Abstract
In some enterobacterial pathogens, but not in Escherichia coli, loss-of-function mutations in the ampD gene are a common route to β-lactam antibiotic resistance. We constructed an assay system for studying mechanism(s) of enterobacterial ampD mutation using the well-developed genetics of E. coli. We integrated the Enterobacter ampRC genes into the E. coli chromosome. These cells acquire spontaneous recombination- and SOS response-independent β-lactam resistance mutations in ampD. This chromosomal system is useful for studying mutation mechanisms that promote antibiotic resistance.
Mutation is a primary cause of bacterial resistance to antibiotics. Some mutations promote resistance directly (e.g., quinolone resistance mutations in genes encoding its Escherichia coli targets, gyrA and gyrB [16]). Other chromosomal mutations can cause mutator phenotypes that increase the likelihood of acquiring a resistance mutation (42). Mutations also ameliorate the otherwise deleterious effects on cell growth and physiology of some antibiotic resistance-conferring mutations (22). Although antibiotic resistance has been studied intensively, the mechanisms that generate resistance mutations are poorly understood.
In addition to spontaneous mutation in exponentially growing cells (growth-dependent mutation), other mutation mechanisms exist that may pertain to antibiotic resistance (34, 36, 37). For example, factors such as antibiotic concentration (23), environmental conditions (12), or other stress-inducing phenomena (1, 34, 36) may enhance or repress mutational machinery that leads to resistance mutations (for a review, see reference 31). Some mutation mechanisms or factors may be more important when the organism is under suboptimal growth conditions, as is probably the case during certain stages of an infection. In this study, we utilize a relatively well-described β-lactam resistance pathway as a model system to begin dissecting the mechanism(s) of antibiotic resistance mutation using the tools of E. coli genetics.
Chromosomally encoded AmpC β-lactamases confer β-lactam antibiotic resistance in many pathogenic and opportunistic bacteria and are ubiquitous in enterobacteria, except for the salmonellae, klebsiellae, Proteus mirabilis, Shigella flexneri, and Shigella dysenteriae (30, 32). Their expression is inducible in all but E. coli and the shigellae (30). In inducible strains, ampC transcription is controlled by the transcriptional activator AmpR (2). AmpR activity is regulated allosterically by two cell wall components, 1,6-anhydromuropeptide and UDP-N-acetylmuramic acid-pentapeptide (UDP-MurNAc-pentapeptide). The first allows, and the second blocks, AmpR transcriptional activator activity at ampC (19). AmpD converts (activator-promoting) 1,6-anhydromuropeptide to (activator-blocking) UDP-MurNAc-pentapeptide, which then binds AmpR and blocks ampC transcription. Thus, loss-of-function mutations in ampD cause 1,6-anhydromuropeptide accumulation and constitutively induced AmpC β-lactamase production (7, 20, 21, 27). ampD missense and nonsense mutations are common in AmpC-mediated, β-lactam-resistant clinical isolates (25, 38). Also, some β-lactam antibiotics can induce expression of ampC by causing an increase in the cytoplasmic concentration of 1,6-anhydromuropeptide (7, 30).
E. coli lacks ampR, and low-level ampC expression results from a promoter embedded in the overlapping fumarate reductase (frdABCD) operon (13). High-level β-lactam resistance, mediated by ampD loss-of-function mutation, can be reconstituted in E. coli when the ampR and ampC genes of other enterobacteria are expressed from a plasmid (28, 35). We have integrated the ampRC genes from Enterobacter cloacae into the E. coli chromosome to assay ampD mutation, as selected by its β-lactam resistance phenotype. Background resistance imparted by the native ampC gene does not interfere with assays involving the reconstituted system. Integrating the ampRC genes into the chromosome improves upon previous plasmid-based ampRC expression systems by allowing genetic analyses not possible previously, first, because many mutant alleles used to study DNA repair, recombination, and mutation cause plasmid instability (e.g., reference 6). Second, the single-copy ampRC locus more closely models the situation in clinically relevant enterobacterial resistance (30). Third, this chromosomal system excludes mutations that confer β-lactam resistance by increasing plasmid (and, therefore, ampC) copy number.
An ampRC expression cassette in the E. coli chromosome was constructed as follows. The strains and plasmids used are shown in Table 1. SMR5222, an E. coli strain carrying the E. cloacae ampRC+ genes in the E. coli chromosome, was constructed using the TGV (transgenic vector) system for integrating linear DNA cassettes into the E. coli chromosome (14). E. cloacae strain MHN1 ampRC+ genes were isolated from plasmid pEc1c (35) by digestion with BamHI and SalI and were ligated into BglII- and XhoI-digested pTGV-light (14) plasmid DNA, creating pJP2. pJP2 was digested with NdeI to generate an ampRC+ fragment flanked on both sides by homology to the E. coli attachment site for phage λ (attλ) for linear transformation in the TGV system (14). SMR5201, a transformant carrying ampRC+ replacing attλ (confirmed by PCR as described elsewhere [14]) was used as a P1 donor to move attλ::ampRC+ into SMR5156 by P1 transduction (as described elsewhere [14]) to create SMR5222. Subsequent strains are isogenic to SMR5222 and were created using standard phage P1 transduction (referenced elsewhere [14]), and the constructions are outlined in Table 1.
TABLE 1.
E. coli strains and plasmids
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| DM49 | lexA3 | 29 |
| FC526 | ΔrecG263::kan | 11 |
| GY8322 | Δ(srlR-recA)306::Tn10 | S. Sommer; ENZ280 (8) carrying the K5353 mini-F plasmid (9) |
| SMR821 | lexA3 malB::Tn9 | 33 |
| SMR1827 | FC40 sulA211 | 33 |
| SMR4562a | rec+attλ+ | 33; genotype identical to FC40 (4) |
| SMR4649 | FC40 sulA211 lexA51 | 33 |
| SMR5078 | recB21 recC22 sbcB15 sbcC201 hsdrK−mK+ (λxis1 cIts857) | 14 |
| SMR5156 | SMR4562 (λxis1 cIts857) | SMR4562 × λSR446 (14) |
| SMR5201 | recB21 recC22 sbcB15 sbcC201 hsdrK−mK+ Δattλ::ampRC | SMR5078 × DNA from pJP2 |
| SMR5222 | SMR4562 Δattλ::ampRC | SMR 5156 × P1 SMR5201 |
| SMR5225 | SMR5222 Δ(srlR-recA)306::Tn10 | SMR5222 × P1 GY8322 |
| SMR5578 | SMR5222 ΔrecG263::kan | SMR5222 × P1 FC526 |
| SMR5652 | SMR5222 ΔrecG263::kan Δ(srlR-recA)306::Tn10 | SMR5578 × P1 GY8322 |
| SMR5701 | SMR1827 (λxis1 cIts857) | SMR1827 × λSR446 |
| SMR5702 | SMR4649 (λxis1 cIts857) | SMR4649 × λSR446 |
| SMR5715 | SMR5222 lexA3 malB::Tn9 | SMR5222 × P1 SMR821 |
| SMR5725 | SMR5222 sulA211 lexA51 | SMR5702 × P1 SMR5201 |
| SMR5749 | SMR5222 sulA211 | SMR5701 × P1 SMR5201 |
| Plasmids | ||
| pEc1c | E. cloacae ampRC+ | 35 |
| pJP2 | pTGV-light ampRC+ | This work |
| pJP19 | pACYC184 ampD+ | This workb |
Full genotype is Δ(lac-pro)XIII thi ara Rifr [F′ α45, lacIq lacI33ΩlacZ].
Plasmid pJP19, carrying the E. coli ampD+ gene and promoter, was created by amplifying ampD+ from E. coli wild-type strain MG1655 (3) chromosomal DNA using primers AmpD no. 1, 5′-GGGTTTTCATGAGAGGCGGCATGTTAAAACTCCAG-3′; and AmpD no. 2, 5′-GGGTTTAAGCTTTCATGTTGTCTCCTTGCTGACCAG-3′. The primers incorporate terminal BspHI and HindIII restriction sites at the 5′ and 3′ ends (respectively) of the amplified fragment. Amplified ampD+ DNA and pACYC184 DNA (5) were digested with BspHI and HindIII, and the ampD+ fragment was ligated into pACYC184. pJP19-mediated ampD expression was confirmed by complementation to β-lactam sensitivity of four independent ampD β-lactam-resistant mutants.
We used the chromosome-based ampRC expression system to determine rates of spontaneous (growth-dependent) ampicillin resistance (Ampr) mutation in otherwise isogenic strains lacking various recombination and SOS genes. Growth-dependent mutation rates were measured in 15 tube fluctuation tests. Fifteen independent cultures for each strain were grown to saturation in 5 or 10 ml of Mueller-Hinton (MH) broth (Difco), shaking at 37°C. Cultures were diluted 10-fold, and 50 μl was plated in duplicate on MH agar plates containing 100 μg/ml ampicillin. The plates were incubated overnight at 37°C, and Ampr colonies were counted. Viable cell counts of the saturated cultures were from dilutions plated on MH incubated overnight at 37°C. Mutation rates were calculated by the method of the median (26).
The recombination and SOS genes examined—recA, recG, recA recG, lexA3(Ind−), sulA, and sulA lexA(Def)—are not required for most growth-dependent mutation; however, they are required for a mechanism of mutation observed under growth-limiting conditions of carbon starvation (for a review, see reference 37; see also references 41 and 43). Moreover, the SOS response controls several mutation-promoting proteins (40) whose possible involvement in β-lactam resistance mutation we wished to test. The β-lactam resistance mutation rate in recombination- and SOS-proficient (rec+) cells is about 1.4 × 10−7 cell−1 generation−1 (Table 2). The strains tested displayed only small differences in growth-dependent mutation rates, indicating that recombination and SOS genes are not important for most growth-dependent β-lactam resistance mutation (Table 2).
TABLE 2.
Rates of β-lactam resistance mutation in E. coli DNA repair-deficient mutants
| Relevant genotypea and exptb | Mutation rate (mutations/cell/ generation, 10−7) | Rate relative to rec+ within each expt | Mean mutation rate (mutations/ cell/generation, 10−7) ± SE | Mean relative to rec+ within each expt ± SE |
|---|---|---|---|---|
| rec+ | ||||
| 1 | 0.460 | 1 | 1.4 ± 0.3 | 1.0 |
| 2 | 2.18 | 1 | ||
| 3 | 2.71 | 1 | ||
| 4 | 1.10 | 1 | ||
| 5 | 1.60 | 1 | ||
| 6 | 0.632 | 1 | ||
| 7 | 1.20 | 1 | ||
| recA | 1.0 ± 0.3 | 0.66 ± 0.1 | ||
| 1 | 0.242 | 0.53 | ||
| 2 | 0.908 | 0.42 | ||
| 3 | 2.72 | 1.0 | ||
| 4 | 0.693 | 0.63 | ||
| 5 | 0.950 | 0.59 | ||
| 6 | 0.277 | 0.44 | ||
| 7 | 1.24 | 1.0 | ||
| recG | 2.0 ± 0.8 | 1.0 ± 0.2 | ||
| 1 | 0.342 | 0.74 | ||
| 2 | 3.20 | 1.5 | ||
| 3 | 2.38 | 0.88 | ||
| recA recG | 1.2 ± 0.5 | 0.65 ± 0.1 | ||
| 1 | 0.235 | 0.51 | ||
| 2 | 1.70 | 0.78 | ||
| 3 | 1.79 | 0.66 | ||
| lexA3 | 0.96 ± 0.2 | 0.79 ± 0.1 | ||
| 5 | 1.48 | 0.93 | ||
| 6 | 0.362 | 0.57 | ||
| 7 | 1.05 | 0.88 | ||
| sulA | 1.0 ± 0.1 | 0.93 ± 0.1 | ||
| 5 | 1.05 | 0.66 | ||
| 6 | 0.665 | 1.1 | ||
| 7 | 1.29 | 1.1 | ||
| sulA lexA5 (Def) | 1.7 ± 0.3 | 1.5 ± 0.04 | ||
| 5 | 2.45 | 1.5 | ||
| 6 | 0.895 | 1.4 | ||
| 7 | 1.69 | 1.4 |
Strains used were SMR5222, rec+ (recombination and SOS response proficient); SMR5225, recA (recombination and SOS response deficient [40]); SMR5578, recG (recombination proficient, elevated for stationary-phase-mutation [11, 15]); SMR5652, recG recA (recombination, SOS, and stationary-phase mutation deficient [11, 15]); SMR5715, lexA3 (recombination proficient, SOS gene induction defective [40]); SMR5749, sulA (allows viability in the presence of a lexA-null mutation [40]); and SMR5725, sulA lexA51(Def) (SOS-induced genes expressed constitutively [40]).
Data preceded by the same experiment number were gathered in parallel.
The following experiments demonstrated that the β-lactam resistance mutations are in ampD. Based on prior observations in an E. coli model and in clinical isolates of enterobacterial pathogens (7, 27), we expected most of the β-lactam resistance mutations to be in ampD. To test this, 20 independent ampicillin resistant mutants (each from a separate independent culture) from the rec+ and recA fluctuation test experiments were examined further. β-Lactam sensitivity was restored to each of the 40 mutants by transforming each with pJP19 (Table 1) carrying the ampD+ gene. This complementation test shows that the ampicillin resistance mutations of the 40 independent mutants are recessive, loss-of-function mutations in ampD.
We determined the ampD sequences for each of the 40 mutants. Little difference was found between the rec+ and recA backgrounds (Fig. 1), suggesting similar mutation mechanism(s) in each. Many different mutations and insertions in ampD conferred AmpC-mediated β-lactam resistance.
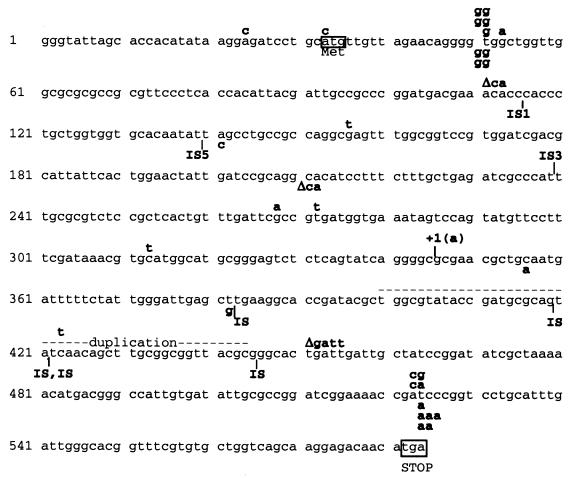
FIG. 1.
Location of ampD mutations. Forty independent Ampr mutants (20 from rec+ strain SMR5222 and 20 from recA strain SMR5225) were screened for ampD mutations by complementation to β-lactam sensitivity using pJP19 (Table 1). Apparent ampD mutations were sequenced (Lone Star Labs, Houston, Tex.) from PCR products using primers AmpD no. 3, 5′-GCGCGTCTCCGCTCACTGTTT-3′; and AmpD no. 4, 5′-GCATGCCATGCACGTTTATCG-3′. The PCR products were generated with primers AmpD no. 1 and AmpD no. 2 (legend to Table 1). The E. coli ampD sequence is given, along with 32 bp upstream of the ATG start codon. Mutations found in rec+ mutants are indicated above, and mutations found in recA mutants are shown below, the sequence. IS, insertion sequence.
Among the eight substitution mutations identified here, two were identified previously from β-lactam resistant isolates. A Trp7Gly substitution in AmpD occurred in both the rec+ and recA strains (Fig. 2) and previously in E. coli ampicillin-resistant mutants of cells expressing the E. cloacae ampRC genes from a plasmid (18). Also, we found Asp164Glu and Asp164Ala in both rec+ and recA. Asp164Glu was found previously in Citrobacter freundii (39). Both previous ampD mutations were shown to cause full derepression of ampC, as ampD-null mutations do (28, 39).
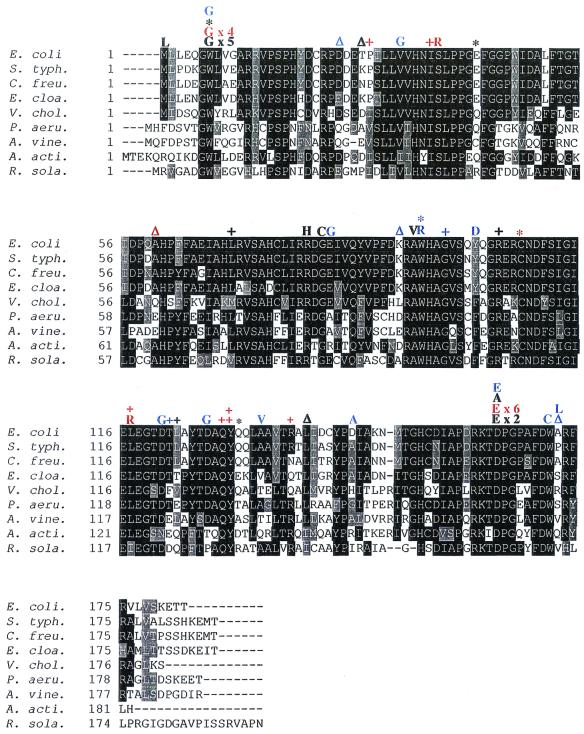
FIG. 2.
Amino acid substitutions, insertions, and deletions identified in the rec+ and recA mutants aligned with the ampD genes from nine enterobacteria. Also included are previously identified ampD mutant proteins from other laboratories (10, 18, 24, 28). rec+ mutations are in black, recA mutations are in red, and previously identified mutations are in blue. Insertions are indicated by a plus sign (+), deletions are indicated by a delta (Δ), and nonsense mutations are indicated by an asterisk (∗). Mutations isolated multiple times show the number of times that each was encountered for each strain (rec+ or recA). Abbreviations: S. typh, Salmonella enterica serovar Typhimurium; C. freu, C. freundii; E. cloa, E. cloacae; V. chol, Vibrio cholerae; P. aeru, Pseudomonas aeruginosa; A. vine, Azotobacter vinelandii; A. acti, Actinobacillus actinomycetemcomitans; and R. sola, Ralstonia solanacearum. Alignment was performed using ClustalW (17) and formatted using BOXSHADE.
Other ampD substitution mutations found include Ser37Arg, Arg80His, Gly82Cys, Ala94Val, and Leu117Arg (Fig. 2). Although these might or might not inactivate ampD fully, substitutions that alter or abolish AmpD function reveal amino acids that are important for AmpD structure and/or function. Conservative substitutions, such as Ala94Val or Asp164Glu, highlight the specific steric and/or chemical requirements of the wild-type amino acids. For example, the intolerance for Ala94Val suggests that the smaller size of alanine is important here, because both amino acids are similarly hydrophobic. The Asp164Glu substitution involves similar charges, suggesting that this amino acid position makes important catalytic or structural contacts disrupted by the larger glutamic acid side chain. Asp164 is probably not simply a surface amino acid, because it has a seemingly stringent size requirement and because alanine at this position is also not tolerated.
Alignments show that ampD is highly conserved among various bacteria (e.g., Fig. 2). Asp164, Ala94, and the other substituted amino acids from our mutation studies are highly conserved among the aligned ampD sequences (Fig. 2) and further highlight their potential structural and/or functional importance.
The variety of loss-of-function mutations observed in this system suggests its utility for studying many kinds of mutation mechanisms. This system may be useful additionally for studying the forward mutation spectra caused by potential damaging agents and environmental factors, because mutations in the small ampD gene are easily selected and sequenced.
Acknowledgments
We thank Mellanie Price for excellent technical support and Mary-Jane Lombardo, P. J. Hastings, Gregory McKenzie, Megan Hersh, Jeanine Pennington, and Rebecca Ponder for critical comments on the manuscript.
This work was supported by a Department of Defense Breast Cancer Research Postdoctoral Fellowship (to J.F.P.) and National Institutes of Health Grant R01-AI43917.
REFERENCES
- 1.Alonso, A., E. Campanario, and J. L. Martinez. 1999. Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology 145:2857-2862. [DOI] [PubMed] [Google Scholar]
- 2.Bartowsky, E., and S. Normark. 1991. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol. Microbiol. 5:1715-1725. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glassner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, A., and A. J. Clark. 1986. Synthesis of linear plasmid multimers in Escherichia coli K-12. J. Bacteriol. 167:327-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietz, H., D. Pfeifle, and B. Wiedemann. 1997. The signal molecule for β-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 41:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dri, A. M., J. Rouviere-Yaniv, and P. L. Moreau. 1991. Inhibition of cell division in hupA hupB mutant bacteria lacking HU protein. J. Bacteriol. 173:2852-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutreix, M., P. L. Moreau, A. Bailone, F. Galibert, J. R. Battista, G. C. Walker, and R. Devoret. 1989. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J. Bacteriol. 171:2415-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrhardt, A. F., C. C. Sanders, J. R. Romero, and J. S. Leser. 1996. Sequencing and analysis of four new Enterobacter ampD alleles. Antimicrob. Agents Chemother. 40:1953-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, P. L., J. M. Trimarchi, and R. A. Maurer. 1996. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics 142:25-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giraud, A., I. Matic, O. Tenaillon, A. Clara, M. Radman, M. Fons, and F. Taddei. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606-2608. [DOI] [PubMed] [Google Scholar]
- 13.Grundström, T., and B. Jaurin. 1982. Overlap between ampC and frd operons on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 79:1111-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gumbiner-Russo, L. M., M.-J. Lombardo, R. G. Ponder, and S. M. Rosenberg. 2001. The TGV transgenic vectors for single-copy gene expression from the Escherichia coli chromosome. Gene 273:97-104. [DOI] [PubMed] [Google Scholar]
- 15.Harris, R. S., K. J. Ross, and S. M. Rosenberg. 1996. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics 142:681-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrera, G., V. Aleixandra, A. Urios, and M. Blanco. 1993. Quinolone action in Escherichia coli cells carrying gyrA and gyrB mutations. FEMS Microbiol. Lett. 106:187-191. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 18.Honoré, N., M.-H. Nicolas, and S. T. Cole. 1989. Regulation of enterobacterial cephalosporinase production: the role of the membrane-bound sensory transducer. Mol. Microbiol. 3:1121-1130. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs, C., J.-M. Frère, and S. Normark. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell 88:823-832. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J.-M. Frère. 1995. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 22.Karunakaran, P., and J. Davies. 2000. Genetic antagonism and hypermutability in Mycobacterium smegmatis. J. Bacteriol. 182:3331-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler, T., M. Michea-Hamzehpour, P. Plesiat, A. L. Kahr, and J. C. Pechère. 1997. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2540-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp, U., B. Wiedemann, S. Lindquist, and S. Normark. 1993. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob. Agents Chemother. 37:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langaee, T. Y., L. Gagnon, and A. Huletsky. 2000. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC β-lactamase expression. Antimicrob. Agents Chemother. 44:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 27.Lindberg, F., S. Lindquist, and S. Normark. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii β-lactamase. J. Bacteriol. 169:1923-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindquist, S., M. Galleni, F. Lindberg, and S. Normark. 1989. Signaling proteins in enterobacterial AmpC β-lactamase regulation. Mol. Microbiol. 3:1091-1102. [DOI] [PubMed] [Google Scholar]
- 29.Little, J. W., S. H. Edmiston, L. Z. Pacelli, and D. W. Mount. 1980. Cleavage of the Escherichia coli LexA protein by the RecA protease. Proc. Natl. Acad. Sci. USA 77:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez, J. L., and F. Baquero. 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44:1771-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurelli, A. T., R. E. Fernández, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenzie, G. J., and S. M. Rosenberg. 2001. Adaptive mutations, mutator DNA polymerases and genetic change strategies of pathogens. Curr. Opin. Microbiol. 4:586-594. [DOI] [PubMed] [Google Scholar]
- 35.Nicolas, M.-H., N. Honore, V. Jarlier, A. Philippon, and S. T. Cole. 1987. Molecular genetic analysis of cephalosporinase production and its role in β-lactam resistance in clinical isolates of Enterobacter cloacae. Antimicrob. Agents Chemother. 31:295-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riesenfeld, C., M. Everett, L. J. Piddock, and B. G. Hall. 1997. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob. Agents Chemother. 41:2059-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg, S. M. 2001. Evolving responsively: adaptive mutation. Nat. Rev. Genet. 2:504-515. [DOI] [PubMed] [Google Scholar]
- 38.Sanders, C. C., and W. E. Sanders, Jr. 1992. β-Lactam resistance in gram-negative bacteria: global trends and clinical impact. Clin. Infect. Dis. 15:824-839. [DOI] [PubMed] [Google Scholar]
- 39.Stapleton, P., K. Shannon, and I. Phillips. 1995. DNA sequence differences of ampD mutants of Citrobacter freundii. Antimicrob. Agents Chemother. 39:2494-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 41.Taddei, F., J. A. Halliday, I. Matic, and M. Radman. 1997. Genetic analysis of mutagenesis in aging Escherichia coli colonies. Mol. Gen. Genet. 256:277-281. [DOI] [PubMed] [Google Scholar]
- 42.Taddei, F., I. Matic, B. Godelle, and M. Radman. 1997. To be a mutator, or how pathogenic and commensal bacteria can evolve rapidly. Trends Microbiol. 5:427-428. [DOI] [PubMed] [Google Scholar]
- 43.Taddei, F., I. Matic, and M. Radman. 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92:11736-11740. [DOI] [PMC free article] [PubMed] [Google Scholar]