Abstract
The de novo formation of multilayered spore walls inside a diploid mother cell is a major landmark of sporulation in the yeast Saccharomyces cerevisiae. Synthesis of the dityrosine-rich outer spore wall takes place toward the end of this process. Bisformyl dityrosine, the major building block of the spore surface, is synthesized in a multistep process in the cytoplasm of the prospores, transported to the maturing wall, and polymerized into a highly cross-linked macromolecule on the spore surface. Here we present evidence that the sporulation-specific protein Dtr1p (encoded by YBR180w) plays an important role in spore wall synthesis by facilitating the translocation of bisformyl dityrosine through the prospore membrane. DTR1 was identified in a genome-wide screen for spore wall mutants. The null mutant accumulates unusually large amounts of bisformyl dityrosine in the cytoplasm and fails to efficiently incorporate this precursor into the spore surface. As a result, many mutant spores have aberrant surface structures. Dtr1p, a member of the poorly characterized DHA12 (drug:H+ antiporter with 12 predicted membrane spans) family, is localized in the prospore membrane throughout spore maturation. Transport by Dtr1p may not be restricted to its natural substrate, bisformyl dityrosine. When expressed in vegetative cells, Dtr1p renders these cells slightly more resistant against unrelated toxic compounds, such as antimalarial drugs and food-grade organic acid preservatives. Dtr1p is the first multidrug resistance protein of the major facilitator superfamily with an assigned physiological role in the yeast cell.
Diploid a/α cells of the budding yeast Saccharomyces cerevisiae undergo a specialized developmental program termed sporulation when transferred to a nitrogen-free medium containing potassium acetate as nonfermentable carbon source. The final product of sporulation is an ascus that consists of four haploid spores surrounded by the ascus wall, the former vegetative cell wall (for a review, see reference 27). Spores are protected from adverse environmental conditions by the spore wall. Especially the surface layers contribute both to the spores' mechanical rigidity and their resistance against chemical and enzymatic attack (3). Spore wall synthesis begins with the formation of the prospore membrane, a bilayered electron-dense structure that starts to form during the second meiotic division on the cytoplasmic side of each of the four spindle pole bodies by fusion of secretory vesicles (9, 13, 23, 31-33). As meiosis progresses, the prospore membrane extends along the outer surface of the nuclear envelope. Septins, among them the sporulation-specific proteins Spr3p and Spr28p, localize at the leading tip of the prospore membrane under control of the Gip1p-Glc7p phosphatase complex and might be involved in its extension and directed growth (14, 16, 44). Other proteins localized to the growing prospore membrane are SspIp, Ady3p, and Don1p, which form the leading edge protein coat (23, 33) and presumably Sps2p (11, 38), but their roles remain to be elucidated. It was originally suggested that the prospore membrane originates from the endoplasmic reticulum (29) but, as shown recently, a specialized sporulation-specific post-Golgi branch of the secretory pathway is involved in the de novo synthesis of this membrane (34). By the end of meiosis II, each of the four prospore membranes have completely engulfed a newly formed haploid nucleus, enclosing also some portions of the mother-cell cytoplasm and organelles. In a final process, spore wall material is deposited in the lumenal space between the two layers of the prospore membrane and the mature spore wall is eventually formed.
In electron micrographs, four layers of the mature spore wall can be distinguished after being stained with OsO4 (24). The two inner layers consist mostly of glucans and mannoproteins, and their chemical composition is very similar to that of the vegetative cell wall (5, 20). Both outer layers have no chemical or morphological equivalent in vegetative cell walls. The second outer layer, which is surrounded by the thin hydrophobic surface layer, consists of chitosan (5). The major component of the surface layer is the amino acid, dityrosine, a highly fluorescent dimer of tyrosine. In the spore surface, dityrosine occurs in its ll-, dl-, and dd-configurations and forms a highly cross-linked, insoluble macromolecule that is completely resistant to proteolytic digestion (3, 7). Dityrosine is synthesized in a two-step process that takes place exclusively in the cytoplasm of the maturing spore, as shown both genetically and biochemically (3, 4). In a first step, free l-tyrosine is chemically modified by Dit1p. The result of this reaction is N-formyl tyrosine. In the next step, two molecules of N-formyl tyrosine are cross-linked by Dit2p, a cytochrome P-450, to form ll-N,N′-bisformyl dityrosine (6). The mechanism by which this soluble precursor is incorporated into the maturing spore wall and epimerized remains to be elucidated.
As part of our long-term project to clarify the mechanism of spore wall formation, we describe here the identification of Dtr1p, encoded by open reading frame YBR180w. Dtr1p, a sporulation-specific member of the major facilitator superfamily involved in multidrug resistance (MFS-MDR) (19, 35), is shown to be the membrane transporter responsible for the translocation of bisformyl dityrosine from the prospores' cytoplasm to the maturing spore wall during spore formation.
MATERIALS AND METHODS
Yeast strains, growth media, and growth conditions.
The strains used in this work are listed in Table 1. Deletion strains were provided by the EUROFAN deletion library (http://mips.gsf.de/proj/yeast/CYGD/db/index.html) in the FY1679 strain background and by the Saccharomyces Genome Deletion Project (http://sequence-www.stanford.edu/group/yeast_deletion_project/deletions3.html) instrain BY4743. Standard yeast genetic methods and growth media were used (42). Unless otherwise noted, cells were grown and sporulated on agar plates at 28°C as described previously (4).
TABLE 1.
S. cerevisiae strains
| Name | Genotype | Source |
|---|---|---|
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0+ +/met15Δ0 ura3Δ0 /ura3Δ0 | EUROSCARF |
| BY4743-Δdtr1 | MATa/α his3Δ1/his3Δ1 leu2Δ0 /leu2Δ0 lys2Δ0/+ +/met15Δ0 ura3Δ0/ura3Δ0 ybr180w::KanMX4/ybr180w::KanMX4 | EUROSCARF |
| FY1679Δ | MATa/α ura3-52/ura3-52 trp1Δ631/+ his3Δ200+ GAL2+/GAL2+ | EUROFAN |
| FY1679-Δdtr1 | MATa/aura3-52/ura3-52 trp1D631/+ his3D200/+ GAL2+/GAL2+ybr180w::KanMX4/ybr180w::KanMX4 | EUROFAN and this work |
| INVsc1 | MATaleu2-3,112 trp 1-289. ura3-52 his3D1 | Invitrogen |
| LP112 | MATa/aade2-1/ade2-1 his3-11,15/his3-11,15 leu2-3,115/leu2-3, 115 trp1-1/trp1-1 ura3-1/ura3-1 canR1-100/canR1-100 | S. Lindquist |
| LP112-a/a | MATa/aade2-1/ade2-1 his3-11,15/his3-11,15 leu2-3,115/leu2-3,115 trp1-1/trp1-1 ura3-1/ura3-1 canR1-100/canR1-100 | S. Lindquist |
| LP112-a/a | MATa/aade2-1/ade2-1 his3-11,15/his3-11,15 leu2-3,115/leu2-3, 115 trp1-1/trp1-1 ura3-1/ura3-1 canR1-100/canR1-100 | S. Lindquist |
| PB39 | MATa/α ura3/ura3 leu2/leu2 his4X/his4B lys2/lys2 cda1-cda2::URA3/cda1-cda2::URA3 ho::LYS2/ho::LYS2 | This work |
| PB75 | Derivative of PB39 and SK1-Δdtr1 | This work |
| SK1 | MATa/α ura3/ura3 leu2/leu2 his4X/his4B lys2/lys2 ho::LYS2/ho::LYS2 | Laboratory stock |
| SK1-Δdtr1 | MATa/α ura3/ura3 leu2/leu2 his4X/his4B lys2/lys2 ho::LYS2/ho::LYS2 ybr180w::KanMX4/ybr180w::KanMX4 | This work |
| W303a | MATaura3-1 trp1-1 leu2-3,112 his3-11 ade2-1 can1-100 | Laboratory stock |
| W303a-Δdtr1 | MATaura3-1 trp1-1 leu2-3,112 his3-11 ade2-1 can1-100 ybr180w::KanMX4 | This work |
| W303-euro | MATa/α ura3-1/ura3-1 trp1-1/trp1-1 leu2-3,112/leu2-3,112 his3-11/his3-11 ade2-1/ade2-1 can1-100/can1-100 | Laboratory stock |
| W303-Δdtr1 | MATa/α ura3-1/ura3-1 trp1-1/trp1-1 leu2-3,112/leu2-3,112 his3-11/his3-11 ade2-1/ade2-1 can1-100/can1-100 ybr180w::KanMX4/ybr180w::KanMX4 | This work |
Mutant screen.
Individual deletion strains were patched on YPD plates (1% yeast extract, 2% peptone, 2% glucose, 2% agar). After 24 h, colonies were transferred to nitrocellulose membranes (Schleicher & Schuell) by replica plating, and the filters were incubated on fresh YPD plates for an additional 12 h. The filters were then transferred to sporulation plates (1% potassium acetate, 0.1% yeast extract, 0.025% glucose, 40 ppm of uracil, 10 ppm of histidine, 60 ppm of leucine, 40 ppm of tryptophan, and 2% Bacto Agar). After completion of sporulation (usually 5 to 6 days in the FY1679 and BY4743 strain backgrounds), cells were removed from the filter and hydrolyzed, and the dityrosine content was determined by reversed-phase high-performance liquid chromatography (RP-HPLC) (2).
Dityrosine analyses.
For analysis of total dityrosine, ca. 2 mg of cells (wet weight) of a sporulated culture were hydrolyzed in 50 μl of 6 N HCl in an open Eppendorf tube at 95°C. After 5 h, the dried hydrolyzate was suspended in 200 μl of water and cleared by centrifugation. Dityrosine analysis was performed on an Agilent 1100 HPLC system and a Waters 470 fluorescence detector set to 285-nm excitation and 425-nm emission wavelengths with a Waters Nova-Pak C18 column (3.9 by 150 mm, 4 μm). Dityrosine was eluted at 7°C under isocratic conditions with 5% (vol/vol) acetonitrile in 0.01 M aqueous trifluoroacetic acid as eluant at a flow rate of 1 ml/min. Under these conditions, baseline separation of the diastereomers ll- and dl-dityrosine was achieved. The ratio of dl- to ll-dityrosine were determined by peak integration. The average elution time of dityrosine was 12 min. Dityrosine-containing spore wall precursors in deproteinated cell extracts (4) were analyzed similarly, except that the column was developed with a linear gradient of acetonitrile in 0.01 M trifluoroacetic acid (0 to 50% CH3CN in 55 min). Standards of dityrosine and its derivatives were prepared as described previously (6).
Construction of plasmids.
Standard techniques were used for plasmid constructions (43). The coding regions of the S. cerevisiae MDR genes mentioned in the text were amplified by PCR from genomic DNA isolated from strain SK1 and cloned into the expression plasmid pYES2 (Invitrogen). For drug susceptibility tests, DTR1 was cloned by the gap repair technique into the centromeric plasmid pFL38, yielding plasmid pYCG_DTR1, as described previously (8). For the localization studies, the complete DTR1 gene (including regulatory regions) was amplified from SK1 DNA and, in a first step, cloned into the centromeric vector pUG35 (kindly provided by J. H. Hegemann), thereby creating a C-terminal fusion with a yeast-enhanced green fluorescent protein (GFP). In a second step, the SacI-KpnI fragment of this vector containing the DTR1-GFP fusion was cloned into the multicopy plasmid YEplac181 (18). The bacterial strain used was Escherichia coli DH5α. The primers are listed in Table 2.
TABLE 2.
Primers used in this studya
| Name | Sequence | Use |
|---|---|---|
| Dtr1-f | 5′-GAGAGAGAGCTCTTAAGGGCTACTGTAACCTGCGTTAT | DTR1-GFP fusion |
| Dtr1-r | 5′-GAGAGAGAGAGGAATTCAAATTCTTTGGCAGTATATT | DTR1-GFP fusion |
| Ybr043-f | 5′-AGAGAGAGGGATCCATGCAAGCCCAAGGTTCACAA | Expression of YBR043c |
| Ybr043-r | 5′-AGAGAGAGTCTAGATTAATCAATTTTGTCGTACAA | Expression of YBR043c |
| Ybr180-f | 5′-GAGAGAGGATCCGAGAATGGGAAGCGAACCGTTTCA | Expression of DTR1 |
| Ybr180-r | 5′-GAGAGATCTAGAAATGCACGTAATTCATAAACT | Expression of DTR1 |
| Yh1035-f | 5′-GAGAGAGAGAGCTCATGGGAACGGATCCCCTTAT | Expression of YHL035c |
| Yh1035-r | 5′-GAGAGAGATCTAGATTATTTCATCATCTTACTTGAT | Expression of YHL035c |
| Yil120-f | 5′-AGAGAGAGGGATCCATGACCAAACAACAAACTTCT | Expression of QDR1 |
| Yil120-r | 5′-AGAGAGAGTCTAGATCACGTCGAAACTTTTTCTGA | Expression of QDR1 |
| Yi1121-f | 5′-AGAGAGAGGGATCCATGGCAGGAGCAACATCAAGT | Expression of YIL121w |
| Yil121-r | 5′-AGAGAGAGCTCGAGTTAATTTACTCCCAGTTCTTG | Expression of YIL121w |
| Yk1209-f | 5′-GAGAGAGAAAGCTTATGAACTTTTTAAGTT | Expression of STE6 |
| Yk1209-r | 5′-GAGAGAGACTCGAGTTAACTGCTTTGGTTGGA | Expression of STE6 |
| Ykr104-f | 5′-GAGAGAGAAAGCTTATGAGTTCAGTAGAGAGAATAA | Expression of YKR104w |
| Ykr104-r | 5′-GAGAGAGATCTAGATTATCTTTTATTATCGAATGAG | Expression of YKR104w |
| Yl1028-f | 5′-GAGAGAGAGGATTCATGTCGGATCATTCTCCCAT | Expression of TPO1 |
| Yl1028-r | 5′-GAGAGAGATCTAGATTAAGCGGCGTAAGCATACT | Expression of TPO1 |
| Ynl065-f | 5′-AGAGAGAGGATCCATGTCACGAAGTAACAGTATAT | Expression of AQR1 |
| Ynl065-r | 5′-AGAGAGAGTCTAGATTAATTATGATTATCGTTCTG | Expression of AQR1 |
Restriction sites used for cloning are underlined.
Northern analysis.
Total RNA was isolated from SK1, and Northern analyses were performed as described previously (3). Probes were labeled with 32P by random priming.
Dityrosine transport in vegetative cells.
INVsc1 cells (Invitrogen) cotransformed with a plasmid harboring DIT1 and DIT2 (4) and a plasmid containing DTR1 (or any other open reading frame coding for an MDR protein) under control of the GAL1 promoter were grown in liquid minimal medium with glucose as the carbon source until reaching an optical density of 0.8, shifted to galactose-containing minimal medium, and incubated for 24 h at 28°C. For the actual dityrosine transport assay, the fully induced cells were centrifuged, washed once with sterile water, and resuspended in fresh galactose medium at a cell density of optical density 0.4. At specific intervals, 1 ml of the cell suspension was removed, and the cells were separated from the medium by centrifugation. The bisformyl dityrosine content of the medium was determined in the acidified supernatant by RP-HPLC with gradient elution. The amount of dityrosine in the cells was estimated after hydrolysis in 6 N HCl by using the isocratic HPLC method. Alternatively, the bisformyl dityrosine content of the cells was determined by analyzing the deproteinated cell extracts by the HPLC gradient elution method.
Microscopy.
For electron microscopy, cells were fixed in glutaraldehyde, stained with acetate-buffered osmium tetroxide, dehydrated with ethanol, and embedded in Epon 812. After the thin sections were stained with uranyl acetate and lead citrate, samples were examined with a Philips EM400 microscope. For fluorescence and differential interference contrast microscopy, a Zeiss Axioskop microscope equipped with an oil-immersion ×100 objective lens was used. DAPI (4′,6′-diamidino-2-phenylindole) and Calcofluor staining was done as described previously (41). Tubulin was visualized by immunofluorescence in formaldehyde-fixed cells as described previously (22) by using a mouse monoclonal anti-α-tubulin antibody (Sigma) as the primary antibody and sheep anti-mouse Cy3 conjugate (Sigma) as the secondary antibody. Images were captured with a video camera and electronically enhanced.
Other techniques.
Multidrug susceptibility assays were performed as described previously (36, 46). They were carried out on MM2 minimal medium agar plates supplemented with the metabolic inhibitors by using a complete tetrad resulting from sporulation of the deleted heterozygous W303 strain. When a susceptibility phenotype was detected in the two dtr1-Δ strains, one wild-type strain (W303a) and one deleted strain (W303a-Δdtr1), belonging to the same tetrad, were transformed either with pYCG_DTR1 or the empty cloning vector, and the effect of the specific metabolic inhibitor on the growth behavior of the transformants was compared on selective MM2-U agar plates. Tests for resistance of spores against heat and organic solvents were performed as described previously (3). Glucosamine levels in hydrolyzates of spore walls were determined by using the Waters Pico-Tag amino acid analysis system (50).
RESULTS
Identification of DTR1.
When sporulated cultures of S. cerevisiae are treated with hydrochloric acid at elevated temperatures, both the ll- and dl-dityrosine-containing macromolecule of the spore surface and soluble spore wall precursors such as bisformyl dityrosine are converted into a mixture of free ll- and dl-dityrosine that can be separated by isocratic RP-HPLC. Hydrolysates of intact cells from a sporulated culture of S. cerevisiae contain ll- and dl-dityrosine in a constant and reproducible ratio that for most laboratory strains is 3:2. Since the epimerization of dityrosine takes place exclusively after incorporation into the maturing spore wall and, on the other hand, all dityrosine-containing soluble spore wall precursors are in the ll-configuration, deviations from the wild-type ratio of dl- to ll-dityrosine are an excellent marker for the identification of defects in the progression of meiosis and/or spore wall formation. This was shown previously with two mutant strains, smk1-Δ and cda2-Δ. In both cases the mutations led to a significant change of the dl/ll ratio (10, 50; unpublished results).
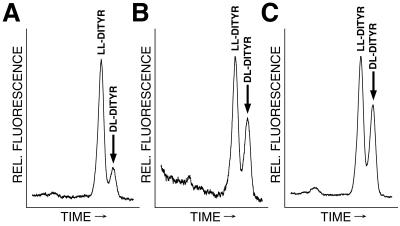
Using this approach, we screened both the EUROFAN deletion library, consisting of 624 single-gene-deletion strains, and the library of the Saccharomyces Genome Deletion Project, which contains a nearly complete set of deletions of all yeast open reading frames (51), for mutants with deviations from the wild-type dl/ll-dityrosine ratio. We identified a total of 15 strains that had lost their ability to form dl-dityrosine altogether and 191 strains that displayed a clear change of the dl/ll ratio. In the strain harboring a deletion of the open reading frame YBR180w, a particularly strong phenotype was observed (Fig. 1A and B). The amount of dl-dityrosine was reduced by 65%, the dl/ll ratio changed from 0.6 in the wild-type strain to 0.2 in the mutant. Otherwise, the mutant strain was at first glance indistinguishable from the corresponding wild type. The sporulation efficiency was not affected, and the asci contained birefringent spores in phase and interference contrast light microscopy. In order to confirm the phenotype and to rule out any strain-specific effects, we repeated the deletion of YBR180w in the strains W303 and SK1. In both strains, the deletion led to an identical phenotype as observed in the original strain backgrounds. A single-copy plasmid containing the complete open reading frame of YBR180w restored the original phenotype (Fig. 1C), indicating that this gene was indeed responsible for the reduced amounts of dl-dityrosine.
FIG. 1.
The disruption of DTR1 leads to a decrease of dl-dityrosine levels in asci. Cells from sporulated cultures of a dtr1-Δ strain (A), the wild type (B), and a dtr1-Δ strain transformed with the cognate clone (C) were hydrolyzed in 6 N HCl to liberate dityrosine from macromolecular structures and separated by isocratic RP-HPLC with fluorescence detection. Only relevant parts of the chromatograms are shown. ll-Dityrosine eluted after 11.3 min; dl-dityrosine eluted after 12.1 min. All strains were FY1679 background.
In silico analysis of YBR180w predicted that this open reading frame codes for a protein of 63,407 Da with 12 membrane-spanning regions. YBR180w, which we named DTR1 (for dityrosine transporter [see below]), was identified by sequence homology as a member of the poorly characterized family of drug:H+ antiporters with 12 predicted transmembrane spans, DHA12 (TC#2.1.3) (19, 37).
dtr1-Δ spores accumulate soluble, unincorporated bisformyl dityrosine.
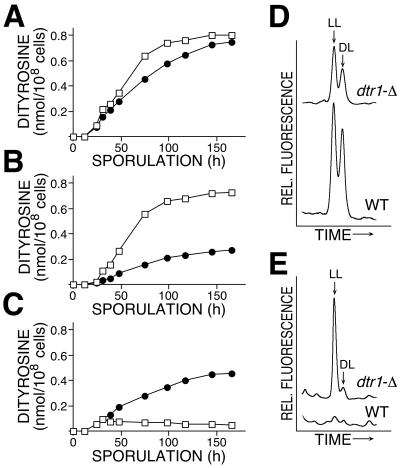
The dramatic reduction of dl-dityrosine in total cell hydrolysates of dtr1-Δ spores could be caused either by a defect in dityrosine epimerization in the maturing spore wall or by the accumulation of unincorporated ll-dityrosine (or its low-molecular-weight derivatives) in the cytoplasmic fraction. To distinguish between these two possibilities, we analyzed the dityrosine content of the cytoplasmic and the wall fraction separately. At specific intervals, cells were removed from the sporulation culture and disintegrated, and the soluble and particulate fractions were separated by centrifugation. After hydrolysis, dityrosine was analyzed and quantified by RP-HPLC. Overall, dityrosine production was comparable in wild-type and dtr1-Δ asci (Fig. 2A), indicating that the synthesis of dityrosine was not affected in the deletion strain and that the observed effects were not merely due to a delay of meiosis or spore formation. A major difference between the wild type and the mutant was observed when the spore wall and cytoplasmic dityrosine were analyzed separately. Incorporation of dityrosine into spore walls followed the same kinetics both in the wild type and the mutant, but the total amount of dityrosine in the mutant spore wall was reduced by more than 60% (Fig. 2B). Despite this significant reduction, the ratio of spore wall dityrosine in the dl and ll configurations remained similar in both strains (Fig. 2D). Since the formation of the insoluble dityrosine-containing macromolecule is closely linked with dityrosine epimerization (4), it can be concluded from this result that the cross-linking reaction itself was not directly affected in dtr1-Δ. Next, we compared dityrosine levels in the cytoplasmic fractions of dtr1-Δ and wild type. During the early phases of sporulation, no difference in the accumulation of soluble dityrosine was noticeable. Only at later stages did the mutant phenotype become obvious. Although the appearance of dityrosine in the cytoplasmic fraction was only transient in the wild type, peaking at 36 h after initiation of sporulation, the mutant strain continued to accumulate soluble dityrosine throughout the process of spore formation (Fig. 2C and E). At the end of sporulation the cytoplasmic fraction of the mutant contained nearly twice the amount of dityrosine than the spore wall fraction. In the wild type, in contrast, >90% of the cytoplasmic dityrosine became incorporated into the spore wall.
FIG. 2.
Comparison of dityrosine synthesis of dtr1-Δ and wild type. (A to C) Time course of dityrosine accumulation. Cells were removed from the sporulation cultures at the indicated times, total cells or fractions of cell homogenates were hydrolyzed in 6 N HCl, and dityrosine was quantitated by RP-HPLC. (A) Dityrosine in total cells; (B) dityrosine incorporated into spore walls; (C) soluble dityrosine in the cytoplasmic fraction. □, Wild type; •, dtr1 deletion mutant. The strain background was FY1679. (D and E) Comparison of the dityrosine content of the wall fraction (D) and the cytoplasmic fraction (E) of cells after the completion of the sporulation program. Strains used for this experiment were of the SK1 background. The relevant parts of the actual HPLC chromatograms are shown; peaks representing ll- and dl-dityrosine are indicated. The corresponding chromatograms are drawn to scale.
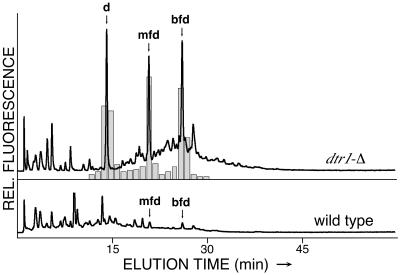
In all of these assays, we analyzed the free amino acid, dityrosine, liberated from higher molecular compounds by acid hydrolysis. Consequently, these experiments did not provide any information about the nature of the dityrosine-containing substances accumulating in the mutant's cytoplasmic fraction. We therefore analyzed deproteinated cell extracts without prior hydrolysis by RP-HPLC with gradient elution. Under these conditions, soluble dityrosine-containing compounds are separated and can be identified by their UV spectra, amino acid analysis, or by comparison with synthetic standards. (6). Figure 3 shows the elution profiles of soluble cell extracts from wild-type and dtr1-Δ cells after 6 days of sporulation and obtained with fluorescence detection. The extract of the wild-type cells contained only trace amounts of bisformyl dityrosine and its degradation product, monoformyl dityrosine. This is typical for wild-type strains at the end of the sporulation program (4). The mutant extract, in contrast, showed three major peaks corresponding to bisformyl dityrosine, monoformyl dityrosine, and free dityrosine, respectively. No other dityrosine-containing compounds were present. We speculate that the deletion of DTR1 primarily caused the accumulation of bisformyl dityrosine. Monoformyl and free dityrosine presumably resulted from the in vivo degradation of bisformyl dityrosine that we occasionally observed in wild-type cells. We therefore assume that their accumulation was only an indirect consequence of the DTR1 deletion. Taking all of these data together, it appears most likely that Dtr1p is involved in the transport of bisformyl dityrosine from the cytoplasm to the maturing spore wall.
FIG. 3.
Analysis of dityrosine-containing compounds accumulating in the cytoplasm of dtr1-Δ spores. Deproteinated extracts of sporulated cells were analyzed by RP-HPLC with gradient elution. Both traces show the signals of the fluorescence detector set to the excitation and emission wavelengths of dityrosine drawn to scale. In addition to free dityrosine (d), the extract of dtr1-Δ contained significant amounts of N-monoformyl dityrosine (mfd) and N,N′-bisformyl dityrosine (bfd) that were identified by coinjection with synthetic standards and by their typical UV spectra. In order to confirm this result, fractions were collected during column development, dried, and hydrolyzed with 6 N HCl. The shaded bars indicate the results of the dityrosine analysis of the hydrolyzed fractions.
A majority of dtr1-Δ spores have an aberrant spore surface.
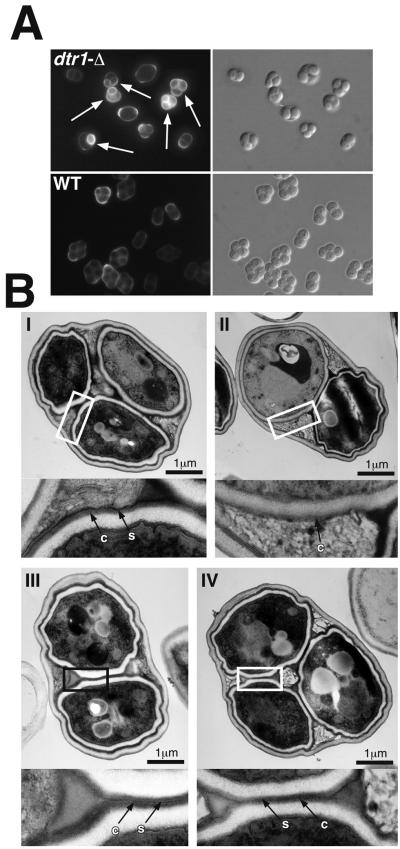
Despite the birefringent wild-type appearance of the spores in light microscopy, a majority of the dtr1-Δ asci contained spores with an altered spore surface. This was apparent both in electron and fluorescence microscopy (Fig. 4). The fluorescent dye Calcofluor white binds to chitin and chitosan and is commonly used to visualize the bud scars of vegetative cells. Wild-type spores are not stained because the dityrosine layer on the spore surface prevents the dye from penetrating into the spore wall and binding to the chitosan layer. Consequently, staining of spores with Calcofluor indicates that their surface layer is not intact. More than 60% of asci of the dtr1-Δ strain contained one or more spores that displayed fluorescence after Calcofluor staining (Fig. 4A). In the corresponding wild-type strain, the percentage was <5%. Electron microscopy of thin sections confirmed that the deletion of DTR1 caused alterations of the dityrosine layer. Figure 4B shows representative examples of the different phenotypes that we observed. The inner layers (electron transparent) and the chitosan layer (electron dense and diffuse) were present in all spores and were not affected by the mutation. This was also confirmed by chemical analysis of the spore walls, which showed that chitosan levels were similar in the wild-type and mutant strains (data not shown). In contrast, the surface layer appeared less electron dense in the majority (60%) of the mutant spores, and in ca. 15% of the spores this layer was not visible at all. To confirm the morphological data, we performed resistance assays, since the resistance of the spores against environmental stress largely depends on the intact spore surface (3). As expected, the mutant spores were more sensitive to heat shock and organic solvents than wild-type spores, and the germination efficiency after treatment with the lytic enzyme Glusulase was reduced from nearly 100% in the wild type to 70% in dtr1-Δ spores (data not shown).
FIG. 4.
Morphological spore wall defects associated with the deletion of DTR1. (A) Fluorescence microscopy with Calcofluor white of dtr1-Δ and wild-type asci. The deletion of DTR1 did not result in a uniform phenotype, instead we observed a mixture of unstained, morphologically wild-type asci and asci containing spores whose chitosan layer became accessible for the fluorescence dye (marked with arrows). Electron microscopy further confirmed the heterogeneity of dtr1-Δ asci. (B) Electron micrographs of representative examples of dtr1-Δ asci. (I) An ascus with the typical appearance of the wild-type spore wall. The dityrosine-containing surface layer (s) is very electron dense and is clearly distinguishable from the underlying chitosan layer (c). 25% of the dtr1-Δ asci showed an unaltered spore wall structure. (II) An ascus containing one spore with a wild-type appearance and one spore lacking the surface layer. Approximately 15% of all asci contained one or more spores with this phenotype. (III and IV) Two examples of asci with an altered surface structure. The surface layer was present but less electron dense than in the wild type and hardly discernible from the chitosan layer. Approximately 60% of the dtr1-Δ asci showed this phenotype. The rectangles indicate the areas shown in the insets at a fourfold magnification.
Expression of DTR1 is sporulation specific.
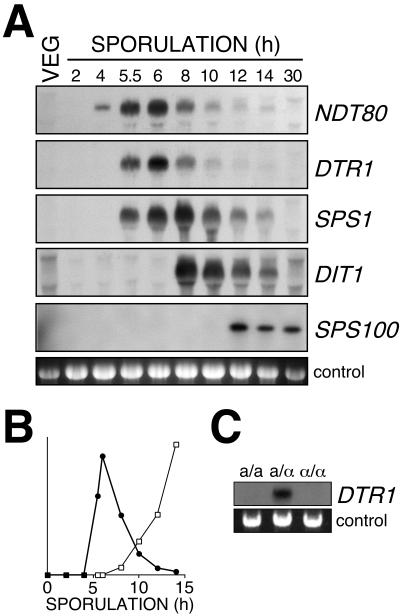
Data obtained in genome-wide studies indicated that expression of DTR1 increases during sporulation (11, 40). In order to verify these data and to correlate DTR1 expression with meiotic events and spore wall formation, we examined total RNA isolated from vegetative cells and cells at different stages of sporulation for presence of DTR1 mRNA by Northern analysis. For this experiment, we used RNA from the fast-sporulating wild-type strain SK1. By using a radiolabeled DTR1 probe, we obtained a signal of ca. 1.7 kb, a finding consistent with the predicted size of the gene. No transcripts were detected in vegetative cells 2, 4, and 5 h after a shift to sporulation medium. At 5.5 h the DTR1 transcript was detected for the first time; maximum accumulation was observed at 6 h. At later time points, expression rapidly declined, and after 10 h only trace amounts of the transcript were detectable (Fig. 5A). The time of maximum DTR1 expression correlated with the onset of dityrosine synthesis (Fig. 5B).
FIG. 5.
Sporulation-specific expression of DTR1. (A) Northern analysis of DTR1 and other sporulation-specific genes. Total RNA isolated from the wild-type strain SK1 at different times of sporulation was hybridized with radiolabeled probes consisting of DNA fragments of the indicated genes. As a control, ethidium bromide-stained rRNA was used. (B) Correlation of DTR1 expression (•) and dityrosine synthesis (□). The relative levels of DTR1 expression was determined by densitometry of the autoradiograph. Dityrosine was analyzed in the hydrolyzates of the cell debris after RNA extraction, thereby ensuring maximum synchronicity between the expression data and the biochemical data. (C) DTR1 is not expressed in asporogenous MATa/a and MATα/α cells. Northern analysis of RNA isolated after 24 h in sporulation medium from three isogenic diploids of the mating type MATa/MATa, MATa/MATα, and MATα/MATα (strain background LP112).
In order to show that transcription of DTR1 is truly sporulation specific and not merely triggered by starvation conditions due to the shift to nitrogen-free sporulation medium, we performed Northern blot analysis with RNAs isolated from isogenic diploid MATa/α, MATa/a, and MATα/α strains under sporulation conditions. The DTR1 transcript was detected exclusively in the sporulating MATa/α strain but not in the asporogeneous MATa/a and MATα/α strains (Fig. 5C).
Sporulation-specific genes are expressed in a strictly hierarchic manner that ensures the ordered progress of the cells through meiosis and spore formation (30). To classify DTR1 expression, we compared the timing of DTR1 mRNA accumulation with several marker genes, namely, NDT80 (delayed early expression), SPS1 (middle expression), DIT1 (mid-late expression), and SPS100 (late expression). We found the maximum accumulation of DTR1 message between 5.5 and 8 h, which is slightly later than NDT80 expression but earlier than SPS1 expression. We therefore propose to classify DTR1 as an “early middle” sporulation-specific gene.
Dtr1p localizes to the prospore membrane.
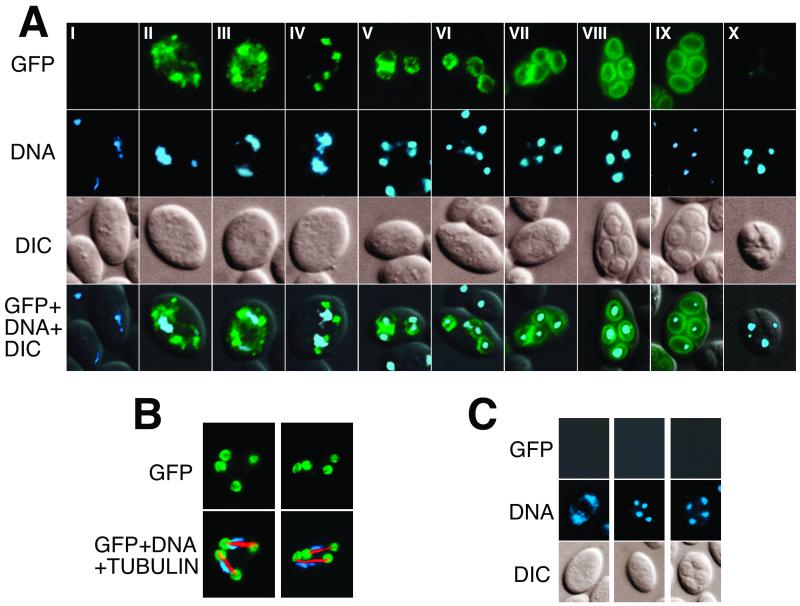
We constructed a C-terminal DTR1-GFP fusion, in order to establish Dtr1p localization during the course of sporulation. The fusion protein was fully functional, since it was able to completely restore the wild-type phenotype of the dtr1-Δ mutant. During the course of this experiment it turned out that the spore wall surface layers interfered strongly with the GFP fluorescence in later stages of sporulation; this was mostly due to dityrosine autofluorescence. We therefore used the strain PB39, a SK1 derivative with a double deletion of the chitin deacetylase genes CDA1 and CDA2. Meiotic and sporulation events are unaffected by the deletion, except that the spores lack the protective surface layers (10), which makes this strain ideal for GFP localization studies during spore formation. The results depicted in Fig. 6 were obtained with this strain transformed with a 2μ plasmid harboring the DTR1-GFP fusion under the control of the native promoter. No GFP-specific signals were detected in vegetative cells and during early meiosis (Fig. 6A, panels I). Toward the end of meiosis I, numerous discrete fluorescing dots, a result most likely representing specific precursors of the prospore membrane, appeared (Fig. 6A, panels II to III). As the cells approached metaphase II, these dots gradually converged into four distinct foci, located at each end of the two bundles of microtubules, as shown by double labeling of formaldehyde-fixed cells with antitubulin antibodies (Fig. 6A, panel IV, and B). It is interesting that in the aldehyde-fixed cells a more detailed structure of these foci was revealed. What appeared as discrete dots in living or ethanol-fixed cells was resolved as cup-shaped structures surrounding the ends of the microtubules (Fig. 6B). As meiosis progressed, the foci expanded to round structures that finally outlined the shape of the immature spores (Fig. 6A, panels V to VIII). Toward the end of spore formation, GFP fluorescence began to fade until it had nearly completely disappeared in mature asci (Fig. 6A, panels IX and X).
FIG. 6.
Localization of GFP-tagged Dtr1p in sporulating cells by fluorescence microscopy. (A) Sporulating cells transformed with a DTR1-GFP-containing multicopy plasmid were ethanol fixed, and the DNA was visualized with DAPI. Representative examples of cells progressing through the sporulation program are shown in chronological order from metaphase of meiosis I to completion of spore formation (subpanels I to X). The pictures of the right column are merges of the GFP, DAPI, and interference contrast signals. (B) Two examples of formaldehyde-fixed cells during meiosis II. To confirm that the localization of the four GFP-fluorescing foci in panel A, subpanel IV, correspond to the prospore membrane adjacent to the meiotic plaque, GFP, tubulin, and DNA were simultaneously detected. The left panels show the location of Dtr1p-GFP; the right panels show the merge of the GFP (green), tubulin (red), and DNA (blue) signals. (C) Cells transformed with an untagged DTR1-containing multicopy plasmid at different stages of meiosis (control).
Given the hydrophobic nature of Dtr1p with 12 membrane-spanning domains, it appears obvious that this protein is an integral membrane component like all other MFS-MDR proteins. Taking this into account, we thus concluded that the GFP-tagged Dtr1p decorated the precursors of the prospore membrane during meiosis I, then the cup-shaped prospore membranes formed by the fusion of these precursors at the sites of the spindle pole bodies (9, 23), and finally the plasma membrane of the immature spores. In contrast to Don1p (23) and Spr3p (16), which localize only to the leading edge, Dtr1p was evenly distributed in the prospore membrane and remained present until the end of spore maturation. These properties make Dtr1p an excellent marker to study the fate of the prospore membrane in mutants with impaired spore wall assembly and morphogenesis and for the isolation and purification of the still poorly characterized prospore membrane.
Dtr1p is a dityrosine transporter.
According to the current model of spore wall synthesis, bisformyl dityrosine is synthesized in the cytoplasm of the prospore and then transported to the maturing spore wall, where it gets incorporated into a macromolecular structure (6). On the way from the cytoplasm to the spore wall, bisformyl dityrosine has to pass through the plasma membrane of the prospore. Based on the data obtained so far, we speculated that Dtr1p might act as dityrosine transporter during spore wall synthesis. The direct experimental proof of this hypothesis in sporulating wild-type cells is difficult, mostly because the processes involved in the formation of the dityrosine-containing macromolecule on the spore surface are poorly understood and cannot be controlled. We therefore tried to verify the postulated function of Dtr1p in a spore wall mutant and in vegetative cells.
Based on our unpublished observation that some spore wall mutants release unincorporated spore wall precursors into the medium when sporulated in liquid medium, we compared in the first experiment the influence of Dtr1p on dityrosine release in a chitin deacetylase-deficient strain that lacks both outer layers of the spore wall (10). Not surprisingly, only trace amounts of dityrosine were detectable in the medium of the wild type and of the dtr1-Δ control, since dityrosine was either incorporated into the maturing spore wall (wild type) or accumulated in the prospores' cytoplasm (dtr1-Δ strain). The medium of the cda1,cda2-Δ strain, on the other hand, contained 85% of the total dityrosine synthesized during sporulation. Presumably as a consequence of the missing spore wall layers, the enzymatic machinery responsible for the polymerization and insolubilization of dityrosine was not functional; therefore, bisformyl dityrosine remained soluble in the ascal cytoplasm and was eventually released into the medium. The additional deletion of DTR1 nearly completely abolished this phenomenon. Spore walls of the cda1,cda2,dtr1-Δ triple deletion were morphologically and chemically indistinguishable from those of the cda1,cda2-Δ strain, since they missed the surface layers and contained only trace amounts of dityrosine (data not shown). Yet the absence of Dtr1p nearly completely abolished dityrosine release into the medium and led to the retention of 85% of the dityrosine within the cells (Fig. 7A).
FIG. 7.
Dityrosine transport by Dtr1p in sporulating (A) and vegetative (B and C) cells. (A) Comparison of the dityrosine content of cells and culture supernatant of wild-type, dtr1-Δ, cda1,cda2-Δ, and dtr1,cda1,cda2-Δ cells after 36 h of sporulation. The dityrosine concentrations are normalized to 1 ml of medium containing 108 cells. Gray bars, dityrosine retained within the asci (no differentiation was made between soluble and spore wall dityrosine); black bars, dityrosine released into the medium. (B) Dityrosine synthesis in vegetative cells ectopically expressing DIT1, DIT2, and DTR1 (□) or DIT1 and DIT2 (•). (Upper panel) Dityrosine released into the medium; (lower panel) dityrosine retained within the cells. (C) RP-HPLC analysis of the culture supernatants of vegetative cells expressing DIT1, DIT2, and DTR1 (upper panel) or DIT1 and DIT2 (lower panel) after 12 h of induction. The peaks corresponding to bisformyl dityrosine are marked with arrows.
The role of Dtr1p as dityrosine transporter was further confirmed in vegetative cells. Normally, vegetative cells are devoid of dityrosine, because DIT1 and DIT2, the two genes responsible for its synthesis, are not expressed. When the sporulation-specific promoter shared by DIT1 and DIT2 is replaced by the GAL1-10 promoter, dityrosine synthesis is induced and bisformyl dityrosine accumulates in the cytoplasmic fraction of the vegetative cells (4). We then asked, what is the fate of bisformyl dityrosine in vegetative cells coexpressing DIT1, DIT2, and DTR1? If our assumption of Dtr1p as a dityrosine transporter was correct, then the ectopic expression of this gene should lead to the measurable secretion of bisformyl dityrosine into the medium, since vegetative cell walls—like the cda1,cda2-Δ mutant spores—do not contain the enzymatic apparatus necessary for dityrosine polymerization and incorporation. As a prerequisite for this experiment, we first determined the localization of Dtr1p in vegetative cells with a C-terminal fusion of Dtr1p with GFP under control of the GAL1 promoter. In induced cells, GFP fluorescence was associated exclusively with the cell envelope, indicating that ectopically expressed Dtr1p was localized in the plasma membrane (data not shown). Fully induced cells transformed with the expression plasmids containing DIT1+DIT2 and DTR1 were transferred to fresh medium, and the dityrosine content of the cells and the medium were measured at different time points. As control, cells containing solely the DIT1+DIT2 plasmid were used. Consistent with the proposed function, DTR1-expressing cells efficiently secreted bisformyl dityrosine into the medium. In the control experiment, dityrosine was retained inside the cells (Fig. 7B and C).
In a similar experiment, we tested whether dityrosine transport by Dtr1p occurs uni- or bidirectionally. Cells ectopically expressing DTR1 were grown in medium containing 0.1 nmol of bisformyl dityrosine/ml. After 24 h, cells were harvested and washed, and their dityrosine content was determined. Despite its presence in the medium, no bisformyl dityrosine (or any other dityrosine derivative) was detectable in these cells (data not shown). This indicates that dityrosine transport by Dtr1p is unidirectional from the inside to the outside of the cell.
Substrate specificity of Dtr1p and related MDR proteins.
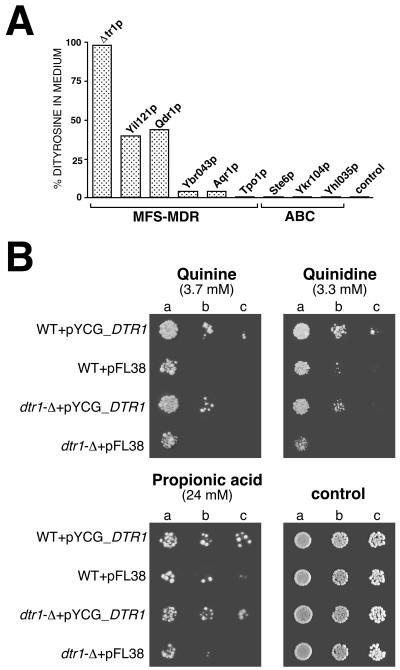
MDR proteins have the ability to extrude a large variety of unrelated, amphiphilic substances from the cell (28). We therefore tested whether (i) dityrosine transport is restricted to Dtr1p and whether (ii) Dtr1p can act as a genuine drug resistance determinant. To answer the first question, eight MDR genes (five representatives of the DHA12 drug efflux family and three members of the ATP-binding cassette [ABC] transporter family) were cloned into expression vectors, and their ability to transport dityrosine was compared to that of Dtr1p. As a control, the empty expression vector was used. Bisformyl dityrosine was most efficiently transported by Dtr1p. Among four DHA12 proteins most closely related to DTR1 (19) and belonging to the same cluster I of Nelissen's classification (35), only Qdr1p (a plasma membrane-localized transporter required for resistance to quinidine and other growth-inhibitory compounds [36]) and Yil121p facilitated dityrosine transport, although the efficiency was reduced by half in comparison to Dtr1p. The medium of cells expressing YBR043c and YNL065w (AQR1), respectively, contained only insignificant amounts of bisformyl dityrosine. No dityrosine secretion was observed in cells expressing TPO1, coding for a polyamine transporter of DHA12 family cluster II (35) localized in the vacuolar membrane (49) and all three ABC transporters tested (STE6, YKR104w, and YHL035c) (Fig. 8A).
FIG. 8.
Substrate specificity of MDR proteins. (A) Transport of bisformyl dityrosine. Dtr1p and MDR proteins belonging to the MFS and ABC transporter families were coexpressed with DIT1/DIT2 in vegetative cells under control of the GAL1 promoter. Bisformyl dityrosine secreted into the medium and dityrosine retained within the cells were determined by RP-HPLC. (B) Comparison of the susceptibility of a wild type and isogenic dtr1-Δ strain to quinine and quinidine at pH 5.5 and to propionic acid at pH 4.0 at the indicated concentrations. The strains contained either the recombinant plasmid pYCG_DTR1 or the empty cloning vector. The cell suspensions used to prepare the spots in lanes b and c were 1:5 and 1:10 dilutions of the suspension used in lane a.
In the complementary experiment, we tested whether Dtr1p can render vegetative cells resistant to drugs and other metabolic inhibitors. Growth tests were performed with wild-type cells and dtr1-Δ cells, transformed either with a DTR1-containing centromeric plasmid or the empty vector, on media supplemented with the toxic compound in question. We observed a slight but consistent increase of resistance in the presence of Dtr1p against the important food-grade organic acid preservatives propionic acid, benzoic acid, and butyric acid, as well as against the antimalarial drugs quinine and quinidine (Fig. 8B). Wild-type cells were slightly less sensitive to these substances than the dtr1-Δ strain, presumably because of a low basal activity of the DTR1 promoter in vegetative cells undetected in the Northern experiments. Increasing the copy number of DTR1 in the cells increased the resistance of the cells, indicating that Dtr1p is indeed responsible for the observed resistance phenotype against these compounds. Detection of the subtle phenotype with the acids was only possible for high inhibitory concentrations, as observed previously (46, 47). The susceptibility of cells against other toxic compounds—such as acetic acid, the antifungal azole derivatives ketoconazole, miconazole, and itraconazole, malachite green, ethidium bromide, benomyl, and 4-NQO—was not consistently affected by Dtr1p.
DISCUSSION
We developed a method for the identification of sporulation mutants in yeast deletion libraries based on the analysis of ll- and dl-dityrosine, the major components of the spore surface by RP-HPLC (2). The rationale of this screen is based on the following two facts: (i) the ratio of dl- to ll-dityrosine is always constant in a given wild-type strain (4) and (ii) the spore wall formation occurs in an ordered pattern, starting with the inner layers (44; unpublished observations). Because of this temporal hierarchy, incorporation and epimerization of bisformyl dityrosine occurs last in spore wall formation and, for the same reason, dityrosine incorporation and epimerization are prone to be affected by perturbations in the deposition of any of the underlying layers, resulting in changes of the dl-/ll-dityrosine ratio. Consequently, analysis of dityrosine and its diastereomers by RP-HPLC is a versatile tool for the identification of mutants with a defect of the surface layer synthesis or with a general deficiency in spore or spore wall formation.
Dtr1p and spore wall maturation.
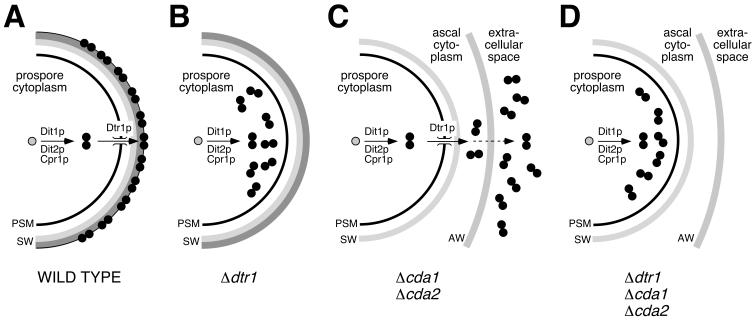
Using this approach, we found in the course of this study that DTR1 (YBR180w) plays an important role during the maturation of the spore surface (summarized in Fig. 9). Homozygous deletion mutants were not affected in their overall dityrosine synthesis, and yet their spore walls contained significantly less dityrosine than corresponding wild-type strains. At the same time, soluble ll-bisformyl dityrosine accumulated in the cytoplasmic fraction of the spores. This resulted in the observed phenotype that hydrolysates of sporulated cultures of dtr1-Δ strains contained mostly ll-dityrosine (Fig. 1). The presence of dityrosine is not restricted to members of the Saccharomycetaceae (39), but it is also found in several other biological systems, such as the in hard fertilization membrane of the sea urchin egg (17) and the cuticula of insects (1). What distinguishes yeast dityrosine is its unique pathway of synthesis. In the sea urchin egg, for instance, dityrosine is formed in a one-step reaction directly in the fertilization membrane (21). In contrast, dityrosine synthesis in S. cerevisiae is far more complex and involves several consecutive steps (6). Dityrosine is not formed in the spore wall itself but in the cytoplasm of the prospores. As a consequence of the spatial separation of synthesis and final destination, yeast cells needed to develop an efficient way to bridge the prospore membrane, which acts as natural barrier between the cytoplasm and maturing spore wall. Based on the phenotype of the dtr1-Δ mutants and the fact that Dtr1p is an MFS-MDR protein (19), we speculated that Dtr1p might be the yet-unidentified transporter of bisformyl dityrosine. Two sets of experiments confirmed our hypothesis. First, we showed that, under sporulation conditions, the deletion of DTR1 prevented the secretion of bisformyl dityrosine into the medium in a spore wall-deficient cda1,cda2-Δ strain. In this experiment we made use of the artifactual release of spore wall precursors into the medium that occurs in this mutant. The fact that the absence of functional Dtr1p counteracted dityrosine release from the asci was a first confirmation of the proposed role of Dtr1p (Fig. 7 and 9). The second experiment was performed in vegetative cells ectopically expressing DIT1 and DIT2, which leads to unscheduled dityrosine synthesis (4). We compared the fate of bisformyl dityrosine produced by these cells in the absence or presence of Dtr1p. Again, we observed retention of bisformyl dityrosine within cells in the absence of Dtr1p, and release into the medium when Dtr1p was present in the plasma membrane.
FIG. 9.
Schematic representations of the fate of bisformyl dityrosine in sporulating wild-type and mutant cells. Bisformyl dityrosine (black circles) is synthesized from free l-tyrosine (gray circles) in the cytoplasm of the prospores in a two-step reaction catalyzed by Dit1p, the cytochrome P-450 Dit2p, and the NADPH-cytochrome P-450 reductase Cpr1p. (A) In the wild type, bisformyl dityrosine is transported through the prospore membrane by Dtr1p and incorporated into the maturing spore wall. (B) The absence of Dtr1p results in the accumulation of bisformyl dityrosine in the prospore cytoplasm and often in an altered structure of the spore surface. (C) In cda1,cda2-Δ cells, bisformyl dityrosine is translocated through the prospore membrane by Dtr1p but not incorporated into the spore wall because of the altered spore wall structure. From the ascal cytoplasm, bisformyl dityrosine eventually diffuses through the ascus wall and accumulates in the medium. (D) The additional deletion of DTR1 in cda1,cda2,dtr1-Δ cells prevents the release into the medium, because bisformyl dityrosine is retained within the prospore. PSM, prospore membrane; SW, spore wall; AW, ascus wall.
The expression profile of DTR1 gave another, although more indirect, hint that Dtr1p plays a role in spore morphogenesis. The time of maximum DTR1 mRNA accumulation coincided with the onset of prospore membrane formation and preceded DIT1 and DIT2 expression, as well as dityrosine accumulation in the spore wall (Fig. 5). No transcripts were detected in vegetative cells, during early phases of sporulation, in mature asci and in nonsporulating a/a or α/α cells, indicating that DTR1 is a genuine sporulation-specific gene. Although no Ndt80p-binding sites were detected in the upstream promoter region, microarray data (11) make it likely that Ndt80p, a key regulator of middle sporulation-specific genes (30), is involved in controlling DTR1 expression. In contrast to DIT1 and DIT2, DTR1 mRNA accumulated prior to the encapsulation of the prospores. As a result, dtr1 behaved recessive and dityrosine profiles of heterozygous dtr1/DTR1 asci were indistinguishable from wild-type asci (data not shown). The reason for Dtr1p's ability to complement mutant spores became fully evident in localization studies with GFP-tagged Dtr1p. GFP fluorescence was first detected in the cytoplasmic precursors of the prospore membrane. Since these secretory vesicles later coalesce at the site of the outer plaque to form the intracellular prospore membrane (23), all four spores are provided with functional Dtr1p. Dtr1p remained present as an integral part of the prospore membrane throughout the phase of its growth and closure, as well as during the later phases of spore wall formation (Fig. 6). Recent publications described proteins that are associated with the prospore membrane. SspIp, Ady3p, and Don1p form the leading edge protein coat at the growing tips of the prospore membrane (33). In contrast to these proteins, Dtr1p apparently does not play a role in the formation of the prospore membrane, since enclosure of the nuclei by the prospore membrane and spore formation were not influenced at all by a deletion of DTR1.
Surprisingly, the deletion of DTR1 did not completely abolish dityrosine transport to the spore wall. The low concentration of dityrosine observed in the spore walls of dtr1-Δ strains could be explained either by passive diffusion of bisformyl dityrosine through the prospore membrane or by unspecific transport by Dtr1p homologues belonging to DHA12 cluster I (see below), some of which are expressed at low levels in sporulating cells (49).
Dtr1p and drug resistance.
As predicted by structural considerations (19, 35, 37), DTR1 confers a slight but consistent resistance to a number of unrelated growth-inhibitory compounds. The resistance phenotypes for DTR1 are close to those observed with other members of DHA12 and DHA14 (14 spanners) families (12, 15, 36, 46, 47), pointing to the prevalence and apparent redundancy of a number of these putative drug transporters. DTR1 also confers a slight resistance to quinine and not only to the stereoisomer quinidine, as is apparently the case with QDR1, AQR1, and TPO4 (12, 15, 36, 46). No effect against ketoconazole and acetic acid toxicity was consistently observed. A number of recent evidences suggests that members of the poorly characterized families of DHA12 and DHA14 transporters may play roles in the yeast cell other than detoxification (36, 46-49). For instance, the plasma membrane proteins Azr1p and Qdr1p, although conferring resistance, could not be clearly implicated in the active export of acetic acid and quinidine, respectively (46, 47). Tpo1p and Tpo4p, polyamine transporters of the vacuolar membrane, were implicated in resistance to a variety of compounds not related to polyamines (12, 15, 45, 49). The observed variety of resistance phenotypes makes it possible that the natural substrates of these transporters and their physiological functions are not yet known.
In the present work we demonstrated that Dtr1p plays an essential role in spore wall synthesis by facilitating the translocation of bisformyl dityrosine through the prospore membrane. This biological role is apparently specific for Dtr1p, since deletions of all other MFS-MDR genes did not influence spore wall maturation (data not shown). Nevertheless, Dtr1p homologues belonging to DHA12 cluster I also facilitated dityrosine transport when coexpressed with DIT1/DIT2 in vegetative cells, although with reduced efficiency. This ability is apparently not shared by Tpo1p, belonging to cluster II, or by the three ABC transporters examined. DTR1 is the first of the MFS-MDR proteins to whom the normal physiological role in the cell was assigned. Highly reminiscent of Ste6p, a member of the ABC transporter family of MDR proteins responsible for the export of the a-factor mating pheromone (25, 26), the primary function of Dtr1p is not related to detoxification, although it is able to alleviate the toxic effects of a wide range of inhibitory compounds in vegetative yeast cells.
Acknowledgments
We thank Michael Breitenbach for many helpful discussions, Fátima Ferreira and Christian Radax for critically reading the manuscript, Albert Thür for the maintenance of the deletion libraries, and Leonor Galvão for carrying out a few preliminary experiments.
This work was supported by Austrian Fonds zur Förderung der Wissenschaftlichen Forschung FWF grant P14735-MOB (to P.B.) and in part by Jubiläumsfonds der Österreichischen Nationalbank grant 6868 (to P. B.) and by the Fundação para a Ciência e a Tecnologia (Ph.D. and postdoctoral fellowships to S.T.).
REFERENCES
- 1.Andersen, S. O. 1964. The cross-links in resilin identified as dityrosine and trityrosine. Biochim. Biophys. Acta 93:213-215. [DOI] [PubMed] [Google Scholar]
- 2.Briza, P., E. Bogengruber, A. Thür, M. Rützler, M. Münsterkötter, I. W. Dawes, and M. Breitenbach. 2002. Systematic analysis of sporulation phenotypes in 624 non-lethal homozygous deletion strains of Saccharomyces cerevisiae. Yeast 19:403-422. [DOI] [PubMed] [Google Scholar]
- 3.Briza, P., M. Breitenbach, A. Ellinger, and J. Segall. 1990. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 4:1775-1789. [DOI] [PubMed] [Google Scholar]
- 4.Briza, P., M. Eckerstorfer, and M. Breitenbach. 1994. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble ll-dityrosine-containing precursor of the yeast spore wall. Proc. Natl. Acad. Sci. USA 91:4524-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briza, P., A. Ellinger, G. Winkler, and M. Breitenbach. 1988. Chemical composition of the yeast ascospore wall: the second outer layer consists of chitosan. J. Biol. Chem. 263:11569-11574. [PubMed] [Google Scholar]
- 6.Briza, P., H. Kalchhauser, E. Pittenauer, G. Allmaier, and M. Breitenbach. 1996. N,N′-bisformyl dityrosine is an in vivo precursor of the yeast ascospore wall. Eur. J. Biochem. 239:124-131. [DOI] [PubMed] [Google Scholar]
- 7.Briza, P., G. Winkler, H. Kalchhauser, and M. Breitenbach. 1986. Dityrosine is a prominent component of the yeast ascospore wall: a proof of its structure. J. Biol. Chem. 261:4288-4294. [PubMed] [Google Scholar]
- 8.Brôco, N., S. Tenreiro, C. A. Viegas, and I. Sá-Correia. 1999. FLR1 gene (ORF YBR008c) is required for benomyl and methotrexate resistance in Saccharomyces cerevisiae and its benomyl-induced expression is dependent on pdr3 transcriptional regulator. Yeast 15:1595-1608. [DOI] [PubMed] [Google Scholar]
- 9.Byers, B. 1981. Cytology of the yeast life cycle, p. 59-96. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Christodoulidou, A., P. Briza, A. Ellinger, and V. Bouriotis. 1999. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 460:275-279. [DOI] [PubMed] [Google Scholar]
- 11.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 12.Delling, U., M. Raymond, and E. Schurr. 1998. Identification of Saccharomyces cerevisiae genes conferring resistance to quinoline ring-containing antimalarial drugs. Antimicrob. Agents Chemother. 42:1034-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, C., and W. S. Saunders. 2001. ADY1, a novel gene required for prospore membrane formation at selected spindle poles in Saccharomyces cerevisiae. Mol. Biol. Cell 12:2646-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Virgilio, C., D. J. DeMarini, and J. R. Pringle. 1996. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology 142:2897-2905. [DOI] [PubMed] [Google Scholar]
- 15.do Valle Matta, M. A., J. L. Jonniaux, E. Balzi, A. Goffeau, and B. van den Hazel. 2001. Novel target genes of the yeast regulator Pdr1p: a contribution of the TPO1 gene in resistance to quinidine and other drugs. Gene 272:111-119. [DOI] [PubMed] [Google Scholar]
- 16.Fares, H., L. Goetsch, and J. R. Pringle. 1996. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 132:399-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foerder, C. A., and B. M. Shapiro. 1977. Release of ovoperoxidase from sea urchin eggs hardens the fertilization membrane with tyrosine crosslinks. Proc. Natl. Acad. Sci. USA 74:4214-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 19.Goffeau, A., J. Park, I. T. Paulsen, J. L. Jonniaux, T. Dinh, P. Mordant, and M. H. Saier, Jr. 1997. Multidrug-resistant transport proteins in yeast: complete inventory and phylogenetic characterization of yeast open reading frames with the major facilitator superfamily. Yeast 13:43-54. [DOI] [PubMed] [Google Scholar]
- 20.Katohda, S., K. Konno, Y. Sasaki, K. Suzuki, and S. Sakamoto. 1984. Isolation and composition of the spore wall of Saccharomyces cerevisiae. Agric. Biol. Chem. 48:895-901. [Google Scholar]
- 21.Kay, E., E. M. Eddy, and B. M. Shapiro. 1982. Assembly of the fertilization membrane of the sea urchin: isolation of a divalent cation-dependent intermediate and its crosslinking in vitro. Cell 29:867-875. [DOI] [PubMed] [Google Scholar]
- 22.Klein, F., T. Laroche, M. E. Cardenas, J. F. Hofmann, D. Schweizer, and S. M. Gasser. 1992. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J. Cell Biol. 117:935-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knop, M., and K. Strasser. 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19:3657-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreger-Van Rij, N. J. 1978. Electron microscopy of germinating ascospores of Saccharomyces cerevisiae. Arch. Microbiol. 117:73-77. [DOI] [PubMed] [Google Scholar]
- 25.Kuchler, K., H. G. Dohlman, and J. Thorner. 1993. The a-factor transporter (STE6 gene product) and cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 120:1203-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuchler, K., R. E. Sterne, and J. Thorner. 1989. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 8:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Lewis, K. 1994. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem. Sci. 19:119-123. [DOI] [PubMed] [Google Scholar]
- 29.Lynn, R. R., and P. T. Magee. 1970. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J. Cell Biol. 44:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moens, P. B. 1971. Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can J. Microbiol. 17:507-510. [DOI] [PubMed] [Google Scholar]
- 32.Moens, P. B., and E. Rapport. 1971. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J. Cell Biol. 50:344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno-Borchart, A. C., K. Strasser, M. G. Finkbeiner, A. Shevchenko, and M. Knop. 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20:6946-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neiman, A. M. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelissen, B., R. De Wachter, and A. Goffeau. 1997. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 21:113-134. [DOI] [PubMed] [Google Scholar]
- 36.Nunes, P. A., S. Tenreiro, and I. Sá-Correia. 2001. Resistance and adaptation to quinidine in Saccharomyces cerevisiae: role of QDR1 (YIL120w), encoding a plasma membrane transporter of the major facilitator superfamily required for multidrug resistance. Antimicrob. Agents Chemother. 45:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulsen, I. T., M. K. Sliwinski, B. Nelissen, A. Goffeau, and M. H. Saier, Jr. 1998. Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 430:116-125. [DOI] [PubMed] [Google Scholar]
- 38.Percival-Smith, A., and J. Segall. 1987. Increased copy number of the 5′ end of the SPS2 gene inhibits sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prillinger, H., W. Schweigkofler, M. Breitenbach, P. Briza, E. Staudacher, K. Lopandic, O. Molnar, F. Weigang, M. Ibl, and A. Ellinger. 1997. Phytopathogenic filamentous (Ashbya, Eremothecium) and dimorphic fungi (Holleya, Nematospora) with needle-shaped ascospores as new members within the Saccharomycetaceae. Yeast 13:945-960. [DOI] [PubMed] [Google Scholar]
- 40.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 41.Pringle, J. R., R. A. Preston, A. E. Adams, T. Stearns, D. G. Drubin, B. K. Haarer, and E. W. Jones. 1989. Fluorescence microscopy methods for yeast. Methods Cell Biol. 31:357-435. [DOI] [PubMed] [Google Scholar]
- 42.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Tachikawa, H., A. Bloecher, K. Tatchell, and A. M. Neiman. 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155:797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teixeira, M. C., and I. Sa-Correia. 2002. Saccharomyces cerevisiae resistance to chlorinated phenoxyacetic acid herbicides involves Pdr1p-mediated transcriptional activation of TPO1 and PDR5 genes. Biochem. Biophys. Res. Commun. 292:530-537. [DOI] [PubMed] [Google Scholar]
- 46.Tenreiro, S., P. A. Nunes, C. A. Viegas, M. S. Neves, M. C. Teixeira, M. G. Cabral, and I. Sá-Correia. 2002. AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 292:741-748. [DOI] [PubMed] [Google Scholar]
- 47.Tenreiro, S., P. C. Rosa, C. A. Viegas, and I. Sá-Correia. 2000. Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast 16:1469-1481. [DOI] [PubMed] [Google Scholar]
- 48.Tomitori, H., K. Kashiwagi, T. Asakawa, Y. Kakinuma, A. J. Michael, and K. Igarashi. 2001. Multiple polyamine transport systems on the vacuolar membrane in yeast. Biochem. J. 353:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomitori, H., K. Kashiwagi, K. Sakata, Y. Kakinuma, and K. Igarashi. 1999. Identification of a gene for a polyamine transport protein in yeast. J. Biol. Chem. 274:3265-3267. [DOI] [PubMed] [Google Scholar]
- 50.Wagner, M., P. Briza, M. Pierce, and E. Winter. 1999. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 151:1327-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]