Abstract
We have previously described an alternative form of RNA polymerase II in yeast lacking the Srb and Med proteins but including Paf1, Cdc73, Hpr1, and Ccr4. The Paf1-RNA polymerase II complex (Paf1 complex) acts in the same pathway as the Pkc1-mitogen-activated protein kinase cascade and is required for full expression of many cell wall biosynthetic genes. The expression of several of these cell integrity genes, as well as many other Paf1-requiring genes identified by differential display and microarray analyses, is regulated during the cell cycle. To determine whether the Paf1 complex is required for basal or cyclic expression of these genes, we assayed transcript abundance throughout the cell cycle. We found that transcript abundance for a subset of cell cycle-regulated genes, including CLN1, HO, RNR1, and FAR1, is reduced from 2- to 13-fold in a paf1Δ strain, but that this reduction is not promoter dependent. Despite the decreased expression levels, cyclic expression is still observed. We also examined the possibility that the Paf1 complex acts in the same pathway as either SBF (Swi4/Swi6) or MBF (Mbp1/Swi6), the partially redundant cell cycle transcription factors. Consistent with the possibility that they have overlapping essential functions, we found that loss of Paf1 is lethal in combination with loss of Swi4 or Swi6. In addition, overexpression of either Swi4 or Mbp1 suppresses some paf1Δ phenotypes. These data establish that the Paf1 complex plays an important role in the essential regulatory pathway controlled by SBF and MBF.
There are multiple forms of RNA polymerase II (RNAP II) in Saccharomyces cerevisiae (8, 18). The holoenzyme form containing the Srb and Med proteins is responsible for transcription of most genes in the yeast genome (reviewed in reference 36). We have described an alternative form of RNAP II, the Paf1 complex, that does not contain Srb or Med proteins but does contain Paf1, Hpr1, Cdc73, and Ccr4, as well as the general initiation factors TFIIB and TFIIF (7, 43, 44). The Paf1 complex is also found associated with Rtf1 and Spt5 (35, 46), factors that are linked to transcriptional elongation. In addition, unlike the Srbs and Meds that are only found in promoter regions, Paf1 complex components are found associated with both promoter and coding regions of genes, indicating that they may play roles at multiple stages of transcription (39).
Loss of Paf1 results in alterations in transcript abundance for a small subset of yeast genes, among which are many cell wall biosynthetic genes controlled by the protein kinase C-mitogen-activated protein (Pkc1-MAP) kinase signaling pathway (7). Consistent with its role in the cell wall biosynthetic pathway, paf1Δ strains are temperature sensitive (ts) and sensitive to cell wall-damaging agents, such as sodium dodecyl sulfate and caffeine (7, 44). Two lines of evidence indicate that the Paf1 complex has multiple functions in yeast in addition to its role in the Pkc1 signaling pathway. First, mutations of Paf1 complex genes have pleiotropic phenotypes consistent with defects in several metabolic pathways, including DNA synthesis, microtubule function, and sterol biosynthesis (4). Second, Paf1 complex mutants affect the expression of many genes other than those required for cell wall biosynthesis (43; M. Chang, J. Fostel, and J. A. Jaehning, unpublished data). Furthermore, the potential homologues of Paf1 in humans, mice, flies, worms, and fission yeast suggest conserved functions for the Paf1 complex throughout the eukaryotic kingdom.
Recently, Koch and coworkers reported that Ctr9 (Cdp1), which is required for full expression of the cell cycle-regulated G1 cyclins CLN1 and CLN2, copurifies with Paf1 and Cdc73 (28). We have confirmed that Ctr9 is part of the Paf1 complex (35), and we have found that isogenic paf1Δ and ctr9Δ strains have identical phenotypes under a wide range of conditions (4). These results, and preliminary experiments comparing whole-genome expression in wild-type versus paf1Δ strains (Chang et al., unpublished), have led us to test the role of the Paf1 complex in expression of a broad range of cell cycle-controlled genes. Published microarray experiments have estimated that from 7 to 13% of yeast genes are cell cycle regulated, depending on experimental conditions and statistical methods used (9, 45, 54). We have found that nearly 30% of genes whose expression decreases in a paf1Δ strain are regulated during the cell cycle, suggesting that the Paf1 complex has a specific function in expression of this important subset of genes.
Two of the major regulators of yeast cell cycle-controlled genes are the transcription factors SBF and MBF, which are activated during G1 by Cdc28/Cln3 (reviewed in reference 34). MBF, a heterodimer of Mbp1 and Swi6, plays an important role in the periodic transcription of genes necessary for DNA replication (27). SBF, a heterodimer of Swi4 and Swi6, controls synthesis of the G1 cyclins CLN1 and CLN2 as well as membrane and cell wall components (15, 23, 31, 38). These transcription factors share the Swi6 transcriptional activation component, with Mbp1 and Swi4 contributing unique DNA binding domains to the complexes (1, 2, 27, 29). Although each factor controls the expression of different genes, they have considerable overlap in function, and in vitro they can cross-recognize each other's DNA targets (47). Consistent with this functional overlap, neither MBP1 nor SWI4 is an essential gene in most yeast strains, but an mbp1 swi4 double mutant is lethal (27). Recently, the in vivo binding sites for Swi4 and Mbp1 were determined on a genomic scale (24). Each factor binds to a subset of yeast genes, with Swi4 more often found upstream of cell wall biosynthetic genes and Mbp1 more often found in association with genes important for DNA replication, strong confirmation of the previous results from gene expression patterns. Consistent with their overlapping functions, both factors are found bound to a significant subset of genes (24).
In addition to its connection to cell cycle regulation via Cdc28, SBF also appears to function downstream of the Pkc1-MAP kinase pathway (31), as does the Paf1 complex (7). The many overlaps between phenotypes and functions of the Paf1 complex with the SBF and MBF factors prompted us to ask whether Paf1 acts through these known transcription factors to control expression of cell cycle-regulated genes. In this work, we demonstrate that Paf1 is necessary throughout the cell cycle for full expression of many periodically expressed genes. We tested the possibility that the Paf1 complex acts through SBF or MBF by comparing cell cycle expression profiles of genes in paf1Δ, swi4Δ, and mbp1Δ strains. Both biochemical and genetic analyses establish that Paf1, acting through both Swi4 and Mbp1, is required for full levels of expression of cell cycle-regulated genes.
MATERIALS AND METHODS
Strains and media.
The strains used in this study are listed in Table 1. Strains YJJ662, YJJ664, YJJ577, YJJ1000, YJJ1232, YJJ1067, YJJ1068, and YJJ1390 are isogenic except for the specific deletion described and were derived from the D273-10B background (48). Strains YJJ755, YJJ756, YJJ1173, and YJJ1239 are isogenic except for the specific deletion described and were derived from the A364a background (19). Strains were grown in yeast extract-peptone-dextrose (YPD) with 4% glucose using standard methods (17).
TABLE 1.
Yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| YJJ662 | MATaleu2Δ1 his3Δ200 ura3-52 | 43 |
| YJJ664 | MATaleu2Δ1 his3Δ200 ura3-52 paf1Δ::HIS3 | 43 |
| YJJ577 | MATα leu2Δ1 his3Δ200 ura3-52 paf1Δ::HIS3 | 43 |
| YJJ755 | MATabar1 his6 his7 leu2 ura3 pep4 prb1 trp1 | R. Sclafani |
| YJJ756 | MATabar1 his6 his7 leu2 ura3 pep4 prb1 trp1 paf1Δ::TRP1 | 7 |
| YJJ1000 | MATaleu2Δ1 his3Δ200 ura3-52 swi4Δ::URA3 | 6 |
| YJJ1067 | MATaleu2Δ1 his3Δ200 ura3-52 mbp1Δ::kanr | J. Betz |
| YJJ1068 | MATα leu2Δ1 his3Δ200 ura3-52 mbp1Δ::kanr | 4 |
| YJJ1173 | MATabar1 his6 his7 leu2 ura3 pep4 prb1 trp1 swi4Δ::URA3 | This work |
| YJJ1232 | MATaleu2Δ1 his3Δ200 ura3-52 swi4Δ::kanr | This work |
| YJJ1233 | MATaleu2Δ1 his3Δ200 ura3-52 swi4Δ::kanr | This work |
| YJJ1239 | MATabar1 his6 his7 leu2 ura3 pep4 prb1 trp1 mbp1Δ::kanr | This work |
| YJJ1390 | MATaleu2Δ1 his3Δ200 ura3-52 swi6Δ::kanr | This work |
Construction of deletion strains.
The swi4Δ::URA3 deletion strain YJJ1173 was constructed using plasmid pJJ1140, which contains a URA3 insertion to disrupt SWI4 (6). Cleavage of pJJ1140 with EcoRI released the swi4Δ::URA3 deletion cassette, which was used to transform strain YJJ755 following standard transformation procedures (17). Transformants were selected as uracil prototrophs and verified by PCR using oligos 230 and 231 (see Table 2 for a list of all primers used), which hybridize upstream and downstream of SWI4, generating a SWI4 PCR product of 2,982 bp and a swi4Δ::URA3 PCR product of 1,542 bp.
TABLE 2.
Oligos used for strain construction and as probes
| Oligo | Use | Sequence |
|---|---|---|
| 230 | swi4Δ verification | 5′-GTTCAAGGTGGGTATGGTAGG-3′ |
| 231 | swi4Δ verification | 5′-TATGCGTTTGCCCTCAAATCC-3′ |
| 287 | swi4Δ strain Kanr replacement | 5′-CCTTCTGTCCCTCTTGCGTAGTTCTAAAAGGTTGATTTATTCGAGAAATCCGTACGCT GCAGGTCGAC-3′ |
| 288 | swi4Δ strain Kanr replacement | 5′-AAAAACTCTGATAATATAGTAAAAATTATTGGTACATTGTGATTAAAATATCGATGA ATTCGAGCTCG-3′ |
| 278 | swi4Δ::Kanr verification | 5′-TCAACAATAATTGCTCTTTGCCG-3′ |
| 331 | swi4Δ::Kanr verification | 5′-GCAACTCAAGCGCAATGAGA-3′ |
| 240 | swi4Δ::Kanr verification | 5′-CTCATCTGTAACATCATTGGCAAC-3′ |
| 380 | mbp1Δ strain Kanr replacement | 5′-CTTAACATTCCGAGACACAACGTAAATCCCAGAAACACAAGCATGCGTACGCTGCAGGTCGAC-3′ |
| 381 | mbp1Δ strain Kanr replacement | 5′-CAGTATATGGATACATGTAAAGTTCCTCTATTTATGTATATTTTAATCGATGAATTCG AGCTCG-3′ |
| 382 | mbp1Δ::Kanr verification | 5′-TCAAGTGCGATTGGTCTGCA-3′ |
| 383 | mbp1Δ::Kanr verification | 5′-GCACTGCTTACTGTTATGTC-3′ |
| 282 | SPC25 probe, 5′ primer | 5′-GCCAGCATAGACGCATTTTCGG-3′ |
| 271 | SPC25 probe, 3′ primer | 5′-TGAATCATCGCCGAATACGAAACGTA-3′ |
| 283 | SAS3 probe, 5′ primer | 5′-GTCATTAACAGCAAACGACGAATCGC-3′ |
| 269 | SAS3 probe, 3′ primer | 5′-CCAGTATCGTTTCTCCTAGCTGCATG-3′ |
| 281 | STP4 probe, 5′ primer | 5′-TGGTATCATCATCTTTTGCATCAAGC-3′ |
| 272 | STP4 probe, 3′ primer | 5′-GGATACACCTCCATATCCAAATTGATC-3′ |
| 284 | PCL7 probe, 5′ primer | 5′-ATGGAGCTAAGTTCACCATCAAAAAAAAC-3′ |
| 273 | PCL7 probe, 3′ primer | 5′-CACTCAAGAACTTCGTGCAAATTGTG-3′ |
| 374 | HO probe, 5′ primer | 5′-CGCAAACGTCACGGCTAAC-3′ |
| 375 | HO probe, 3′ primer | 5′-TCAAACTGTAAGATTCCGCCAC-3′ |
| 84 | 18S rRNA probe | 5′-GCTTATACTTAGACATGCAT-3′ |
| 211 | RNR1 probe | 5′-ACACATTTCACAAGCTTCTGGG-3′ |
| 213 | FAR1 probe | 5′-TTCTATCAAACTAGAATGCGGGTGTT-3′ |
| 214 | RPS9A probe | 5′-CCGCCTTCCTTGCAGCATTTCTTCTA-3′ |
| 216 | CYB5 probe | 5′-CCACTACCTTTACTTTGGTTTTCAGA-3′ |
| 217 | FKS1 probe | 5′-CCAATGAATCTCCAATGTGTTTGTGG-3′ |
| 265 | HTB1 probe | 5′-CAGTGGAAGTGGAAGTCTTTTTAGCG-3′ |
| 268 | PIR3 probe | 5′-GAGCCAATTCTACCCTTCCTATCGGT-3′ |
| 344 | CLN1 promoter, 5′ | 5′-CGAAAACACCGCGCGTAAAG-3′ |
| 396 | swi6Δ strain Kanr replacement, 5′ | 5′-AAAAAGAATAATAAAGGGGAACACAGTATAATTCTCGAGAGGCGTACGCTGCAGG TCGAC-3′ |
| 397 | swi6Δ strain Kanr replacement, 3′ | 5′-AGTGCCTATATATCCCATCTGAGTACGTAAAAATTTTGTAAATCGATGAATTCGAG CTCG-3′ |
| 410 | swi6Δ::Kanr verification, 5′ | 5′-GGACCGTTTGGATAAGATCATTCACC-3′ |
| 411 | swi6Δ::Kanr verification, 3′ | 5′-GGTGGAAGGACAGGCTTAAGATCTTGTAG-3′ |
| 588 | CLN1 promoter, 3′ | 5′-CCAACTCGAGTCTAGAAGTGGAGTGGTGGTAGTGC-3′ |
| 597 | FKS1 promoter, 5′ | 5′-AAGGTACCTCTAGAATGTACGGGCAATCAGAATCTG-3′ |
| 598 | FKS1 promoter, 3′ | 5′-AAGGTACCGGTCTGACCGTTGTATGAAAGAC-3′ |
| 599 | GLK1 promoter, 5′ | 5′-AAGGTACCTCTAGAGCTTTAACGTATTCAGGAGCCATG-3′ |
| 600 | GLK1 promoter, 3′ | 5′-AAGGTACCGTATTAGTGGTGGTGTTGTGGTTTAC-3′ |
The swi4::kanr strain YJJ1232 was constructed using a PCR-based method. Primers were designed such that the internal section would be homologous to the kanr gene of plasmid pFA6a-kanMX4 (50), while the external section was homologous to the yeast SWI4 gene. Oligos 287 and 288 were used in a standard PCR amplification (3) with pFA6a-kanMX4 as the template. The product of this PCR amplification was used to transform strain YJJ662 using standard methods (17). Kanamycin-resistant transformants were verified using PCR with oligos 278 and 331 (5′ and 3′ SWI4 primers outside of deleted region) or oligos 278 and 240 (5′ SWI4 and 3′ primer for kanamycin region). Oligos 278 plus 240 do not generate a PCR product with SWI4 DNA template but do generate a 732-bp PCR product with swi4Δ::kanr. Oligos 278 and 331 generate a product of 3,509 bp with SWI4 DNA and a product of 1,731 bp with swi4Δ::kanr.
The mbp1Δ::kanr strain YJJ1239 was also constructed using a PCR-based method as described above, with oligos 380 and 381, plasmid pFA6a-kanMX4, and transformation into YJJ755. Kanamycin-resistant transformants were verified using PCR with oligos 382 and 383, which generate an MBP1 PCR product of 2,760 bp and a mbp1Δ::kanr PCR product of 1,770 bp. Using oligos 382 and 240 (in the kanamycin resistance gene) generates no product with MBP1 and a 705-bp product with mbp1Δ::kanr.
The swi6Δ::kanr strain YJJ1390 was constructed as above using PCR primers 396 and 397 with plasmid pFA6a-kanMX4. This PCR product was transformed into strain YJJ662, and kanamycin-resistant transformants were verified using PCR with oligos 410 and 411. These oligos generate a SWI6 PCR product of 2,854 bp and a swi6Δ PCR product of 1,847 bp.
Cell cycle synchronization.
Cells were synchronized with α-factor as previously described (5) using strains YJJ755 (wild type), YJJ756 (paf1Δ), YJJ1173 (swi4Δ), and YJJ1239 (mpb1Δ), all carrying the bar1 mutation to facilitate synchronization (5). Cells were grown at 30°C to a density of 40 Klett units (1.5 × 107 cells/ml), and α-factor (catalog no. T6901; Sigma) was added to a concentration of 20 nM. Cells were incubated in α-factor for one doubling (1.5 h, or 3 h for paf1Δ), and progression of the arrest was measured microscopically. To remove the α-factor, cells were collected in a sterile 0.8-μm-pore-size filter (90 mm; Osmonics) in a Millipore filter apparatus and washed with 15 volumes of YPD warmed to 30°C. Cells were resuspended in 400 ml of fresh YPD warmed to 30°C, Pronase (Calbiochem) was added to a concentration of 50 μg/ml, and the culture was incubated with shaking at 200 rpm at 30°C. Aliquots were taken every 15 min for RNA isolation, budding index profiles, and flow cytometry. For RNA isolation, cells from duplicate 10-ml samples were pelleted, washed in diethyl pyrocarbonate-treated distilled H2O (dH2O), pelleted again, quick-frozen in liquid nitrogen, and stored at −80°C. For determination of the budding index, 180 μl of culture was mixed with 20 μl of formaldehyde and stored at 4°C until microscopic analysis of budding stages. For each time point, at least 200 cells were categorized as unbudded, or small, medium, or large budded. For flow cytometry, 820 μl of culture was sonicated for 30 s (10 s for paf1Δ strains) and pelleted in a microcentrifuge. Cells were resuspended in 300 μl of dH2O and fixed by adding 700 μl of ethanol while vortexing. Samples were stored at 4°C until processing according to the method described by Nash et al. (37).
RNA isolation and analysis.
RNA was isolated from both synchronized and asynchronous cells by either the glass bead method (3) or the hot phenol method (41) with equivalent results. Synchronized cells were prepared as described above. Asynchronous cells were grown to early log phase and harvested at a density of 60 Klett units (2 × 106 cells/ml). RNA was fractionated by formaldehyde gel electrophoresis according to standard methods (3). For each sample, 10 μg of total RNA was loaded. RNA was visualized using SYBRGold dye (S-11494; Molecular Probes) and transferred to Zeta-Probe GT membranes (Bio-Rad). Membranes were hybridized to specific 32P-labeled probes using conditions suggested by the manufacturer. Probes were either 5′-end-labeled oligonucleotide probes designed to hybridize to the 3′ ends of specific mRNAs, or probes generated by PCR amplification of specific genes from yeast genomic DNA (Table 2). The CLN1 probe was a gel-purified 500-bp NcoI-EcoRI fragment from pJJ961, which contains the 1.2-kb CLN1 coding region in plasmid pRS313 (Robert Sclafani, University of Colorado Health Sciences Center). PCR-generated probes were gel purified and then labeled using the random prime method (RadPrime kit; Invitrogen). Hybridizations and washes were performed at either 64°C for random prime-labeled probes or between 38 and 45°C for oligonucleotide probes (listed in Table 2).
Quantitation was performed on blots by exposing them to a storage phosphor screen and scanning the screens using a Storm imaging system (Molecular Dynamics). An RNA loading control involved hybridizing and normalizing all blots to an oligonucleotide complementary to the yeast 18S rRNA. Band densities were quantified using ImageQuant (Molecular Dynamics) for the Macintosh.
Genetic analyses.
Mating YJJ577 and YJJ1232 generated a paf1Δ/PAF1 swi4Δ/SWI4 heterozygous diploid. The diploid was sporulated, and tetrads were dissected onto a YPD plate containing 1 M sorbitol using standard methods (17). The genotypes of spore colonies were determined by patching to plates that would distinguish the markers, i.e., the paf1Δ spores were HIS3 and the swi4Δ were kanr. Wild-type cells were his3Δ200 and sensitive to kanamycin. Mating YJJ664 and YJJ1068 generated a paf1Δ/PAF1 mbp1Δ/MBP1 heterozygous diploid. Diploids were sporulated and dissected, and spores were genotyped as above; the paf1Δ was HIS3 and the mbp1Δ was kanr. The wild-type spores were his3Δ200 and sensitive to kanamycin, while the double mutants were HIS3 kanr. Mating YJJ577 and YJJ1390 generated a paf1Δ/PAF1 swi6Δ/SWI6 heterozygous diploid. Diploids were sporulated, dissected, and genotyped as above; the paf1Δ was HIS3 and the swi6Δ was kanr.
Plasmids YEp24, YEp352, B327, and BK72 were kindly provided by M. Snyder (Yale University). B327 is vector YEp352 plus the Swi4 gene, while BK72 is vector YEp24 plus the Mbp1 gene. Both YEp24 and YEp352 are 2μm plasmids, so Swi4 and Mbp1 are functionally overexpressed when carried on these plasmids. The plasmids were transformed into wild type (YJJ662) and paf1Δ (YJJ664) strains and maintained on SD-URA medium (17). For spot assays, strains were grown under selection, serially diluted, and plated to YPD, YPD containing 6 mM caffeine, or YPD containing 100 mM hydroxyurea (HU). Spots were made by applying 4 μl of cell suspensions that were diluted to 107, 106, 105, 104, and 103 cells/ml. The experiments were repeated with two independently isolated clones for each plasmid with similar results.
Luciferase assays.
Luciferase constructs were made using the plasmid pMTLuc provided by D. Reines (Emory University). This plasmid is derived from the vector pRS316 and contains the firefly luciferase gene without a yeast TATA or upstream activation sequence (42). There is a unique KpnI site just upstream of the luciferase gene available for cloning, and we constructed pJJ1322, a version of the plasmid that contains a unique upstream XhoI site. Using template DNA from strain YJJ662 we amplified the CLN1 promoter using primers 344 and 588 (−38 to −680), the FKS1 promoter using primers 597 and 598 (−1 to −716), and the GLK1 promoter using primers 599 and 600 (−19 to −837) (Table 2). Each PCR product was cleaved and cloned into the appropriate vector to create pJJ1346, the CLN1 promoter in pJJ1322; pJJ1333, the FKS1 promoter in pMTLuc; and pJJ1339, the GLK1 promoter in pMTLuc. The vectors and promoter reporter plasmids were transformed into yeast strains YJJ662, YJJ664, YJJ1233, and YJJ1067 using standard transformation procedures (17). Luciferase assays were performed according to the Promega luciferase assay system. Cells were lysed by adding approximately 100 μl of glass beads to 1 optical density at 260 nm unit of cells in 150 μl of Promega cell culture lysis buffer and vortexed at high speed for 2 min. The assays were performed on a Dynex Technologies model MLX luminometer using a 96-well plate format. Units of activity were normalized to the protein concentration measured by using protein assay reagents from Bio-Rad.
RESULTS
Paf1 is required for full expression of a variety of cell cycle-regulated genes.
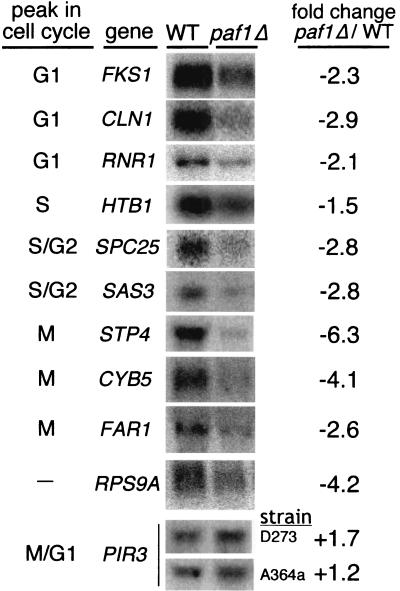
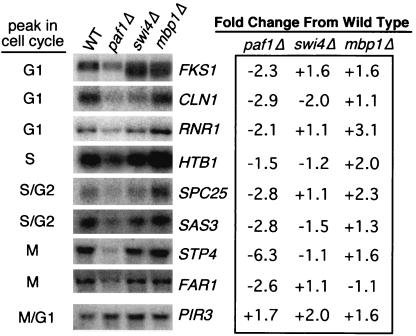
We initiated these studies based on results from differential display and microarray analyses that demonstrated that many genes dependent on Paf1 are cell cycle regulated (7, 43; Chang et al., unpublished). To further examine the role of the Paf1 complex in expression of these cycling genes, we first validated some of the microarray results by analyzing mRNA levels in asynchronous cultures. We probed RNA from wild-type and paf1Δ strains for several cell cycle-regulated genes, as shown in Fig. 1. These experiments were consistent with the microarray results, yielding a magnitude of decreased expression from two- to sixfold. The paf1Δ mutation does not seem to affect a specific cell cycle stage; instead, we saw diminished expression of genes that peak in G1 (FKS1, CLN1, and RNR1), S (HTB1), G2 (SPC25 and SAS3), and M (STP4, CYB5, and FAR1).
FIG. 1.
Deletion of the PAF1 gene causes a decrease in mRNA levels for many cell cycle-regulated genes. Asynchronous cultures of wild type (YJJ662) and paf1Δ (YJJ664) cells were grown to a Klett density of 60 (2 × 106 cells/ml), total RNA was harvested, and 10 μg of RNA per lane was fractionated and used to detect mRNAs for specific cell cycle-regulated genes as described in Materials and Methods. The cycle peak designations are from the microarray analysis of Spellman et al. (45; http://genome-www.stanford.edu/cellcycle/). The fold decrease of each mRNA in the paf1Δ strain compared to that in the wild type was calculated from an average of at least two experiments after normalization to 18S rRNA. RPS9A is an example of a gene that decreases in paf1Δ but is not cell cycle regulated. PIR3 is an example of a cell cycle-regulated gene that does not decrease in a paf1Δ strain. Results for two different strain backgrounds are shown for PIR3. The other mRNAs shown in this figure were from the D273-10b background. Similar results were obtained from both strains for the other mRNAs shown in this figure.
Not all genes whose expression is decreased in paf1Δ are cell cycle regulated; for example, RPS9A does not cycle, but its expression is reduced more than fourfold in paf1Δ (Fig. 1). In addition, not all cell cycle-regulated genes depend on Paf1: the abundance of the M/G1 phase-regulated PIR3 gene is actually slightly elevated in paf1Δ strains (Fig. 1). We have established that the paf1Δ effects are not an artifact of the yeast strain background used. For example, notice that PIR3 transcript abundance in Fig. 1 is very similar in both the A364a background, used in all of the cell cycle synchronization experiments described below, and the D273-10b background. Both strains were used in the asynchronous cell experiments, yielding very similar results for the different genes analyzed.
Wild-type and paf1Δ strains can both be arrested by α-factor and released into a synchronous cell cycle.
Unlike many genes important for cell cycle regulation, the abundance of PAF1 mRNA and Paf1 protein does not vary during the cell cycle (9, 45; M. Chang, unpublished data). Another feature of many critical cell cycle regulatory genes is that their mutation results in impairment of a particular cycle stage. However, other than its slow growth phenotype (3 h doubling time versus 1.5 h for wild type), a paf1Δ mutation does not lead to cells accumulating at a unique cell cycle stage, as determined by measuring budding index and DNA content (by flow cytometry) of asynchronous populations (data not shown). In addition, shifting a ts paf1Δ strain to its nonpermissive temperature does not result in a uniform terminal arrest phenotype as found with classic cdc mutations (40). Mutation of Paf1 complex components also does not lead to mating defects (43), indicating an unimpaired sensitivity to the mating pheromone α-factor. We therefore used α-factor to synchronize isogenic wild-type and paf1Δ strains in order to analyze gene expression patterns during an entire cell cycle. The paf1Δ strain was efficiently arrested by the same concentration of α-factor used with the wild-type strain and could be released into a synchronous cell cycle (Fig. 2).
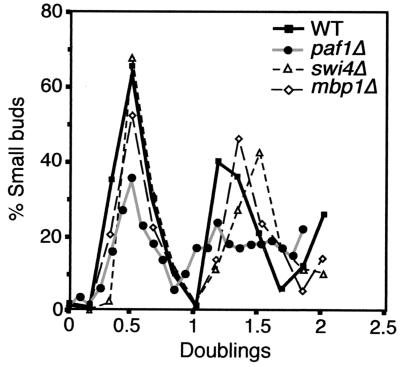
FIG. 2.
Synchronized wild-type, paf1Δ, swi4Δ, and mbp1Δ strains have similar budding profiles. Strains were synchronized with α-factor as described in Materials and Methods. Samples were taken at 15-min intervals, and the number of small budded cells in each sample was determined. Data are graphed as the percentage of cells with small buds versus doublings, rather than time, to directly compare the budding indices of the slow-growing paf1Δ strain to the other strains. Doublings for the wild type (YJJ755), swi4Δ (YJJ1173), and mbp1Δ (YJJ1239) were calculated by dividing the time, in hours, of each time point by 1.5 h. Doublings for paf1Δ (YJJ756) were calculated by dividing the time, in hours, of each time point by 3 h.
After release from the α-factor arrest, samples were isolated every 15 min for RNA preparation, flow cytometry, and budding index determination. The percentages of unbudded, small, medium, and large budded cells were determined for each time point. Compensating for the longer doubling time of the paf1Δ strain by plotting the data relative to doublings, rather than time after release, revealed that the profiles for all classes of cells are remarkably similar between the wild type and paf1Δ (small budded cells are plotted in Fig. 2 as an example). Although the fraction of cells recovering from pheromone treatment of the paf1Δ strain is a little lower and the synchrony is not as precise as for the wild-type strain, especially in the second cycle, the paf1Δ strain is clearly passing through the stages of the cell cycle. As was found for the asynchronous cultures, we did not detect any particular stage where the paf1Δ strain spends more time relative to the wild type. The conclusion that the paf1Δ strain is passing through a relatively normal cell cycle is confirmed by the expression profiles of periodic genes, as shown in the next section.
mRNA abundance is reduced throughout the cell cycle in paf1Δ cells, but expression is still cyclic.
The paf1Δ mutation affects the expression of genes that peak at different times throughout the cell cycle. Is this the result of an overall decrease in expression at all stages, or is expression of these genes no longer cyclic? To distinguish between these two possibilities, we isolated RNA from the synchronized wild-type and paf1Δ strains described in the previous section. These samples were probed for many of the same cell cycle-regulated transcripts and controls used for the asynchronous analysis shown in Fig. 1. All signals were normalized to that of a probe for 18S rRNA and plotted relative to doubling time after pheromone release, as shown in Fig. 3. In each case, the synchronized wild-type and paf1Δ samples were run on the same gel so that signal intensities could be directly compared. Compared to published data on expression patterns after release from α-factor, the profiles we obtained from wild-type cells were very similar to the microarray results of Spellman et al. (45) in terms of peak stage and magnitude of cycling.
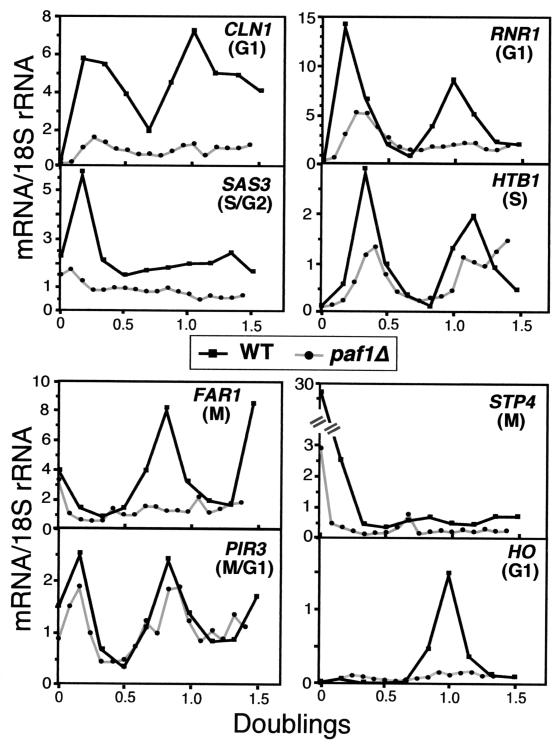
FIG. 3.
mRNA abundance for cycling genes is decreased throughout the cell cycle in the paf1Δ strain. Fractionated total RNA from synchronized wild-type (YJJ755) and paf1Δ (YJJ756) cells was analyzed with probes (see Table 2) that detect periodic genes that peak in the indicated parts of the cell cycle. PIR3 expression peaks in M/G1 but is not affected by lack of Paf1. Each graph shows the normalized amount of mRNA for that gene over a period of 1.5 doublings for wild-type and paf1Δ strains. The mRNA/18S rRNA units are arbitrary phosphorimager numbers and are not comparable between probes. However, each individual panel represents equal amounts of RNA from wild-type and paf1Δ cells fractionated on the same gel for direct comparison.
For nearly all of the genes analyzed, mRNA abundance in paf1Δ was reduced significantly throughout the cell cycle relative to levels observed in the wild-type strain (Fig. 3). As seen by other investigators studying critical cell cycle regulatory factors (23), we observed greater paf1Δ-dependent decreases in the synchronous cells than we did for the asynchronous samples. For example, CLN1, RNR1, and FAR1 were reduced 2.9-, 2.1-, and 2.6-fold in the asynchronous samples (Fig. 1) but were reduced 5.2-, 4.4-, and 4.7-fold at the peak of expression in the synchronized samples. S/G2-phase-regulated SAS3 and M-phase-regulated STP4 transcripts were decreased 2.8- and 6.3-fold in asynchronous cells and 4.1- and 13-fold at their peak in synchronized cells. Expression of HTB1 decreased only slightly, 2.2-fold, in the synchronized cells, but still to a greater extent than the 1.5-fold decrease seen in the asynchronous experiments (Fig. 1). We did not measure HO expression in asynchronous populations, but in synchronized cells HO expression was down more than 10-fold in paf1Δ compared to the wild type.
The expression of two control genes was also analyzed throughout the cell cycle. RPS9A is not cell cycle regulated but is dependent on Paf1 for full expression (Fig. 1). We observed that the approximately fourfold reduction in expression of RPS9A seen in the asynchronous cells was maintained during passage through the cell cycle (Fig. 4 and data not shown). PIR3 is cell cycle regulated but is not dependent on Paf1 in asynchronous cells (Fig. 1). PIR3 cycling in paf1Δ is virtually indistinguishable from its pattern in the wild type (Fig. 3). This result confirms both that, apart from the longer doubling time, the paf1Δ cells are cycling normally and that only a subset of cycling genes depends on Paf1.
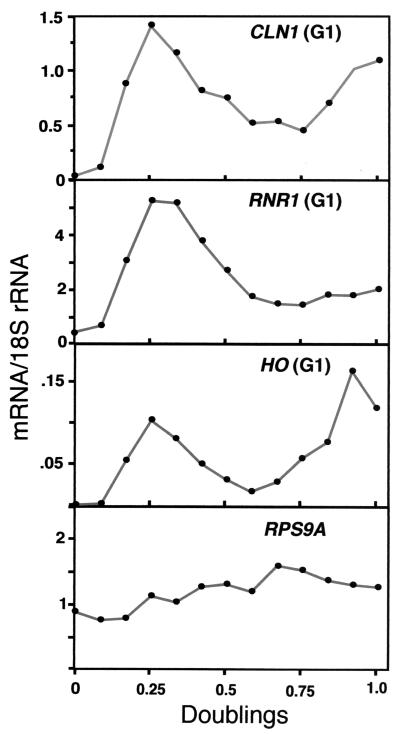
FIG. 4.
Cell cycle-regulated genes still cycle in paf1Δ. Representative data from Fig. 3 showing the cell cycle profiles from paf1Δ (YJJ756) cells of three G1-phase genes (CLN1, RNR1, and HO) normalized to 18S rRNA are shown here on an expanded scale. Profile of the non-cell-cycle-regulated gene RPS9A is shown for comparison.
Some genes, for example HTB1, still clearly demonstrate a cyclical profile in a paf1Δ strain. However, for other genes, like CLN1 and HO, where expression is dramatically reduced relative to that in the wild-type strains, it is difficult to see in the data in Fig. 1 whether the residual expression is still cyclic. To look in more detail at the cycling patterns, the mRNA abundance profiles for CLN1, RNR1, and HO in the paf1Δ strain were plotted independently (Fig. 4). This view of the data makes it clear that the cyclic pattern is still evident. All of the genes showed cyclic patterns in paf1Δ when plotted in this way (data not shown). As a control, we show the profile of the noncyclic gene RPS9A in the paf1Δ strain (Fig. 4). Unlike the cell cycle-regulated transcripts, RPS9A is present at a constant level throughout the cell cycle. These data clearly demonstrate that while the amount of mRNA for several cell cycle-regulated genes is diminished in paf1Δ, expression is still cyclic.
Gene expression patterns of neither swi4Δ nor mbp1Δ exactly duplicate those seen in paf1Δ.
If the Paf1 complex is acting through the known cell cycle transcription factors Swi4 and Mbp1, then we would predict that removal of one or the other, or possibly both, factor(s) might mimic the effects we have described for the absence of Paf1. The phenotypes of the single-mutant swi4Δ and mbp1Δ strains in the genetic backgrounds used in these studies are not nearly as severe as that of a paf1Δ strain (4). However, as previously described for other genetic backgrounds (12), we found that a swi4Δ mbp1Δ double mutant is lethal. Therefore, the Paf1 complex could be communicating with either, or both, of these factors to activate transcription of target genes. To test this possibility we analyzed mRNA levels of cell cycle-regulated genes in asynchronous cultures of isogenic swi4Δ and mbp1Δ strains and compared them to the levels in paf1Δ (Fig. 5).
FIG. 5.
Gene expression profiles of swi4Δ and mbp1Δ strains are not identical to that of a paf1Δ strain. Asynchronous cultures of wild type (YJJ662), paf1Δ (YJJ664), swi4Δ (YJJ1000), and mbp1Δ (YJJ1067) cells were grown to a Klett density of 60 (2 × 106 cells/ml). Total RNA was harvested, 10 μg of RNA per lane was fractionated, and specific mRNAs were detected using probes for cell cycle-regulated genes and normalized to 18S rRNA. The fold change of each mRNA in paf1Δ, swi4Δ, and mbp1Δ strains compared to that for the wild type is an average of at least two experiments. PIR3 is a cell cycle-regulated gene that does not decrease in a paf1Δ strain (see Fig. 1). Note that the asynchronous samples shown in this figure were from the D273-10b genetic background; similar results were obtained in the A364a background.
Consistent with the modest phenotypes of the swi4Δ and mbp1Δ strains, we did not observe dramatic effects on transcript abundance in these strains. However, in agreement with previous reports (38), we found that CLN1 expression is reduced in the absence of Swi4 but is not affected by loss of Mbp1 (Fig. 5). Note that the twofold reduction of CLN1 mRNA in the absence of Swi4 is not as severe as the nearly threefold reduction seen in the absence of Paf1. Neither swi4Δnor mbp1Δ results in a pattern of gene expression identical to that of paf1Δ. For example, FKS1 expression decreased over twofold in paf1Δ yet increased in both swi4Δ and mbp1Δ, and STP4 expression decreased over sixfold in paf1Δ yet did not change in swi4Δ and increased slightly in mbp1Δ. The abundance of RNR1, reported to depend on Mbp1 for correct expression during the cell cycle (25), actually increased over threefold in the mbp1Δ strain, was unaffected by swi4Δ, and decreased over twofold in paf1Δ in the asynchronous cells.
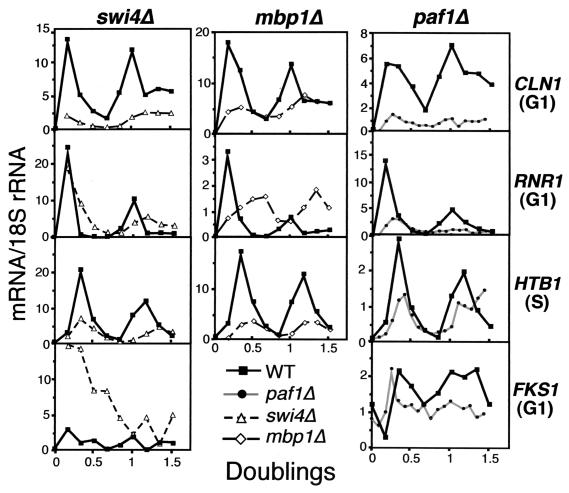
To further compare expression of cell cycle-regulated genes in paf1Δ versus swi4Δ and mbp1Δ, we synchronized swi4Δ and mbp1Δ strains with α-factor and analyzed transcripts at 15-min intervals after release from arrest. The doubling time of the swi4Δ and mbp1Δ strains was 1.5 h, indistinguishable from that of the wild type, and passage through the cell cycle as measured by budding index of the swi4Δ and mbp1Δ mutants was also very similar to that of the wild type (Fig. 2). In all cases, RNAs from mutant and wild-type cells were separated on the same gels and probed together, so transcript abundance could be compared directly (Fig. 6).
FIG. 6.
The gene expression profiles of swi4Δ and mbp1Δ strains do not mimic the cycling profiles seen in a paf1Δ strain. RNA from synchronized wild-type (YJJ755), paf1Δ (YJJ756), swi4Δ (YJJ1173), and mbp1Δ (YJJ1239) cells was fractionated and probed for the indicated transcripts. The graphs show the amount of normalized mRNA for the indicated genes over a period of 1.5 doublings for wild-type versus paf1Δ, swi4Δ, and mbp1Δ strains. The mRNA/18S rRNA units are arbitrary phosphorimager counts and are not comparable between probes. However, each individual panel represents equal amounts of RNA from wild type and the indicated mutant strains fractionated on the same gels for a direct comparison.
As shown in Fig. 6, although mutation of SWI4 and MBP1 had definite effects on expression of the cell cycle-regulated genes CLN1, RNR1, HTB1, and FKS1, none of the changes were superimposable with the pattern observed in the paf1Δ strain. For example, CLN1 expression was reduced in all three mutants. Timing of CLN1 expression was similar to that of the wild type in paf1Δ and mbp1Δ, but abundance was reduced significantly more in paf1Δ (more than fivefold) than in mbp1Δ (more than threefold) throughout the cell cycle. In contrast, CLN1 expression in swi4Δ was both significantly reduced and altered in timing. RNR1 expression was shifted in time in both swi4Δ and mbp1Δ, but not paf1Δ. Consistent with the results seen in the asynchronous cells (Fig. 5), the abundance of the RNR1 transcript was not significantly reduced in swi4Δ, but it decreased more than threefold in paf1Δ. None of the mutations seemed to affect the timing of HTB1 expression, but both swi4Δ (3.7-fold) and mbp1Δ (4.6-fold) reduced the abundance of this transcript significantly more than did the paf1Δ mutation (2.2-fold). Finally, the FKS1 transcript that was previously shown to be dependent on Swi4 for cyclic expression (23), demonstrated both a change in cycling and a dramatic (nearly fivefold) increase in expression in the swi4Δ strain; neither effect was seen in paf1Δ.
Genetic evidence suggests that Paf1 functions in both the Swi4- and Mbp1-controlled pathways.
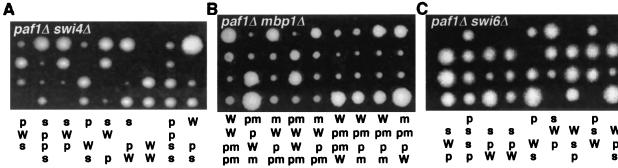
Combining a mutation in PAF1 with mutations in MBP1 or SWI4 could potentially give us additional information about whether they are functioning in the same, or possibly parallel, pathways to regulate expression of cell cycle genes. If the double mutant combinations are similar in phenotype to the most severe single mutant, in this case the paf1Δ strain, then the two genes may be functioning in the same pathway. However, an enhanced phenotype would indicate that the two proteins might have functions in independent pathways (16). This analysis is complicated by the fact that Swi4 and Mbp1 perform partially overlapping essential functions, and the fact that the factors interact with Swi6 which provides a transcriptional activation function to both DNA binding proteins (reviewed in reference 34). Despite these caveats, we found that loss of Paf1 is equivalent to loss of Mbp1 in that a paf1Δ swi4Δ double mutant was inviable, and a paf1Δ mbp1Δ combination was viable and not significantly impaired relative to the paf1Δ strain (Fig. 7 and data not shown).
FIG. 7.
paf1Δ swi4Δ and paf1Δ swi6Δ, but not paf1Δ mbp1Δ, are synthetically lethal. Heterozygous diploids containing paf1Δ and swi4Δ, paf1Δ and swi6Δ, or paf1Δ and mbp1Δ mutations were sporulated, and tetrads were dissected onto YPD plates containing 1 M sorbitol and incubated at 30°C. Tetrads were also dissected onto YPD plates and onto YPD plates at low temperature (23°C) with similar results. Representative tetrads are shown. Spore genotypes are indicated below each tetrad: W, wild type; p, paf1Δ; s, swi4Δ or swi6Δ as appropriate; m, mbp1Δ; pm, paf1Δ mbp1Δ. Genotypes were scored as described in Materials and Methods.
Despite the fact that loss of both Swi4 and Mbp1 is lethal, a mutant lacking Swi6, which is the associated transcription activation component of both factors, is viable (1). However, swi6Δ swi4Δ is lethal, while a swi6Δ mbp1Δ mutant is viable (27). The viability of a swi6Δ mbp1Δ mutant may be due to the ability of Swi4 to activate transcription in the absence of Swi6 (11). We found that, like a swi4Δ swi6Δ mutant, a paf1Δ swi6Δ mutant is not viable (Fig. 7). Paf1 may therefore be required for the essential residual transcription activity of Swi4 in the absence of Swi6.
Overexpression of Swi4 or Mbp1 suppresses some, but not all, paf1Δ phenotypes.
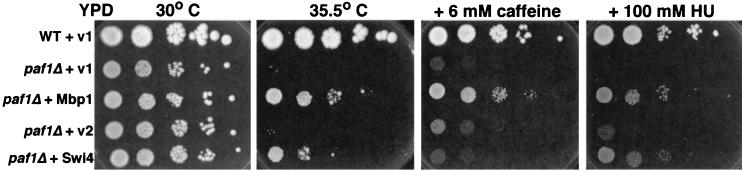
If the Paf1 complex is required for full expression of Swi4- and Mbp1-dependent genes, then overexpression of one or both of the factors might suppress paf1Δ phenotypes. We found that increased expression of Mbp1 suppressed the ts phenotype of paf1Δ at 35.5°C (Fig. 8). Overexpression of Mbp1 also suppressed the cell wall defects of paf1Δ on caffeine and the sensitivity to HU caused by reduced expression of RNR1 (Fig. 8). Overexpression of Swi4 also suppressed the ts defect and sensitivity to HU but was no better than vector alone for suppression of caffeine sensitivity (Fig. 8). Not all paf1Δ phenotypes are corrected by increased expression of Mbp1 or Swi4; for example, neither factor can correct the inability of paf1Δ to grow on 30 μg of hygromycin/ml (data not shown). The ability of both transcription factors to suppress some paf1Δ phenotypes strongly supports the model that they are involved in the same pathway as the Paf1-RNAP II complex.
FIG. 8.
Both Mbp1 and Swi4, when overexpressed, can partially suppress paf1Δ. Spot assays were performed as described in Materials and Methods. WT + v1, YJJ662 transformed with YEp24; paf1Δ + v1, YJJ664 transformed with YEp24; paf1Δ + Mbp1, YJJ664 transformed with BK72; paf1Δ + v2, YJJ664 transformed with YEp352; paf1Δ + Swi4, YJJ664 transformed with B327. Spots from left to right were made by applying 4 μl of a cell suspension containing 107, 106, 105, 104, and 103 cells/ml. Similar results were obtained with two independent sets of transformants.
CLN1 and FKS1 promoter/reporter constructs are not sensitive to loss of Paf1.
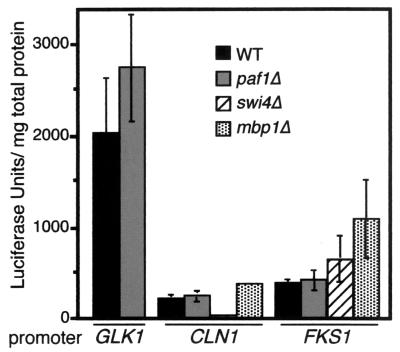
The Paf1 complex is associated with both initiation factors (TFIIB and TFIIF) and factors linked to elongation (Rtf1 and Spt5). To determine which step in transcription of the cell cycle-regulated genes is dependent on Paf1, we made luciferase constructs containing promoters for two genes whose expression is reduced two- to threefold in paf1Δ, FKS1, and CLN1. Both promoters are bound by SBF in vivo (24). In addition, we made a GLK1 promoter construct as a negative control, because abundance of the GLK1 transcript is not decreased in paf1Δ (data not shown). As shown in Fig. 9, expression from this strong promoter is not diminished in paf1Δ. If Paf1 is acting at initiation, then luciferase activity driven by CLN1 and FKS1 promoters should be decreased in paf1Δ compared to that in the wild type. In contrast, we observed that expression from neither the CLN1 nor FKS1 promoter/reporter constructs is decreased in paf1Δ versus wild-type cells. However, the CLN1 promoter is sensitive to loss of Swi4, demonstrating a 5- to 10-fold decrease in swi4Δ. In addition, expression from the FKS1 promoter is slightly increased in swi4Δ and mbp1Δ, matching the small increases in FKS1 expression we observed by RNA analysis (Fig. 5). We conclude that, although full CLN1 transcript abundance is dependent on both Paf1 and Swi4, these factors are acting at different stages of transcription.
FIG. 9.
Output of promoter/reporter constructs does not correlate with measurements of RNA abundance. Constructs containing the GLK1, FKS1, and CLN1 promoters driving expression of the luciferase gene were transformed into wild-type (YJJ662) and paf1Δ (YJJ664) strains. The FKS1 and CLN1 constructs were also transformed into swi4Δ (YJJ1233) and mbp1Δ (YJJ1067) strains. Extracts were made and luciferase activity was measured as described in Materials and Methods. The bars represent the average and standard error derived from two independent experiments, with the activity of each construct measured in quadruplicate in each assay. Data are presented as relative luciferase units per milligram of total protein from the extract.
DISCUSSION
Layers of overlapping controls act to protect the expression of genes critical for cell cycle regulation. The complexity of these promoters and regulatory factors is perhaps an obvious consequence of the importance of maintaining proper regulation of cell division. In this work we have added a new player to the roster of cell cycle regulatory factors. We have shown that a functional form of the yeast Paf1-RNAP II complex is required for normal levels of expression of many cell cycle-regulated genes, including the G1 cyclin CLN1 and the essential RNR1 gene encoding ribonucleotide reductase. The genes analyzed in this study included examples of periodic expression that peak in the G1 (CLN1, RNR1, FKS1, and HO), S (HTB1), S/G2 (SPC25 and SAS3), and M (STP4, CYB5, and FAR1) phases of the cell cycle. In the absence of Paf1, transcript abundance from these genes is reduced from 2- to 6-fold in asynchronous cells, and from 2- to 13-fold during a synchronous cell cycle (Fig. 1 and 3). Paf1 therefore does not seem to interfere with a unique stage of the cell cycle, nor does its loss result in arrest in a particular phase.
Our results are consistent with those of Koch and coworkers, who found that another component of the Paf1 complex, Ctr9, is required for full expression of the G1 cyclin gene CLN2 (28). Mutation of either CTR9 or PAF1 results in identical phenotypes under a variety of growth conditions, including temperature sensitivity (4, 28). When arrested ctr9Δ or paf1Δ cells are released from α-factor at the nonpermissive temperature of 37°C, Koch found that the normal appearance of the CLN2 transcript in G1 is blocked in both strains (28). We have extended this analysis of cells cultured under lethal conditions to show that when paf1Δ cells are synchronized under conditions where they are still viable (30°C), CLN1 expression, and the expression of many other cell cycle-regulated genes, is reduced dramatically throughout the cell cycle (Fig. 3).
CLN1 and CLN2 are coordinately regulated in G1 (9, 45, 53), and their expression requires many of the same transcription factors (reviewed in reference 34). Of particular interest is the report that the TATA-binding protein-associated factor Taf145 is essential for G1 cyclin expression (51). However, a subsequent microarray analysis demonstrated that the reduction in CLN1 and CLN2 expression is not a primary effect of Taf145 depletion. Instead, there is first a dramatic (sevenfold) reduction in CTR9 expression within 45 min of Taf145 shutoff, apparently followed by the loss of CLN1 and CLN2 expression (22). Therefore, Taf145 is required for expression of Ctr9 and a functional form of the Paf1 complex. Without the Paf1 complex there are defects in G1 cyclin expression.
Connections between Paf1 and known cell cycle regulatory factors.
Loss of Paf1 affects genes expressed throughout the cell cycle, but Paf1 is not required for proper expression of all periodic genes. For example, the M/G1 phase-expressed PIR3 gene is expressed normally in both asynchronous and synchronized cultures of a paf1Δ strain (Fig. 1 and 3). When comparing the pattern of genes that depend on Paf1 to the expression patterns for known transcription factors, we see statistically significant, but by no means complete, overlap with genes known to be targets of the SBF and MBF cell cycle regulatory factors (24, 34). For example, mutations in Paf1 affect the SBF targets CLN1 and the HO gene (Fig. 3), as well as CLN2 (28), but Paf1 does not appear to be required for the coordinately expressed CLB5 gene (Chang et al., unpublished). Similarly, Paf1 is required for full expression of the MBF target RNR1, but not POL1. These data must, however, be considered preliminary since they are based on analyses of RNA from asynchronous cultures. We have found, as reported by other investigators (23, 27), that many effects on the expression of cell cycle-regulated genes are much more clearly seen in samples from synchronized cells. For example, we found that CLN1 expression is reduced about threefold in asynchronous paf1Δ cells (Fig. 1), but the effect is greater than fivefold in synchronized cells (Fig. 3).
Ideally, an accurate comparison of the overlaps between genes affected by loss of Paf1 and loss of Swi4 or Mbp1, the selective DNA-binding components of SBF and MBF, respectively, awaits a complete genomic assay using multiple samples from synchronous cultures. Lacking that comprehensive information, we compared a small number of genes throughout the cell cycle in isogenic wild-type, paf1Δ, swi4Δ, and mbp1Δ strains. We found that the effects of neither the swi4Δ nor the mbp1Δ mutation exactly matched that of paf1Δ (Fig. 6). In general, the effect of the paf1Δ mutation was to reduce abundance without altering the timing of the residual cyclic expression. In contrast, we found that the swi4Δ mutation more often affected the timing of expression, as observed previously by Igual et al. (23). In some cases the effect of swi4Δ was to reduce expression, as seen with CLN1 and HTB1, but loss of Swi4 also lead to a dramatic increase of FKS1 expression. Mutation of Mbp1 also results in complex patterns: we found that expression of CLN1 and HTB1 is reduced, but cycling is normal, and RNR1 expression is both reduced and shifted in time in synchronized cells. The lack of a perfect overlap in expression patterns is undoubtedly due to the fact that when Mbp1 is missing, Swi4 can perform many of its functions and vice versa (reviewed in reference 34). In addition, Swi4 possesses a Swi6-independent transcription activation function that may result in unique patterns of expression (11).
Finally, our observation that loss of Paf1 acts at a different transcriptional stage than Swi4 or Mbp1 helps to explain both the nonidentical effects on gene expression and the suppression of some paf1Δ phenotypes by overexpression of Swi4 or Mbp1. Perhaps the increased abundance of the transcriptional activators drives increased activation that partially overcomes the defects in later stages of transcription caused by lack of Paf1.
The observed similarities in effects on gene expression seen in paf1Δ, swi4Δ, and mbp1Δ strains are consistent with our genetic analysis of interactions between mutations in the PAF1, SWI4, MBP1, and SWI6 genes (Fig. 7). Loss of Paf1 behaves genetically like loss of Mbp1 in that it is lethal in combination with loss of Swi4. It also acts like loss of Swi4 in its lethality in combination with loss of Swi6. Loss of Paf1 results in reduced levels of CLN1 (and other genes), and loss of Swi4 or Swi6 also reduces CLN1 levels at a different transcriptional stage. Therefore, combining these mutations would be predicted to reduce expression of CLN1 (or other essential genes) to a lethal level.
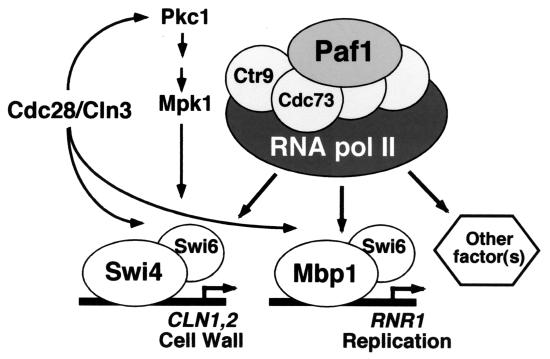
Other transcription factors that are important for cell cycle regulation include the essential Mcm1 protein that acts through the early cell cycle box (ECB) to control expression of several genes, including CLN3, FAR1, and CDC47 at M/G1 (30, 33). Although we have shown that FAR1 expression is significantly reduced in a paf1Δ strain (Fig. 1 and 3), microarray data indicate that other ECB-regulated genes, including CLN3 and CDC47, are not Paf1 dependent (Chang et al., unpublished). However, these data derived from asynchronous samples are subject to the same caveats described above. In addition, because they significantly reduce the expression of CLN1 and CLN2, paf1Δ and ctr9Δ are lethal in combination with cln3Δ (28). These data are consistent with the model shown in Fig. 10, in which a major function of the Paf1 complex is downstream of Cdc28/Cln3 at the level of stimulating the second wave of G1 cyclin synthesis as well as genes important for cell wall biosynthesis and DNA replication.
FIG. 10.
The Paf1 complex is required for full expression of many SBF- and MBF-regulated genes. The model, described further in the Discussion, summarizes the genetic and molecular analyses in this work. Mutation of PAF1 results in reduced expression of Swi4 targets like CLN1 and Mbp1 targets like RNR1. The synthetic lethality of paf1Δ in combination with swi4Δ or swi6Δ indicates that the Paf1 complex has essential functions that are redundant with these factors. High-copy suppression of paf1Δ phenotypes by SWI4 and MBP1 is consistent with these factors acting downstream of the Paf1 complex, or at a different stage of transcription.
Since the phenotype of a paf1Δ mutant is much more severe than the phenotypes of either mbp1Δ or swi4Δ single mutants, it is clear that there must be other genes dependent on the Paf1 complex (Fig. 10). Potential targets include genes regulated by Rlm1, Xbp1, Stb1 and Fkh1 and -2. Rlm1 is a MADS box transcription factor thought to act downstream of the Pkc1 pathway (14, 52). Because combining mutations in RLM1 and PAF1 does not lead to an enhanced phenotype (4), these two factors may function in the same pathway. However, the relatively small number of genes controlled by Rlm1 (26) make it unlikely that this factor is a major contributor to the defects seen in a paf1Δ strain. Xbp1 is a transcriptional repressor related to Swi4 and Mbp1 that is involved in CLN1 gene expression during meiosis (32). The Stb1 protein interacts directly with the Swi6 transcriptional activator portion of SBF and MBF (20). Stb1 is a phosphorylation target of Cdc28/Cln, and in the absence of Cln3 the loss of Stb1 leads to defects in progression through G1 (20). Finally, the forkhead transcription factors Fkh1 and Fkh2 play an important role in cell cycle regulation as well as in control of cell morphology and gene silencing (21, 55). It is possible that the Paf1 complex is acting at genes controlled by one or more of these factors.
Multiple layers of cell cycle control.
Although it is clear that MBF and SBF are among the most important players in cell cycle regulation, even their roles have been difficult to define due to the apparently redundant controls for each periodic promoter. For example, distinct DNA-binding sequences have been identified for Mbp1 (ACGCGTNA) and Swi4 (CACGAAA), yet the two proteins can cross-recognize each other's DNA elements in vitro and apparently in vivo (reviewed in reference 34). In fact, in the CLN1 promoter the MBF sites are controlled by Swi4, but not by Mbp1 (27, 38). In addition, MBF sites in the RNR1 promoter are responsive to mutation of MBP1, but not of SWI4 (13), and when the same sequences are assayed in a reporter construct, mutation of SWI4 affects expression (49). In this regard it is interesting that we observed that overexpression of either Swi4 or Mbp1 suppresses the HU sensitivity of paf1Δ caused by reduced expression of Rnr1 (Fig. 8). The recent genomic analysis of Swi4 and Mbp1 in vivo binding sites is a first step towards resolving this issue (24). There are sites where only one or the other factor is bound, plus many genes where both factors reside. However, information about where, for example, Swi4 is bound when Mbp1 is absent is still not available. Even when all of the obvious MBF and SBF sites have been removed from the CLN1 promoter, expression is still cyclic (38). It is therefore perhaps not surprising that mutations in PAF1 do not exactly resemble mutations in either SWI4 or MBP1. As shown in Fig. 10, the Paf1 complex probably acts at genes controlled by both of these factors, plus others, at a subset of the promoters under their control.
Recently, the Nasmyth lab reported a temporal dissection of the events at the HO, CLN1, and CLN2 promoters leading to transcriptional activation in G1 (10, 11). They found that binding of transcription factor Swi5 leads to the recruitment of chromatin remodeling complexes, which facilitates binding of SBF. Bound SBF recruits the Srb-mediator complex in the absence of RNAP II. Recruitment of RNAP II occurs in a subsequent step that requires activation of Cdc28. It will be interesting to determine whether the Paf1-RNAP II complex is playing a unique role at these promoters that are subject to such complex controls.
Acknowledgments
We thank members of the Jaehning lab, including J. L. Betz, C. Mueller, and K. Penheiter, for sharing results prior to publication and for their comments on the manuscript. We are grateful to C. Phernetton for her help with the high-copy suppression studies and to J. Fostel for the microarray experiments. D. Reines is gratefully acknowledged for the luciferase reporter construct and for invaluable advice about the assays. We also thank our colleagues D. Bentley, A. Gutierrez-Hartmann, M. Huang, R. Sclafani, and P. Dohrmann for reagents, input into the experimental design, and their comments on the manuscript.
This work was supported by a grant from the National Institutes of Health (GM38101) to J.A.J.
REFERENCES
- 1.Andrews, B. J., and I. Herskowitz. 1989. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature 342:830-833. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, B. J., and L. A. Moore. 1992. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc. Natl. Acad. Sci. USA 89:11852-11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., et al. (ed.). 1987. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 4.Betz, J. L., T. M. Washburn, S. E. Porter, C. L. Mueller, and J. A. Jaehning. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis and lipid and nucleic acid metabolism. Mol. Genet. Genom., in press. [DOI] [PubMed]
- 5.Breeden, L. L. 1997. Alpha-factor synchronization of budding yeast. Methods Enzymol. 283:332-341. [DOI] [PubMed] [Google Scholar]
- 6.Chang, M. 1998. Identification and characterization of a novel RNA polymerase II complex in Saccharomyces cerevisiae. Ph.D. thesis. University of Colorado Health Sciences Center, Denver.
- 7.Chang, M., D. French-Cornay, H. Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, M., and J. A. Jaehning. 1997. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 25:4861-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway, L. Wodicka, T. G. Wolfsberg, A. E. Gabrielian, D. Landsman, D. J. Lockhart, and R. W. Davis. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2:65-73. [DOI] [PubMed] [Google Scholar]
- 10.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 11.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 12.Dirick, L., T. Bohm, and K. Nasmyth. 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dirick, L., T. Moll, H. Auer, and K. Nasmyth. 1992. A central role for SWI6 in modulating cell cycle Start-specific transcription in yeast. Nature 357:508-513. [DOI] [PubMed] [Google Scholar]
- 14.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin, and I. Herskowitz. 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 16:4924-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarente, L. 1993. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 9:362-366. [DOI] [PubMed] [Google Scholar]
- 17.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press Inc., San Diego, Calif.
- 18.Hampsey, M., and D. Reinberg. 1999. RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev. 9:132-139. [DOI] [PubMed] [Google Scholar]
- 19.Hartwell, L. H. 1967. Macromolecule synthesis in temperature-sensitive mutants of yeast. J. Bacteriol. 93:1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, Y., M. Costanzo, L. Moore, R. Kobayashi, and B. J. Andrews. 1999. Regulation of transcription at the Saccharomyces cerevisiae Start transition by Stb1, a Swi6-binding protein. Mol. Cell. Biol. 19:5267-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollenhorst, P. C., M. E. Bose, M. R. Mielke, U. Muller, and C. A. Fox. 2000. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics 154:1533-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 23.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001-5013. [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 25.Johnston, L. H., and A. L. Johnson. 1995. The DNA repair genes RAD54 and UNG1 are cell cycle regulated in budding yeast but MCB promoter elements have no essential role in the DNA damage response. Nucleic Acids Res. 23:2147-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 27.Koch, C., T. Moll, M. Neuberg, H. Ahorn, and K. Nasmyth. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261:1551-1557. [DOI] [PubMed] [Google Scholar]
- 28.Koch, C., P. Wollmann, M. Dahl, and F. Lottspeich. 1999. A role for Ctr9p and Paf1p in the regulation of G1 cyclin expression in yeast. Nucleic Acids Res. 27:2126-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowndes, N. F., A. L. Johnson, and L. H. Johnston. 1991. Coordination of expression of DNA synthesis genes in budding yeast by a cell-cycle regulated trans factor. Nature 350:247-250. [DOI] [PubMed] [Google Scholar]
- 30.MacKay, V. L., B. Mai, L. Waters, and L. L. Breeden. 2001. Early cell cycle box-mediated transcription of CLN3 and SWI4 contributes to the proper timing of the G1-to-S transition in budding yeast. Mol. Cell. Biol. 21:4140-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madden, K., Y. J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781-1784. [DOI] [PubMed] [Google Scholar]
- 32.Mai, B., and L. Breeden. 1997. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol. Cell. Biol. 17:6491-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McInerny, C. J., J. F. Partridge, G. E. Mikesell, D. P. Creemer, and L. L. Breeden. 1997. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 11:1277-1288. [DOI] [PubMed] [Google Scholar]
- 34.Mendenhall, M. D., and A. E. Hodge. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1191-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller, C. L., and J. A. Jaehning. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22:1971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 37.Nash, R., G. Tokiwa, S. Anand, K. Erickson, and A. B. Futcher. 1988. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7:4335-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Partridge, J. F., G. E. Mikesell, and L. L. Breeden. 1997. Cell cycle-dependent transcription of CLN1 involves swi4 binding to MCB-like elements. J. Biol. Chem. 272:9071-9077. [DOI] [PubMed] [Google Scholar]
- 39.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 40.Pringle, J. R., and L. H. Hartwell. 1981. The Saccharomyces cerevisiae cell cycle, p. 97-142. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), Molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi, X., M. Chang, A. J. Wolf, C. H. Chang, A. A. Frazer-Abel, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 17:1160-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi, X., A. Finkelstein, A. J. Wolf, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, I. A., P. B. McIntosh, P. Pala, M. K. Treiber, S. Howell, A. N. Lane, and S. J. Smerdon. 2000. Characterization of the DNA-binding domains from the yeast cell-cycle transcription factors Mbp1 and Swi4. Biochemistry 39:3943-3954. [DOI] [PubMed] [Google Scholar]
- 48.Ulery, T. L., D. A. Mangus, and J. A. Jaehning. 1991. The yeast IMP1 gene is allelic to GAL2. Mol. Gen. Genet. 230:129-135. [DOI] [PubMed] [Google Scholar]
- 49.Verma, R., J. Smiley, B. Andrews, and J. L. Campbell. 1992. Regulation of the yeast DNA replication genes through the Mlu I cell cycle box is dependent on SWI6. Proc. Natl. Acad. Sci. USA 89:9479-9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 51.Walker, S. S., W. C. Shen, J. C. Reese, L. M. Apone, and M. R. Green. 1997. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell 90:607-614. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe, Y., G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:2615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wittenberg, C., K. Sugimoto, and S. I. Reed. 1990. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell 62:225-237. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, L. P., R. Prentice, and L. Breeden. 2001. Statistical modeling of large microarray data sets to identify stimulus-response profiles. Proc. Natl. Acad. Sci. USA 98:5631-5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]