Abstract
A highly sensitive and specific PCR-based method of monitoring 16S rRNA genes of Pseudomonas stutzeri was developed for searching P. stutzeri DNA in environmental samples. This monitoring was combined with a reliable and sensitive method for isolating P. stutzeri colony formers from soil and sediment, depending on their utilization of ethylene glycol, starch, and maltose. With these techniques, P. stutzeri populations (n = 2 to 170) were obtained from five of six sites giving positive PCR signals (including three marine sediment and two soil samples). The phylogenetic positions of isolates from the five sites, based on their 16S ribosomal DNA sequences, indicated that the environmental isolates were affiliated with different genomovars of P. stutzeri. Using the broad-host-range plasmid pNS1 with kanamycin and gentamicin resistance determinants as the transforming DNA, naturally transformable strains were identified among the isolates from all sites. For one population from soil, the genetic relationship of the 120 members was determined by randomly amplified polymorphic DNA-PCR with three PCR primers. Among the population members which are taxonomically closely related as determined by 16S sequence comparisons of group representatives, a rather high genetic diversity and a characteristic clustering into subgroups were found. Remarkably, within the population, nontransformability and different levels of transformability (a frequency between about 10−9 and 10−4 per cell) were often associated with distinct genetic subgroups. It is concluded that transformability is widespread among environmental P. stutzeri strains and that its specific level is a heritable trait that may vary strongly within a local population.
Pseudomonas stutzeri is a highly diverse species of great physiological and ecological versatility and widespread geographic distribution. Members of this species have been shown to be involved in nitrification and denitrification processes (48) as well as in the degradation of environmental pollutants (2, 36). Most of the well-characterized strains of P. stutzeri are clinical isolates (32, 35, 40). Recently, P. stutzeri has been isolated from nonhuman vertebrate feces (19), papermaking chemicals (45), and drinking water (30). Strains were also recovered from environmental habitats (3, 35). The potential for natural genetic transformation was found only in a few P. stutzeri strains which were mostly clinical isolates (7, 24), and the type strain (ATCC 17588) is not transformable (7). Strains ZoBell (ATCC 14405), JM300 (DSM 10701), 19SMN4, and DNSP21 are the only nonclinical transformable isolates of P. stutzeri. Several studies of transformability with ZoBell and JM300 have been reported (24, 25, 27, 41). With JM300, evidence was provided that natural transformation can occur in the soil by the addition of DNA and with DNA released in soil by bacteria (39).
In this communication, we describe improved methods for the detection and isolation of P. stutzeri strains. The methods were applied to environmental samples and used to isolate several local populations. Screening of population members for their potential for natural transformation indicated that only some of the isolates are transformable. Moreover, the level of transformability varied widely among the members of one local population and was apparently correlated with clusters of closely related strains.
MATERIALS AND METHODS
Media, plasmid DNA, and isolation of genomic DNA.
For the isolation of P. stutzeri from environmental samples, SW-LB medium plates were used (containing, per liter, 10 g of Bacto Tryptone, 5 g of yeast extract, and 15 g of agar [all from Difco Laboratories, Detroit, Mich.] in artificial seawater containing, per liter, 24 g of NaCl, 10.5 g of MgSO4·7H2O, and 0.11 g of NaHCO3), with 50 mg of cycloheximide added for suppression of fungal growth. Minimal medium (26) prepared with artificial seawater contained either 0.5% (wt/vol) of ethylene glycol, starch, or maltose as a sole carbon source (abbreviated SW-ME, SW-MS, and SW-MM, respectively). The plasmid pNS1 coding for gentamicin resistance (Gmr) (the aacC1 gene from Tn1696) and kanamycin resistance (Kmr) (the nptII gene from Tn5) was constructed as follows. The 1.3-kb NsiI-AlwNI fragment from pBBR1 MCS-2 (23) carrying nptII was cloned into the single BsaI site of pBBR1 MCS-5 carrying aacC1 to give pNS1. Plasmid pUCP-Gm was derived from pUCP19 (38) with the aacC1 gene from Tn1696 on a 700-bp fragment cloned into the ScaI site.
Genomic DNA from P. stutzeri (ATCC 17588T), Pseudomonas alcaligenes (ATCC 14909T), Pseudomonas mendocina (ATCC 25411T), Pseudomonas corrugata (ATCC 29736T), and Pseudomonas balearica (DSM 6083T) was prepared according to Marmur (28). Plasmid pNS1 was isolated from Escherichia coli K-12 SF8 using the Plasmid Midi kit (Qiagen, Hilden, Germany).
Extraction of total DNA from environmental samples.
Material from soil or sediment (200 mg) was resuspended in 1.4 ml of extraction buffer (250 mM NaCl, 100 mM EDTA [pH 8.0], 2% sodium dodecyl sulfate) and vortexed thoroughly for 15 s. After addition of 0.1 ml of a 5 M guanidine thiocyanate solution (Sigma, Deisenhofen, Germany) in 0.1 M Tris-HCl, pH 7.5, the suspension was vortexed for 15 s and incubated for 70 min with a sonification step (3 min in a Sonifier 250; Branson Ultrasonics, Danbury, Conn.) after 10 min. The solids were sedimented (15 min at 14,000 × g). The DNA in the supernatant was precipitated with isopropanol, washed with 70% ethanol, and dissolved in 100 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). For further purification, 50 μl of DNA solution was thoroughly mixed with 200 μl of acid-washed polyvinylpolypyrrolidone (PVPP; Sigma) and incubated for 5 min at 20°C. The DNA was separated from PVPP by centrifugation (Ultrafree MC Centrifugal filter unit; Millipore Corporation, Bedford, Mass.) for 60 s at 14,000 × g. The filtrate was mixed with a buffer (50 mM MOPS [morpholinepropanesulfonic acid], 750 mM NaCl [pH 7.5]) to give a final volume of 1 ml and loaded onto a minicolumn from the Qiagen Plasmid DNA Purification kit. After being washed seven times with 2-ml portions of buffer QC (Qiagen), the DNA was eluted according to the manufacturer’s instructions, precipitated with isopropanol, washed with 70% ethanol, and dissolved in 50 μl of TE buffer.
Isolation of environmental strains and growth conditions.
From a suspension of about 0.5 g of soil or sediment in 5 ml of liquid SW-LB, 200-μl portions were plated on SW-LB. After incubation for 16 h at 28°C, the colonies were replica plated on SW-ME, SW-MS, and SW-MM and incubated (for 5 days at 28°C). Colonies growing on SW-ME and additionally on SW-MM and/or SW-MS were streaked at least once on SW-ME and then at least once on SW-LB for single-colony isolation. Cells of the purified isolates were identified as P. stutzeri by PCR with samples of overnight cultures. Cells from overnight culture in SW-LB (28°C) were stored in 10% glycerol at −80°C.
PCR with a primer pair for specific amplification of a region of P. stutzeri 16S ribosomal DNA (rDNA).
PCR analysis with genomic DNA was performed with 20-μl reaction mixtures containing 1 μl of DNA solution (about 2 ng), primers fps158 (5"-GTGGGGGACAACGTTTC-3") and rps743minus4 (5"-TCAGTGTCAGTATTAGC-3") at 1 μM, the four deoxyribonucleoside triphosphates (dNTPs) at 50 μM each (Pharmacia, Freiburg, Germany), 3 mg of bovine serum albumin (Behringwerke AG, Marburg, Germany) per ml, and 0.5 U of REDTaq DNA polymerase (Sigma) in the supplied reaction buffer. The cycling program was 5 min at 94°C; 40 cycles of 2 min at 94°C, 2 min at 57°C, 3 min at 72°C; and a final step of 10 min at 72°C (Robocycler; Stratagene, Amsterdam, The Netherlands). PCR with samples of overnight cultures was carried out similarly to PCR with purified DNA except that rps743 (5"-CACCTCAGTGTCAGTATTAGC-3") was used as a reverse primer and the annealing temperature was 63°C. Generally, 2 μl of washed overnight culture was used.
Amplification and sequencing of 16S rDNA genes and determination of phylogenetic position.
Amplification of 16S rRNA genes was performed with 150 μl of reaction buffer with 6 μl of DNA (prepared with the GeneReleaser kit) (Eurogentec, Seraing, Belgium), 5% dimethyl sulfoxide, primers fD1 and rD1 at 1 μM each (47), the four dNTPs at 50 μM each (Pharmacia), and 1.0 U of Taq DNA polymerase (catalogue no. M2868; Promega, Mannheim, Germany). The cycling program was 5 min at 92°C; 30 cycles of 2 min at 92°C, 2 min at 58°C, and 3 min at 70°C; and a final step at 70°C for 10 min (Perkin-Elmer 480 DNA ThermoCycler). The PCR product was sequenced using primers GTATTACCGCGGCTGCTGGC and CAGCAGCCGCGGTAATAC (both spanning E. coli numbering positions 517 to 536 in forward and backward directions) and primer CTCCTACGGGAGGCAGCAG (E. coli numbering positions 339 to 357). Alignment of sequences was performed with CLUSTAL X, version 1.64b (44) using default parameters (gap opening, 10.00; gap extension, 0.05; delay divergent sequences, 40%; DNA transition weight, 0.50). The alignment was corrected manually. Determination of Jukes-Cantor distances and cluster analysis using a neighbor-joining algorithm were performed with TREECON software (46).
Plate transformation assay.
This assay was performed essentially as described (13). An aliquot (40 μl) from a fresh overnight culture of an isolate in SW-LB was mixed with 1,000 ng of pNS1 DNA (or other DNA) to a final volume of 50 μl and spotted onto an SW-LB agar plate. After incubation (normally 24 h at 37°C if not stated otherwise), the piece of agar with the spot of cells was transferred to a glass tube with 1 ml of SW-LB. The tubes were vigorously vortexed to resuspend the cells from the agar (final cell titer, ∼1010 cells/ml). When the cells did not resuspend easily, the cell material was scraped off with a scalpel from the agar and resuspended in a sterile Potter-Elvehjem microhomogenizer. From the suspension, 200 μl was plated for transformants on SW-LB with 5 μg of gentamicin/ml (or 50 μg of kanamycin/ml) and incubated (3 days at 37°C). Cell material from the colonies was streaked for single-colony growth on SW-LB with kanamycin (or gentamicin) to verify the presence of pNS1. The total viable count on SW-LB plates was determined. Control experiments were performed identically except that plasmid DNA was omitted. Transformation frequencies are given as the number of Kmr or Gmr transformants per viable count. The frequency of spontaneous Kmr or Gmr mutants was ≤10−9 to ≤10−10.
RAPD-PCR.
The reactions for randomly amplified polymorphic DNA (RAPD)-PCR were carried out with 25-μl volumes containing 1 μl of DNA prepared with the GeneReleaser kit (Eurogentec) from a fresh overnight culture; one of the following primers (0.2 μM): (i) 5"-CGAGCTTCGCGTACCACCCC-3", (ii) 5"-GTTTCGCTCGATGCGCTACC-3", or (iii) 5"-CGGCACACTGTTCCTCGACG-3"; Taq DNA polymerase (1 U, M2868; Promega); and dNTPs (100 μM each; Pharmacia) under a drop of mineral oil (Sigma) in reaction buffer (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, and 0.2 mg of bovine serum albumin per ml). Four cycles at 94, 40, and 70°C for 5 min each were run, followed by 30 cycles at 94 and 55°C for 1 min each and at 70°C for 2 min, with a final primer extension cycle at 70°C for 5 min with an MJ Research PTC-100 cycler (Biozym Diagnostik, Hessisch Oldendorf, Germany). The PCR products were separated by agarose gel (1.3%) electrophoresis. A single master mixture including all PCR components except DNA polymerase and template DNA was used for all isolates. Several control experiments were performed, including separate RAPD-PCR runs on the same and/or separate overnight cultures of the same strain and electrophoresis of RAPD-PCR products with different agarose gels to ensure the reproducibility of the RAPD patterns. A size marker (Ladder Mix; MBI Fermentas, St. Leon-Rot, Germany) was used as a reference in all gels. The software Gene ImagIR, version 3.52 (Scanalytics, Inc., Fairfax, Va.) was used to analyze the RAPD pattern, and the results were exported into TREECON software (46) to generate an unweighted pair group method with arithmetic mean dendrogram from a distance matrix (29).
Nucleotide sequence accession numbers.
The accession numbers of the 16S sequences of the environmental isolates reported here are AJ312175, AJ270452, AJ312158, AJ312160, AJ270456, AJ270451, AJ312165, AJ319662, AJ312161, AJ312162, AJ312164, AJ312166, AJ312167, AJ312168, AJ312169, and AJ312171 (EMBL database). The accession numbers of the 16S sequences of the reference strains are U26415, AJ006108, U22427, U25280, AF063219, AJ006105, AJ005167, U26419, U26261, U25431, U26420, X98607, AJ006106, AJ006103, U26416, U26414, U26262, U25432, AJ006104, U58660, U26418, U26417, AF054936, AJ006107, Z76659, Z76668, Z76651, and J01695.
RESULTS
Detection of P. stutzeri DNA in environmental samples by PCR.
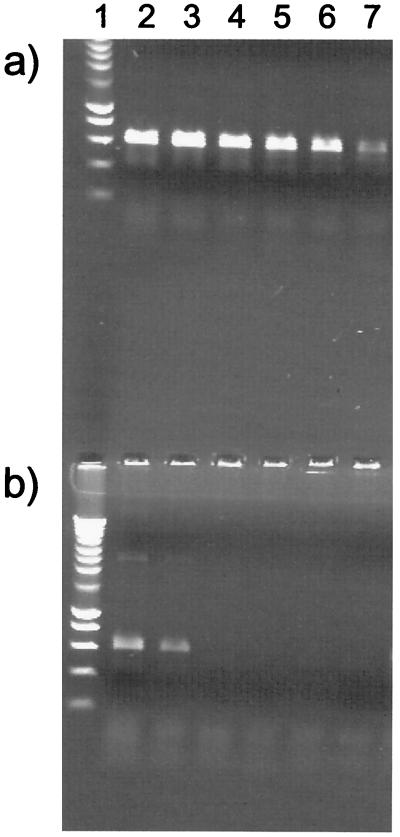
We established a PCR protocol for the analysis of environmental samples in the presence of P. stutzeri DNA. The protocol is based on the highly specific amplification of a region of the 16S rRNA gene of P. stutzeri. A primer pair for this purpose consisting of fps158 and rps743 (Fig. 1) was recently described (3). We increased the specificity of the protocol for P. stutzeri relative to other bacteria (Fig. 1) by shortening the primer rps743 at the 5" end by four nucleotides (primer rps743minus4), which was expected to increase the destabilizing effect of mismatches at the 3" site during primer binding to 16S rRNA genes of bacteria other than P. stutzeri. Further, we used an adapted PCR protocol which allows for an particularly efficient amplification of DNA isolated from the environment (which often contains traces of humic substances known to inhibit PCR), owing to the presence of a very high concentration of bovine serum albumin (34). Under these conditions, an amplification product with the specific primers was obtained with DNA amounts corresponding to only 5 genome equivalents of the P. stutzeri type strain (Fig. 2). The specificity of the PCR amplification with the new primer pair was tested with DNA of various Pseudomonas species, including P. alcaligenes, P. mendocina, P. corrugata, and P. balearica, which are most similar to P. stutzeri at their primer binding sites (Fig. 1). With DNA from these species, 5 × 104 genome equivalents (the P. alcaligenes type strain) (Fig. 2) or more than 5 × 105 genome equivalents (the P. mendocina, P. corrugata, and P. balearica type strains) (data not shown) were required to generate a false-positive signal. The primers depicted in Fig. 1 match perfectly the 16S sequences of all P. stutzeri strains shown in Fig. 3 with the exception of DSM 10701 and ATCC 17641, which have a single mismatch in fps158. Thus, they encompass all seven genomovars (4, 37). Environmental samples from several locations in different habitats were collected (marine sediments, agricultural soils, and other soils), and total DNA was extracted and purified. With this DNA, a PCR product of the expected size was obtained from six of nine samples (Table 1). The absence of a PCR product with the DNA of three of the samples was not due to PCR-inhibiting substances, since the addition of DNA from the P. stutzeri type strain (5 genome equivalents per assay) generated the expected PCR product (Table 1). The results of the PCR monitoring suggested that in samples giving the PCR signal, P. stutzeri cells were present and isolates would be obtained. This was the case in five of six samples (see below).
FIG. 1.
Position of mismatches in the P. stutzeri annealing sites for primers fps158 and rps743 in the 16S rDNA sequences of several type strains from related species. A dot represents an identical base. The nucleotide positions (E. coli) of primer annealing sites were 142 to 158 (fps158) and 743 to 763 (rps743).
FIG. 2.
Specificity and sensitivity of the primer pair fps158 and rps743minus4 with chromosomal DNA of the P. stutzeri type strain (a) and the P. alcaligenes type strain (b). The amounts of template DNA were approximately 5 × 105 (lane 2), 5 × 104 (lane 3), 5 × 103 (lane 4), 5 × 102 (lane 5), 50 (lane 6), and 5 genome equivalents (lane 7). The PCR product was 625 bp. Lane 1 contains molecular weight markers (Smart Ladder; Eurogentec).
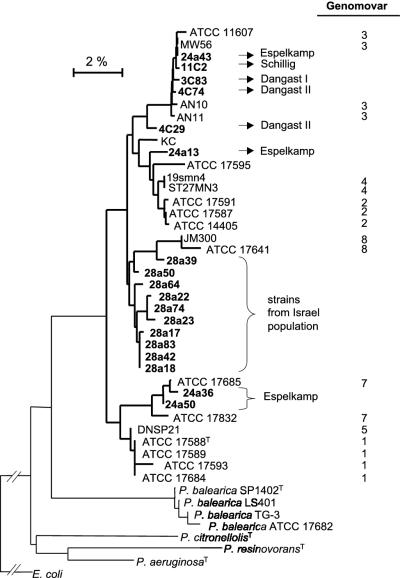
FIG. 3.
Tree based on 16S rDNA, reflecting the phylogenetic relationship of the type strain and selected isolates of P. stutzeri. The tree is based on neighbor-joining analysis of Jukes-Cantor distances. All strains in the dendrogram are P. stutzeri, unless stated otherwise. The bar corresponds to 0.02 Jukes-Cantor substitution per nucleotide.
TABLE 1.
Detection of P. stutzeri in environmental samples
| Habitat | Locationa | PCR product in total DNAb |
P. stutzeri isolatesc
|
|||
|---|---|---|---|---|---|---|
| Total no. of isolates | No. tested/no. identified by PCR | No. of isolates/g of sample material | No. of colonies/total coloniesd | |||
| North Sea sediment | Dangast I (G) | +, (+) | 139 | 139/139 | ∼600 | 1/∼1,160 |
| Dangast II (G) | +, (+) | 170 | 17/17 | ∼2,073 | 1/∼430 | |
| Schillig (G) | +, (+) | 2 | 2/2 | ∼24 | 1/∼9,100 | |
| Hooksiel (G) | −, (+) | 0 | NA | NA | NA | |
| Soil contaminated with mineral oil | Filling station, Espelkamp (G) | +, (+) | 118 | 20/20 | ∼1,370 | 1/∼3,330 |
| Agricultural soil | Kiel I (G) | −, (+) | 0 | NA | NA | NA |
| Kiel II (G) | +, (+) | 0 | NA | NA | NA | |
| Quedlinburg (G) | −, (+) | 0 | NA | NA | NA | |
| Soil | Tel Aviv airport area (I) | +, (+) | 120 | 10/10 | ∼1,800 | 1/∼1,600 |
All samples were from Germany (G) or Israel (I). The location name refers to the town located at the sampling place. Dangast I and Dangast II were sampled about 2 m apart.
+, a specific PCR product was obtained; (+), a specific PCR product was obtained when five genome equivalents of P. stutzeriT DNA were added to the PCR assay; no PCR product obtained.
NA, not applicable.
On rich medium agar plates.
Isolation of P. stutzeri from environmental samples.
The environmental samples were subjected to an isolation procedure for viable P. stutzeri cells. The isolation procedure included (i) plating of sample material on rich medium agar, (ii) replica plating of total colonies on three minimal medium plates with ethylene glycol, maltose, or starch, respectively, as the sole carbon source, and (iii) identification of colonies grown on plates with ethylene glycol (which were also present on maltose and/or starch medium) by PCR analysis with the specific PCR primers. In this way, P. stutzeri was isolated from five of six samples giving a P. stutzeri-specific PCR signal (Table 1). Colonies identified as P. stutzeri by PCR were of a yellow to brownish color and showed the dry and wrinkled morphology on ethylene glycol medium which was previously attributed to fresh P. stutzeri isolates (35). Colonies with a different color or colony morphology did not yield the expected PCR signal. The 139 colonies identified as P. stutzeri on SW-ME plates inoculated from the Dangast I sample all proved to be P. stutzeri by specific PCR analysis. Thus, P. stutzeri colonies were recognized rather easily and reliably. This encouraged routine isolation of P. stutzeri, during which not all of the 549 isolates obtained were verified by PCR (Table 1). Five populations consisting of 139, 170, 118, 120, and 2 isolates were obtained (Table 1). The colonies within each population varied considerably in color (different nuances between yellow and brown at different intensities) and colony morphology (different degrees of wrinkling or indenting of colonies). There was no class of colony morphology or color that was typical of the P. stutzeri isolates from a site. The frequency of P. stutzeri cells was up to 2.1 × 103 per g of sample material. The isolation procedure turned out to be highly efficient, since one P. stutzeri colony was detected among 9,100 colonies of other bacteria (Table 1). P. stutzeri could not be isolated from the Kiel II sample, although the DNA recovered from the sample gave a signal in the monitoring by PCR. Perhaps the PCR resulted from dead cells or from DNA generating a false-positive signal. As well, P. stutzeri could not be isolated from samples without a specific PCR product.
Phylogenetic positions of selected isolates.
Phylogenetic comparison of the 16S sequences (approximately 1,450 nucleotides) of randomly chosen strains of the five populations with 16S sequences of P. stutzeri reference strains representing the sequence diversity characteristic of this species confirmed that the isolates belong to P. stutzeri (Fig. 3). It has previously been shown that positioning in a phylogenetic tree based on 16S sequences correlates well with the grouping of strains into genomic subgroups termed genomovars (4, 37, 40). Based on this, strains 24a43 (Espelkamp), 11C2 (Schillig), 3C83 (Dangast I), and 4C74 (Dangast II) (sampling sites are given in parentheses) belong to genomovar 3 and strains 24a36 and 24a50 from Espelkamp belong to genomovar 7 (Fig. 3). Based solely on 16S sequence comparisons, strains 4C29 (Dangast II) and 24a13 (Espelkamp) and the strains of the Israel population cannot be assigned to any of the known genomovars. The more detailed analysis of the Israel population by 16S sequence comparison of RAPD cluster representatives (see below and Fig. 4) showed that the majority of strains (>85%) belonged to a genomically closely related group probably constituting a separate genomovar. Another group of strains (represented by 28a39 and 28a50) (Fig. 3 and 4) needs further analysis to clear up its genomovar grouping.
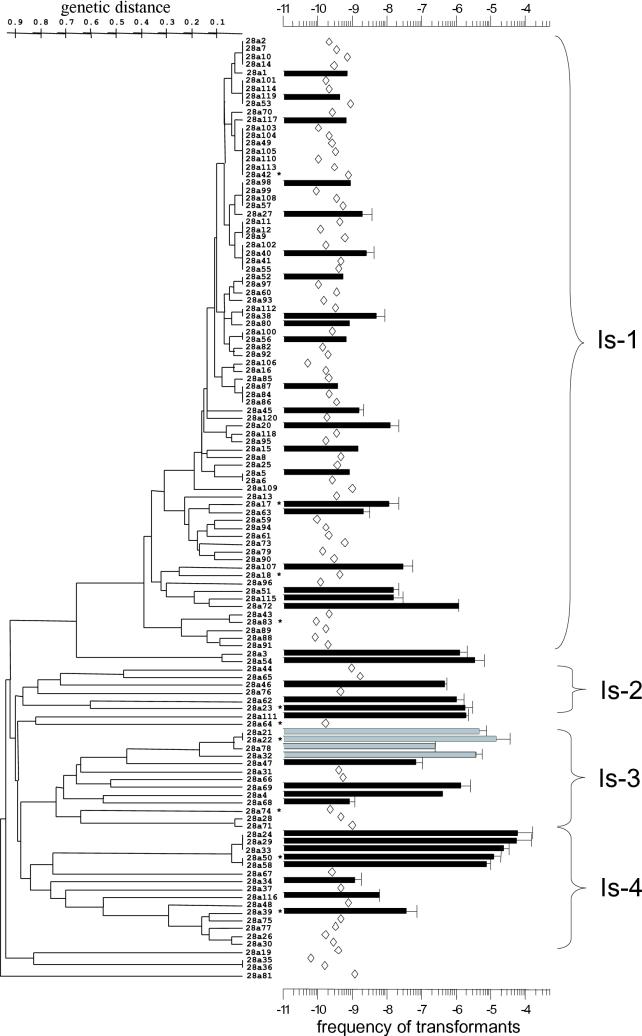
FIG. 4.
The 120 strains of a local soil population from Israel are grouped according to a phenetic analysis based upon RAPD data obtained with three primers. The scale on the left gives the genetic distance. The 16S sequence (approximately 1,450 nucleotides) was determined for the strains marked with asterisks. The scale on the right gives the transformation frequency determined with pNS1-DNA (Kmr transformants). Black bars indicate that Kmr clones were obtained and were verified as transformants by the simultaneous acquisition of Gmr. Gray bars indicate that the transformants were not stable (see the text). Open diamonds show that Kmr clones were not obtained and therefore indicate the limit of detection. The data are the means of two to three experiments (given with deviation from the mean or standard deviation). The clusters Is-1 to Is-4 are discussed in the text.
Natural transformation of P. stutzeri isolates by plasmid DNA.
P. stutzeri was considered to be transformable by plasmid DNA only when the plasmid contained an insert of chromosomal P. stutzeri DNA (8). Later it was found that a broad-host-range plasmid (RSF1010 carrying a streptomycin resistance determinant) without a chromosomal insert transformed P. stutzeri JM300 (6). Transformation of JM300 by the broad-host-range plasmid pNS1 carrying determinants for Gmr and Kmr was as efficient as that by RSF1010 and followed a one-hit mechanism (i.e., a single pNS1 molecule sufficed to generate a transformant). This allowed us to assess the transformability of isolates quantitatively by using plasmid DNA and circumvented the need for the isolation of an antibiotic-resistant mutant of each isolate and preparation of its DNA before the transformability of the isolate was determined. Randomly chosen isolates from each population (five to seven each) and the two isolates from the Schillig sample site were tested with pNS1 DNA for transformability in the quantitative plate assay. In this assay, an inoculum of about 3 × 108 cells is allowed to grow in one spot on a nonselective broth plate for several generations in the presence of transforming DNA. During this time, the cells pass through the DNA uptake-competent stage and finally reach the stationary phase (about 2 × 109 to 1 × 1010 cells). The results with the 27 isolates indicated that transformable strains were present in each population (Table 2). Altogether, about one-third of the strains was considered nontransformable. The Kmr strains obtained after treatment with pNS1 were verified as transformants by the simultaneous acquisition of the second plasmid resistance determinant, Gmr (data not shown). The two isolates from the Schillig site had a rather similar transformation frequency. Remarkably, among the members of each of the other populations, the transformability differed by up to 3 or 4 orders of magnitude (Table 2). It appears that transformability is a rather variable trait among environmental P. stutzeri isolates. The transformation levels of the isolates, however, were constant even after several transfers on fresh agar plates.
TABLE 2.
Frequency of transformants with 1 μg of plasmid pNS1 per transformation assay
| Sampling site and strain | Frequency |
|---|---|
| Dangast I | |
| 3C35 | 1.8 × 10−7 ± 4.0 × 10−8 |
| 3C42 | 3.1 × 10−6 ± 2.9 × 10−6 |
| 3C44 | 1.5 × 10−5 ± 9.0 × 10−6 |
| 3C48 | 1.2 × 10−9 ± 1.0 × 10−10 |
| 3C53 | ≤3.1 × 10−9 |
| 3C83 | ≤4.8 × 10−10 |
| Dangast II | |
| 4C33 | ≤1.4 × 10−9 |
| 4C52 | ≤7.9 × 10−10 |
| 4C99 | ≤8.4 × 10−9 |
| 4C29 | 3.2 × 10−8 ± 2.5 × 10−8 |
| 4C74 | 7.2 × 10−7 ± 6.9 × 10−7 |
| Israel | |
| 28a14 | ≤3.0 × 10−10 |
| 28a39 | 3.7 × 10−8 ± 3.6 × 10−8 |
| 28a40 | 2.5 × 10−9 ± 1.9 × 10−9 |
| 28a45 | 1.6 × 10−9 ± 6.0 × 10−10 |
| 28a50 | 1.2 × 10−5 ± 7.8 × 10−6 |
| 28a59 | ≤9.6 × 10−11 |
| 28a72 | 1.2 × 10−6 ± 0 |
| Espelkamp | |
| 24a22 | 4.4 × 10−8 ± 1.6 × 10−8 |
| 24a26 | 3.0 × 10−6 ± 9.0 × 10−7 |
| 24a30 | 1.7 × 10−8 ± 7.0 × 10−9 |
| 24a45 | 1.3 × 10−5 ± 4.0 × 10−6 |
| 24a43 | 7.9 × 10−6 ± 7.1 × 10−6 |
| 24a36 | 4.8 × 10−9 ± 3.7 × 10−9 |
| 24a50 | ≤7.0 × 10−9 |
| Schillig | |
| 11C1 | 3.0 × 10−6 |
| 11C2 | 1.2 × 10−6 |
The level of transformability correlates with clusters of related strains within a population.
RAPD analysis is a powerful method for examining the genetic relationship between the members of a population (5, 18, 40). We used three primers for the RAPD analysis of the 120 members of the population from sampling sites in Israel, which provided altogether 15 ± 4 (mean ± standard error) bands per strain. From this data, the dendrogram shown in Fig. 4 was generated, showing the strains for which 16S sequences were determined. As discussed above, they indicate that the strains of the population constitute a genomically closely related group (Fig. 3). According to the RAPD data, the majority of the isolates (n = 78) are relatively closely related (with a distance of less than about 0.4 on the genetic dissimilarity scale) (Fig. 4) and form the main cluster, designated Is-1, of the population. Most of the remaining strains were grouped into several clusters (Is-2 to Is-4) which are separated from each other and the few nongrouped strains by branch distances of up to 0.9 on the genetic dissimilarity scale. Relatively few strains were identical in their RAPD pattern, suggesting considerable genetic diversity in the population.
By application of our quantitative test for natural transformation to all members of the population (Fig. 4), three observations were made. First, about one-third of the population members were considered transformable by the test in which the limit of detection was at a transformation frequency between about 10−9 and 10−10. Second, transformable strains were present within each major cluster of the population. The transformation frequency of the cluster Is-1 strains was about 10−8 to 10−7 (except for strain 28a72, which had a frequency of 10−6) and was 100- to 1,000-fold lower than that of the five strains of the Is-4 subcluster, including strain 28a50 (almost 10−4). The majority of transformable strains in clusters Is-2 and Is-3 had intermediate levels of transformability. Third, genetically closely related strains positioned side by side in the dendrogram often had similar levels of transformability. This was also seen with a subcluster of four isolates of Is-3 (Fig. 2) which were rather highly transformable but in which Kmr was lost upon subsequent transfer of transformants to fresh selective plates, perhaps as a result of plasmid instability (Gmr was lost in parallel). The distribution of the transformation levels within the population followed rather closely the pattern of genotypic relatedness. However, there were also cases in which a nontransformable strain appeared in a cluster of transformable strains (as in Is-2) or a transformable strain appeared within a group of nontransformable strains (as in Is-4) (Fig. 2).
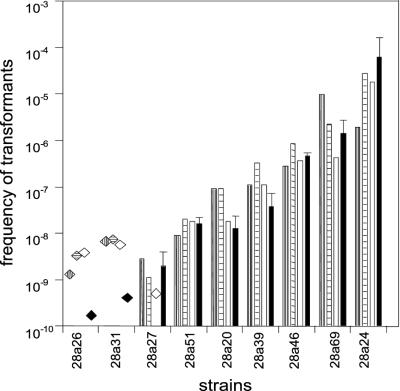
Several different experiments were performed which verified that the different transformation levels of the strains were real and did not result from any specific characteristics of the transformation procedure or of pNS1. First, the same pattern of transformation frequencies with the strains was obtained when the primary selection was for Gmr instead of Kmr (as shown in Fig. 4). Thus, the selection for a different antibiotic resistance marker did not affect the measurement of transformability. Second, different kinetics of competence development of the strains during plate transformation that could influence their transformation frequencies were not observed in nine strains showing high to low or no levels of transformation (Fig. 5). With each of these strains, the characteristic transformation frequency was obtained when the plate transformation was terminated after 6, 12, or 24 h. Third, the transformation level pattern shown in Fig. 4 and 5 did not result from different plasmid replicon establishment in the strains because the same pattern of high to low or no levels of transformation that were obtained with pNS1 were similarly obtained with pUCP-Gm (Fig. 6). This plasmid differs from the pBBR-derived pNS1 in having a Pseudomonas-specific origin of replication in addition to a ColE1-type origin (38). With a third plasmid derived from RP4 (21), pRK415-Km, the transformation frequencies were generally about 100-fold lower than those shown in Fig. 6. Therefore, transformation of strains with transformability of 10−6 or lower with pNS1 fell below detection level. The strains classified as nontransformable with pNS1 were also found to be nontransformable with plasmids pUCP-Gm and pRK415-Km. Finally, by applying a transformation test with chromosomal DNA (24), the high or intermediate transformability of several strains observed with pNS1 plasmid DNA was also obtained with the chromosomal DNA fragments (J. Sikorski and W. Wackernagel, unpublished data).
FIG. 5.
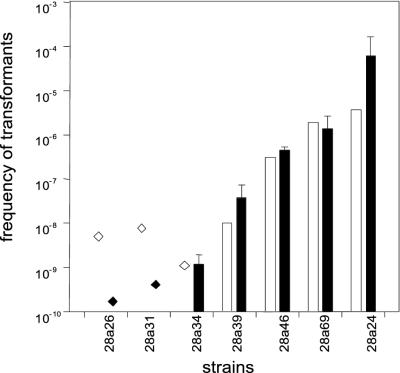
Kinetics of the development of transformation frequencies during plate transformation of nine members of the Israel population having different transformation-level phenotypes. The transforming plasmid was pNS1 and selection was for Gmr. Plate transformation was terminated after 6 h (vertically striped bars), 12 h (horizontally striped bars), and 24 h (open bars). The data for each strain from Fig. 4 are included (black bars). The transformation frequencies of strains 28a26 and 28a31, given as striped or open diamonds, are seemingly higher than the corresponding data from Fig. 4 (black diamonds). However, the data shown in Fig. 5 are from a single experiment in which no transformants were found, whereas the data in Fig. 4 were accumulated from three independent determinations in which no transformants were obtained.
FIG. 6.
The frequency of transformation obtained by plate transformation with pUCP-Gm DNA (open symbols) of seven members of the Israel population having different transformation phenotypes. Selection was for Gmr. The data with pNS1 from Fig. 4 are included (black symbols). For transformation frequencies of strains 26a26 and 28a31, see the legend to Fig. 5.
The results suggest that transformability and its level are heritable traits which may strongly vary in a local population.
DISCUSSION
We have developed a method of specific and sensitive detection of 16S rRNA genes of P. stutzeri in DNA from environmental samples by PCR. The method includes improved specificity of the previously described procedure (2) by primer adaptation and has additionally been optimized in its sensitivity for application with DNA isolated from the environment. With this method, a specific PCR product was generated from only 5 genome equivalents of P. stutzeri in DNA preparations from environmental samples and covered at least six of the seven genomovars. Recently, Grüntzig et al. (17) described a very sensitive real-time PCR method based on one primer pair for the nirS gene (one gene copy could be amplified) for detecting the presence of P. stutzeri DNA in environmental samples. However, the primer pair did not cover the amplification of strains from genomovars 4, 5, and 7. We further developed a method for the effective isolation of viable P. stutzeri cells from environmental samples. Isolation of P. stutzeri strains is usually based on their potential for denitrification, a technique which is laborious and time-consuming. We made use of the ability of P. stutzeri to grow on rich medium and to utilize ethylene glycol, starch, and maltose as carbon sources. Employing replica plating allowed us to isolate P. stutzeri, even when only a few colony formers were present per gram of sample and when an up to 104-fold excess of colony formers of other bacterial species was present. P. stutzeri was isolated from three marine sediments and from two soils, one of which was contaminated with mineral oil. We were not able to isolate P. stutzeri from the three agricultural soils sampled in this study, which may be explained by specific agrotechnical conditions in these soils. A similar observation was reported by Grüntzig et al. (17). From four sites with P. stutzeri present, populations of more than 100 members were recovered, which opens the possibility of comparative population genetic studies. In this context, it is encouraging for future studies of the abundance of P. stutzeri in the environment that it was possible to isolate viable P. stutzeri cells by our plating method at five of six sites where PCR amplification suggested the presence of P. stutzeri (Table 1). The 16S rRNA sequences obtained from several isolates from the various sites positioned them into several groups within the phylogenetic tree of P. stutzeri, of which at least two groups are established genomovars (Fig. 3).
A test of natural transformation by pNS1 DNA of 27 randomly chosen strains from our isolates from the five environmental sites suggested that a large fraction of the P. stutzeri isolates can develop DNA uptake competence (Table 2). The levels of transformability of the isolates were very different (by up to 4 orders of magnitude). Previously, of 12 members of a worldwide collection of P. stutzeri strains, 6 were found to be transformable (24), and the transformation frequencies varied over 3 orders of magnitude. Whereas in that study representatives of all genomovars were included, thus covering the huge genomic diversity within this species by one to three strains per genomic group, the Israel soil strains allowed the study of the diversity of the transformation phenotype among a high number of genomically closely related strains. Among the 120 members of the Israel soil strains, the abundance of transformability was about 30%. Levels of transformation differed by 4 orders of magnitude (Fig. 4). The different transformation levels obtained with plasmid pNS1 DNA and two different antibiotic resistance determinants were confirmed for a limited number of strains with two other replicons and also with chromosomal DNA. Our results support the notion that transformability is not generally associated with strains belonging to P. stutzeri. Moreover, the data from the soil strains from Israel indicate that within a local population a variety of transformation-level phenotypes can exist, ranging from presumably nontransformable through intermediate to high levels of transformability. The transformation phenotypes were often correlated with the genomic relatedness of the isolates.
How can the strong variability of the transformation phenotype within a local population be explained? Due to the many proteins involved in the DNA uptake process, the transformability of bacteria is a polygenic phenotype (10). In addition, we can distinguish between primary and secondary transformation genes. Primary genes are those which are directly involved in DNA uptake and are therefore essential for transformation. For P. stutzeri, eight genes have presently been identified that are necessary for the biogenesis of type IV pili (pil genes), which are absolutely required for uptake of DNA into the periplasm of P. stutzeri (13, 16; S. Graupner and W. Wackernagel, unpublished data). That 8 genes are involved is probably a strong underestimation, because more than 34 genes involved in type IV pilus biogenesis and function in Pseudomonas aeruginosa have been identified so far (1, 9). Additional gene functions are needed for the translocation of DNA from the periplasm into the cytoplasm. For P. stutzeri, two such genes have been identified so far, one of which is comA (14); the other is comF (Graupner and Wackernagel, unpublished). In other transformable species, further genes for DNA translocation through the cytoplasmic membrane have already been identified (10). Null mutations in any of the above genes knock out transformation. Secondary transformation genes may be considered those genes which act indirectly on DNA uptake and translocation processes, e.g., by affecting cell wall structure and function. Inactivation of these genes may affect transformation to various extents. In P. stutzeri the exbB gene presumably functions in energy transfer from the cytoplasmic membrane to the periplasm and outer membrane and would be such a gene, since null mutants were still about 10% transformable compared to the wild type (14). The comL gene of Neisseria gonorrhoeae is involved in murein metabolism, and a mutation in it decreased transformation and cell size (12). In Acinetobacter, the level of polysaccharide capsule formation influences the level of transformation (20). In P. stutzeri the knockout of gene pilAII, which is very similar to the gene for the structural pilus protein, increased transformability about 20-fold, indicating that the gene normally acts as a suppressor of transformation (15). Altogether, the many genes either necessary for transformation or modulating its effectiveness constitute a considerable part of the genome and therefore provide a large target for mutational alterations. The primary and presumably many secondary transformation genes are not essential. If mutability is high in a population, it is very likely that transformation and its level would be affected as was observed. A relatively frequent formation of mutations in the local population studied here would be in accordance with the high genetic diversity of the population members that was detectable by the RAPD analysis (Fig. 4). A frequent occurrence of mutations would also explain the high variability in the morphology and color of colonies among the population members. In the development of the local population, any rather recent mutational alterations of transformation genes would be visible because closely related strains would have the same transformation phenotype. This was indeed observed. It has been suggested that during starvation-stationary phase, bacteria increase their genetic diversity by decreasing mismatch repair activity and increasing mutation formation by the SOS response (11, 42, 43). The environmental habitats of bacteria provide mostly starvation conditions (22, 31). Our finding of strong diversity within a local population fits with the recent conclusions drawn from the analysis of a worldwide collection of P. stutzeri strains (33, 40), which culminated in the conclusion that P. stutzeri is an extremely diverse species (33). These researchers argued that species diversity results from local diversity of niche-adapting subpopulations. With P. stutzeri the strong adaptive diversity of subpopulations would result from frequent mutation and, to a lesser extent, from recombination, since the global population structure was largely clonal (33).
If the assumption that mutability of P. stutzeri cells is rather high in the habitat and frequently results in a change of the level of transformability or its total loss is correct, it is interesting that the ability for transformation has not already been extinguished. In this context it may be recalled that transformability is assumed to have a high evolutionary potential, since it has been maintained within species and among organisms throughout the bacteria and archaea (10, 25). How any mutationally inactivated genes are reactivated, i.e., by reverse mutations or recombinational repair after horizontal gene transfer, is not known.
Acknowledgments
We thank Jorge Lalucat and Michael G. Lorenz for supplying bacterial strains, Petra Meier for providing experimental details prior to publication, and Johann de Vries for helpful suggestions.
This work was supported by the Bundesministerium für Bildung, Wissenschaft, Forschung, und Technologie and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Alm, R. A., and J. S. Mattick. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89-98. [DOI] [PubMed] [Google Scholar]
- 2.Baggi, G., P. Barbieri, E. Galli, and S. Tollari. 1987. Isolation of a Pseudomonas stutzeri strain that degrades o-xylene. Appl. Environ. Microbiol. 53:2129-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennasar, A., C. Guasp, and J. Lalucat. 1998. Molecular methods for the detection and identification of Pseudomonas stutzeri in pure culture and environmental samples. Microb. Ecol. 35:22-33. [DOI] [PubMed] [Google Scholar]
- 4.Bennasar, A., R. Rosselló-Mora, J. Lalucat, and E. R. Moore. 1996. 16S rRNA gene sequence analysis relative to genomovars of Pseudomonas stutzeri and proposal of Pseudomonas balearica sp. nov. Int. J. Syst. Bacteriol. 46:200-205. [DOI] [PubMed] [Google Scholar]
- 5.Boerlin, P., E. Bannerman, F. Ischer, J. Rocourt, and J. Bille. 1995. Typing Listeria monocytogenes: a comparison of random amplification of polymorphic DNA with 5 other methods. Res. Microbiol. 146:35-49. [DOI] [PubMed] [Google Scholar]
- 6.Bruns, S., K. Reipschläger, M. G. Lorenz, and W. Wackernagel. 1992. Characterization of natural transformation of the soil bacteria Pseudomonas stutzeri and Acinetobacter calcoaceticus by chromosomal and plasmid DNA, p. 115-126. In M. J. Gauthier (ed.), Gene transfers and environment. Springer-Verlag, Berlin, Germany.
- 7.Carlson, C. A., L. S. Pierson, J. J. Rosen, and J. L. Ingraham. 1983. Pseudomonas stutzeri and related species undergo natural transformation. J. Bacteriol. 153:93-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson, C. A., S. M. Steenbergen, J. J. Rosen, and J. L. Ingraham. 1984. Natural transformation of Pseudomonas stutzeri by plasmids that contain cloned fragments of chromosomal deoxyribonucleic acid. Arch. Microbiol. 140:134-138. [Google Scholar]
- 9.Darzins, A., and M. A. Russell. 1997. Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system—a review. Gene 192:109-115. [DOI] [PubMed] [Google Scholar]
- 10.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 11.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96:4023-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fussenegger, M., D. Facius, J. Meier, and T. F. Meyer. 1996. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol. Microbiol. 19:1095-1105. [DOI] [PubMed] [Google Scholar]
- 13.Graupner, S., V. Frey, R. Hashemi, M. G. Lorenz, G. Brandes, and W. Wackernagel. 2000. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J. Bacteriol. 182:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graupner, S., and W. Wackernagel. 2001. Identification and characterization of novel competence genes comA and exbB involved in natural genetic transformation of Pseudomonas stutzeri. Res. Microbiol. 152:451-460. [DOI] [PubMed] [Google Scholar]
- 15.Graupner, S., and W. Wackernagel. 2001. Pseudomonas stutzeri has two closely related pilA genes (type IV pilus structural protein) with opposite influences on natural genetic transformation. J. Bacteriol. 183:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graupner, S., N. Weger, M. Sohni, and W. Wackernagel. 2001. Requirement of novel competence genes pilT and pilU of Pseudomonas stutzeri for natural transformation and suppression of pilT deficiency by a hexahistidine tag on the type IV pilus protein PilAI. J. Bacteriol. 183:4694-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grüntzig, V., S. C. Nold, J. Zhou, and J. M. Tiedje. 2001. Pseudomonas stutzeri nitrite reductase gene abundance in environmental samples measured by real-time PCR. Appl. Environ. Microbiol. 67:760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez, J., A. Fayos, M. A. Ferrus, and R. J. Owen. 1995. Random amplified polymorphic DNA fingerprinting of Campylobacter jejuni and C. coli isolated from human faeces, seawater and poultry products. Res. Microbiol. 146:685-696. [DOI] [PubMed] [Google Scholar]
- 19.Hubalek, Z., Z. Pacova, J. Halouzka, I. Sedlacek, M. Dlouhy, and M. Honza. 1998. Selective isolation of Pseudomonas stutzeri from vertebrate faeces on Rambach agar. Zentbl. Bakteriol. 288:343-349. [DOI] [PubMed] [Google Scholar]
- 20.Juni, E., and A. Janik. 1969. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J. Bacteriol. 98:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 22.Kolter, R., D. A. Siegele, and A. Tormo. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855-874. [DOI] [PubMed] [Google Scholar]
- 23.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 24.Lorenz, M. G., and J. Sikorski. 2000. The potential for intraspecific horizontal gene exchange by natural genetic transformation: sexual isolation among genomovars of Pseudomonas stutzeri. Microbiology 146:3081-3090. [DOI] [PubMed] [Google Scholar]
- 25.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenz, M. G., and W. Wackernagel. 1991. High frequency of natural genetic tranformation of Pseudomonas stutzeri in soil extract supplemented with a carbon/energy and phosphorus source. Appl. Environ. Microbiol. 57:1246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenz, M. G., and W. Wackernagel. 1990. Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA. Arch. Microbiol. 154:380-385. [DOI] [PubMed] [Google Scholar]
- 28.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 29.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papapetropoulou, M., J. Iliopoulou, G. Rodopoulou, J. Detorakis, and O. Paniara. 1994. Occurrence and antibiotic-resistance of Pseudomonas species isolated from drinking water in southern Greece. J. Chemother. 6:111-116. [DOI] [PubMed] [Google Scholar]
- 31.Postgate, J. 1989. A microbial way of death. New Scientist 1665:43-46. [Google Scholar]
- 32.Rainey, P. B., I. P. Thompson, and N. J. Palleroni. 1994. Genome and fatty acid analysis of Pseudomonas stutzeri. Int. J. Syst. Bacteriol. 44:54-61. [DOI] [PubMed] [Google Scholar]
- 33.Rius, N., M. C. Fuste, C. Guasp, J. Lalucat, and J. G. Loren. 2001. Clonal population structure of Pseudomonas stutzeri, a species with exceptional genetic diversity. J. Bacteriol. 183:736-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romanowski, G., M. G. Lorenz, and W. Wackernagel. 1993. Use of polymerase chain reaction and electroporation of Escherichia coli to monitor the persistence of extracellular plasmid DNA introduced into natural soils. Appl. Environ. Microbiol. 59:3438-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossello, R. A., E. Garcia-Valdes, J. Lalucat, and J. Ursing. 1991. Genotypic and phenotypic diversity of Pseudomonas stutzeri. Syst. Appl. Microbiol. 14:150-157. [Google Scholar]
- 36.Rosselló-Mora, R. A., J. Lalucat, and E. García-Valdés. 1994. Comparative biochemical and genetic analysis of naphthalene degradation among Pseudomonas stutzeri strains. Appl. Environ. Microbiol. 60:966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosselló-Mora, R. A., J. Lalucat, and E. R. B. Moore. 1996. Strain JM300 represents a new genomovar within Pseudomonas stutzeri. Syst. Appl. Microbiol. 19:596-599. [Google Scholar]
- 38.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 39.Sikorski, J., S. Graupner, M. G. Lorenz, and W. Wackernagel. 1998. Natural genetic transformation of Pseudomonas stutzeri in a non-sterile soil. Microbiology 144:569-576. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski, J., R. Rossello-Mora, and M. G. Lorenz. 1999. Analysis of genotypic diversity and relationships among Pseudomonas stutzeri strains by PCR-based genomic fingerprinting and multilocus enzyme electrophoresis. Syst. Appl. Microbiol. 22:393-402. [DOI] [PubMed] [Google Scholar]
- 41.Stewart, G. J., and C. D. Sinigalliano. 1991. Exchange of chromosomal markers by natural transformation between the soil isolate, Pseudomonas stutzeri JM300, and the marine isolate, Pseudomonas stutzeri strain ZoBell. Antonie Leeuwenhoek 59:19-25. [DOI] [PubMed] [Google Scholar]
- 42.Taddei, F., J. A. Halliday, I. Matic, and M. Radman. 1997. Genetic analysis of mutagenesis in aging Escherichia coli colonies. Mol. Gen. Genet. 256:277-281. [DOI] [PubMed] [Google Scholar]
- 43.Taddei, F., M. Vulic, M. Radman, and I. Matic. 1997. Genetic variability and adaptation to stress. EXS 83:271-290. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaisanen, O. M., A. Weber, A. Bennasar, F. A. Rainey, H. J. Busse, and M. S. Salkinoja-Salonen. 1998. Microbial communities of printing paper machines. J. Appl. Microbiol. 84:1069-1084. [DOI] [PubMed] [Google Scholar]
- 46.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 47.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]