosal immunization. Springer-Verlag and Landes Bioscience, Austin, Tex.
- 5.Chatel, J.-M., P. Langella, K. Adel-Patient, J. Commissaire, J. M. Wal, and G. Corthier. 2001. Induction of local immune response after intranasal and oral inoculation of mice with Lactococcus lactis producing bovine beta-lactoglobulin. Clin. Diagn. Lab. Immunol. 8:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheville, N. F. 2000. Development, testing and commercialization of a new brucellosis vaccine for cattle. Ann. N. Y. Acad. Sci. 916:147-153. [DOI] [PubMed] [Google Scholar]
- 7.Corner, L. A., and G. G. Alton. 1981. Persistence of Brucella abortus strain 19 infection in adult cattle vaccinated with reduced doses. Res. Vet. Sci. 31:342-344. [PubMed] [Google Scholar]
- 8.Debarbieux, L., and J. Beckwith. 1998. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc. Natl. Acad. Sci. USA 95:10751-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieye, Y., S. Usai, F. Clier, and J. C. Piard. 2001. Design of a protein targeting system for lactic acid bacteria. J. Bacteriol. 183:4157-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drouault, S., G. Corthier, S. D. Ehrlich, and P. Renault. 1999. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 65:4881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enouf, V., P. Langella, J. Commissaire, J. Cohen, and G. Corthier. 2001. Bovine rotavirus nonstructural protein 4 produced by Lactococcus lactis is antigenic and immunogenic. Appl. Environ. Microbiol. 67:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded proteins in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, T. J. 1984. Ph.D. thesis. University of Cambridge, Cambridge, England.
- 15.Gilbert, C., K. Robinson, R. W. Le Page, and J. M. Wells. 2000. Heterologous expression of an immunogenic pneumococcal type 3 capsular polysaccharide in Lactococcus lactis. Infect. Immun. 68:3251-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilloteau, L. A., K. Laroucau, N. Vizcaino, I. Jacques, and G. Dubray. 1999. Immunogenicity of recombinant Escherichia coli expressing the omp31 gene of Brucella melitensis in BALB/C mice. Vaccine 17:353-361. [DOI] [PubMed] [Google Scholar]
- 17.Hirt, H., S. L. Erlandsen, and G. M. Dunny. 2000. Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance Asc10 in Lactococcus lactis and Streptococcus gordonii contributes to cell hydrophobicity and adhesion to fibrin. J. Bacteriol. 182:2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreeger, T. J., M. W. Miller, M. A. Wild, P. H. Elzer and S. C. Olsen. 2000. Safety and efficacy of Brucella abortus strain RB51 vaccine in captive pregnant elk. J. Wildl. Dis. 36:477-483. [DOI] [PubMed] [Google Scholar]
- 19.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezen, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 20.Langella, P., and Y. Le Loir. 1999. Heterologous protein secretion in Lactococcus lactis: a novel antigen delivery system. Braz. J. Med. Biol. Res. 32:191-198. [DOI] [PubMed] [Google Scholar]
- 21.Langella, P., Y. Le Loir, S. D. Ehrlich, and A. Gruss. 1993. Efficient plasmid mobilization by pIP501 in Lactococcus lactis subsp. lactis. J. Bacteriol. 175:5806-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Loir, Y., A. Gruss, S. D. Ehrlich, and P. Langella. 1994. Direct screening of recombinants in gram-positive bacteria using the secreted staphylococcal nuclease as a reporter. J. Bacteriol. 176:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Loir, Y., A. Gruss, S. D. Ehrlich, and P. Langella. 1998. A nine-residue synthetic propeptide enhances secretion efficiency of heterologous proteins in Lactococcus lactis. J. Bacteriol. 180:1895-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Loir, Y., S. Nouaille, J. Commissaire, L. Brétigny, A. Gruss, and P. Langella. 2001. Signal peptide and propeptide optimization for protein secretion in Lactococcus lactis. Appl. Environ. Microbiol. 67:4119-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norton, P. M., H. W. Brown, and R. W. Le Page. 1994. The immune response to Lactococcus lactis: implications for its use as a vaccine delivery vehicle. FEMS Microbiol. Lett. 120:249-256. [DOI] [PubMed] [Google Scholar]
- 26.Norton, P. M., H. W. Brown, J. M. Wells, A. M. Macpherson, P. W. Wilson, and R. W. Le Page. 1996. Factors affecting the immunogenicity of tetanus toxin fragment C expressed in Lactococcus lactis. FEMS Immunol. Med. Microbiol. 14:167-177. [DOI] [PubMed] [Google Scholar]
- 27.Norton, P. M., J. M. Wells, H. W. Brown, A. M. Macpherson, and R. W. Le Page. 1997. Protection against tetanus toxin in mice nasally immunized with recombinant Lactococcus lactis expressing tetanus toxin fragment C. Vaccine 15:616-619. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira, S. C., and G. A. Splitter. 1994. Subcloning and expression of the Brucella abortus L7/L12 ribosomal gene and T-lymphocyte recognition of the recombinant protein. Infect. Immun. 62:5201-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira, S. C., and G. A. Splitter. 1996. Immunization of mice with recombinant L7/L12 ribosomal protein confers protection against Brucella abortus infection. Vaccine 14:959-962. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira, S. C., J. S. Harms, M. Banai, and G. A. Splitter. 1996. Recombinant Brucella abortus proteins that induce proliferation and gamma-interferon secretion by CD4+ T cells from Brucella-vaccinated mice and delayed-type hypersensitivity in sensitized guinea pigs. Cell. Immunol. 172:262-268. [DOI] [PubMed] [Google Scholar]
- 31.Onate, A. A., R. Vemulapalli, E. Andrews, G. G. Schurig, S. Boyle, and H. Folch. 1999. Vaccination with live Escherichia coli expressing Brucella abortus Cu/Zn superoxide dismutase protects mice against virulent B. abortus. Infect Immun. 67:986-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piard, J.-C., R. Jimenez-Diaz, V. A. Fischetti, S. D. Ehrlich, and A. Gruss. 1997. The M6 protein of Streptococcus pyogenes and its potential as a tool to anchor biologically active molecules at the surface of lactic acid bacteria. Adv. Exp. Med. Biol. 418:545-550. [DOI] [PubMed] [Google Scholar]
- 33.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Que, Y. A., J. A. Haefliger, P. Francioli, and P. Moreillon. 2000. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect. Immun. 68:3516-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson, K., L. M. Chamberlain, K. M. Schofield, J. M. Wells, and R. W. Le Page. 1997. Oral vaccination of mice against tetanus with recombinant Lactococcus lactis. Nat. Biotechnol. 15:653-657. [DOI] [PubMed] [Google Scholar]
- 36.Roop, R. M., II, T. W. Fletcher, N. M. Sriranganathan, S. M. Boyle, and G. G. Schurig. 1994. Identification of an immunoreactive Brucella abortus HtrA stress response protein homolog. Infect. Immun. 62:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Shortle, D. 1983. A genetic system for analysis of staphylococcal nuclease. Gene 22:181-189. [DOI] [PubMed] [Google Scholar]
- 39.Simonen, M., and I. Palva. 1993. Protein secretion in Bacillus species. Microbiol. Rev. 57:109-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steidler, L., W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers, and E. Remaut. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289:1352-1355. [DOI] [PubMed] [Google Scholar]
- 41.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Asseldonk, M., W. M. de Vos, and G. Simons. 1993. Functional analysis of the Lactococcus lactis usp45 secretion signal in the secretion of a homologous proteinase and a heterologous alpha-amylase. Mol. Gen. Genet. 240:428-434. [DOI] [PubMed] [Google Scholar]
- 43.van Metre, D. C., G. A. Kennedy, S. C. Olsen, G. R. Hansen, and D. R. Ewalt. 1999. Brucellosis induced by RB51 vaccine in a pregnant heifer. J. Am. Vet. Med. Assoc. 215:1491-1493. [PubMed] [Google Scholar]
- 44.Vemulapalli, R., H. Yongqun, S. Cravero, S. Nammalwar, S. M. Boyle, and G. G. Schurig. 2000. Overexpression of protective antigen as a novel approach to enhance vaccine efficacy of Brucella abortus strain RB51. Infect. Immun. 68:3286-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vemulapalli, R., Yongqun He, S. M. Boyle, N. Sriranganathan, and G. G. Schurig. 2000. Brucella abortus strain RB51 as a vector for heterologous protein expression and induction of specific Th1 type immune responses. Infect. Immun. 68:3290-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitini, E., S. Alvarez, M. Medina, M. Medici, M. V. de Budeguer, and G. Perdigon. 2000. Gut mucosal immunostimulation by lactic acid bacteria. Biocell 24:223-232. [PubMed] [Google Scholar]
- 47.Wells, J. M., P. W. Wilson, P. M. Norton, M. J. Gasson, and R. W. Le Page. 1993. Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol. Microbiol. 8:1155-1162. [DOI] [PubMed] [Google Scholar]
- 48.Wells, J. M., P. M. Norton, and R. W. F. Le Page. 1995. Progress in the development of mucosal vaccines based on Lactococcus lactis. Int. Dairy J. 5:1071-1079. [Google Scholar]
- 49.Young, E. J. 1995. An overview of human brucellosis. Clin. Infect. Dis. 21:283-289. [DOI] [PubMed] [Google Scholar]