Abstract
The G1 cyclin Cln3 is a key activator of cell-cycle entry in budding yeast. Here we show that Whi3, a negative G1 regulator of Cln3, interacts in vivo with the cyclin-dependent kinase Cdc28 and regulates its localization in the cell. Efficient interaction with Cdc28 depends on an N-terminal domain of Whi3 that is also required for cytoplasmic localization of Cdc28, and for proper regulation of G1 length and filamentous growth. On the other hand, nuclear accumulation of Cdc28 requires the nuclear localization signal of Cln3, which is also found in Whi3 complexes. Both Cln3 and Cdc28 are mainly cytoplasmic during early G1, and become nuclear in late G1. However, Whi3-deficient cells show a distinct nuclear accumulation of Cln3 and Cdc28 already in early G1. We propose that Whi3 constitutes a cytoplasmic retention device for Cln3–Cdc28 complexes, thus defining a key G1 event in yeast cells.
Keywords: cell cycle, Cdc28, cytoplasmic retention, Cln3, Saccharomyces cerevisiae
Introduction
While a single cyclin-dependent kinase (Cdk), termed Cdc28, is sufficient to drive the cell cycle of the budding yeast Saccharomyces cerevisiae, at least nine cyclins act with some redundancy at different stages of the cycle: Cln1, Cln2 and Cln3 in the G1 phase; Clb5 and Clb6 in the S phase; and Clb1, Clb2, Clb3 and Clb4 in the G2 and M phases (Futcher, 1996; Mendenhall and Hodge, 1998). Binding of regulatory cyclin subunits to Cdc28 induces a conformational change that is required for kinase activity. As Cdc28 executes diverse events through the cell cycle, cyclin moieties are presumed to provide the required specificity by participating in substrate recognition (Morgan, 1995). On the other hand, most cyclins are expressed only at particular times during the cell cycle, thus imposing temporal constraints to each cyclin–Cdc28 complex. An exception to this rule is Cln3, a cyclin with a key role in triggering cell-cycle entry that is nonetheless present at rather constant levels during late G1 and the G1/S transition (Tyers et al, 1993).
The G1 cyclin Cln3 is the most upstream effector of cell-cycle entry. In combination with Cdc28, Cln3 activates the expression of a large set of G1/S genes that are regulated by the transcriptional factors SBF and MBF (Andrews and Herskowitz, 1989; Nasmyth and Dirick, 1991; Lowndes et al, 1992; Tyers et al, 1993; Dirick et al, 1995; Stuart and Wittenberg, 1995; Spellman et al, 1998). Cln3 contains a functional bipartite nuclear localization signal (NLS) at the C-terminus, and seems to perform its essential functions as a nuclear protein (Edgington and Futcher, 2001; Miller and Cross, 2001). Transcriptional induction by Cln3–Cdc28 involves the loading of RNA polymerase II onto SBF-containing promoters in late G1 (Cosma et al, 2001). Nutrients, which are among the most important trophic factors for yeast cells, control cell-cycle entry by regulating Cln3 expression at different levels (Huble et al, 1993; Barbet et al, 1996; Gallego et al, 1997; Hall et al, 1998; Newcomb et al, 2002). However, when nutritional and other external signals are kept constant, additional mechanisms must exist to restrict Cln3–Cdc28-dependent activation of gene expression to late G1, perhaps after some cell size and/or growth requirements have been met. Cell division is coupled to cell growth in budding yeast by multiple mechanisms (Jorgensen et al, 2002), and CLN3 itself had been first identified as a mutation that conferred a small cell size phenotype (Sudbery et al, 1980). Similarly, WHI3 was isolated as a gene involved in cell size regulation (Nash et al, 2001), and we have shown recently that Whi3 is a cytoplasmic protein that exerts a negative role on Cln3 activity in G1 (Garí et al, 2001).
Here we show that Whi3 is associated to Cdc28 in vivo, and restricts its cellular distribution to the cytoplasm. Whi3 contains an N-terminal domain required for (1) interaction with Cdc28, (2) cytoplasmic retention of Cdc28, (3) proper regulation of cell-cycle entry, and (4) initiation of developmental programs in G1 such as filamentous growth. We also show that both Cdc28 and Cln3 are cytoplasmic in early G1 cells and accumulate in the nucleus in late G1, while they are already nuclear in early G1 cells deficient for Whi3. Our results suggest that Whi3 constitutes a cytoplasmic retention device for Cln3–Cdc28 complexes, thus defining a key G1 event in the regulatory network that controls cell-cycle entry in yeast cells.
Results
Characterization of Whi3 protein complexes
We had shown in a previous work that Whi3 is a cytoplasmic protein that plays an important role in regulating Cln3 activity in G1 (Garí et al, 2001). To better understand this negative regulation, we attempted to identify other proteins that could interact with Whi3. By differential centrifugation of yeast cell extracts through glycerol gradients, we found Whi3 in high-molecular-weight complexes (Figure 1A). The size distribution of these complexes was extremely wide, most of them being larger than ribosomes, which suggested that Whi3 may form part of a large cytoplasmic structure such as the cytoskeleton or the endoplasmic reticulum.
Figure 1.
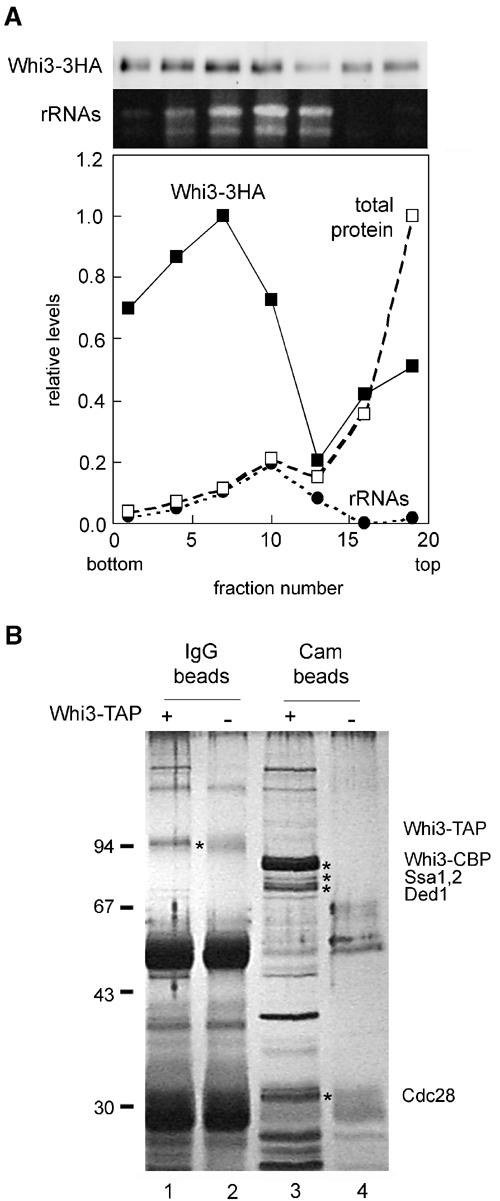
Characterization of Whi3 high-molecular-weight complexes. (A) Distribution of Whi3 in a glycerol gradient. Whi3-3HA, total protein and ribosomal RNA levels measured for each fraction are plotted as relative values. (B) TAP of the Whi3 complex. Cell extracts obtained from a Whi3-TAP-tagged strain (lanes 1 and 3) and an untagged strain (lanes 2 and 4) were used to purify first the Whi3-TAP complexes onto IgG beads (lanes 1 and 2). Eluates obtained after cleavage of the protein A moiety of the TAP tag, which released Whi3-CBP, were then purified onto calmodulin beads (lanes 3 and 4). Silver-stained bands were analyzed by mass spectrometry and some tentatively identified polypeptides are indicated.
Next we used a tandem-affinity purification (TAP) strategy (Rigaut et al, 1999), and built a Whi3-TAP fusion expressed at endogenous levels at the chromosomal locus to identify proteins that could uphold stable interactions to Whi3. Cells carrying this fusion were only slightly affected in cell size compared to wild type, indicating that fusion to TAP did not alter Whi3 function significantly. Figure 1B shows that Whi3 seems to interact specifically with a large number of different polypeptides. By applying mass spectrometry to trypsin-digested mixtures of some of these polypeptides, we found candidates for the Ssa1,2 chaperones, which have been involved in the formation of cyclin–Cdk complexes (Yaglom et al, 1996; Honey et al, 2001), and the translation initiation factor Ded1, which belongs to an RNA-helicase family of proteins needed to unfold long 5′UTRs (Chuang et al, 1997). In addition, a ca. 33-kDa protein was identified as a putative candidate for Cdc28, the yeast Cdk involved in cell-cycle regulation. Thus, although none of these candidate proteins could be predicted to act as simple negative regulators of the Cln3 cyclin, the presence of the yeast Cdk suggested that Whi3 could have a direct negative function on the activity of Cln3–Cdc28 complexes.
Whi3 interacts with Cdc28
To confirm the ability of Whi3 to interact with Cdc28, we used a regular coimmunoprecipitation approach with a fully functional Whi3-3HA fusion protein (Garí et al, 2001). Figure 2A shows that Whi3-3HA is specifically detected in immunoprecipitates obtained with a polyclonal anti-Cdc28 antibody. The association of Whi3 with Cdc28 does not require the RNA-recognition motif (RRM) present in the C-terminus of Whi3 (Figure 2B), making unlikely the possibility that the Whi3–Cdc28 interaction is mediated by nascent Cln3 polypeptides translated from Whi3-bound CLN3 mRNA molecules. Moreover, Figure 2B shows that the anti-Cdc28 antibody was able to immunoprecipitate Whi3 as efficiently as the mitotic cyclin Clb2, one of the proteins that is known to interact more strongly with Cdc28 (Surana et al, 1991; Honey et al, 2001). Assuming that the efficiency of immunoprecipitation by the anti-Cdc28 antibody is not affected by the association between Cdc28 and Whi3, quantification of these two proteins in both cell extracts and Cdc28 immunoprecipitates indicated that at least 10% of the total Whi3 protein in the cell may be associated with Cdc28. Overexpression of a GST fusion to the whole Whi3 protein proved to be deleterious for cell viability and very unstable (not shown). However, overexpression of a deletion of Whi3 that lacks the RRM neither causes a G1 arrest nor affects cell viability (Nash et al, 2001). Thus, to perform the reverse interaction analysis we used a GST-Whi3ΔRRM fusion, and detected the specific presence of Cdc28 in the corresponding glutathione beads (Figure 2C). These results confirm that, either directly or indirectly, Whi3 interacts with Cdc28 in vivo.
Figure 2.
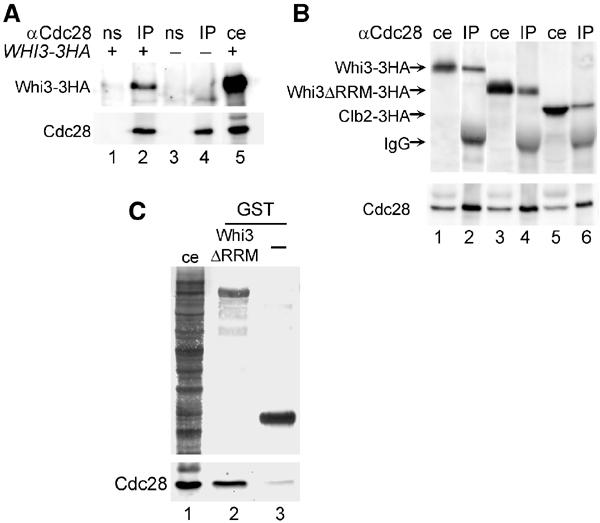
Whi3 interacts with Cdc28 in vivo. (A) Whi3-3HA-tagged (lanes 1 and 2) and untagged (lanes 3 and 4) cell extracts were immunoprecipitated with an anti-Cdc28 antibody (IP, lanes 2 and 4) or nonspecific serum (ns, lanes 1 and 3), and bound proteins were analyzed by Western blot with either anti-HA (top) or anti-Cdc28 (bottom) antibodies. Lane 5 contains a sample of the Whi3-3HA cell extract (ce) as control. (B) Cell extracts containing 3HA-tagged Whi3 (lanes 1 and 2), Whi3ΔRRM (lanes 3 and 4) and Clb2 (lanes 5 and 6) proteins were immunoprecipitated with the anti-Cdc28 antibody. The corresponding immunoprecipitates (IP, lanes 2, 4 and 6) and cell extracts (ce, lanes 1, 3 and 5) were analyzed as in (A). (C) GST-Whi3ΔRRM (lane 2) or GST (lane 3, as control) proteins were expressed in a wild-type yeast strain and purified onto glutathione beads. Samples of bound proteins were analyzed in Coomassie-blue-stained gels (top) or by Western blot with the anti-Cdc28 antibody (bottom). Lane 1 contains a cell extract (ce) sample of the strain used.
To determine the functional relevance of the interaction between Whi3 and Cdc28, we obtained a set of mutants by serial deletion of the WHI3 open reading frame. Among the different deletions constructed (Figure 3A), only one of them showed reduced levels in Cdc28 immunoprecipitates (Figure 3B), suggesting that the N-terminal domain spanning amino acids 121–220 of Whi3 plays a unique and important role in the interaction with Cdc28. More importantly, this Cdc28-recruitment region (CRR) was also found to be essential for other key functional properties of Whi3, that is the whi3ΔCRR mutation was unable to complement a whi3 null mutant regarding defects in cell size (Figure 3A and C), and invasive or filamentous growth (Figure 3D and E). All other deletions obtained did not cause any significant effect in the Whi3–Cdc28 interaction (Figures 2B and 3B) and, aside from the whi3ΔRRM mutation, they were perfectly capable of complementing the aforementioned defects of the whi3 null mutant (Figure 3). As the RNA-binding ability of Whi3ΔCRR was not affected compared to the wild-type protein (data not shown), all these results suggest that the interaction between Whi3 and Cdc28 may be per se a key aspect of Whi3 function.
Figure 3.
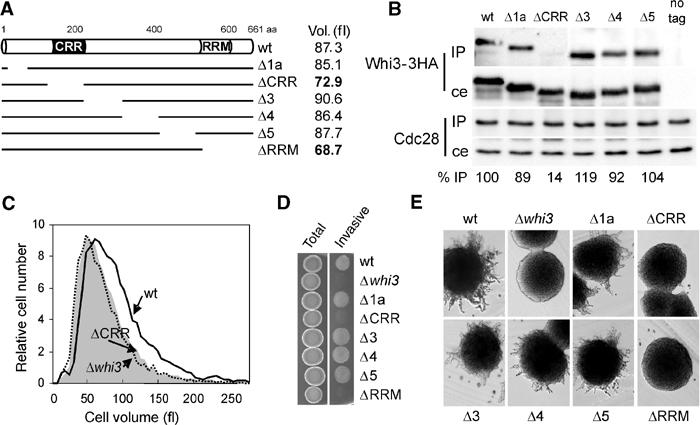
Functional analysis of the interaction between Whi3 and Cdc28. (A) Scheme depicting the Whi3 deletions described under Materials and methods. The average cell volume of exponentially growing cells containing the different deletions of Whi3 is also shown. (B) Cell extracts obtained from strains expressing the 3HA-tagged versions of Whi3 depicted in (A) were immunoprecipitated with the anti-Cdc28 antibody. The presence of Cdc28 and Whi3-3HA proteins in the corresponding immunoprecipitates (IP) and cell extracts (ce) was analyzed as described in Figure 2A. Samples obtained from an untagged strain are shown as control (no tag). Immunoprecipitation efficiencies are shown at the bottom as percentages relative to the value obtained for wild-type Whi3. (C) Volume distributions of cells carrying a wild-type WHI3 gene (wt), the whi3ΔCRR deletion that removes amino acids 121–220 (ΔCRR) or a null allele (Δwhi3). (D) Invasive growth in YEPD plates of derivative haploid Σ1278 strains carrying the 3HA-tagged versions of Whi3 depicted in (A). (E) Filamentation in SLAD plates of derivative diploid Σ1278 strains carrying the 3HA-tagged versions of Whi3 depicted in (A).
The serial deletion analysis also identified an additional domain between amino acids 61 and 120 that may be important for proper expression or stability of the Whi3 protein, since the overall levels of the corresponding deleted protein (Whi3Δ1b) showed a 10-fold reduction in whole-cell extracts compared to wild type (not shown), and was not analyzed further. As WHI3 gene dosage is very important for proper function (Nash et al, 2001), the fact that Whi3Δ1b was not able to complement the whi3 null mutant could be directly due to the low levels attained by this mutant protein (data not shown).
Whi3 is required for cytoplasmic localization of Cdc28
We next asked about the nature of the functional role of Whi3 on Cdc28. By using immunofluorescence and subcellular fractionation methods, Cdc28 had been found to be partially associated with a cytoplasmic matrix (Wittenberg et al, 1987). On the other hand, by using a fusion to four copies of GFP, Cdc28 was found to be enriched in the nucleus throughout the cell cycle (Maekawa et al, 2003), which suggests that nucleocytoplasmic partitioning is a characteristic of Cdc28. Since the essential role of Cln3–Cdc28 complexes seems to be confined to the nucleus (Edgington and Futcher, 2001; Miller and Cross, 2001), we hypothesized that Whi3 could have an effect on the nucleocytoplasmic distribution of Cdc28. First, we determined by immunofluorescence the localization of a functional Cdc28-3HA protein (Deshaies and Kirschner, 1995) in exponentially growing cells. Many wild-type cells showed a rather homogeneous distribution of Cdc28 in the cell with no strong nuclear accumulation or exclusion. However, most budded cells showed a distinct accumulation of Cdc28-3HA in the nucleus (data not shown), suggesting that preferential import of Cdc28 to the nucleus could be a cell-cycle-regulated event in budding yeast (see below). To test further whether Whi3 could act as a cytoplasmic retention factor for Cdc28 in G1 cells, we analyzed by immunofluorescence the localization of Cdc28-3HA in elutriated wild-type and whi3 mutant cells. Figure 4A shows that Whi3-deficient G1 cells showed a brighter nuclear signal for Cdc28-3HA compared to wild-type G1 cells (Figure 4A). As the overall Cdc28-3HA levels were very similar in both strains (Figure 4B), these results suggest that Whi3 could play a role in regulating the nucleocytoplasmic partitioning of Cdc28 in G1. Although we found that a functional Cdc28-sGFP fusion was already slightly enriched in the nucleus of elutriated G1 cells of the wild-type strain, the nuclear signal became much brighter during cell-cycle entry (data not shown). In addition, elutriated G1 cells of the Δwhi3 mutant strain showed a modest but reproducible increase in the intensity of the nuclear signal of Cdc28-sGFP when compared to the wild-type strain (data not shown). Supporting the idea that the Whi3–Cdc28 interaction is important for retaining Cdc28 in the cytoplasm, elutriated G1 cells expressing Whi3ΔCRR, which lacks the N-terminal domain required for efficient interaction with Cdc28 (Figure 3), also showed a clear nuclear accumulation of Cdc28 (Figure 4A). Finally, overexpression of Whi3 at levels that cause a G1 arrest (Garí et al, 2001; Nash et al, 2001) imposed an exclusion of Cdc28 from the nucleus in most cells (Figure 4C). We also found that Cdc28 accumulated in the nucleus in elutriated G1 cells expressing a Whi3ΔRRM protein (Figure 4A), which lacks the RRM. However, the Whi3ΔRRM protein was able to bind Cdc28 as efficiently as the wild-type Whi3 protein (Figure 2B), which suggests the possibility that Whi3 recruitment of the CLN3 mRNA (Garí et al, 2001) may also be important to modulate the intracellular localization of Cdc28.
Figure 4.
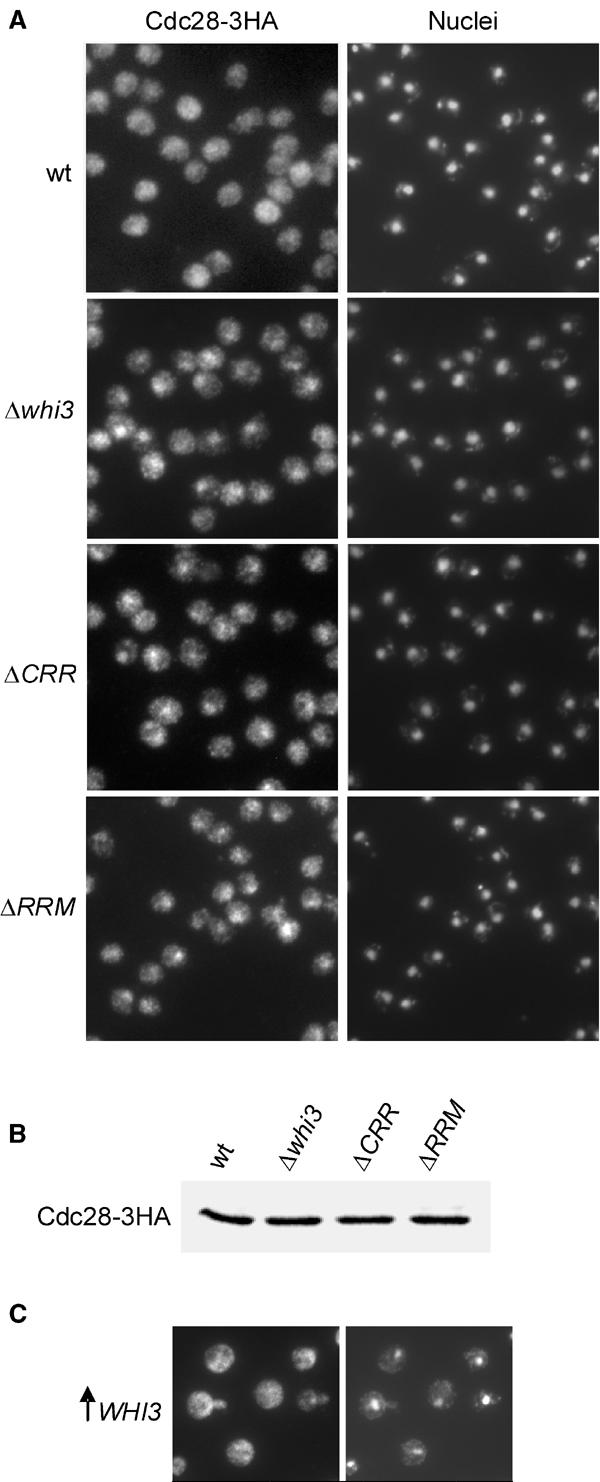
Whi3 restricts the presence of Cdc28 to the cytoplasm. (A) Cdc28-3HA-tagged cells otherwise wild type (wt), deficient for Whi3 (Δwhi3), containing a Whi3 protein that lacks the Cdc28-recruitment region (ΔCRR), or the RNA-recognition motif (ΔRRM) were fixed with formaldehyde and elutriated to obtain small G1 cells, which were then analyzed by immunofluorescence with an anti-HA antibody. Corresponding pictures of DAPI-stained nuclei are also shown. (B) Cdc28-3HA was detected by Western blot using equal amounts of total protein extracts from strains used in (A). (C) Cdc28-3HA-tagged cells overexpressing Whi3 (↑ Whi3) were analyzed by immunofluorescence as in (A).
In summary, all these results suggest that the Whi3–Cdc28 interaction is part of a cytoplasmic retention mechanism for Cdc28, which would also provide Whi3 with a direct negative function on the activity of Cln3–Cdc28 complexes. If this were the case, the Cln3 protein might be found, although only transiently, in trimeric complexes with Cdc28 and Whi3. Also, Cln3 could play a role in driving the accumulation of Cdc28 in the nucleus. The next two sections describe experiments designed to test these ideas.
Whi3, Cdc28 and G1 cyclins form multimeric complexes
To investigate the possibility that Whi3 and G1 cyclins coexist in complexes with Cdc28, a functional Cln3-6FLAG protein (data not shown) was expressed in a wild-type strain carrying the WHI3-3HA construct, and cell extracts were obtained and immunoprecipitated with a monoclonal anti-FLAG antibody. As shown in Figure 5A, a small but reproducible amount of Whi3 was detected in the Cln3-6FLAG immunoprecipitate, suggesting that Whi3 may coexist with G1 cyclins in Cdc28 complexes. The relative levels of Whi3 in Cln3-6FLAG immunoprecipitates were much lower compared to the Whi3 levels obtained in Cdc28 immunoprecipitates, suggesting that the existence of these multimeric complexes would only be transient, perhaps due to the intrinsic instability of Cln3 (Yaglom et al, 1996). Whi3 was not detected in immunoprecipitates with the anti-FLAG antibody from cells expressing Cln3ΔCB-6FLAG (Figure 5A), which carries a deletion of the fifth helix of the cyclin box (CB) required for binding to Cdc28 (Huang et al, 1997). Although we cannot rule out a direct interaction between the Cln3 cyclin box and Whi3, this result suggests that the Cln3 cyclin would require Cdc28 to interact with Whi3, and that a direct interaction between Cln3 and Whi3 may not take place.
Figure 5.
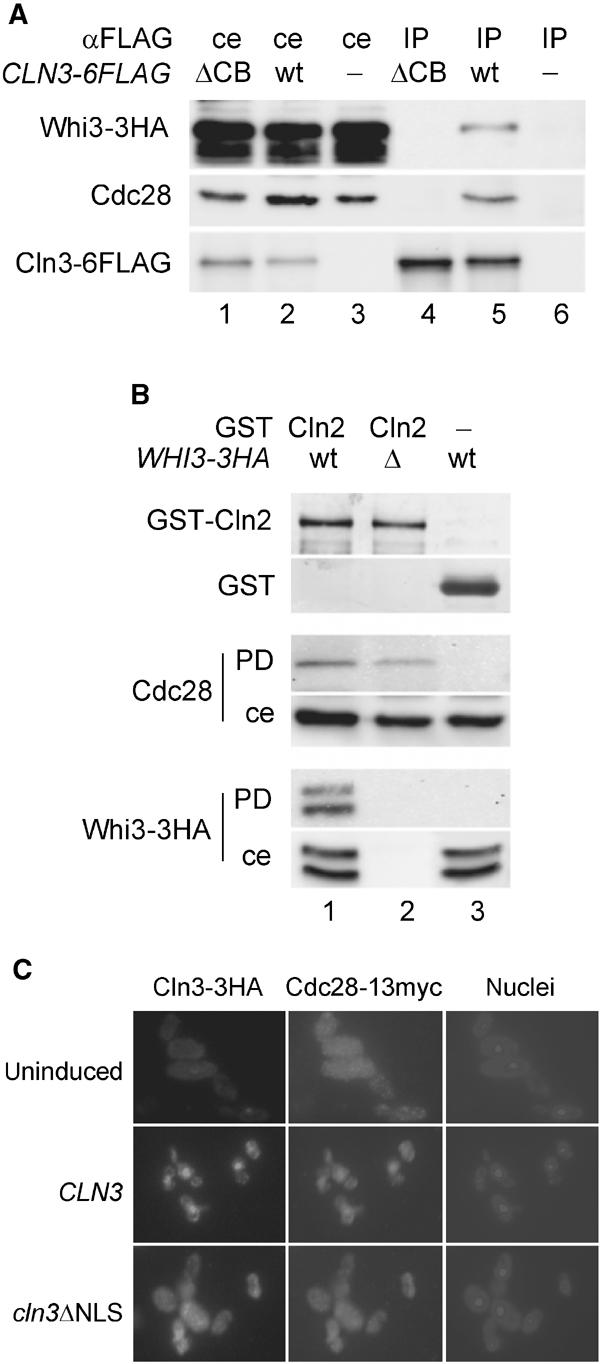
Physical and functional interactions between Cln3, Cdc28 and Whi3. (A) Cln3 is also found in complexes with Whi3. Cell extracts from a Whi3-3HA-tagged strain expressing a 6FLAG-tagged mutant protein that lacks the cyclin box (ΔCB, lanes 1 and 4), a 6FLAG-tagged wild-type Cln3 protein (wt, lanes 2 and 5) or carrying an empty vector (lanes 3 and 6) were immunoprecipitated with an anti-FLAG antibody. Cell extracts (ce) and immunoprecipitates (IP) were analyzed by Western blot to detect Whi3-3HA, Cdc28 and Cln3-6FLAG proteins with anti-HA (top), anti-Cdc28 (middle) or anti-FLAG (bottom) antibodies, respectively. (B) Reconstitution assays. Δcln1 Δcln2 GAL1p-CLN3 cells either expressing a WHI3-3HA fusion (wt, lanes 1 and 3) or carrying a deletion of the WHI3 gene (Δ, lane 2) were depleted from G1 cyclins by promoter shut-off. Cell extracts were then used to reconstitute active Cdc28 complexes with exogenously added GST-Cln2 (lanes 1 and 2) or GST (lane 3) as control. Samples of glutathione beads used were analyzed in Coomassie-blue-stained gels (top) to detect GST-Cln2 and GST. Pulled-down proteins (PD) as well as cell extract samples (ce) were analyzed by Western blot with anti-Cdc28 and anti-HA antibodies. (C) Cdc28 requires the Cln3 NLS to accumulate in the nucleus. Cells expressing a Cdc28-13myc fusion that lacked an endogenous CLN3 gene (Δcln3) were transformed with plasmids containing GAL1p-driven 3HA fusions to a wild-type CLN3 gene or a deletion that removes the NLS of Cln3 (cln3ΔNLS). Cells were analyzed by immunofluorescence with anti-HA and anti-myc antibodies before (uninduced) or after 30 min of galactose addition to trigger CLN3 or cln3ΔNLS expression. Corresponding pictures of DAPI-stained nuclei are also shown.
To sustain the existence of these trimeric complexes, we decided to test whether Whi3 can be found in reconstituted G1 cyclin–Cdc28 complexes. The reconstitution assay designed by Deshaies and Kirschner (1995) uses a GST-Cln2 fusion that, in contrast to Cln3, can be expressed at high levels in Escherichia coli. Despite their different localization patterns in the cell, Cln2 and Cln3 are assumed to play similar biochemical roles as cyclin moieties of Cdc28 (Stuart and Wittenberg, 1995; Levine et al, 1996). Thus, we used purified GST-Cln2 bound to glutathione beads to test its ability to bind Cdc28 and Whi3-3HA from cell extracts that had been depleted by promoter shut-off of endogenous G1 cyclins. Figure 5B shows that reconstituted complexes of GST-Cln2 and Cdc28 also contain Whi3-3HA. When made relative to the respective amounts in the total cell extract, levels of Cdc28 and Whi3-3HA bound to GST-Cln2 were very similar. These results reinforce the notion that Whi3, Cdc28 and a G1 cyclin can coexist in multimeric complexes.
The Cln3 NLS is required for nuclear accumulation of Cdc28
Since Cln3 contains a bipartite NLS within the last 25 amino acids of the C-terminus signal (Edgington and Futcher, 2001; Miller and Cross, 2001), we asked whether Cdc28 import to the nucleus could be mediated by its interaction with Cln3, and whether it would depend on the Cln3 NLS. In the absence of Cln3-3HA, a functional Cdc28-13myc fusion was found to be homogeneously distributed in the cell (Figure 5C). In contrast, most of the Cdc28 signal was located at the nucleus in 30 min after inducing Cln3-3HA expression. However, a cln3ΔNLS construct that lacked the bipartite NLS of Cln3 and produced a cytoplasmic protein was unable to cause an accumulation of Cdc28 in the nucleus (Figure 5C). Thus, Cdc28 would need to bind Cln3 in order to be recognized, through the Cln3 bipartite NLS, as a cargo by the nuclear import machinery and driven to the nucleus by a Ran-dependent mechanism (Miller and Cross, 2001). Comparable results were obtained in Whi3-deficient cells (not shown), suggesting that the absence of Whi3 is not sufficient to promote nuclear accumulation of Cdc28, and that cytoplasmic retention by Whi3 and nuclear import mediated by Cln3 are independent.
We have found that endogenously expressed Cln3-3HA accumulates in the nucleus of cells arrested in late G1 by alpha factor (our unpublished observations), where Cln3 is the only Cdc28-specific cyclin present at normal levels. Under these late G1 arrest conditions, Cdc28-sGFP accumulated in the nucleus only in the presence of Cln3, but not in the presence of a Cln3ΔNLS mutant protein (data not shown), reinforcing the notion that endogenous levels of Cln3 may cause an accumulation of Cdc28 in the nucleus.
Whi3 restricts to late G1 the nuclear accumulation of both Cln3 and Cdc28
A high proportion of budded cells from a wild-type strain showed a strong nuclear accumulation of Cdc28, while in most unbudded cells Cdc28 was found to be more homogeneously distributed in the cell (see above). This observation suggests that the mechanisms that retain Cdc28 in the cytoplasm or drive it to the nucleus might be differently regulated during the cell cycle. As it is required for Cdc28 entry into the nucleus, the Cln3 cyclin would be an obvious candidate for such a specific cell-cycle regulatory step. However, whether Cln3 (or Cdc28) accumulates in the nucleus in a cell-cycle-regulated manner was still unknown. As we already had set up a highly sensitive immunofluorescence method to detect Cln3-3HA expressed at endogenous levels from its chromosomal promoter sequences (Garí et al, 2001), we decided to analyze the subcellular localization of both Cln3 and Cdc28 proteins in synchronized cultures of wild-type and Whi3-deficient cells obtained by elutriation. To this end, 3HA-tagged fusions expressed at endogenous levels from their natural 5′ sequences at their chromosomal loci were used, and the extent of their nuclear localization was quantified by pixel correlation analysis of respective immunofluorescence and DAPI images. In wild-type cells, Cln3 showed a punctuate cytoplasmic distribution in early G1, accumulated in the nucleus in late G1, and reached a maximal nuclear signal around Start, which quickly disappeared during S phase (Figure 6A). Nuclear localization of Cln3-3HA in late G1 was confirmed by confocal microscopy (not shown). Cdc28 behaved in a very similar way to Cln3, although it remained mainly nuclear during the S phase. In contrast, Whi3-deficient cells showed a distinct nuclear accumulation of both Cln3 and Cdc28 already in early G1, which persisted until Start (Figure 6B). Similarly to wild-type cells, while Cdc28 remained mainly in the nucleus, the Cln3 signal again became punctuate in the cytoplasm during the S phase entry in Whi3-deficient cells. Thus, both Cln3 and Cdc28 accumulate in the nucleus in late G1 in a precisely regulated manner. This regulatory event shows a clear requirement for Whi3, which supports the notion of Whi3 being part of a cell-cycle-regulated retention device for Cln3–Cdc28 complexes.
Figure 6.
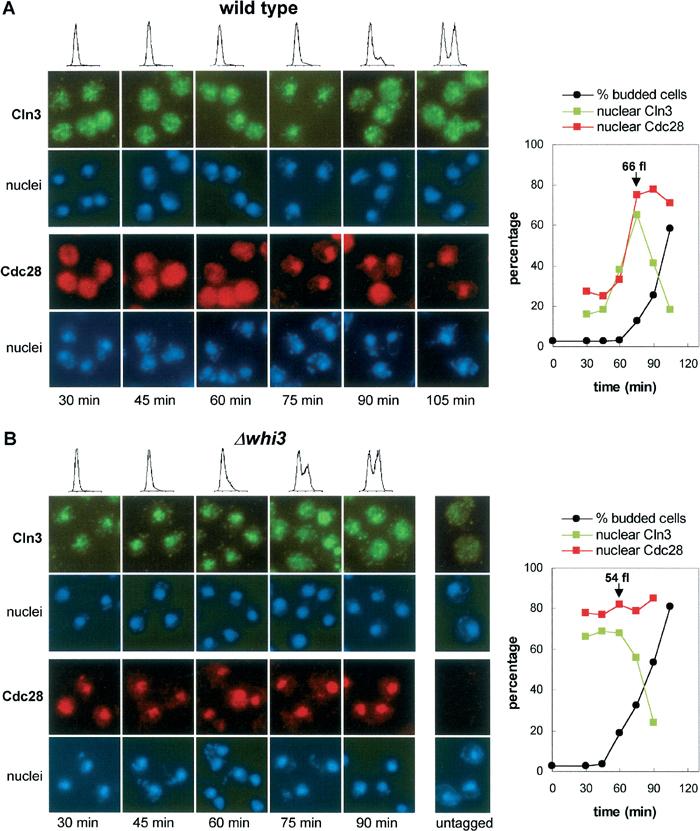
Cln3 and Cdc28 are both restricted to the cytoplasm by Whi3 during early G1, and accumulate in the nucleus in late G1. (A) Small G1 wild-type cells expressing Cln3-3HA or Cdc28-3HA at endogenous levels were obtained by elutriation and grown in YPD at 30°C, and samples were taken at the indicated time points for immunofluorescence with an anti-HA antibody. The corresponding pictures of DAPI-stained nuclei are also shown. The nuclear signal in both Cln3 and Cdc28 immunofluorescence images was quantified by pixel correlation analysis as described under Materials and methods, and the percentages of nuclear pixels obtained are plotted. Budding indexes determined for each time point are also plotted, and corresponding DNA content distributions are shown. Under these conditions, wild-type cells initiated budding and S phase at 75 min, with an average cell volume of 66 fl. (B) Small G1 Whi3-deficient (Δwhi3) cells expressing Cln3-3HA or Cdc28-3HA at endogenous levels were obtained and analyzed as in (A). Under these conditions, Whi3-deficient cells initiated budding and S phase at 60 min, with an average cell volume of 54 fl. A sample of untagged cells is also shown.
Whi3 contributes to the G1 arrest caused by nitrogen limitation
In addition to its role in regulating G1 length in mitotic cells, Whi3 has been shown to be essential for yeast cells to take other developmental options in G1 such as filamentous growth, meiosis and mating (Mösch and Fink, 1997; Garí et al, 2001; Nash et al, 2001; also see Figure 3). The proposal of Whi3 as a retention device for Cln3–Cdc28 complexes in G1 may provide a mechanism that could contribute to better coordinate cell growth and cell-cycle entry, and to establish a tighter decision mechanism within a narrow range of Cln3 levels. To test this hypothesis, we evaluated the ability of wild-type and Whi3-deficient cells to arrest properly in G1 under different limiting concentrations of the nitrogen source, which have been shown to readily modulate Cln3 synthesis at a translational level (Gallego et al, 1997). In addition, poor nitrogen sources and nitrogen starvation are key nutritional conditions for filamentous growth and meiosis, respectively. Figure 7A shows that wild-type cells arrested in G1 or entered the cell cycle within a narrow range of ammonium concentrations. In contrast, the whi3 deletant strain showed a more relaxed behavior with respect to G1 arrest, with higher percentages of cycling cells at any limiting concentration of ammonium sulfate. Moreover, the abnormalities caused by Whi3 loss required Cln3, as cln3 and double cln3 whi3 null mutant strains behaved as the wild type (Figure 7A). These observations suggest that Whi3-dependent retention of Cln3–Cdc28 in the cytoplasm may help set a threshold for the activity of this G1 cyclin–Cdk complex to coordinate growth in the G1 phase and cell-cycle entry.
Figure 7.
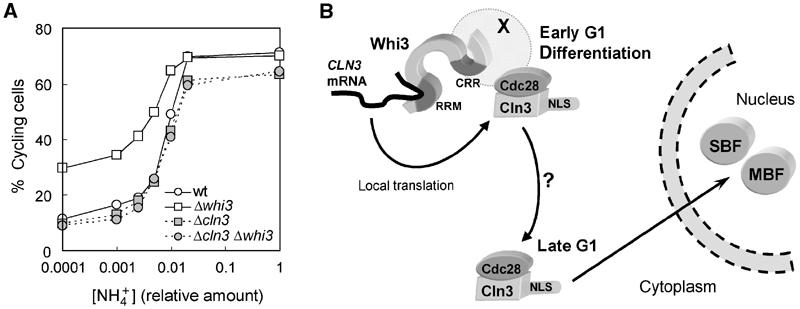
Whi3 sets a threshold for cell-cycle entry. (A) Wild-type (wt), Δwhi3, Δcln3 and double Δwhi3 Δcln3 mutant cells growing exponentially were transferred to media with different ammonium-limiting concentrations for 4 h. Fractions of cycling cells (in S, G2 and M phases) were deduced from DNA content distributions and are plotted as a function of the initial ammonium concentration. (B) A model for the cytoplasmic retention device is presented based upon physical and functional interactions between Whi3, Cdc28 and the CLN3 mRNA. Whi3 interacts in vivo with Cdc28 even in the absence of G1 cyclins, suggesting that Whi3 would recruit Cdc28 already in early G1 cells. As Whi3 also binds the CLN3 mRNA, newly synthesized Cln3 polypeptides would form complexes and activate those Cdc28 molecules that interact with Whi3. As Whi3 is a protein present in large cytoplasmic structures, newly assembled Cln3–Cdc28 complexes would become trapped and unable to reach essential targets in the nucleus, that is the transcriptional factors SBF and MBF. However, and as the cell grows during G1, an unknown event would release Cln3–Cdc28 complexes in the cytoplasm, which would be rapidly imported to the nucleus by the Cln3 NLS to activate SBF- and MBF-dependent transcription in late G1. In addition, Whi3 could set a threshold for Cln3 levels to arrest promptly the cell cycle in G1 when Cln3 synthesis rates decrease under nutritionally compromised conditions, thus helping yeast cells to take different fate options only after the budding mitotic cycle has been fully deactivated.
Discussion
We have shown that Whi3 interacts with Cdc28 in vivo and plays a key role in retaining both Cdc28 and Cln3 in the cytoplasm until late G1. We have identified an N-terminal domain in Whi3 that is required for efficient interaction with and cytoplasmic localization of Cdc28. This CRR is also essential for key aspects of Whi3 function such as G1 length regulation and filamentous growth, suggesting that the interaction between Whi3 and Cdc28 is part of a cytoplasmic retention mechanism with an important role in regulating G1 events.
Whi3 had been identified as a putative RNA-binding protein involved in cell size regulation (Nash et al, 2001). In addition, we had shown that Whi3 binds the CLN3 mRNA and plays an important negative role in all functional aspects of Cln3 activity, that is regulation of G1 length, and modulation of cell fate options such as mating, meiosis and filamentous growth (Garí et al, 2001). Here we propose that Whi3 recruits Cdc28 and retains Cln3–Cdc28 complexes in the cytoplasm during G1. We have detected Whi3 in complexes with Cln3, showing that these multimeric complexes do exist, although perhaps only transiently. Moreover, the presence of Whi3 in Cln3 immunoprecipitates required the cyclin box of Cln3 that is needed for binding to Cdc28, suggesting that Cdc28 acts as a linking bridge between Whi3 and Cln3. Supporting the notion that Whi3 might localize Cln3–Cdc28 complex formation, we have identified in Whi3-TAP complexes a putative candidate for the Ssa1,2 chaperones. Although the presence of these proteins may reflect unspecific interactions created during the affinity-purification procedures, Ssa1,2 have been involved in the formation of active cyclin–Cdk complexes (Yaglom et al, 1996; Honey et al, 2001). In addition, the model requires that Whi3 should recruit Cdc28 prior to the synthesis of the Cln3 cyclin and the concomitant activation of Cdc28. We have found that Whi3 binds Cdc28 independently of the presence of cyclins (our unpublished data). Finally, two different fusions of the Cln3 NLS to GFP expressed at different levels from the tet promoter produced proteins that accumulated in the nucleus independently of Whi3 (our unpublished data), suggesting that retention is not due to a direct masking effect of Whi3 on the Cln3 NLS. In summary, these data suggest that recruitment of Cdc28 may be important for Whi3 to retain newly formed Cln3–Cdc28 complexes in the cytoplasm.
Figure 7B outlines our current model for Whi3 function, where both the RNA-binding domain (RRM) and the CRR are required. The Whi3 RRM would bind the CLN3 mRNA and restrict Cln3 synthesis to a local molecular environment, while the Whi3 CRR would be responsible for retaining some Cdc28 molecules in the same molecular environment, so newly synthesized Cln3 polypeptides would bind preferentially the retained Cdc28 molecules. By using a double 3HA-tagged strain for Whi3 and Cdc28 we have estimated that Cdc28 molecules are five times more abundant than Whi3 molecules in G1 cells (our unpublished results). In other words, only a fraction of Cdc28 may be found in complexes with Whi3. Thus, when Whi3 lacks the RRM domain, CLN3 mRNA translation would take place away from Cdc28–Whi3 complexes, so newly synthesized Cln3 polypeptides would readily bind to free excess Cdc28 molecules, thus escaping the retention mechanism. Accordingly, a 3- to 4-fold overproduction of Whi3ΔRRM does not cause any cell-cycle defect (Garí et al, 2001). On the other hand, if Whi3 lacks the CRR domain, no Cdc28 molecules would be subject to retention, so newly synthesized Cln3 polypeptides would also bind free Cdc28 molecules and escape the retention mechanism. This model fits with the fact that both domains are equally important for Whi3 function.
We do not know whether the interaction between Cdc28 and Whi3 is direct or indirect. Our attempts to detect this interaction in vitro with purified proteins have so far been unsuccessful. However, similarly to the interaction between Cdc28 and G1 cyclins (Deshaies and Kirschner, 1995; Gerber et al, 1995), the possibility that specific chaperones and/or other proteins are required to induce a direct interaction should still be considered. Cdc28 had been found in the cytoplasm partially associated with a large structural matrix (Wittenberg et al, 1987). Although Whi3 shows a behavior very similar when using the same differential centrifugation conditions used by these authors, we do not know whether the fraction of Cdc28 bound to Whi3 is the same as they defined as being associated to a cytoplasmic matrix.
Here we show that Whi3 is important for yeast cells to arrest efficiently in G1 under nutritionally compromised conditions, suggesting that Whi3 may play a role in coordinating cell growth in G1 and cell-cycle progression. When Cln3 is at low levels due to limited trophic signals, newly synthesized Cln3 polypeptides could be efficiently retained on Cdc28 molecules bound to Whi3, and prevent these complexes from reaching their nuclear targets, namely SBF and MBF, thus holding the cell in G1. As Cln3 synthesis rates are extremely dependent on nutrient availability (Barbet et al, 1996; Gallego et al, 1997; Polymenis and Schmidt, 1997; Newcomb et al, 2002), such a mechanism would narrow the trophic signal interval at which the cell would commit to a new cycle. In agreement with this hypothesis, a moderate Cln3 overexpression is able to override the negative regulation exerted by Whi3 in G1 (Garí et al, 2001; Nash et al, 2001).
How the retention potential of Whi3 is modulated to allow the nuclear accumulation of Cln3–Cdc28 in late G1 is still an open question. Although we have not observed changes in the affinity of Whi3 for Cdc28 during entry into the cell cycle, we cannot discard the possibility that the sustained presence of Cln3–Cdc28 complexes bound to Whi3 during G1 might affect their respective functions, favor a transient release of the G1 cyclin–Cdk complex, and allow the nuclear accumulation of Cln3–Cdc28 in late G1 (Figure 6B). However, Whi3 did not inhibit Cdc28 in in vitro kinase assays, and neither Cln3–Cdc28 nor Cln2–Cdc28 was able to phosphorylate Whi3 in vitro (unpublished data). Alternatively, Whi3 could also locally recruit some other factors that might modulate Cln3–Cdc28 complex formation and/or retention in late G1. Here we have shown that the RNA-binding domain of Whi3 is essential for cytoplasmic retention of Cdc28, but not for the interaction. On the other hand, we have found in Whi3-TAP complexes a putative candidate for Ded1, an essential translation initiation factor (Chuang et al, 1997). These data suggest that Whi3 could recruit some mRNAs to localize their translation. In this regard, a three-hybrid screen using Whi3 as RNA-binding bait yielded a large number of RNA sequences with one or more copies of the GCAU tetranucleotide (DJ SenGupta, personal communication). Remarkably, the CLN3 mRNA contains two regions with five tetranucleotides each, the first within a fragment of 150 nt at the 5′UTR, and the second within 200 nt at the 3′ region of the ORF, which we had found to be important for efficient binding to the Whi3 RRM in vitro (Garí et al, 2001). However, as the presence of such a tetranucleotide is obviously not restricted to the CLN3 mRNA, the RRM of Whi3 could also cause local synthesis of other proteins in addition to Cln3 to modulate the retention of Cln3–Cdc28 complexes in the cytoplasm during G1.
The Whi3-mediated step may help define the nuclear accumulation of Cln3–Cdc28 as a key G1 event. In this respect, we can now subdivide the G1 phase into three different molecular events, which find a clear parallelism when comparing higher eukaryotes and budding yeast. The early G1 period could be defined by the sustained expression of G1 cyclins, Cln3 in yeast (Tyers et al, 1993) and D-type cyclins in higher eukaryotes (reviewed by Sherr, 1995), which would take place at transcriptional or translational levels (Aktas et al, 1997; Gallego et al, 1997; Polymenis and Schmidt, 1997; Weber et al, 1997; Muise-Helmericks et al, 1998; Newcomb et al, 2002) depending on the nature of the proliferation signal, and would be especially noticeable when cells initiate the cell cycle from a G0 or resting state. The mid-G1 period would be defined by the nuclear accumulation of the corresponding G1 cyclin–Cdk. Here we have shown that Cln3–Cdc28 accumulates in the nucleus during G1 in a similar manner to what is described for cyclin D–Cdk4 in higher eukaryotes (Baldin et al, 1993). However, there is still no evidence for a homolog of Whi3 or an analogous cytoplasmic retention mechanism that could regulate the localization of cyclin D–Cdk4 complexes during G1 in higher eukaryotes. Export of cyclin D1 to the cytoplasm during the S phase involves phosphorylation by GSK-3β and depends on Crm1 in higher eukaryotes (Diehl et al, 1998; Alt et al, 2000). We have found that Cln3 also becomes cytoplasmic during the S phase, suggesting that an export mechanism may also operate in yeast cells. Finally, transcriptional activation of G1/S genes would take place in the late G1 period, mainly mediated by Cdk-dependent phosphorylation of pRB in higher eukaryotic cells (Weinberg, 1995), and by the Cdc28-dependent recruitment of the basal complex onto SBF- and MBF-containing promoters in yeast cells (Cosma et al, 2001). In this regard, although Whi3-deficient cells show a clear accumulation of Cln3 and Cdc28 in the nucleus early in G1, they still display a measurable G1 phase, indicating the existence of additional fences in the nucleus to prevent premature activation of G1/S gene expression. Unraveling the complexity of molecular mechanisms operating at different moments during the G1 phase will allow us to understand how eukaryotic cells integrate a plethora of quantitative signals to take a single and binary decision: to enter or not the cell cycle.
Materials and methods
Strains, plasmids and growth conditions
Yeast parental strains (CML128, W303 and Σ1278), standard growing conditions, and special media for nitrogen limitation, and to induce filamentation or invasive growth were as described (Mösch and Fink, 1997; Gallego et al, 1997; Garí et al, 2001). Strains used in G1 cyclin depletion experiments derive from BF305 (MATa, GALp-CLN3, Δcln1∷HIS3, Δcln2∷TRP1, leu2, ura3, ade1, arg5, 6, met14) (Tyers et al, 1993). Chromosomal gene disruptions, C-terminal fusions to several different epitope tags (3HA, 6FLAG, 13myc and TAP) and sGFP, as well as gene fusions to the GAL1 promoter were performed by gene transplacement methods as described (Gallego et al, 1997). As the presence of WHI3 is deleterious to E. coli cells, the C-terminal 3HA-tagged gene including promoter sequences had to be split into two vectors for mutagenesis, the whole gene or appropriate deletions being rebuilt by homologous recombination within overlapping sequences into an appropriate centromeric vector during transformation of Δwhi3 yeast cells. Productive recombination events were assessed by Western blot analysis. Serial deletions of Whi3 were obtained by PCR-based mutagenesis essentially as described (Gallego et al, 1997), removing amino acids 21–72 in Whi3Δ1a, 73–120 in Whi3Δ1b, 121–220 in Whi3ΔCRR, 221–320 in Whi3Δ3, 321–440 in Whi3Δ4, 421–520 in Whi3Δ5 and 541–661 in Whi3ΔRRM. The cln3ΔNLS construct carries a deletion that removes the last 55 amino acids of Cln3 containing a bipartite NLS (Edgington and Futcher, 2001; Miller and Cross, 2001), cln3ΔCB lacks the fifth helix (amino acids 188–199 in Cln3) of the cyclin box (Huang et al, 1997), and cln3nt carries a point mutation that eliminates the start codon (Garí et al, 2001). The GST-Cln2 fusion is as described (Deshaies and Kirschner, 1995). Details of strain and plasmid constructions are available upon request.
Affinity purification and characterization of Whi3 complexes
Whi3 complexes were purified from 2 l cell cultures grown in YPD at OD600=2 by the TAP procedure developed by Rigaut et al (1999). Final material bound to calmodulin beads was analyzed by SDS–PAGE. Desired bands were cut from silver-stained gels and trypsin-digested mixtures were analyzed by MALDI mass spectrometry (Shevchenko et al, 1996). Whi3 complexes were also characterized in glycerol gradients. Briefly, Whi3-3HA cell extracts were spun through a 10–40% glycerol gradient at 15 krpm for 15 h in an SW21 Beckman rotor, under identical buffer conditions as used for the TAP procedure. Fractions were collected from the bottom, and analyzed for the presence of Whi3-3HA by Western blotting, and ribosomal RNA by agarose electrophoresis.
Interaction assays
Cdc28 was immunoprecipitated with a rabbit anti-Cdc28 antibody (a gift from C. Mann), and Cln3-6FLAG with a mouse anti-FLAG antibody (clone M2, Eastman Kodak), as described by Tyers et al (1993). The same buffer conditions were used to purify GST-Whi3ΔRRM onto glutathione beads (Pharmacia). Reconstitution of active Cln2–Cdc28 complexes was assayed by the method developed by Deshaies and Kirschner (1995).
Immunofluorescence
The localization of 3HA- and 13myc-tagged proteins was carried out by immunofluorescence (Colomina et al, 1999) with a rat anti-HA antibody (clone 3F10, Roche) or a mouse anti-myc antibody (clone 9E10, Roche) followed by incubation with anti-rat Alexa488-labeled or anti-mouse Alexa546-labeled antibodies (Molecular Probes). Endogenous levels of Cln3-HA were detected by a signal-amplification method. Briefly, after incubation with rat anti-HA antibody, slides were sequentially incubated with goat anti-rat, rabbit anti-goat and goat anti-rabbit antibodies, all labeled with Alexa488 (Molecular Probes). Digital images were obtained with the aid of a 16-bit cooled CCD camera (Ultrapix SK1600, Life Science Resources), which was mounted on a Nikon Eclipse E600 microscope equipped with epifluorescence illumination and the required filters. To quantify nuclear localization of Cln3 and Cdc28 in Figure 6, respective immunofluorescence images were first binarized above a fixed threshold that eliminated all pixels with an intensity value lower than 25% of the brighter pixels in each image. Corresponding DAPI images were also binarized to eliminate any dim pixels outside the nuclear compartment. Colocalization was quantified in a minimum of 100 cells with the Ultraview (Perkin-Elmer) software as the percentage of pixels in the immunofluorescence image that matched pixels in the DAPI image. Nuclear localization of Cln3-3HA in late G1 cells was assessed with the aid of an Olympus FV500-IX81 confocal microscope.
Miscellany
Northern methods and probes used, as well as Western conditions to detect specific tagged proteins were carried out as described previously (Gallego et al, 1997). TAP-tagged proteins were detected with a peroxidase-labeled rabbit anti-peroxidase antibody soluble complex (Sigma) as described (Rigaut et al, 1999). Small G1 cells were obtained by elutriation (Tyers et al, 1993). Cell volume distributions were obtained in a Z2 Coulter Counter, and DNA content distributions and budding indexes were determined as described (Gallego et al, 1997).
Acknowledgments
We thank Sònia Rius and Anna Valls for their technical assistance, Emilio Camafeita for the analysis by MALDI mass spectrometry, Bruce Futcher, Bertrand Sèraphin and Pamela Silver for providing strains and plasmids, and Carl Mann for the anti-Cdc28 antibody. We also thank Neus Colomina for her help in some experiments, Yuhui Liu for helpful discussions and technical advice, and Joan Comella for critical comments on the manuscript. This work was funded by the Ministry of Science and Technology of Spain, Fundació La Caixa and FEDER. HW and EV received fellowships from AECI and Generalitat de Catalunya, respectively. EG is a researcher of the Ramon y Cajal program.
References
- Aktas H, Cai H, Cooper GM (1997) Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the Cdk inhibitor p27KIP1. Mol Cell Biol 17: 3850–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt JR, Cleveland JL, Hannink M, Diehl JA (2000) Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev 14: 3102–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews BJ, Herskowitz I (1989) The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature 342: 830–833 [DOI] [PubMed] [Google Scholar]
- Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G (1993) Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev 7: 812–821 [DOI] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN (1996) TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, Chang TH (1997) Requirement of the DEAD-Box protein Ded1p for messenger RNA translation. Science 275: 1468–1471 [DOI] [PubMed] [Google Scholar]
- Colomina N, Gari E, Gallego C, Herrero E, Aldea M (1999) G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J 18: 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Panizza S, Nasmyth K (2001) Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell 7: 1213–1220 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Kirschner M (1995) G1 cyclin-dependent activation of p34CDC28 (Cdc28p) in vitro. Proc Natl Acad Sci USA 92: 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ (1998) Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev 12: 3499–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L, Bohm T, Nasmyth K (1995) Roles and regulation of Cln–Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J 14: 4803–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington NP, Futcher B (2001) Relationship between the function and the location of G1 cyclins in S. cerevisiae. J Cell Sci 114: 4599–4611 [DOI] [PubMed] [Google Scholar]
- Futcher B (1996) Cyclins and the wiring of the yeast cell cycle. Yeast 12: 1635–1646 [DOI] [PubMed] [Google Scholar]
- Gallego C, Garí E, Colomina N, Herrero E, Aldea M (1997) The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J 16: 7196–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garí E, Volpe T, Wang H, Gallego C, Futcher B, Aldea M (2001) Whi3 binds the mRNA of the G1 cyclin CLN3 to modulate cell fate in budding yeast. Genes Dev 15: 2803–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber MR, Deshaies RJ, Herskowitz I, Morgan DO (1995) Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proc Natl Acad Sci USA 92: 4651–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DD, Markwardt DD, Parviz F, Heideman W (1998) Regulation of the Cln3–Cdc28 kinase by cAMP in Saccharomyces cerevisiae. EMBO J 17: 4370–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey S, Schneider BL, Schieltz DM, Yates JR, Futcher B (2001) A novel multiple affinity purification tag and its use in identification of proteins associated with a cyclin–CDK complex. Nucleic Acids Res 29: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KN, Odinsky SA, Cross FR (1997) Structure–function analysis of the Saccharomyces cerevisiae G1 cyclin Cln2. Mol Cell Biol 17: 4654–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huble L, Bradshaw-Rouse J, Heideman W (1993) Connections between the Ras-cyclic AMP pathway and G1 cyclin expression in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol 13: 6274–6282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz B-J, Tyers M (2002) Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395–400 [DOI] [PubMed] [Google Scholar]
- Levine K, Huang K, Cross FR (1996) Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol 16: 6794–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes NF, Johnson AL, Breeden L, Johnston LH (1992) SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature 357: 505–508 [DOI] [PubMed] [Google Scholar]
- Maekawa H, Usui T, Knop M, Schiebel E (2003) Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J 22: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE (1998) Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1191–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ME, Cross FR (2001) Mechanisms controlling subcellular localization of the G1 cyclins Cln2p and Cln3p in budding yeast. Mol Cell Biol 21: 6292–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO (1995) Principles of CDK regulation. Nature 374: 131–134 [DOI] [PubMed] [Google Scholar]
- Mösch HU, Fink JR (1997) Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 145: 671–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muise-Helmericks RC, Grimes HL, Bellacosa A, Malstrom SE, Tsichlis PN, Rosen N (1998) Cyclin D expression is controlled post-transcriptionally via a phosphatidylinositol 3-kinase/Akt-dependent pathway. J Biol Chem 273: 29864–29872 [DOI] [PubMed] [Google Scholar]
- Nash RS, Volpe T, Futcher B (2001) Isolation and characterization of WHI3, a size-control gene of Saccharomyces cerevisiae. Genetics 157: 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Dirick L (1991) The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell 66: 995–1013 [DOI] [PubMed] [Google Scholar]
- Newcomb LL, Hall DD, Heideman W (2002) AZF1 is a glucose-dependent positive regulator of CLN3 transcription in Saccharomyces cerevisiae. Mol Cell Biol 22: 1607–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenis M, Schmidt EV (1997) Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev 11: 2522–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 10: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Sherr CJ (1995) D-type cyclins. Trends Biochem Sci 20: 187–190 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C (1995) CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev 9: 2780–2794 [DOI] [PubMed] [Google Scholar]
- Sudbery PE, Goodey AR, Carter BL (1980) Genes which control cell proliferation in the yeast Saccharomyces cerevisiae. Nature 288: 401–404 [DOI] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher B, Nasmyth K (1991) The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 65: 145–161 [DOI] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B (1993) Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J 12: 1955–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Hu W, Jefcoat SC, Raben DM Jr, Baldassare JJ (1997) Ras-stimulated extracellular signal-related kinase 1 and RhoA activities coordinate platelet-derived growth factor-induced G1 progression through the independent regulation of cyclin D1 and p27. J Biol Chem 272: 32966–32971 [DOI] [PubMed] [Google Scholar]
- Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Wittenberg C, Richardson SL, Reed SI (1987) Subcellular localization of a protein kinase required for cell cycle initiation in Saccharomyces cerevisiae: evidence for an association between the CDC28 gene product and the insoluble cytoplasmic matrix. J Cell Biol 105: 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaglom JA, Goldberg AL, Finley D, Sherman MY (1996) The molecular chaperone Ydj1 is required for the p34CDC28-dependent phosphorylation of the cyclin Cln3 that signals its degradation. Mol Cell Biol 16: 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]


