Abstract
Five missense mutations of the winged-helix FOXC1 transcription factor, found in patients with Axenfeld-Rieger (AR) malformations, were investigated for their effects on FOXC1 structure and function. Molecular modeling of the FOXC1 forkhead domain predicted that the missense mutations did not alter FOXC1 structure. Biochemical analyses indicated that, whereas all mutant proteins correctly localize to the cell nucleus, the I87M mutation reduced FOXC1-protein levels. DNA-binding experiments revealed that, although the S82T and S131L mutations decreased DNA binding, the F112S and I126M mutations did not. However, the F112S and I126M mutations decrease the transactivation ability of FOXC1. All the FOXC1 mutations had the net effect of reducing FOXC1 transactivation ability. These results indicate that the FOXC1 forkhead domain contains separable DNA-binding and transactivation functions. In addition, these findings demonstrate that reduced stability, DNA binding, or transactivation, all causing a decrease in the ability of FOXC1 to transactivate genes, can underlie AR malformations.
Introduction
The forkhead/winged-helix family of transcription factors is required for a variety of developmental processes, including embryogenesis and tissue-specific cell differentiation, as well as for other biologically important events, such as tumorigenesis (Kaufmann and Knochel 1996). These transcription factors contain a monomeric, 110-amino-acid DNA-binding domain, first identified as a region of homology between the Drosophila melanogaster protein fork head and rat hepatocyte nuclear factor 3 proteins (Weigel and Jackle 1990). Forkhead domains are evolutionarily conserved and exist in a wide range of species, from yeast to human (Kaufmann and Knochel 1996). This DNA-binding motif is a variant of the helix-turn-helix motif and consists of three α helices and two large loops that form “wing” structures, hence the name “winged-helix.”
FOXC1 (forkhead box [MIM 601090]) is a member of this winged-helix family of transcription factors (Larsson et al. 1995). Mutations in FOXC1 (formerly known as “FREAC3” and “FKHL7”) underlie Axenfeld-Rieger (AR) anterior eye-segment defects mapping to chromosome 6p25 (Mears et al. 1998; Nishimura et al. 1998; Mirzayans et al. 2000). Patients with AR malformations who have FOXC1 mutations typically show a spectrum of ocular findings, including iris hypoplasia, a prominent Schwalbe line, iris adhesions, and goniodysgenesis. The maldevelopment of the iridocorneal angle, through which the aqueous humor must pass, can result in increased intraocular pressure. Elevated intraocular pressure increases the potential for atrophy of the optic nerve and for retinal ganglion cell death, thereby raising the risk of development of glaucoma. These abnormalities in iridocorneal-angle development are thought to arise from a defect in the migration and/or differentiation of mesenchymal cells that contribute to the anterior segment of the eye (Kume et al. 1998).
Patients with AR malformations who have FOXC1 mutations may also present with nonocular findings including cardiac defects and dental dysgenesis (Swiderski et al. 1999; Winnier et al. 1999; Mirzayans et al. 2000). The systemic phenotypes suggest that FOXC1 has a broad role in the developmental process. FOXC1 is expressed in fetal human tissues and is widely expressed in adult human tissues (Pierrou et al. 1994; Mears et al. 1998; Nishimura et al. 1998). The closely related murine gene, Foxc1, is also expressed embryonically in a wide range of tissues, including the periocular mesenchyme, cornea, and developing skeletal and organ structures such as the heart and kidney (Hiemisch et al. 1998; Kume et al. 1998, 2000). Foxc1−/− homozygous mutant mice die at birth, with hydrocephalus and skeletal and eye defects, including an absent anterior chamber and open or absent eyelids (Kume et al. 1998; Hong et al. 1999; Kidson et al. 1999). Similar studies of Foxc1+/− heterozygotes have shown that these mice have anterior eye-segment defects similar to those found in human patients with FOXC1 mutations, including iris hypoplasia, a displaced Schwalbe line, and iridocorneal-angle dysgenesis (Smith et al. 2000).
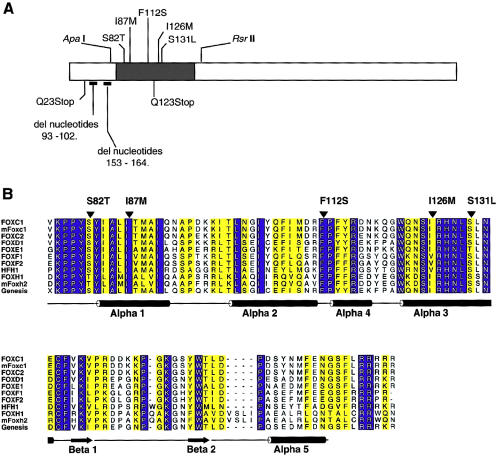
Two nonsense mutations and two deletions, all resulting in frameshift mutations, have been reported in FOXC1 (fig. 2A; Kume et al. 1998; Mears et al. 1998; Nishimura et al. 1998; Mirzayans et al. 2000), producing truncated products that are likely to be rapidly degraded by cellular machinery. There also have been five reported missense mutations of FOXC1 found in patients with AR malformations (fig. 2A; Mears et al. 1998; Nishimura et al. 1998), which provide a valuable tool for initiation of a functional dissection of FOXC1.
Figure 2.
A, Schematic of the FOXC1 protein. The blackened rectangle represents the forkhead domain, and the missense mutations studied are indicated above the forkhead domain. Mutations presented below the ideogram result in truncated products and were not tested. Positions of restriction-enzyme sites used in subcloning are indicated above the ideogram. B, Multiple-sequence alignment of the forkhead domain of human FOXC1 and related FOX proteins. The sequences shown in single-letter amino acid codes are those of human FOXC1 (AF048693), mouse Foxc1 (NM008592), human FOXC2 (Y08223), human FOXD1 (U13222), human FOXE1 (U89995), human FOXF1 (U13219), human FOXF2 (U13220), human HFH1 (AF153341), human FOXH1 (AF076292), mouse Foxh2 (AF110506), and rat Genesis/Foxd3 (NM012183), respectively (NCBI Databases) . Amino acid residues showing absolute identity among these proteins are shown in white against a blue background; those positions with conservative substitutions are shown against a yellow background. The positions of the α-helices and β-strands, as defined in the NMR structure of Genesis, are schematically represented below the alignment. Positions of mutations and corresponding amino acid changes in FOXC1 are indicated by the blackened arrowheads above the alignment. ALSCRIPT was used to format the alignment.
Missense mutations often modify the function of a transcription factor; for example, PITX2 (MIM 601542) is a member of the paired bicoid family of transcription factors and has been shown to underlie anterior eye-segment defects mapping to chromosome 4q25 (Semina et al. 1996; Alward et al. 1998; Kulak et al. 1998). Missense mutations reduce the ability of PITX2 to bind DNA and to transactivate reporter genes, with the severity of the defects corresponding to the residual binding capacity of the PITX2 mutant proteins (Kozlowski and Walter 2000). One of the missense mutations of PITX2 has the additional effect of reducing the localization of PITX2 to the nucleus. Similarly, in Oct1 (MIM 602607), a POU-class transcription factor, a single missense mutation can alter the protein-protein interactions that are required for Oct1 to activate genes (Lai et al. 1992). We therefore investigated the way in which the naturally occurring missense mutations within FOXC1 adversely affect FOXC1 structure and/or function. We used molecular modeling, cell biology, and biochemistry to assay the effects that missense mutations of FOXC1 have on the ability of mutant FOXC1 proteins to adopt wild-type conformations, localize to the nucleus, bind DNA, and activate gene expression.
Patients, Material, and Methods
Reports on Patients
All five families with FOXC1 missense mutations presented with AR malformations. The clinical features of these patients have been reported elsewhere (Mears et al. 1998; Nishimura et al. 1998; Swiderski et al. 1999; Winnier et al. 1999).
Threading Analysis
Threading experiments were performed by the method of Bryant and Lawrence (1993), with detailed derivations and methodology provided therein. Each query sequence was threaded through the atomic coordinates of the solution structure of the forkhead domain, of the rat forkhead protein Genesis, bound to its target DNA (Jin et al. 1999). Five core segments (CS) were defined on the basis of the nuclear magnetic resonance (NMR) structure: CS1 spanned residues 10–20, CS2 spanned residues 24–34, CS3 spanned residues 47–56, CS4 spanned residues 60–63, and CS5 spanned residues 73–97 (the numbering is based on the sequence of Genesis [pdb:2HDC A], as presented in fig. 2B). For each possible alignment, individual pairwise residue interactions were determined on the basis of chemical type and distance intervals, the tables for which have been published by Bryant and Lawrence (1993). Through use of these values, a conformational energy, ΔGR|M, defined as the expected work required for substitution of a specific sequence R for a random sequence with the same composition, in the context of folding-motif M, was then calculated for each alignment (table 1). Z-scores (ZR|M) and chance-occurrence probabilities (ER|M) were calculated to compare conformational energies for different alignments. Chance-occurrence probabilities give the odds that a random sequence of the same length and amino acid composition would yield a threading energy as low as the query sequence R. Calculations of energies and of statistical significance were performed with C and S-PLUS subroutines (Becker et al. 1988). Critical interactions are defined as those having a pairwise residue interaction energy <−1 kcal/mol.
Table 1.
Statistics for Optimal Threads of Forkhead Domain–Containing Proteins
| Protein | ΔGR|Ma | ZR|Mb | ER|M |
| FOXC1 | 102.16 | 5.28 | 1.06×10-3 |
| S82T | 103.04 | 5.37 | 5.56×10-5 |
| I87M | 100.58 | 5.23 | 1.56×10-3 |
| F112S | 105.12 | 5.51 | 3.70×10-4 |
| I126M | 99.02 | 5.15 | 5.39×10-3 |
| S131L | 104.94 | 5.37 | 8.28×10-4 |
| Genesisc | 95.69 | 5.03 | 9.43×10-3 |
Sums of contact potentials are expressed as ΔGR|M , the energy associated with nonlocal, nonbonded interactions (Bryant and Lawrence 1993).
Z-scores, in SD units, represent the variance from the mean of ΔGR|M.
Statistics on Genesis represent the “self thread”—that is, the alignment of the Genesis sequence against its own structure.
Plasmid Construction and Mutagenesis
FOXC1 cDNA (a gift from P. Carlsson) was repaired at the 3′ end so that the FOXC1 cDNA encoded the entire FOXC1 open reading frame. The FOXC1 cDNA was subcloned into EcoRI–XbaI sites in the pcDNA4 His/Max B™ plasmid (Invitrogen). The high G-C content of FOXC1 precluded mutagenesis of the entire cDNA (data not shown), so an additional cloning step was required. An ApaI–RsrII FOXCI fragment (fig. 2A) encompassing the forkhead domain was cloned into pBluescript and was mutagenized with the Quickchange™ mutagenesis kit (Stratagene) and with appropriate primers, according to the manufacturer's protocol, with the addition of 5% dimethyl sulfoxide. The resulting protein change, with the nucleotide change in square brackets and the primer sequences in parentheses, are as follows: S82T [G245C] forward (5′-GGTGAAGCCGCCCTATACCTACATCGCGCTCATCACC-3′) and reverse (5′-GGTGATGAGCGCGATGTAGGTATAGGGCGGCTTCACC-3′); I87M [C261G] forward (5′-GCTACATCGCGCTCATGACCATGGCCATCCAG-3′) and reverse (5′-CTGGATGGCCATGGTCATGAGCGCGATGTAGC-3′); F112S [T335C] forward (5′-CCAGTTCATCATGGACCGCTCCCCCTTCTACCGGG-3′) and reverse (5′-CCCGGTAGAAGGGGGAGCGGTCCATGATGAACTGG-3′); I126M [C378G] forward (5′-GGCTGGCAGAACAGCATGCGCCACAACCTCTCG-3′) and reverse (5′-CGAGAGGTTGTGGCGCATGCTGTTCTGCCAGCC-3′); and S131L [C392T] forward (5′-CCGCCACAACCTCTTGCTCAACGAGTGCTTCG-3′) and reverse (5′-CGAAGCACTCGTTGAGCAAGAGGTTGTGGCGG-3′).
Potential mutant constructs were sequenced with an automated sequencer (Li-COR). Subclones that were confirmed to contain the required mutation were subcloned back into the FOXC1 pcDNA His/Max vector, and these final clones were sequenced.
Cell-Transfection and -Cotransfection Assays
One-hundred-millimeter plates of COS-7 cells grown in Dulbecco’s modified Eagle’s medium and 10% fetal bovine serum were transfected, at 80% confluency, with 2 μg of plasmid DNA, through use of FuGene6™ transfection reagent (Roche) according to the manufacturer's protocol. For FOXC1/Lac Z cotransfection assays, 1 μg each of pcDNA4 His/Max Lac Z and of either pcDNA4 His/Max FOXC1 or pcDNA4 His/Max FOXC1 I87M were cotransfected into COS-7 cells. After 48 h, proteins were extracted and analyzed by western analysis.
Protein Extraction, Western Analysis, and Immunofluorescence
Forty-eight hours after transfection, cells were washed with PBS and were harvested by scraping. Cells were pelleted; were resuspended in lysis buffer (20 mM HEPES pH 7.6, 150 mM NaCl, 0.5 mM DTT, 25% glycerol, 2.5 mM phenylmethylsulfonyl fluoride, 10 μg of Aprotinin/μl, 9 μg of Leupeptin/μl, and 10 μg of Pepstatin A/μl) at 4°C; and were lysed by gentle sonication on ice. After centrifugation at 13,000 g for 5 min at 4°C, supernatants were transferred to a microfuge tube and were resolved by SDS-PAGE. Western analysis and immunofluorescence were performed as described by Kozlowski and Walter (2000).
Northern Analysis
Forty-eight hours after transfection, COS-7 cells were washed with PBS, and RNA was extracted through use of TRIzol (Gibco/BRL) reagent. The RNA was size-separated on a 1 × 3-(N-morpholino) propane sulfonic acid pH 7.0, 0.66 M formaldehyde (BDH) agarose gel and was transferred to Hybond (Amersham). The Hybond blot was hybridized with [32P]-dCTP–labeled pcDNA4 His/Max B linearized with EcoRI. A second hybridization of the blot was performed with a [32P]-dCTP–labeled PCR product from the 5′ FOXC1 (nucleotides 20–233). Control hybridization for loading amounts was performed with [32P]-dCTP–labeled S26 ribosomal-protein RNA (Vincent et al. 1993).
Electrophoretic Mobility–Shift Assay (EMSA)
COS-7 cell extracts containing recombinant FOXC1 were equalized for amounts of recombinant FOXC1 protein, by western analysis. Reactions were brought up to volume through use of untransfected COS-7 cell extract. Protein extracts were incubated with 1.3 mM DTT, 5 μg of sheared salmon-sperm DNA, 1 μg of poly dIdC (Sigma), and 10,000 cpm of [32P]-dCTP–labeled double-stranded DNA containing the FOXC1-binding site (forward, 5′-GATCCAAAGTAAATAAACAACAGA-3′; and reverse, 5′-GATCTCTGTTGTTTATTTACTTTG-3′). Reactions were incubated at room temperature for 15 min, after which 3 μl of 50% glycerol was added. Six-percent polyacrylamide Tris-glycine-EDTA gels were prerun for 15 min, and the products of the reactions were subjected to electrophoresis at room temperature for 50 min. Binding-specificity EMSAs were performed as described above, with the addition of the [32P]-dCTP–labeled double-stranded DNA oligonucleotides listed in table 2.
Table 2.
The FOXC1 Binding Site and Variant Oligonucleotides
|
Sequencea |
||
| Oligonucleotide | Forward | Reverse |
| FOXC1 site | GATCCAAAGTAAATAAACAACAGA | GATCTCTGTTGTTTATTTACTTTG |
| Variant: | ||
| 1 | GATCCAAACTAAATAAACAACAGA | GATCTCTGTTGTTTATTTAGTTTG |
| 2 | GATCCAAAGAAAATAAACAACAGA | GATCTCTGTTGTTTATTTTCTTTG |
| 3 | GATCCAAAGTTAATAAACAACAGA | GATCTCTGTTGTTTATTAACTTTG |
| 4 | GATCCAAAGTATATAAACAACAGA | GATCTCTGTTGTTTATATACTTTG |
| 5 | GATCCAAAGTAATTAAACAACAGA | GATCTCTGTTGTTTAATTACTTTG |
| 6 | GATCCAAAGTAAAAAAACAACAGA | GATCTCTGTTGTTTTTTTACTTTG |
| 7 | GATCCAAAGTAAATTAACAACAGA | GATCTCTGTTGTTAATTTACTTTG |
| 8 | GATCCAAAGTAAATATACAACAGA | GATCTCTGTTGTATATTTACTTTG |
| 9 | GATCCAAAGTAAATAATCAACAGA | GATCTCTGTTGATTATTTACTTTG |
The 9-bp core sequence of the in vitro–derived FOXC1 site (Pierrou et al. 1994) and of the variants is underlined; the converted base in each oligonucleotide is shown in boldface italics. Purines were converted to pyrimidines with an equivalent number of hydrogen bonds, and pyrimidines were converted to purines with an equivalent number of hydrogen bonds.
Dual-Luciferase Assay
The dual-luciferase assay combines two reporter enzymes into an integrated, single-tube, dual-reporter assay format. The system provides sequential quantification of both Photinus pyralis luciferase (reporter) and Renilla reniformis luciferase (control) and takes advantage of the luciferases’ different substrate requirements, making it possible to differentiate between their respective bioluminescent reactions.
The cytomegalovirus promoter of pGL3 (Promega) was replaced with a herpes simplex–virus thymidine kinase (TK) promoter, from pRLTK (Promega), which was cloned into the BglII-HindIII sites of pGL3. Six copies of the FOXC1-binding sites (underlining indicates the core, 9-bp FOXC1-binding site: forward, 5′-CTAGCCAAAGTAAATAAACAACAGCAAAGTAAATAAACAACAGG-3′; and reverse, 5′-CTAGCCTGTTGTTTATTTACTTTGCTGTTGTTTATTTACTTTGG-3′) were then cloned into the EcoRI- NheI sites, 5′ to the TK promoter. HeLa cells were then transfected with 50 ng of the pGL3 TK construct, 1 ng of the Renilla control vector, and 500 ng of a given FOXC1 pcDNA4 His/Max construct. Transfected cells were grown for 48 h. The dual-luciferase assays were performed with the Promega Dual Luciferase Assay kit, according to the manufacturer’s protocol (Promega). Reactions were replicated a minimum of three times.
Results
Threading Experiments on the Forkhead Domain of FOXC1
To study FOXC1 and understand the effects that missense mutations have on FOXC1 structure, molecular models of the FOXC1 forkhead domain were generated through use of threading algorithms developed by Bryant and Lawrence (1993). Threading methods such as the one employed here can be used to predict whether a given protein sequence has the potential to adopt the known three-dimensional structure of a wild-type protein. This computational technique identifies the alignments, between a query sequence and a given folding motif, that are most likely to represent stable conformational states, on the basis of implied pairwise and hydrophobic interactions of residues that are nonlocal in the sequence (Bryant and Lawrence 1993; Fetrow and Bryant 1993).
The three-dimensional structure of the rat Genesis protein bound with its target DNA (2HDC, chain A; Jin et al. 1999) was used as a template for the threading analysis of FOXC1. The positions of the α-helices in the NMR structure of Genesis were used to define the core segments within this motif. The core segments, therefore, represent the secondary-structural regions of the protein and exclude most of the flexible regions. All possible placements of the core segments along the query sequence were considered in light of the constraints of sequence length, core segment length, and limits of loop length. Threading-contact energies were corrected for sequence-composition bias, by random shuffling of the aligned residues, to generate composition-corrected threading scores (i.e., ZR|M). To evaluate the statistical significance of computed threading scores, 100 random permutations of the query sequence were generated, and the alignment-optimization procedure was then repeated on these now-shuffled sequences. On the basis of this collection of scores, ER|M was calculated. The results with the most favorable conformational energies (i.e., those with the lowest ΔGR|M values) were selected for further study. A summary of the threading results is shown in table 1. The calculated conformational energies of the “self-thread” of Genesis and FOXC1 are only slightly different from one another (by ∼6 kcal/mol). More important, the probability values (i.e., ER|M) for both structures meet the statistical criteria of being ⩾.05 (5%), corresponding to 95% confidence in the threading prediction. As such, it can be concluded that there is a statistically significant alignment of the sequence of FOXC1 with the structure of Genesis.
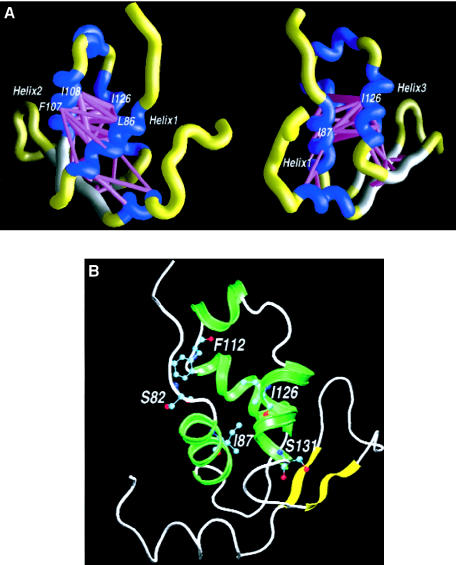
Figure 1A shows the energy scaffolds generated in this threading experiment for the forkhead domain of FOXC1. The energy scaffolds provide a method for visualization of the important intramolecular interactions taking place within a protein. Here, the winged-helical bundle can be seen to be held together by numerous hydrophobic interactions, represented by the thick, magenta-colored cylinders. Several of the highly conserved, large hydrophobic residues in this protein are involved in the maintenance of these interactions within the hydrophobic core, which occur primarily between the conserved residues at the helical interfaces. The energy scaffolds indicate that the most favorable hydrophobic interactions observed in the threading model of the FOXC1 forkhead domain involve Leu 86 and Ile 87 in helix 1; Ile 104, Phe 107, and Ile 108 in helix 2; Ile 126 and Leu 130 in helix 3; and Phe 151 and Trp152 in the last β-strand. The amino acid residues involved in these critical pairwise interactions are conserved absolutely between FOXC1 and Genesis.
Figure 1.
A, Energy scaffolds for the forkhead domain of FOXC1. The α-carbon backbone of the protein is depicted as a curving “worm.” Within the backbone, segments of the forkhead domain that comprise the core folding motifs are shown in blue, whereas the intervening loop regions are shown in yellow. Helices and loop regions are as defined in figure 2B. Pairwise residue-interaction energies between core residues (Bryant and Lawrence 1993) are shown by the width and coloring of the connected α-carbon positions on the protein backbone. Indicated interactions are limited to those with pairwise residue-interaction energies ⩽−1 kcal/mol (“critical” pairwise interactions). Thick, magenta-colored cylinders denote the most favorable interactions; cylinders of intermediate thickness denote interactions with lower, less-favorable pairwise energies. Scaffolds were generated by the graphics program GRASP (Nicholls et al. 1991). The amino acid numbering corresponds to that in the multiple-sequence alignment presented in figure 2B. The second view is 70° counterclockwise around the Y-axis, with respect to the first view. B, Molecular model of the forkhead domain of FOXC1. A ribbon model of the backbone of FOXC1 is shown, with the side chains of the mutated amino acid residues shown in a “ball-and-sticks” representation. The missense mutations in FOXC1 shown here are S82T at the N-terminal end of helix 1, I87M in helix 1, F112S at the C-terminal end of helix 2, and I126M and S131L in helix 3. The numbering in the figure corresponds to that of the human FOXC1 sequence.
To further assess the structural consequences of FOXC1 mutations, threading analyses were performed on all of the five missense mutations analyzed in this study. All of these five missense mutations produced threading scores similar to that of the wild-type FOXC1. None of the mutations significantly altered any critical pairwise residue interactions or destabilized the forkhead domain of FOXC1 (table 1), suggesting that these missense mutations do not significantly alter the structure of FOXC1. Since these missense mutations have only subtle structural significance, the consequences that these five mutations have on FOXC1 function were investigated.
Expression of FOXC1
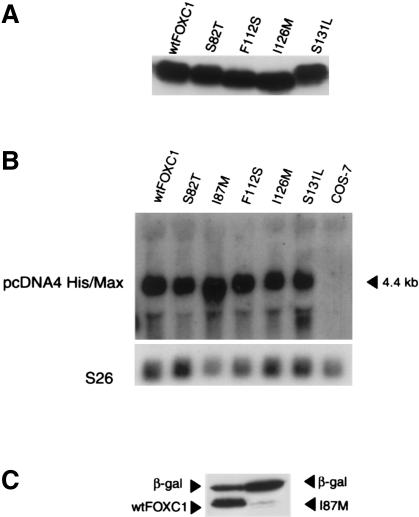
All five missense mutations occur within the forkhead domain of FOXC1 (fig. 2A). COS-7 cells were transfected with the vectors encoding wild-type and mutated FOXC1. Whole-cell extracts of the transfected COS-7 cells were resolved by SDS-PAGE and immunoblot analysis. Detection of the N-terminal, vector-encoded, Xpress epitope demonstrated a stable product, ∼65 kD in size, for wild-type FOXC1 and for FOXC1 containing four of the five missense mutations (fig. 3A). The fusion proteins have the molecular weights expected from the 4-kD Xpress epitope and have the predicted 61-kD FOXC1.
Figure 3.
A, Western blot analysis of both recombinant FOXC1 and FOXC1 containing missense mutations. Transfected COS-7 whole-cell extracts were resolved by SDS-PAGE and by the N-terminal Xpress epitope detected by immunoblotting with a mouse anti-Xpress monoclonal antibody. B, Northern analysis of COS-7 extracts transfected with FOXC1 pcDNA4 His/Max wild-type and missense mutation constructs. [32P]-labeled EcoRI-linearized pcDNA4 His/Max vector is hybridized to the Xpress epitope, detecting an appropriately sized product at 4.4 kb. S26 cDNA was used as a loading control. C, FOXC1 I87M, which reduces FOXC1 levels. COS-7 cells were transiently cotransfected with LacZ pcDNA4 His/Max and either FOXC1 pcDNA4 His/Max or FOXC1 I87M pcDNA4 His/Max. Transfected COS-7 whole-cell extracts were resolved by SDS-PAGE and by the N-terminal Xpress epitope detected by immunoblotting. Only small amounts (<5% of wild type) of the FOXC1 I87M mutant protein were detected.
Reduced Protein Levels Resulting from the FOXC1 I87M Mutation
When cell extracts were made from COS-7 cells transfected with the FOXC1 constructs, <5% of wild-type amounts of the FOXC1 I87M mutant could be isolated. To determine whether this problem with protein isolation could be the result of a cryptic mutation in the remainder of the FOXC1 pcDNA4 plasmid, five additional, different clones from independent mutagenesis events of the I87M FOXC1 pcDNA4 His/Max were transfected into COS-7 cells. All of these independent FOXC1 I87M clones also produced <5% of wild-type amounts of protein (data not shown). Northern analysis was performed to test whether the FOXC1 cDNA containing the I87M mutation was able to produce mRNA. As seen in figure 3B, when mRNA is extracted from COS-7 cells transfected with equivalent amounts of recombinant plasmid DNA, the amount of mRNA between the FOXC1 variants appears to be equal. To confirm that the FOXC1 I87M missense mutation reduced the levels of FOXC1, LacZ pcDNA4 His/Max was cotransfected in COS-7 cells with either I87M FOXC1 or the wild-type FOXC1 construct, and protein was extracted. Both of the transfected COS-7 plates produced β-galactosidase from the LacZ gene, indicating that the cells were able to produce protein. However, the I87M FOXC1 variant was present only in small amounts as compared with wild-type FOXC1 (fig. 3C). It would appear, therefore, that, while the I87M FOXC1 variant is being introduced into cells, mRNA is being produced, and the protein machinery of the cell is competent to synthesize new proteins, FOXC1 containing the I87M mutation nevertheless shows markedly reduced levels.
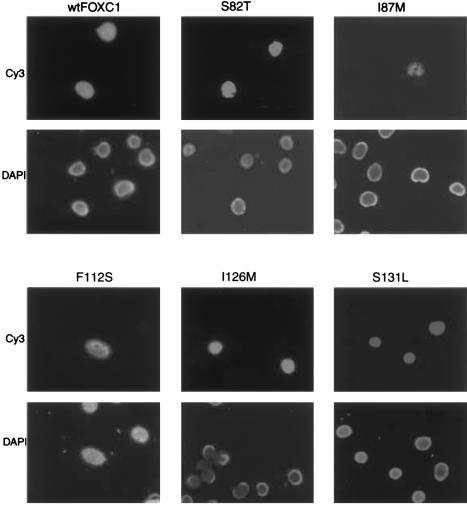
Localization of FOXC1 and Mutant Variants of FOXC1 to the Nuclei of COS-7 Cells
FOXC1 is a DNA-binding protein (Pierrou et al. 1994) and, as such, should localize to the nucleus. To determine where FOXC1 localized within cells, COS-7 cells were transiently transfected with the FOXC1 expression vector, and immunofluorescence was performed against the vector-encoded Xpress epitope. Immunofluorescence against the Xpress-tagged recombinant FOXC1 indicated that FOXC1 is localized to the nucleus (fig. 4). The effects that the missense mutations have on FOXC1 localization were tested in a similar manner. COS-7 cells were transiently transfected with the pcDNA4 His/Max FOXC1 missense-mutation vectors. All FOXC1 proteins containing missense mutations were localized to the nuclei, demonstrated by the colocalization of the Xpress epitope of the recombinant FOXC1 proteins by DAPI staining of nuclei (fig. 4). Interestingly, FOXC1 containing the I87M mutation could still be localized to the cell nucleus by immunofluorescence. The immunofluorescent signal was weaker and was present in a smaller percentage (<1%) of cells within the visual field, compared with the signal in the other missense mutations and in wild-type FOXC1 (fig. 4). This weak immunofluorescence is likely due to the reduced levels of the FOXC1 I87M mutant protein. As a result of the instability of this protein, the FOXC1 I87M mutation was not tested further.
Figure 4.
Immunofluorescence of Xpress epitope–tagged FOXC1 proteins. COS-7 cells were transiently transfected with the FOXC1 pcDNA4 His/Max constructs. In the upper six panels, the Xpress epitope–tagged recombinant FOXC1 proteins are localized to the nucleus, indicated by Cy3 fluorescence. The lower six panels indicate the position of the nuclei, by staining with DAPI. Note that the Cy3 fluorescence in the I87M transfection is weaker than it is in the other mutations.
EMSAs
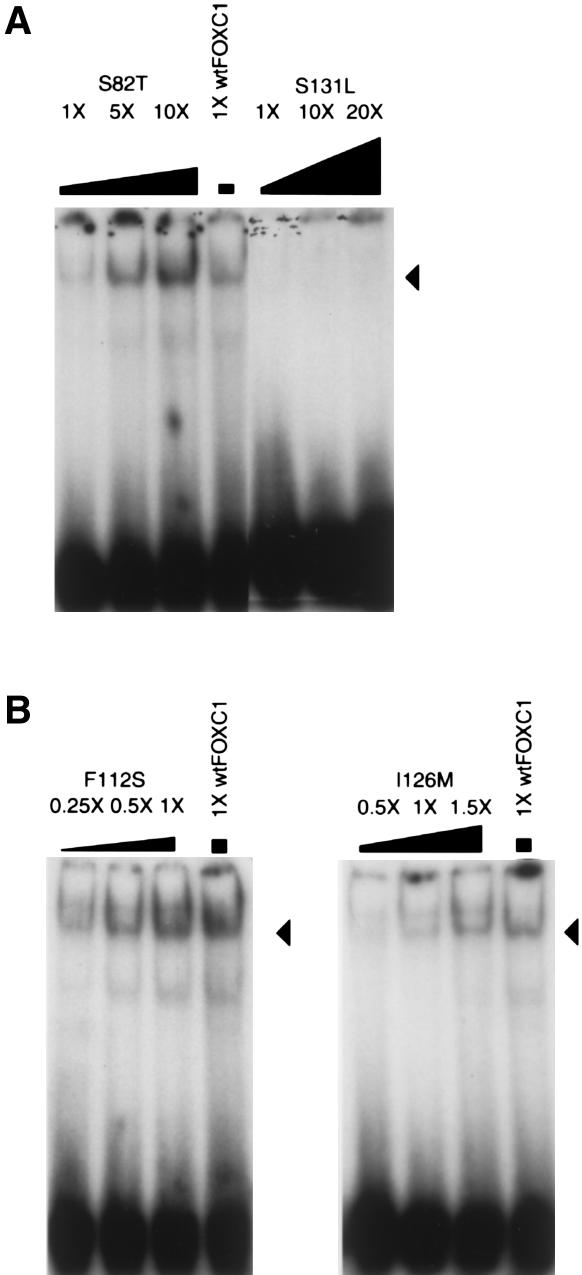
EMSAs demonstrated that the FOXC1 forkhead domain preferentially forms DNA-protein complexes with an in vitro–derived oligonucleotide, the FOXC1 site (table 2; Pierrou et al. 1994). This oligonucleotide was used in EMSA experiments to investigate whether FOXC1 missense mutations affect binding to the FOXC1 site. The S82T missense mutation showed an approximately three- to fivefold reduction in binding to the FOXC1 binding site when equal amounts of wild-type and S82T FOXC1 protein were tested (fig. 5A). The S131L mutation showed a more pronounced reduction in binding to the FOXC1 binding site. At 20 times the amount of protein, binding of the S131L mutant was still reduced to levels well below that in wild-type FOXC1 (fig. 5A).
Figure 5.
FOXC1 missense mutations that affect the DNA-binding ability of FOXC1. EMSA using the [32P]-labeled FOXC1 site was incubated with FOXC1 wild-type and missense mutations containing COS-7 protein extracts. Recombinant FOXC1 missense proteins were equalized to recombinant wild-type FOXC1 by western blotting. The position of the FOXC1-DNA complex is indicated by the blackened arrowheads. A, FOXC1 containing the S82T missense mutation, showing reduced affinity for the FOXC1 site. FOXC1 containing the S131L missense mutations shows an ∼20-fold reduction in affinity for the FOXC1 site. B, FOXC1 containing the F112S and I126M mutations, showing DNA binding that is at wild-type or near–wild-type FOXC1-protein levels.
In contrast to the reduced binding capacity of the FOXC1 S82T and S131L missense mutations, the FOXC1 F112S and I126M missense mutants bound the FOXC1 site at near–wild-type FOXC1-protein levels. The affinity that the FOXC1 F112S mutation has for the FOXC1 binding site is 1–2 times that of wild-type FOXC1, whereas the binding capacity of FOXC1 I126M is 0.5–1.0 times that of wild-type FOXC1 (fig. 5B).
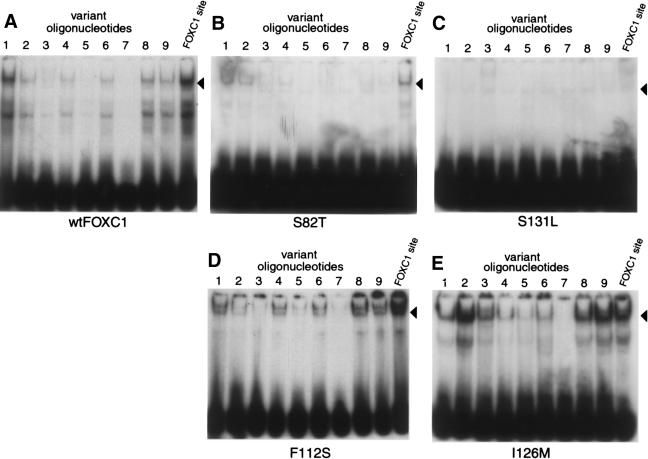
Altered Binding Specificity of the FOXC1 I126M Mutation
To determine whether the FOXC1 missense mutations altered the DNA-binding affinity of FOXC1, variant oligonucleotides of the FOXC1 binding site were constructed (table 2). Of special interest were the FOXC1 F112S and I126M missense mutants, which bound to the FOXC1 site at near–wild-type levels. EMSAs were performed using the variant FOXC1 binding sites. The FOXC1 F112S missense mutant showed an affinity for the variant oligonucleotides that was equivalent to the wild-type FOXC1 affinity for these nucleotides (fig. 6A and D). The FOXC1 I126M mutant, however, did show altered specificity compared with the wild-type FOXC1 protein. The affinity that FOXC1 I126M had for the variant FOXC1 binding-site oligonucleotides 2 (alteration of the core binding site is underlined; GAAAATAAA), 3 (GTTAATAAA), 8 (GTAAATATA), and 9 (GTAAATAAT) was higher than the affinity that the wild-type FOXC1 protein had for these variant binding sites (fig. 6A and E). The affinity of FOXC1 I126M for variant FOXC1 binding sites 2 and 9 was also higher than its affinity for the FOXC1 site. FOXC1 S82T showed only a weak affinity for oligonucleotides 1 and 2, whereas the FOXC1 S131L mutant bound none of the variant oligonucleotides (fig. 6B and C).
Figure 6.
Effect of FOXC1 missense mutations on DNA-binding specificity. EMSAs were performed with [32P]-labeled variant sites (see table 2) incubated with COS-7 protein extracts containing either wild-type or mutant FOXC1. Recombinant proteins were equalized to wild-type FOXC1 by western blotting. The position of the FOXC1-DNA complex is indicated by the blackened arrowheads. The FOXC1 I126M protein shows an increased affinity for variant oligonucleotides 2, 3, 8, and 9, compared with wild-type FOXC1’s affinity for these oligonucleotides. Also, compared with its affinity for the FOXC1 site, I126M shows an increased affinity for variant oligonucleotides 2 and 9.
Transactivation Assays
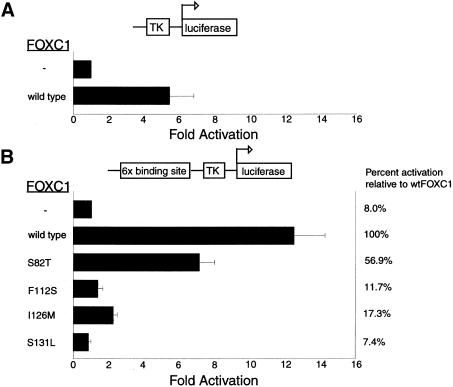
FOXC1 is thought to act as a transcription factor; therefore, the ability of FOXC1 to regulate expression of a reporter gene was tested. The herpes simplex–virus thymidine kinase (TK) promoter was positioned upstream of a luciferase reporter. HeLa cells were cotransfected with the TK-luciferase reporter construct and either pcDNA4 His/Max or FOXC1 pcDNA4 His/Max. FOXC1 was found to stimulate transcription from the TK promoter alone, showing luciferase activity to be increased 5.3-fold over that in an empty vector control (fig. 7A). The effect that the FOXC1 binding site had on FOXC1 transactivation ability was then tested. Six copies of the FOXC1 binding site were positioned upstream of the herpes simplex–virus TK promoter, again used to activate transcription of a luciferase reporter. When the FOXC1 binding sites were inserted upstream of the TK promoter, activation of the luciferase gene by FOXC1 was increased ∼12.5-fold over basal levels for the TK promoter alone (fig. 7B). The effect that the missense mutations had on FOXC1 function was then tested. The FOXC1 S82T mutant showed luciferase activity to be increased 7.1-fold (56.9% of wild-type levels) over basal levels, whereas the FOXC1 S131L mutant showed no transactivation of the luciferase reporter (7.4% of wild-type levels, compared with 8% for the empty vector alone; fig. 7B). These transactivation results are in good agreement with the FOXC1 DNA-binding studies.
Figure 7.
Disruption of the transactivation of a luciferase reporter construct, by FOXC1 missense mutations. The thick blackened bars represent the mean of the values, and the Y error bar represents the SD of the values. In each panel, the upper bar, denoted by a minus sign (−), represents a vector-only control. In panel A, a schematic of the TK promoter–luciferase gene reporter construct is shown above the histogram; transfected HeLa cells were assayed for luciferase activity, as described in the Patients, Material, and Methods section. In panel B, a schematic of the FOXC1 site-TK promoter-luciferase gene reporter construct is shown above the histogram.
Of particular interest were the FOXC1 F112S and I126M mutants. Although these mutants did bind the FOXC1 site at affinities similar to those of the wild-type FOXC1 protein (fig. 5B), the transactivation of these mutants was severely reduced. The FOXC1 F112S mutant was able to transactivate expression of luciferase at only 11.7% of the level of wild-type activation. Similarly, the FOXC1 I126M missense mutant was able to transactivate expression of luciferase at only 17.3% of wild-type FOXC1 levels. These data indicate that, although the FOXC1 F112S and I126M mutant proteins bind the FOXC1 binding site at near–wild-type levels, their abilities to activate expression of a reporter gene are markedly reduced.
Discussion
A three-dimensional structure of FOXC1 was modeled on the basis of the resolved Genesis structure reported by Jin et al. (1999). The model of the FOXC1 forkhead domain was used to predict the effects that naturally occurring missense mutations, identified in patients with ocular phenotypes, would have on the structure of the FOXC1 protein. Structural predictions were used in parallel with biochemical investigations of the molecular consequences of these disease-causing alterations of FOXC1. By comparing the structure, stability, nuclear localization, DNA-binding capacity and specificity, and transactivation potential, of wild-type versus mutant FOXC1, we have elucidated the molecular defect underlying each missense mutation.
The isoleucine residue at position 87 is located within the first helix (fig. 1B) and is one of the main participants in the formation of the hydrophobic core. I87 is involved in favorable hydrophobic interactions both with I99, I104, and I108, in helix 2, and with I126, L130, and L132, in helix 3 (fig. 1A). None of these favorable hydrophobic interactions are disrupted by the substitution, at position 87, of a methionine (a hydrophobic amino acid) for the isoleucine. Although the mechanisms underlying this reduced stability are unknown, experimentally the FOXC1 I87M mutation reduces levels of FOXC1 protein (fig. 3). Protein instability may thus represent the mechanism by which the I87M mutation of FOXC1 causes disease in patients. The small amount of I87M FOXC1 that is detected by immunofluorescence (fig. 4) and recovered in protein extracts likely is due to the overexpression of the recombinant protein in the cell-culture system used here.
The FOXC1 S82T mutation occurs in a putative nuclear-localization signal (NLS) of the forkhead domain. This region of the forkhead domain is proposed to be part one of a bipartite NLS found within the forkhead domain (Kaufmann and Knochel 1996) and thus could interfere with proper nuclear localization of the protein. Although the serine at amino acid 82 of the forkhead domain is highly conserved throughout the forkhead family, the presence of a threonine at this position does occur naturally, albeit rarely, within the forkhead family. FAST1 and Fast2 have a threonine instead of a serine at amino acid 82, and both proteins function in a manner consistent with nuclear localization (Labbe et al. 1998; Zhou et al. 1998; Yeo et al. 1999). In this light, it is not surprising that the S82T change does not disrupt the NLS and that the FOXC1 S82T protein correctly localizes to the cell nucleus. In fact, in the present study, all five missense mutants correctly localized to the cell nuclei. Our data thus indicate that incorrect subcellular localization of FOXC1 does not underlie impaired FOXC1 function in the patients in whom these missense mutations were found.
The missense mutation S82T resides at the N-terminal end of the first helix of FOXC1 (fig. 2B). The serine residue at position 82 is located outside the hydrophobic core, on the hydrophilic face of FOXC1. As expected, substitution of this serine residue by a threonine residue did not significantly change the threading scores, meaning that the now-mutated sequence is still able to form the predicted FOXC1 forkhead structure (table 1). However, experimentally the S82T mutant FOXC1 protein has a defect in DNA-binding capacity. To produce wild-type levels of DNA binding, three to five times the amount of FOXC1 S82T protein is required (fig. 5A), in agreement with the reduction in transactivation (fig. 7B). This would indicate that the FOXC1 S82T mutation affects the DNA-binding capacity, not the transactivation capacity.
The serine residue at position 131, found in the third helix, is on the hydrophilic face of the FOXC1 forkhead model. Since this position is not involved in hydrophobic-core formation, substitution of a serine residue by a leucine residue did not significantly alter the threading scores shown in table 1. The mutated sequence still shows a statistically significant match to its target structure. On the basis of its position within FOXC1, this mutation is most likely involved in DNA binding. Experimentally, the S131L missense mutation markedly reduces the ability of FOXC1 to bind the FOXC1 site (fig. 5A), and this is consistent with the structural predictions. Therefore, the reduced affinities that both the FOXC1 S82T and S131L mutant proteins have for the FOXC1 binding site are likely to underlie impaired FOXC1 function in patients with these mutations.
FOXC1 F112S and I126M missense mutants showed near–wild-type FOXC1 levels in binding to the FOXC1 binding sites but showed severe reductions in transactivation capacity. Previously, the FOXC1 forkhead domain had been thought to be responsible for nuclear localization and DNA binding (Pierrou et al. 1994; Mears et al. 1998; Nishimura et al. 1998). It is clear from the experiments in the present study that amino acid changes within the forkhead domain also can perturb the transactivation potential of FOXC1. These data also indicate that, within the FOXC1 forkhead domain, transactivation and DNA-binding capabilities are two distinct and separable processes.
The loss of transactivation activity of the FOXC1 F112S and I126M mutant proteins may be attributable to an inability to initiate transcription. In the λ repressor, a DNA-binding protein with a helix-turn-helix structure like that of FOXC1, there are positive control mutations that affect the activation ability but have no effect on binding capacity (Guarente et al. 1982; Hochschild et al. 1983). These missense mutations in the λ repressor affect the interaction between the λ repressor and RNA polymerase, rather than affecting the protein’s interaction with DNA (Hawley and McClure 1983; Li et al. 1994). The F112S and I126M missense mutations in FOXC1 may represent positive control mutants in the forkhead family of transcription factors.
On the basis of their position within the three-dimensional structure of the forkhead domain of FOXC1, the F112S and I126M missense mutations of FOXC1 can be further distinguished from one another. The F112S mutation is located outside the hydrophobic core, in the loop region between helices 2 and 4 (fig. 1B). This position is not involved in hydrophobic-core formation; hence, replacement at this position did not interfere with the overall pattern of intermolecular interactions. (It is important to note that this computational method takes into account only residues involved in pairwise contacts with other residues. Thus, residues with their side chains pointed outward presumably are not involved in such interactions, and, therefore, the net effect of mutations at these positions cannot be assessed directly.) Biochemical studies showing that recombinant F112S binds DNA as effectively as does the wild-type protein—but that it does not transactivate reporter constructs that contain the same DNA target sequence—can be explained by the predicted location of this mutant residue, away from the DNA-binding face of FOXC1. The F112S mutation in FOXC1 most likely interferes with protein-protein interactions that are necessary for the transactivation of downstream genes.
The isoleucine at position 126 is within helix 3 and is another important residue in the formation of the hydrophobic core. The I126M mutation did not significantly change the threading scores in table 1, since the substitution is by another similar hydrophobic amino acid. The reduction in transactivation by recombinant I126M probably is due to subtle changes in intermolecular interactions, resulting in a more rigid molecule. The I126M mutation may render FOXC1 unable to alter the conformation of target DNA, thus preventing transcription initiation.
The fact that the FOXC1 F112S and I126M mutant proteins are able to bind to the FOXC1 binding site indicates that the winged-helix structure that acts to bind DNA sequences is fundamentally intact. Additionally, the binding specificities that FOXC1 F112S and I126M mutant proteins have with respect to the variant oligonucleotides (fig. 6 and table 2) indicate that the binding of these FOXC1 mutant proteins is sequence specific. The NMR structural analyses of Jin et al. (1999) predicted that helix 3 of forkhead proteins recognizes the major groove of the DNA, implicating this region of the protein in DNA-site recognition. Our experimental data, demonstrating that the I126M mutation in helix 3 of FOXC1 results in a mutant protein with altered DNA-site preference, are in excellent agreement with this prediction.
In the mouse mutant dysgenic lens (dyl), Foxe3, the gene responsible for the dyl phenotype, has two missense mutations, F93L and F98S, both in the forkhead domain. The F112S mutation of FOXC1 corresponds to the same position in the forkhead domain as does the F98S mutation found in dyl mice (Blixt et al. 2000). In both cases, the phenylalanine is replaced by a serine. The dyl mutation, like the F112S mutation of FOXC1, may therefore result in an inability of Foxe3 to transactivate genes required for proper lens formation. The F93L substitution, also found in the dyl allele of Foxe3, may also exert an effect in addition to the effect of the F98S substitution.
Data from patients with FOXC1 mutations (Mears et al. 1998; Nishimura et al. 1998), as well as data from mouse models with Foxc1 alterations (Kume et al. 1998; Mears et al. 1998; Kidson et al. 1999; Smith et al. 2000), present strong evidence for a haploinsufficiency model for FOXC1. In mice, a null-heterozygous mutation of Foxc1 results in anterior-segment defects of the eye, similar to those found in patients with AR malformations. In humans, missense mutations that still produce full-length FOXC1 with reduced activity cannot be distinguished, at a phenotypic level, from null or nonsense mutations. The S82T FOXC1 missense mutation provides strong evidence for a model of haploinsufficiency for FOXC1. The S82T missense mutation shows almost 60% of wild-type transactivation activity, yet the phenotypic consequences are the same as those in patients with the I87M mutation, which produces <5% of wild-type amounts of FOXC1-protein. In fact, the ocular phenotypic differences between patients with any of the missense mutations were the same as the ocular phenotypic differences between patients with the same missense mutations. Within a family with a given missense mutation of FOXC1, there is often a spectrum of phenotypic consequences (Mears et al. 1998; Nishimura et al. 1998).
Although the penetrance of FOXC1 defects within the eye is high (Mears et al. 1998; Nishimura et al. 1998), there are likely to be environmental factors and/or modifier genes that result in variable expressivity. Phenotypic variability could reflect a level of chance in developmental events, which is related to the timing, location, and level of expression of developmentally important downstream targets of FOXC1. Regulation of FOXC1 levels is therefore critical for proper development. A recent report by other researchers indicates that individuals with three copies of FOXC1 show anterior eye-segment defects (Lehmann et al. 2000). This report, combined with the present study's data on partial activity of one of the FOXC1 missense mutations, lead us to conclude that the regulation of FOXC1 levels is extremely stringent, with >78% of wild-type levels (normal allele activity plus FOXC1 S82T activity) but <150% of wild-type levels (activity of three alleles) of FOXC1 being required for correct FOXC1 function.
Mutations in FOXC1 and PITX2 result in similar ocular phenotypes. Although genotype-phenotype correlations have been established for defects in PITX2 (Kozlowski and Walter 2000), it appears that no such relationship can be established for FOXC1. Mutant PITX2 proteins that retain partial function result in anterior eye-segment defects that are milder than those found in patients with no PITX2 function, but this clearly is not the case for FOXC1. Aberrant ocular development arising from PITX2 mutations may follow a mechanism/pathway different than that of ocular defects arising from FOXC1 mutations.
The upstream elements and downstream targets of FOXC1 remain unknown. Possible target genes include those involved in cellular processes such as cell-cell adhesion, cell migration, and cell differentiation (Kume et al. 1998; Smith et al. 2000). Tight regulation of FOXC1, possibly within the aforementioned cellular pathways, appears to be critical for normal eye development.
Acknowledgments
We would like to thank Dr. Peter Carlsson for the FOXC1 cDNA, Mr. James Friedman for technical assistance, Ms. Margaret Hughes for tissue culture, and Drs. Moira Glerum and Alan Underhill (Department of Medical Genetics) and the Ocular Genetics Lab for critical reading of the manuscript and for helpful comments. This work was supported by the Alberta Heritage Foundation for Medical Research (AHFMR) and the Canadian Institutes for Health Research (CIHR). R.A.S. is supported by an AHFMR studentship. M.A.W. is an AHFMR Senior Scholar and a CIHR Investigator.
Note added in proof.—A very recent study by Nishimura et al. (2001) found an additional eight novel mutations: one missense mutation, five frameshift mutations, and two duplications.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- National Center for Biotechnology Information (NCBI) Databases, http://www.ncbi.nlm.nih.gov/Database/index.html (for human FOXC1 [AF048693], mouse Foxc1 [NM008592], human FOXC2 [Y08223], human FOXD1 [U13222], human FOXE1 [U89995], human FOXF1 [U13219], human FOXF2 [U13220], human HFH1 [AF153341], human FOXH1 [AF076292], mouse Foxh2 [AF110506], and rat Genesis/Foxd3 [NM012183])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FOXC1 [MIM 601090], PITX2 [MIM 601542], Oct1 [MIM 602607], and FOXC2 [MIM 602402])
References
- Alward WL, Semina EV, Kalenak JW, Heon E, Sheth BP, Stone EM, Murray JC (1998) Autosomal dominant iris hypoplasia is caused by a mutation in the Rieger syndrome (RIEG/PITX2) gene. Am J Ophthalmol 125:98–100 [DOI] [PubMed] [Google Scholar]
- Becker R, Chambers J, Wilks A (1988) The new S language—a programming environment for data analysis and graphics. Wadsworth, Pacific Grove, CA [Google Scholar]
- Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P (2000) A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev 14:245–254 [PMC free article] [PubMed] [Google Scholar]
- Bryant SH, Lawrence CE (1993) An empirical energy function for threading protein sequence through the folding motif. Proteins 16:92–112 [DOI] [PubMed] [Google Scholar]
- Fetrow J, Bryant SH (1993) New programs for protein tertiary structure prediction. Biotechnology 11:479–484 [DOI] [PubMed] [Google Scholar]
- Guarente L, Nye JS, Hochschild A, Ptashne M (1982) Mutant lambda phage repressor with a specific defect in its positive control function. Proc Natl Acad Sci USA 79:2236–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley DK, McClure WR (1983) The effect of a lambda repressor mutation on the activation of transcription initiation from the lambda PRM promoter. Cell 32:327–333 [DOI] [PubMed] [Google Scholar]
- Hiemisch H, Monaghan AP, Schutz G, Kaestner KH (1998) Expression of the mouse Fkh1/Mf1 and Mfh1 genes in late gestation embryos is restricted to mesoderm derivatives. Mech Dev 73:129–132 [DOI] [PubMed] [Google Scholar]
- Hochschild A, Irwin N, Ptashne M (1983) Repressor structure and the mechanism of positive control. Cell 32:319–325 [DOI] [PubMed] [Google Scholar]
- Hong HK, Lass JH, Chakravarti A (1999) Pleiotropic skeletal and ocular phenotypes of the mouse mutation congenital hydrocephalus (ch/Mf1) arise from a winged helix/forkhead transcription factor gene. Hum Mol Genet 8:625–637 [DOI] [PubMed] [Google Scholar]
- Jin C, Marsden I, Chen X, Liao X (1999) Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J Mol Biol 289:683–690 [DOI] [PubMed] [Google Scholar]
- Kaufmann E, Knochel W (1996) Five years on the wings of fork head. Mech Dev 57:3–20 [DOI] [PubMed] [Google Scholar]
- Kidson SH, Kume T, Deng K, Winfrey V, Hogan BL (1999) The forkhead/winged-helix gene, Mf1, is necessary for the normal development of the cornea and formation of the anterior chamber in the mouse eye. Dev Biol 211:306–322 [DOI] [PubMed] [Google Scholar]
- Kozlowski K, Walter MA (2000) Variation in residual PITX2 activity underlies the phenotypic spectrum of anterior segment developmental disorders. Hum Mol Genet 9:2131–2139 [DOI] [PubMed] [Google Scholar]
- Kulak SC, Kozlowski K, Semina EV, Pearce WG, Walter MA (1998) Mutation in the RIEG1 gene in patients with iridogoniodysgenesis syndrome. Hum Mol Genet 7:1113–1117 [DOI] [PubMed] [Google Scholar]
- Kume T, Deng K, Hogan BL (2000) Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development 127:1387–1395 [DOI] [PubMed] [Google Scholar]
- Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL (1998) The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell 93:985–996 [DOI] [PubMed] [Google Scholar]
- Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L (1998) Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell 2:109–120 [DOI] [PubMed] [Google Scholar]
- Lai JS, Cleary MA, Herr W (1992) A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeo domain. Genes Dev 6:2058–2065 (erratum: Genes Dev 6:2663 [1992]) [DOI] [PubMed] [Google Scholar]
- Larsson C, Hellqvist M, Pierrou S, White I, Enerbäck S, Carlsson P (1995) Chromosomal localization of six human forkhead genes, freac-1 (FKHL5), -3 (FKHL7), -4 (FKHL8), -5 (FKHL9), -6 (FKHL10), and -8 (FKHL12). Genomics 30:464–469 [DOI] [PubMed] [Google Scholar]
- Lehmann OJ, Ebenezer ND, Jordan T, Fox M, Ocaka L, Payne A, Leroy BP, Clark BJ, Hitchings RA, Povey S, Khaw PT, Bhattacharya SS (2000) Chromosomal duplication involving the forkhead transcription factor gene FOXC1 causes iris hypoplasia and glaucoma. Am J Hum Genet 67:1129–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Moyle H, Susskind MM (1994) Target of the transcriptional activation function of phage lambda cI protein. Science 263:75–77 [DOI] [PubMed] [Google Scholar]
- Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo W, Collins C, Marshall J, Gould DB, Pearce W, Carlsson P, Enerbäck S, Morissette J, Bhattacharya S, Hogan B, Raymond V, Walter MA (1998) Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet 63:1316–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayans F, Gould DB, Heon E, Billingsley GD, Cheung JC, Mears AJ, Walter MA (2000) Axenfeld-Rieger syndrome resulting from mutation of the FKHL7 gene on chromosome 6p25. Eur J Hum Genet 8:71–74 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281–296 [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Searby CC, Alward WL, Walton D, Craig JE, Mackey DA, Kawase K, Kanis AB, Patil SR, Stone EM, Sheffield VC (2001) A spectrum of FOXC1 mutations suggests gene dosage as a mechanism for developmental defects of the anterior chamber of the eye. Am J Hum Genet 68:364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC (1998) The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet 19:140–147 [DOI] [PubMed] [Google Scholar]
- Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P (1994) Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J 13:5002–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC (1996) Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet 14:392–399 [DOI] [PubMed] [Google Scholar]
- Smith RS, Zabaleta A, Kume T, Savinova OV, Kidson SH, Martin JE, Nishimura DY, Alward WLM, Hogan BLM, John SWM (2000) Haploinsufficiency of the transcription factors FOXC1 and FOXC2 results in aberrant ocular development. Hum Mol Genet 9:1021–1032 [DOI] [PubMed] [Google Scholar]
- Swiderski RE, Reiter RS, Nishimura DY, Alward WL, Kalenak JW, Searby CS, Stone EM, Sheffield VC, Lin JJ (1999) Expression of the Mf1 gene in developing mouse hearts: implication in the development of human congenital heart defects. Dev Dyn 216:16–27 [DOI] [PubMed] [Google Scholar]
- Vincent S, Marty L, Fort P (1993) S26 ribosomal protein RNA: an invariant control for gene regulation experiments in eucaryotic cells and tissues. Nucleic Acids Res 21:1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Jackle H (1990) The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell 63:455–456 [DOI] [PubMed] [Google Scholar]
- Winnier GE, Kume T, Deng K, Rogers R, Bundy J, Raines C, Walter MA, Hogan BLM, Conway SJ (1999) Roles for the winged helix transcription factors MF1 and MFH1 in cardiovascular development revealed by nonallelic noncomplementation of null alleles. Dev Biol 213:418–431 [DOI] [PubMed] [Google Scholar]
- Yeo CY, Chen X, Whitman M (1999) The role of FAST-1 and Smads in transcriptional regulation by activin during early Xenopus embryogenesis. J Biol Chem 274:26584–26590 [DOI] [PubMed] [Google Scholar]
- Zhou S, Zawel L, Lengauer C, Kinzler KW, Vogelstein B (1998) Characterization of human FAST-1, a TGF beta and activin signal transducer. Mol Cell 2:121–127 [DOI] [PubMed] [Google Scholar]