Abstract
Infections have been the major cause of disease throughout the history of human populations. With the introduction of antibiotics, it was thought that this problem should disappear. However, bacteria have been able to evolve to become antibiotic resistant. Nowadays, a proficient pathogen must be virulent, epidemic, and resistant to antibiotics. Analysis of the interplay among these features of bacterial populations is needed to predict the future of infectious diseases. In this regard, we have reviewed the genetic linkage of antibiotic resistance and bacterial virulence in the same genetic determinants as well as the cross talk between antibiotic resistance and virulence regulatory circuits with the aim of understanding the effect of acquisition of resistance on bacterial virulence. We also discuss the possibility that antibiotic resistance and bacterial virulence might prevail as linked phenotypes in the future. The novel situation brought about by the worldwide use of antibiotics is undoubtedly changing bacterial populations. These changes might alter the properties of not only bacterial pathogens, but also the normal host microbiota. The evolutionary consequences of the release of antibiotics into the environment are largely unknown, but most probably restoration of the microbiota from the preantibiotic era is beyond our current abilities.
INTRODUCTION
The mechanisms involved in the virulence (defined as the relative capacity of a microbe to cause damage in a host [72, 73]) of pathogenic bacteria as well as those determining antibiotic resistance are important and widely studied topics in clinical microbiology. However, they have rarely been analyzed and integrated as we intend to do in the present review.
In terms of evolution and ecology, antibiotic resistance and virulence determinants share some basic characteristics. Since these determinants have been acquired by horizontal gene transfer from other organisms, many are examples of what has been termed “evolution in quantum leaps” (153). Also, most determinants serve to escape the action of antibacterial defense systems that have developed either by natural (host anti-infectious mechanisms) or cultural (antibiotics) evolution (123) to prevent infections. Both the natural anti-infective defenses and antibiotic treatments lead to stringent conditions for bacterial growth. In biology, any limiting condition for the majority is a golden opportunity for the minority. Those bacteria that are capable of surviving and multiplying under these conditions will gain access to organic spaces in which competition with other microorganisms is avoided (exclusive environments).
We might then assume that both virulence and antibiotic resistance are formally similar adaptive mechanisms selected to survive under stress conditions (either host invasion or antibiotic treatment). From an ecological point of view, both infective conditions and antibiotic treatments are evolutionary bottlenecks that tend to reduce microbial biodiversity, so that only a very specific subset of bacteria are capable of colonizing the host under those conditions (Fig. 1 ). There are several bacterial species that are able to grow at 37°C and are tolerant to the oxygen tension present in different parts of the human body. The fact that environmental microorganisms that are unable to produce disease in the healthy host frequently infect immunocompromised patients indicates that many organisms are ecologically compatible with the physicochemical conditions within the human body. The human body and its physicochemical conditions are then an ecological space that can be colonized by several microorganisms (182), frequently with an environmental origin (307). In the normal host, this potential for colonization is limited by the immune system, which actively impedes colonization of the human body by opportunistic pathogens. In the immunodepressed host, only antibiotic treatment maintains a small colonizable space in the human body (see below).
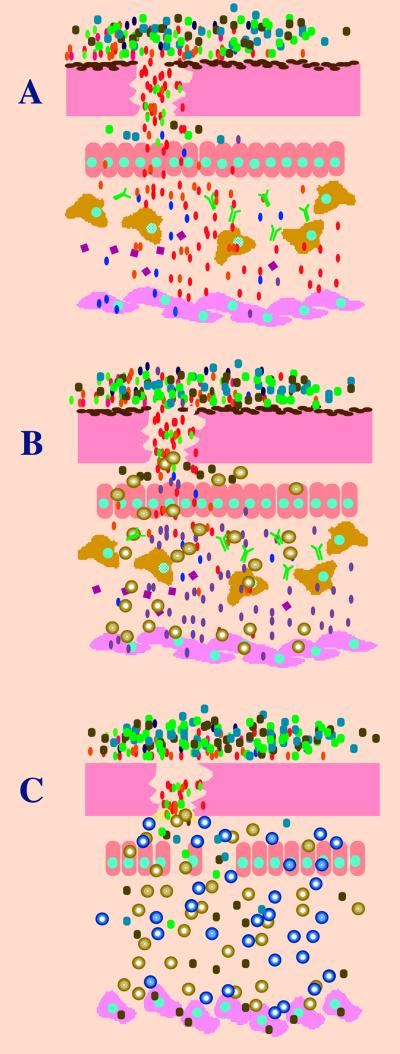
FIG. 1.
Infection and antibiotic treatment are both stringent growing conditions. Several bacteria are able to grow at the temperature and oxygen tension of and using the nutrients present in the human body. However, only some are able to produce infection; this is shownin panel A. Several bacterial species (ovals are pathogens, circles are environmental ones) coexist outside the host. Some species are able to displace the commensal flora, traverse through different epithelia, resist the action of macrophages, antibodies, defensins (with squares), and all the anti-infectious mechanisms of the human body, to finally reach the target cells where the disease is produced. At any of these sequential bottlenecks, only some bacteria are selected, and their population is further amplified, so that, from the high variability of bacteria that could potentially produce the disease, only some are really pathogenic. In panel B, the same situation is analyzed but under antibiotic treatment (spheres). In this case, only a small proportion of the cells belonging to the infectious species (the antibiotic-resistant ones; blue ovals with a red line) are able to produce infection, so that the antibiotic-treated host is a more stringent ecosystem for the growth of bacteria. However, in the case of a debilitated person (panel C), the situation is somewhat different. In these patients, the indigenous microflora might be removed because of antibiotic use, the epithelial integrity may be impaired (intubated patients), the immune system can be abolished (immunocompromised patients), and even the target cells can change. Under these circumstances, some environmental species can produce infection (opportunistic pathogens), because growth conditions are less stringent. However, at least in the developed world, those patients are usually under heavy antibiotic treatment, frequently with a combination of antibiotics (golden and blue spheres), so that only those species with an intrinsically antibiotic-resistant phenotype can produce the infection. Under these circumstances, the main selective force is antibiotics, so that antibiotic treatment becomes a risk factor for some pathogens.
We want then to go one step beyond. Is there any evolutionary relationship between resistance and virulence? If modern medicine has limited the spread and maybe the evolution of bacterial pathogens, this has been done at the expense of increasing antibiotic resistance. Apparently, a decrease in the size of pathogenic populations and an increase in the number of antibiotic-resistant microorganisms (378) have characterized the evolution of infectious diseases. In pure theory, when the number of pathogens decrease to a critical value, antibiotics should be less required, and restoration of antibiotic susceptibility could be expected to occur. We know, however, that this is not true. The widespread dissemination of antibiotic resistance among bacterial populations (275) has maintained or even increased the number of harmful bacteria involved in infections. In fact, and in spite of previous claims, infectious diseases are among the most prominent health problem nowadays (419), in part as the consequence of the increasing number of antibiotic resistance phenotypes, which make bacterial infections untreatable by current therapeutic protocols (277, 329).
If new antibiotics and protocols are required for fighting infectious diseases, we must first understand the relationship between virulence, transmissibility, and antibiotic resistance. Thus, two essential topics are first reviewed in this work: the effect of acquisition of antibiotic resistance (and antibiotic treatment) on bacterial virulence, and analysis of whether pathogenicity and antibiotic resistance might prevail as linked phenotypes in the world of the future, if pathogens become antibiotic resistant as a consequence of intense antibiotic selective pressure.
GENETIC LINKAGE OF RESISTANCE AND VIRULENCE
Virulent bacteria have acquired their phenotype through a long evolutionary course in close contact with their natural hosts. Most virulence determinants are either located in chromosomal gene clusters (pathogenicity islands) or harbored in genetic accessory elements such as plasmids and phages. This suggests that evolution from an avirulent way of life to pathogenicity frequently implies the acquisition of foreign pieces of DNA (125, 153, 157). Nevertheless, for the organism to be a real pathogen, these pathogenicity factors should be inserted in an organism ecologically compatible with the potential host. Moreover, in some cases it is not an acquisition but a deletion (virulence-associated “black holes”) which is needed to become a pathogen (254, 271, 359).
Indeed, any change in lifestyle has a biological cost, as functions needed in one habitat may cause a burden in another habitat and could therefore be counterselected. Acquisition of a virulence phenotype might then require the acquisition of some different pathogenicity islands and the loss of some chromosomal DNA regions. Therefore, the construction of a pathogen by the acquisition of specific pathogenic elements in ecologically compatible host-adapted bacterial genomes has probably occurred over a long evolutionary time.
In this regard, the record of infectious diseases (mainly epidemic ones) in human history and bioarchaeological analysis of the paleontological record indicate that we have been in contact with pathogens similar to those currently producing infections for a long time. In this way, it has been demonstrated that infectious diseases such as syphilis, tuberculosis, and other infections due to bacteria, viruses, and parasites were common in prehistoric men (420), particularly in the Neolithic age, when agriculture and farming ensured close contact between humans and between humans and animals. In some cases, however, such evolution has occurred in historical times, as the evolution from Yersinia pseudotuberculosis to Y. pestis, which was shortly followed by the first pandemic plagues (2). Obviously, the evolutionary outcome of pathogenic organisms tends to be limited in modern times by the anti-infectious repertoire developed by humans that includes hygienic measures, epidemiological controls, vaccines, and, significantly, antibiotics.
Conversely, acquisition and further spread of antibiotic resistance genes among pathogenic bacteria is a phenomenon that has occurred just in the last 50 years as a consequence of extensive antibiotic use for human therapy and animal farming. At first glance, pathogenicity and resistance should be unlinked phenomena. However, several examples indicate that this is not the situation for several bacterial pathogens. Antibiotic resistance and virulence genes can be linked (and then coselected) in the same replicon, or eventually a single determinant can be involved in both virulence and resistance. Some examples of these situations will be reviewed.
Common Mechanisms Involved in Virulence and Resistance
In this section, we analyze whether a mechanism which has been selected on the basis of a bacterial virulence phenotype might also be relevant for antibiotic resistance and vice versa.
Let us first examine the effect of the spatial localization of bacteria in the human body. Several virulent bacteria base their pathogenic characteristics in an intracellular way of life during infection (125). Internalization may be required for the induction of inflammatory cytokines (166) and produce tissue damage through the induction of either necrotic (130) or apoptotic (167, 410) responses. A remarkable example is the apoptosis of macrophages induced by pathogens such as Shigella, which both avoid the antibacterial effect of those cells and trigger inflammation (427, 428). In some well-characterized examples, bacteria are able to travel from cell to cell without any significant contact with the extracellular milieu (43, 273), thus avoiding the immune system as well as contact with antibiotics that do not enter mammalian cells (12, 172). This provides the first clue as to how well-characterized virulence phenotypes can be related to antibiotic resistance.
Mammalian cells are poorly permeable to or easily exclude several families of antibiotics (12, 166). Even if the antibiotic enters the mammalian cells, intracellular growth might induce a transient antibiotic-resistant phenotype, a situation that has been described for Legionella pneumophila (35). The location within the host may alter in an unspecific way the susceptibility of the bacterial organism to antibiotics. Haemophilus influenzae grown in animals undergoes modifications in penicillin-binding proteins, as peptidoglycan metabolism is directly affected by the environment (93), and Salmonella peptidoglycan is dramatically altered for bacteria growing intracellularly (308). Although the effect of these changes on antibiotic susceptibility has not been analyzed extensively, it is possible that those metabolic changes may alter the activity of antibiotics such as beta-lactams against bacteria growing during infection. It is then possible that intracellular localization may allow bacteria to maintain a phenotype of antibiotic resistance.
In this respect, treatment with some antibiotics might select for intracellular clones. This could be the case for species in which some clones are able to invade mammalian cells and others are noninvasive; one such species is Pseudomonas aeruginosa (126). On the contrary, some antibiotics, such as macrolides and fluoroquinolones, are actively accumulated by human cells (262). In this case, intracellular bacterial localization might eventually increase the antibiotic effect against bacterial pathogens, leading to better eradication and favoring the less invasive strains.
A similar situation of phenotypic antibiotic resistance can be observed for bacteria growing in biofilms. Bacterial biofilms are frequent in persistent infections (85), such as those associated with cystic fibrosis (354), chronic bronchitis, osteomyelitis (152), and foreign-body-associated infections. Bacteria growing in biofilms are more resistant than those under a planktonic way of life to the action of phagocytic cells' antibacterial activity (261) as well as the action of antibiotics (16, 83, 149, 370, 404). As in the previous case, antibiotics might select biofilm-forming organisms, thereby increasing the prevalence of chronic infections. Other mechanisms of virulence could prevent the action of antibiotics against bacteria. For instance, toxins leading to local necrosis or abscess formation certainly will decrease the local availability of antibiotics due to reduction in antibiotic arrival to the foci or because of local inactivation of the drugs by altered pH, free proteins, or DNA. In these cases, the mechanisms of pathogenicity may finally serve as mechanisms for antibiotic resistance.
A final example of a virulence mechanism with a role in antibiotic resistance is presented by Bordetella pertussis, the etiological agent of whooping cough. The cell wall of the virulent strains of this microorganism is infrequently susceptible to autolysis triggered by beta-lactam antibiotics; only avirulent B. pertussis strains are known to be lysed. It was demonstrated that this phenotypic tolerance and the antibiotic-induced autolytic activity are controlled by the vir locus, which determines phase transition in B. pertussis, a key element in the virulence properties of this bacterial species (389).
In previous examples, we have seen that a “virulent way of life” can contribute to an antibiotic resistance phenotype. Could antibiotic resistance determinants also contribute to bacterial virulence? Some examples suggest that this could be the case. Recently, one area of intense study of antibiotic resistance is the analysis of multidrug resistance (MDR) efflux pumps (280, 293). These determinants are able to extrude an ample range of substances that include antibiotics, solvents, dyes, and quorum-sensing signals (295, 423). Extrusion by the same pump of compounds as diverse as erythromycin, quinolones, beta-lactams, and ethidium bromide is a rule more than an exception.
Might these MDR determinants extrude compounds involved in the host defense be contributing to bacterial virulence? The answer is yes. For instance, adaptation to growth in the presence of bile salts is a prerequisite for any pathogen to colonize the intestinal tract. It has been reported that Escherichia coli extrudes bile salts (382) through the acrAB system, which was first characterized as an MDR determinant (6, 230); the same occurs in Salmonella enterica serotype Typhimurium. In fact, it has been suggested that extrusion of bile salts present in the intestinal ecosystem to which these bacteria are adapted is the function for which those MDR determinants were selected (230). Analysis of S. enterica serotype Typhimurium mutants with high susceptibility to bile salts has demonstrated that this higher susceptibility was a consequence of inactivation of the MDR determinant acrB. This inactivation leads to a reduced ability to colonize the intestinal tract in a murine infection model (199), which indicates that acrAB is involved in both antibiotic resistance and virulence.
Some MDR systems, such as Mtr from Neisseria gonorrhoeae (158) and SapA (291) from S. enterica serotype Typhimurium, can actively extrude both defensins and antibiotics, thus contributing to reducing antibiotic susceptibility as well as to increasing the number of virulence determinants of the organisms. In fact, it has been described that resistance to defensins is required for a full virulence phenotype in the case of Salmonella (154). The mechanisms of action of defensins resemble at least in part that of peptidic antibiotics currently used in clinical practice (180, 391). Thus, it is conceivable that they might share the same mechanisms for resistance. This situation has been described for polymixin B. The sensitivities to polycationic peptides were compared between two groups of Yersinia enterocolitica isolates, one with an environmental origin and the other from infections (38). Data indicated that pathogenic strains of Y. enterocolitica were resistant to defensins and also to polymixin B, showing a clear cross-linkage between antibiotic resistance and virulence as a consequence of the expression of a resistance determinant.
Not only MDR pumps but also biocide efflux determinants may play a role in bacterial virulence. It has been reported that the QacA pump of Staphylococcus aureus, involved in resistance to several organic cations (biocides included), has a role in resistance to thrombin-induced platelet microbicidal protein (197). Thus, the presence of the pump contributes to the survival of strains carrying this determinant at sites of endothelial damage as well as in experimental endovascular infections. We want to stress here that the mechanisms of extrusion of antibiotics are similar to those translocating some proteins involved in pathogenicity. The hemolysin export apparatus in Escherichia coli comprises the outer membrane channel trimmer protein TolC that is also involved in the AcrA-B multidrug efflux transport complex (279). The periplasmic proteins of both systems, AcrA and HlyD, have a similar structure and are able to interact with TolC (278).
In all previously discussed examples, a mechanism selected for bacterial virulence can also produce an antibiotic resistance phenotype. Selection for the most virulent strains might select for antibiotic resistance, and conversely, selection for antibiotic resistance might select the most virulent microorganisms. However, the opposite situation can also be found: antibiotic resistance may decrease bacterial virulence (see below). For instance, the KatG catalase-peroxidase activity is important for the survival of Mycobacterium tuberculosis in the host (216). Mutations that eliminate this activity prevent the activation of isoniazid and are the major cause of resistance to this drug in Mycobacterium spp. (316, 425). It might then be predicted that isoniazid-resistant Mycobacterium mutants might be less virulent than wild-type isoniazid-susceptible strains, at least in the case of M. tuberculosis (264) and Mycobacterium bovis (414). However, when isoniazid-resistant isolates were analyzed, such reduced virulence was not found. It turned out that M. tuberculosis pathogenic isolates accumulate compensatory mutations in the gene encoding the alkyl peroxidase AphC, which can substitute for KatG for survival inside the host (350).
Costs of and Compensations for Virulence and Resistance
It is largely assumed that acquisition of novel genetic determinants may have a cost for the bacterial host. That may happen because of partial incompatibility of previous and acquired lifestyles, or because of the extra energy required to maintain the genetic vectors carrying the new genes. The acquisition of plasmids or antibiotic resistance-virulence genes might have an effect on bacterial fitness (8, 13, 426). As previously stated, pathogenicity islands, acquired through horizontal transfer of large gene arrangements, have a major role in bacterial virulence (157). Also, large vir plasmids are needed for some pathogens, such as Shigella (337, 406) and Yersinia (306), to express a pathogenic phenotype.
We might speculate that acquisition of these determinants implied a cost in bacterial fitness when they were included in the genome of the previously nonvirulent bacteria. However, acquisition of resistance genes by pathogenic bacteria has occurred over the last 50 years, but acquisition and further evolution of structures as pathogenicity islands occurred thousands of years ago. Thus, the evolutionary time needed to acquire and optimize compensatory mutations to alleviate the effect on fitness of the acquisition of pathogenicity determinants has been much longer. Also, for plasmids carrying vir determinants, the costs of the acquisition of such determinants might be compensated for by mutations in other loci. Of note, it has been shown that the vir plasmid can be easily lost in vitro in Shigella (342). However, growth conditions in vivo should preserve the plasmid's presence during infection, otherwise Shigella would not be virulent anymore.
Acquisition of vir plasmids ensures the ability to colonize a different biological compartment, thus evading competition with other bacterial species and eventually reducing the potential fitness cost imposed by the new genetic element. The same has probably occurred for enteroinvasive E. coli isolates (335, 336). Recent work suggests that the invasive ability of those isolates has evolved in different chromosomal backgrounds, probably through the spread of plasmid-borne invasion genes, and the maintenance of invasive phenotypes in separate lineages suggests that this ability confers a selective advantage to invasive strains (247). Although the Ewald hypothesis (119, 120) is still controversial, it can be speculated that sanitation procedures, vaccination, and widespread antibiotic use, impeding host-to-host spread of vir plasmid-based pathogens, may reduce the overall pathogenic power of microorganisms.
Due to the rapid acquisition and fixation of compensatory mutations by bacteria, experiments to evaluate the biological cost of the expression of novel antibiotic resistance and virulence determinants are difficult to perform. Antibiotic resistance and virulence can be acquired either by horizontal transfer of antibiotic resistance genes (97) or by mutation (241). The presence of new plasmids and transposons in the bacterial genome has a relevant cost for the recipient bacteria (13). However, it has been demonstrated that under these circumstances, the cost is compensated for in few generations as the consequence of genetic change by the host, getting the specific plasmid-bacterial strain association more fit than the previous non-plasmid-containing bacterium (61).
Also, in the case of mutations leading to antibiotic resistance, some examples demonstrate the possibility of fitness reduction, which may also reduce bacterial virulence (see last example in previous section). For instance, current evidence supports the idea that highly fluoroquinolone-resistant Salmonella strains could be counterselected in the field (145). A substantial reduction of the virulent characteristics has been also described for antibiotic-resistant S. enterica serotype Typhimurium isolates (48). Although this may occur for some bacteria, it does not mean that it will always occur. In fact, an outbreak of quinolone-resistant S. enterica serotype Typhimurium DT104 has recently been described (268), and an increase in quinolone-resistant Campylobacter jejuni infections has been reported in Minnesota (356).
The emergence of compensatory mutations rapidly alleviates the biological cost of antibiotic resistance in S. enterica serotype Typhimurium isolates (45), and this could be the situation for the aforementioned outbreaks of quinolone-resistant bacteria. Noteworthy, the compensatory mutations that are selected in vivo and in vitro are different, reflecting the different growing conditions for bacteria during infection compared with microorganisms growing in vitro (49). This indicates that the metabolic requirements needed for infection are different from those for surviving in the environment. Therefore, acquisition of the characteristics required for infection and antibiotic resistance might make them less proficient for surviving in the environment and to be transmitted among different hosts.
It seems then that acquisition of novel virulence and antibiotic resistance traits has a cost in bacterial fitness, but the cost is rapidly compensated for due to the high plasticity of bacterial genomes and the huge populations of bacteria from which compensatory mutants can be selected (59, 209, 213). However, this situation may not always occur. Possibly, only those mechanisms that can be compensated for by mutations in other loci are selected because these are the only ones that can avoid fitness reduction. As stated by other authors (13), the effect of antibiotic resistance (and in a higher grade of virulence determinants) on bacterial fitness has been properly addressed in only a few studies. This topic needs to be investigated so that the biological potential of novel virulent and antibiotic-resistant bacteria may be predicted.
Evolution and Dissemination of Genes Involved in Virulence and Resistance
As previously stated, both antibiotic resistance genes and virulence determinants have been acquired by horizontal gene transfer in most cases (285). One intriguing question that has not been completely resolved resides in the origin of these determinants.
It is widely accepted by the plasmids and transposons that carry antibiotic resistance genes may have originated in antibiotic-producing organisms in order to avoid the deleterious effect of the antibiotic on themselves (39, 98, 239, 290, 407). Eventually, they could have further evolved in organisms in an ecological consortium with antibiotic producers. More recent works indicate that this hypothesis about the origin of resistance is only half true (11, 98). For instance, it is difficult to believe that chromosomal beta-lactamases (71, 217) as well as some aminoglycoside-inactivating enzymes (4, 201, 233) and a plethora of MDR determinants (280, 293) which are present in all isolates of a given bacterial species originated in the antibiotic producers. In fact, all these determinants must have a functional role other than antibiotic resistance, and this phenotype will only be a consequence of their primary physiological function (see reference 11 for a review of this concept).
In the case of virulence determinants, the current paradigm indicates that pathogenicity islands acquired through horizontal gene transfer are frequently responsible for the pathogenic properties of virulent bacterial species (125, 153, 157). Different from antibiotic resistance genes, the pathogenicity islands are more difficult to explain, because pathogenic bacterial ancestors carrying such gene clusters have not been detected. It has been proposed that pathogenicity islands might be relevant for the biodegradative properties of microorganisms (including decomposition of dead bodies), to kill living cells (to obtain food and reduce competition), and to live inside eukaryotic cells (such as amoeba, protozoa, plants, and animals) in natural environments (161).
Antibiotic resistance genes and virulence determinants might then play a different role in the original organisms from which they were transferred to pathogenic bacteria. However, once those determinants have been selected in the heterologous host, they confer a selective advantage and are fixed in the pathogenic bacterial populations, where they can evolve further and eventually be transferred to other bacterial species.
Mutation and recombination.
Two different processes account for evolution of antibiotic resistance-virulence phenotypes: gene mutation and gene recombination (horizontal gene transfer included). The role of mutation in antibiotic resistance is well known (241). In the case of virulence, fewer examples of such a role have been described. However, mutational activation-inactivation of intrinsic genes might also contribute to a virulent phenotype in the case of opportunistic pathogens. As an example of cryptic virulence determinants activatable by mutations, laboratory isolates of E. coli are hemolytic (105) because of mutations in the hns (147) and fnr (315) genes. Whether this is just a laboratory curiosity and might actually have a role in the course of nosocomial infections by E. coli is a matter of speculation.
Some virulence determinants of P. aeruginosa, such as alginate production (60) and cytotoxicity, are downregulated. However, mutants in which the expression of such determinants is derepressed are frequently found during infection (240). This indicates that mutation probably plays a major role in the emergence of the different virulent phenotypes shown by P. aeruginosa clinical isolates. Pathogenic P. aeruginosa isolates can be broadly classified into two groups, those with a cytoinvasive phenotype and those with a cytotoxic phenotype. However, both kinds of isolates contain the genes required for invading epithelial cells. Invasive and cytotoxic strains differ in the expression of the genes under the control of an activator called ExsA (116). Mutations in the gene exsA, which encodes the activator, lead to not a cytotoxic but only an invasive phenotype (116, 126). Although both types of bacteria are virulent, they occupy different environments (inside and outside epithelial cells), so that one or the other phenotype might be selected in vivo by, for the moment, unknown processes.
Recombination also has an important role in the evolution of antibiotic resistance determinants. Progressive clustering of genes, leading to an operon structure, may have occurred to optimize and regulate the expression of ancient genes producing a resistance (frequently low-level resistance) phenotype (206). In a similar way, the construction of a pathogenicity island requires the recruitment of different genes that recombine to produce a single genetic element containing several genes with a common role, virulence. In other cases, intragenic recombination is essential to produce a resistant phenotype. The most conspicuous example of recombination in chromosomal DNA leading to antibiotic resistance is beta-lactam resistance produced by recombination of penicillin-binding-protein genes in Neisseria gonorrhoeae and Streptococcus pneumoniae (79, 235).
Similar recombinatorial processes may have influenced the evolution of virulence. However, unlike for antibiotic resistance, few examples of recent evolution of bacteria to a virulent phenotype have been described. One might be recombination between different sets of genes, which causes the rearrangements that can be observed in the capsular antigens of S. pneumoniae (78-80, 276) and can be considered a mechanism of defense against host immunity. Another could be the emergence and further dissemination of enterohemorrhagic E. coli O157:H7 strains (131). This E. coli serogroup has emerged as a relevant pathogen in the last 20 years. Enterohemorrhagic E. coli isolates contain virulence plasmids and pathogenicity islands similar to those found in Shigella spp. (70, 237). Although it was a matter of speculation whether these genetic determinants were acquired recently by E. coli, the recent sequencing of the genome of an E. coli O157:H7 strain (297) demonstrated that it contains as many as 1,387 new genes in comparison with the previously sequenced nonpathogenic laboratory strain E. coli K-12 (52).
We must be aware that acquisition of pathogenicity determinants by previously nonpathogenic organisms might occur in a similar way as it happens with antibiotic resistance determinants. For instance, the acquisition of virulence plasmids by bacteria forming part of the human indigenous flora and thus already well adapted for surviving inside their host (such as E. coli) might lead to the emergence of novel pathogens. In a similar way, the acquisition of virulence determinants by environmental microorganisms should produce novel pathogenic bacteria. Although the evolution of novel pathogens is possible, the selective pressure in favor of the selection of virulence determinants is not as strong as in the case of antibiotic resistance determinants. Therefore, an explosion of novel pathogens, such as has occurred with antibiotic-resistant bacteria, will most likely not happen in the near future, not only because the selective pressure for a pathogenic phenotype is less strong than for an antibiotic resistance phenotype, but because selection for pathogenicity in today's world might decrease with sanitation (119, 120) if the Ewald hypothesis is true.
A critical point to discuss here is that virulence and/or antibiotic treatment might increase the rate of bacterial variation. In the case of infection, this situation might contribute to increased mutation rates (310, 326) and even to the selection of hypermutator strains (see below), thus enhancing antibiotic resistance mutation (67, 208, 287, 380). In a similar way, antibiotic challenge might also produce hypermutable phenotypes (9, 318, 320), increasing the possibility of mutants overexpressing virulence determinants. Mechanisms favoring hypermutation may also facilitate recombination (253). Moreover, in vivo transfer of plasmids carrying antibiotic resistance genes and/or virulence determinants and recombination are probably favored during infection due to host signals that enhance gene transfer (260).
In this respect, the effect of antibiotics in inducing the transfer of plasmid and transposons has been demonstrated in vitro (387). It has been described that the same genes required to initiate infection of human macrophages by Legionella pneumophila are involved in plasmid mobilization (344). Recent results in our laboratory also suggest that bacterial expression of factors involved in cell-to-cell DNA transfer (likely antibiotic resistance plasmids) in some organisms, such as S. pneumoniae, may be triggered by inflammatory products (M. R. Baquero, unpublished results). It can then be suggested that bacteria are expected to evolve more rapidly inside the host and under antibiotic selective pressure, so that an infected patient under antibiotic therapy may act as an evolutionary accelerator.
Plasmids.
It is well known that plasmids are major vectors for the dissemination of both antibiotic resistance and virulence determinants among bacterial populations. It is also clear that the presence of virulence and antibiotic resistance determinants in the same genetic element will produce coselection of both types of determinants. This applies as well for genes present in transposons, phages, and cassettes, which are discussed below. For bacteria carrying those linked determinants, the selection of an infective population will select for antibiotic resistance, and antibiotic selective pressure will select the virulence trait. Table 1 shows some published examples regarding this type of gene linkage. Among them, antibiotic resistance plasmids carrying, alone and in combination, genes encoding the synthesis of bacteriocins (272), siderophores (82, 168, 401), cytotoxins (34, 132), and adhesion factors (202) have been described.
TABLE 1.
Examples of virulence determinants encoded by antibiotic resistance plasmids
General plasmid-encoded transfer functions (present in all transferable resistance plasmids) may be relevant by themselves for bacterial virulence. One of these functions is related to the traT genes, involved in plasmid conjugation (374). The other function is the production of pheromones (77), also involved in conjugation of plasmids and transposons in gram-positive bacteria. Conjugative plasmids have a relevant role in the dissemination of antibiotic resistance. Plasmid conjugation is encoded in the tra region. It has been demonstrated that expression of traT genes might be involved in bacterial resistance to serum (181, 270), biofilm development (141), and phagocytosis (3), therefore contributing to the virulent properties of bacteria carrying conjugative (frequently antibiotic resistance) plasmids.
The plasmid transfer proteins are sometimes chromosomally encoded, but they can have a role in both plasmid transfer and virulence. This could be the case for Legionella pneumophila. It has been described that the dot virulence genes encode a large putative membrane complex that functions as a secretion system that is able to transfer plasmid DNA from one cell to another (402). Mutations in the dot genes reduced both virulence and plasmid transfer. In the case of gram-positive bacteria, aggregation substances encoded by pheromone plasmids are involved in the clumping required for an efficient transfer of DNA by conjugation. It has also been described that Enterococcus faecalis aggregation substance promotes resistance to killing by human neutrophils in spite of phagocytosis and neutrophil activation (314) and enhances pathogenicity in a rabbit model (340) and thus, as said by R. Wirth, is “more than a plasmid collection mechanism” (416).
The analysis of plasmids from the preantibiotic era demonstrated that there have not been major changes in the families of plasmids present in pathogenic bacterial populations, but just recruitment of antibiotic resistance genes by formerly “antibiotic-susceptible” plasmids (95, 171, 179). Since plasmids encoding adaptive traits (eventually involved in the pathogenic lifestyle) certainly preceded antibiotic resistance, it could be suspected that many current resistance plasmids contain genes with a role in bacterial colonization and virulence. In fact, this has probably been the origin of plasmids containing virulence determinants and antibiotic resistance genes such as those listed in Table 1. We do not know, however, if this association is the rule for antibiotic resistance plasmids, because only a very small fraction of these determinants have been entirely sequenced.
Transposons.
Transposons are also relevant for the dissemination of antibiotic resistance genes, either by integration in transferable plasmids or by direct conjugation and further integration in the bacterial chromosome. Also, transposons containing virulence determinants have been described. One example is the aerobactin operon (46, 142, 244). Aerobactin is a siderophore produced by several bacterial species (74). It has been proposed that aerobactin is a virulence factor with relevance for iron acquisition at the site of infection (103). Aerobactin genes have been found in the chromosomes of different bacterial species and in several different plasmids, a situation which indicates easy mobilization. The aerobactin operon forms part of a transposable element (102), and it has been proposed that it is part of a pathogenicity island in Shigella flexneri (403). Other virulence determinants found in transposable elements are the E. coli enterotoxin STII gene (170), the Shiga toxin operon, which has been found in a putative composite transposon in Shigella dysenteriae 1 (257), and the toxic shock toxin, carried by a family of mobile pathogenicity islands in Staphylococcus aureus (218).
Little is known about the association between antibiotic resistance and virulence determinants in the same transposon. However, there is no reason for this lack of association. Indeed, like other complex gene arrangements (see below), transposons frequently have a mosaic structure in which highly recombinogenic regions are included (56, 90, 205), so that the acquisition of novel traits is a common occurrence in transposon evolution (256).
Phages.
The presence of virulence determinants in phages infecting different bacterial species has been described (Table 2). Also, a phage origin has been proposed for at least some pathogenicity islands (see below). Bacteriophage-associated transduction of antibiotic resistance determinants has been described as well (50, 51, 341, 390). Nevertheless, the presence of antibiotic resistance genes together with virulence determinants in the same phage has not been reported. A possible explanation for this phenomenon is the size requirements for phage DNAs. Phage DNA needs to accommodate inside the head of the phage particle, so that its length must fit a fixed range of sizes. In such circumstances, the gain of some genes must be accompanied by the loss of others. Because of this, the possibility of combining antibiotic resistance genes and virulence determinants in the same bacteriophage is lower than in the case of plasmids, which have less stringent requirements for the incorporation of novel DNA fragments.
TABLE 2.
Examples of virulence determinants encoded by phages
Gene cassettes.
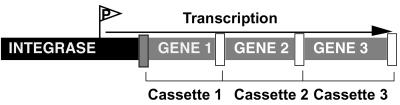
In the last few years, several works have shown the presence of gene cassettes capable of integrating novel genes from different DNA sources, offering possibilities for the horizontal dissemination-linked antibiotic resistance-virulence genes. The most relevant family of these gene cassettes is the integron family (328, 377). Integrons are the primary systems for the capture of antibiotic resistance genes in gram-negative bacteria and have also been described in gram-positive bacteria. Integrons are formed by gene cassettes located downstream of a recombinase-encoding conserved sequence that includes a strong promoter (Fig. 2). This organization allows the formation of large arrays of gene cassettes which can eventually be transferred as a whole between different replicons.
FIG. 2.
Structure of an integron. Integrons are site-specific recombination elements that mediate the acquisition and spread of genes among bacterial populations. Integrons are formed by an integrase gene followed by one primary att recombination site (dark grey box) and several cassettes, each including one gene and one 59-bp recombination site (white square). The transcription of the system is controlled by a strong promoter located upstream of the integron. This structure favors the arrangement of genes in tandem, which are transferred as single elements among bacterial populations.
Most of the integrons described to date are formed by antibiotic resistance genes (328). However, several different genes, including not only antibiotic resistance but also virulence determinants, have been found in integrons. Prototypic of composite integrons containing virulence genes are the VCR cassettes found in the chromosome of Vibrio cholerae. VCRs (from Vibrio cholerae repeated sequences) are a family of 123- to 126-bp sequences of imperfect dyad symmetry, highly repeated in the chromosome of V. cholerae (up to 60 copies). Analysis of VCR clusters demonstrated that the gene-VCR organization is identical to that of the antibiotic resistance cassettes present in integrons (255). Also, the formation of the VCR islands likely occurred by means of an integrase-mediated process. This indicates that VCR islands might have a role in gene capture similar to the proposed role of integrons in the acquisition of antibiotic resistance genes. It cannot be dismissed that some genes in VCRs may play a role in antibiotic resistance, perhaps to still undiscovered antibiotics.
Another type of gene cassette with a key role in the establishment of the infective process are pathogenicity islands. These DNA regions found in the chromosomes and plasmids of bacterial pathogens are composed of clusters of genes (typically from 15 to 25, although more are possible) which have been acquired by horizontal gene transfer (157). It has been shown that several of these gene arrangements have been introduced into tRNA genes through a phage-mediated transfer mechanism (75). However, a role for insertion sequences has also been suggested in other cases. This is the case for the pathogenicity island required for growth of Y. pestis in iron-deprived environments (162). This island is flanked by direct repeats of IS100, an insertion sequence which is present in the chromosome and plasmid of this bacterial species and thus may contribute to recombination of genes from different replicons.
The presence of sequences with a role in DNA mobility either inside or flanking pathogenicity islands is quite common (157). However, few of these gene clusters have been demonstrated to be mobile. One example is the aforementioned family of pathogenicity islands carrying the gene encoding toxic shock toxin in Staphylococcus aureus (218). Stabilization of pathogenicity islands in the bacterial genome probably requires inactivation of the functions involved in mobility. In fact, the “mobility elements” present in pathogenicity islands frequently carry a large number of mutations, often leading to stop codons (21), which abolish the expression of such functions. Antibiotic resistance determinants have not been detected in pathogenicity islands, but that is an open possibility.
The transfer of gene clusters (contained in either plasmids, transposons, bacteriophages, or gene cassettes) is extremely relevant to bacterial adaptation to novel environments because it allows “bacterial evolution in quantum leaps” (153). As previously stated, both bacterial virulence and antibiotic resistance can be considered strategies to explore and colonize novel environments in which bacterial competitors are scarce. Bacteria will make use of all the tools (mutation, gene transfer, recombination) which allow the high plasticity shown by bacterial genomes (176, 211, 285, 349). In the treated host, adaptation of pathogens will require both antibiotic resistance elements and virulence determinants, so that both adaptive mechanisms must evolve together to produce the explosive spread of virulent and antibiotic-resistant microorganisms that we are now facing.
Phenotypic Adaptation in Virulence and Antibiotic Resistance
Over the course of an infection, bacteria must face several different environments (Fig. 1), so that phenotypic adaptation to all of them is an important trait for the final pathogenicity result. In this respect, both virulence and antibiotic resistance determinants can be environmentally regulated. The signaling network that regulates the expression of bacterial virulence and antibiotic resistance determinants during infection is a hot topic in current research in microbial pathogenesis.
Environmental signaling in virulence and antibiotic resistance.
Bacteria are constantly sensing the environment in order to respond to changes in its composition. In the case of bacterial pathogens, constant cross talk occurs between the microorganisms and their hosts (86) which modulates the expression of several genes in both the bacterial and host cells. Physiological signals such as calcium concentration (372), low pH (128, 296), high temperature (194, 296), and low iron concentration (88, 399) among others, trigger the expression of several virulence genes. However, little is known about whether such signals might also trigger the expression of antibiotic resistance genes.
Nevertheless, we know that the antibiotic susceptibility of bacteria is modulated by several factors which include growth phase (164), pH (89, 203, 226), carbon dioxide (226), temperature (311, 312), bile salts (230), and low iron (305). It is then clear that certain conditions during infection might induce the expression of both antibiotic resistance and virulence genes, and a common regulation of both types of determinants might then occur. Most probably this regulation does not always involve the same regulatory network, but in some cases virulence and antibiotic resistance genes can be part of the same regulon. In this regard, it has recently been described that the MarA protein (from multiple antibiotic resistance) of E. coli regulates the expression of more than 60 chromosomal genes. These genes include not only MDR determinants (156), but also genes with a potential role in virulence, such as those involved in oxidative stress and iron metabolism (33).
In general, a linkage between antibiotic resistance and virulence gene regulation might occur in global regulons, which modulates the expression of stress genes (see below). This type of regulation, coupled with phenotypic resistance of bacteria growing in biofilms, inside cells, and even in resting cells which are resistant to antibiotics, might produce situations of in host resistance at the site of infection that is impossible to predict by routine laboratory susceptibility tests (reviewed in reference 242).
Antibiotics as effectors of bacterial virulence.
Common models of bacterial virulence analyze host-pathogen interactions under a defined set of conditions that usually do not include antibiotic therapy. Although the first stages of an infection usually occur without treatment, once a diagnosis is available, most infectious diseases evolve under antibiotic treatment. We want to stress here that antibiotics are not only bacterial killers, but also modulators of bacterial and even host cell transcription of different genes (238, 347). In such a way, antibiotics modulate the interaction between host cells and bacteria (289). It has also been suggested that they may have an important role as modulators of the interactions among natural environmental bacterial populations (100). For instance, quinolones alter DNA supercoiling (5), and the expression of many genes, including those involved in virulence, is dependent on DNA supercoiling (44, 112).
More recently, the role of a natural quinolone on quorum sensing in Pseudomonas aeruginosa has been demonstrated (258, 299). Quinolone treatment may alter the expression of several bacterial genes. In fact, a decrease in the expression of virulence determinants upon exposure to subinhibitory concentrations of quinolones has been described in some cases (62, 361). In contrast, ciprofloxacin and other SOS-inducing antimicrobial agents cause Shiga toxin-encoding bacteriophage induction and enhanced Shiga toxin production from E. coli O157:H7 both in vitro and in animal models (187, 424). Therefore, quinolones clearly affect the expression of virulence determinants, but their role as either enhancers or inhibitors of bacterial virulence depends on the model analyzed and must be established for each pathogenic bacterium. On the other hand, resistance to quinolones as a consequence of mutations in bacterial topoisomerases in some cases alters DNA supercoiling (5, 22, 367), leading to changes in the level of expression of several bacterial proteins, including virulence determinants. For instance, treatment of quinolone-resistant S. aureus with subinhibitory concentrations of quinolones increases the expression of fibronectin-binding proteins, contributing in part to their emergence and maintenance in clinical settings (47).
Other antibiotics besides quinolones can affect the expression of virulence determinants in different ways. For instance, Suerbaum demonstrated that low antibiotic concentrations increased serum sensitivity in E. coli strains protected by the K1 antigen (373), and low macrolide concentrations appeared to reduce the adhesive properties of some gram-negative rods (269). Also, antibiotic treatment may trigger the release of bacterial products, including lipopolysaccharide vesicles (188) and endotoxin (355), that interact with host cells, changing the virulence characteristics of pathogenic bacteria.
Exposure to antibiotics can produce global changes in the metabolism of bacteria, as is possibly the case with Mycobacterium tuberculosis. Exposure to low concentrations of antibiotics induces expression of the alternate sigma factor sigF, which is involved in the regulation of several genes, including those involved in virulence (263). In light of these data, a role of chemotherapy in the persistence of this important pathogen has been suggested. Recently, it has been shown that tetracyclines are iron scavengers (151). Iron deprivation induces the expression of several virulence-associated proteins in bacteria (224, 243), and it is known that free iron is scarce during infection (409). In this way, tetracyclines might trigger the expression of several bacterial genes just as a consequence of their iron-chelating activity.
The influence of subinhibitory concentrations of antibiotics on virulence is then dependent on the bacterial species-antibiotic combination. The response of resistant strains to antibiotics will be different from that of susceptible ones. For instance, if subinhibitory concentrations of antibiotics reduce the expression of virulence determinants, resistance to drugs (even low-level resistance [29]) would restore the original virulence levels. Major efforts must be made to analyze the effect of antibiotics on the metabolism of antibiotic-resistant and -susceptible bacteria. One such approach is the recently published analysis of isoniazid-induced alterations in M. tuberculosis gene expression (7, 415). The authors showed that treatment with this drug produces a dramatic change in the expression of several bacterial genes, some of them related to the drug's mode of action, but others with no apparent relationship to the antibiotic killing mechanism, which may eventually influence the outcome of infection. The current availability of tools for whole-genome expression analysis may allow a better understanding of the role of antibiotics on regulation of bacterial metabolism and thus potentially virulence.
Cross talk between virulence and antibiotic resistance regulons.
The environmental regulation of the expression of antibiotic resistance and virulence determinants has already been discussed. The possible cross talk of both types of regulons will now be reviewed. The most relevant example to emerge in the last few years is the effect of quorum-sensing signals on bacterial virulence and the effect of antibiotic resistance on quorum sensing.
Several bacterial species are able to determine the local concentration of bacterial cells, a process known as quorum sensing (37). Quorum-sensing signals trigger the expression of several genes, some of which are involved in bacterial virulence (37, 292, 294). Quite interestingly, it has recently been described that mutations leading to multidrug resistance in P. aeruginosa as a consequence of overproduction of the MexAB-OprM efflux pump also affect the quorum-sensing response (117). This response could be due to a direct effect on the extrusion of quorum signals by this MDR pump in P. aeruginosa (295). More recently, it has also been described that overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa (192).
Since quorum-sensing signals trigger the expression of several virulence-associated genes, such as those involved in production of elastase, rhamolipid, pyocianin, proteases (63), and type III secretion system, antibiotic resistance might strongly influence virulence through the differential processing of quorum-sensing signals in antibiotic-resistant and antibiotic-susceptible bacterial populations. Indeed, recent work in our laboratory has shown that MDR P. aeruginosa mutants are impaired in their virulence properties (334).
Conversely, elements with a role in virulence might also be involved in the regulation of antibiotic resistance. As previously stated, iron deprivation induces expression of the MDR efflux pump MexAB-OprM. Also, the expression of MDR determinants can be induced by salicylate (251). Salicylate is a relevant signal molecule in plant-bacteria interactions (351). It is also an intermediate in the synthesis of different siderophores (104, 346) and is itself a siderophore in Pseudomonas spp. (346). Bacterial siderophores are relevant virulence factors that are produced during infection (243) as a consequence of the small amount of free available iron that is present in the human body (409). Since P. aeruginosa might produce salicylate during infection and this compound induces the expression of MDR determinants, salicylate might thus induce a phenotype of antibiotic resistance for bacteria growing under iron-deprived conditions, such as occur during infection (409).
Mutations in the genes encoding some histidine kinases, which are involved in signal transduction mechanisms in Streptococcus pneumoniae, cause a phenotype of tolerance to the killing ability of glycopeptide antibiotics (281), and these mutants are also less virulent when tested in animal models (384). Deletion of the ciaR gene, which is involved in regulation of competence and cefotaxime resistance in S. pneumoniae, led to a 1,000-fold attenuation of bacterial growth in vivo. Deletion of the vncSR gene pair, which is involved in tolerance to glycopeptide antibiotics in this bacterial species (281), also produces growth attenuation, although the effect is smaller. These signal transduction systems are thus pleiotropic regulators that may influence antibiotic susceptibility, virulence, and even horizontal gene transfer. Therefore, antibiotics may select for variants unable to respond to a number of environmental signals. If that is the case, the environmental signals that induce pathogenicity traits may eventually be altered.
Signal transduction systems also have a role in antibiotic resistance in P. aeruginosa. phoP-phoQ mutants showing increased resistance to aminoglycosides, cationic peptides, and polymyxin have been described (231, 232). PhoP-PhoQ is a two-component regulatory system that has been studied mainly in S. enterica serovar Typhimurium. In this bacterial species, PhoP-PhoQ regulates the expression of at least 40 genes in response to magnesium concentrations (360) and has an important role in both virulence (139, 266) and resistance to polymyxin (155). Although a role of P. aeruginosa PhoP-PhoQ in virulence has not yet been demonstrated, it is highly expressed under magnesium starvation (232), which indicates a role similar to that previously described in the case of S. enterica serovar Typhimurium. PhoP-PhoQ may then coordinately regulate both virulence and antibiotic resistance in response to the magnesium concentration.
We have addressed some examples of cross talk between antibiotic resistance and virulence regulons. However, genes encoding both types of determinants might also be part of the same regulon. As previously stated, this is the case for the Mar regulon in E. coli and S. enterica serovar Typhimurium, which is involved in multiple antibiotic resistance (156), superoxide resistance (106, 177), organic solvent tolerance (18), and bile salts resistance (230), among others. The same situation probably occurs for several other MDR determinants, so that the possibility that the physiological signals that bacteria receive during infection might trigger a global response including the regulation of both antibiotic resistance and virulence genes should be carefully analyzed.
THE CROSSROADS OF VIRULENCE, EPIDEMICITY, AND ANTIBIOTIC RESISTANCE
The pathogenic relevance of a bacterium relies mainly on two different properties, virulence and epidemicity (the ability to produce epidemics). A compromise between the two properties is required to cause a major disease because a nonvirulent bacterium obviously will not produce any disease, whereas a nontransmissible bacterium might cause a dangerous disease but only in a limited number of people. Since bacteria are under antibiotic pressure during infection (see above), highly efficient pathogens must be not only virulent and epidemic but antibiotic resistant as well. The problem from the microorganisms' perspective is whether or not the acquisition of one of these traits could have a detrimental effect on the expression of the others. In such a case, an “optimization strategy” will be required. We will review whether the acquisition of these abilities is detrimental to the expression of the others and, conversely, reinforces the possibility of acquisition of a full proinfective repertoire, virulence, epidemicity, and antibiotic resistance.
Are Antibiotic-Resistant Bacteria More or Less Virulent?
When antibiotic resistance occurs as a consequence of a genetic alteration (241) in a housekeeping gene (for instance, those involved in ribosome function, cell wall construction, biosynthetic pathways, and the DNA replication machinery), conventional wisdom suggests that an evolutionarily optimized mechanism has deviated from its functional optimum. Therefore, resistance should have a “direct” cost in bacterial fitness, and resistant organisms should exhibit suboptimal behavior. The location of antibiotic resistance determinants in accessory genetic elements (plasmids, transposons, integrons) probably reduces this “direct” cost, but at the expense of the “indirect” cost of carriage of the elements themselves (8). In both cases, antibiotic resistance should have a fitness cost (13), which might reduce bacterial virulence.
Although it is difficult to find examples of the effect of antibiotic resistance on the virulence of clinical bacterial isolates, epidemiological data support the idea that, in some cases, antibiotic-resistant organisms may show a decrease in pathogenicity. For instance, in urinary tract infections, fluoroquinolone-resistant E. coli are easily found in cystitis but are isolated at very low frequencies as causative agents of pyelonephritis (54). Recent studies also suggest that penicillin-resistant S. pneumoniae strains may be less pathogenic than susceptible ones, at least in animal models (20, 234), and isolates from cases of bacteremia are frequently less resistant than those isolated from mucosal infections and even in carriers (150, 175). In this case, a possible explanation is that the capsulation involved in serum resistance eventually interferes with acquisition of foreign DNA encoding resistant penicillin-binding proteins.
Nevertheless, the pathogenic process might provide an advantage for the bacteria, such as permitting access to exclusive habitats. For instance, a reduction in the rate of protein synthesis because of a ribosomal resistance mutation may be deleterious for a bacterial organism competing with the wild-type susceptible strain in a rich medium, where the effect of such a deficiency is maximized, while under slow-growth conditions, the difference would be minimal. The expression of genes involved in pathogenicity is frequently regulated by stationary-phase signals (160, 207), so that the slow growth due to a reduction in protein synthesis might eventually trigger the expression of genes involved in pathogenesis. In this case, the growth defect produced by antibiotic resistance could be compensated for by the increased virulence of the pathogen, and bacteria could easily accumulate compensatory mutations which restore their original pathogenic abilities totally or in part (213).
Although antibiotic resistance usually reduces bacterial fitness, the opposite situation might also occur. For instance, the bleomycin resistance gene contained in transposon Tn5 confers improved survival and growth advantage on Escherichia coli (55). The increase in the virulence properties of antibiotic-resistant bacteria is a disturbing possibility, because in these cases, revertants that become susceptible by means of mutations and loss of antibiotic resistance plasmids may be at a selective disadvantage relative to the ancestor resistant population, so that the resistant population will prevail even in the absence of antibiotic selective pressure.
In general, antibiotic resistance obviously increases the overall pathogenic potential of the organism in the treated patient, because the susceptible organisms are killed by the antibiotics and the infection does not progress. For instance, the mortality rate of S. pneumoniae meningitis in South African children is significantly higher for antibiotic-resistant strains (133, 134). However, in the absence of antibiotics, antibiotic resistance may influence pathogenicity in different ways, depending on the antibiotic resistance determinants involved and the possible accumulation of compensatory mutations.
Are Antibiotic-Resistant Bacteria More or Less Epidemic?
The term epidemic is usually restricted to widespread outbreaks of pathogenic and/or resistant microorganisms. Thus, the actual dimension of the link between transmissibility and resistance may be difficult to evaluate without extensive studies on the population biology of microbes associated with humans. Although the epidemiology of nonvirulent and nonresistant organisms remains to be evaluated, some important observations (163) indicate that in countries such as Sweden, where antibiotic-mediated selection is low, epidemics of antibiotic-susceptible but pathogenic bacteria may be, as in the preantibiotic era, the usual case. Studies of commensal bacteria in wild animals also suggest that the gut flora of wild populations with little human contact (and thus no contact with antibiotics) shows very low levels of antibiotic resistance (288), whereas those from the guts of animals that have been in contact with humans show higher levels of antibiotic resistance (143, 144). On the other hand, the most relevant increases in antibiotic resistance at the local and global levels are due to the spread of a relatively small number of resistant bacterial clones, such as with the “international” resistant clones of S. pneumoniae (79, 165, 225), methicillin-resistant S. aureus (135, 225, 393), and vancomycin-resistant Enterococcus faecium (28, 84). The extreme rapidity with which some of these resistant organisms spread in remote countries suggests that their colonizing potential is nearly optimal for a broad range of hosts.
Several reasons can be given in favor of the existence of an evolutionary link between antibiotic resistance and host-to-host transmission. The diminution in bacterial diversity due to antibiotic treatment will result in overgrowth of resistant bacterial populations that are in the minority under normal competitive circumstances. Antibiotic treatment is then a bottleneck that is crossed by the few antibiotic-resistant bacteria that are present in the population. However, once this bottleneck is crossed, the best colonizers among the remaining antibiotic-resistant bacteria will have an advantage for recolonizing the host. If the challenge is repeated several times, it could be expected that a certain within-host evolution of bacteria to optimize colonization will occur (412). Considering that all members of the host population are very similar in their ability to be colonized by each particular organism, success in colonizing the host will be reflected in a corresponding success in between-host transmission ability. Large numbers of resistant cells within the host should facilitate host-to-host transfer, particularly to other treated hosts. This perspective may have some exceptions. Some bacterial populations are strictly linked to particular environments, including a dependence on other local bacterial populations (41). That situation is not infrequent in mucosa-associated communities. In these cases, the antibiotic-resistant population may remain confined and even disappear under antibiotic therapy just because the other members of the community are eradicated.
The possibility that a resistant population will be enriched under antibiotic treatment thus requires the ability of this species to independently exploit the available habitat. This ability is common among organisms capable of crossing ecological barriers, including the more epidemic ones. In serial-passage experiments with mixed populations, the strain with the greatest number of cells in the transferred inoculum has a selective advantage. The driving force may be within-host competition and selection for increased parasite growth rate (114). In this regard, the ecological abundance of specific clones in natural populations of Staphylococcus aureus might be linked to the virulence of such clones (99). The clones should then have been selected because they are good colonizers as well as being highly transmissible and highly virulent (99, 221). This statement is true in general, except perhaps for organisms with high virulence, such as Mycobacterium tuberculosis, for which a small inoculum may be sufficient to cause the illness.
Epidemicity ensures constant high multiplication rates that may be needed for the acquisition of resistance. Therefore, these bacteria are simultaneously more prone to becoming resistant, to be selected for by antibiotics, to increase their absolute numbers, and consequently to be transmitted efficiently. There is an obvious corollary: antibiotics should be prescribed (57) in a way that ensures eradication of the bacterial pathogen (as said by Paul Ehrlich, “hit hard and hit early”), as any survival gives the organism an opportunity to evolve and spread in a more efficient way.
The relationship between epidemicity and pathogenicity may also help to modulate the development of resistance. Within-host growth usually induces the expression of virulence determinants in several pathogens, thereby increasing their virulence. Increased between-host bacterial transfer may also increase virulence. It is also well known that serial passage of a given organism among susceptible hosts frequently increases its pathogenic power in the new host, although it is attenuated for the former host (114). Bacterial virulence might then be increased in densely clustered host groups (camps, schools, hospitals), with close contact and high transmission rates, particularly in the early stages of the spread. Antibiotics preferentially exert their selective pressure on clinically ill patients, who are treated more frequently than carriers. If extinction of the pathogen does not occur as a result of treatment, those epidemic strains will tend to be subjected to increased antibiotic pressure, and antibiotic resistance will be more easily achieved.
Although we think that antibiotic resistance usually helps microorganisms in their transmissibility, this is not the case for all. For instance, comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest that in some cases, a counterselection of highly fluoroquinolone-resistant strains might occur in the field (145). This indicates that fluoroquinolone-resistant Salmonella spp. probably have a strong reduction in fitness outside of the host, so that they cannot be maintained in the environment. Alternatively, it could be that the compensatory mutations accumulated during infection (49) are deleterious for bacteria growing in the environment (see above). The same probably occurs for opportunistic pathogens such as P. aeruginosa, Burkholderia cepacia, and Stenotrophomonas maltophilia. These bacterial species are intrinsically resistant to several antibiotics (307), but, during therapy, mutants with higher levels of resistance and displaying resistance to other antibiotics are selected (140, 378, 392, 422).
It is noteworthy that cross infection is not frequent for these opportunistic pathogens, and the resistant clones selected during therapy are not frequently disseminated. In the case of fluoroquinolone-resistant Salmonella strains, this suggests that these resistant opportunistic pathogens are probably impaired in their dissemination in natural environments (mainly water and soil). Of course, as we want to stress throughout this review, this is not a black and white situation, and outbreaks of nosocomial infections due to the transmission of resistant clones have been described. Relevant examples of this situation are vancomycin-resistant E. faecium (28), the selection of such antibiotic-resistant bacteria in avoparcin-exposed farms (321), and some outbreaks of infections with multidrug-resistant Acinetobacter baumanii (191, 228).
Are Virulent Bacteria More Resistant to Antibiotics?
Because virulent organisms are able to produce clinical symptoms of infection in human and animal hosts and should therefore be exposed more frequently to antimicrobial drugs, the risk of development of resistance is expected to be higher. That is expected to produce a link between higher virulence and higher resistance. Despite this prediction, the opposite is frequently the case. Pathogens such as Neisseria meningitidis, Salmonella enterica serovar Typhi, Shigella dysenteriae, Bordetella pertussis, Leptospira icterohaemorragica, and Brucella melitensis are rarely resistant to antibiotics. Some reasons can be given for this phenomenon.
First, as stated previously, the possible cost (in fitness) of antibiotic resistance should be tolerable by the bacterial host. Usually this tolerance involves the acquisition of cost-compensating mutations that must be tolerated as well. This is not a critical problem for many opportunistic pathogens that are able to survive in various alternative environments. But in several highly specialized pathogens, the genetic structure cannot be extensively modified by mutation without risk of local extinction as a consequence of an increased number of lethal mutations (14). Second, in contrast to less-specialized pathogens, some of these highly virulent organisms have probably evolved in protected environments less open to competition and therefore less exposed to natural antibiotics. Third, the absolute number of many pathogens is reduced compared to their commensal counterparts, so that its chances of being exposed to antibiotics and acquiring antibiotic resistance should be lower. This view has been proposed as an explanation for the differences in the ability to develop mutational resistance to penicillins in S. pneumoniae and the viridans group streptococci versus S. pyogenes (28). Fourth, the particular niche occupied by highly pathogenic organisms may be excluded or almost excluded from the action of antimicrobial agents (see below), so that they are not subjected to antibiotic selective pressure.
Are Epidemic Bacteria More Resistant to Antibiotics?
Epidemic bacteria are considered here as those able to spread in an efficient way between hosts in a short period of time (note that we do not mention whether epidemic bacteria are pathogens or not). This property is also frequently expressed as organisms having a “high reproductive rate” (R0) (19, 283). By definition, epidemic bacteria are frequently encountered in a large number of individuals and thus reach high population sizes throughout the host community. Because these populations frequently face variable environments in terms of antibiotic selective pressure (patients under therapy and not, different therapeutic regimens), the chances for acquisition of antibiotic resistance increase (32). Additionally, bacteria with a high dispersal (migration) rate are in contact with more types of microorganisms occupying different ecological habitats from which antibiotic resistance genes can be acquired by horizontal transfer. These characteristics make epidemic bacteria more prone to acquire antibiotic resistance. Once an antibiotic resistance phenotype is acquired, an epidemic microorganism can be widely disseminated. In addition, antibiotic selective pressure in treated patients will convert such an organism into a predominant clone in this bacterial species (Fig. 3). This is probably the cause of the worldwide distribution of antibiotic-resistant clones of species such as S. pneumoniae (see above).
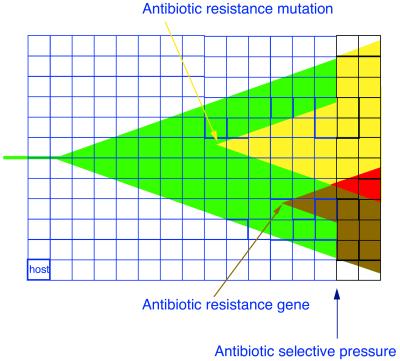
FIG. 3.
Epidemic bacteria can easily acquire an antibiotic-resistant phenotype. An epidemic bacterial strain (green) spreading between hosts (each individual blue square) increases its population size, facilitating the emergence of antibiotic-resistant mutations (yellow arrow) and the acquisition of resistance genes by horizontal transfer (brown arrow). Acquisition of resistance is immediately amplified by the epidemicity of the strain, even in the absence of antibiotic selection (resistant variants, yellow and brown). Interactions between different mechanisms of resistance may occur, eventually leading to multiresistance (red). If antibiotics are almost ubiquitously present in the hosts, as in an intensive care unit (black squares), the spread of the resistant bacteria is favored.
Epidemicity implies that the microorganism can colonize a large number of hosts, so that the overall number of bacterial cells to which selection can be applied is also large. Thus, the probability of selecting an antibiotic-resistant mutant is higher than in the case of nonepidemic microorganisms. On the other hand, if an epidemic organism becomes resistant in one host and is transferred to other hosts, the use of antibiotics in each treated host ensures the removal of the susceptible bacterial populations and its replacement by the epidemic population. At this stage, the antibiotic-resistant microorganisms increase in number, and all the possible variants within this population compete for establishment in the novel hosts. As a result of this competition, the best colonizers of the epidemic, antibiotic-resistant microorganism are now selected. Selection of epidemic and antibiotic-resistant bacteria is thus based on a feedback mechanism which links the two properties, unless the cost of antibiotic resistance severely impairs the effectiveness of host-to-host spread (see above). This cost will depend on the specific bacterial species-antibiotic resistance mechanisms. In this way, studies which integrate virulence properties and the behavior of antibiotic-resistant microorganisms in the environment are needed to establish the best protocols for avoiding the spread of virulent antibiotic-resistant bacteria.
RESISTANCE AND VIRULENCE: AN INTEGRATED VIEW
The human host has acquired several different mechanisms in the course of evolution for fighting infections, including the immune system, fever, production of antimicrobial peptides (136, 174, 397), and iron-scavenging (376), among others (210). Another mechanism of defense against colonization and further infection is the presence of an indigenous microflora which has coevolved with its host (41). This defense constitutes the very first biological barrier against subsequent colonization and infection with external potentially pathogenic microorganisms. Very recently in terms of evolutionary time, a new line of defense has been added to the armamentarium of humans in their fight against infection, that being antibiotic therapy.
Based on the effect that they produce on infectious bacteria, pharmaceutically produced antibiotics play the same anti-infectious role as antimicrobial compounds produced by either the host or its allied microbiota. All these types of substances are able to inhibit bacterial growth and thus constitute a part of the defense of the human host. The only difference between these substances resides in the fact that conventional (industrial) antibiotics are a product of the so-called “cultural evolution” (123), whereas the other mechanisms for defense against infections are the result of the natural “genetic evolution” of the human species. In opportunistic infections, where the genetic defense strategy is severely impaired, the only line of defense that remains is the antibiotic cultural defense. With this approach, antibiotic resistance is hard to distinguish from other mechanisms involved in microbial pathogenicity.
Resistance as a Colonization Factor in the Treated Host
Acquisition of an antibiotic resistance phenotype will have two effects on bacteria during colonization and infection of the treated host. First, resistance is required for evading cultural host defenses (in this case, antibiotic treatment). Therefore, it is equivalent to other systems for evading other host defenses, such as production of siderophores (88), resistance to oxidative stress (68, 106, 284), resistance to defensins (136, 174), and resistance to bile salts (382, 396) for intestinal pathogens. These characteristics are needed, although not sufficient, for the development of infection. Second, resistance in the presence of antibiotics in the treated host will favor colonization. As stated previously, the very first line of defense against infections is the indigenous microflora (41), which form a colonization barrier against exogenous microorganisms. However, this microflora is severely affected by antibiotic therapy. Therefore, antibiotic therapy debilitates the defenses of the host by altering the protective biological defenses. Only if the infecting microorganism is resistant to antibiotics can it make use of this advantage for better colonization of the host.
We might thus conclude that antibiotic treatment promotes further colonization and infection by antibiotic-resistant bacteria which otherwise may not be able to effectively compete with indigenous microflora. In this way, antibiotic resistance is a relevant colonization factor for the microorganisms interacting with the treated patient. In fact, epidemiological studies have demonstrated that antibiotic therapy is a relevant risk factor for colonization by antibiotic-resistant bacteria such as S. pneumoniae, methicillin-resistant Staphylococcus aureus, Stenotrophomonas maltophilia (108), Acinetobacter baumannii (307), and Campylobacter jejuni (356).
Opportunistic Infections
Opportunistic infections occur in individuals whose natural defenses against bacterial infection are severely impaired (42, 182, 307). In the preantibiotic era, these people were mainly infected by microorganisms belonging to their own microflora. Currently, even when the genetic defense has collapsed, the second line of defense, antibiotics, keeps these hosts free from infection, but obviously only if antibiotic-susceptible organisms are involved. As research on antibiotics has been focused for years on frequent human pathogens, many antibiotics are still active on microbes associated with humans. Because of this, many opportunistic pathogens today come from the environment, with one of their more salient characteristics being high intrinsic resistance to a broad range of antibiotics (307). Indeed, it is this ability that allows opportunistic pathogens to invade the compromised host under antibiotic therapy, since environmental opportunistic pathogens do not need to harbor specific pathogenicity determinants for humans (as the natural defense has been broken), but only antibiotic resistance.
In fact, opportunistic pathogens are unable to produce infections in nondebilitated patients. Therefore, environmental and clinical isolates of opportunistic pathogens such as P. aeruginosa (127, 323) and Stenotrophomonas maltophilia (40) are genetically indistinguishable. Analysis of two collections of P. aeruginosa isolates from environmental and clinical settings has demonstrated that they were functionally equivalent in traits relevant for both their pathogenic (host-oriented) and biodegradative (environment-oriented) abilities (10).
The healthy host is quite an “extreme” environment in terms of stringent growing conditions for microorganisms. Although few bacteria have evolved to efficiently colonize this environment, many more are potentially able to grow using the resources of human body (for instance, those involved in corpse decomposition and putrefaction). In other words, the dead body implies a rupture of some ecological constraints of the living one. Also, because conditions for bacterial growth are less stringent in the ill host (Fig. 4), a greater variety of bacteria can eventually produce an infection. However, when antibiotic treatment is begun, there is a new ecological constraint on the growth of the bacteria, the antibiotic, so that under these conditions only antibiotic-resistant bacteria will be able to maintain the infection. Finally, because opportunistic pathogens are not the result of evolution of an infectious bacterial species in contact with the host, the term “emergent pathogens” has been inappropriately used for their description. These organisms, having acquired the ability to colonize different habitats, can infect the sick host. Their selection is the result of cultural human evolution, since antibiotic treatment has selected a previously naturally resistant bacterial species. The replacement of traditional opportunistic pathogens with novel, more resistant ones has occurred over the last few years (307). Thus, antibiotic resistance linked to antibiotic therapy might be considered the primary factor influencing the selection of such novel antibiotic-resistant opportunistic pathogens.
FIG. 4.
Colonization space of opportunistic pathogens. Colonization space can be defined as the combination of the different physicochemical parameters (including the space) in which an organism can survive. In this regard, stringent growth conditions (for instance, absence of oxygen) reduce the ecological space. As discussed, conditions for infection are quite stringent, so that only a few bacteria can colonize the host's ecological space (dark gray square) compared to bacteria that colonize the environment and, eventually, the dead host (lightly shaded square). In the case of the sick host, however, stringency for colonization is lower, so that some environmental bacteria can now colonize this larger ecological space (intermediately shaded square in panels b and c). These bacteria with an environmental origin are opportunistic pathogens. Once infected with these bacteria, the host is usually treated with antibiotics. Again, antibiotic treatment restricts the ecological space so that only antibiotic-resistant bacteria can grow under these conditions (oval in C). In all cases, small ovals indicate human-associated bacteria and small circles indicate environmental bacteria. White, antibiotic-susceptible bacteria; black, antibiotic-resistant bacteria.
Does Virulence Affect Exposure to Antibiotics?
As previously stated, virulent organisms producing clinical symptoms are more frequently treated with antibiotics than nonvirulent ones, and therefore virulence should increase antibiotic exposure. Antibiotic treatment will affect both the infecting bacteria and indigenous microflora. However, pathogenic bacteria will be treated as soon as they are detected (and their presence suspected from clinical symptoms), and thus antibiotic selective pressure is exerted quite frequently on these organisms. Indigenous microflora are challenged with antibiotics only as an unwanted side effect of the therapy directed against the pathogens, so that most time is maintained without antibiotics. Nevertheless, several bacterial pathogens may behave as commensals over long periods of time.
Many bacterial infections are controlled by the host in many cases without clinical symptoms (210). In this case, even pathogenic organisms may not be frequently challenged by antibiotics. In some cases, host lesions induced by pathogenic microbes may occur but with few if any clinical symptoms (subacute infections). In these cases, these organisms with “low virulence” will also be less exposed to antibiotics. One might suggest that antibiotic therapy could be considered a cost and a penalty associated with virulence. If this were the case, it is conceivable that some pathogens could evolve to low pathogenicity after prolonged use of antibiotics in human populations. In general, close relatives to pathogenic bacteria are now members of the normal flora. Are they the result of the evolution from pathogen to harmless variant to escape host defense mechanisms?
In particularly severe infections, the host may quickly die, reducing bacterial exposure to antibiotics. Once the host dies, the microorganisms may either die as well (if they need to grow inside the host) and be released into the environment, where the antibiotic pressure is low. It is thus possible that the most virulent bacteria are perhaps less exposed to antibiotics than nonvirulent ones. We can conclude that bacteria with intermediate levels of bacterial virulence have a greater probability of being exposed and developing resistance to antibiotics than less virulent and highly virulent organisms (Fig. 5), in the first instance because they are not always detected and thus not treated, and in the second, because the host dies without giving them enough time for the development of antibiotic-resistant variants.
FIG. 5.
Infectious bacteria under risk of antibiotic treatment. As stated in the text, in some cases infections are extremely acute (black line), so that the host is damaged (and even killed) before an antibiotic treatment is implemented. Other infections present no symptoms and have subclinical manifestations, so that they are not treated even if they occur for long periods of time (light gray line). Only infectious bacteria that produce clinically relevant symptoms and are present long enough to be treated (dark gray line) are under antibiotic selective pressure.
Finally, epidemic organisms may evade a short antibiotic exposure because, by definition, they move to another host (usually nontreated) more easily than a nonepidemic microorganism. Thus, mechanisms facilitating between-host transfer may in some cases reduce exposure to antibiotics, particularly if the organism is not highly virulent. In the long run, however, epidemic organisms will confront higher levels of antibiotics, as the hosts are treated after the infection is detected (see above).
In host spatial location and exposure to antibiotics.
The location of microorganisms in tissues during infection is an important factor in their exposure to antibiotics. In some cases, organisms are refractory to treatment even if they are classified as susceptible by standard in vitro tests (242). Virulent organisms may invade spaces in which the achievable concentration of antibiotic is low, such as in otitis, sinusitis, and meningitis (186). Also, the pharmacodynamic properties of antibiotics influence in a critical way the inhibitory and killing effects of these drugs. For instance, if a given concentration of a beta-lactam is not maintained in contact with the organism over a critical period of time, the antibacterial effect (predicted by the AUC/MIC ratio and time above the MIC) will be reduced.
In other cases, there is a critical concentration of antibiotic (Cmax) that is affected by changes in the medium during infection. For example, low pH is characteristic of inflammatory fluids as well as some intracellular compartments (353). Several antibiotics are less active in acidic conditions, so that the environment of the infection site may contribute to lowering the antibacterial effect of antibiotics (193, 226). As previously discussed, several bacterial pathogens can invade host cells and are thus less exposed to antibiotics (395). Biofilms (see above) also produce an antibiotic resistance phenotype due at least in part to the structure of the biofilm, which precludes the efficient transfer of antibiotic to all the bacteria present in the biofilm (83, 370). More recently, it has been described that resting cells are as resistant as bacteria growing in biofilms, and the effect of biofilms on antibiotic susceptibility might be due to a stationary-like growth phase of the bacteria growing in the biofilms (364)
Obligate intracellular pathogens such as Chlamydia, Lawsonia, and Rickettsia spp. are infrequently resistant to antibiotics. For these pathogens, the MICs of antibiotics such as tetracycline and erythromycin that are distributed either extracellularly or in intracellular compartments that are different from those in which bacteria are located have remained constant over decades (259) This suggests that some intracellular compartments serve as refuges against antibiotics. It is obvious that intracellular location also prevents easy acquisition of resistance elements such as plasmids and transposons, from other organisms. These elements may occasionally be present in bacteria, as Coxiella and Listeria spp., which do not have an absolute requirement for an intracellular location for growing. Other obligate intracellular pathogenic organisms such as Brucella spp. are infrequently resistant to antibiotics and rarely contain plasmids.
Comparison of the genomes of different bacterial species has demonstrated that in organisms with small genomes, horizontal gene transfer is very infrequent (15, 285). It has been suggested that because of the extreme specialization of the small genome of intracellular organisms, any new gene or gene modification may disrupt the delicate equilibrium of vital genes, leading to extinction of the microorganism (113, 286). In other words, resistance genes might reduce the viability of intracellular organisms. Moreover, the altered metabolism of these organisms might reduce their ability to utilize resistance genes. For instance, the energy needed for expression of resistance to tetracyclines and macrolides by efflux mechanisms may not be accessible in organisms dependent on host cell energy sources (259). In general, intracellular bacteria might tolerate only mutational changes that lead to energy-independent mechanisms of resistance and not reduce intracellular fitness.
The possibility that some bacteria could have developed mechanisms of intracellular invasion in the environment, as an escape from natural antibiotics, cannot be discarded. Facultative intracellular pathogens such as Legionella spp. have environmental protozoa as natural hosts, and the pathogenicity of these organisms in humans may only reflect an accidental change leading to the invasion of another type of eukaryotic cell (161). Something similar may have occurred in Listeria. Interestingly, acquired antibiotic resistance is a very unusual event in both genera, which indicates that intracellular location is a effective way to escape from antibiotics.
In summary, we want to stress that the location of an organism in a particular body site is influenced by its colonizing and pathogenic abilities. Therefore, these properties determine the cell-antibiotic interaction. On the other hand, this interaction is critical for the eventual development of antibiotic resistance.
Normal microbiota versus pathogenic bacteria.
The normal microbiota is the first line of defense against infections due to its role as a colonization barrier (41). However, once colonization occurs, normal bacterial flora might also protect virulent bacteria from antibiotic action. Protection of a susceptible population by a resistant one has been described in vitro for beta-lactam antibiotics (30). Beta-lactamase-producing bacteria rapidly inactivate beta-lactam antibiotics. If a mixed population of beta-lactamase producers and nonproducers is exposed to beta-lactams, the nonproducers will begin to die. However, once the antibiotic is inactivated by the beta-lactamase producers, the remaining susceptible bacteria that have not been killed by the beta-lactam can grow. This effect is frequently seen in the laboratory, when ampicillin-susceptible colonies exhibit satellite growth around an ampicillin-resistant clone in an ampicillin-containing plate.
It has been demonstrated that minute amounts of beta-lactamase-producing strains can protect fully susceptible strains from the action of beta-lactams (30). During infection, the same situation might occur in vivo. Antibiotic-resistant populations, either as normal colonizers or in the case of coinfection by different bacteria, can then protect virulent, susceptible bacteria from antibiotic action. This effect has been claimed as relevant when beta-lactam therapy failed to eradicate fully susceptible bacteria from colonized environments (66).
Normal microbiota can also contribute to the acquisition of antibiotic resistance determinants by bacterial pathogens because in vivo transfer of antibiotic resistance determinants has been reported (248). Thus, acquisition of antibiotic resistance genes by normal, nonpathogenic microbiota is a concern because the final outcome of antibiotic resistance in bacterial pathogens might be compromised. Indeed, the possibility of maintaining hospitals with a reduced proportion of resistant bacteria depends on the local reduction the antibiotic selection, but also on the dilution of the resistant organisms of the hospital caused by the admission of patients from the community with susceptible normal flora (222).
POPULATION STRUCTURE
The population structure of clinically relevant bacteria is an important epidemiological topic (212). If only one or a few clones of a bacterial species are relevant for infection and antibiotic resistance, efforts must concentrate on these clones. Also, preventive strategies can be more easily designed if the prevalence and spread of the various clones involved in disease are known. A more comprehensive analysis of the infection must involve the study of bacterial populations from both the ecological and evolutionary perspectives. This analysis requires the use of molecular biology tools for analyzing the structure and evolution of bacterial pathogens.
Bacterial Variation Facing Stressful Environments
As was stated above, bacterial adaptation is essentially the consequence of mutation and gene recombination. Mutational events are particularly important when a large population of cells is confronted with critical, abrupt, and nonspecific changes in the environment, eventually permitting rapid but not always optimal adaptation. In fact, selection of bacteria with a hypermutator phenotype (see below) has been suggested to be a good adaptive strategy to sudden environmental changes (381), and mutational adaptation requires large bacterial populations (381), as this evolutionary strategy implies the death of most members of the population (246). On the other hand, recombination allows the acquisition of several different traits in a single step (153).
In this regard, horizontal transfer of genes and even large chromosomal regions is frequent in all bacterial species (101, 176), contributing to the acquisition of several physiological functions in a single step (see above). However, recombination between different strains of the same bacterial species occurs frequently in only a subset of bacterial species (1, 79, 122) that are highly specialized to grow in constant and frequently in unique habitats (such as Helicobacter pylori, Neisseria meningitidis, Neisseria gonorrhoeae, Haemophilus influenzae, and S. pneumoniae). In these cases, recombination between members of the same species is important for combining the beneficial mutations present in the entire bacterial population and eliminate the deleterious ones. In fact, it has been stated that a high recombination rate could be selected when several mutations (present in different members of the population) are needed for adaptation (118).
Bacterial adaptation can increase under conditions of stress such as infection and antibiotic selective pressure. In the last few years, it has been shown that bacteria have stress-regulated systems which allow them to mutate at a higher rate under stress conditions. Two different situations are relevant for increased mutation rates in vivo in bacterial populations.
Stress-induced transient increases in mutation rates.
Bacteria increase their mutation rate as a consequence of stress, such as starvation (185, 348), antibiotic challenge (9, 318), and possibly during infections; this process is termed adaptive mutation (129, 348). A situation of stress triggers a bacterial response that involves changes in the expression of several proteins. These changes are a consequence of the recognition by the bacteria of “stress signals” that activate a global response based in the concomitant transcriptional regulation of several genes (343, 383, 421). Some aspects of these responses are specific to the type of stress involved, but other changes are common to different stress situations.
Little is known about the bacterial responses to pathogenic and antibiotic-induced bacterial stresses. However, it is conceivable that these types of stress trigger responses similar to those in other bacterial stress situations. For instance, treatment with antibiotics at bacteriostatic concentrations provokes a stationary phase-like response, and it has been demonstrated that subinhibitory concentrations of antibiotics targeting the ribosome induce the synthesis of either heat shock or cold shock proteins, depending on whether the A site of the ribosome was empty or occupied (45). In any case, adaptive mutation to stress is a regulated phenomenon by means of which bacteria can increase their mutation rate severalfold (327). The primary factors in this process are stress-responsive, error-prone DNA polymerases V (umuCD) and IV (dinB), whose expression transiently increase the rate of bacterial mutation (310). Eventually, bacterial stress over the pathogenic process may increase the probability of emergence of antibiotic-resistant mutants; conversely, antibiotic stress may increase bacterial adaptation to the host.
Hypermutable (mutator) strains.
Hypermutation can be the consequence of a regulated increase in mutation rates (see below) or the presence of subpopulations of “mutator” bacteria within “normal” bacterial populations (252, 386). Some works have demonstrated the presence of such mutator populations in infecting bacteria (67, 208, 287). These cells have mutation rates that can be increased as much as 10,000-fold, usually as the consequence of inherited defects on the methyl-directed mismatch repair system (380). These strains should have increased abilities to survive antibiotic challenge and antibacterial host defenses. Among strains lacking the methyl-directed mismatch repair system, the ability to recombine (homologous recombination) should increase (253), which may facilitate the acquisition of both antibiotic resistance and bacterial virulence in the same organism.
Mutator strains are supposed to be less fit than wild-type strains outside the stress situation (adaptive bottleneck). Induction of a transient hypermutation under stress serves to generate mutants which can escape from transient stress situations, whereas stable mutator strains are fixed in the populations only when bacteria are subjected to frequent bottlenecks. This situation probably occurs in chronic infections, such as those associated with cystic fibrosis. In fact, P. aeruginosa isolates from cystic fibrosis patients frequently present a mutator phenotype, a characteristic feature that does not occur in P. aeruginosa isolates obtained from blood and sputum cultures of non-cystic fibrosis patients (287).
Recently, the fitness of mutator E. coli strains has been analyzed in an in vivo model system (146). A high mutation rate was shown to be beneficial because it allowed faster adaptation. However, the benefit disappeared once adaptation was achieved, because mutator strains accumulated mutations that, although neutral in the analyzed model system, can be deleterious in a secondary environment. Thus, mutator strains may have short- and long-term advantages that justify high competitiveness of mutator bacteria during infection, but low competitiveness during transmission and recolonization of other hosts.
In summary, stressful growth conditions in the host, particularly under antibiotic therapy, are important elements for the evolution of bacterial populations, influencing gene transfer and increasing mutation rates. Stress associated with the pathogenic process and antibiotic exposure not only increases bacterial variation, but also strongly selects particular bacterial variants. In this sense, pathogenesis and antibiotic exposure can be considered “accelerators” (31) of bacterial evolution.
Clonal Structure
Molecular epidemiology has demonstrated that a few clones may account for the worldwide dissemination of virulent and antibiotic-resistant bacteria for several bacterial species. This view of the genetic structure of bacterial species is known as the clone concept (345, 368). Further analysis has demonstrated that the degree of clonality varies depending on the species analyzed. In that way, pathogenic species such as E. coli and Salmonella spp. are highly clonal, whereas species such as Neisseria gonorrhoeae present a very high rate of recombination and have a panmictic (species that are not subdivided into discrete phylogenetic lineages [385]) genetic structure (357). Even for microorganisms with a predominantly clonal structure, analysis of complete genome sequences has demonstrated that horizontal transfer due to recombination is frequent in bacterial populations (204).
The high rate of recombination in several bacterial species might put in question the validity of the epidemiological analysis of clinically relevant bacteria under the clonal structure approach. However, even for bacteria for which recombination occurs very frequently, this approach can be extremely useful. When analyzing the genetic structure of these bacterial populations, evolution can be traced over time, following the routes of transmission. For highly epidemic species such as Neisseria meningitidis, clones emerge that disseminate further worldwide. Vaccination against the most frequent serotypes may produce the replacement with less common ones (see below). Also, the replacement of antibiotic-susceptible with antibiotic-resistant clones has occurred, and these replacements are easily followed by using molecular typing methods (225). In this case, a transient “quasi-clonal” population can be observed at a fixed time depending on the geographic localization (148).
This situation is the consequence of the emergence of clonal lineages which are more proficient in their epidemicity, virulence, and antibiotic resistance than others. The more successful lineages will then predominate. However, once they become the predominant population, they will produce novel divergent lineages as a consequence of recombination with other clones and may then expand in different environments. The presence worldwide of different environmental habitats for bacterial populations guarantees this divergence, as has been demonstrated in different model systems (313).
The clonal hypothesis is also relevant for understanding the evolution of genetic elements that are transmitted in bacterial populations. In fact, the variability of antibiotic resistance genes with clinical relevance is not very high, which indicates a “clonal” origin for several. Thus, most antibiotic resistance genes currently acquired by horizontal transfer were originated by transfer of an ancestor gene to a single or a few lineages of bacteria and then disseminated among bacterial populations. For instance, it has been demonstrated that the gene mecA, responsible for resistance to methicillin in S. aureus, was acquired only once by this bacterial species (196). Something similar has probably occurred for widely distributed genes such as plasmid-borne beta-lactamase genes of the TEM group (71).
Even for highly recombinogenic bacterial species, a clonal origin of their genetic resistance determinants can eventually be traced. An example is the evolution of the genes encoding the synthesis of penicillin-binding proteins in Neisseria and Streptococcus spp. The analysis of pbp genes from different isolates has demonstrated that they are very uniform among penicillin-susceptible strains of N. meningitidis, N. gonorrhoeae, and S. pneumoniae, whereas those from penicillin-resistant strains consist of a mosaic structure that arose from the horizontal transfer of blocks of DNA from other, more naturally penicillin-resistant, commensal species by genetic transformation (79, 365). Analysis of beta-lactam-resistant populations has demonstrated that these novel pbp genes have occasionally been selected, as single gene “clones,” and afterwards, either the antibiotic-resistant bacterial clone or the pbp “resistant gene clone” disseminates and evolves further among the bacterial population.
The same notions applied for analyzing the evolutionary biology of bacterial populations can be applied when analyzing accessory genetic elements (mainly plasmids and transposons). The most successful plasmid will be one with a good compromise between plasmid “epidemicity,” defined as the ability to transfer between different species (number of different species or strains to which the plasmid can be transferred), “infectivity,” defined as the ability to infect a specific host (frequency of entrance in the bacteria); and presence of selective traits (see above). As discussed above, antibiotic resistance may reduce bacterial fitness (13), and the same occurs with the acquisition of plasmids (8). If the fitness reduction is too high, the resistant bacteria will be unable to compete with the wild-type susceptible ones. Thus, most probably, we only detect those antibiotic resistance genes that do not produce an unaffordable fitness reduction on bacteria.
We think that there are probably many more antibiotic resistance genes than those currently detected in bacterial populations. As stated previously, accessory genetic elements of clinically relevant bacterial populations also follow the rules of the clonal hypothesis. The same vir plasmids are found in all members of a single Yersinia species and are quite similar when one species is compared to another (306). This indicates both the clonal origin for these determinants and their successful maintenance in Yersinia populations for very long periods of time. Also, dissemination of epidemic antibiotic resistance plasmids is very frequent, and the acquisition of novel genetic determinants by a clonally originated plasmid has been documented (31, 98). In fact, analysis of bacterial collections from the preantibiotic and postantibiotic eras demonstrated that they contain plasmids belonging to the same families, although the ones from the preantibiotic era do not contain antibiotic resistance genes (95, 171). These results indicate the ability of plasmids to evolve as evolutionary units through the acquisition of antibiotic resistance determinants.
The clonal hypothesis is then a useful tool for analyzing relationships between bacterial strains as well as mobile elements and even between genes involved in clinically relevant phenotypes. The use of this analysis of the lineages of such different elements can give a complete picture of the evolution of bacterial pathogens in response to novel selective pressure (such as antibiotic treatment). This approach is also relevant from the epidemiological point of view and can offer new light for understanding the future evolution of bacterial pathogens in terms of virulence, epidemicity, and antibiotic resistance.
The application of the clonal hypothesis to the study of both organisms and genes will eventually serve to explore the hypothesis of possible convergent evolution of antibiotic resistance and bacterial pathogenicity, eventually leading to increasing numbers of organisms that are both resistant and virulent (see below).
Resistance and virulence decrease bacterial diversity.
Virulent bacteria must face several environmental changes over the course of infection. To infect the host, bacteria must enter the organism, traverse through different epithelia, avoid the action of different host defense mechanisms against infection, and finally reach the target organ(s), where they produce the disease (Fig. 1). It is intuitively appealing that the challenge of bacterial populations by these successive selective bottlenecks should reduce genetic diversity, at least in the short term.
Changing environments are very stringent conditions for evolutionary adaptation, so that only a few organisms can survive those changes. As stated in the legend to Fig. 1, this is the case for the process of infection. The diversity of commensal bacteria in humans is extremely high, which shows that many different bacteria are adapted to grow on each of the different host surfaces (intestine, skin, oropharynx). On the contrary, the diversity of virulent bacteria is much lower (357). Virulent bacteria need to traverse and eventually colonize different environments as well as survive stringent and changing conditions (see Fig. 1). The requirement for multiplication under all these stringent conditions might explain the low diversity of very specialized pathogens. A release from some of the stringent conditions required for growth in the healthy host will increase the possible diversity of potential pathogens. In this regard, a diverse array of opportunistic pathogens are able to infect the immunosuppressed and severely ill host, which presents a much less stringent environment (see above).
A stringent selection also occurs for antibiotic resistance. In this case, rapid changes in the environment, produced by antibiotic exposure, rapidly lead to lethal conditions of selection. Only resistant clones will survive, replicate, and subsequently colonize the previously complex ecosystem. In fact, it is well known that many antibiotic treatments reduce the bacterial diversity of the human-associated microflora. Selection is even stronger with conditions of combined and successive antibiotic therapy, because bacteria are challenged by simultaneous and consecutive lethal bottlenecks. Therefore, only a few types of bacteria will survive, and diversity will be strongly reduced as a consequence of the treatment.
Taxonomic Implications of Changes in Resistance and Virulence
The development of antibiotic resistance and the emergence of new pathogens have two implications for the taxonomy of microorganisms.
Misclassification as new species.
The phenotypic changes associated with the acquisition of an antibiotic-resistant phenotype may alter the characteristics of the well-known human pathogens, producing mistakes in the classification of these microorganisms. For instance, optochin-resistant S. pneumoniae (300) could be easily classified with the viridans streptococci (58), and penicillin-resistant pneumococci manifest major changes in peptidoglycan structure (138). If classic approaches to phenotypic identification are used without knowledge that these bacteria are antibiotic-resistant derivatives of pneumococci, a new species defined as “an antibiotic-resistant gram-positive organism, very close to S. pneumoniae but with some different features, such as resistance to beta-lactams and a characteristic peptidoglycan structure” could have been described.
Emergence of novel microbial species.
The emergence of novel virulent species may occur by the acquisition of pathogenicity islands, which changes the pathogenic characteristics of the bacterial species. For instance, we know now that Y. pestis, the cause of plague, is a recently emerged clone of the much less virulent Y. pseudotuberculosis (2). Also, acquisition of a new host specificity could lead to novel species, such as occurs with the human immunodeficiency virus (for instance, from chimpanzees to humans [137]) and several mycobacteria (from ruminants to humans [17, 53]). In fact, it has been suggested that speciation of M. tuberculosis occurred 15,000 to 20,000 years ago (183) at the time of ruminant domestication.
Finally, new bacterial species (and strains) for the moment unknown might emerge linked to novel diseases. For instance, enterohemorrhagic E. coli (131) has been recognized as a relevant health problem just in the last two decades. However, the large divergences found upon comparison of the sequence of the whole genome of the enterohemorrhagic E. coli strain O157:H7 (297) with the nonpathogenic laboratory strain K-12 (52) demonstrates that these strains diverged a long time ago.
FUTURE EVOLUTION IN AN ECOLOGICAL PERSPECTIVE
Major Factors
The future evolution of bacterial virulence is expected to be shaped (Table 3) by at least a dozen major factors.
TABLE 3.
Factors that might influence the future evolution of virulence and antibiotic resistance
| Factor | Outcome |
|---|---|
| Sanitation and hygienic measures | Reduction in the number of infections |
| Preservation (eventually bioremediation) of ecosystems | Maintenance of microbial diversity |
| Human demographic changes | Increased number of host-pathogen contacts |
| Environmental changes | Changes in the geographic localization of microorganisms |
| Commercial interchanges | Wide spread of microorganisms |
| Increased number of elderly people | Increased number of opportunistic infections |
| Aggressive medical technologies | Increased number of opportunistic infections |
| Intensive farming | Reduction in microbial diversity; selection of antibiotic-resistant bacteria by antibiotics and food additives |
| Diseases that alter host response to infection | Increased number of opportunistic infections |
| Novel antimicrobial strategies | Better treatment of infections |
| New vaccination technologies | Prevention of infections |
| Bioterrorism/biological war | Spread of old and/or novel pathogens |
Sanitation and hygienic measures (mainly drinking water and sewage management).
The rise in the world population coupled with demographic changes (megacities, displacement of the population from the countryside to the cities, increased numbers of refugees) produces an increased requirement for noncontaminated water reservoirs. Control strategies must be implemented to guarantee the quality of water and food resources used in agriculture and animal and human consumption in order to prevent the emergence and dissemination of potentially relevant novel diseases.
Preservation and eventual bioremediation of normal ecosystems in human, animal, and environmental microbiota.
Industrial contamination produces severe changes in natural ecosystems. Heavy metals, pesticides, solvents, and even antibiotics are among the most conspicuous contaminants. The effect of this type of contamination on indigenous bacterial populations has been little analyzed; however, the above-mentioned deleterious compounds may have a profound impact on natural microbial populations, including the selection of antibiotic-resistant variants (11), increased recombination, and, in general, qualitative changes (388) in the microbial species and clones that colonize different habitats. Bioremediation of contaminated environments may help to reduce the potential effect of pollution on microbial ecosystems (11).
Number of contacts between microbes from infected and noninfected hosts (human and animal) and between infected people and the environment.
The demographic structure of the human population (338) increasingly facilitates urban crowding, favoring host-to-host contact and the dissemination of infectious diseases among the population.
Global environmental changes.
Global environmental changes, including increases in warming and UV exposure, altering the biogeographical limits of bacterial species and thus mixing them with new hosts might serve to accelerate bacterial evolution and increase the number of diseases with previous geographic restrictions. For instance, global warming might increase the prevalence of tropical infectious diseases in areas such as the United States and the European Union.
Human sociological movements.
Human sociological movements, including commercial interchanges (330), emigration, travel, war, and establishment of underdeveloped social islands in developed societies might increase contact between people infected with pathogens with a previous closely restricted geographical allocation (see the example of Yanomami Indians in reference 6).
Increase in susceptible hosts.
The increase in the number of elderly people, immunosuppressed patients, and children, particularly if they are densely clustered (day care centers, nursing homes), will increase the role of opportunistic pathogens in infectious diseases.
New technologies.
New aggressive medical technologies for diagnosis and therapy (not anti-infectious) might reduce the incidence of pathologies such as cancer and diseases of the elderly at the cost of increasing the problem of infections due to opportunistic pathogens (42, 378).
Intensive farming.
Intensive farming, which reduces the diversity of animals (the same animal breeds are used in different farms worldwide), might also reduce the diversity of microorganisms colonizing such animals, producing an ecological alteration in the populations of microbes that can come into contact with humans (94). Also, the use of antibiotics for farming purposes might increase the emergence or spread of antibiotic resistance (301, 417).
Viral and virus-like diseases.
Viral and virus-like diseases affecting humans and animals, such as AIDS, might evolve and alter the host response against infections.
New antimicrobial strategies.
The discovery and use of new antimicrobial strategies, including antibiotics, antivirals, conventional vaccines, pro- and prebiotics, antipathogenic therapies, and modulators of biological response modifiers in the infected host, may be useful for fighting infectious diseases.
Vaccination.
New technologies in vaccination (in some cases food-associated; see below) and gene or sequence therapy may be useful for reducing susceptibility to some infections (309).
Bioterrorism.
Eventually, bioterrorism will affect antibiotic resistance and virulence, with dangerous and unpredictable consequences (190).
Interestingly, all these factors will also influence antibiotic resistance. Over the last two decades, resistance to antibiotics has crossed the limits of the patient and hospital to become a global environmental phenomenon in which infectious diseases of wildlife and domestic animals might have a deep impact on the emergence and further spread of new human pathogens (94). Everything that promotes bacterial spread among different natural ecosystems and among different hosts (protozoa, animals, plants, and humans) will also potentially affect the spread of resistance. A common theme resulting from all these factors is the possible progressive reduction in bacterial diversity by alteration of the complexity of current ecosystems (see above).
In this regard, intensive farming procedures may have consequences for the future evolution of virulence and resistance. In fact, farming procedures reduce microbial diversity (388). The collapse in the diversity of animal and plant natural variants (races) within agronomically relevant species has already started, because only those variants with high productivity rates are being used worldwide. In fact, most crops are obtained from the seeds produced by a few companies. The result of animal cloning procedures, by which the offspring will be genetically identical to the progenitor, will produce an even larger reduction in the biodiversity of farming animals.
Genetic diversity among hosts is probably a natural barrier to infections, since multiple-passage experiments have shown that adaptation to a new host frequently involves attenuation for the former one (114). From this perspective, a reduction in host diversity will allow bacterial organisms infecting a particular host to spread with greater efficiency to other closely related hosts (identical in the case of clonal farming), eventually increasing virulence. The effect of a possible change of host (for instance, from animals to humans) may result in bacterial attenuation for the former animal host with an unexpected increase in virulence for the new human host (114). The economic and health consequences of these possible changes in bacterial transmission have not been evaluated. The possible outcome is an increase in certain bacterial populations that will alter their potential virulence as a result of interactions with new hosts and then become targets for antibiotic intervention.
If antibiotics are used over the next 50 years as they have been used in the previous 50 years, there is no doubt that the number of replicons and genetic elements carrying antibiotic resistance genes will increase continuously on Earth, and the cost of harboring these genes and vectors will probably be minimized (214, 285), particularly in the more common bacterial populations, thus ensuring their perpetuation in the microbial world.
Strategies against Virulence May Reduce Antibiotic Resistance
Antivirulence strategies tend to reduce the number of bacterial pathogens and may act on the expression of pathogenic mechanisms. In some cases, if the pathogenic mechanism is essential for the lifestyle of the bacteria, the elimination of the pathogenic factor will reduce the overall presence of the pathogen. For instance, nonsystemic toxins, such as necrotic toxins and hemolysins, contribute to the successful growth of the bacterial pathogen inside the host, so that antitoxin vaccination will not only reduce the toxic effect of the toxin, but also contribute to eradicating the bacterial species. In this regard, strategies against virulence may reduce antibiotic resistance, because lower number of pathogenic bacteria implies less antibiotic exposure, a smaller number of antibiotic-resistant mutants, and fewer possibilities of acquisition of antibiotic resistance genes from other bacterial pathogens.
Reducing host-to-host transmission: hygiene and vaccination.
Any strategy that leads to a reduction in the number of infections in the host population will reduce the probability of emergence of antibiotic resistance for two reasons: a reduction in the number of bacteria under antibiotic selective pressure, and a reduction in the number of selective environments (different hosts). This results in a decreased probability of selecting different antibiotic-resistant mutants (32).
In this regard, hygiene and vaccination are among the best systems for avoiding infections and resistance. The use of conjugate vaccines against those S. pneumoniae serotypes that are more frequently resistant to antibiotics have significantly decreased the rate of penicillin resistance in closed communities (91, 92). As predicted by mathematical models, this decrease is related to the selection of nontargeted and poorly targeted serotypes (219, 220). Nevertheless, in some instances, the more susceptible serotypes may also be the more invasive (163). In this case, if these serotypes are not included in the vaccine, vaccination could increase pathogenicity. Vaccination against Haemophilus influenzae type b reduces resistance is this species, as resistance is found more frequently in this serotype (175). In this particular case, both pathogenicity and resistance have dropped simultaneously, as no clear replacement by other types has been documented to date.
In general, the vaccine-mediated reduction in the number of pathogenic organisms and infected hosts reduces the likelihood that resistance will evolve in these bacteria, as acquisition of resistance by mutation and horizontal transfer is a density-dependent phenomenon (see above). Obviously, the principle applies not only to human but also to animal infections and may have an important impact on the development of resistance in the food industry, where antibiotics are frequently used to prevent infections.
In the next few years, we will probably face an explosion in the number of potentially effective novel vaccines. Such a trend is based on the growing use of genomic information together with in silico prediction of useful antigens and massive production of recombinant proteins to be used for immunization of animal models (303). This has been called reverse vaccinology (317) and will doubtless offer a great number of antigenic candidates for the development of a new generation of vaccines. We must insist, however, that when vaccines are developed, the local microbial ecology should be considered.
We have mentioned that antibiotic-susceptible but more virulent bacterial types may emerge because a vaccine is directed against resistant organisms. But it is also true that some vaccines may eventually be more effective in eliminating some antibiotic-susceptible types and therefore select for antibiotic resistance. This possibility should be considered when a vaccine is introduced into a given human habitat (163, 220). A more optimistic perspective is to consider that prolonged use of antibiotics has reduced the diversity of some bacterial pathogens, so that some pathogenic species may be less able to adapt to novel challenges. If an effective vaccine is available for reducing the number of antibiotic-resistant clones, some pathogens in some parts of the world may become locally extinct (see below).
Antivirulence-directed therapeutic approaches.
Antivirulence is considered here as interventions directed against virulent organisms and/or the expression of the virulence determinants. One of the best-known side effects of antibiotic treatment is the replacement of the antibiotic-susceptible commensal flora with resistant organisms. The release of antibiotics in the environment, as well as the use of antibiotics as growth promoters in animal feed, may have a profound impact on bacterial populations. One theoretical possibility to avoid this side effect is to develop antibiotics directed exclusively against virulent organisms. A “magic bullet” is one that only kills virulent bacteria in such a way as to preserve the antibiotic-susceptible phenotype of nonpathogenic bacteria. In this way, commensal bacteria will remain a colonization barrier against infection and will not acquire antibiotic resistance genes. Acquisition of antibiotic resistance genes by nonpathogenic bacteria is detrimental for two reasons. First, these bacteria will constitute a reservoir of antibiotic resistance genes (and antibiotic resistance vectors) that may be transferred to virulent bacteria. Second, antibiotic-resistant bacteria can protect the susceptible ones (eventually pathogenic) from the action of antibiotics (see above).
Although this magic bullet will obviously be desirable, it is not easy to find a lethal target present only in virulent bacteria. Some types of antibiotics act and have been designed in a way that antibiotic resistance will automatically reduce bacterial fitness and consequently the pathogenicity of the resistant organism. For instance, naturally occurring albomycins (124), metalloporphyrins (371), and synthetic derivatives of Fe3+ siderophores with antibacterial activity (301) are taken up across the bacterial outer membrane by transport systems for Fe3+ siderophores and heme transport systems. Indeed, some antibiotics have been developed that include a siderophore moiety linked to the antibiotic molecule (65, 111, 265). Those antibiotics make use of the siderophore transport system for entry into bacteria. Resistant bacteria can arise as a consequence of mutations in this transport system. However, loss of the iron transport system is detrimental for bacterial growth at the site of infection, so that treatment with this type of antibiotic may severely affect bacterial survival, either by the antibiotic effect itself and by allowing host defenses to cope with the infection due to a mechanism of resistance that has an excessive biological cost for the bacteria.
Vaccination directed against virulence determinants has also been proposed as an effective antivirulence approach to prevention of infectious diseases. Such as all strategies not based on antibiotic treatment, this may help reduce the emergence and spread of antibiotic-resistant bacteria. One example of this strategy is the development of novel vaccines against S. aureus. In this organism, the expression of virulence factors is triggered (25) by the RNA III activating peptide (RAP). This peptide is continuously secreted by bacteria, and its activity resembles some other quorum-sensing signals which are also inducers of the expression of virulence determinants (37, 189). Vaccination with RAP protects mice from S. aureus pathology (24). This indicates that anti-quorum-sensing strategies might be useful to fight infection.
A different approach for antivirulence vaccination was used by Mason et al. (250). A transgenic potato containing the gene for E. coli heat-labile enterotoxin was obtained (250, 379) and used to feed mice. Feeding with such potatoes produced higher levels of antitoxin antibodies than the whole-bacteria vaccine. Thus, this opens the possibility of developing transgenic plants containing several virulence factors that could be used as multisubunit edible vaccines (249).
Strategies Directed against Resistance May Reduce Bacterial Virulence
It has been suggested that the search for and further use of antibiotics active only against antibiotic-resistant bacteria might reverse the problem of antibiotic resistance. The hypothesis of antibiotics able to select susceptible organisms was suggested some years ago (27). For instance, to fight the spread of beta-lactamase-producing bacteria, it may be possible to develop an antibiotic drug whose activation depends on the breakage of a beta-lactam ring. Because only beta-lactamase-positive resistant bacteria would be susceptible to such an antibiotic, beta-lactam-susceptible organisms will be selected. This and other approaches are currently under investigation. Obviously, any new antibiotic able to eliminate (because of the absence of cross-drug resistance) resistant organisms to other drugs has at least the possibility of eradicating resistance. This effect is only possible if the resistant organisms are in a minority with respect to susceptible ones, so that recolonization after therapy will occur from within the host and from other hosts with susceptible bacteria.
However, until now, the best (and maybe only) strategy against resistance is a good policy for the prescription and use of antibiotics (169). Rotation in antibiotic use has been suggested (322) and even implemented, in some cases, with an improvement in antibiotic resistance rates (282). However, the levels of resistance never drop to zero, so that rotation of antibiotics is useful only transiently (332). Antibiotics must be used only when needed (215, 223), and the consumption of antibiotics for purposes other than the treatment of infectious diseases must be avoided as much as possible. For instance, the ban on the use of avoparcin in animal feed has curbed the development of vancomycin resistance in the European Union (23). It has been discussed whether the ban would be followed by a reduction in the productivity and an increase in the morbidity and mortality of the animals as a consequence of an increasing number of infections (23, 115). However, neither a reduction in productivity nor an increase in the number of infections has been observed after discontinuing the use of antimicrobial growth promoters (115), probably because hygienic measures were implemented at the same time as the ban (23).
Strategies against the emergence and further spread of antibiotic resistance genes in bacterial populations will obviously reduce virulence because infections will be easily treated and the disease will disappear. We cannot forget that during the 1980s, it was common to believe that infections would not be a relevant clinical problem in the future of mankind. However, it became clear in the 1990s that this was not the case. Some infections became untreatable by conventional chemotherapy, and this produced a reemergence of the problem of infectious diseases (378). This renaissance has not been a consequence of the introduction of novel bacteria with a higher virulence potential, but mainly as a result of emergence of the same families of bacteria which now are resistant to antibiotics. Because antibiotics are the very last line of defense incorporated along human evolution in its fight against infections, breaking down this defense will restore the problem of infectious diseases to the situation of the preantibiotic era. Therefore, antiresistance strategies will clearly diminish the overall virulence of bacterial pathogens, because bacteria will remain susceptible to treatment so that they will not produce disease. To achieve this goal, novel antibiotics are required. (419)
Drug diversification.
Although several antibiotics are currently being used in therapy, all belong to a few structural families, so that resistance to one antibiotic frequently produces resistance to other members belonging to the same family. Novel antibiotics belonging to different structural families than those currently in use are then needed in order to fight against antibiotic-resistant bacteria
Two approaches can be combined in the search for novel antibiotics. One is the classical search for novel inhibitors of lethal bacterial targets either by using natural sources or by screening of synthetic compounds. The other is the search for inhibitors of resistance systems, with the aim of recovering a susceptible phenotype in a previously resistant population. Examples of the latter strategy are the beta-lactamase inhibitors (236) currently used in clinical practice, and the inhibitors of MDR pumps which are under development by some pharmaceutical companies.
Concerning the search of inhibitors of classical targets, major efforts are currently under way in order to use the knowledge derived from the sequencing of whole microbial genomes (325). The advantages derived from the use of combinatorial chemistry (109) and rational design by computerized molecular modeling of crystal structures of potential targets (331), the use of new methods for the screening of natural compounds (69), and the increasingly refined methods of high-throughput screening (64, 87) will be useful for developing novel families of antibiotics in the future. Nevertheless, we must still wait for the introduction of new antibiotics developed by using these methodologies.
In the meantime, new fluoroquinolones such as moxifloxacin (26), gemifloxacin (229), and gatifloxacin (298), new tetracyclines such as glycylcycline (375), streptogramins such as the combination quinupristin/dalfopristin (200), macrolide-related agents such as ketolides (107), and a new family of antimicrobials, oxazolidinones (110), have been launched and/or in the last phases of development. Some new families and resistance inhibitors are under development. These include the novel inhibitors of class C beta-lactamases such as boronic acids (411) and inhibitors of MDR efflux pumps (227, 319, 369)
The diversity of clones within a given bacterial species is severely reduced by antibiotics. In some instances, the species may maintain a certain degree of diversity when the resistance determinant disseminates by horizontal transfer into different clones, and if a mechanism of diversification is available, for instance, capsular transformation in S. pneumoniae. Nevertheless, even in the case of species able to exchange DNA, a progressive simplification of the population structure is expected to occur. In S. pneumoniae, prolonged challenge with beta-lactams has selected a relatively small number of “international resistant clones,” certainly at the expense of reducing the number of susceptible clones. It has been discussed that this reduction in diversity may lead to the potential extinction of some highly virulent but antibiotic-susceptible clones. In this sense, the overuse of antibiotics may have reduced the overall pathogenicity of certain organisms. In any case, the antibiotic-driven reduction in diversity of the bacterial clones may cause extinction of some of these bacterial pathogens, using either vaccines against the reduced number of resistant clones or new antibiotics to which the resistant bacteria are susceptible, or both strategies simultaneously. Implementation of barriers to transmission will increase the possibility of reaching extinction. It seems that a low-diversity population structure will reduce the possibility of maintaining a pathogen under these new pressures.
INFLUENCE ON EVOLUTION OF THE HOST
The sentence “I am myself and my circumstances” from the Spanish philosopher Ortega means, in biological terms, that not only genetics but also the environment is relevant to phenotype. In the case of humans, it might also mean that the human body is formed by 10% mammalian cells and 90% prokaryotic cells (our normal microbiota) (41), each of which can influence the evolution of the other. The human host is also surrounded by a plethora of environmental microorganisms, some of which may interact transiently with the human body. Therefore, it is clear that the ability of bacteria to grow and damage human tissues has most probably had an effect on human evolution. For instance, the build-up of the extremely complex system of immunological responses involved in the defense against infection that was selected during the evolution of animals was necessary (and, so, selectable) only because of the existence of virulent microorganisms.
In this regard, it has been suggested that infectious diseases might be the major evolutionary force driving the high diversity displayed by the major histocompatibility complex (178). Body temperature, which is higher than the mean environmental temperature, limits the growth of microorganisms in humans to those that can grow in a narrow range around 37°C. In fact, it has been demonstrated that pathological hypothermia favors infection by environmental bacteria such as Pseudomonas putida (76). We thus speculate that body temperature is a very first line of defense that precludes infection by environmental microorganisms. It is thus possible that this anti-infective function together with several other selective pressures has contributed to select 37°C (a temperature that requires that constant metabolic activity be maintained) as the human body temperature. These examples illustrate that virulent microorganisms might have had a relevant effect on host evolution, as suggested by Haldane more than 50 years ago (159).
Since microorganisms can overcome human anti-infective defenses and kill the human host, infection might select the most resistant (in terms of infection) genetic backgrounds in the host population (309). In such a way, the prevalence of specific infectious microorganisms in different geographic allocations might be a driving force in human evolution. A very well known example of this viewpoint is the prevalence of sickle cell anemia in regions with a high rate of malaria infections (198). Sickle-shaped hemoglobin is protective against malaria. Thus, infection by Plasmodium falciparum has produced a bias in the number of sickle cell heterozygotes. The percentage of sickle cell homozygous individuals did not show such a relevant increase because these people have a high mortality risk caused by complications of sickle cell disease.
A more recent example is the prevalence of cystic fibrosis in Caucasian populations, which is higher than predicted by genetic analysis. Cystic fibrosis is a genetic disease caused by the lack of activity of a chlorine channel (CFTR). CFTR is also the receptor for some bacterial pathogens, such as Salmonella enterica serovar Typhi. Therefore, it has been suggested that cystic fibrosis could prevent infections by this bacterial species (302). In such instances, an increased risk of infection by S. enterica serovar Typhi might select populations with a bias in the percentage of cystic fibrosis patients.
In a similar way, it has been found that resistance and susceptibility to virulent M. tuberculosis is a complex genetic trait (195), so that different genetic backgrounds make people more and less susceptible to developing clinical tuberculosis once they have been infected with M. tuberculosis. It has been described that Yanomami Indians of the Brazilian Amazon have diminished cell-mediated immune responses against M. tuberculosis compared with control individuals of European extraction living within the same region. Yanomami Indians remained isolated until the mid-1960s and have not been in contact with M. tuberculosis until recently. As stated by the authors of that work, M. tuberculosis infections exerted a powerful selective pressure, “resulting in the elimination of a significant proportion of highly susceptible individuals over their reproductive age” (362). This selection occurred before the availability of antituberculosis treatment in the 1950s and might be occurring now if the treatment is not effective due to the emergence of antibiotic-resistant M. tuberculosis.
The same occurs for other infectious diseases. We must to keep in mind that the contact between Europeans and native populations in America produced dramatic effects on the Native Americans due to the dissemination of “European” infectious diseases among them. Whereas Europeans had been in close contact with these infectious agents, with a resulting selection against the most susceptible individuals occurring a long time ago, the first encounter between the native population and the European “alien” infectious agents caused the death of a great number of people. It has also been suggested that infections in the New World have been an important force for selecting the mixed race (mestizo) produced by the mating between Spaniards and Indians. As suggested by the authors, mestizos should be immunologically more capable of defending themselves against the different infectious agents brought over from the Old World (274) and maintaining their lower susceptibility to New World infections, so that they should be more adapted than Europeans and Native Americans to the new conditions produced by the introduction of novel pathogens in the environment.
These examples illustrate that infections have been important driving forces in human evolution. We have to keep in mind that several genetic changes might have been selected over human evolution just because they are protective against infections. However, we have only detected those changes which, like cystic fibrosis and sickle cell disease, are associated with a relevant genetic disorder. More recently, the study of infection-resistant phenotypes by classic human genetic analysis (309) is shedding new light on the potential role of infectious diseases as driving forces in host evolution. The use of antibiotics and the development of antibiotic resistance mechanisms might also have an effect on host evolution, because they will change the risk for infection.
To illustrate, we can speculate that the introduction of effective treatments (antibiotics, prophylaxis, vaccines) against S. enterica serovar Typhi might eventually reduce (in the long term) the prevalence of cystic fibrosis to the percentage of population predicted by purely genetic criteria. Also, as stated previously, effective antituberculosis treatment will allow the highly susceptible population to survive, because M. tuberculosis would not exert any selective pressure. Antibiotic treatment will then have the effect of removing selective pressure due to infections, and antibiotic resistance will have the effect of returning such selective pressure.
We want to stress here that, in any case, the time scale for a detectable effect of antibiotic treatment on human evolution is a lot longer than the time scale needed for bacterial evolution due to the extremely different lifetimes of the two types of organisms (from minutes to hours for bacteria, several decades for humans), so that a direct effect of antibiotics in human evolution is unsuitable unless antibiotics are maintained as the main choice for treatment of infections over several centuries. However, the most rapid and evident effect of antibiotic treatment will be on the composition of normal microflora because antibiotic treatment will select for resistant variants not only in the treated virulent microorganisms, but in the commensal microflora also (see below).
Changes in Normal Host Microbiota
Might virulent and/or antibiotic-resistant bacteria change host microflora? It is clear that a bacterial pathogen needs to successfully compete with normal host microflora in order to produce an infection, so that a temporal displacement of commensal bacteria might occur over the very first steps of infection. On the other hand, virulent bacteria might either select “resistant” hosts (see above), in which they can multiply without producing any relevant disease, and attenuated bacterial variants might also be selected. In any of these cases, the previously virulent bacteria might finally be incorporated as a commensal microorganism into the normal host microflora. Antibiotic treatment will also displace normal host microflora, allowing the recolonization by exogenous antibiotic-resistant bacteria as well as by antibiotic-resistant mutants belonging to normal microflora. This effect will probably be buffered in the long run, because human populations are not always under antibiotic treatment, so that the risk for selection of resistant variants will be lower for commensal microflora than for virulent microorganisms (see above).
Nevertheless, it cannot be ruled out that because of the huge consumption of antimicrobials in the world, some bacterial members which form the normal host microbiota will be replaced by others. For instance, highly sporulating gram-positive bacilli may replace (because spores are antibiotic resistant) less-sporulating organisms. Some Lactobacillus species are intrinsically less resistant to antibiotics than others and will be progressively selected. In general, bacteria with lower growth rates (less susceptible to most antibiotics) may also be selected. It can be predicted that those changes will produce effects, for the moment largely unknown, on human health.
Even for the same bacterial species, a replacement of human-adapted strains by animal-adapted strains might occur. For instance, natural populations of E. coli are organized in an ecotypic structure where adaptation to the host plays an important role (363). As discussed before, the use of antimicrobials as growth promoters selects antibiotic-resistant clones in the commensal microbial flora of farming animals (394, 418). Those clones that are not good colonizers of humans might replace human-adapted antibiotic-susceptible clones in the case of antibiotic treatment, thereby breaking down the close association between bacterial clones and their host. It is true that current bacterial diversity will probably offer “almost equivalent” resistant organisms to replace those that may be under danger of extinction. Nevertheless, most probably our intestinal flora will change, and therefore our microbial heritage for the future of humankind (the “historical host” in the future) will be adulterated. Again, the evolutionary consequences are totally unknown.
Microbial evolution is not a reversible process. The release of huge quantities of antibiotics on the Earth, together with other environmental toxic compounds, and the changes in the population structures of human (338) and animal hosts have profoundly changed the interactions of microbes and humans. The possibility of wide restoration of a wild antibiotic-susceptible bacterial flora in nature is beyond our current abilities.
Acknowledgments
Thanks are given to Fernando Rojo, Juan Carlos Galán, Marisa Morosini, Rafael Cantón, José Claudio Pérez-Díaz, Alejandro Dinamarca, and Jesús Blázquez for inspiring discussions on the topics reviewed in the present work as well as for fruitful criticisms and comments on draft versions of the manuscript. Thanks are also given to two anonymous referees for their excellent work reviewing and editing this article.
Work in our laboratories is supported by grants BIO2001-1081, QLRT-2000-01339, CAM08.2/0020.1/2001, and QLK2-CT-2001-00873.
Footnotes
This work is dedicated to the memory of our good friend Cristina Negri.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguero, M. E., L. Aron, A. G. DeLuca, K. N. Timmis, and F. C. Cabello. 1984. A plasmid-encoded outer membrane protein, TraT, enhances resistance of Escherichia coli to phagocytosis. Infect. Immun. 46:740-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ainsa, J. A., E. Perez, V. Pelicic, F. X. Berthet, B. Gicquel, and C. Martin. 1997. Aminoglycoside 2′-N-acetyltransferase genes are universally present in mycobacteria: characterization of the aac(2′)-Ic gene from Mycobacterium tuberculosis and the aac(2′)-Id gene from Mycobacterium smegmatis. Mol. Microbiol. 24:431-441. [DOI] [PubMed] [Google Scholar]
- 5.Aleixandre, V., G. Herrera, A. Urios, and M. Blanco. 1991. Effects of ciprofloxacin on plasmid DNA supercoiling of Escherichia coli topoisomerase I and gyrase mutants. Antimicrob.. Agents Chemother. 35:20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 7.Alland, D., I. Kramnik, T. R. Weisbrod, L. Otsubo, R. Cerny, L. P. Miller, W. R. J. Jacobs, and B. R. Bloom. 1998. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 95:13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alldrick, A. J., and J. T. Smith. 1983. R-plasmid effects on bacterial multiplication and survival. Antonie Van Leeuwenhoek 49:133-142. [DOI] [PubMed] [Google Scholar]
- 9.Alonso, A., E. Campanario, and J. L. Martinez. 1999. Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology UK 145:2857-2862. [DOI] [PubMed] [Google Scholar]
- 10.Alonso, A., F. Rojo, and J. L. Martinez. 1999. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1:421-430. [DOI] [PubMed] [Google Scholar]
- 11.Alonso, A., P. Sanchez, and J. L. Martinez. 2001. Environmental selection of antibiotic resistance genes. Environ. Microbiol. 3:1-10. [DOI] [PubMed] [Google Scholar]
- 12.Anderson, R., G. Joone, and C. E. van Rensburg. 1986. An in vitro investigation of the intracellular bioactivity of amoxicillin, clindamycin, and erythromycin for Staphylococcus aureus. J. Infect. Dis. 153:593-600. [DOI] [PubMed] [Google Scholar]
- 13.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 14.Andersson, D. I., and D. Hughes. 1996. Muller's ratchet decreases fitness of a DNA-based microbe. Proc. Natl. Acad. Sci. USA 93:906-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersson, S. G., and C. G. Kurland. 1998. Reductive evolution of resident genomes. Trends Microbiol. 263:263-268. [DOI] [PubMed] [Google Scholar]
- 16.Anwar, H., M. Dasgupta, K. Lam, and J. W. Costerton. 1989. Tobramycin resistance of mucoid Pseudomonas aeruginosa biofilm grown under iron limitation. J. Antimicrob.. Chemother. 24:647-655. [DOI] [PubMed] [Google Scholar]
- 17.Aranaz, A., E. Liebana, E. Gomez-Mampaso, J. C. Galan, D. Cousins, A. Ortega, J. Blazquez, F. Baquero, A. Mateos, G. Suarez, and L. Dominguez. 1999. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int. J. Syst. Bacteriol. 3:1263-1273. [DOI] [PubMed] [Google Scholar]
- 18.Asako, H., H. Nakajima, K. Kobayashi, M. Kobayashi, and R. Aono. 1997. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl. Environ. Microbiol. 63:1428-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Austin, D. J., M. J. Bonten, R. A. Weinstein, S. Slaughter, and R. M. Anderson. 1999. Vancomycin-resistant enterococci in intensive-care hospital settings: transmission dynamics, persistence, and the impact of infection control programs. Proc. Natl. Acad. Sci. USA 96:6908-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azoulay-Dupuis, E., V. Rieux, M. Muffat-Joly, J. P. Bedos, E. Vallee, C. Rivier, R. Isturiz, C. Carbon, and P. Moine. 2000. Relationship between capsular type, penicillin susceptibility, and virulence of human Streptococcus pneumoniae isolates in mice. Antimicrob.. Agents Chemother. 44:1575-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bach, S., C. Buchrieser, M. Prentice, A. Guiyoule, T. Msadek, and E. Carniel. 1999. The high-pathogenicity island of Yersinia enterocolitica Ye8081 undergoes low-frequency deletion but not precise excision, suggesting recent stabilization in the genome. Infect. Immun. 67:5091-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagel, S., V. Hullen, B. Wiedemann, and P. Heisig. 1999. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob.. Agents Chemother 43:868-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bager, F., F. M. Aarestrup, and H. C. Wegener. 2000. Dealing with antimicrobial resistance—the Danish experience. Can. J. Anim. Sci. 80:223-228. [Google Scholar]
- 24.Balaban, N., T. Goldkorn, R. T. Nhan, L. B. Dang, S. Scott, R. M. Ridgley, A. Rasooly, S. C. Wright, J. W. Larrick, R. Rasooly, and J. R. Carlson. 1998. Autoinducer of virulence as a target for vaccine and therapy against Staphylococcus aureus. Science 280:438-440. [DOI] [PubMed] [Google Scholar]
- 25.Balaban, N., and R. P. Novick. 1995. Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 92:1619-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balfour, J. A., and L. R. Wiseman. 1999. Moxifloxacin. Drugs 57:363-373. [DOI] [PubMed] [Google Scholar]
- 27.Baquero, F. 1995. Is man winning the battle against the microbe? Clinician 13:1-8. [Google Scholar]
- 28.Baquero, F. 1997. Gram-positive resistance: challenge for the development of new antibiotics. J. Antimicrob.. Chemother. 39(Suppl. A):1-6. [DOI] [PubMed] [Google Scholar]
- 29.Baquero, F., M. C. Negri, M. I. Morosini, and J. Blazquez. 1997. The antibiotic selective process: concentration-specific amplification of low-level resistant populations. Ciba Found. Symp. 207:93-105. [DOI] [PubMed] [Google Scholar]
- 30.Baquero, F., M. F. Vicente, and J. C. Perez-Diaz. 1985. beta-lactam coselection of sensitive and TEM-1 beta-lactamase-producing subpopulations in heterogeneous Escherichia coli colonies. J. Antimicrob. Chemother. 15:151-157. [DOI] [PubMed] [Google Scholar]
- 31.Baquero, F., and J. Blazquez. 1997. Evolution of antibiotic resistance. Trends Ecol. Evolut. 12:482-487. [DOI] [PubMed] [Google Scholar]
- 32.Baquero, F., M. C. Negri, M. I. Morosini, and J. Blazquez. 1998. Antibiotic-selective environments. Clin. Infect. Dis. 27:S5-S11. [DOI] [PubMed] [Google Scholar]
- 33.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barja, J. L., Y. Santos, I. Huq, R. R. Colwell, and A. E. Toranzo. 1990. Plasmids and factors associated with virulence in environmental isolates of Vibrio cholerae non-O1 in Bangladesh. J. Med. Microbiol. 33:107-114. [DOI] [PubMed] [Google Scholar]
- 35.Barker, J., H. Scaife, and M. R. Brown. 1995. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob. Agents Chemother. 39:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barondess, J. J., and J. Beckwith. 1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346:871-874. [DOI] [PubMed] [Google Scholar]
- 37.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 38.Bengoechea, J. A., R. Diaz, and I. Moriyon. 1996. Outer membrane differences between pathogenic and environmental Yersinia enterocolitica biogroups probed with hydrophobic permeants and polycationic peptides. Infect. Immun. 64:4891-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benveniste, R., and J. Davies. 1973. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 70:2276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berg, G., N. Roskot, and K. Smalla. 1999. Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 37:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berg, R. D. 1996. The indigenous gastrointestinal microflora. Trends Microbiol. 4:430-435. [DOI] [PubMed] [Google Scholar]
- 42.Bergogne Berezin, E., D. Decre, and M. L. Joly Guillou. 1993. Opportunistic nosocomial multiply resistant bacterial infections-their treatment and prevention. J. Antimicrob. Chemother. 32(Suppl. A):39-47 [DOI] [PubMed] [Google Scholar]
- 43.Bernardini, M. L., J. Mounier, H. d'Hauteville, M. Coquis-Rondon, and P. J. Sansonetti. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 86:3867-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhriain, N. N., C. J. Dorman, and C. F. Higgins. 1989. An overlap between osmotic and anaerobic stress responses: a potential role for DNA supercoiling in the coordinate regulation of gene expression. Mol. Microbiol. 3:933-942. [DOI] [PubMed] [Google Scholar]
- 45.Bianchi, A. A., and F. Baneyx. 1999. Stress responses as a tool to detect and characterize the mode of action of antibacterial agents. Appl. Environ. Microbiol. 65:5023-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bindereif, A., and J. B. Neilands. 1983. Cloning of the aerobactin-mediated iron assimilation system of plasmid ColV. J. Bacteriol. 153:1111-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bisognano, C., P. E. Vaudaux, D. P. Lew, E. Y. Ng, and D. C. Hooper. 1997. Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 41:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bjorkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bjorkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 50.Blahova, J., K. Kralikova, V. Krcmery, and P. Jezek. 1999. Transduction of antibiotic resistance in Pseudomonas aeruginosa: Relationship between lytic and transducing activity of phage isolate AP-423. Acta Virol. 43:395-398. [PubMed] [Google Scholar]
- 51.Blahova, J., K. Kralikova, V. Krcmery, A. Mikovicova, and N. Bartonikova. 1998. Two high-frequency-transduction phage isolates from lysogenic strains of Pseudomonas aeruginosa transducing antibiotic resistance. Acta Virol. 42:175-179. [PubMed] [Google Scholar]
- 52.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 53.Blazquez, J., L. E. Espinosa de los Monteros, S. Samper, C. Martin, A. Guerrero, J. Cobo, J. Van Embden, F. Baquero, and E. Gomez-Mampaso. 1997. Genetic characterization of multidrug-resistant Mycibacterium bovis strains from a hospital outbreak involving human immunodeficiency virus-positive patients. J. Clin. Microbiol. 35:1390-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blazquez, R., A. Menasalvas, I. Carpena, C. Ramirez, C. Guerrero, and S. Moreno. 1999. Invasive disease caused by ciprofloxacin-resistant uropathogenic Escherichia coli. Eur. J. Clin. Microbiol. Infect. Dis. 18:503-505. [DOI] [PubMed] [Google Scholar]
- 55.Blot, M., B. Hauer, and G. Monnet. 1994. The Tn5 bleomycin resistance gene confers improved survival and growth advantage on Escherichia coli. Mol. Gen. Genet. 242:595-601. [DOI] [PubMed] [Google Scholar]
- 56.Bonafede, M. E., L. L. Carias, and L. B. Rice. 1997. Enterococcal transposon Tn5384: evolution of a composite transposon through cointegration of enterococcal and staphylococcal plasmids. Antimicrob. Agents Chemother. 41:1854-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonhoeffer, S., M. Lipsitch, and B. R. Levin. 1997. Evaluating treatment protocols to prevent antibiotic resistance. Proc. Natl. Acad. Sci. USA 94:12106-12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borek, A. P., D. C. Dressel, J. Hussong, and L. R. Peterson. 1997. Evolving clinical problems with Streptococcus pneumoniae: increasing resistance to antimicrobial agents, and failure of traditional optochin identification in Chicago, Illinois, between 1993 and 1996. Diagn. Microbiol. Infect. Dis. 29:209-214. [DOI] [PubMed] [Google Scholar]
- 59.Bottger, E. C., B. Springer, M. Pletschette, and P. Sander. 1998. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat. Med. 4:1343-1344. [DOI] [PubMed] [Google Scholar]
- 60.Boucher, J. C., J. Martinez Salazar, M. J. Schurr, M. H. Mudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouma, J. E., and R. E. Lenski. 1988. Evolution of a bacteria/plasmid association. Nature 335:351-352. [DOI] [PubMed] [Google Scholar]
- 62.Braga, P. C., M. T. Sala, and M. Dal Sasso. 1999. Pharmacodynamic effects of subinhibitory concentrations of rufloxacin on bacterial virulence factors. Antimicrob. Agents Chemother. 43:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Broach, J. R., and J. Thorner. 1996. High-throughput screening for drug discovery. Nature 384:14-16. [PubMed] [Google Scholar]
- 65.Brochu, A., N. Brochu, T. I. Nicas, T. R. J. Parr, A. A. J. Minnick, E. K. Dolence, J. A. McKee, M. J. Miller, M. C. Lavoie, and F. Malouin. 1992. Modes of action and inhibitory activities of new siderophore-beta-lactam conjugates that use specific iron uptake pathways for entry into bacteria. Antimicrob. Agents Chemother. 36:2166-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brook, I. 1998. Microbial factors leading to recurrent upper respiratory tract infections. Pediatr. Infect. Dis. J. 17:62-67. [DOI] [PubMed] [Google Scholar]
- 67.Bucci, C., A. Lavitola, P. Salvatore, L. Del Giudice, D. R. Massardo, C. B. Bruni, and P. Alifano. 1999. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol. Cell 3:435-445. [DOI] [PubMed] [Google Scholar]
- 68.Buchmeier, N., S. Bossie, C. Y. Chen, F. C. Fang, D. G. Guiney, and S. J. Libby. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bull, A. T., A. C. Ward, and M. Goodfellow. 2000. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol. Mol. Biol. Rev. 64:573-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Casadevall, A., and L. A. Pirofski. 1999. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect. Immun. 67:3703-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casadevall, A., and L. A. Pirofski. 2000. Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect. Immun. 68:6511-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cercenado, E., F. Baquero, A. Delgado-Iribarren, and J. L. Martinez. 1986. Epidemiology of aerobactin production in Enterobacteriaceae. Ann. Inst. Pasteur Microbiol. 137B:297-303. [DOI] [PubMed] [Google Scholar]
- 75.Cheetham, B. F., and M. E. Katz. 1995. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18:201-208. [DOI] [PubMed] [Google Scholar]
- 76.Chiu, C., H., T. Y. Lin, and J. L. Wu. 1998. Hypothermia predisposing to Pseudomonas putida sepsis in a child with panhypopituitarism. J. Formos. Med. Assoc. 97:286-288. [PubMed] [Google Scholar]
- 77.Clewell, D. B. 1993. Bacterial sex pheromone-induced plasmid transfer. Cell 73:9-12. [DOI] [PubMed] [Google Scholar]
- 78.Coffey, T. J., M. Daniels, M. C. Enright, and B. G. Spratt. 1999. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology UK 145:2023-2031. [DOI] [PubMed] [Google Scholar]
- 79.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 80.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Paton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 81.Colonna, B., M. Bernardini, G. Micheli, F. Maimone, M. Nicoletti, and M. Casalino. 1988. The Salmonella wien virulence plasmid pZM3 carries Tn1935, a multiresistance transposon containing a composite IS1936-kanamycin resistance element. Plasmid 20:221-231. [DOI] [PubMed] [Google Scholar]
- 82.Colonna, B., L. Ranucci, P. A. Fradiani, M. Casalino, A. Calconi, and M. Nicoletti. 1992. Organization of aerobactin, hemolysin, and antibacterial resistance genes in lactose-negative Escherichia coli strains of serotype O4 isolated from children with diarrhea. Infect. Immun. 60:5224-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Coquet, L., G. A. Junter, and T. Jouenne. 1998. Resistance of artificial biofilms of Pseudomonas aeruginosa to imipenem and tobramycin. J. Antimicrob. Chemother. 42:755-760. [DOI] [PubMed] [Google Scholar]
- 84.Cormican, M. G., and R. N. Jones. 1996. Emerging resistance to antimicrobial agents in gram-positive bacteria. Enterococci, staphylococci and nonpneumococcal streptococci. Drugs 51(Suppl. 1):6-12 [DOI] [PubMed] [Google Scholar]
- 85.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 86.Cotter, P. A., and J. F. Miller. 1996. Triggering bacterial virulence. Science 273:1183-1184. [DOI] [PubMed] [Google Scholar]
- 87.Cox, B., J. C. Denyer, A. Binnie, M. C. Donnelly, B. Evans, D. V. Green, J. A. Lewis, T. H. Mander, A. T. Merritt, M. J. Valler, and S. P. Watson. 2000. Application of high-throughput screening techniques to drug discovery. Prog. Med. Chem. 37:83-133. [DOI] [PubMed] [Google Scholar]
- 88.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Culebras, E., J. L. Martinez, F. Baquero, and J. C. Perez-Diaz. 1996. pH modulation of aminoglycoside resistance in Staphylococcus epidermidis harbouring 6′-N-aminoglycoside acetyltransferase. J. Antimicrob. Chemother. 37:881-889. [DOI] [PubMed] [Google Scholar]
- 90.Culebras, E., and J. L. Martinez. 1999. Aminoglycoside resistance mediated by the bifunctional enzyme 6′-N-aminoglycoside acetyltransferase-2”-O-aminoglycoside phosphotransferase. Front. Biosci. 4:1-8. [DOI] [PubMed] [Google Scholar]
- 91.Dagan, R., and D. Fraser. 2000. Conjugate pneumococcal vaccine and antibiotic-resistant Streptococcus pneumoniae: herd immunity and reduction of otitis morbidity. Pediatr. Infect. Dis. J. 19:S79-87. [DOI] [PubMed] [Google Scholar]
- 92.Dagan, R., N. Givon-Lavi, L. Shkolnik, P. Yagupsky, and D. Fraser. 2000. Acute otitis media caused by antibiotic-resistant Streptococcus pneumoniae in southern Israel: implication for immunizing with conjugate vaccines. J. Infect. Dis. 181:1322-1329. [DOI] [PubMed] [Google Scholar]
- 93.Dargis, M., P. Gourde, D. Beauchamp, B. Foiry, M. Jacques, and F. Malouin. 1992. Modification in penicillin-binding proteins over in vivo development of genetic competence of Haemophilus influenzae is associated with a rapid change in the physiological state of cells. Infect. Immun. 60:4024-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Daszak, P., A. A. Cunningham, and A. D. Hyatt. 2000. Emerging infectious diseases of wildlife-threats to biodiversity and human health. Science 287:443-449. [DOI] [PubMed] [Google Scholar]
- 95.Datta, N., and V. M. Hughes. 1983. Plasmids of the same Inc groups in Enterobacteria before and after the medical use of antibiotics. Nature 306:616-617. [DOI] [PubMed] [Google Scholar]
- 96.Datta, S., A. Pal, S. Basu, and P. C. Banerjee. 1997. Involvement of a 70-kb plasmid of the epidemic Shigella dysenteriae type 1 (Dt66) strain in drug-resistance, lipopolysaccharide synthesis, and virulence. Microb. Drug Resist. 3:351-357. [DOI] [PubMed] [Google Scholar]
- 97.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375-382. [DOI] [PubMed] [Google Scholar]
- 98.Davies, J. E. 1997. Origins, acquisition and dissemination of antibiotic resistance determinants. Ciba Found. Symp. 207:15-27. [PubMed] [Google Scholar]
- 99.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. [DOI] [PubMed] [Google Scholar]
- 100.de la Cruz, F., J. M. García-Lobo, and J. Davies. 2001. Antibiotic resistance: on how bacterial populations respond to a simple evolutionary force. In K. Lewis et al. (ed.), bacterial resistance to antimicrobials: mechanisms, genetics, medical practice and public health. Marcel Dekker, New York, N.Y.
- 101.de La Cruz, I., and I. Davies. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8:128-133. [DOI] [PubMed] [Google Scholar]
- 102.de Lorenzo, V., M. Herrero, and J. B. Neilands. 1988. IS1-mediated mobility of the aerobactin system of pColV-K30 in Escherichia coli. Mol. Gen. Genet. 213:487-490. [DOI] [PubMed] [Google Scholar]
- 103.de Lorenzo, V., and J. L. Martinez. 1988. Aerobactin production as a virulence factor: a reevaluation. Eur. J. Clin. Microbiol. Infect. Dis. 7:621-629. [DOI] [PubMed] [Google Scholar]
- 104.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. R. Barry. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.del Castillo, F. J., S. C. Leal, F. Moreno, and F. del Castillo. 1996. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol. Microbiol. 25:107-115. [DOI] [PubMed] [Google Scholar]
- 106.Demple, B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53-57. [DOI] [PubMed] [Google Scholar]
- 107.Denis, A., C. Agouridas, J. M. Auger, Y. Benedetti, A. Bonnefoy, F. Bretin, J. F. Chantot, A. Dussarat, C. Fromentin, S. G. D'Ambrieres, S. Lachaud, P. Laurin, O. Le Martret, V. Loyau, N. Tessot, J. M. Pejac, and S. Perron. 1999. Synthesis and antibacterial activity of HMR 3647 a new ketolide highly potent against erythromycin-resistant and susceptible pathogens. Bioorg. Med. Chem. Lett. 9:3075-3080. [DOI] [PubMed] [Google Scholar]
- 108.Denton, M., N. J. Todd, and J. M. Littlewood. 1996. Role of antipseudomonal antibiotics in the emergence of Stenotrophomonas maltophilia in cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 15:402-405. [DOI] [PubMed] [Google Scholar]
- 109.Desnottes, J. F. 1996. New targets and strategies for the development of antibacterial agents. Trends Biotechnol. 14:134-140. [DOI] [PubMed] [Google Scholar]
- 110.Diekema, D. I., and R. N. Jones. 2000. Oxazolidinones: a review. Drugs 59:7-16. [DOI] [PubMed] [Google Scholar]
- 111.Dolence, E. K., A. A. Minnick, C. E. Lin, M. J. Miller, and S. M. Payne. 1991. Synthesis and siderophore and antibacterial activity of N5-acetyl-N5-hydroxy-L-ornithine-derived siderophore-beta-lactam conjugates: iron-transport-mediated drug delivery. J. Med. Chem. 34:968-978. [DOI] [PubMed] [Google Scholar]
- 112.Dorman, C. J., N. N. Bhriain, and C. F. Higgins. 1990. DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature 344:789-792. [DOI] [PubMed] [Google Scholar]
- 113.Dykhuizen, D. E., and G. Baranton. 2001. The implications of low rate of horizontal transfer in Borrelia. Trends Microbiol. 9:344-350. [DOI] [PubMed] [Google Scholar]
- 114.Ebert, D. 1998. Experimental evolution of parasites. Science 282:1432-1435. [DOI] [PubMed] [Google Scholar]
- 115.Emborg, H., A. K. Ersb/oll, O. E. Heuer, and H. C. Wegener. 2001. The effect of discontinuing the use of antimicrobial growth promoters on the productivity in the Danish broiler production. Prev. Vet. Med. 50:53-70. [DOI] [PubMed] [Google Scholar]
- 116.Evans, D. J., D. W. Frank, V. Finck Barbancon, C. Wu, and S. M. Fleiszig. 1998. Pseudomonas aeruginosa invasion and cytotoxicity are independent events, both of which involve protein tyrosine kinase activity. Infect. Immun. 66:1453-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Evans, K., L. Passador, R. Srikumar, E. Tsang, J. Nezezon, and K. Poole. 1998. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 180:5443-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Evans, R. 1986. Niche expansion in bacteria: can infectious gene exchange affect the rate of evolution? Genetics 113:775-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ewald, P. W. 1987. Transmission modes and evolution of the parasitism-mutualism continuum. Ann. N.Y. Acad. Sci. 503:295-306. [DOI] [PubMed] [Google Scholar]
- 120.Ewald, P. W. 1998. The evolution of virulence and emerging diseases. J. Urban Health 75:480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Faruque, S. M., Asadulghani, A. R. Alim, M. J. Albert, K. M. Islam, and J. J. Mekalanos. 1998. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect. Immun. 66:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 123.Feldman, M. W., and K. N. Laland. 1996. Gene-culture coevolutionary theory. Trends Ecol. Evolut. 11:453-457. [DOI] [PubMed] [Google Scholar]
- 124.Ferguson, A. D., V. Braun, H. P. Fiedler, J. W. Coulton, K. Diederichs, and W. Welte. 2000. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 9:956-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fleiszig, S. M., J. P. Wiener Kronish, H. Miyazaki, V. Vallas, K. E. Mostov, D. Kanada, T. Sawa, T. S. Yen, and D. W. Frank. 1997. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect. Immun. 65:579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Foght, J. M., D. W. Westlake, W. M. Johnson, and H. F. Ridgway. 1996. Environmental gasoline-utilizing isolates and clinical isolates of Pseudomonas aeruginosa are taxonomically indistinguishable by chemotaxonomic and molecular techniques. Microbiology 142:2333-2340. [DOI] [PubMed] [Google Scholar]
- 128.Foster, J. W. 1999. When protons attack: microbial strategies of acid adaptation. Curr. Opin. Microbiol. 2:170-174. [DOI] [PubMed] [Google Scholar]
- 129.Foster, P. L. 1993. Adaptive mutation: the uses of adversity. Annu. Rev. Microbiol. 47:467-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Francois, M., V. Le Cabec, M. A. Dupont, P. J. Sansonetti, and I. Maridonneau-Parini. 2000. Induction of necrosis in human neutrophils by Shigella flexneri requires type III secretion, IpaB and IpaC invasins, and actin polymerization. Infect. Immun. 68:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 132.Franklin, A., Soderlind, and R. Mollby. 1981. Plasmids coding for enterotoxins, K88 antigen and colicins in porcine Escherichia coli strains of O-group 149. Med. Microbiol. Immunol. 170:63-72. [DOI] [PubMed] [Google Scholar]
- 133.Friedland, I. R., and K. P. Klugman. 1991. Recurrent penicillin-resistant pneumococcal meningitis after chloramphenicol therapy. Pediatr. Infect. Dis. J. 10:705-707. [DOI] [PubMed] [Google Scholar]
- 134.Friedland, I. R., and K. P. Klugman. 1992. Failure of chloramphenicol therapy in penicillin-resistant pneumococcal meningitis. Lancet 339:405-408. [DOI] [PubMed] [Google Scholar]
- 135.Gales, A. C., R. N. Jones, M. A. Pfaller, K. A. Gordon, and H. S. Sader. 2000. Two-year assessment of the pathogen frequency and antimicrobial resistance patterns among organisms isolated from skin and soft tissue infections in Latin American hospitals: results from the SENTRY antimicrobial surveillance program, 1997-98. SENTRY Study Group. Int. J. Infect. Dis. 4:75-84. [DOI] [PubMed] [Google Scholar]
- 136.Ganz, T., and R. I. Lehrer. 1998. Antimicrobial peptides of vertebrates. Curr. Opin. Immunol. 10:41-44. [DOI] [PubMed] [Google Scholar]
- 137.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, L. O., M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:385-386. [DOI] [PubMed] [Google Scholar]
- 138.Garcia Bustos, J., and A. Tomasz. 1990. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc. Natl. Acad. Sci. USA 87:5415-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Garcia Vescovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 140.Garrison, M. W., D. E. Anderson, D. M. Campbell, K. C. Carroll, C. L. Malone, J. D. Anderson, R. J. Hollis, and M. A. Pfaller. 1996. Stenotrophomonas maltophilia: Emergence of multidrug-resistant strains during therapy and in an in vitro pharmacodynamic chamber model. Antimicrob. Agents Chemother. 40:2859-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 142.Gibson, F., and D. I. Magrath. 1969. The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim. Biophys. Acta 192:175-184. [DOI] [PubMed] [Google Scholar]
- 143.Gilliver, M. A., M. Bennett, M. Begon, S. M. Hazel, and C. A. Hart. 1999. Enterobacteria—antibiotic resistance found in wild rodents. Nature 401:233-234. [DOI] [PubMed] [Google Scholar]
- 144.Gilliver, M. A., M. Bennett, M. Begon, S. M. Hazel, and C. A. Hart. 2001. Antibiotic resistance—how wild are wild mammals? Nature 409:38. [DOI] [PubMed] [Google Scholar]
- 145.Giraud, E., A. Brisabois, J. L. Martel, and E. ChaslusDancla. 1999. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob. Agents Chemother. 43:2131-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Giraud, A., I. Matic, O. Tenaillon, A. Clara, M. Radman, M. Fons, and F. Taddei. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606-2628. [DOI] [PubMed] [Google Scholar]
- 147.Gomez-Gomez, J. M., J. Blazquez, F. Baquero, and J. L. Martinez. 1996. HNS mutant unveils the presence of a latent haemolytic activity in Escherichia coli K-12. Mol. Microbiol. 19:909-910. [DOI] [PubMed] [Google Scholar]
- 148.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 149.Gracia, E., A. Lacleriga, M. Monzon, J. Leiva, C. Oteiza, and B. Amorena. 1998. Application of a rat osteomyelitis model to compare in vivo and in vitro the antibiotic efficacy against bacteria with high capacity to form biofilms. J. Surg. Res. 79:146-153. [DOI] [PubMed] [Google Scholar]
- 150.Gratten, M., and J. Montgomery. 1991. The bacteriology of acute pneumonia and meningitis in children in Papua New Guinea: assumptions, facts and technical strategies. PNG Med. J. 34:185-198. [PubMed] [Google Scholar]
- 151.Grenier, D., M. P. Huot, and D. Mayrand. 2000. Iron-chelating activity of tetracyclines and its impact on the susceptibility of Actinobacillus actinomycetemcomitans to these antibiotics. Antimicrob. Agents Chemother. 44:763-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gristina, A. G., M. Oga, L. X. Webb, and C. D. Hobgood. 1985. Adherent bacterial colonization in the pathogenesis of osteomyelitis. Science 228:990-993. [DOI] [PubMed] [Google Scholar]
- 153.Groisman, E. A. 1996. Pathogenicity islands: Bacterial evolution in quantum leaps. Cell 87:791-794. [DOI] [PubMed] [Google Scholar]
- 154.Groisman, E. A., C. Parra-Lopez, M. Salcedo, C. J. Lipps, and F. Heffron. 1992. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11939-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hachler, H., S. P. Cohen, and S. B. Levy. 1991. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 173:5532-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 158.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177:4162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Haldane, J. B. 1949. Disease and evolution. Ricerca Sci. Suppl. 19:68-76. [Google Scholar]
- 160.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 161.Harb, O. S., L. Gao, and Y. A. Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 162.Hare, J. M., A. K. Wagner, and K. A. McDonough. 1999. Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol. Microbiol. 31:291-303. [DOI] [PubMed] [Google Scholar]
- 163.Henriques, B., M. Kalin, A. Ortqvist, B. O. Liljequist, M. Almela, T. J. Marrie, M. A. Mufson, A. Torres, M. A. Woodhead, S. B. Svenson, and G. Kallenius. 2000. Molecular epidemiology of Streptococcus pneumoniae causing invasive disease in 5 countries. J. Infect. Dis. 182:833-839. [DOI] [PubMed] [Google Scholar]
- 164.Herbert, D., C. N. Paramasivan, P. Venkatesan, G. Kubendiran, R. Prabhakar, and D. A. Mitchison. 1996. Bactericidal action of ofloxacin, sulbactam-ampicillin, rifampin, and isoniazid on logarithmic- and stationary-phase cultures of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 40:2296-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hermans, P. W., M. Sluijter, S. Dejsirilert, N. Lemmens, K. Elzenaar, A. van Veen, W. H. Goessens, and R. de Groot. 1997. Molecular epidemiology of drug-resistant pneumococci: toward an international approach. Microb. Drug Resist. 3:243-251. [DOI] [PubMed] [Google Scholar]
- 166.Hersh, D., J. Weiss, and A. Zychlinsky. 1998. How bacteria initiate inflammation: aspects of the emerging story. Curr. Opin. Microbiol. 1:43-48. [DOI] [PubMed] [Google Scholar]
- 167.Hilbi, H., A. Zychlinsky, and P. J. Sansonetti. 1997. Macrophage apoptosis in microbial infections. Parasitology. 115(Suppl.):S79-S87. [DOI] [PubMed] [Google Scholar]
- 168.Hirsh, D. C., C. Kirkham, and W. D. Wilson. 1993. Linkage of serum resistance, aerobactin production, and resistance to antimicrobial agents on conjugal plasmids in some strains of Escherichia coli isolated from septic foals. Am. J. Vet. Res. 54:878-881. [PubMed] [Google Scholar]
- 169.Hoiby, N. 2000. Ecological antibiotic policy. J. Antimicrob. Chemother. 46:59-62. [PubMed] [Google Scholar]
- 170.Hu, S. T., and C. H. Lee. 1988. Characterization of the transposon carrying the STII gene of enterotoxigenic Escherichia coli. Mol. Gen. Genet. 214:490-495. [DOI] [PubMed] [Google Scholar]
- 171.Hughes, V. M., and N. Datta. 1983. Conjugative plasmids in bacteria of the ′pre-antibiotic' era. Nature 302:725-726. [DOI] [PubMed] [Google Scholar]
- 172.Hulten, K., R. Rigo, I. Gustafsson, and L. Engstrand. 1996. New pharmacokinetic in vitro model for studies of antibiotic activity against intracellular microorganisms. Antimicrob. Agents Chemother. 40:2727-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hussey, G., J. Hitchcock, D. Hanslo, G. Coetzee, E. Van Schalkwyk, J. Pitout, and H. Schaaf. 1994. Serotypes and antimicrobial susceptibility of Haemophilus influenzae. J. Antimicrob. Chemother. 34:1031-1036. [DOI] [PubMed] [Google Scholar]
- 174.Huttner, K. M., and C. L. Bevins. 1999. Antimicrobial peptides as mediators of epithelial host defense. Pediatr. Res. 45:785-794. [DOI] [PubMed] [Google Scholar]
- 175.Jacobs, M. R., H. J. Koornhof, R. M. Robins-Browne, C. M. Stevenson, Z. A. Vermaak, I. Freiman, G. B. Miller, M. A. Witcomb, M. Isaacson, J. I. Ward, and R. Austrian. 1978. Emergence of multiply resistant pneumococci. N. Engl. J. Med. 299:735-740. [DOI] [PubMed] [Google Scholar]
- 176.Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jair, K. W., R. G. Martin, J. L. Rosner, N. Fujita, A. Ishihama, and R. E. J. Wolf. 1995. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J. Bacteriol. 177:7100-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jeffery, K. J., and R. M. Bangham. 2000. Do infectious diseases drive MHC diversity?. Microbes Infect. 2:1335-1341. [DOI] [PubMed] [Google Scholar]
- 179.Jones, C., and J. Stanley. 1992. Salmonella plasmids of the preantibiotic era. J. Gen. Microbiol. 138:189-197. [DOI] [PubMed] [Google Scholar]
- 180.Kagan, B. L., T. Ganz, and R. I. Lehrer. 1994. Defensins: a family of antimicrobial and cytotoxic peptides. Toxicology 87:131-149. [DOI] [PubMed] [Google Scholar]
- 181.Kanukollu, U., S. Bieler, S. Hull, and R. Hull. 1985. Contribution of the traT gene to serum resistance among clinical isolates of Enterobacteriaceae. J. Med. Microbiol. 19:61-67. [DOI] [PubMed] [Google Scholar]
- 182.Kaplan, J. E., G. Roselle, and K. Sepkowitz. 1998. Opportunistic infections in immunodeficient populations. Emerg. Infect. Dis. 4:421-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kapur, V., T. S. Whittam, and J. M. Musser. 1994. Is Mycobacterium tuberculosis 15,000 years old? J. Infect. Dis. 170:1348-1349. [DOI] [PubMed] [Google Scholar]
- 184.Karaolis, D. K., S. Somara, D. R. J. Maneval, J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 185.Karunakaran, P., and J. Davies. 2000. Genetic antagonism and hypermutability in Mycobacterium smegmatis. J. Bacteriol. 182:3331-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kearney, B. P., and F. T. Aweeka. 1999. The penetration of antiinfectives into the central nervous system. Neurol. Clin. 17:883-900. [DOI] [PubMed] [Google Scholar]
- 187.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 2000. Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 6:458-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kirikae, T., F. Kirikae, S. Saito, K. Tominaga, H. Tamura, Y. Uemura, T. Yokochi, and M. Nakano. 1998. Biological characterization of endotoxins released from antibiotic-treated Pseudomonas aeruginosa and Escherichia coli. Antimicrob. Agents Chemother. 42:1015-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kleerebezem, M., L. E. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 190.Klietmann, W. F., and K. L. Ruoff. 2001. Bioterrorism: implications for the clinical microbiologist. Clin. Microbiol. Rev. 14:364-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Koeleman, J. G., G. A. Parlevliet, L. Dijkshoorn, P. H. Savelkoul, and C. M. Vandenbroucke-Grauls. 1997. Nosocomial outbreak of multiresistant Acinetobacter baumannii on a surgical ward: epidemiology and risk factors for acquisition. J. Hosp. Infect. 37:113-123. [DOI] [PubMed] [Google Scholar]
- 192.Kohler, T., C. van Delden, L. K. Curty, M. Michea-Hamzehpour, and J. C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Köning, C., H. P. Simmen, and J. Blaser. 1993. Effect of pathological changes of pH, pO2 and pCO2 on the activity of antimicrobial agents in vitro. Eur. J. Clin. Microbiol. Infect. Dis. 12:519-526. [DOI] [PubMed] [Google Scholar]
- 194.Konkel, M. E., and K. Tilly. 2000. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2:157-166. [DOI] [PubMed] [Google Scholar]
- 195.Kramnik, I., W. F. Dietrich, P. Demant, and B. R. Bloom. 2000. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 97:8560-8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. N. Maslow, A. McGeer, D. E. Low, and R. P. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259:227-230. [DOI] [PubMed] [Google Scholar]
- 197.Kupferwasser, L. I., R. A. Skurray, M. H. Brown, N. Firth, M. R. Yeaman, and A. S. Bayer. 1999. Plasmid-mediated resistance to thrombin-induced platelet microbicidal protein in staphylococci: role of the qacA locus. Antimicrob. Agents Chemother. 43:2395-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kwiatkowski, D. 2000. Genetic susceptibility to malaria getting complex. Curr. Opin. Genet. Dev. 10:320-324. [DOI] [PubMed] [Google Scholar]
- 199.Lacroix, F. J., A. Cloeckaert, O. Grepinet, C. Pinault, M. Y. Popoff, H. Waxin, and P. Pardon. 1996. Salmonella typhimurium acrB-such as gene: identification and role in resistance to biliary salts and detergents and in murine infection. FEMS Microbiol. Lett. 135:161-167. [DOI] [PubMed] [Google Scholar]
- 200.Lamb, H. M., D. P. Figgitt, and D. Faulds. 1999. Quinupristin/dalfopristin: a review of its use in the management of serious gram-positive infections. Drugs 58:1061-1097. [DOI] [PubMed] [Google Scholar]
- 201.Lambert, T., M. C. Ploy, F. Denis, and P. Courvalin. 1999. Characterization of the chromosomal aac(6′)-Iz gene of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 43:2366-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Laporta, M. Z., M. L. Silva, I. C. Scaletsky, and L. R. Trabulsi. 1986. Plasmids coding for drug resistance and localized adherence to HeLa cells in enteropathogenic Escherichia coli O55:H− and O55:H6. Infect. Immun. 51:715-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Laub, R., Y. J. Schneider, and A. Trouet. 1989. Antibiotic susceptibility of Salmonella spp. at different pH values. J. Gen. Microbiol. 135:1407-1416. [DOI] [PubMed] [Google Scholar]
- 204.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lawrence, J. G., H. Ochman, and D. L. Hartl. 1992. The evolution of insertion sequences within enteric bacteria. Genetics 131:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lazazzera, B. A. 2000. Quorum sensing and starvation: signals for entry into stationary phase. Curr. Opin. Microbiol. 3:177-182. [DOI] [PubMed] [Google Scholar]
- 208.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 209.Lenski, R. E. 1997. The cost of antibiotic resistance-from the perspective of a bacterium. Ciba Found. Symp. 207:131-151. [DOI] [PubMed] [Google Scholar]
- 210.Levin, B. R., and R. Antia. 2001. Why we don't get sick: the within-host population dynamics of bacterial infections. Science 292:1112-1115. [DOI] [PubMed] [Google Scholar]
- 211.Levin, B. R., and C. T. Bergstrom. 2000. Bacteria are different: Observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc. Natl. Acad. Sci. USA 97:6981-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Levin, B. R., M. Lipsitch, and S. Bonhoeffer. 1999. Evolution and disease—population biology, evolution, and infectious disease: Convergence and synthesis. Science 283:806-809. [DOI] [PubMed] [Google Scholar]
- 213.Levin, B. R., V. Perrot, and N. Walker. 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154:985-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Levin, B. R. 1993. The accessory genetic elements of bacteria: existence conditions and (co)evolution. Curr. Opin. Genet. Dev. 3:849-854. [DOI] [PubMed] [Google Scholar]
- 215.Levin, B. R., M. Lipsitch, V. Perrot, S. Schrag, R. Antia, L. Simonsen, N. M. Walker, and F. M. Stewart. 1997. The population genetics of antibiotic resistance. Clin. Infect. Dis. 24(Suppl. 1):S9-S16. [DOI] [PubMed] [Google Scholar]
- 216.Li, Z., C. Kelley, F. Collins, D. Rouse, and S. Morris. 1998. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistence in mice and guinea pigs. J. Infect. Dis. 177:1030-1035. [DOI] [PubMed] [Google Scholar]
- 217.Lindberg, F., and S. Normark. 1986. Contribution of chromosomal beta-lactamases to beta-lactam resistance in enterobacteria. Rev Infect. Dis. 8:292-304. [DOI] [PubMed] [Google Scholar]
- 218.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527-543. [DOI] [PubMed] [Google Scholar]
- 219.Lipsitch, M. 1997. Vaccination against colonizing bacteria with multiple serotypes. Proc. Natl. Acad. Sci. USA 94:6571-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lipsitch, M. 1999. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg. Infect. Dis. 5:336-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lipsitch, M. 2001. Bacterial population genetics and disease. Science 292:59-60. [DOI] [PubMed] [Google Scholar]
- 222.Lipsitch, M., C. T. Bergstrom, and B. R. Levin. 2000. The epidemiology of antibiotic resistance in hospitals: Paradoxes and prescriptions. Proc. Natl. Acad. Sci. USA 97:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lipsitch, M., and B. R. Levin. 1997. The population dynamics of antimicrobial chemotherapy. Antimicrob. Agents Chemother. 41:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Livermore, D. M. 2000. Epidemiology of antibiotic resistance. Intensive Care Med. 26(Suppl. 1):14-21. [DOI] [PubMed] [Google Scholar]
- 226.Livermore, D. M., and J. E. Corkill. 1992. Effects of CO2 and pH on inhibition of TEM-1 and other beta-lactamases by penicillanic acid sulfones. Antimicrob. Agents Chemother. 36:1870-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lortholary, O., J. Y. Fagon, A. B. Hoi, M. A. Slama, J. Pierre, P. Giral, R. Rosenzweig, L. Gutmann, M. Safar, and J. Acar. 1995. Nosocomial acquisition of multiresistant Acinetobacter baumannii: risk factors and prognosis. Clin. Infect Dis. 20:790-796. [DOI] [PubMed] [Google Scholar]
- 229.Lowe, M. N., and H. M. Lamb. 2000. Gemifloxacin. Drugs 59:1137-1147. [DOI] [PubMed] [Google Scholar]
- 230.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 231.Macfarlane, E. L., A. Kwasnicka, and R. E. Hancock. 2000. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology UK 146:2543-2554. [DOI] [PubMed] [Google Scholar]
- 232.Macfarlane, E. L., A. Kwasnicka, M. M. Ochs, and R. E. Hancock. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305-316. [DOI] [PubMed] [Google Scholar]
- 233.Macinga, D. R., and P. N. Rather. 1999. The chromosomal 2′-N-acetyltransferase of Providencia stuartii: physiological functions and genetic regulation. Front. Biosci. 4:132-140. [DOI] [PubMed] [Google Scholar]
- 234.Magnusdottir, A. B., A. Hermansson, and A. Melhus. 2000. Experimental study of the virulence of Streptococcus pneumoniae with reduced susceptibility to penicillin. Int. J. Pediatr. Otorhinolaryngol. 55:1-9. [DOI] [PubMed] [Google Scholar]
- 235.Maiden, M. C. J. 1998. Horizontal genetic exchange, evolution, and spread of antibiotic resistance in bacteria. Clin. Infect. Dis. 27:S12-S20. [DOI] [PubMed] [Google Scholar]
- 236.Maiti, S. N., O. A. Phillips, R. G. Micetich, and D. M. Livermore. 1998. Beta-lactamase inhibitors: agents to overcome bacterial resistance. Curr. Med. Chem. 5:441-456. [PubMed] [Google Scholar]
- 237.Makino, K., K. Ishii, T. Yasunaga, M. Hattori, K. Yokoyama, C. H. Yutsudo, Y. Kubota, Y. Yamaichi, T. Iida, K. Yamamoto, T. Honda, C. G. Han, E. Ohtsubo, M. Kasamatsu, T. Hayashi, S. Kuhara, and H. Shinagawa. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 5:1-9. [DOI] [PubMed] [Google Scholar]
- 238.Marchal, S., J. L. Merlin, P. Colosetti, and C. Finance. 1999. Influence of the fluoroquinolone ofloxacin on the intrinsic expression of multidrug resistance phenotype in HCT-8 human colon carcinoma cells. Oncol. Res. 11:375-381. [PubMed] [Google Scholar]
- 239.Marshall, C. G., G. Broadhead, B. K. Leskiw, and G. D. Wright. 1997. d-Ala-d-Ala ligases from glycopeptide antibiotic-producing organisms are highly homologous to the enterococcal vancomycin-resistance ligases VanA and VanB. Proc. Natl. Acad. Sci. USA 94:6480-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Martinez, J. L., and F. Baquero. 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44:1771-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Martinez, J. L., J. Blazquez, and F. Baquero. 1994. Non-canonical mechanisms of antibiotic resistance. Eur. J. Clin. Microbiol. Infect. Dis. 13:1015-1022. [DOI] [PubMed] [Google Scholar]
- 243.Martinez, J. L., A. Delgado-Iribarren, and F. Baquero. 1990. Mechanisms of iron acquisition and bacterial virulence. FEMS Microbiol. Rev. 6:45-56. [DOI] [PubMed] [Google Scholar]
- 244.Martinez, J. L., M. Herrero, and V. de Lorenzo. 1994. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutA genes. J. Mol. Biol. 238:288-293. [DOI] [PubMed] [Google Scholar]
- 245.Martinez, J. L., and J. C. Perez-Diaz. 1990. Cloning of the determinants for microcin D93 production and analysis of three different D-type microcin plasmids. Plasmid 23:216-225. [DOI] [PubMed] [Google Scholar]
- 246.Martínez, J. L. 1995. On simplicity and complexity as different evolutionary strategies. E vol. Theory 10:317-320. [Google Scholar]
- 247.Martinez, M. B., T. S. Whittan, E. A. McGraw, J. Rodrigues, and L. R. Trabulsi. 1999. Clonal relationship among invasive and non-invasive strains of enteroinvasive Escherichia coli serogroups. FEMS Microbiol. Lett. 172:145-151. [DOI] [PubMed] [Google Scholar]
- 248.Martinez-Suarez, J. V., J. L. Martinez, M. J. Lopez de Goicoechea, J. C. Perez-Diaz, F. Baquero, M. Meseguer, and J. Linares. 1987. Acquisition of antibiotic resistance plasmids in vivo by extraintestinal Salmonella spp. J. Antimicrob. Chemother. 20:452-453. [DOI] [PubMed] [Google Scholar]
- 249.Mason, H. S., and C. J. Arntzen. 1995. Transgenic plants as vaccine production systems. Trends Biotechnol. 13:388-392. [DOI] [PubMed] [Google Scholar]
- 250.Mason, H. S., T. A. Haq, J. D. Clements, and C. J. Arntzen. 1998. Edible vaccine protects mice against Escherichia coli heat-labile enterotoxin (LT): potatoes expressing a synthetic LT-B gene. Vaccine 16:1336-1343. [DOI] [PubMed] [Google Scholar]
- 251.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Matic, I., M. Radman, F. Taddei, B. Picard, C. Doit, E. Bingen, E. Denamur, and J. Elion. 1997. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277:1833-1834. [DOI] [PubMed] [Google Scholar]
- 253.Matic, I., F. Taddei, and M. Radman. 2000. No genetic barriers between Salmonella enterica serovar Typhimurium and Escherichia coli in SOS-induced mismatch repair-deficient cells. J. Bacteriol. 182:5922-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 256.McDonald, J. F. 1993. Evolution and consequences of transposable elements. Curr. Opin. Genet. Dev. 3:855-864. [DOI] [PubMed] [Google Scholar]
- 257.McDonough, M. A., and J. R. Butterton. 1999. Spontaneous tandem amplification and deletion of the shiga toxin operon in Shigella dysenteriae 1. Mol. Microbiol. 34:1058-1069. [DOI] [PubMed] [Google Scholar]
- 258.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.McOrist, S. 2000. Obligate intracellular bacteria and antibiotic resistance. Trends Microbiol. 8:483-486. [DOI] [PubMed] [Google Scholar]
- 260.Mel, S. F., and J. J. Mekalanos. 1996. Modulation of horizontal gene transfer in pathogenic bacteria by in vivo signals. Cell 87:795-798. [DOI] [PubMed] [Google Scholar]
- 261.Meluleni, G. J., M. Grout, D. J. Evans, and G. B. Pier. 1995. Mucoid Pseudomonas aeruginosa growing in a biofilm in vitro are killed by opsonic antibodies to the mucoid exopolysaccharide capsule but not by antibodies produced over chronic lung infection in cystic fibrosis patients. J. Immunol. 155:2029-2038. [PubMed] [Google Scholar]
- 262.Meyer, A. P., C. Bril-Bazuin, H. Mattie, and P. J. van den Broek. 1993. Uptake of azithromycin by human monocytes and enhanced intracellular antibacterial activity against Staphylococcus aureus. Antimicrob. Agents Chemother. 37:2318-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Michele, T. M., C. Ko, and W. R. Bishai. 1999. Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob. Agents Chemother. 43:218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Middlebrook, G., and M. L. Cohn. 1953. Some observations on the pathogenicity of isoniazid-resistant variants of tubercle bacilli. Science 118:297-299. [DOI] [PubMed] [Google Scholar]
- 265.Miller, M. J., J. A. McKee, A. A. Minnick, and E. K. Dolence. 1991. The design, synthesis and study of siderophore-antibiotic conjugates. Siderophore mediated drug transport. Biol. Met. 4:62-69. [DOI] [PubMed] [Google Scholar]
- 266.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Molbak, K., D. L. Baggesen, F. M. Aarestrup, J. M. Ebbesen, J. Engberg, K. Frydendahl, P. Gerner-Smidt, A. M. Petersen, and H. C. Wegener. 1999. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype typhimurium DT104. N. Engl. J. Med. 341:1420-1425. [DOI] [PubMed] [Google Scholar]
- 269.Molinari, G., C. A. Guzman, A. Pesce, and G. C. Schito. 1993. Inhibition of Pseudomonas aeruginosa virulence factors by subinhibitory concentrations of azithromycin and other macrolide antibiotics. J. Antimicrob. Chemother. 31:681-688. [DOI] [PubMed] [Google Scholar]
- 270.Moll, A., P. A. Manning, and K. N. Timmis. 1980. Plasmid-determined resistance to serum bactericidal activity: a major outer membrane protein, the traT gene product, is responsible for plasmid-specified serum resistance in Escherichia coli. Infect. Immun. 28:359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Morosini, M. I., J. A. Ayala, F. Baquero, J. L. Martinez, and J. Blazquez. 2000. Biological cost of AmpC production for Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 44:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Morzejko, E., E. Panek, J. Skala, T. M. Lachowicz, and S. Cebrat. 1989. Genetic properties of plasmids isolated from pathogenic strain of Citrobacter freundii. Acta Microbiol. Pol. 38:159-170. [PubMed] [Google Scholar]
- 273.Mounier, J., A. Ryter, M. Coquis-Rondon, and P. J. Sansonetti. 1990. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infect. Immun. 58:1048-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Naranjo, P. 1992. Epidemic hecatomb in the New World. Allergy Proc. 13:237-241. [DOI] [PubMed] [Google Scholar]
- 275.Neu, H. C. 1992. The crisis in antibiotic resistance. Science 257:1064-1073. [DOI] [PubMed] [Google Scholar]
- 276.Nesin, M., M. Ramirez, and A. Tomasz. 1998. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J. Infect. Dis. 177:707-713. [DOI] [PubMed] [Google Scholar]
- 277.Niederman, M. S. 2001. Impact of antibiotic resistance on clinical outcomes and the cost of care. Crit. Care Med. 29:N114-120. [DOI] [PubMed] [Google Scholar]
- 278.Nikaido, H. 2000. How do exported proteins and antibiotics bypass the periplasm in Gram-negative bacterial cells? Trends Microbiol. 8:481-483. [DOI] [PubMed] [Google Scholar]
- 279.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1:516-523. [DOI] [PubMed] [Google Scholar]
- 281.Novak, R., B. Henriques, E. Charpentier, S. Normark, and E. Tuomanen. 1999. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature 399:590-593. [DOI] [PubMed] [Google Scholar]
- 282.Nowak, R. 1994. Hungary sees an improvement in penicillin resistance. Science 264:364. [DOI] [PubMed] [Google Scholar]
- 283.Nowak, M. A., and R. M. May. 1994. Superinfection and the evolution of parasite virulence. Proc. R. Soc. Lond. B Biol. Sci. 255:81-89. [DOI] [PubMed] [Google Scholar]
- 284.Nunoshiba, T., T. deRojas Walker, J. S. Wishnok, S. R. Tannenbaum, and B. Demple. 1993. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc. Natl. Acad. Sci. USA 90:9993-9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 286.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1098. [DOI] [PubMed] [Google Scholar]
- 287.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 288.Osterblad, M., K. Norrdahl, E. Korpimaki, and P. Huovinen. 2001. Antibiotic resistance—how wild are wild mammals? Nature 409:37-38. [DOI] [PubMed] [Google Scholar]
- 289.Pallister, C. J., and R. J. Lewis. 2000. Effects of antimicrobial drugs on human neutrophil-microbe interactions. Br. J. Biomed. Sci. 57:19-27. [PubMed] [Google Scholar]
- 290.Pang, Y., B. A. Brown, V. A. Steingrube, R. J. J. Wallace, and M. C. Roberts. 1994. Tetracycline resistance determinants in Mycobacterium and Streptomyces species. Antimicrob. Agents Chemother. 38:1408-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Parra-Lopez, C., M. T. Baer, and E. A. Groisman. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12:4053-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Pearson, J. P., M. Feldman, B. H. Iglewski, and A. Prince. 2000. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect. Immun. 68:4331-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 297.Perna, N. T., G. Plukett, I. I. I., V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Pósfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davies, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 298.Perry, C. M., J. A. Barman Balfour, and H. M. Lamb. 1999. Gatifloxacin. Drugs 58:683-696. [DOI] [PubMed] [Google Scholar]
- 299.Pesci, E. C., J. B. J. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Phillips, G., R. Barker, and O. Brogan. 1988. Optochin-resistant Streptococcus pneumoniae. Lancet ii:281. [DOI] [PubMed] [Google Scholar]
- 301.Piddock, L. J. 1996. Does the use of antimicrobial agents in veterinary medicine and animal husbandry select antibiotic-resistant bacteria that infect man and compromise antimicrobial chemotherapy? J. Antimicrob. Chemother. 38:1-3. [DOI] [PubMed] [Google Scholar]
- 302.Pier, G. B., M. Grout, T. Zaidi, G. Meluleni, S. S. Mueschenborn, G. Banting, R. Ratcliff, M. J. Evans, and W. H. Colledge. 1998. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature 393:79-82. [DOI] [PubMed] [Google Scholar]
- 303.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B Meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 304.Plunkett, G. R., D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Portnoy, D. A., H. Wolf-Watz, I. Bolin, A. B. Beeder, and S. Falkow. 1984. Characterization of common virulence plasmids in Yersinia species and their role in the expression of outer membrane proteins. Infect. Immun. 43:108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Quinn, J. P. 1998. Clinical problems posed by multiresistant nonfermenting gram-negative pathogens. Clin. Infect. Dis. 27:S117-S124. [DOI] [PubMed] [Google Scholar]
- 308.Quintela, J. C., M. A. de Pedro, P. Zollner, G. Allmaier, and F. Garcia-del Portillo. 1997. Peptidoglycan structure of Salmonella typhimurium growing within cultured mammalian cells. Mol. Microbiol. 23:693-704. [DOI] [PubMed] [Google Scholar]
- 309.Qureshi, S. T., E. Skamene, and D. Malo. 1999. Comparative genomics and host resistance against infectious diseases. Emerg. Infect. Dis. 5:36-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Radman, M. 1999. Enzymes of evolutionary change. Nature 401:866-867. [DOI] [PubMed] [Google Scholar]
- 311.Rahmati Bahram, A., J. T. Magee, and S. K. Jackson. 1995. Growth temperature-dependent variation of cell envelope lipids and antibiotic susceptibility in Stenotrophomonas (Xanthomonas) maltophilia. J. Antimicrob. Chemother. 36:317-326. [DOI] [PubMed] [Google Scholar]
- 312.Rahmati Bahram, A., J. T. Magee, and S. K. Jackson. 1997. Effect of temperature on aminoglycoside binding sites in Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 39:19-24. [DOI] [PubMed] [Google Scholar]
- 313.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 314.Rakita, R. M., N. N. Vanek, K. Jacques-Palaz, M. Mee, M. M. Mariscalco, G. M. Dunny, M. Snuggs, W. B. Van Winkle, and S. I. Simon. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect. Immun. 67:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Ralph, E. T., J. R. Guest, and J. Green. 1998. Altering the anaerobic transcription factor FNR confers a hemolytic phenotype on Escherichia coli K12. Proc. Natl. Acad. Sci. USA 95:10449-10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 317.Rappuoli, R. 2000. Reverse vaccinology. Curr. Opin. Microbiol. 3:445-450. [DOI] [PubMed] [Google Scholar]
- 318.Ren, L., M. S. Rahman, and M. Z. Humayun. 1999. Escherichia coli cells exposed to streptomycin display a mutator phenotype. J. Bacteriol. 181:1043-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Renau, T. E., R. Leger, E. M. Flamme, J. Sangalang, M. W. She, R. Yen, C. L. Gannon, D. Griffith, S. Chamberland, O. Lomovskaya, S. J. Hecker, V. J. Lee, T. Ohta, and K. Nakayama. 1999. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 42:4928-4931. [DOI] [PubMed] [Google Scholar]
- 320.Riesenfeld, C., M. Everett, L. J. Piddock, and B. G. Hall. 1997. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob. Agents Chemother. 41:2059-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Robredo, B., K. V. Singh, F. Baquero, B. E. Murray, and C. Torres. 1999. From vanA Enterococcus hirae to vanA Enterococcus faecium: a study of feed supplementation with avoparcin and tylosin in young chickens. Antimicrob. Agents Chemother 43:1137-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Roccanova, L., and P. R. Rappa. 2000. Antibiotic rotation. Science 287:803. [DOI] [PubMed] [Google Scholar]
- 323.Romling, U., J. Wingender, H. Muller, and B. Tummler. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Roosenberg, J. M. n., Y. M. Lin, Y. Lu, and M. J. Miller. 2000. Studies and syntheses of siderophores, microbial iron chelators, and analogs as potential drug delivery agents. Curr. Med. Chem. 7:159-197. [DOI] [PubMed] [Google Scholar]
- 325.Rosamond, J., and A. Allsop. 2000. Harnessing the power of the genome in the search for new antibiotics. Science 287:1973-1976. [DOI] [PubMed] [Google Scholar]
- 326.Rosenberg, S. M. 1997. Mutation for survival. Curr. Opin. Genet. Dev. 7:829-834. [DOI] [PubMed] [Google Scholar]
- 327.Rosenberg, S. M. 2001. Evolving responsively: adaptive mutation. Nat. Rev. Genet. 2:504-515. [DOI] [PubMed] [Google Scholar]
- 328.Rowe-Magnus, D. A., and D. Mazel. 1999. Resistance gene capture. Curr. Opin. Microbiol. 2:483-488. [DOI] [PubMed] [Google Scholar]
- 329.Rowland, K. E., and J. D. Turnidge. 2000. The impact of penicillin resistance on the outcome of invasive Streptococcus pneumoniae infection in children. Aust. N. Z. J. Med. 30:441-449. [DOI] [PubMed] [Google Scholar]
- 330.Ruiz, G. M., T. K. Rawlings, F. C. Dobbs, L. A. Drake, T. Mullady, A. Huq, and R. R. Colwell. 2000. Global spread of microorganisms by ships. Nature 408:49-50. [DOI] [PubMed] [Google Scholar]
- 331.Russell, R. B., and D. S. Eggleston. 2000. New roles for structure in biology and drug discovery. Nat. Struct. Biol. 7(Suppl.):928-930. [DOI] [PubMed] [Google Scholar]
- 332.Salyers, A. A., and C. F. Amabile-Cuevas. 1997. Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41:2321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Sameshima, T., H. Ito, I. Uchida, H. Danbara, and N. Terakado. 1993. A conjugative plasmid pTE195 coding for drug resistance and virulence phenotypes from Salmonella naestved strain of calf origin. Vet. Microbiol. 36:197-203. [DOI] [PubMed] [Google Scholar]
- 334.Sanchez, P., J. F. Linares, B. Ruiz-Diez, E. Campanario, A. Navas, F. Baquero, and J. L. Martinez. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother., in press. [DOI] [PubMed]
- 335.Sansonetti, P. J., H. d'Hauteville, C. Ecobichon, and C. Pourcel. 1983. Molecular comparison of virulence plasmids in Shigella and enteroinvasive Escherichia coli. Ann. Microbiol. Paris 134A:295-318. [PubMed] [Google Scholar]
- 336.Sansonetti, P. J., H. d'Hauteville, S. B. Formal, and M. Toucas. 1982. Plasmid-mediated invasiveness of “Shigella-such as” Escherichia coli. Ann. Microbiol. Paris 133:351-355. [PubMed] [Google Scholar]
- 337.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Sattar, S. A., J. Tetro, and V. S. Springthorpe. 1999. Impact of changing societal trends on the spread of infections in American and Canadian homes. Am. J. Infect. Control 27:S4-S21. [DOI] [PubMed] [Google Scholar]
- 339.Scaletsky, I. C., M. S. Gatti, J. F. da Silveira, I. M. DeLuca, E. Freymuller, and L. R. Travassos. 1995. Plasmid coding for drug resistance and invasion of epithelial cells in enteropathogenic Escherichia coli 0111:H. Microb. Pathog. 18:387-399. [DOI] [PubMed] [Google Scholar]
- 340.Schlievert, P. M., P. J. Gahr, A. P. Assimacopoulos, M. M. Dinges, J. A. Stoehr, J. W. Harmala, H. Hirt, and G. M. Dunny. 1998. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 66:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Schmieger, H., and P. Schicklmaier. 1999. Transduction of multiple drug resistance of Salmonella enterica serovar Typhimurium DT104. FEMS Microbiol. Lett. 170:251-256. [DOI] [PubMed] [Google Scholar]
- 342.Schuch, R., and A. T. Maurelli. 1997. Virulence plasmid instability in Shigella flexneri 2a is induced by virulence gene expression. Infect. Immun. 65:3686-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Segal, G., and E. Z. Ron. 1998. Regulation of heat-shock response in bacteria. Ann. N. Y. Acad. Sci. 851:147-151. [DOI] [PubMed] [Google Scholar]
- 344.Segal, G., and H. A. Shuman. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol. Microbiol. 30:197-208. [DOI] [PubMed] [Google Scholar]
- 345.Selander, R. K., and B. R. Levin. 1980. Genetic diversity and structure in Escherichia coli populations. Science 210:545-547. [DOI] [PubMed] [Google Scholar]
- 346.Serino, L., C. Reimmann, H. Baur, M. Beyeler, P. Visca, and D. Haas. 1995. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol. Gen. Genet. 249:217-228. [DOI] [PubMed] [Google Scholar]
- 347.Shakibaei, M., C. Forster, H. J. Merker, and R. Stahlmann. 1995. Ofloxacin alters expression of integrins on chondrocytes from mouse fetuses in vitro. Drugs 49(Suppl. 2):293-295 [DOI] [PubMed] [Google Scholar]
- 348.Shapiro, J. A. 1984. Observations on the formation of clones containing araB-lacZ cistron fusions. Mol. Gen. Genet. 194:79-90. [DOI] [PubMed] [Google Scholar]
- 349.Shapiro, J. A. 1993. Natural genetic engineering of the bacterial genome. Curr. Opin. Genet. Dev. 3:845-848. [DOI] [PubMed] [Google Scholar]
- 350.Sherman, D. R., K. Mdluli, M. J. Hickey, T. M. Arain, S. L. Morris, C. E. R. Barry, and C. K. Stover. 1996. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 272:1641-1643. [DOI] [PubMed] [Google Scholar]
- 351.Shirasu, K., H. Nakajima, V. K. Rajasekhar, R. A. Dixon, and C. Lamb. 1997. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9:261-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Siegrist, H. H., B. R. Birch, and A. E. Jacob. 1987. Detection of a large haemolysin plasmid carrying multiple antibiotic resistance markers in Streptococcus faecalis. Microb. Pathog. 2:155-158. [DOI] [PubMed] [Google Scholar]
- 353.Simmen, H. P., and J. Blaser. 1993. Analysis of pH and pO2 in abscesses, peritoneal and drainage fluid in the presence and the absence of bacterial infection over and following abdominal surgery. Am. J. Surg. 166:24-27. [DOI] [PubMed] [Google Scholar]
- 354.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 355.Sjölin, J., G. Goscinski, M. Lundholm, J. Bring, and I. Odenholt. 2000. Endotoxin release from Escherichia coli after exposure to tobramycin: dose-dependency and reduction in cefuroxime-induced endotoxin release. Clin. Microbiol. Infect. Dis. 6:74-81. [DOI] [PubMed] [Google Scholar]
- 356.Smith, K. E., J. M. Besser, C. W. Hedberg, F. T. Leano, J. B. Bender, J. H. Wicklund, B. P. Johnson, K. A. Moore, and M. T. Osterholm. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. Investigation Team. N. Engl. J. Med. 340:1525-1532. [DOI] [PubMed] [Google Scholar]
- 357.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Reference deleted.
- 359.Sokurenko, E. V., D. L. Hasty, and D. E. Dykhuizen. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 360.Soncini, F. C., E. Garcia Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Sonstein, S. A., and J. C. Burnham. 1993. Effect of low concentrations of quinolone antibiotics on bacterial virulence mechanisms. Diagn Microbiol. Infect. Dis. 16:277-289. [DOI] [PubMed] [Google Scholar]
- 362.Sousa, A. O., J. I. Salem, F. K. Lee, M. C. Vercosa, P. Cruaud, B. R. Bloom, P. H. Lagrange, and H. L. David. 1997. An epidemic of tuberculosis with a high rate of tuberculin anergy among a population previously unexposed to tuberculosis, the Yanomami Indians of the Brazilian Amazon. Proc. Natl. Acad. Sci. USA 94:13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Souza, V., M. Rocha, A. Valera, and L. E. Eguiarte. 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Spratt, B. G., L. D. Bowler, Q. Y. Zhang, J. Zhou, and J. M. Smith. 1992. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J. Mol. Evol. 34:115-125. [DOI] [PubMed] [Google Scholar]
- 366.Stanley, T. L., C. D. Ellermeier, and J. M. Slauch. 2000. Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar Typhimurium survival in Peyer's patches. J. Bacteriol. 182:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Steck, T. R., R. J. Franco, J. Y. Wang, and K. Drlica. 1993. Topoisomerase mutations affect the relative abundance of many Escherichia coli proteins. Mol. Microbiol. 10:473-481. [DOI] [PubMed] [Google Scholar]
- 368.Stenderup, J., and F. Orskov. 1983. The clonal nature of enteropathogenic Escherichia coli strains. J. Infect. Dis. 148:1019-1024. [DOI] [PubMed] [Google Scholar]
- 369.Stermitz, F. R., P. Lorenz, J. N. Tawara, L. A. Zenewicz, and K. Lewis. 2000. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. USA 97:1433-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Stewart, P. S. 1994. Biofilm accumulation model that predicts antibiotic resistance of Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 38:1052-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Stojiljkovic, I., V. Kumar, and N. Srinivasan. 1999. Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol. 31:429-442. [DOI] [PubMed] [Google Scholar]
- 372.Straley, S. C., G. V. Plano, E. Skrzypek, P. L. Haddix, and K. A. Fields. 1993. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol. Microbiol. 8:1005-1010. [DOI] [PubMed] [Google Scholar]
- 373.Suerbaum, S., H. Leying, B. Meyer, and W. Opferkuch. 1990. Influence of beta-lactam antibiotics on serum resistance of K1-positive blood culture isolates of Escherichia coli. Antimicrob. Agents Chemother. 34:628-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Sukupolvi, S., and C. D. O'Connor. 1990. TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. Microbiol. Rev. 54:331-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Sum, P. E., F. W. Sum, and S. J. Projan. 1998. Recent developments in tetracycline antibiotics. Curr. Pharm. Des. 4:119-132. [PubMed] [Google Scholar]
- 376.Sunder Plassmann, G., S. I. Patruta, and W. H. Horl. 1999. Pathobiology of the role of iron in infection. Am. J. Kidney Dis. 34:25-29. [DOI] [PubMed] [Google Scholar]
- 377.Sundstrom, L. 1998. The potential of integrons and connected programmed rearrangements for mediating horizontal gene transfer. APMIS Suppl. 84:37-42. [DOI] [PubMed] [Google Scholar]
- 378.Swartz, M. N. 1994. Hospital-acquired infections: diseases with increasingly limited therapies. Proc. Natl. Acad. Sci. USA 91:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Tacket, C. O., H. S. Mason, G. Losonsky, J. D. Clements, M. M. Levine, and C. J. Arntzen. 1998. Immunogenicity in humans of a recombinant bacterial antigen delivered in a transgenic potato. Nat. Med. 4:607-609. [DOI] [PubMed] [Google Scholar]
- 380.Taddei, F., M. Radman, J. Maynard-Smith, B. Toupance, P. H. Gouyon, and B. Godelle. 1997. Role of mutator alleles in adaptive evolution. Nature 387:700-702. [DOI] [PubMed] [Google Scholar]
- 381.Tenaillon, O., F. Taddei, M. Radman, and I. Matic. 2001. Second-order selection in bacterial evolution: selection acting on mutation and recombination rates in the course of adaptation. Res. Microbiol. 152:11-16 [DOI] [PubMed] [Google Scholar]
- 382.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Thieringer, H. A., P. G. Jones, and M. Inouye. 1998. Cold shock and adaptation. Bioessays 20:49-57. [DOI] [PubMed] [Google Scholar]
- 384.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 385.Tibayrenc, M. 1996. Towards a unified evolutionary genetics of microorganisms. Annu. Rev. Microbiol. 50:401-429. [DOI] [PubMed] [Google Scholar]
- 386.Torkelson, J., R. S. Harris, M. J. Lombardo, J. Nagendran, C. Thulin, and S. M. Rosenberg. 1997. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 16:3303-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Torres, O. R., R. Z. Korman, S. A. Zahler, and G. M. Dunny. 1991. The conjugative transposon Tn925: enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in Bacillus subtilis and E. faecalis. Mol. Gen. Genet. 225:395-400. [DOI] [PubMed] [Google Scholar]
- 388.Torsvik, V., F. L. Daae, R. A. Sandaa, and L. Ovreas. 1998. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 64:53-62. [DOI] [PubMed] [Google Scholar]
- 389.Tuomanen, E., J. Schwartz, and S. Sande. 1990. The vir locus affects the response of Bordetella pertussis to antibiotics: phenotypic tolerance and control of autolysis. J. Infect. Dis. 162:560-563. [DOI] [PubMed] [Google Scholar]
- 390.Ubukata, K., M. Konno, and R. Fuji. 1975. Transduction of drug resistance to tetracycline, chloramphenicol, macrolides, lincomycin and clindamycin with phages induced from Streptococcus pyogenes. J. Antibiot. 28:681-688. [DOI] [PubMed] [Google Scholar]
- 391.Vaara, M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Valdezate, S., A. Vindel, F. Baquero, and R. Canton. 1999. Comparative in vitro activity of quinolones against Stenotrophomonas maltophilia. Eur. J. Clin. Microbiol. Infect. Dis. 18:908-911. [DOI] [PubMed] [Google Scholar]
- 393.van Belkum, A., W. van Leeuwen, R. Verkooyen, S. C. Sacilik, C. Cokmus, and H. Verbrugh. 1997. Dissemination of a single clone of methicillin-resistant Staphylococcus aureus among Turkish hospitals. J. Clin. Microbiol. 35:978-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Van den Bogaard, A. E., and E. E. Stobberingh. 2000. Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob.. Agents 14:327-335. [DOI] [PubMed] [Google Scholar]
- 395.Van der Auwera, P., G. Prinz, and G. Petrikkos. 1991. Activity of intracellular antibiotics. Infection. 19(Suppl. 4):216-223. [DOI] [PubMed] [Google Scholar]
- 396.van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.van Wetering, S., P. J. Sterk, K. F. Rabe, and P. S. Hiemstra. 1999. Defensins: key players and bystanders in infection, injury, and repair in the lung? J. Allergy Clin. Immunol. 104:1131-1138. [DOI] [PubMed] [Google Scholar]
- 398.Vartivarian, S., E. Anaissie, G. Bodey, H. Sprigg, and K. Rolston. 1994. A changing pattern of susceptibility of Xanthomonas maltophilia to antimicrobial agents: implications for therapy. Antimicrob. Agents Chemother. 38:624-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34:399-413. [DOI] [PubMed] [Google Scholar]
- 400.Vica Pacheco, S., O. Garcia Gonzalez, and G. L. Paniagua Contreras. 1997. The lom gene of bacteriophage lambda is involved in Escherichia coli K12 adhesion to human buccal epithelial cells. FEMS Microbiol. Lett. 156:129-132. [DOI] [PubMed] [Google Scholar]
- 401.Vidotto, M. C., J. M. Cacao, C. R. Goes, and D. S. Santos. 1991. Plasmid coding for aerobactin production and drug resistance is involved in virulence of Escherichia coli avian strains. Braz. J. Med. Biol. Res. 24:677-685. [PubMed] [Google Scholar]
- 402.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-886. [DOI] [PubMed] [Google Scholar]
- 403.Vokes, S. A., S. A. Reeves, A. G. Torres, and S. M. Payne. 1999. The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol. Microbiol. 33:63-73. [DOI] [PubMed] [Google Scholar]
- 404.Vrany, J. D., P. S. Stewart, and P. A. Suci. 1997. Comparison of recalcitrance to ciprofloxacin and levofloxacin exhibited by Pseudomonas aeruginosa bofilms displaying rapid-transport characteristics. Antimicrob. Agents Chemother. 41:1352-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 406.Watanabe, H., and A. Nakamura. 1985. Large plasmids associated with virulence in Shigella species have a common function necessary for epithelial cell penetration. Infect. Immun. 48:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Webb, V., and J. Davies. 1994. Accidental release of antibiotic-resistance genes. Trends Biotechnol. 12:74-75. [DOI] [PubMed] [Google Scholar]
- 408.Weeks, C. R., and J. J. Ferretti. 1984. The gene for type A streptococcal exotoxin (erythrogenic toxin) is located in bacteriophage T12. Infect. Immun. 46:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Weinberg, E. D. 1984. Iron withholding: a defense against infection and neoplasia. Physiol. Rev. 64:65-102. [DOI] [PubMed] [Google Scholar]
- 410.Weinrauch, Y., and A. Zychlinsky. 1999. The induction of apoptosis by bacterial pathogens. Annu. Rev. Microbiol. 53:155-187. [DOI] [PubMed] [Google Scholar]
- 411.Weston, G. S., J. Blazquez, F. Baquero, and B. K. Shoichet. 1998. Structure-based enhancement of boronic acid-based inhibitors of AmpC beta-lactamase. J. Med. Chem. 41:4577-4586. [DOI] [PubMed] [Google Scholar]
- 412.Wilkinson, D. M. 1999. Bacterial ecology, antibiotics and selection for virulence. Ecol. Lett. 2:207-209. [Google Scholar]
- 413.Willems, R. J., W. Homan, J. Top, M. van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
- 414.Wilson, T. M., G. W. de Lisle, and D. M. Collins. 1995. Effect of inhA and katG on isoniazid resistance and virulence of Mycobacterium bovis. Mol. Microbiol. 15:1009-1015. [DOI] [PubMed] [Google Scholar]
- 415.Wilson, M., J. DeRisi, H. H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Wirth, R. 1994. The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism? Eur. J. Biochem. 222:235-246. [DOI] [PubMed] [Google Scholar]
- 417.Witte, W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996-997. [DOI] [PubMed] [Google Scholar]
- 418.Witte, W. 2000. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob.. Agents 16:S19-S24. [DOI] [PubMed] [Google Scholar]
- 419.World Health Organization. 2000. Overcoming antibiotic resistance. World Health Organization Report in Infectious Diseases 2000. World Health Organization, Geneva, Switzerland.
- 420.Wright, J. M. 1971. Syphilis and Neanderthal man. Nature 229:409. [DOI] [PubMed] [Google Scholar]
- 421.Yura, T., K. Nakahigashi, and M. Kanemori. 1996. Transcriptional regulation of stress-inducible genes in prokaryotes. EXS 77:165-181. [DOI] [PubMed] [Google Scholar]
- 422.Zabner, R., and J. P. Quinn. 1992. Antimicrobials in cystic fibrosis: emergence of resistance and implications for treatment. Semin. Respir. Infect. 7:210-217. [PubMed] [Google Scholar]
- 423.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]
- 424.Zhang, X. P., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. K. Acheson. 2000. Quinolone antibiotics induce shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]
- 425.Zhang, Y., B. Heym, B. Allen, D. Young, and S. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]
- 426.Zund, P., and G. Lebek. 1980. Generation time-prolonging R plasmids: correlation between increases in the generation time of Escherichia coli caused by R plasmids and their molecular size. Plasmid 3:65-69. [DOI] [PubMed] [Google Scholar]
- 427.Zychlinsky, A., M. C. Prevost, and P. J. Sansonetti. 1992. Shigella flexneri induces apoptosis in infected macrophages. Nature 358:167-169. [DOI] [PubMed] [Google Scholar]
- 428.Zychlinsky, A., and P. J. Sansonetti. 1997. Apoptosis as a proinflammatory event: what can we learn from bacteria-induced cell death? Trends Microbiol. 5:201-204. [DOI] [PubMed] [Google Scholar]