Abstract
The gram-negative plant-pathogenic bacterium Xanthomonas campestris pv. vesicatoria is the causative agent of bacterial spot disease in pepper and tomato plants, which leads to economically important yield losses. This pathosystem has become a well-established model for studying bacterial infection strategies. Here, we present the whole-genome sequence of the pepper-pathogenic Xanthomonas campestris pv. vesicatoria strain 85-10, which comprises a 5.17-Mb circular chromosome and four plasmids. The genome has a high G+C content (64.75%) and signatures of extensive genome plasticity. Whole-genome comparisons revealed a gene order similar to both Xanthomonas axonopodis pv. citri and Xanthomonas campestris pv. campestris and a structure completely different from Xanthomonas oryzae pv. oryzae. A total of 548 coding sequences (12.2%) are unique to X. campestris pv. vesicatoria. In addition to a type III secretion system, which is essential for pathogenicity, the genome of strain 85-10 encodes all other types of protein secretion systems described so far in gram-negative bacteria. Remarkably, one of the putative type IV secretion systems encoded on the largest plasmid is similar to the Icm/Dot systems of the human pathogens Legionella pneumophila and Coxiella burnetii. Comparisons with other completely sequenced plant pathogens predicted six novel type III effector proteins and several other virulence factors, including adhesins, cell wall-degrading enzymes, and extracellular polysaccharides.
Xanthomonas campestris pv. vesicatoria (also designated Xanthomonas axonopodis pv. vesicatoria [101] or Xanthomonas euvesicatoria [46]) is a gram-negative, rod-shaped γ-proteobacterium with a high genomic G+C content. Members of the genus Xanthomonas represent an omnipresent group of plant-pathogenic bacteria which infect most economically important crop plants and cause a broad variety of diseases (54). X. campestris pv. vesicatoria, the causative agent of bacterial spot disease on pepper (Capsicum spp.) and tomato (Lycopersicon spp.) plants, enters the plant tissue through stomata and wounds. Bacterial colonization of plant intercellular spaces is locally restricted and induces macroscopically visible disease symptoms, so-called water-soaked lesions that later become necrotic (91). The disease results in defoliation and severely spotted fruits, both of which cause massive yield losses. Bacterial spot disease occurs worldwide but is most pernicious in regions with a warm and humid climate.
Pathogenicity of X. campestris pv. vesicatoria depends on a type III protein secretion system (TTSS) (11, 17), which is highly conserved among plant and animal pathogenic bacteria (24, 97). In X. campestris pv. vesicatoria, the TTSS is encoded by the chromosomal hrp gene cluster (hypersensitive response and pathogenicity) (11) and translocates effector proteins into the plant cell (96). Once inside the plant cytoplasm, the effectors modulate host cell processes, such as suppression of the plant basal defense mechanisms, for the benefit of the pathogen (1, 13). Some effectors, termed avirulence proteins, are recognized by the host plant. This leads to the induction of the plant defense, including a rapid, locally restricted cell death reaction, the hypersensitive response, which ultimately arrests bacterial growth (1). So far, only a small number of type III effectors have been identified in X. campestris pv. vesicatoria, and their molecular functions are widely unknown (17, 19, 70, 82).
The expression of the Hrp TTSS is controlled in X. campestris pv. vesicatoria in planta by two key regulatory proteins, HrpG and HrpX (107, 109). The OmpR family regulator HrpG is activated by an unknown mechanism and controls the expression of a genome-wide regulon, including hrp and effector genes and several TTSS-unrelated genes (71). Many, but not all, HrpG-regulated genes are also regulated by HrpX, an AraC-type regulator which is controlled by HrpG. Since some HrpX-regulated genes contain a conserved sequence motif in their promoter region (plant-inducible promoter [PIP] box; TTCGC-N15-TTCGC), it is speculated that HrpX controls their expression by binding to this promoter element (35, 107).
Recently, genome analyses and comparative genomics have been used to identify novel virulence factors. Genome sequencing of the phytopathogenic bacteria Xanthomonas axonopodis pv. citri, Xanthomonas campestris pv. campestris (26), Xanthomonas oryzae pv. oryzae (53), Pseudomonas syringae pv. tomato DC3000 (14), Erwinia carotovora subsp. atroseptica (8), and Ralstonia solanacearum (87) proved to be a milestone for the identification of putative effectors and other candidate pathogenicity factors by bioinformatic approaches (19). All of these pathogens have different lifestyles and host specificities. For example, X. axonopodis pv. citri causes citrus canker, whereas X. campestris pv. campestris is the causative agent of black rot, a systemic disease on a wide range of crucifers. Xanthomonas oryzae pv. oryzae causes bacterial blight on rice, which leads to massive yield losses. R. solanacearum, which belongs to the β-proteobacteria, has an unusually wide host range and, as X. campestris pv. vesicatoria does, also infects tomato. The identification of the complete repertoire of virulence factors of these bacteria and their biological functions is a prerequisite to understanding the pathogen-plant interaction, e.g., different host range and infection strategies.
Here, we describe the complete genome sequence of X. campestris pv. vesicatoria strain 85-10, which is pathogenic for pepper but not for tomato plants (pepper race 2) (20, 63). Strain 85-10 was chosen because it is a well-established model for bacterium-plant interactions. For instance, type III secretion and several avirulence genes have been studied in this strain in great detail (17, 19). By comparison with other plant-pathogenic bacteria, we have gained novel insights into the genome evolution of xanthomonads and detected unique features of strain 85-10, such as its specific set of candidate virulence determinants.
MATERIALS AND METHODS
Whole-genome sequencing.
Genomic DNA from X. campestris pv. vesicatoria 85-10 (20) was extracted from cells grown at 30°C in NYG liquid cultures (25) using the Genomic-tip 100/G kit (QIAGEN GmbH, Hilden, Germany). DNA shotgun libraries with insert sizes of 1 kb and 2 to 3 kb were constructed in pGEM-T (Promega GmbH, Mannheim, Germany), and 8-kb fragments were cloned into pTrueBlue-rop (MoBiTec GmbH, Göttingen, Germany) by MWG Biotech AG (Ebersberg, Germany). Plasmid clones were end sequenced on ABI 3700 sequencers (PE Applied Biosystems) by MWG Biotech AG. Base calling was done using TraceTuner (PE Applied Biosystems). High-quality reads were defined by a minimal length of 250 bp, with an averaging quality value of ≥20 in a sliding window of 30 bp. Finally, 70,042 high-quality reads, a total of 49,029 (5.66 X), 14,183 (1.64 X), and 6,830 (0.79 X) end sequences (X's indicate genome equivalents) from libraries with 1-kb, 2- to 3-kb, and 8-kb inserts, respectively, were established.
Sequence assembly and validation.
Base calling, quality control, and elimination of vector DNA sequences of the sequences were performed using the software package BioMake (Bielefeld University, unpublished) as described previously (47). Sequences were assembled using the PHRAP assembly tool (www.phrap.org), resulting in 139 contigs. For finishing of the genome sequence, the Consed/Autofinish software package (38, 39) was used, supplemented by the in-house tool BACCardI (5).
For gap closure and assembly validation, a fosmid library with inserts of approximately 35 to 38 kb was constructed using the EpiFOS fosmid library production kit (Epicenter, Madison, WI). A total of 768 fosmid clones were end sequenced on ABI 3730xl and ABI 377 sequencers by the Max Planck Institute for Developmental Biology (Tübingen, Germany) and IIT GmbH (Bielefeld, Germany), respectively. Remaining gaps were closed by sequencing on shotgun and fosmid clones carried out by IIT GmbH on LI-COR 4200L (LI-COR Inc., Lincoln, NE) and ABI 377 sequencers. Additionally, gaps were closed on fosmids using an ABI 3730xl DNA analyzer by the Max Planck Institute for Developmental Biology. To obtain a high-quality genome sequence, all regions of the consensus sequence were polished to at least phred40 quality by primer walking. Collectively, 1,731 sequencing reads were added to the primary shotgun assembly for the finishing and polishing of the genomic sequence. Repetitive elements, i.e., insertion sequence (IS) elements and rRNA operons, were sequenced completely by primer walking on fosmids and, in some cases, on shotgun clones. For assembly validation, fosmid end sequences were mapped onto the genome sequence employing BACCardI (5).
Genome analysis and annotation.
The genome was annotated using GenDB 2.0 (62). Briefly, a combined gene prediction strategy (58) was applied on the assembled sequences using Glimmer (88) and CRITICA (4). Putative ribosomal binding sites and tRNA genes were identified with RBSfinder (94) and tRNAscan-SE (57), respectively. Prior to the manual annotation of each predicted gene, an automatic functional annotation was computed based on different analyses. Similarity searches were performed against different databases, including SWISS-PROT and TrEMBL (9), KEGG (48), Pfam (6), TIGRFAM (41), and InterPro (65). Additionally, SignalP (67), helix-turn-helix (32), and TMHMM (52) were applied. Finally, each gene was functionally classified by assigning a clusters of orthologous groups (COG) number and corresponding COG category (98) and gene ontology numbers (42). Putative membrane transporters were compared against the transporter classification (TC) database for functional assignment (16). Potential substrates of the twin-arginine translocation (TAT) secretion pathway were predicted using TATFind 1.2 (30).
Genomic comparison.
For comparative analyses, the annotated genome sequences of the following bacteria were imported into GenDB: X. campestris pv. campestris ATCC 33913 (GenBank accession no. AE008922) (26), X. axonopodis pv. citri 306 and plasmids pXAC33 and pXAC64 (GenBank accession nos. AE008923, AE008924, and AE008925, respectively) (26), X. oryzae pv. oryzae KACC10331 (GenBank accession no. AE013598) (53), Erwinia carotovora subsp. atroseptica SCRI1043 (GenBank accession no. BX950851) (8), Xylella fastidiosa 9a5c (GenBank accession no. AE003849) (90), Pseudomonas syringae pv. tomato DC3000 (GenBank accession nos. AE016853 to AE016855) (14), and Ralstonia solanacearum GMI1000 (GenBank accession nos. AL646052 and AL646053) (87). Homology searches were conducted on the nucleotide and amino acid sequence level using BLAST (2). Comparisons of chromosomal sequences were carried out with the Artemis Comparison Tool (ACT) (www.sanger.ac.uk /Software/ACT).
Detection of regions with atypical G+C content.
Genomic regions with atypical G+C content were identified using a “sliding window” technique with a window size of 2,000 bp and a step size of 1,000 bp. For this, the G+C content was treated as a Gaussian distribution, and regions with differences of at least 1.5 standard deviations from the mean were calculated.
Database submission.
The nucleotide sequences of the chromosome of X. campestris pv. vesicatoria strain 85-10 and its four plasmids, pXCV2, pXCV19, pXCV38, and pXCV183, were submitted to GenBank (accession numbers: AM039948, AM039949, AM039950, AM039951, and AM039952). Sequence files giving more annotation details are available in the supplementary material at https://www.genetik.uni-bielefeld.de/GenoMik/genomeproj.html.
RESULTS AND DISCUSSION
General features of the X. campestris pv. vesicatoria genome.
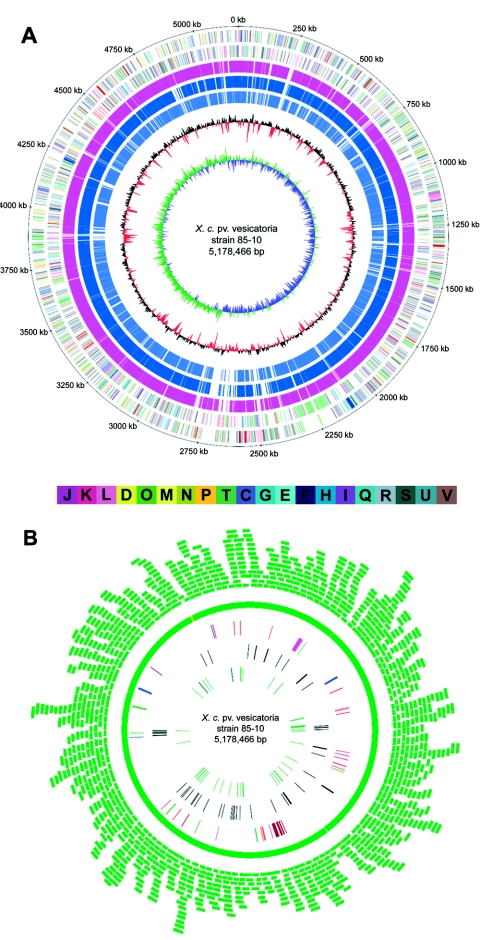
The X. campestris pv. vesicatoria strain 85-10 genome sequence was established by employing a whole-genome shotgun approach. In the final consensus sequence, each base matched at least phred40 quality. The assembly of the high-quality sequence was validated by a complete fosmid map (Fig. 1).
FIG. 1.
Circular representations of the X. campestris pv. vesicatoria strain 85-10 chromosome displaying relevant genome features (A and B) and validation of the sequence assembly by a fosmid map (B). (A) From the outer to the inner concentric circle: circle 1, genomic position in kilobases; circles 2 and 3, predicted CDS on the forward (outer wheel) and the reverse (inner wheel) strands colored according to the assigned COG classes; circles 4, 5, and 6, CDS with homologs in the chromosomes of X. axonopodis pv. citri (pink), X. campestris pv. campestris (dark blue), and X. oryzae pv. oryzae (light blue), respectively; circle 7, the G+C content showing deviations from the average (64.75%); circle 8, GC skew. The bar below the circles represents the COG colors for the functional groups (C, energy production and conversion; D, cell cycle control, mitosis, and meiosis; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolite biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking and secretion; V, defense mechanisms). (B) Fosmid map of the X. campestris pv. vesicatoria chromosome. Each green arc represents a single fosmid clone mapped onto the assembled sequence; circle 1, coverage of the X. campestris pv. vesicatoria chromosome with fosmid clones (green indicates that the chromosome is covered by more than one fosmid clone, and yellow indicates that the chromosome is covered by one fosmid clone.); circle 2, genes coding for the different secretion systems and the flagellar gene cluster (red, Sec and TAT pathway; black, putative type I secretion system; blue, type II secretion systems; magenta, type III secretion system; green, type IV system components; brown, flagellar system); circle 3, complete IS elements; circle 4, tRNA genes (green) and the two rRNA operons (blue).
X. campestris pv. vesicatoria contains a single, circular chromosome of 5,178,466 bp (Fig. 1) and four extrachromosomal elements (Table 1). While the presence of plasmids pXCV2 (1,852 bp), pXCV38 (38,116 bp), and pXCV183 (182,572 bp) was known previously (unpublished data), the genome project revealed the existence of a fourth plasmid, which was designated pXCV19 (19,146 bp) (Table 1).
TABLE 1.
Features of the chromosome and plasmids of X. campestris pv. vesicatoria strain 85-10
| Feature | Chromosome | Plasmid
|
|||
|---|---|---|---|---|---|
| pXCV2 | pXCV19 | pXCV38 | pXCV183 | ||
| Size, bp | 5,178,466 | 1,852 | 19,146 | 38,116 | 182,572 |
| G+C content, % | 64.75 | 56.59 | 59.76 | 60.73 | 60.49 |
| CDS | |||||
| Predicted no. of CDS | 4,487 | 2 | 22 | 43 | 172 |
| Function assigned | 2,965 | 1 | 12 | 26 | 76 |
| Conserved hypothetical protein | 658 | 4 | 14 | 21 | |
| Hypothetical protein | 864 | 1 | 6 | 3 | 75 |
| % of genome coding | 87.13 | 80.72 | 82.86 | 84.81 | 85.52 |
| Average length, bp | 1,009 | 767 | 722 | 754 | 910 |
| Maximal length, bp | 11,130 | 1,059 | 2,976 | 2,976 | 5,259 |
| % ATG initiation codons | 83.42 | 100 | 68.18 | 90.70 | 84.30 |
| % GTG initiation codons | 12.70 | 31.82 | 9.30 | 13.37 | |
| % TTG initiation codons | 3.77 | 2.33 | |||
| Ribosomal RNA operons | 2 | ||||
| Transfer RNA | 54 | 1 | 1 | ||
| Insertion sequence elements | 58 | 2 | 1 | 5 | |
The G+C content averages 64.75% for the chromosome and varies between 56.59% and 60.73% for the plasmids. The origin of replication is clearly detectable by a bias of G toward the leading strand (GC skew) (76). The dnaA gene is located in this region, and, therefore, its start codon has been defined as the zero point of the chromosome (Fig. 1). The GC skew analysis of the chromosome indicates a bidirectional replication mechanism, but the chromosome is not clearly divided into two equal replichores (Fig. 1A). The location of the predicted replication terminus appears to be skewed from the 180° position opposite to oriC, resulting in an approximately 370-kb size difference between the replichores.
X. campestris pv. vesicatoria contains a total of 4,726 predicted coding sequences (CDS) (Table 1). The chromosome shows an average coding capacity of 87.13%, which is typical for most bacteria. There is no significant asymmetry in the distribution of CDS on the chromosome between the leading strand (2,223 CDS; 49.5%) and the lagging strand (2,264 CDS; 50.5%). Based on the manual annotation, biological roles were assigned to 3,080 of the 4,726 CDS. The remaining 1,646 CDS comprise 697 conserved hypothetical CDS and 949 CDS of unknown function (Fig. 1A; Table 1).
X. campestris pv. vesicatoria contains two rRNA operons organized in the order 16S-23S-5S and located in a region of approximately 500 kb (between 4,600,000 bp and 5,100,000 bp) on the left replichore (Fig. 1B). Altogether, 56 genes for tRNAs representing all 20 amino acids were identified. A total of 54 tRNA genes are located on the chromosome (Fig. 1B), whereas two genes for tRNAs both recognizing the AUA codon are exclusively carried on the plasmids pXCV19 and pXCV183 (Table 1).
Horizontal gene transfer and genome plasticity.
The genome of X. campestris pv. vesicatoria contains 66 IS elements. A total of 58 IS elements are located on the chromosome, and 8 are located on the four plasmids (Table 1; see supplementary Table 1 at https://www.genetik.uni-bielefeld.de/GenoMik/genomeproj.html). A total of 48 elements belong to the IS3 family, 10 elements belong to the IS5 family, and 4 elements belong to the IS6 family. Four IS elements could not be grouped with a known family. In X. campestris pv. vesicatoria, the IS3 family member IS476/IS1477 (20 copies) is the most abundant element. IS476 has been characterized in X. campestris pv. vesicatoria and shown to inactivate the avirulence gene avrBs1 (50). The IS3 family is also highly abundant in X. axonopodis pv. citri, whereas in X. campestris pv. campestris, most IS elements belong to the IS3 and IS5 families.
An indication of high genomic plasticity is the presence of atypical DNA regions, for example, regions that differ in their G+C contents from the average of the genome. Most likely, these regions were acquired by horizontal gene transfer (31). The chromosomal CDS range in their G+C contents from 40.1% to 75.1%, with an average of 65.1%. More than 85% of the CDS have a G+C content between 60% and 70%. Analysis of the 30 CDS with the lowest G+C contents (40.1% to 50.4%) revealed that 50% are ORFans, i.e., genes that are restricted to a particular genome and that possess no known homologs. With one exception, these ORFans are short (average length, 475 bp). A total of 21 ORFans are found in the vicinity of IS elements. In contrast, there are no ORFans among the 30 CDS with the highest G+C contents (72.3% to 75.1%). The small size and low G+C content of the ORFans in X. campestris pv. vesicatoria are similar to observations made in Escherichia coli (28).
To identify alien nucleotide sequences, the chromosome was analyzed for regions with significant deviations from the genomic mean in G+C content (see Materials and Methods). These regions vary in size between 2 kb and 26 kb and contain 50 out of 58 IS elements and 15 tRNA genes. The presence of IS elements and tRNA genes is typical for genomic islands (31). Interestingly, the two largest regions (3,870,000 to 3,899,000 bp and 3,190,000 to 3,215,000 bp) carry parts of a type IV secretion system (virB6 or virB9, virB8, and virD4) and components of the type 4 pilus (i.e., pilE and fimT). The location of parts of the type IV apparatus within a genomic region of atypical G+C content, an indicator for horizontal gene transfer, has also been observed for Wolinella succinogenes, a member of the ɛ-proteobacteria (3). Interestingly, another ∼15-kb atypical region carries the complete xanthan biosynthesis cluster gumA to gumM and is flanked on one side by IS1477. An ∼14-kb region harboring genes with homology to the filamentous phage φLf (∼2,784,000 to 2,798,000 bp) could also be identified by this approach. Additionally, smaller atypical regions encode putative virulence determinants, such as type III effector proteins and adhesins (see below).
In summary, features such as the high number of mobile elements and deviations in G+C content suggest a highly flexible genome. This is advantageous for pathogen evolution driven by the need for continuous adaptation to the host in order to evade or suppress coevolving host defense mechanisms. This idea is corroborated by the presence of a large number of virulence determinants (see below).
Comparison of the X. campestris pv. vesicatoria sequence to genomes of other plant-pathogenic bacteria.
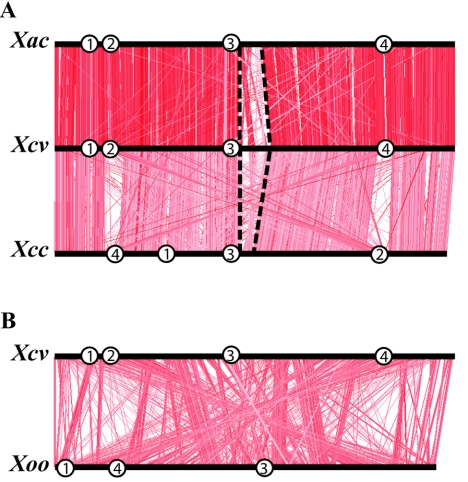
Previously, the complete genomic sequences of three different xanthomonads, X. axonopodis pv. citri, X. campestris pv. campestris, and X. oryzae pv. oryzae, causal agents of citrus canker, the systemic black rot disease of crucifers, and rice blight, respectively, were determined (26, 53). All four xanthomonads have similar chromosome sizes; however, they differ in their plasmid contents. X. campestris pv. vesicatoria harbors four plasmids, and X. axonopodis pv. citri carries two plasmids, whereas X. campestris pv. campestris and X. oryzae pv. oryzae lack any plasmids. A comparison of the chromosomal sequences of the four xanthomonads revealed that X. campestris pv. vesicatoria is most closely related to X. axonopodis pv. citri (Fig. 1A; Fig. 2). Since the genome structure of X. oryzae pv. oryzae is completely different from the three other xanthomonads (Fig. 2B), we focused our comparative analyses of the genomic organization on X. axonopodis pv. citri and X. campestris pv. campestris. Most genes are syntenic to each other among these three xanthomonads; however, striking differences are found between the flagellar gene cluster and the replication terminus of the chromosomes (Fig. 1A; Fig. 2A). Integration events in this region are probably the reason for the asymmetric replichores (see above).
FIG. 2.
Comparison of the chromosomal sequences of (A) X. axonopodis pv. citri (Xac), X. campestris pv. vesicatoria (Xcv), and X. campestris pv. campestris (Xcc) and (B) X. campestris pv. vesicatoria (Xcv) and X. oryzae pv. oryzae (Xoo) using ACT. Red lines indicate DNA sequences longer than 400 bp with at least 80% identity. The darker the color, the higher the DNA identity. Numbers mark the locations of different gene clusters on the chromosomes, which encode (1) the hrp type III secretion system, (2) the xcs type II secretion system, (3) the flagellar system, and (4) the xps type II secretion system. Dashed lines highlight the less-conserved region in the middle of the X. campestris pv. vesicatoria, X. axonopodis pv. citri, and X. campestris pv. campestris chromosomes.
The DNA sequences of X. campestris pv. vesicatoria and X. campestris pv. campestris chromosomes differ in the locations of some large gene clusters (Fig. 2A). For instance, two gene clusters coding for different type II secretion systems and their flanking regions are colinear between X. campestris pv. vesicatoria and X. axonopodis pv. citri, whereas their positions are exchanged in the chromosome of X. campestris pv. campestris compared to the other two xanthomonads (Fig. 2A). The same is true for the chromosomal location of the hrp gene cluster, which is found at the same position in X. campestris pv. vesicatoria and X. axonopodis pv. citri but at a different location in X. campestris pv. campestris (Fig. 2A). Thus, the chromosomes of X. campestris pv. vesicatoria and X. axonopodis pv. citri are not completely colinear with X. campestris pv. campestris (Fig. 2A).
Using a cutoff E value of 10−30, comparisons of the predicted protein sequences of the four xanthomonads revealed that 2,999 CDS (66.8%) of the X. campestris pv. vesicatoria chromosome are orthologs of genes in X. axonopodis pv. citri, X. campestris pv. campestris, and X. oryzae pv. oryzae, thus representing the conserved chromosomal backbone of these four species. A total of 184 CDS (4.1%), 87 CDS (1.9%), and 45 CDS (1.0%) were considered orthologs of genes present in only X. axonopodis pv. citri, X. campestris pv. campestris, and X. oryzae pv. oryzae, respectively. A total of 548 CDS (12.2%) are unique to X. campestris pv. vesicatoria, most of which encode hypothetical and conserved hypothetical proteins.
We also compared X. campestris pv. vesicatoria with the following other plant-pathogenic bacteria: X. fastidiosa, P. syringae pv. tomato, R. solanacearum and E. carotovora subsp. atroseptica. Because of the low overall DNA sequence identity, we limited the comparisons to predicted protein sequences. Using the same cutoff E value as above, there are 1,714 CDS (38.2%) in X. campestris pv. vesicatoria which are homologous to predicted proteins of X. fastidiosa, 1,780 CDS (39.7%) of P. syringae pv. tomato, 1,600 CDS (35.7%) of R. solanacearum (chromosome and megaplasmid), and 1,434 CDS (32.0%) of E. carotovora subsp. atroseptica.
Four plasmids of X. campestris pv. vesicatoria strain 85-10.
Many Xanthomonas strains carry plasmids, but little is known about their function and relevance for pathogenicity (102). X. campestris pv. vesicatoria strain 85-10 harbors four different plasmids (Table 1). The smallest plasmid, pXCV2, is basically identical to pXV64 of X. campestris pv. vesicatoria strain Xv64 (106). The presence of plasmid pXCV19 was detected only due to the efforts of the genome sequencing project and contains almost exclusively genes necessary for plasmid partitioning and conjugation. pXCV38 shows similarities to the X. axonopodis pv. citri plasmid pXAC33; however, pXCV38 lacks the type III effector genes pthA1 and pthA2. Instead, pXCV38 contains nine genes encoding a putative type IV secretion system of the Vir/Tra type (XCVc0028 to XCVc0033, XCVc0035, XCVc0041, and XCVc0042) which probably serve for conjugal transfer.
Intriguingly, the largest plasmid in strain 85-10, pXCV183, encodes a putative type IV secretion system which is most similar to the Icm/Dot system of human pathogens (21). We could identify 14 of the 22 components found in Legionella pneumophila and Coxiella burnetii (21, 111) (see supplementary Table 2 at https://www.genetik.uni-bielefeld.de/GenoMik/genomeproj.html). We alsofound candidate genes which could fulfill the function of the missing components. This is the first report of a putative Icm/Dot-like type IV secretion system in a plant-pathogenic bacterium. The essential role of Icm/Dot type IV secretion for the virulence of Coxiella and Legionella species (21, 111) suggests that this system might contribute to the virulence of Xanthomonas.
TABLE 2.
Putative pathogenicity factors and their no. of genes for X. campestris pv. vesicatoria strain 85-10
| Factor by functional group | No. of genes |
|---|---|
| Secretion system | |
| Sec and TAT pathway | 19 |
| Type I secretion system | 4 |
| Type II secretion system | 31 |
| Type III secretion system | 29 |
| Type IV secretion system | 35 |
| Type V autotransporter | 4 |
| Others | 4 |
| Flagellum | 44 |
| Secreted proteins | |
| TAT substrates | 66 |
| Host cell wall-degrading enzymes | 26 |
| Type III effectors | 20 |
| Detoxification | 65 |
| Surface structures and adhesion | 120 |
| Quorum sensing | 13 |
One interesting question concerns the segregational stability of the plasmids, which appears to be maintained by different mechanisms. pXCV183 contains a putative postsegregational killing system (Fig. 3) of the ζ toxin (XCVd0099) ɛ antitoxin (XCVd0100) type (59), which is also present on pXAC64. Additionally, the presence of an isoleucin tRNA gene on plasmids pXCV183 and pXCV19, which is missing in the chromosome, might contribute to plasmid stability. This feature is unique for X. campestris pv. vesicatoria relative to X. axonopodis pv. citri.
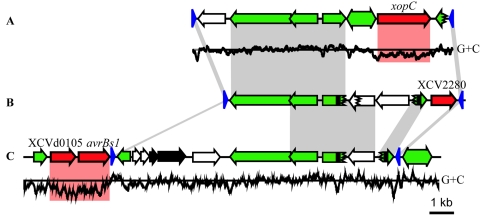
FIG. 3.
Comparison between three effector integron regions of the X. campestris pv. vesicatoria genome. Schematic overview of the chromosomal (A) xopC, (B) XCV2280, and (C) the avrBs1 region on plasmid pXCV183. Gray shaded regions indicate DNA identities of at least 80%, and red marks indicate CDS with low G+C content. Single-headed arrows represent CDS and the direction of transcription. Red arrows refer to putative effector genes. Green CDS are related to mobile genetic elements. Green double-headed arrows represent IS elements, black arrows represent a putative postsegregational killing system, and white arrows represent genes of unknown function. Blue triangles refer to 62-bp inverted repeats. Black rectangles correspond to direct repeats. A jagged line in a CDS indicates frame shifts. The G+C content of the xopC and avrBs1 region was calculated for 100-bp windows (65% on average) and is displayed at the bottom.
Genetic information storage and processing.
All basic genes for information storage and processing (i.e., DNA replication, recombination, repair, transcription, and translation) are present. Interestingly, there are three genes encoding histone-like nuclear structuring proteins (H-NS). Mutations in these genes often have pleiotropic effects, and a regulatory function of these genes in virulence has been implicated (80).
Most of the transcriptional regulators in X. campestris pv. vesicatoria belong to one of the following families: LysR (40 genes), AraC (18 genes), TetR (17 genes), LacI (10 genes), and MarR (10 genes). In addition, X. campestris pv. vesicatoria contains several putative two-component signal transduction systems. Approximately half of the 68 response regulators possess a predicted DNA binding motif as an output domain, while the rest contain other output domains, such as diguanylate cyclase (GGDEF domain), cyclic diguanylate (c-diGMP) phosphodiesterase (EAL domain), metal-dependent phosphohydrolase (HD domain), and CheB-like methylesterase. Altogether, 29 GGDEF and 14 EAL domains were found. These protein domains are responsible for the synthesis and turnover of the secondary messenger c-diGMP. Originally, this signal molecule was identified as an allosteric regulator of the cellulose synthase of Glucoacetobacter xylinum (86). In several bacteria, cellulose serves as an extracellular matrix involved in biofilm formation (84). Interestingly, X. campestris pv. vesicatoria also possesses a cellulose synthase operon (XCV3640 to XCV3644) with a c-diGMP binding subunit (XCV3642). However, this catalytic subunit is probably not functional due to an internal stop codon (XCV3643 to XCV3644).
Although histidine kinases and methyl-accepting proteins (see below) are the main sensors of extracellular signals, they are by no means the only ones present in X. campestris pv. vesicatoria. As known for most bacteria, PAS (31 proteins) and GAF (17 proteins) domains are the most common cytoplasmic signaling domains found in X. campestris pv. vesicatoria (99).
Finally, the genome of X. campestris pv. vesicatoria encodes 15 RNA polymerase σ factors (69), including 1 primary σ factor (RpoD), 1 heat-shock factor (RpoH), 1 flagellar-specific factor (FliA), 10 alternate extracytoplasmic function-type factors, and 2 σ54 family factors (RpoN). Interestingly, one of the alternate sigma factors (XCV1276) carries a C-terminal extension of approximately 200 amino acids which is unique to X. campestris pv. vesicatoria, X. campestris pv. campestris, and X. oryzae pv. oryzae.
The high number and modular diversity of regulatory proteins relative to those of other bacteria, e.g., E. coli (36), suggest that xanthomonads are able to cope with complex and changing environmental conditions. Future studies will reveal whether any of these proteins are components of the regulatory cascade which controls expression of virulence factors like the TTSS.
Quorum sensing, chemotaxis, and motility for the adaptation to environmental conditions.
Quorum sensing is a mechanism by which bacteria regulate the expression of certain genes in response to their population density (103). Only a few xanthomonads appear to produce the typical autoinducer molecule l-homoserine lactone, and there is no indication that X. campestris pv. vesicatoria does so (22). Two different alternative autoinducers have been well studied in X. campestris pv. campestris. The diffusible signal factor (DSF), an α,β-unsaturated fatty acid (105), is involved in the regulation of the synthesis of extracellular enzymes, exopolysaccharides, and cyclic glucans. Eight genes (rpfA to H) which are involved in DSF production and perception are also present in X. campestris pv. vesicatoria, thus providing circumstantial evidence for the existence of a DSF regulatory system in X. campestris pv. vesicatoria. A second diffusible factor (DF), a butyrolactone, is involved in the regulation of pigment (xanthomonadin) and exopolysaccharide synthesis in X. campestris pv. campestris (77). Next to the xanthomonadin biosynthesis gene cluster in X. campestris pv. vesicatoria is an operon that might correspond to the pigB locus of X. campestris pv. campestris, which has been implicated in the synthesis of DF (77). Both quorum-sensing systems participate in the regulation of exopolysaccharide, an important virulence factor in X. campestris pv. campestris (75, 78). Based on the conserved gene content, these systems are probably functional in X. campestris pv. vesicatoria.
X. campestris pv. vesicatoria carries a single unipolar flagellum. We identified a 147-kb region in the chromosome (116 genes, XCV1929 to XCV2044) which is almost exclusively devoted to chemotaxis and flagellar biosynthesis. The most interesting feature of this large chromosomal region is the presence of 14 tandemly repeated genes encoding methyl-accepting chemotaxis proteins. Similar gene clusters were also found in X. axonopodis pv. citri (10 mcp genes), X. campestris pv. campestris (10 mcp genes), and X. oryzae pv. oryzae (7 mcp genes). Intriguingly, this region appears to be highly dynamic since its size and gene order differ among the four Xanthomonas strains analyzed. The following other chemotaxis-related genes are also present in several copies which are dispersed in the chromosome: 10 more mcp genes, 3 cheA homologs, 2 cheB homologs, 4 cheR homologs, and 6 cheW homologs. It will be interesting to elucidate which environmental stimuli are sensed by this complex chemotactic system.
Bacterial motility is not limited to swimming. Type 4 pili enable movement by retraction and mediate bacterial adhesion to plant tissue. Immunofluorescence studies showed that purified type 4 pili of X. campestris pv. hyacinthi attached to stomata of hyacinth leaves, suggesting a role for these surface structures in the first stages of yellow disease (100). In X. campestris pv. vesicatoria strain 85-10, several operons are predicted to encode components of type 4 pili. Although type 4 pilins are very diverse in their primary structure, they can be identified by the presence of a prepilin peptidase-processing site followed by a transmembrane α-helix. Using this criterion, 15 type 4-related pilin genes were found, 8 of which are part of the two different type II protein secretion systems (see below). So far, the function of type 4 pili in pathogenicity of X. campestris pv. vesicatoria is not clear (74).
Surface structures involved in bacterial interactions.
Besides type 4 pili, several other surface structures might be implicated in the adherence of gram-negative bacterial pathogens. X. campestris pv. vesicatoria encodes several proteins which could act as adhesins (XCV1860, XCV1861, XCV2103, XCV3670, XCV3672, XCV4203, and XCV4444), such as homologs of the Yersinia proteins YapH and YadA (43), HecA from Erwinia chrysanthemi (83), and the filamentous hemagglutinin from Bordetella pertussis (56). In E. chrysanthemi, HecA is encoded next to the TTSS cluster and plays a role in virulence (51). Homologs of hecA are also present in the genomes of Xylella spp. and R. solanacearum (83). The nonfimbrial adhesin YadA forms a sheath-like structure on the surface of Yersinia and mediates adherence to epithelial cells and autoagglutination (43). Homologs of YadA have been found in several plant and animal pathogenic bacteria (73), e.g., in X. oryzae pv. oryzae, where XadA was shown to be an important virulence factor (79).
Gram-negative bacteria exhibit complex sets of polysaccharides on their surfaces, which often contribute to their pathogenic interactions with plant and animal cells (29). The typical exopolysaccharide of the genus Xanthomonas is xanthan, the structure and biosynthesis of which have been well studied in X. campestris pv. campestris (7). The gum gene cluster which controls xanthan biosynthesis in X. campestris pv. campestris is conserved in X. campestris pv. vesicatoria (XCV2776 to XCV2789). Mutant studies in X. campestris pv. campestris demonstrated that xanthan gum is not essential for pathogenicity but contributes to bacterial aggressiveness against the host (49).
Lipopolysaccharides from Xanthomonas play a role as elicitors of plant defense reactions (61, 66). Lipopolysaccharide biosynthesis in X. campestris pv. vesicatoria strain 85-10 appears to follow the ABC transporter-dependent pathway (encoded by wzm and wzt). Next to the genes for the ABC transporter and the lipopolysaccharide core biosynthesis (rmd and gmd), several genes for O-antigen synthesis (wxc) are predicted. Interestingly, the wxc genes have counterparts in X. campestris pv. campestris but not in X. axonopodis pv. citri or X. oryzae pv. oryzae. However, all of the glycosyltransferase genes required for O-antigen modification in X. campestris pv. campestris (104) are missing in X. campestris pv. vesicatoria. Instead, two deviant glycosyltransferase genes (wbdA1 and wbdA2) were found within this gene cluster, and they encode homologs of mannosyltransferases (see supplementary Table 3 at https://www.genetik.uni-bielefeld.de/GenoMik/genomeproj.html). Thus, it seems that the O-antigen of X. campestris pv. vesicatoria is basically a polymannan, in contrast to the xylosylated polyrhamnan of X. campestris pv. campestris (64).
TABLE 3.
Characteristics of perfect PIP boxes in the chromosome of X. campestris pv. vesicatoria strain 85-10
| Positiona | Distanceb | Gene | Gene product |
|---|---|---|---|
| 821742 | 119 | pgl | Polygalacturonase |
| 569831 | 153 | trpE | Anthranilate synthase component I |
| 532702 | 43 | avrRxvc | Avirulence protein AvrRxv |
| 490643 | 130 | xopAd | Xanthomonas outer protein A |
| 477427 | 81 | hrpB1d | HrpB1 protein |
| 477412 | 69 | hrcUd | HrcU protein |
| 475194 | 855 | hpaCd | HpaC protein |
| 473817 | 248 | hrcQd | HrcQ protein |
| 470920 | 917 | hrpD6d | HrpD6 protein |
| 362283 | 945 | gatA | Glu-tRNAGln amidotransferase A subunit |
| 244787 | 719 | XCV0208 | Putative secreted protein |
| 62238 | 64 | avrBs2 | Avirulence protein AvrBs2 |
| 5169633 | 191 | XCV4480 | TonB-dependent outer membrane receptor (C-terminal fragment) |
| 5020263 | 942 | XCV4364 | Xylosidase |
| 4824118 | 922 | yapH | Filamentous hemagglutinin-related protein |
| 3910091 | 62 | XCV3425 | Predicted aminopeptidase |
| 3634784 | 713 | gcvH | Glycine cleavage H protein |
| 3191886 | 273 | XCV2805 | IS1477 transposase |
| 2894866 | 229 | XCV2568 | Putative secreted protein |
| 2568565 | 140 | pgl | 6-Phosphogluconolactonase |
| 2088821 | 976 | XCV1851 | ISXccl transposase |
| 1631628 | 349 | XCV1453 | Conserved hypothetical protein |
| 1371078 | 158 | dnaE1 | DNA polymerase III alpha chain protein |
Position of the first bp of the PIP box (TTCGC-N15-TTCGC) within the chromosomal sequence. PIP boxes in antisense direction and more than 1,000 bp distant from a CDS start were excluded.
Distance in base pairs between the end of the PIP box and the start codon.
AvrRxv is not regulated by HrpG (23).
Surprisingly, a novel gene cluster of 11 genes (XCV1723 to XCV1733), which encodes a putative polysaccharide polymerase and a chain length regulator, both characteristic for heteropolymer polysaccharide biosynthesis, was identified. In addition, this cluster harbors three predicted glycosyltransferases and enzymes modifying sugar nucleotides or transferring amino or methyl groups. Homologs of this gene cluster were also found in the genomes of X. axonopodis pv. citri, X. campestris pv. campestris, and X. oryzae pv. oryzae. It is not clear which carbohydrate is produced by this novel gene cluster.
General transport systems.
Candidate transport proteins have been classified according to the TC database (16). Most transport systems belong to the class of secondary transporters, such as proteins of the major facilitator superfamily (TC 2.A.1; 38 proteins) and the resistance-nodulation-cell division superfamily (TC 2.A.6; 15 proteins). Some secondary transporters are encoded in the close vicinity of accessory components (i.e., membrane fusion proteins, TC 8.A.1) which might be important for efflux of drugs, heavy metal cations, oligosaccharides, or proteins. Altogether there are 17 complete secondary transporter-type export systems, 10 of which also contain an outer membrane factor (TC 1.B.17). Often, secondary transporters lead to multidrug resistance (68). For Erwinia amylovora, it was shown that multidrug efflux contributes to virulence (15). Predominance of secondary transporters is in accordance with the fact that X. campestris pv. vesicatoria has an aerobic lifestyle.
Additionally, genes for 28 primary transporters (traffic ATPases; TP 3.A.1) were identified. Eight of these are associated with a gene encoding a periplasmic binding protein, indicating that these systems are responsible for import. One traffic ATPase (XCV1571 and XCV1573) is encoded next to genes for a membrane fusion protein (XCV1570) and an outer membrane factor (XCV1569). We believe that this gene cluster encodes a type I protein secretion system (see below) which exports several predicted extracellular proteases and peptidases.
Pathogenicity determinants of X. campestris pv. vesicatoria.
Bacterial secretion systems are important for the interaction of pathogens with the host (18). X. campestris pv. vesicatoria contains genes for all known protein transport systems in gram-negative bacteria, namely the Sec, signal recognition particle, and TAT pathways; at least one putative type I, two type II, one type III, and two putative type IV secretion systems of different types; four type V autotransporters; and two two-partner secretion systems (Table 2; supplementary Table 3). The presence of all known secretion systems was highlighted before for only the plant pathogens E. carotovora subsp. atroseptica (8) and R. solanacearum (37).
The Sec pathway is important for the export of many proteins into the periplasmic space (Table 2; see supplementary Table 3). The TAT system (tatA-tatC) offers an alternative route to the periplasm for folded proteins (81), but so far the function of this pathway has not been elucidated for Xanthomonas. Predicted candidate substrates of the TAT pathway are 66 proteins (Table 2; see supplementary Table 4 at https://www.genetik.uni-bielefeld.de/GenoMik/genomeproj.html), e.g., putative xylosidases and cellulases.
TABLE 4.
Characteristics of known and predicted type III effector proteins of X. campestris pv. vesicatoria strain 85-10
| Gene no. | Gene name | Comment(s)a | G+C (%) | Homologb in:
|
Reference(s) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Xac | Xcc | Xoo | Pst | Rs | Eca | |||||
| XCVd0104 | avrBs1 | Unknown function | 42.23 | − | + | − | − | − | − | 85 |
| XCV0052 | avrBs2 | Putative glycerophosphoryl-diester phosphodiesterase | 63.59 | + | + | + | − | − | − | 95 |
| XCV0471 | avrRxv | YopJ/AvrRxv family, putative cysteine protease | 52.32 | − | (+) | − | − | + | − | 110 |
| XCV0581 | xopB | Homology to HopD1 (P. syringae pv. tomato) | 55.54 | − | − | − | + | − | − | 71 |
| XCV2435 | xopC | Unknown function | 47.50 | − | − | − | − | (+) | − | 70 |
| XCV0437 | xopD | SUMO cysteine protease; C48 family | 54.76 | − | + | − | − | − | − | 44, 72 |
| XCV0414 | xopF1 | Unknown function | 65.47 | (+) | (+) | + | − | − | − | 82 |
| XCV2942 | xopF2 | Unknown function | 64.72 | (+) | (+) | + | − | − | − | 82 |
| XCV2156 | xopJ | YopJ/AvrRxv family, putative cysteine protease | 56.86 | − | (+) | − | − | + | − | 70 |
| XCV2944 | xopN | Unknown function | 63.44 | + | + | + | − | − | − | 82 |
| XCV1055 | xopO | Homology to HopK1 and AvrRps4 (P. syringae) | 52.04 | − | − | − | + | − | − | 82 |
| XCV1236 | xopP | Unknown function | 61.66 | + | + | + | − | + | − | 82 |
| XCV4438 | xopQ | HopQ1-1 family protein, putative inosine-uridine nucleoside N-ribohydrolase | 68.88 | + | + | + | + | + | − | 82 |
| XCV0572 | xopX | Unknown function | 65.95 | + | + | + | − | − | − | 60 |
| XCVd0105c | Homology to HopAO1 (P. syringae pv. tomato); putative tyrosine phosphatase | 49.39 | − | + | − | + | − | − | ||
| XCV0294c | Homology to HopX2 (P. syringae pv. maculicola) | 63.34 | + | + | − | + | + | − | ||
| XCV1298c | Homology to HopH1 (P. syringae pv. tomato) | 52.02 | − | + | + | + | + | − | ||
| XCV2280c | Homology to HopX2 (P. syringae pv. maculicola) | 60.72 | + | + | − | + | + | − | ||
| XCV3786c | Homology to HopK1 (P. syringae pv. tomato) | 60.36 | (+) | − | − | + | − | − | ||
| XCV4428c | avrRxoI | Homology to AvrRxoI (X. oryzae pv. oryzicola) | 50.77 | − | − | − | − | − | − | 112 |
Putative function and homology to known type III effector proteins from Pseudomonas syringae or other Xanthomonas spp. For Pseudomonas effectors, the unified nomenclature was used (55).
Homologs were determined using BLAST algorithms. −, absence of a homolog; +, presence of a homolog; (+), partial homology or a disrupted homolog in the corresponding genome (Xac, X. axonopodis pv. citri; Xcc, X. campestris pv. campestris; Xoo, X. oryzae pv. oryzae; Pst, P. syringae pv. tomato; Rs, R. solanacearum; Eca, E. carotovora subsp. atroseptica).
Putative type III effector, not experimentally verified.
Type II secretion systems of pathogenic bacteria secrete a vast number of proteins, including possible virulence determinants, into plant intercellular space (89). X. campestris pv. vesicatoria, like X. axonopodis pv. citri and X. campestris pv. campestris (26), harbors two type II secretion systems, whereas X. oryzae pv. oryzae encodes only one (Fig. 2). Remarkably, the gene for a prepilin leader peptidase is lacking. Instead, this task might be fulfilled by the type 4 pilus assembly leader peptidase (XCV3355) (45). In X. campestris pv. vesicatoria strain 85-10, likely substrates include cellulases (nine candidates), β-glucosidases (five candidates), pectate lyases (four candidates), polygalacturonases (three candidates), and xylanases (five candidates) (Table 2; see supplementary Table 3). These proteins probably exhibit plant cell wall-degrading activity. The nature and number of these candidates are similar in all four xanthomonads. Additionally, we found a disrupted homolog of Pat-1 from the gram-positive tomato pathogen Clavibacter michiganensis subsp. michiganensis (XCV4424 and XCV4425). This gene, which encodes a putative secreted serine protease that is essential for wilt induction on tomato (33), is not present in X. axonopodis pv. citri and X. campestris pv. campestris. However, a nondisrupted homolog is present in X. oryzae pv. oryzae (XOO0986), suggesting a possible role of this gene in virulence.
In addition to the well-studied secretion systems, X. campestris pv. vesicatoria contains homologs of raxST, raxA, raxB, and raxC (XCV1244 to XCV1246 and XCV3591), which are involved in AvrXa21 activity in X. oryzae pv. oryzae. The nature of AvrXa21 is unknown, but it leads to specific recognition of bacteria in rice plants expressing the Xa21 resistance gene (27). RaxA and RaxB show similarity to a double-glycine leader peptide-dependent secretion system and might, in concert with RaxC, secrete the so-far-unknown substrate (27).
The main virulence determinant of X. campestris pv. vesicatoria is the TTSS (17, 19), which is encoded by the hrp pathogenicity gene cluster (11) located in a 35.3-kb chromosomal genomic island (459,555 bp to 494,868 bp). Interestingly, the regions flanking the 16.2-kb core cluster (hrcC-hpaB) encode accessory components and substrates of the TTSS and are not conserved among the different xanthomonads (19, 53, 72).
Most promoters in the hrp gene cluster contain PIP boxes (TTCGC-N15-TTCGC), the supposed binding motif for the transcriptional regulator HrpX (35, 107). In addition to 8 known PIP boxes, we identified 15 novel boxes within a reasonable distance upstream of the CDS (Table 3). The genome contains 189 imperfect PIP boxes (TTCG-N16-TTCG); however, no clear association with pathogenicity-related genes is evident. Future work will elucidate how many of these genes are regulated by HrpG and HrpX and might play a role in pathogenicity. However, it is known already that not all HrpX-regulated genes possess this motif in their promoter regions (71). The genome sequence enables us to determine the complexities of HrpG- and HrpX-dependent regulation of gene expression by transcriptome analyses.
TTSS of plant pathogens translocate a large number of effector proteins into the plant cells (1). So far, 14 type III effectors of X. campestris pv. vesicatoria strain 85-10 are known (19, 60, 70, 82). Analysis of the X. campestris pv. vesicatoria genome sequence led to the identification of six new candidate effectors (XCVd0105, XCV0294, XCV1298, XCV2280, XCV3786, and XCV4428) (Table 4), most of which have homologs in other xanthomonads and different P. syringae strains but not in E. carotovora subsp. atroseptica (Table 4). Type III effectors of P. syringae were designated according to the new unified nomenclature (55). It appears that homologs of HopX2 (formerly HopPmaB), HopH1 (formerly HopPtoH), HopQ1-1 (formerly HopPtoQ), and XopP are distributed most widely among plant-pathogenic bacteria. For almost all effectors, homologous proteins can be found in other pathogenic bacteria. The only exception is XopC, which is unique for X. campestris pv. vesicatoria (70). It should be noted that in contrast to other X. campestris pv. vesicatoria strains, strain 85-10 does not contain homologs of the well-studied AvrBs3 family of effectors (17). Xanthomonads also lack effectors of the YopT family, which is found in many animal and plant-pathogenic bacteria, such as P. syringae and R. solanacearum. In summary, the set of effectors differs between several species and even between closely related bacteria like the four xanthomonads (Table 4). The specific effector set of a given strain is certainly important for its interaction with the plant, thus potentially determining host range.
Ten of the 20 candidate and proven effector genes of X. campestris pv. vesicatoria show significantly lower G+C contents, indicative for acquisition by horizontal gene transfer (31). This idea is corroborated by the fact that some of the predicted effector genes are located in the vicinity of mobile genetic elements. It is noteworthy that we identified two novel effector integrons similar to a previously described element (Fig. 3) (70). All three integrons are flanked by inverted repeats that were first identified next to genes coding for effectors of the AvrBs3 family (10, 70). One of the newly identified integrons is located on the chromosome and is associated with a gene (XCV2280) encoding a homolog of the effector HopX2 from P. syringae (Table 4) (40). The other integron is located on the plasmid pXCV183 and harbors the putative postsegregational killing system. This integron is flanked by a type III effector locus, which encodes AvrBs1 and a putative tyrosine phosphatase homologous to the effector HopAO1 (formerly HopPtoD2) from P. syringae pv. tomato DC3000 (12, 34). We were surprised that these are the only two effector genes present on the plasmids of X. campestris pv. vesicatoria strain 85-10. Similar effector integrons are also present in the X. axonopodis pv. citri chromosome and plasmid pXAC64 (70).
In addition to protein secretion systems, other factors might be important for the pathogen-plant interaction. We identified 65 proteins with possible functions in defense of X. campestris pv. vesicatoria against antimicrobial substances (Table 2; see supplementary Table 3). Interestingly, genes coding for a copper-efflux system of X. campestris pv. vesicatoria (XCV3747 and XCV3748) were first described to be carried on the same plasmid as AvrBs1 (92), but in strain 85-10, they are located on the chromosome. The known streptomycin resistance of this strain is encoded on a transposon (93). Additionally, we identified genes coding for catalases (five candidates), peroxidases (five candidates), and superoxide dismutases (four candidates), which might function in detoxification of reactive oxygen species produced during the plant defense reaction (see supplementary Table 3).
Conclusions.
In this study, we present the complete genome sequence of X. campestris pv. vesicatoria, which revealed novel insights into the genome plasticity and pathogenicity of this important plant pathogen. All xanthomonads carry a high number of mobile elements; however, their set differs in each strain. Several regions of atypical G+C content are associated with both IS elements and tRNA genes and contain putative pathogenicity determinants which might have been acquired by horizontal gene transfer. The presence of four plasmids, two of which carry a tRNA gene not present on the chromosome, was unexpected. The role of the plasmids in pathogenicity is currently being addressed by plasmid curing.
Intriguingly, X. campestris pv. vesicatoria possesses all known types of protein secretion systems. Previous studies revealed that the TTSS is essential for pathogenicity. Here, several novel type III effectors were predicted and these will be analyzed in the future. In addition, the role of the other secretion systems in the interaction with the plant will be examined with a focus on cell wall-degrading enzymes and adhesins. One of the highlights of our genome analysis is the finding of a candidate Icm/Dot-like type IV secretion system in X. campestris pv. vesicatoria.
We were surprised by the number of genes devoted to regulation, a process that is poorly understood in X. campestris pv. vesicatoria. The identification of two quorum-sensing systems and of a complex and apparently dynamic chemotactic system will stimulate the study of regulation in the context of pathogenicity.
Finally, the first identification of genes possibly involved in adhesion will open new directions in the study of the complete arsenal of virulence factors in X. campestris pv. vesicatoria.
Acknowledgments
We gratefully thank all the people involved in this project.
This work was supported by the GenoMik initiative of the German Federal Ministry of Education and Research (BMBF) and grants to Ulla Bonas, Alfred Pühler, and the competence network center Bielefeld (BMBF grant numbers 031U213D and 0313105). D. H. Nies and G. Mittenhuber were supported by an HWP grant.
REFERENCES
- 1.Alfano, J. R., and A. Collmer. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baar, C., M. Eppinger, G. Raddatz, J. Simon, C. Lanz, O. Klimmek, R. Nandakumar, R. Gross, A. Rosinus, H. Keller, P. Jagtap, B. Linke, F. Meyer, H. Lederer, and S. C. Schuster. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. USA 100:11690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badger, J. H., and G. J. Olsen. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512-524. [DOI] [PubMed] [Google Scholar]
- 5.Bartels, D., S. Kespohl, S. Albaum, T. Druke, A. Goesmann, J. Herold, O. Kaiser, A. Pühler, F. Pfeiffer, G. Raddatz, J. Stoye, F. Meyer, and S. C. Schuster. 2005. BACCardI—a tool for the validation of genomic assemblies, assisting genome finishing and intergenome comparison. Bioinformatics 21:853-859. [DOI] [PubMed] [Google Scholar]
- 6.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, A., F. Katzen, A. Pühler, and L. Ielpi. 1998. Xanthan gum biosynthesis and application: a biochemical/genetic perspective. Appl. Microbiol. Biotechnol. 50:145-152. [DOI] [PubMed] [Google Scholar]
- 8.Bell, K. S., M. Sebaihia, L. Pritchard, M. T. Holden, L. J. Hyman, M. C. Holeva, N. R. Thomson, S. D. Bentley, L. J. Churcher, K. Mungall, R. Atkin, N. Bason, K. Brooks, T. Chillingworth, K. Clark, J. Doggett, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, H. Norbertczak, D. Ormond, C. Price, M. A. Quail, M. Sanders, D. Walker, S. Whitehead, G. P. Salmond, P. R. Birch, J. Parkhill, and I. K. Toth. 2004. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 101:11105-11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeckmann, B., A. Bairoch, R. Apweiler, M. C. Blatter, A. Estreicher, E. Gasteiger, M. J. Martin, K. Michoud, C. O'Donovan, I. Phan, S. Pilbout, and M. Schneider. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonas, U., J. Conrads-Strauch, and I. Balbo. 1993. Resistance in tomato to Xanthomonas campestris pv. vesicatoria is determined by alleles of the pepper-specific avirulence gene avrBs3. Mol. Gen. Genet. 238:261-269. [DOI] [PubMed] [Google Scholar]
- 11.Bonas, U., R. Schulte, S. Fenselau, G. V. Minsavage, B. J. Staskawicz, and R. E. Stall. 1991. Isolation of a gene-cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant-Microbe Interact. 4:81-88. [Google Scholar]
- 12.Bretz, J. R., N. M. Mock, J. C. Charity, S. Zeyad, C. J. Baker, and S. W. Hutcheson. 2003. A translocated protein tyrosine phosphatase of Pseudomonas syringae pv. tomato DC3000 modulates plant defence response to infection. Mol. Microbiol. 49:389-400. [DOI] [PubMed] [Google Scholar]
- 13.Brown, I., J. Mansfield, and U. Bonas. 1995. hrp genes in Xanthomonas campestris pv. vesicatoria determine ability to suppress papillae deposition in pepper mesophyll cells. Mol. Plant-Microbe Interact. 8:825-836. [Google Scholar]
- 14.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burse, A., H. Weingart, and M. S. Ullrich. 2004. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant-Microbe Interact. 17:43-54. [DOI] [PubMed] [Google Scholar]
- 16.Busch, W., and M. H. Saier. 2004. The IUBMB-endorsed transporter classification system. Mol. Biotechnol. 27:253-262. [DOI] [PubMed] [Google Scholar]
- 17.Büttner, D., and U. Bonas. 2002. Getting across—bacterial type III effector proteins on their way to the plant cell. EMBO J. 21:5313-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Büttner, D., and U. Bonas. 2002. Port of entry—the type III secretion translocon. Trends Microbiol. 10:186-192. [DOI] [PubMed] [Google Scholar]
- 19.Büttner, D., L. Noël, F. Thieme, and U. Bonas. 2003. Genomic approaches in Xanthomonas campestris pv. vesicatoria allow fishing for virulence genes. J. Biotechnol. 106:203-214. [DOI] [PubMed] [Google Scholar]
- 20.Canteros, B. J. 1990. Diversity of plasmids and plasmid-encoded phenotypic traits in Xanthomonas campestris pv. vesicatoria. Ph.D. thesis. University of Florida, Gainesville.
- 21.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 23.Ciesiolka, L. D., T. Hwin, J. D. Gearlds, G. V. Minsavage, R. Saenz, M. Bravo, V. Handley, S. M. Conover, H. Zhang, J. Caporgno, N. B. Phengrasamy, A. O. Toms, R. E. Stall, and M. C. Whalen. 1999. Regulation of expression of avirulence gene avrRxv and identification of a family of host interaction factors by sequence analysis of avrBsT. Mol. Plant-Microbe Interact. 12:35-44. [DOI] [PubMed] [Google Scholar]
- 24.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 25.Daniels, M. J., C. E. Barber, P. C. Turner, M. K. Sawczyc, R. J. W. Byrde, and A. H. Fielding. 1984. Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J. 3:3323-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Sluys, N. F. Almeida, L. M. Alves, A. M. Do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. De Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade Dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 27.da Silva, F. G., Y. Shen, C. Dardick, S. Burdman, R. C. Yadav, A. L. de Leon, and P. C. Ronald. 2004. Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol. Plant-Microbe Interact. 17:593-601. [DOI] [PubMed] [Google Scholar]
- 28.Daubin, V., and H. Ochman. 2004. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 14:1036-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Haeze, W., and M. Holsters. 2004. Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 12:555-561. [DOI] [PubMed] [Google Scholar]
- 30.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschröder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 32.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn- helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18: 5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dreier, J., D. Meletzus, and R. Eichenlaub. 1997. Characterization of the plasmid encoded virulence region pat-1 of phytopathogenic Clavibacter michiganensis subsp. michiganensis. Mol. Plant-Microbe Interact. 10:195-206. [DOI] [PubMed] [Google Scholar]
- 34.Espinosa, A., M. Guo, V. C. Tam, Z. Q. Fu, and J. R. Alfano. 2003. The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol. Microbiol. 49:377-387. [DOI] [PubMed] [Google Scholar]
- 35.Fenselau, S., and U. Bonas. 1995. Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol. Plant-Microbe Interact. 8:845-854. [DOI] [PubMed] [Google Scholar]
- 36.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genin, S., and C. Boucher. 2004. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42:107-134. [DOI] [PubMed] [Google Scholar]
- 38.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 39.Gordon, D., C. Desmarais, and P. Green. 2001. Automated finishing with autofinish. Genome Res. 11:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 41.Haft, D. H., B. J. Loftus, D. L. Richardson, F. Yang, J. A. Eisen, I. T. Paulsen, and O. White. 2001. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 29:41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris, M. A., J. Clark, A. Ireland, J. Lomax, M. Ashburner, R. Foulger, K. Eilbeck, S. Lewis, B. Marshall, C. Mungall, J. Richter, G. M. Rubin, J. A. Blake, C. Bult, M. Dolan, H. Drabkin, J. T. Eppig, D. P. Hill, L. Ni, M. Ringwald, R. Balakrishnan, J. M. Cherry, K. R. Christie, M. C. Costanzo, S. S. Dwight, S. Engel, D. G. Fisk, J. E. Hirschman, E. L. Hong, R. S. Nash, A. Sethuraman, C. L. Theesfeld, D. Botstein, K. Dolinski, B. Feierbach, T. Berardini, S. Mundodi, S. Y. Rhee, R. Apweiler, D. Barrell, E. Camon, E. Dimmer, V. Lee, R. Chisholm, P. Gaudet, W. Kibbe, R. Kishore, E. M. Schwarz, P. Sternberg, M. Gwinn, L. Hannick, J. Wortman, M. Berriman, V. Wood, N. de la Cruz, P. Tonellato, P. Jaiswal, T. Seigfried, and R. White. 2004. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 32:258-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hotson, A., R. Chosed, H. Shu, K. Orth, and M. B. Mudgett. 2003. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50:377-389. [DOI] [PubMed] [Google Scholar]
- 45.Hu, N. T., P. F. Lee, and C. Chen. 1995. The type IV pre-pilin leader peptidase of Xanthomonas campestris pv. campestris is functional without conserved cysteine residues. Mol. Microbiol. 18:769-777. [DOI] [PubMed] [Google Scholar]
- 46.Jones, J. B., G. H. Lacy, H. Bouzar, R. E. Stall, and N. W. Schaad. 2004. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 27:755-762. [DOI] [PubMed] [Google Scholar]
- 47.Kaiser, O., D. Bartels, T. Bekel, A. Goesmann, S. Kespohl, A. Pühler, and F. Meyer. 2003. Whole genome shotgun sequencing guided by bioinformatics pipelines—an optimized approach for an established technique. J. Biotechnol. 106:121-133. [DOI] [PubMed] [Google Scholar]
- 48.Kanehisa, M., S. Goto, S. Kawashima, and A. Nakaya. 2002. The KEGG databases at GenomeNet. Nucleic Acids Res. 30:42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katzen, F., D. U. Ferreiro, C. G. Oddo, M. V. Ielmini, A. Becker, A. Pühler, and L. Ielpi. 1998. Xanthomonas campestris pv. campestris gum mutants: effects on xanthan biosynthesis and plant virulence. J. Bacteriol. 180:1607-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kearney, B., and B. J. Staskawicz. 1990. Characterization of IS476 and its role in bacterial spot disease of tomato and pepper. J. Bacteriol. 172: 143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim, J. F., J. H. Ham, D. W. Bauer, A. Collmer, and S. V. Beer. 1998. The hrpC and hrpN operons of Erwinia chrysanthemi EC16 are flanked by plcA and homologs of hemolysin/adhesin genes and accompanying activator/transporter genes. Mol. Plant-Microbe Interact. 11:563-567. [DOI] [PubMed] [Google Scholar]
- 52.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 53.Lee, B. M., Y. J. Park, D. S. Park, H. W. Kang, J. G. Kim, E. S. Song, I. C. Park, U. H. Yoon, J. H. Hahn, B. S. Koo, G. B. Lee, H. Kim, H. S. Park, K. O. Yoon, J. H. Kim, C. H. Jung, N. H. Koh, J. S. Seo, and S. J. Go. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leyns, F., M. De Cleene, J. Swings, and J. De Ley. 1984. The host range of the genus Xanthomonas. Bot. Rev. 50:305-355. [Google Scholar]
- 55.Lindeberg, M., J. Stavrinides, J. H. Chang, J. R. Alfano, A. Collmer, J. L. Dangl, J. T. Greenberg, J. W. Mansfield, and D. S. Guttman. 2005. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:275-282. [DOI] [PubMed] [Google Scholar]
- 56.Locht, C., R. Antoine, and F. Jacob-Dubuisson. 2001. Bordetella pertussis, molecular pathogenesis under multiple aspects. Curr. Opin. Microbiol. 4:82-89. [DOI] [PubMed] [Google Scholar]
- 57.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McHardy, A. C., A. Goesmann, A. Pühler, and F. Meyer. 2004. Development of joint application strategies for two microbial gene finders. Bioinformatics 20:1622-1631. [DOI] [PubMed] [Google Scholar]
- 59.Meinhart, A., J. C. Alonso, N. Strater, and W. Saenger. 2003. Crystal structure of the plasmid maintenance system epsilon/zeta: functional mechanism of toxin zeta and inactivation by epsilon 2 zeta 2 complex formation. Proc. Natl. Acad. Sci. USA 100:1661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Metz, M., D. Dahlbeck, C. Q. Morales, B. Al Sady, E. T. Clark, and B. J. Staskawicz. 2005. The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana. Plant J. 41:801-814. [DOI] [PubMed] [Google Scholar]
- 61.Meyer, A., A. Pühler, and K. Niehaus. 2001. The lipopolysaccharides of the phytopathogen Xanthomonas campestris pv. campestris induce an oxidative burst reaction in cell cultures of Nicotiana tabacum. Planta 213:214-222. [DOI] [PubMed] [Google Scholar]
- 62.Meyer, F., A. Goesmann, A. C. McHardy, D. Bartels, T. Bekel, J. Clausen, J. Kalinowski, B. Linke, O. Rupp, R. Giegerich, and A. Pühler. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minsavage, G. V., D. Dahlbeck, M. C. Whalen, B. Kearny, U. Bonas, B. J. Staskawicz, and R. E. Stall. 1990. Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria-pepper interactions. Mol. Plant-Microbe Interact. 3:41-47. [Google Scholar]
- 64.Molinaro, A., A. Evidente, S. Fiore, N. S. Iacobellis, R. Lanzetta, and M. Parrilli. 2000. Structure elucidation of the O-chain from the major lipopolysaccharide of the Xanthomonas campestris strain 642. Carbohydr. Res. 325:222-229. [DOI] [PubMed] [Google Scholar]
- 65.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, D. Barrell, A. Bateman, D. Binns, M. Biswas, P. Bradley, P. Bork, P. Bucher, R. R. Copley, E. Courcelle, U. Das, R. Durbin, L. Falquet, W. Fleischmann, S. Griffiths-Jones, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, R. Lopez, I. Letunic, D. Lonsdale, V. Silventoinen, S. E. Orchard, M. Pagni, D. Peyruc, C. P. Ponting, J. D. Selengut, F. Servant, C. J. Sigrist, R. Vaughan, and E. M. Zdobnov. 2003. The InterPro database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31:315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Newman, M. A., M. J. Daniels, and J. M. Dow. 1995. Lipopolysaccharide from Xanthomonas campestris induces defense-related gene expression in Brassica campestris. Mol. Plant-Microbe Interact. 8:778-780. [DOI] [PubMed] [Google Scholar]
- 67.Nielsen, H., and A. Krogh. 1998. Prediction of signal peptides and signal anchors by a hidden Markov model, p. 122-130. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB 6). AAAI Press, Menlo Park, Calif. [PubMed]
- 68.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 69.Nies, D. H. 2004. Incidence and function of sigma factors in Ralstonia metallidurans and other bacteria. Arch. Microbiol. 181:255-268. [DOI] [PubMed] [Google Scholar]
- 70.Noël, L., F. Thieme, J. Gäbler, D. Büttner, and U. Bonas. 2003. XopC and XopJ, two novel type III effector proteins from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 185:7092-7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noël, L., F. Thieme, D. Nennstiel, and U. Bonas. 2001. cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 41:1271-1281. [DOI] [PubMed] [Google Scholar]
- 72.Noël, L., F. Thieme, D. Nennstiel, and U. Bonas. 2002. Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nummelin, H., M. C. Merckel, J. C. Leo, H. Lankinen, M. Skurnik, and A. Goldman. 2004. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 23:701-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ojanen-Reuhs, T., N. Kalkkinen, B. Westerlund-Wikstrom, J. van Doorn, K. Haahtela, E. L. Nurmiaho-Lassila, K. Wengelnik, U. Bonas, and T. K. Korhonen. 1997. Characterization of the fimA gene encoding bundle-forming fimbriae of the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 179:1280-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Osbourn, A. E., B. R. Clarke, and M. J. Daniels. 1990. Identification and DNA sequence of a pathogenicity gene of Xanthomonas campestris pv. campestris. Mol. Plant-Microbe Interact. 3:280-285. [DOI] [PubMed] [Google Scholar]
- 76.Pedersen, A. G., L. J. Jensen, S. Brunak, H. H. Staerfeldt, and D. W. Ussery. 2000. A DNA structural atlas for Escherichia coli. J. Mol. Biol. 299:907-930. [DOI] [PubMed] [Google Scholar]
- 77.Poplawsky, A. R., and W. Chun. 1997. pigB determines a diffusible factor needed for extracellular polysaccharide slime and xanthomonadin production in Xanthomonas campestris pv. campestris. J. Bacteriol. 179:439-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poplawsky, A. R., and W. Chun. 1998. Xanthomonas campestris pv. campestris requires a functional pigB for epiphytic survival and host infection. Mol. Plant-Microbe Interact. 11:466-475. [DOI] [PubMed] [Google Scholar]
- 79.Ray, S. K., R. Rajeshwari, Y. Sharma, and R. V. Sonti. 2002. A high-molecular-weight outer membrane protein of Xanthomonas oryzae pv. oryzae exhibits similarity to non-fimbrial adhesins of animal pathogenic bacteria and is required for optimum virulence. Mol. Microbiol. 46: 637-647. [DOI] [PubMed] [Google Scholar]
- 80.Rimsky, S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7:109-114. [DOI] [PubMed] [Google Scholar]
- 81.Robinson, C., and A. Bolhuis. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2:350-356. [DOI] [PubMed] [Google Scholar]
- 82.Roden, J. A., B. Belt, J. B. Ross, T. Tachibana, J. Vargas, and M. B. Mudgett. 2004. A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. USA 101:16624-16629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rojas, C. M., J. H. Ham, W. L. Deng, J. J. Doyle, and A. Collmer. 2002. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. USA 99:13142-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Römling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205-212. [DOI] [PubMed] [Google Scholar]
- 85.Ronald, P. C., and B. J. Staskawicz. 1988. The avirulence gene avrBs1 from Xanthomonas campestris pv. vesicatoria encodes a 50-kDa protein. Mol. Plant-Microbe Interact. 1:191-198. [PubMed] [Google Scholar]
- 86.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 87.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 88.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sandkvist, M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. Briones, M. R. Bueno, A. A. Camargo, L. E. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. Costa, C. M. Costa-Neto, L. L. Coutinho, M. Cristofani, E. Dias-Neto, C. Docena, H. El-Dorry, A. P. Facincani, A. J. Ferreira, V. C. Ferreira, J. A. Ferro, J. S. Fraga, S. C. Franca, M. C. Franco, M. Frohme, L. R. Furlan, M. Garnier, G. H. Goldman, M. H. Goldman, S. L. Gomes, A. Gruber, P. L. Ho, J. D. Hoheisel, M. L. Junqueira, E. L. Kemper, J. P. Kitajima, J. E. Krieger, E. E. Kuramae, F. Laigret, M. R. Lambais, L. C. Leite, E. G. Lemos, M. V. Lemos, S. A. Lopes, C. R. Lopes, J. A. Machado, M. A. Machado, A. M. Madeira, H. M. Madeira, C. L. Marino, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 91.Stall, R. E. 1995. Xanthomonas campestris pv. vesicatoria, p. 167-184. In R. P. Singh, U. S. Singh, and K. Kohmoto (ed.), Pathogenesis and host-parasite specificity in plant diseases, vol. I. Prokaryotes. Pergamon, Elsevier Science Inc., Tarrytown, New York. [Google Scholar]
- 92.Stall, R. E., D. C. Loschke, and J. B. Jones. 1986. Linkage of copper resistance and avirulence loci on a self-transmissible plasmid in Xanthomonas campestris pv. vesicatoria. Phytopathology 76:240-243. [Google Scholar]
- 93.Sundin, G. W., and C. L. Bender. 1995. Expression of the strA-strB streptomycin resistance genes in Pseudomonas syringae and Xanthomonas campestris and characterization of IS6100 in X. campestris. Appl. Environ. Microbiol. 61:2891-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Suzek, B. E., M. D. Ermolaeva, M. Schreiber, and S. L. Salzberg. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17:1123-1130. [DOI] [PubMed] [Google Scholar]
- 95.Swords, K. M., D. Dahlbeck, B. Kearney, M. Roy, and B. J. Staskawicz. 1996. Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2. J. Bacteriol. 178:4661-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Szurek, B., O. Rossier, G. Hause, and U. Bonas. 2002. Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol. Microbiol. 46:13-23. [DOI] [PubMed] [Google Scholar]
- 97.Tampakaki, A. P., V. E. Fadouloglou, A. D. Gazi, N. J. Panopoulos, and M. Kokkinidis. 2004. Conserved features of type III secretion. Cell. Microbiol. 6:805-816. [DOI] [PubMed] [Google Scholar]
- 98.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ulrich, L. E., E. V. Koonin, and I. B. Zhulin. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 13:52-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Doorn, J., P. M. Boonekamp, and B. Oudega. 1994. Partial characterization of fimbriae of Xanthomonas campestris pv. hyacinthi. Mol. Plant-Microbe Interact. 7:334-344. [DOI] [PubMed] [Google Scholar]
- 101.Vauterin, L., J. Rademaker, and J. Swings. 2000. Synopsis on the taxonomy of the genus Xanthomonas. Phytopathology 90:677-682. [DOI] [PubMed] [Google Scholar]
- 102.Vivian, A., J. Murillo, and R. W. Jackson. 2001. The roles of plasmids in phytopathogenic bacteria: mobile arsenals? Microbiology 147:763-780. [DOI] [PubMed] [Google Scholar]
- 103.Von Bodman, S. B., W. D. Bauer, and D. L. Coplin. 2003. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41:455-482. [DOI] [PubMed] [Google Scholar]
- 104.Vorhölter, F. J., K. Niehaus, and A. Pühler. 2001. Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: a cluster of 15 genes is involved in the biosynthesis of the LPS O-antigen and the LPS core. Mol. Genet. Genomics 266:79-95. [DOI] [PubMed] [Google Scholar]
- 105.Wang, L. H., Y. He, Y. Gao, J. E. Wu, Y. H. Dong, C. He, S. X. Wang, L. X. Weng, J. L. Xu, L. Tay, R. X. Fang, and L. H. Zhang. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51:903-912. [DOI] [PubMed] [Google Scholar]
- 106.Weng, S. F., Y. F. Fan, Y. H. Tseng, and J. W. Lin. 1997. Sequence analysis of the small cryptic Xanthomonas campestris pv. vesicatoria plasmid pXV64 encoding a Rep protein similar to gene II protein of phage I2-2. Biochem. Biophys. Res. Commun. 231:121-125. [DOI] [PubMed] [Google Scholar]
- 107.Wengelnik, K., and U. Bonas. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178:3462-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wengelnik, K., O. Rossier, and U. Bonas. 1999. Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 181:6828-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wengelnik, K., G. Van den Ackerveken, and U. Bonas. 1996. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol. Plant-Microbe Interact. 9:704-712. [DOI] [PubMed] [Google Scholar]
- 110.Whalen, M. C., J. F. Wang, F. M. Carland, M. E. Heiskell, D. Dahlbeck, G. V. Minsavage, J. B. Jones, J. W. Scott, R. E. Stall, and B. J. Staskawicz. 1993. Avirulence gene avrRxv from Xanthomonas campestris pv. vesicatoria specifies resistance on tomato line Hawaii 7998. Mol. Plant-Microbe Interact. 6:616-627. [DOI] [PubMed] [Google Scholar]
- 111.Zamboni, D. S., S. McGrath, M. Rabinovitch, and C. R. Roy. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49:965-976. [DOI] [PubMed] [Google Scholar]
- 112.Zhao, B., E. Y. Ardales, A. Raymundo, J. Bai, H. N. Trick, J. E. Leach, and S. H. Hulbert. 2004. The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1. Mol. Plant-Microbe Interact. 17:771-779. [DOI] [PubMed]